Significance
We characterize in humans a coordinated network of brain activity describing neurobehavioral correlates of reward anticipation. The network involves nodes in striatal and cortical brain regions, which are preferentially associated with distinct externalizing behaviors—hyperactivity and alcohol consumption—suggesting that the heterogeneity of reward-related behaviors might be accounted for by different association patterns of nodes and their connecting links. In a genome-wide association study of the striatal node with subsequent functional validation in Drosophila, we identify molecular genetic mechanisms involving vacuolar protein sorting-associated protein 4A (VPS4A) in dopamine regulation, reward anticipation, and hyperactivity. Our approach might facilitate the identification of causal neural mechanisms, important for the identification of previously unidentified targets and the establishment of neurobehaviorally informed end points for clinical trials.
Keywords: fMRI, neural network, VPS4A, dopamine receptor, GWAS
Abstract
Dysfunctional reward processing is implicated in various mental disorders, including attention deficit hyperactivity disorder (ADHD) and addictions. Such impairments might involve different components of the reward process, including brain activity during reward anticipation. We examined brain nodes engaged by reward anticipation in 1,544 adolescents and identified a network containing a core striatal node and cortical nodes facilitating outcome prediction and response preparation. Distinct nodes and functional connections were preferentially associated with either adolescent hyperactivity or alcohol consumption, thus conveying specificity of reward processing to clinically relevant behavior. We observed associations between the striatal node, hyperactivity, and the vacuolar protein sorting-associated protein 4A (VPS4A) gene in humans, and the causal role of Vps4 for hyperactivity was validated in Drosophila. Our data provide a neurobehavioral model explaining the heterogeneity of reward-related behaviors and generate a hypothesis accounting for their enduring nature.
Successful behavioral adaptation requires effective reward processing that determines whether a desired goal is approached and maintained. Reward processing can be separated into behavioral anticipation or reward expectancy as a consequence of learning and behavioral and subjective responses to rewarding outcomes (1). In humans, dysfunctional reward processing (in particular, dysfunctional reward anticipation) has been implicated in various externalizing disorders, including attention-deficit hyperactivity disorder (ADHD) (2) and addiction (3). Brain regions involved in reward anticipation include the ventral tegmental area, the medial forebrain bundle, and the nucleus accumbens/ventral striatum (VS; including the ventral caudate-putamen) as well as the ventromedial and insular cortices (4). More recently, observations have been reported to link reward processing in humans with cortical activation (5), including the primary somatosensory (6), primary visual (V1) (7), and auditory (8) cortices. Dopamine is the principal neurotransmitter regulating reward processing, particularly through the mesocorticolimbic pathway (9), the neuronal projection from the ventral tegmental area to the VS and prefrontal cortex. A general feature of striatal information processing is the control by reward-related dopamine signals of direct and indirect cortical inputs from different neurotransmitter systems, including noradrenaline, glutamate, and GABA as well as acetylcholine, endogenous opioids, and cannabinoids (10). As a consequence, striatal dopaminergic activity integrates cortical and subcortical inputs with reward response. In addition to direct and indirect regulation by heteroceptors, dopamine release is regulated by presynaptic autoreceptors of the D2 family, in particular D2 dopamine receptors (DRD2) that are coupled to inhibitory G proteins, modulate ion channel activity, and/or inhibit adenylyl cyclase. Postsynaptic dopamine receptors include DRD1, which activates the cAMP pathway and is colocalized with glutamatergic NMDA receptors in the postsynaptic density, and they are thought to contribute to the glutamate–dopamine cross-talk (11).
Despite neurobiological and molecular evidence indicating extensive corticostriatal integration in reward processing, most human neuroimaging studies have limited their investigations to regions of interest analyses of very selected brain structures, namely the VS and the orbitofrontal cortex. There is, as yet, no comprehensive analysis to investigate a coordinated network of brain activity during reward anticipation in large human datasets or study its genetic basis. Given that the behavioral heterogeneity associated with dysfunctional reward processes is too extensive to be easily explained by differences in brain activities in these regions of interest alone, such a network-based analysis might help to explain the neural underpinnings of common and distinct neuropsychological deficits associated with reward-related mental disorders.
Results
Functional Brain Network of Reward Anticipation.
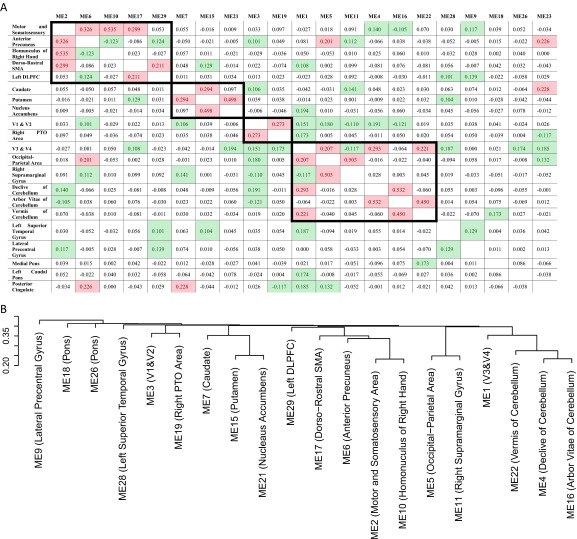
We investigated the pattern of brain activation during reward anticipation in the IMAGEN sample (12) by measuring the blood oxygen level-dependent (BOLD) response in functional neuroimaging [functional MRI (fMRI)] analyses of 1,544 14-y-old adolescents (Table 1) using the high win vs. no win contrast of the monetary incentive delay (MID) task (Materials and Methods and SI Materials and Methods). We applied a hypothesis-free brain-wide weighted voxel coactivation network analysis (WVCNA) (13) and obtained 1,397 modules of brain activation during reward anticipation. Of these, 21 modules fulfilling stringent methodological requirements were selected for additional analysis (SI Materials and Methods and Fig. S1). The modules included subcortical reward-processing areas, including the striatum and cortical areas in (but not limited to) the frontal, parietal, and occipital lobes (Tables S1 and S2). We examined relationships among these modules by generating functional connections (i.e., a partial correlation matrix) (SI Materials and Methods and Fig. S2A). Subsequent hierarchical clustering identified four nodes involved in reward anticipation (Fig. 1 and Fig. S2B). Node 1 consisted of the caudate nucleus, putamen, and nucleus accumbens (striatum), node 2 included occipital areas (V1/V2) involved in early visual processing, node 3 included somatosensory and motor areas, and node 4 involved occipital, parietal, and cerebellar areas. Their corresponding first principle components were used in the following analyses.
Table 1.
Summary of descriptive statistics for the IMAGEN sample
| Sample information | Descriptive statistics |
| fMRI full sample size | n = 1,544 (53% female) |
| Age, y (range) | 14.41 (12.56–16.04) |
| Affective go/no go task (n = 1,333) | |
| Positive stimuli | 12.3 (8.09) |
| Negative stimuli | 14.0 (7.42) |
| SWM (n = 1,506) | |
| Between search error | 19.3 (14.0) |
| Delay discounting task (n = 1,518) | |
| Small reward | 0.0381 (0.0516) |
| Medium reward | 0.0276 (0.0404) |
| Large reward | 0.0176 (0.0342) |
| SDQ (n = 1,534) | |
| Hyperactivity (parent-rated) | 2.86 (2.56) |
| ESPAD (n = 1,495) | |
| Lifetime alcohol consumption | 17.2 (34.3) |
| TCI-R (n = 1,521) | |
| Impulsiveness | 26.74 (4.80) |
Statistics are mean (SD) unless noted otherwise. ESPAD, European School Survey Project on Alcohol and Drugs; SDQ, Strengths and Difficulties Questionnaire; SWM, spatial working memory; TCI-R: Temperament and Character Inventory, Revised Version. Distributions of variables were shown in Fig. S7.
Fig. S1.
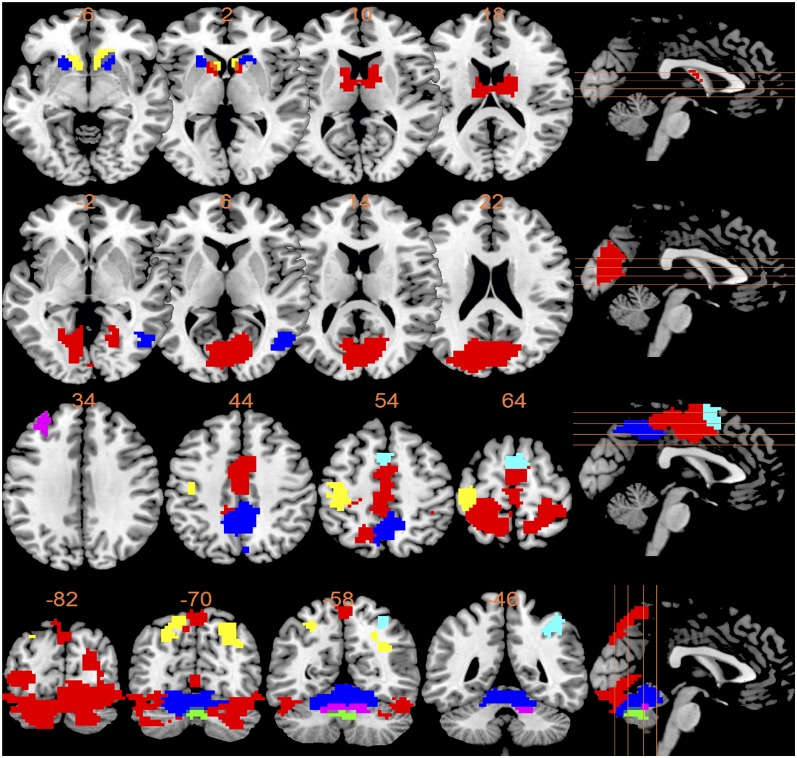
Top 30 modules from the WVCNA. (A) Occipital cortex regions. Red (module 1): Brodmann areas 18 and 19. Blue (module 3): Brodmann areas 17–19. (B) Sensory and motor cortex regions. Red (module 2): Brodmann areas 3–7. Blue (module 6): Brodmann area 7. Yellow (module 10): Brodmann areas 3 and 4. (C) Subcortical regions. Red (module 7): caudate. Blue (module 15): putamen. Yellow (module 21): nucleus accumbens. (D) Parietal cortex regions. Red (module 5): Brodmann area 7. (Right) Blue (module 11): Brodmann area 40. Yellow (module 19): Brodmann areas 37 and 39. (E) Cerebellum regions. Red (module 4): anterior lobe and declive of posterior lobe. Blue (module 16): arbor vitae. Yellow (module 22): tuber and declive vermis. (F) Frontal cortex regions. Red (module 17): Brodmann area 6. (Left) Blue (module 29): Brodmann area 9. (G) Ungrouped modules I. Red (module 9): Brodmann areas 4 and 6. Blue (module 23): Brodmann areas 23 and 31. (Left) Yellow (module 28): Brodmann areas 22 and 38. (H) Ungrouped modules II. Red (module 18): medial pons. Blue (module 26): left caudal pons. (I) Noise modules. Red (module 12), blue (module 13), and yellow (module 24). (J) Negative BOLD response modules I. Red (the whole-brain activation map): positive BOLD response. Blue (module 8), yellow (module 14), pink (module 20), light blue (module 25), and green (module 30). (K) Negative BOLD response modules II. Red (the whole-brian activation map): positive BOLD Response. Blue (module 27).
Table S1.
Tables for fMRI-weighted coactivation network analyses: Anatomical regions
| Modules | Anatomical (Brodmann) area involved | Main area |
| 1 | Brodmann 7, 17–19, and 37 | Brodmann 7, 18, and 19 (dorsal precuneus and associated vision area V3, V4) |
| 2 | Brodmann 1–7, 24, and 31 | Brodmann 1–3 and 5–7 (superior parietal lobule, medial frontal gyrus, and postcentral gyrus) |
| 3 | Brodmann 7, 17–19, 23, 30, and 31 | Brodmann 18 and 19 (primary and secondary vision area V1 and V2) |
| 4 | Cerebellum (culmen of anterior lobe and declive of posterior lobe) | Declive of posterior lobe |
| 5 | Brodmann 7, 19, and 39 | Brodmann 7 (occipital/parietal area) |
| 6 | Brodmann 5 and 7 | Brodmann 7 (anterior part of precuneus) |
| 7 | Caudate body, caudate head | Caudate |
| 9 | Brodmann 3, 4, and 6 | Brodmann 4 and 6 (lateral precentral gyrus) |
| 10 | Brodmann 1–4, 6, and 40 (left only) | Brodmann 3 and 4 (left superior precentral gyrus and left superior postcentral gyrus) |
| 11 | Brodmann 7 and 40 (right only) | Brodmann 40 (right inferior parietal lobule) |
| 15 | Caudate head, putamen | Putamen |
| 16 | Arbor vitae | |
| 17 | Brodmann 6 | Brodmann 6 (superior/medial frontal gyrus) |
| 18 | Pons | |
| 19 | Brodmann 19, 37, and 39 (right only) | Brodmann 19, 37, and 39 (right middle temporal gyrus and right middle occipital gyrus) |
| 21 | Nucleus accoumbens | Nucleus accumbens |
| 22 | Cerebellar vermis (posterior lobe) | Tuber and declive vermis |
| 23 | Brodmann 23, 30, and 31 | Brodmann 23 and 31 (posterior cingulate) |
| 26 | Pons | |
| 28 | Brodmann 22, 38, and 47 (left only) | Brodmann 22 (left superior temporal gyrus) |
| 29 | Brodmann 9 and 10 (left only) | Brodmann 9 (left dorsolateral prefrontal cortex) |
The anatomical regions related to each module were generated from TalairachClient (www.talairach.org/).
Table S2.
Tables for fMRI-weighted coactivation network analyses: Size and Montreal Neurological Institute coordinates
| Modules | Stereotaxic coordinates given in MNI space of subcluster 1 | Size of subcluster 1 | Stereotaxic coordinates given in MNI space of subcluster 2 | Size of subcluster 2 | Size of core voxels | Size of the origin module | ||||
| x/Sagittal | y/Axial | z/Coronal | x/Sagittal | y/Axial | z/Coronal | |||||
| 1 | 4.36 | −81.52 | −10.78 | 1,886 | 0.54 | −68.15 | 60.21 | 176 | 2,062 | 2,952 |
| 2 | −18.62 | −49.90 | 65.40 | 501 | 0.10 | −14.79 | 56.84 | 371 | 872 | 1,497 |
| 21.22 | −47.70 | 67.26 | ||||||||
| 3 | −2.15 | −77.98 | 17.67 | 1,003 | 1,003 | 1,356 | ||||
| 4 | −0.32 | −62.32 | −15.88 | 705 | 705 | 924 | ||||
| 5 | −23.69 | −68.71 | 47.45 | 52 | 31.66 | −67.42 | 47.83 | 42 | 94 | 393 |
| 6 | 1.97 | −52.23 | 52.27 | 171 | 171 | 380 | ||||
| 7 | −10.77 | −3.76 | 18.24 | 179 | 179 | 353 | ||||
| 12.40 | −4.32 | 19.28 | ||||||||
| 9 | −54.91 | −10.11 | 33.86 | 54 | 58.60 | −5.77 | 25.52 | 30 | 84 | 298 |
| 10 | −40.06 | −25.10 | 61.28 | 129 | 129 | 272 | ||||
| 11 | 41.01 | −47.38 | 53.61 | 42 | 42 | 219 | ||||
| 15 | −20.65 | 13.12 | −6.27 | 19 | 13.52 | 16.08 | −2.32 | 10 | 29 | 166 |
| 16 | 3.88 | −59.15 | −22.02 | 57 | 57 | 144 | ||||
| 17 | 1.18 | 7.49 | 67.63 | 26 | 0.85 | 12.50 | 56.69 | 14 | 40 | 139 |
| 18 | 0.09 | −15.94 | −30.92 | 46 | 46 | 139 | ||||
| 19 | 43.43 | −69.32 | 8.13 | 18 | 50.26 | −61.35 | 8.98 | 12 | 30 | 135 |
| 21 | 12.49 | 21.69 | −6.99 | 27 | −12.01 | 19.24 | −8.00 | 14 | 41 | 126 |
| 22 | 1.71 | −69.70 | −30.44 | 31 | −0.81 | −61.79 | −32.00 | 17 | 48 | 124 |
| 23 | 0.70 | −34.86 | 28.07 | 47 | 47 | 120 | ||||
| 26 | −14.99 | −27.59 | −45.51 | 48 | −16.38 | −23.51 | −40.09 | 27 | 75 | 113 |
| 28 | −55.37 | 14.93 | −6.74 | 43 | 43 | 112 | ||||
| 29 | −33.67 | 43.68 | 28.68 | 14 | 14 | 110 | ||||
The subclusters of each module are generated through the hierarchical clustering on the corresponding stability matrix that measures the frequency of being regrouped into the same cluster for each pair of voxels. The Montreal Neurological Institute (MNI) coordinates of the central voxel are calculated as the weighted averages of MNI coordinates of all of the voxels in the corresponding subcluster. The weight for each voxel is calculated as the average of its stability matrix (SI Materials and Methods) entries belonging to the corresponding subcluster. The sizes of subclusters and original modules are also shown as the numbers of voxels involved, where the dimension of each voxel is 3 × 3 × 3 mm.
Fig. S2.
Hierarchical clustering analysis of functional connection matrix. (A) The functional connection matrix was calculated as pairwise partial correlations between all 21 valid modules, with all of the rest of the modules as control variables. For each module, the module names are listed in row 1, and the involved brain areas are listed in column 1. Cells with partial correlations that survived the Bonferroni correction for multiple testing are highlighted in green, and values over 0.20 are highlighted in red. Functional clusters/nodes established from hierarchical clustering as shown in B were grouped and highlighted with bold borders. (B) Tree of hierarchical clustering (in four clusters/nodes): node 1 (modules 7, 15, and 21), node 2 (modules 3 and 19), node 3 (modules 2, 6, 10, 17, and 29), and node 4 (modules 1, 4, 5, 11, 16, and 22). Module 23 (posterior cingulate cortex) was excluded from the tree because of its high correlation with multiple nodes. DLPFC, dorsal/lateral prefrontal cortex; ME, module eigenvoxel; PTO, parietal/temporal/occipital; SMA, supplementary motor area.
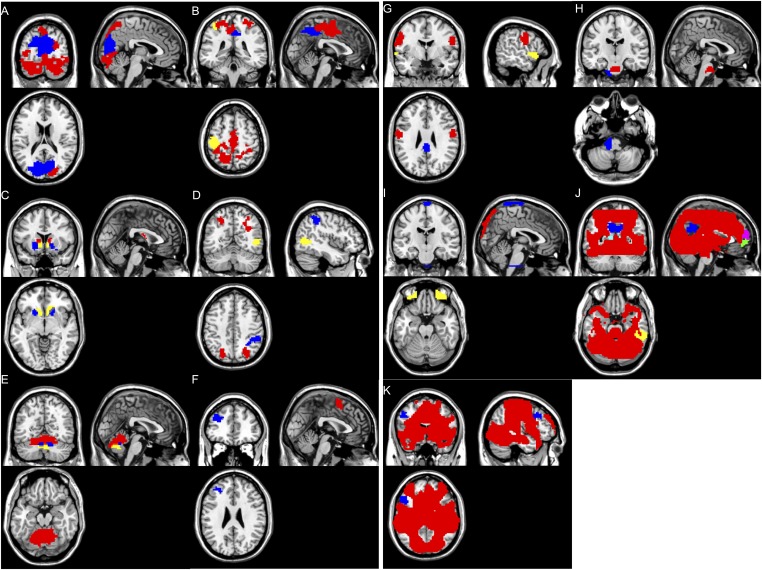
Fig. 1.
Illustration of fMRI clusters/nodes. (Row 1) Node 1: module 7, caudate (red); module 15, putamen (blue); and module 21, nucleus accumbens (yellow), with multisplicing axial view at Montreal Neurological Institute (MNI) coordinate z equal to −6, 2, 10, and 18. (Row 2) Node 2: module 3, visual area V1 and V2 (red) and module 19, the right parietal/temporal/occipital area (blue), with multisplicing axial view at MNI coordinate z equal to −2, 6, 14, and 22. (Row 3) Node 3: module 2, primary somatosensory and motor areas (red); module 6, anterior precuneus (blue); module 10, left precentral and postcentral gyrus (yellow); module 17, dorsorostral supplementary motor area (light blue); and module 29, left dorsolateral prefrontal cortex (purple), with multisplicing axial view at MNI coordinate z equal to 34, 44, 54, and 64. (Row 4) Node 4: module 1, visual area V3 and V4 (red); module 4, cerebellum anterior lobe and declive of posterior lobe (blue); module 5, superior parietal lobe (yellow); module 11, right supramarginal gyrus (light blue); module 16, arbor vitae (purple); and module 22, cerebellum vermis (green), with multisplicing sagittal view at MNI coordinate y equal to −82, −70, −58, and −46.
Characteristics of the Functional Brain Network.
We assessed associations of these nodes with neuropsychological tests (Table 1) related to reward processing through Cambridge Neuropsychological Test Automated Battery (Cantab) (www.cambridgecognition.com), including (i) the affective go/no go task, which measures selective attentional bias to affective stimuli, and (ii) the spatial working memory task, which is akin to an optimal foraging task for reward (14) and has been associated with ADHD (15), and the delay discounting task (Monetary Choice Questionnaire), a measure of delayed gratification and impulsiveness (16), taking into account that damage to the rat nucleus accumbens impairs delayed reward discounting (17). Because activity in different nodes may not be independent, we identified the predominant node driving the association with performance in these tests by carrying out partial correlation analyses controlling for the effects of all remaining nodes (Table 2 and Table S3). Predominant association was defined as a P < 0.1 after partial correlation analysis. The predominant association in striatal node 1 was with fewer errors in the spatial working memory task (R = −0.12; Pcorrected = 0.0001; df = 1,495) and reduced delay discounting of small (R = −0.08; Pcorrected = 0.0461; df = 1,507) and medium gains (R = −0.08; Pcorrected = 0.0209; df = 1,507). Activation of node 2 (V1/V2) revealed predominant association with fewer omissions of responses under negative (R = −0.09; Pcorrected = 0.0090; df = 1,323) stimuli in the affective go/no go task. Node 3 (somatosensory/motor) was predominantly associated with less delay discounting of large rewards (R = −0.08; Pcorrected = 0.0190; df = 1,507). In node 4, we detected no predominant association with any of the neuropsychological measures. Other associations that were significant after permutation (Table 2) but were not predominant after partial correlation analysis are not described here. Although all four identified nodes are part of a reward anticipation network, their different anatomical localization as well as their distinct neuropsychological characteristics may suggest that these nodes represent functional correlates of a coordinated process underlying reward anticipation (6, 7).
Table 2.
Results of the association analyses between fMRI nodes and neuropsychological tests
| Outcomes (df) | Node 1 | Node 2 | Node 3 | Node 4 |
| AGN omission positive stimuli (1,323) | n.s. | 0.0090 (−0.09)*† | n.s. | n.s. |
| AGN omission negative stimuli (1,323) | n.s. | 0.0014 (−0.11)* | 0.0026 (−0.11)* | 0.0038 (−0.10)* |
| SWM between search errors (1,495) | 0.0001 (−0.12)*† | 0.0003 (−0.11)* | 0.0007 (−0.09)* | 0.0004 (−0.11)* |
| Delay discounting (small reward; 1,507) | 0.0461 (−0.08)*† | n.s. | n.s. | n.s. |
| Delay discounting (medium reward; 1,507) | 0.0209 (−0.08)*† | n.s. | n.s. | n.s. |
| Delay discounting (large reward; 1,507) | n.s. | n.s. | 0.0190 (−0.08)*† | n.s. |
The P values corrected for multiple testing were calculated based on 10,000 permutation analyses. AGN, affective go/no go; n.s., no significance; SWM, spatial working memory.
Results were provided as P value (partial correlation, i.e., the effect size).
A node was classed as predominant if its P value after controlling for all other nodes was smaller than 0.10 (Table S3). Study sites, gender, and handedness were controlled.
Table S3.
Tables for fMRI-weighted coactivation network analyses: Results of the predominance analysis for neuropsychological tests
| Outcomes (df) | Controlled P values | |||
| Node 1 | Node 2 | Node 3 | Node 4 | |
| AGN omission positive stimuli (1,323) | 4.57 × 10−1 | 4.60 × 10−2* | 5.37 × 10−1 | 9.56 × 10−1 |
| AGN omission negative stimuli (1,323) | 4.71 × 10−1 | 1.70 × 10−1 | 2.58 × 10−1 | 6.41 × 10−1 |
| SWM between trial errors (1,495) | 6.67 × 10−2* | 2.18 × 10−1 | 4.49 × 10−1 | 5.16 × 10−1 |
| Delay discounting (small reward; 1,507) | 2.12 × 10−2* | 4.26 × 10−1 | 1.98 × 10−1 | 3.08 × 10−1 |
| Delay discounting (medium reward; 1,507) | 6.30 × 10−2* | 6.32 × 10−1 | 1.92 × 10−1 | 1.61 × 10−1 |
| Delay discounting (large reward; 1,507) | 2.56 × 10−1 | 9.91 × 10−1 | 6.12 × 10−2* | 3.38 × 10−1 |
For each neuropsychological measurement, the controlled P values of each node were calculated by controlling the remaining nodes. A predominance node was then claimed if its controlled P value was smaller than 0.10. Study sites, gender, and handedness were always controlled for all of the tests conducted. AGN, affective go/no go; SWM, spatial working memory.
The node was classed as predominant if controlled P value was <0.1.
Functional Network and Externalizing Behaviors.
We next explored the relation of the fMRI nodes with indicators of psychopathology by searching for associations with behavioral outcomes relevant for ADHD and addictive behavior (Table 1). These outcomes included measures of hyperactivity from the Strengths and Difficulties Questionnaire (18) and lifetime alcohol consumption from the European School Survey Project on Alcohol and Drugs (19) (Table 3 and Table S4). We found that lower activation in striatal node 1 was associated with higher parent-rated hyperactivity (R = −0.07; Pcorrected = 0.0310; df = 1,523). This association was only observed in boys (R = −0.10; P = 6.53 × 10−3; df = 712). A similar but weaker association was also observed in the small win vs. no win contrast of anticipation phase (SI Materials and Methods) (R = −0.05; P = 0.0439; df = 1,427 in the full sample and R = −0.06; P = 0.104; df = 670 in boys), suggesting that the association strength is proportional to the strength of reward stimuli. The occipital cortical node 2 showed the most significant association with reduced lifetime alcohol consumption (R = −0.09; Pcorrected = 0.0038; df = 1,484). However, alcohol consumption was not only dependent on one node alone but also related to a link between nodes 1 and 2. It was associated with both caudate nucleus (R = 0.09; P = 6.16 × 10−4; df = 1,483) in node 1 and V1/V2 activation (R = 0.08; P = 1.57 × 10−3; df = 1,483) in node 2, which were significantly correlated (R = 0.11; Pcorrected = 9.43 × 10−3; df = 1,514) (Fig. S2A).
Table 3.
Results of the association analyses between fMRI nodes and psychopathological assessment
| Outcomes (df) | Node 1 | Node 2 | Node 3 |
| ADHD symptoms (1,523) | 0.0310 (−0.07)*† | n.s. | n.s. |
| Alcohol usage (1,484) | n.s. | 0.0038 (−0.09)*† | 0.0432 (−0.07)* |
Results were provided as P value (partial correlation, i.e., the effect size).
A node was classed as predominant if its P value after controlling for all other nodes was smaller than 0.10 (Table S4). Study sites, gender, and handedness were controlled.
Table S4.
Tables for fMRI-weighted coactivation network analyses: Results of predominance analysis for psychopathological assessment
| Outcomes (df) | Controlled P values | ||
| Node 1 | Node 2 | Node 3 | |
| ADHD symptoms (1,523) | 7.13 × 10−2* | 5.12 × 10−1 | 5.28 × 10−1 |
| Alcohol use (1,484) | 8.19 × 10−1 | 4.28 × 10−2* | 9.67 × 10−1 |
ADHD symptoms were assessed using the SDQ. Lifetime alcohol consumption was calculated using the European School Survey Project on Alcohol and Drugs. For each neuropsychological measurement, the controlled P values of each node were calculated by controlling the remaining nodes. A predominance node was then claimed if its controlled P value was smaller than 0.10. Study sites, gender, and handedness were always controlled for all of the tests conducted.
The node was classed as predominant if controlled P value was <0.1.
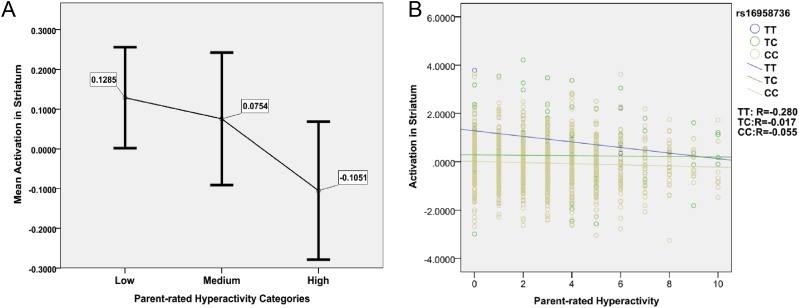
To further assess the relation of activation in node 1 with hyperactivity, we selected extreme cases with clinically relevant hyperactivity scores ≥7 (n = 113; 76 boys), cases with mild hyperactivity scores =4 (n = 179; 91 boys), and cases with no indication of hyperactivity (scores =0) as controls (n = 256; 80 boys; www.sdqinfo.com). In extreme cases vs. controls, we found a twofold increase in the effect size of the correlation between hyperactivity and node 1 activation (R = −0.12; P = 0.0197; df = 358) compared with a quantitative analysis in the full IMAGEN sample (R = −0.069). Similar to the full sample, the association in extreme cases vs. controls was only observed in boys (R = −0.22; P = 7.20 × 10−3; df = 146). We observed a monotonically decreased mean activation in node 1 with higher hyperactivity, providing no evidence for a U-shaped model between hyperactivity and BOLD response (20) (Fig. S3A). There was a highly significant association between impulsivity (as assessed with the Cloninger’s Temperament and Character Inventory, Revised Version) (Table 1) (21) and parent-rated hyperactivity (P = 3.44 × 10−11; df = 1,523), but only a modest explanation of variance (R2 = 0.029).
Fig. S3.
Evaluating the U-shaped model and genetic moderator model between ADHD symptoms and striatal activation. (A) The vertical axis indicates the mean activations in striatum, where the data have been standardized with mean equal to zero and SD equal to 1. The horizontal axis indicates three categories of parent-rated hyperactivity (the severity scores from 0 to 10, where 0 indicates no symptoms), where individuals scored 0 are marked as low to represent the control group, individuals scored 4 are marked as median, and individuals scored 7–10 are marked as high to represent the clinical relevant group. For each group, an error bar is drawn to illustrate the mean, which is shown as the number next to the error bar, and the 95% confidence interval is indicated by the horizontal bar at each end. The figure illustrates that the mean activation in striatum monotonically decreases with higher parent-rated hyperactivity score and therefore, does not support the inverted U-shaped model suggested by Plichta and Scheres (20). Meanwhile, the high group shows significantly lower striatum activation than the low group (P = 0.0197; df = 358). (B) The association between activations in striatum and patent-rated hyperactivity was evaluated in three subsamples in terms of genotypes of rs16958736 of the VPS4A gene, where CC represents homozygous cytosine, TC represent heterozygous thymine cytosine, and TT represents homozygous thymine. The vertical axis represents the activation in striatum, where the data have been standardized in the full sample with mean equal to zero and SD equal to 1. The horizontal axis represents the parent-rated hyperactivity ranging from 0 to 10 (i.e., from low to high).
Genome-Wide Association Study of Reward Sensitivity.
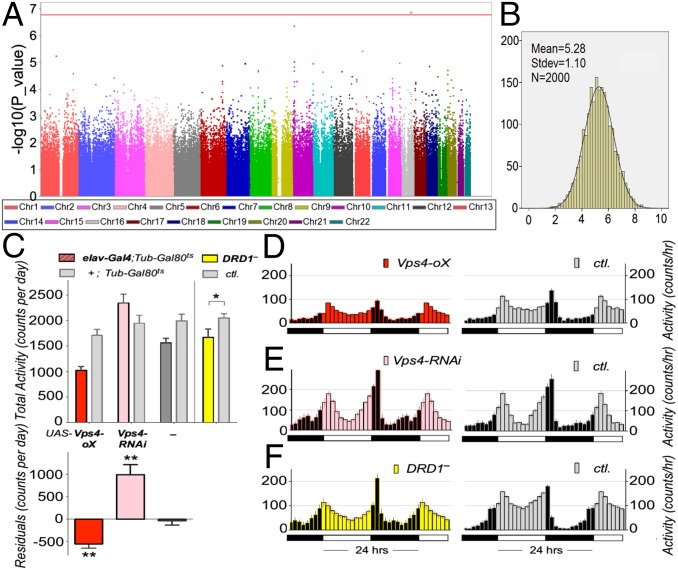
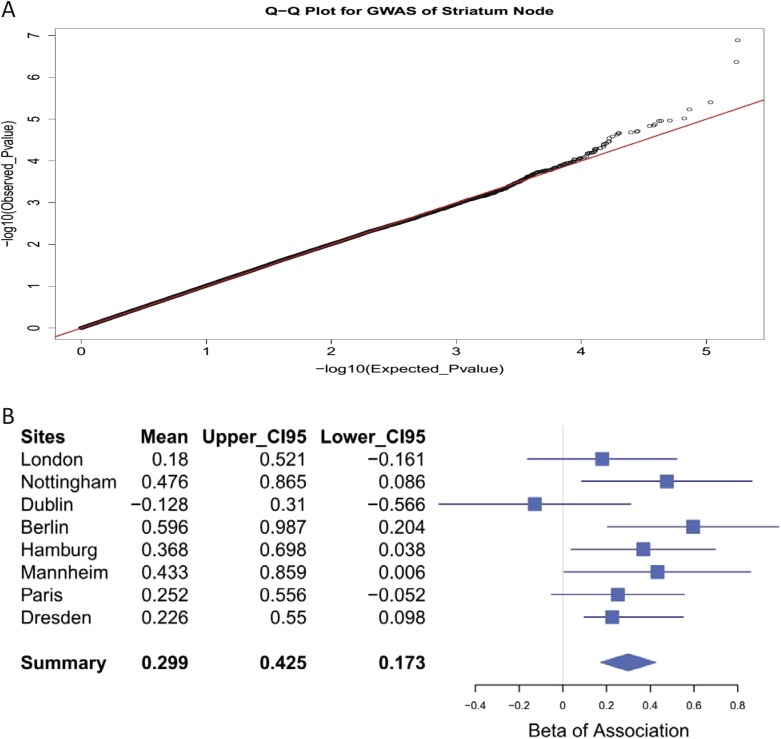
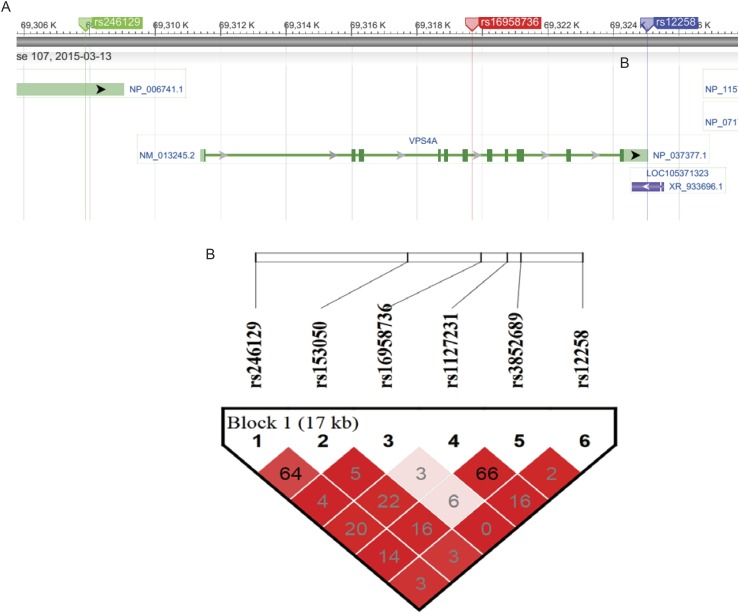
The involvement of the striatum in reward anticipation is well-established, and striatal node 1 was associated with both neuropsychological indicators of dysfunctional reward processing and behavioral symptoms of hyperactivity. Therefore, we carried out a genome-wide association study (GWAS) of node 1 BOLD response during reward anticipation in the IMAGEN sample (n = 1,403). We detected a signal in the sixth intron of the vacuolar protein sorting-associated protein 4A (VPS4A) gene locus, the C/T SNP rs16958736. The major C allele was associated with decreased activation in the striatal node 1 (R = 0.14; P = 1.30 × 10−7) (Fig. 2A and Fig. S4A). Although the VPS4A signal does not reach the commonly used threshold for genome-wide significance (P = 5.00 × 10−8), it remains significant if corrected for the number of independent tests (22), where the 0.05 significance threshold was detected at P = 1.71 × 10−7. It is, thus, a strongly suggestive candidate. A similar but weaker association was also observed in the small win vs. no win contrast of anticipation phase (R = 0.06; P = 0.0366). VPS4A encodes an ATPase involved in trafficking of G protein-coupled receptors, including dopamine receptors (23). VPS4A genotypes did not alter the direction of the correlation between BOLD response in node 1 and hyperactivity (Fig. S3B). The stability of the association of VPS4A with node 1 was supported by both the consistent directional associations across recruitment sites (seven of eight; R = 0.13; P = 3.26 × 10−6 from metaanalysis) and normally distributed t statistics from bootstrapping analyses (R = 0.14; P = 1.53 × 10−7; mean t statistic) (Fig. 2B, SI Materials and Methods, and Fig. S4B). To assess the genetic information of the entire VPS4A locus, we conducted a haplotype analysis, and VPS4A was associated with the striatal node 1 (Table S5) (η2 = 0.02; P = 1.58 × 10−4; omnibus test) as a single haplotype block (SI Materials and Methods and Fig. S5). This association was driven by a positive association of haplotype 4 (R = 0.12; frequency = 0.033; P = 1.33 × 10−5; df = 1,392), and it was mainly observed in boys (R = 0.14; P = 2.29 × 10−4; df = 653) and observed less in girls (R = 0.09; P = 0.019; df = 730).
Fig. 2.
Results for the VPS4A gene. (A) Manhattan plot of GWAS of the striatal node. The red line indicates the 5% genome-wide significance level based on the number of independent tests (22). SNP rs16958736 (Chr16: 69353587 in hg19) in the sixth intron of VPS4A was significant. (B) Histogram of bootstrapping results of the association between node 1 (the striatum) and SNP rs16958736; t statistics follow normal distribution, suggesting no hidden substructure and highly stable results. The mean t statistic of 5.28 (from 2,000 bootstrapping iterations) is equivalent to P = 1.53 × 10−7 (two-tailed test; df = 1,393) (SI Materials and Methods). (C) Locomotion phenotypes in Drosophila mutant strains. (Upper) Total daily locomotion activity of Drosophila expressing UAS transgenes for Vps4 in the nervous system specifically with the elav-Gal4 driver. In both males and females, expression of elav-Gal4 (dark gray bar; group marked by —) reduced activity by 20%. (Lower) We, thus, corrected for this in the experimental elav-Gal4;Tub-Gal80ts flies by dividing their total activity by 0.8, and then, we plotted the difference of the corrected experimental elav-Gal4;Tub-Gal80ts activity from their control +;Tub-Gal80ts activity. This figure shows that Vps4 overexpression (oX) significantly decreased (Cohen’s d = 1.02; t = 5.68; df = 31) and that knockdown (RNAi) significantly increased locomotion activity (Cohen’s d = 1.03; t = 4.51; df = 19). DRD1 mutant flies (yellow) showed hypolocomotion compared with their genetic background-matched control (Cohen’s d = 0.26; t = 2.08; df = 62). *P < 0.05; **P < 0.01. (D–F) Average activity plots of flies from C. Mutants (D) Vps4 overexpression, (E) Vps4 knockdown, and (F) DRD1 mutant are in Left, and their controls are in Right, respectively. Note that, for genetic reasons, we had to use females for Vps4 overexpression, which show a less diurnal activity pattern. ctl., Control.
Fig. S4.
Assessing the reliability of the GWAS finding. (A) The quantile–quantile (Q–Q) plot shows an almost perfect match between the observed and expected P values, which indicates that the applied significance level is reliable. (B) The forest plots of associations between node 1 and SNP rs16958736 in each research site: the regression coefficients (i.e., β or mean) and the corresponding upper and lower 95% confidence intervals (CI95s) of the association between the activation in node 1 and SNP rs16958736 for research sites are listed in Left. In the forest plot in Right, the means or β-values of each research site are shown as blue cubes, and the ranges between upper and lower CI95s are shown as the blue lines. The integrated mean or β as well as its upper and lower CI95s were calculated from the metaanalysis by using the inversion of squared SEs as weight, and the result is illustrated as the blue diamond, where the mean is indicated vertically and the range between upper and lower CI95s is indicated by horizontally. The corresponding P value of the summarized result was 3.26 × 10−6.
Table S5.
Haplotype analysis of VPS4A: Haplotype phases for VPS4A and their associations with node 1 (the striatum) and hyperactivity in the IMAGEN
| Haplotype | Frequency | rs246129 | rs153050 | rs16958736 | rs1127231 | rs3852689 | rs12258 | Striatal node | Hyperactivity in boys |
| Hap1 | 0.0699 | G | C | C | G | G | A | 6.30 × 10−2 | |
| Hap2 | 0.0490 | G | C | T | G | T | G | 1.38 × 10−3* | |
| Hap3 | 0.191 | G | C | C | G | T | G | 3.16 × 10−1 | |
| Hap4 | 0.0326 | G | C | T | A | G | G | 1.33 × 10−5‡ | 2.16 × 10−2† |
| Hap5 | 0.287 | A | T | C | A | G | G | 9.11 × 10−1 | |
| Hap6 | 0.0563 | G | T | C | A | G | G | 4.73 × 10−1 | |
| Hap7 | 0.0291 | A | C | C | A | G | G | 9.11 × 10−1 | |
| Hap8 | 0.272 | G | C | C | A | G | G | 2.39 × 10−1 | |
| Omnibus | 1.64 × 10−04‡ |
The frequency of each haplotype is provided. A, T, G, and C indicate nucleic acid adenine, thymine, guanine, and cytosine, respectively. An omnibus test (F test) was conducted to measure the overall influence of VPS4A haplotypes on the activation in node 1 (i.e., the comparison of models with/without the contribution of all haplotypes). The most significant haplotype phase (i.e., Hap4) with node 1 was then tested for association with hyperactivity in boys, where the hyperactivity and node 1 were observed in significant association.
Significance at level 0.01.
Significance at level 0.05.
Significance at level 0.001.
Fig. S5.
The structure and linkage disequilibrium (LD) plot of VPS4A genes. (A) The structure of the VPS4A gene was generated from the National Center for Biotechnology Information website (www.ncbi.nlm.nih.gov), where the first (rs246129) and last (rs12258) SNPs included in the following haplotype analyses are indicated in green and blue, respectively, and the main genetic finding rs16958736 is highlighted in red within the sixth intron of the VPS4A gene. (B) Haplotype blocks were generated through method solid spine of the LD. The whole VPS4A gene was involved in a single haplotype block. The white to red color scale measures the adjusted LD (D′) from low to high, whereas the number scale measures the squares of correlation coefficients (R2) between alleles from paired SNPs. The D′ is calculated as the LD divided by its theoretical maximum, and thus, D′ ranges between zero and one.
Haplotype Analysis of VPS4A with Hyperactivity.
Haplotype 4 of VPS4A was significantly associated with hyperactivity in boys (R = 0.08; P = 0.0216; df = 935), but there was no association of rs16958736, indicating that, although the VPS4A gene is involved in the regulation of reward sensitivity and hyperactivity, this SNP is likely to be a marker for an undetected causal genetic variation. Because there is no functional neuroimaging sample of comparable magnitude with the MID task, we were forced to restrict replication to VPS4A and hyperactivity in two independent samples (Table S6). In the Saguenay sample (24) of 481 adolescent boys (SI Materials and Methods), we found a significant association of VPS4A with the ADHD symptoms in both the overall haplotypes (η2 = 0.04; P = 0.0239; omnibus test) and haplotype 4 (R = 0.09; Pone tailed = 0.0200; df = 478). In the Avon Longitudinal Study of Parents and Children sample (25) (SI Materials and Methods), we confirmed the association of VPS4A haplotypes with hyperactivity in 2,550 13-y-old boys (η2 = 0.01; P = 0.0271; omnibus test) but not for haplotype 4.
Table S6.
Haplotype analysis of VPS4A: Frequency of haplotype phases for VPS4A in males from the IMAGEN, the Saguenay, and the ALSPAC
| Haplotype | Phases | Frequency | ||
| IMAGEN | Saguenay | ALSPAC | ||
| Hap1 | GCCGGA | 0.070 | 0.0433 | 0.0693 |
| Hap2 | GCTGTG | 0.049 | 0.0672 | 0.0629 |
| Hap3 | GCCGTG | 0.191 | 0.164 | 0.185 |
| Hap4 | GCTAGG | 0.033 | 0.0486 | 0.0475 |
| Hap5 | ATCAGG | 0.287 | 0.230 | 0.274 |
| Hap6 | GTCAGG | 0.056 | 0.0305 | 0.058 |
| Hap7 | ACCAGG | 0.029 | 0.0290 | 0.0375 |
| Hap8 | GCCAGG | 0.272 | 0.376 | 0.259 |
The same haplotype phases with frequency larger than 1% have been detected in all three datasets. The frequency of each haplotype is provided. A, T, G, and C indicate nucleic acid adenine, thymine, guanine, and cytosine, respectively. An omnibus test (F test) was conducted to measure the overall influence of VPS4A haplotypes on the activation in node 1 (i.e., the comparison of models with/without the contribution of all haplotypes). The most significant haplotype phase (i.e., Hap4) with node 1 was then tested for association with hyperactivity in boys, where the hyperactivity and node 1 were observed in significant association.
Table S7.
Author list for the IMAGEN Consortium
| First name | Last name | Institution |
| Lisa | Albrecht | Charite |
| Mercedes | Arroyo | Cambridge University |
| Eric | Artiges | INSERM |
| Semiha | Aydin | Physikalisch-Technische Bundesanstalt |
| Christine | Bach | Central Institute of Mental Health |
| Tobias | Banaschewski | Central Institute of Mental Health |
| Alexis | Barbot | Commissariat à l'Energie Atomique |
| Gareth | Barker | Institute of Psychiatry, Psychology and Neuroscience |
| Arun | Bokde | Trinity College Dublin |
| Zuleima | Bricaud | INSERM |
| Uli | Bromberg | University of Hamburg |
| Ruediger | Bruehl | Physikalisch-Technische Bundesanstalt |
| Christian | Büchel | University of Hamburg |
| Anna | Cattrell | Institute of Psychiatry, Psychology and Neuroscience |
| Patricia | Conrod | Institute of Psychiatry, Psychology and Neuroscience |
| Katharina | Czech | Charite |
| Jeffrey | Dalley | Cambridge University |
| Sylvane | Desrivieres | Institute of Psychiatry, Psychology and Neuroscience |
| Tahmine | Fadai | University of Hamburg |
| Herta | Flor | Central Institute of Mental Health |
| Vincent | Frouin | Commissariat à l'Energie Atomique |
| Jürgen | Gallinat | University Medical Center Hamburg-Eppendorf |
| Hugh | Garavan | Trinity College Dublin |
| Fanny | Gollier Briand | INSERM |
| Penny | Gowland | University of Nottingham |
| Bert | Heinrichs | Deutsches Referenzzentrum für Ethik |
| Andreas | Heinz | Charite |
| Thomas | Hübner | Technische Universität Dresden |
| Albrecht | Ihlenfeld | Physikalisch-Technische Bundesanstalt |
| Alex | Ing | Institute of Psychiatry, Psychology and Neuroscience |
| Bernd | Ittermann | Physikalisch-Technische Bundesanstalt |
| Tianye | Jia | Institute of Psychiatry, Psychology and Neuroscience |
| Jennifer | Jones | Trinity College Dublin |
| Eleanor | Kennedy | Institute of Psychiatry, Psychology and Neuroscience |
| Dirk | Lanzerath | Deutsches Referenzzentrum für Ethik |
| Mark | Lathrop | McGill University and Genome Quebec Innovation Centre |
| Claire | Lawrence | University of Nottingham |
| Hervé | Lemaitre | INSERM |
| Katharina | Lüdemann | Charite |
| Christine | Macare | Institute of Psychiatry, Psychology and Neuroscience |
| Karl | Mann | Central Institute of Mental Health |
| Adam | Mar | Cambridge University |
| Jean-Luc | Martinot | INSERM |
| Eva | Mennigen | Technische Universität Dresden |
| Fabiana | Mesquita de Carvahlo | Institute of Psychiatry, Psychology and Neuroscience |
| Kathrin | Müller | Technische Universität Dresden |
| Frauke | Nees | Central Institute of Mental Health |
| Charlotte | Nymberg | Institute of Psychiatry, Psychology and Neuroscience |
| Marie-Laure | Paillere | INSERM |
| Tomas | Paus | University of Toronto |
| Zdenka | Pausova | University of Toronto |
| Jean-Baptiste | Poline | University of California, Berkeley and Commissariat à l'Energie Atomique |
| Luise | Poustka | Central Institute of Mental Health |
| Erin | Quinlan | Institute of Psychiatry, Psychology and Neuroscience |
| Jan | Reuter | Charite |
| Stephan | Ripke | Technische Universität Dresden |
| Trevor | Robbins | Cambridge University |
| Gabriel | Robert | Institute of Psychiatry, Psychology and Neuroscience |
| Sarah | Rodehacke | Technische Universität Dresden |
| Barbara | Ruggeri | Institute of Psychiatry, Psychology and Neuroscience |
| Barbara | Ruggeri | Institute of Psychiatry, Psychology and Neuroscience |
| Dirk | Schmidt | Technische Universität Dresden |
| Sophia | Schneider | University of Hamburg |
| Florian | Schubert | Physikalisch-Technische Bundesanstalt |
| Michael | Smolka | Technische Universität Dresden |
| Wolfgang | Sommer | Central Institute of Mental Health |
| Rainer | Spanagel | Central Institute of Mental Health |
| Claudia | Speiser | GABO:Milliarium MbH & Co. KG |
| Tade | Spranger | Deutsches Referenzzentrum für Ethik/Institut of Science and Ethics |
| Alicia | Stedman | University of Nottingham |
| Dai | Stephens | University of Sussex |
| Nicole | Strache | Charite |
| Andreas | Ströhle | Charite |
| Maren | Struve | Central Institute of Mental Health |
| Naresh | Subramaniam | Cambridge University |
| Amir | Tahmasebi | University of Toronto |
| David | Theobald | Cambridge University |
| Nora | Vetter | Technische Universität Dresden |
| Helene | Vulser | INSERM |
| Bernadeta | Walaszek | Physikalisch-Technische Bundesanstalt |
| Robert | Whelan | Trinity College Dublin |
| Steve | Williams | Institute of Psychiatry, Psychology and Neuroscience |
| Bing | Xu | Institute of Psychiatry, Psychology and Neuroscience |
| Juliana | Yacubian | University of Hamburg |
| Veronika | Ziesch | Technische Universität Dresden |
Gene Manipulation in Drosophila.
Vps4 is the highly conserved, sole ortholog of two mammalian VPS4 genes (26), and the fly protein has 74% identity and 86% similarity to human VPS4A. We neuronally overexpressed Drosophila Vps4 and found that these flies were hypoactive (P < 0.01; Cohen’s d = 1.02; t = 5.68; df = 31), whereas flies with neuronal Vps4 knockdown showed significant hyperactivity (P < 0.01; Cohen’s d = 1.03; t = 4.51; df = 19) (Fig. 2 C–E). In rodents, the Vps4b paralogue is associated with locomotor activity, dysregulation of the dopamine system, and altered alcohol reward sensitivity (27). Because flies do not have noradrenaline, their catecholaminergic function is restricted to dopamine. Drosophila dopamine receptors—including DRD1—show closest homology to both human DRD and alpha adrenergic receptors (ADRA) and are correlated with locomotor activity (28). We confirmed this by testing Drosophila DRD1 (Drd1; also known as Dop1R1) mutants that were hypoactive compared with control (P < 0.05; Cohen’s d = 0.26; t = 2.08; df = 62) (Fig. 2 C and F). Because in Drosophila locomotion, Vps4 overexpression resulted in the same phenotype as loss of DRD1 function, we were interested in a more detailed investigation of the coexpression patterns of VPS4A and catecholamine genes.
Coregulation Patterns Between VPS4A and Catecholamine Receptors in Humans and Mice.
We measured coexpressions of VPS4A with major pre- and postsynaptic dopamine and noradrenaline receptors in frontocortical postmortem human brain data from BrainCloud (n = 248) (SI Materials and Methods) (29). VPS4A showed a negative correlation with postsynaptic activating DRD1 (R = −0.22; P = 6.48 × 10−4) and a positive correlation with presynaptic inhibitory DRD2S (R = 0.39; P = 1.85 × 10−10). Similarly, in mouse striatum (n = 31) (SI Materials and Methods) (30), we observed a negative correlation of Vps4a and Drd1 expression (R = −0.48; P = 6.80 × 10−3) and a positive correlation of Vps4a and Drd2 expression (R = 0.53; P = 2.07 × 10−3). VPS4A was also significantly correlated with presynaptic ADRA2C expression in both human (R = 0.56; P = 1.40 × 10−21) and mouse data (R = 0.49; P = 5.70 × 10−3).
SI Materials and Methods
Participants.
One thousand five hundred forty-four adolescents (mean age = 14.44 y old; SD = 0.42; range = 12.88–16.44 y old) from the baseline assessment of the IMAGEN sample were included in the WVCNA. The IMAGEN is a longitudinal imaging genetics study in 2,000 healthy adolescents of Caucasian origin. Detailed descriptions of this study have previously been published (12). Of these 1,544 participants, 1,403 (mean age = 14.44 y old; SD = 0.42; range = 12.88–16.44 y old) had genotypic data and were included in the genome-wide association analyses between the fMRI clusters and genotypes. The corresponding dfs of the remaining analyses are provided along with the P values. Gender and imaging sites were included as covariates in all of these analyses. Whenever the fMRI data were included, handedness was additionally controlled.
fMRI Data Acquisition and Analysis.
Structural MRI and fMRI data were acquired at eight IMAGEN assessment sites with 3-T MRI scanners of different manufacturers (Siemens, Philips, General Electric, and Bruker). The scanning variables were specifically chosen to be compatible with all scanners. The same scanning protocol was used in all sites. In brief, high-resolution T1-weighted 3D structural images were acquired for anatomical localization and coregistration with the functional time series. BOLD functional images were acquired with a gradient echo, echo planar imaging sequence. For the MID task, 300 vol were acquired for each participant, and each volume consisted of 40 slices aligned to the anterior commission/posterior commission line (2.4-mm slice thickness and 1-mm gap). The echo time was optimized (echo time = 30 ms; repetition time = 2,200 ms) to provide reliable imaging of subcortical areas.
fMRI data were analyzed with SPM8 (Statistical Parametric Mapping; www.fil.ion.ucl.ac.uk/spm). Spatial preprocessing included slice time correction to adjust for time differences caused by multislice imaging acquisition, realignment to the first volume in line, nonlinearly warping to the Montreal Neurological Institute space [based on a custom echo planar imaging template (53 × 63 × 46 voxels) created out of an average of the mean images of 400 adolescents], resampling at a resolution of 3 × 3 × 3 mm3, and smoothing with an isotropic Gaussian kernel of 5-mm FWHM.
At the first level of analysis, changes in the BOLD response for each subject were assessed by linear combinations at the individual subject level for each experimental condition, and each trial (i.e., reward anticipation high gain) was convolved with the hemodynamic response function to form regressors that account for potential noise variance (e.g., head movement) associated with the processing of reward anticipation. Estimated movement parameters were added to the design matrix in the form of 18 additional columns (three translations, three rotations, three quadratic and three cubic translations, and three translations each with a shift of ±1 repetition time). To analyze the anticipation phase, we contrasted brain activation during anticipation of high win (here signaled by a circle) vs. anticipation of no win (here signaled by a triangle). The single-subject contrast images were then taken to the population-based weighted network analysis.
MID Task for fMRI.
Participants performed a modified version of the MID task to examine neural responses to reward anticipation and reward outcome. The task consisted of 66 10-s trials. In each trial, participants were presented with one of three cue shapes (cue; 250 ms) denoting whether a target (white square) would subsequently appear on the left or right side of the screen and whether 0, 2, or 10 points could be won in that trial. After a variable delay (4,000–4,500 ms) of fixation on a white cross-hair, participants were instructed to respond with a left/right button press as soon as the target appeared. Feedback on whether and how many points were won during the trial was presented for 1,450 ms after the response. Using a tracking algorithm, task difficulty (i.e., target duration varied between 100 and 300 ms) was individually adjusted, such that each participant successfully responded on ∼66% of trials. Participants had first completed a practice session outside the scanner (∼5 min), during which they were instructed that, for each five points won, they would receive one food snack in the form of small chocolate candies. Based on prior research suggesting reliable associations between ADHD symptoms and fMRI BOLD responses measured during reward anticipation, this study used the contrast anticipation of high win vs. anticipation of no win but also, included information from anticipation of small win vs. anticipation of no win when necessary. Only successfully hit trials were included here.
Population-Based Weighted Neuroimaging Network Analysis.
As illustrated in the whole-brain activation map (Fig. S6C) of the anticipation phase of high win vs. no win contrast for the MID task, most parts of the brain are activated. Therefore, it is almost impossible to properly identify activation nodes from another either visually or based on an arbitrary threshold. Meanwhile, the commonly used method [principle component analysis (PCA) and independent component analysis] is not expected to achieve a satisfactory resolution because of the complexity of the high coactivation patterns among brain regions from a task-based fMRI (i.e., no independence among activated brain regions should be hypothesized). A possible solution would be to use predefined functional regions from previous literature. However, the definition of a functional region is normally arbitrary and ambiguous, and it lacks consensus. To cope with the difficulty of defining meaningfully activated regions, we implement the weighted correlation network analysis, which is purely data-driven and initially applied to capture the patterns from a scale-free network (for example, a gene coexpression network or a functional brain network) (45). The robustness of this method has also been proved before (13).
Fig. S6.
The preprocessing and stability assessment of the WVCNA. (A) The plots show the fitness of the scale-free model with different power parameters applied, and the power with the best fitness (power = 8) will be adopted to form the adjacency matrix. (B) The horizontal axis represents the sequence of modules, and the vertical axis represents the corresponding R2 values. In the blue line, each peak represents the original R2 of the corresponding module. In the red line, the averaged R2 is cumulatively calculated as the average of R2 values from module 1 to the current module. The overall average R2 is 0.88. Modules are ordered in decreasing size. (C) The positive t statistic of each voxel is presented with a color scale from red (low t statistic) to white (high t statistic).
The R package WGCNA (42) was implemented to perform WVCNA of the anticipation high win vs. no win contrast of the MID task. Preprocessing steps involved removing null voxels and potential participant outliers from contrast data based on low intersample correlations. The final dataset involved 1,544 participants and 92,119 voxels. Using the scale-free topology criterion, the soft threshold parameter was set to eight (Fig. S6A).
We used an unsigned correlation network, which is based on the absolute value of the pairwise correlation coefficients between voxels. We also explored the use of a signed correlation network but found that it led to very similar results. The corresponding adjacency matrix was then calculated as the power-weighted unsigned correlation matrix. To generate fMRI modules based on average linkage hierarchical clustering, we used a dissimilarity measure based on the topological overlap measure. To cut branches of the resulting clustering tree, we used the dynamic tree cut method.
In total, 1,397 stable modules were generated, and we selected the 30 most stable modules based on the bootstrapping process as described below. Among these modules, modules 12, 13, and 24 (Fig. S1I) were identified as noise (i.e., detected outside of brain area), and modules 8, 14, 20, 25, 27, and 30 (Fig. S1 J and K) were excluded, because they showed a negative BOLD response compared with the whole-brain activation map as illustrated in Fig. S6C. The remaining 21 modules were further examined and labeled using the TalairachClient (www.talairach.org/) to identify the related regions (Tables S1 and S2), and they were visualized in SPM8 (www.fil.ion.ucl.ac.uk/spm/) and MRIcron (www.mccauslandcenter.sc.edu/mricro/). The R package Rniftilib (https://r-forge.r-project.org/R/?group_id=427) was used to help in visualization of the modules. The first eigenvector of each module (i.e., the module eigenvoxel) is generated from singular value decomposition and implemented as the BOLD response of the corresponding module in the following association analyses.
Bootstrapping Process and the Establishment of Core Voxels for Modules.
Command sample() of the R program was used to conduct the random resampling with replacement (i.e., the bootstrapping process) of 1,544 baseline individuals with fMRI data. The resampled individuals were then analyzed in the package WGCNA to establish a bootstrapped coactivation network. This process was iterated 50 times, and the probability that a pair of voxels was regrouped together per iteration was calculated from the stability matrix of each module. The hierarchical clustering was then applied on the stability matrices (i.e., as the similarity matrices, with a cutoff at 0.20, which is equivalent to an 80% chance of regrouping into the same module during each bootstrapping process). For each stability matrix, the largest subcluster was always chosen, and the second largest one was also considered if it was either larger than 100 voxels or larger than one-half of the first subcluster. The subclusters, hence, represent the core voxels of the corresponding module (i.e., the core module). The eigenvoxel of each core module was then calculated through singular value decomposition and compared with the eigenvoxel of the corresponding original module to assess the stability through their shared variance (i.e., the higher R2 that we have, the more stable the investigated module). Fig. S6B illustrates the R2 values of the top 200 modules in size, and the increased variations of R2 values as well as the decreased average R2 can be observed along with the decrease of the module sizes, which indicates that the smaller modules are overall less stable than the larger ones. Meanwhile, the average R2 shows convergence at about 0.88 when module size is small, and therefore, we set a cutoff for the average R2 at 0.94 (i.e., the mean of the theoretical maximum 1.0 and the observed convergence 0.88), which resulted in the top 30 modules in size being selected.
R2 values between the core and original modules as the measurement of stability are preferred rather than simply the percentage of matching shared voxels across bootstrapping processes, because it is the eigenvoxels that meaningfully represent the BOLD response of modules, and therefore, they are the only measurement that matters for all of the following analyses.
Partial Correlation Matrix and the Clustering of Modules.
Of these 21 stable modules, paired correlations were fairly high, which is because of the fact that task-related brain activations were virtually stimulated by the same event (i.e., the anticipation phase of reward in the MID task). We, therefore, adopted the partial correlation strategy to capture the real functional connections between any pair of 21 stable modules by controlling for the contributions from all of the remaining modules as well as for gender, imaging site, and handedness. The thus-established functional connection matrix (Fig. S2A) was then used to generate the distance matrix [i.e., (1 − abs(functional connection matrix))/2] for hierarchical clustering, where abs indicates absolute value. This process resulted in four nodes representing clusters of correlated brain activation (Fig. 1 and Fig. S2B).
We then calculated the first principal component (FPC) for each of these four functional nodes to represent the amount of variability in BOLD response that they explain. The FPCs explained 80.67% variance of the first node, 85.79% variance of the second node, 71.16% variance of the third node, and 68.35% variance of the fourth node.
In the small win vs. no win contrast of anticipation phase, the same modules for caudate, putamen, and nucleus accumbens acquired from the high win vs. no win contrast were extracted, and their FPC (77.78% variance explained) was used to represent the striatal activation in the corresponding analyses.
Neuropsychological Testing and Measures of Personality.
Participants underwent neuropsychological testing that included the Cambridge Neuropsychological Test Automated Battery (CANTAB) (www.cambridgecognition.com) and the Delay Discounting Test [Monetary Choice Questionnaire by Kirby and Marakovic (16)]. The CANTAB is widely used for assessing various neurocognitive functions and processes, such as executive function/cognitive control, attention, and decision-making. Measures from the CANTAB and the work by Kirby and Marakovic (16), which have previously been related to reward processing, were selected to assess emotional processing (the affective go/no go task) and executive function/cognitive control (spatial working memory task). The descriptive statistics of the above outcome variables are listed in Table 1, and the corresponding distributions are illustrated in Fig. S7 A–F.
Fig. S7.
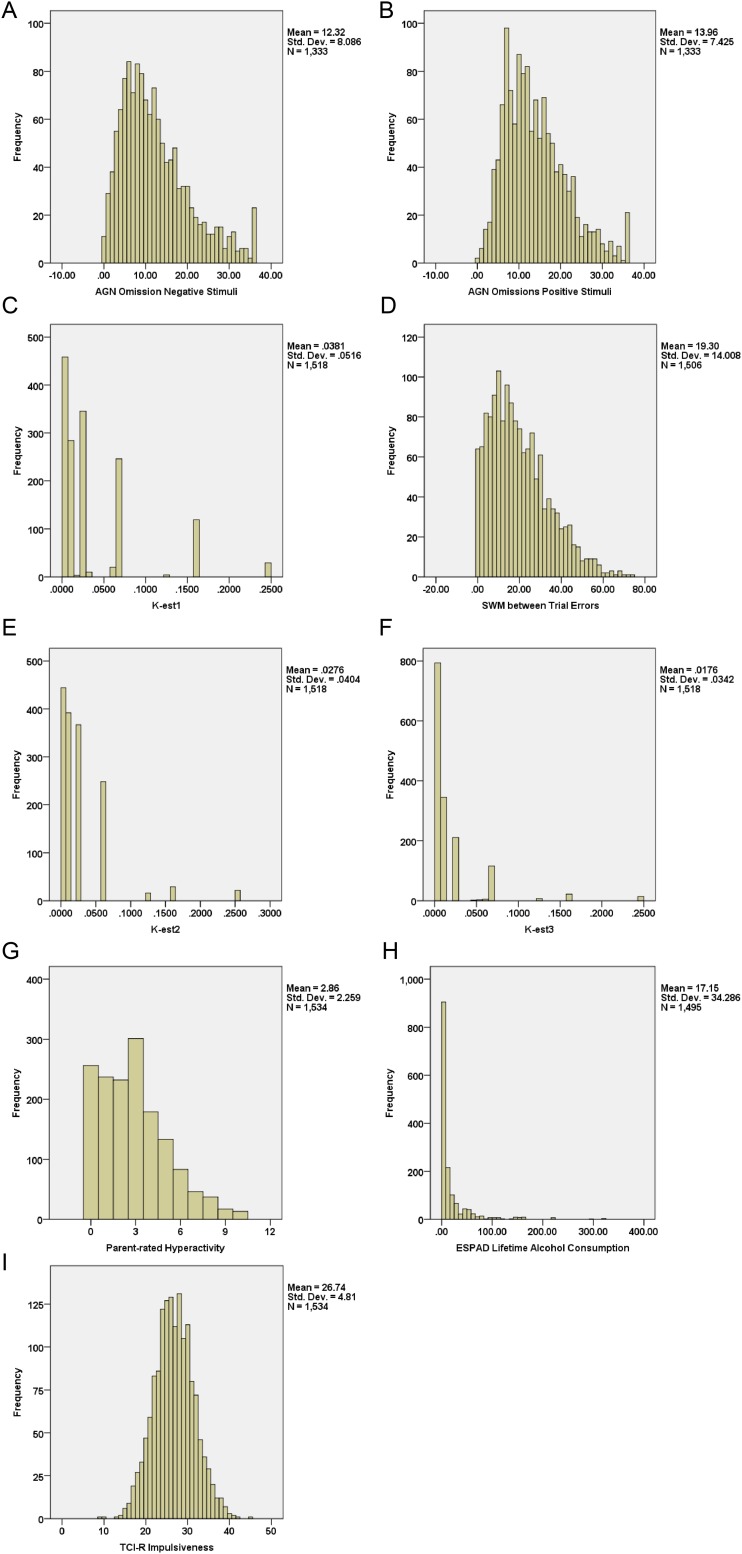
Histograms illustrating the distributions of the main variables. Main neuropsychological variables were the CANTAB affective go/no-go (AGN) task number of errors of omission to (A) positive and (B) negative stimuli and (C) spatial working memory (SWM) task number between trial errors. Main questionnaire variables were (D) k-est1 = estimated delay discounting rates for small long delay rewards, (E) k-est2 = estimated delay discounting rates for medium long delay rewards, and (F) k-est3 = estimated delay discounting rates for large long delay reward from the Monetary Choice Questionnaire, (G) the hyperactivity total scale scores from the SDQ, and (H) lifetime alcohol consumption from the European School Survey Project on Alcohol and Drugs (ESPAD). (I) In addition, we also included the measurement of impulsiveness from Cloninger’s Temperament and Character Inventory, Revised Version (TCI-R). For each variable, the mean, SD, and sample size were provided.
An adapted version of the affective go/no go task consisting of 13 blocks in 18 trials was used here. Participants saw a word, either a target or a distractor (in 50% of the trials), flashed onto the center of the screen for 300 ms, with an interstimulus interval of 900 ms. Target words were of positive (e.g., hopeful or alive), anxiety-related (e.g., stress or sinister), or negative (e.g., rejection or ashamed) content. Distractor words were of neutral content (e.g., contrast or primary). Participants were given a target category and asked to indicate whether the word presented matched the target category by pressing the touch pad located in front of them and inhibit the response whenever a distractor is shown. Mean latency (i.e., time taken to respond to positive or negative targets) and number of omissions (i.e., failure to respond to positive or negative target stimuli) were examined in this study. Higher latency and numbers of omissions are indicative of greater emotional bias. The task lasted ∼8 min.
The spatial working memory task is a self-ordered search task assessing the participant’s ability to retain spatial information and manipulate working memory content. Participants were required to search through boxes for a blue hidden token by touching each box one by one on the screen. Participants were instructed that only one token was hidden in each search, and after a token was found, the next token will be hidden in another box; a box previously found to have a hidden token will not contain another token in the following searches. Therefore, participants should continue searching in the rest of the boxes until hidden tokens had been found in all boxes. The number of boxes increased throughout the task: three, four, six, and eight boxes were presented four times each (i.e., four trials for each set of boxes). The color and position of the boxes were altered from trial to trial to minimize the use of fixed search strategies (e.g., always starting with a red box). An error was recorded whenever participants revisited boxes in which tokens have been found previously (labeled between-search error and used to evaluate working memory performance). The task lasted ∼8 min, but there was no time limit for each search. A demonstration of this task is available from the CANTAB website (www.cambridgecognition.com/tests/working-memory-swm).
The Monetary Choice Questionnaire investigates delay discounting performance that measures reward-related impulsivity (16). Subjects are asked to choose from either an immediate smaller reward or a larger reward in the future (16). Participants are presented a fixed set of 27 choices between smaller, immediate rewards and larger, delayed rewards. Sample items are “would you prefer £54 today or £55 in 117 d?” and “would you prefer £55 today or £75 in 61 d?” Questions are grouped into three groups of nine items each depending on the value of reward: small (£25–£35), medium (£50–£60), and large (£75–£85) rewards. Delay discounting is the reduction of the value of a future reward as the delay to that reward increases. The hyperbolic discount parameter (k) was calculated for each participant for high, medium, and low rewards; k can be thought of as an indication for impulsiveness (increasing from k-est1 to k-est3), whereby higher values refer to higher levels of impulsiveness.
Assessment of Problem Behaviors.
The Strength and Difficulties Questionnaire (SDQ) (18) is a brief 25-item behavioral screening tool probing hyperactivity, emotional symptoms, conduct problems, peer problems, and prosocial behavior for 3–16 y olds. In this study, we chose parent-rated hyperactivity, and it has been shown that externalizing problems (e.g., hyperactivity) are more reliably assessed by parents than children themselves (46). Although named as hyperactivity, this sum score of five items actually measures both inattention and hyperactivity and therefore, is commonly used as a screening tool of ADHD in adolescents.
Lifetime drinking phenotypes (i.e., frequency of drinking, quantity of drinking on a typical day, and frequency of binge drinking) were defined using the European School Survey Project on Alcohol and Drugs Questionnaire (19). These phenotypes were then used to calculate the lifetime alcohol consumption index using the method introduced by Stahre et al. (47) with minor adaptions for the European School Survey Project on Alcohol and Drugs Questionnaire. The descriptive statistics of the above outcome variables are listed in Table 1, and the corresponding distributions are illustrated in Fig. S7 G and H.
Predominant fMRI Nodes.
Because all fMRI nodes responded to the same stimuli, they were highly correlated to each other. To remove shared variance and identify the predominant association of a node with a behavioral measure, we carried out partial correlation analyses. First, each node was tested for association with the phenotype of interest by controlling all of the rest nodes. Second, if one and only one node reached trend-level significance (i.e., P < 0.1), this node would be labeled as predominant.
Measurement of Impulsivity.
The Novelty-Seeking Scale of the revised version of Cloninger’s Temperament and Character Inventory, Revised Version (21), a 51-item self-report questionnaire, was applied to assess dimensions of temperament. Items are answered on a five-point scale (i.e., definitely false to definitely true). Scores on each subscale (i.e., exploratory excitability, extravagance, disorderliness, and impulsiveness) are combined in a total novelty-seeking score. The impulsiveness subscale was selected as a measure of impulsivity to test for its association with parents-rated hyperactivity from the SDQ. The descriptive statistics of the Temperament and Character Inventory, Revised Version impulsiveness are listed in Table 1, and the corresponding distribution is illustrated in Fig. S7I.
Genotyping.
DNA purification and genotyping were performed by the Centre National de Génotypage. DNA was extracted from whole-blood samples (∼10 mL) preserved in BD Vacutainer EDTA Tubes (Becton, Dickinson and Company) using the Gentra Puregene Blood Kit (QIAGEN Inc.) according to the manufacturer’s instructions. Genotype information was collected at 582,982 markers using the Illumina HumanHap610 Genotyping BeadChip (Illumina) as part of a previous GWAS (12).
SNPs with call rates of <98%, minor allele frequency <1%, or deviation from the Hardy–Weinberg equilibrium (P < 1.00 × 10−4) were excluded from the analyses. Individuals with an ambiguous sex code, excessive missing genotypes (failure rate >2%), and outlying heterozygosity (heterozygosity rate of 3 SDs from the mean) were also excluded. Identity by state similarity was used to estimate cryptic relatedness for each pair of individuals using PLINK software (pngu.mgh.harvard.edu/∼purcell/plink/). Closely related individuals with identity by descent >0.1875 were eliminated from the subsequent analysis. Population stratification for the GWAS data was examined by PCA using EIGENSTRAT software (genetics.med.harvard.edu/reich/Reich_Lab/Software.html). The four HapMap populations were used as reference groups in the PCA, and individuals with divergent ancestry (from Utah residents with ancestry from northern and western Europe, i.e., CEU) were also excluded.
Association Analyses Between fMRI Modules, Genetic Data, and Phenotypes.
The Efficient Mixed Model Association Expedited (EMMAX) was implemented to perform the GWAS for the eigenvoxels of selected fMRI modules (genetics.cs.ucla.edu/emmax/). The effective number of independent test for the GWAS was calculated by the genetic type 1 error calculator as 292,884.5 of 506,932 SNPs, and it yielded a Bonferroni corrected genome-wide significance level of 5% as 1.71 × 10−7 (22). The Manhattan and the quantile–quantile (Q–Q) plots for the log10-transformed P values of the striatum node (node 1) from the GWAS are illustrated in Fig. 2A and Fig. S4A. The Q–Q plot shows an almost perfect match between the observed and expected P values, which indicates that the applied significance level is reliable.
Haplotype blocks were generated and illustrated through software Haploview (https://sourceforge.net/projects/haploview/) using the solid spine of the linkage disequilibrium method with a parameter of 0.80. Haploview was also used to generate the Manhattan plot of GWAS results from EMMAX. The haplotype phases were estimated through the software PLINK (pngu.mgh.harvard.edu/∼purcell/plink/), where the probabilities of haplotype phases (i.e., the dosage value) were always included unless otherwise specified.
Linear regression was implemented to detect the association between the eigenvoxels of fMRI modules and phenotypes. All of the association analyses, except for the GWAS, were conducted in R program unless otherwise specified.
Throughout the paper, the uncorrected P values are reported unless specified (for example, in case of multiple comparisons, either a corrected P value or a threshold at a significance level of 0.05 after correction is provided). Two-tailed tests are always applied unless otherwise specified.
Internal Replication of the GWAS Finding.
To investigate the stability of the association between the activation in node 1 and the VPS4A SNP rs16958736, we have carried out two internal replications.
First, we tested the same association in each eight research sites separately in PLINK. The regression coefficients (β) and their corresponding SEs were then used to conduct the metaanalysis. The R package rmeta (https://cran.r-project.org/web/packages/rmeta/rmeta.pdf) was then used to generate the forest plot.
Second, a resampling of the original data with replacement (a bootstrapping process) was conducted to generate a new sample with the same sample size as the original data. The process was replicated 1,403 times, and all of the t statistics were summarized in a histogram to illustrate the stability of the association of interest.
Saguenay Sibling Sample and the Validation of VPS4A Findings.
The procedures for recruitment have been described previously (24). Recruitment and all assessments took place in the Saguenay–Lac Saint Jean region of Quebec, Canada between 2003 and 2012. Adolescents and their maternal and paternal grandparents were of white Caucasian French–Canadian ancestry born within the region. Adolescents had to be between the ages of 12 and 18 y of age and have at least one sibling of the same age group. The study was approved by the ethics committee at Montreal Neurological Institute, McGill University, and details of the sample have been described elsewhere (24).
The ADHD in Saguenay sample was assessed by the DISK (diagnostic interview schedule with children) predictive scales (48). The same haplotype block of VPS4A was constructed in 1,028 Saguenay samples through PLINK based on the genotyped and imputed genetic data, where the same haplotype phases were acquired with similar frequencies to those of the IMAGEN sample (Table S6). The association between VPS4A haplotypes and ADHD symptoms in 496 boys was calculated based on the omnibus test in R by comparing linear models with and without all eight haplotypes.
Hap4 of VPS4A, the main finding in the IMAGEN sample, was selected to test for association with ADHD symptoms through a mixed model from the EMMAX, where the identity by descent matrix, based on record, was implemented to correct for the relatedness between siblings. Because of the limitation of the EMMAX, the best estimated haplotype phases were used here instead of its dosage value.
In all above analyses, maternal exposure to cigarette smoking and age of participants were included as the covariates.
Avon Longitudinal Study of Parents and Children Sample and the Validation of VPS4A Findings.
The Avon Longitudinal Study of Parents and Children (ALSPAC) included data on 13,971 babies with expected delivery dates between April of 1991 and December of 1992. The study was approved by the ethics committee at the University of Bristol, and detailed information can be found elsewhere (25).
In the ALSPAC sample, the SDQ was used to assess ADHD at 9, 11, 13, and 16 y old. In this paper, the data at 13 y old are of particular interest, because they are the best match of our 14-y-old individuals. The parent-rated hyperactivity was used for the following association analyses.
The same haplotype block was constructed in 8,358 ALSPAC individuals, and the same haplotype phases were acquired with similar frequencies to those from the IMAGEN (Table S6). The association between VPS4A haplotypes and parent-rated hyperactivity was calculated based on the omnibus test in R by comparing linear models with and without all eight haplotypes. The first 10 principle components were included in the regression models as control variables for the population stratification.
After matching for individuals with nonmissing data of hyperactivity at 13 y old, covariates, and genetic data, only 5,132 individuals were left, of which 2,550 are boys.
Drosophila Experiments.
Locomotion activity was measured in the Drosophila Activity Monitor System (Trikinetics) at 25 °C in 12:12-h light–dark cycles for 3 d. Activity was measured in 30-min bins and aggregated into 1-h bins (Fig. 2 D–F). Flies were entrained to the light–dark regimen for at least 3 d. Males were used when possible, but for Vps4 overexpression, we assayed females (because both Vps4[EP] and elav-Gal4 are on the X chromosome), which show less marked crepuscular behavior than males (i.e., do not take a siesta) as previously reported (43). The various transgenes were obtained from the Bloomington Stock Center (Vps4[EP.G524] for overexpression and [TRiP.HM04061] for knockdown). Experimental (elav-Gal4;Tub-Gal80[ts];UAS-transgene) and control (+;Tub-Gal80[ts];UAS-transgene) flies were in the same hybrid genetic background (Berlin/transgene), reared at room temperature, and shifted to 29 °C for 2 d before activity measurements to increase transgene expression. The DRD1 receptor loss-of-function mutant used was allele f02676, and it was outcrossed to w Berlin (44).
For Vps4 experiment, the expression of elav-Gal4 (dark gray bar in the group marked — in Fig. 2C, Upper) reduced activity by 20% in both males and females. To compensate for this effect, we divided all elav-Gal4;Tub-Gal80ts locomotion activity levels by 0.8, from which the residuals were calculated by subtracting the corresponding +;Tub-Gal80ts locomotion activity levels (Fig. 2C, Lower). The third residual bar (group marked — in Fig. 2C, Upper) shows that the above normalization procedure properly corrects for reduced activity level introduced by elav-Gal4.
BrainCloud Database.
The BrainCloud database contains 269 postmortem subjects with both genome-wide DNA and RNA data (29). Although the RNA data were collected from the prefrontal cortex but not from the subcortical striatum area, they should still properly reflect the general coexpression patterns in the human brain; 21 individuals with RNA integrity <7.0 were removed, and in the following coexpression analysis, age, gender, and RNA integrity were used as covariates. We choose to look at the G protein-coupled receptors of dopamine and noradrenaline, which have been closely linked to ADHD (49). Among these receptors, dopamine receptors DRD1, DRD2, and ADRA2C were selected, because they were reported to be the main receptors expressed in striatum area (50, 51). For DRD2, two probes were included to target two different isoforms (DRD2S and DRD2L), where the former probe targets the 3′ UTR of both isoforms, whereas the latter probe targets the sixth exon that was only included in the long isoform. Because no coexpression was observed between DRD2L and VPS4A (R = 0.023; P = 0.704), we only focused on DRD2S, which expresses presynaptically, in the following analyses.
Mouse Expression Data (Human Brain Project/Rosen Striatum M430v2 Probe Dependent Nearest Neighbors Clean Dataset).
The caudate nucleus of forebrain was extracted from 33 lines of mice. Approximately 250 brain samples were taken from both males and females. The platform Affymetrix Mouse Genome 430 2.0 short oligomer microarrays were used for the measure of mRNA expression. Two strains of mouse were removed during the quality control process. Detailed information of these data can be found in ref. 30, and the data are available at www.genenetwork.org/home.html. When multiple probes were available for the same gene, we only focused on the one targeting 3′ UTR to keep consistent with human data.
Discussion
We describe a coordinated neural network, which is activated on response to anticipated reward. This network involves a core subcortical node in the striatum (node 1) as well as accessory cortical nodes in the visual association cortex (node 2) and somatosensory cortex (node 3). In our population-based sample, BOLD responses in these nodes were preferentially associated with either ADHD symptoms (node 1) or lifetime alcohol consumption (node 2). Increased BOLD response in node 1 was also associated with better short-term working memory and reduced delay discounting, which together with its localization, may suggest a contribution to the initiation and monitoring of goal-directed behaviors (5). This node is likely to work in concert with cortical nodes 2 and 3 (6, 7) to execute motivated, planned behaviors. BOLD response in the occipital visual node 2 was correlated with affective processing and stimulus expectancy (7). Its link with the striatal reward circuit in node 1 was associated with alcohol consumption. These findings may emphasize the joint modulation of reward-related function and dysfunction by attentional affective and motivational factors. The sensorimotor areas, such as the supplementary motor area, in node 3 were not predominantly associated with externalizing behavior, and they may be viewed as output modules that are driven by the valence and arousal of the rewarding stimuli and instigate motor responses but also, may act back on the striatal reward node 1 (5). This hypothesis was supported by the correlation of BOLD response in node 3 with delay discounting, predominantly to large rewards. Thus, we hypothesize that node 1 may act together with nodes 2 and 3 associated with perception, cognition, and motor control to process reward. Specificity of the reward anticipation network for these different reward-related cognitive and externalizing behavioral symptoms may be provided by distinct network configurations, which are reflected in different association patterns of nodes and their connecting links. Reward-related disorders, including alcohol use disorders and ADHD, have strong comorbid relations. Comorbidity between ADHD and alcohol abuse in adults of 12.9% (31) and 61–64% in adolescence (32) suggests relatedness of the neural mechanisms underlying these disorders, which to date, are diagnosed separately and treated differently.
It is a limitation of this manuscript that some of the proposed functions of the nodes identified were partly based on reverse inference (33). Although we are aware of the potential logical fallacy associated with this method, its application was inevitable to help interpret our findings from data-driven analyses. Nevertheless, we tried to minimize the risk for inaccurate interpretations by increasing prior probability through linking tasked-based data with neuropsychological and behavioral measures that are directly related to the psychological construct interrogated in the task setting (i.e., reward processing) (34). In addition, reward anticipation involves a number of additional psychological functions, such as higher attention, salience attribution, valuation, and arousal, which are inherent in reward anticipation and may have contributed to the observed associations.
We found a negative correlation of hyperactivity and BOLD response in the striatal node 1, which is consistent with previous studies in both clinical (2, 20) and nonclinical samples (35). However, it has been suggested that, in population-based samples, impulsivity measures show mostly positive correlation with VS activation during reward anticipation, whereas in clinical studies of ADHD patients, often a negative correlation is observed (20). Plichta and Scheres (20) have proposed three models explaining these relations, which assume an inverted U shape, (genetic) moderators, or unrelatedness to account for the observed inconsistencies. In the IMAGEN dataset, we did not find evidence for either a U-shaped distribution (Fig. S3A) or genetic moderation by VPS4A genotypes (Fig. S3B) of the correlation of hyperactivity and VS activation. However, in a previous investigation, we have observed genetic moderation by monoamine oxidase A (MAOA) genotype of the interplay of inferior frontal gyrus and VS activation on ADHD symptoms (2). The third model, which assumes unrelatedness, accounts for the possibility “that the blunted VS response as observed in individuals with ADHD is related to additional disease-related factors and not trait impulsivity per se” (20). We found highly significant association of hyperactivity and trait impulsivity, but 97% of the variance of hyperactivity could not be explained by the Temperament and Character Inventory, Revised Version impulsivity measure. Although these analyses are limited by the fact that the extreme cases were selected from a population-based sample as opposed to a clinical ADHD sample, they indicate the presence of additional stochastic factors, which contribute to the emergence of ADHD symptoms. Despite their statistical significance, the effect sizes between fMRI nodes and neuropsychological tasks and behaviors were smaller than those detected in clinical samples (20). Our data were population-based and not selected for differences in hyperactivity and/or impulsivity, which would be the case for clinical ADHD case control studies. Comparing participants with high ADHD symptoms vs. controls, we observed a doubling of the effect size. It is also important to note that behavioral constructs, such as hyperactivity, are not optimized to reflect neural processes, thus limiting the degree of correlation with measures of brain activation.
VPS4A is well-conserved across species (26) and highly expressed in the brain (ds.biogps.org/?dataset=GSE1133&gene=27183). It encodes a member of the ATPases associated with diverse cellular activities (AAA) family involved in the late steps of the generating endosomal multivesicular bodies (MVBs) by promoting membrane invagination and scission from the limiting endosome membrane (36). MVBs are then delivered to lysosomes for protein degradation or recycled to the plasma membrane (37). In neurons, MVBs are involved in trafficking of neurotransmitter receptors after endocytosis, including dopamine and noradrenaline G protein-coupled receptors in the striatum (23). Although we did not find an association of rs16958736 with VPS4A expression, we observed significant correlations of VPS4A expression and expressions of dopamine and noradrenaline receptors in both human frontal cortex and mouse striatum. These brain regions were investigated as exemplary given that dopaminergic and noradrenergic gene expression and extracellular responses (e.g., to alcohol treatment) are comparable between striatum and other brain areas (38, 39). Increased expression of VPS4A is correlated with decreased expression of postsynaptic DRD1 and increased expression of presynaptic DRD2 and ADRA2C, thus possibly altering reward sensitivity and level of activity (27, 28). The observed coregulation patterns are consistent with the result of the manipulation of Vps4 and Drd1 in Drosophila, where Vps4 and Drd1 have opposing effects on activity. They also conform to the predominant association of the striatal node 1 with ADHD symptoms and ADHD symptoms with VPS4A. Together, these data give rise to the hypothesis that VPS4A affects activity and reward processing by regulating brain catecholaminergic systems, including D1 and D2 dopaminergic and α2C adrenergic receptors. Nevertheless, detailed molecular and physiologic studies are required to further investigate possible functional relations between VPS4A and signal transductions of catecholamine.
Our approach to neurobehaviorally characterize reward anticipation can potentially be extended to other behavioral domains and help contribute to a reclassification of mental disorders (40). It may facilitate the identification of causal neural mechanisms important for the identification of previously unidentified targets and repurposing of existing drugs as well as the establishment of neurobehaviorally informed end points for clinical trials (41).
Materials and Methods
Details are in SI Materials and Methods.
MID Task.
During the anticipation phase of the MID task, cues are shown to indicate that no reward, a small reward, or a large reward might be won during the trial. Participants are then shown a target stimulus and asked to respond to the target to gain the reward. We analyzed the contrast comparing the BOLD signal during anticipation of large (or small) rewards with the BOLD signal during anticipation of no rewards; the only difference between the two conditions was the presented target stimuli.
WVCNA.
The R package WGCNA (42) was implemented to perform the WVCNA of the contrasts of the MID task. The final dataset involved 1,544 participants and 92,119 voxels after removing null data and outliers. Using the scale-free topology criterion, the soft threshold parameter was set to eight (Fig. S6A). The stabilities of generated modules were assessed through bootstrapping (Fig. S6B).
GWAS and Haplotype Analysis.
The Efficient Mixed Model Association Expedited was implemented to perform the GWAS for the eigenvoxels of selected fMRI modules (genetics.cs.ucla.edu/emmax/). Haplotype blocks were generated and illustrated through the software Haploview (https://sourceforge.net/projects/haploview/) using the solid spine of the linkage disequilibrium method with parameter 0.80. Haploview was also used to generate the Manhattan plot of GWAS results from Efficient Mixed Model Association Expedited (Fig. 2A). The haplotype phases were estimated through software PLINK (pngu.mgh.harvard.edu/∼purcell/plink/).
Drosophila Experiments.
Locomotion activity was measured in the Drosophila Activity Monitor System (Trikinetics) at 25 °C in 12:12-h light–dark cycles for 3 d. Activity was measured in 30-min bins and aggregated into 1-h bins (Fig. 2 D–F). Males were used where possible, but for Vps4 overexpression, we assayed females (because both Vps4[EP] and elav-Gal4 are on the X chromosome), which show less marked crepuscular behavior than males (i.e., do not take a siesta) as previously reported (43). Experimental (elav-Gal4;Tub-Gal80[ts];UAS-transgene) and control (+;Tub-Gal80[ts];UAS-transgene) flies were in the same hybrid genetic background (Berlin/transgene). The DRD1 receptor loss-of-function mutant used was allele f02676, and it was outcrossed to w Berlin (44).
Ethical Approval.
The IMAGEN Study was approved by local ethics research committees at each research site: King’s College London, University of Nottingham, Trinity College Dublin, University of Heidelberg, Technische Universität Dresden, Commissariat à l'Energie Atomique et aux Energies Alternatives, and University Medical Center. Informed consent was sought from all participants and a parent/guardian of each participant.
Acknowledgments
This work received support from the following sources: the European Union-funded FP6 Integrated Project IMAGEN (Reinforcement-related behaviour in normal brain function and psychopathology) LSHM-CT- 2007-037286; the FP7 Projects IMAGEMEND (602450; Imaging Genetics for Mental Disorders) and Multidisciplinary Approaches to Translational Research in Conduct Syndromes (603016); Innovative Medicine Initiative Project EU-AIMS 115300-2; Medical Research Council Grants 93558 (Developmental pathways into adolescent substance abuse) and MR/N000390/1 (Consortium on Vulnerability to Externalizing Disorders and Addictions); the Swedish funding agency FORMAS (Forskningsrådet för Miljö, Areella näringar och Samhällsbyggande); the National Institute for Research Biomedical Research Centre at South London and Maudsley National Health Service Foundation Trust and King’s College London; Bundesministerium für Bildung und Forschung Grants 01GS08152, 01EV0711, eMED SysAlc01ZX1311A, and Forschungsnetz AERIAL (Addiction: Early Recognition and Intervention Across the Lifespan); NIH Grant R01 MH085772-01A1 (Axon, Testosterone and Mental Health during Adolescence); and NIH Consortium Grant U54 EB020403 supported by a cross-NIH alliance that funds Big Data to Knowledge Centres of Excellence. The Drosophila experiments were supported by NIH Grants T32 DA007290 (to D.A.G.), R01AA019526 (to A.R.), and R21AA022404 (to A.R.).
Footnotes
Conflict of interest statement: T.B. has served as an adviser or consultant to Eli Lilly, Hexal Pharma, Medice, Novartis, Otsuka, Oxford Outcomes, PCM Scientific, Shire, and Vifor Pharma; he has received conference attendance support, conference support, or speaking fees from Eli Lilly, Medice, Novartis, and Shire, and he is involved in clinical trials conducted by Eli Lilly, Shire, and Vifor Pharma. G.J.B. has received honoraria for teaching from GE Healthcare and has served as a consultant for IXICO. A.L.W.B. has received funding from Science Foundation Ireland. T.R. has served as a consultant for Cambridge Cognition, Eli Lilly, Lundbeck, Otsuka, Shire, and Teva; he has received research support from Eli Lilly, GlaxoSmithKline, and Lundbeck; editorial honoraria from Elsevier and Springer-Verlag; educational speaking honoraria from Merck, Sharp, and Dohme; and royalties from Cambridge Cognition. The other authors report no financial relationships with commercial interests.
This article is a PNAS Direct Submission.
Data deposition: IMAGEN data are available from a dedicated database: https://imagen.cea.fr.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1503252113/-/DCSupplemental.
Contributor Information
Collaborators: Lisa Albrecht, Mercedes Arroyo, Eric Artiges, Semiha Aydin, Christine Bach, Tobias Banaschewski, Alexis Barbot, Gareth Barker, Arun Bokde, Zuleima Bricaud, Uli Bromberg, Ruediger Bruehl, Christian Büchel, Anna Cattrell, Patricia Conrod, Katharina Czech, Jeffrey Dalley, Sylvane Desrivieres, Tahmine Fadai, Herta Flor, Vincent Frouin, Jürgen Gallinat, Hugh Garavan, Fanny Gollier Briand, Penny Gowland, Bert Heinrichs, Andreas Heinz, Thomas Hübner, Albrecht Ihlenfeld, Alex Ing, Bernd Ittermann, Tianye Jia, Jennifer Jones, Eleanor Kennedy, Dirk Lanzerath, Mark Lathrop, Claire Lawrence, Hervé Lemaitre, Katharina Lüdemann, Christine Macare, Karl Mann, Adam Mar, Jean-Luc Martinot, Eva Mennigen, Fabiana Mesquita de Carvahlo, Kathrin Müller, Frauke Nees, Charlotte Nymberg, Marie-Laure Paillere, Tomas Paus, Zdenka Pausova, Jean-Baptiste Poline, Luise Poustka, Erin Quinlan, Jan Reuter, Stephan Ripke, Trevor Robbins, Gabriel Robert, Sarah Rodehacke, Barbara Ruggeri, Barbara Ruggeri, Dirk Schmidt, Sophia Schneider, Florian Schubert, Michael Smolka, Wolfgang Sommer, Rainer Spanagel, Claudia Speiser, Tade Spranger, Alicia Stedman, Dai Stephens, Nicole Strache, Andreas Ströhle, Maren Struve, Naresh Subramaniam, Amir Tahmasebi, David Theobald, Nora Vetter, Helene Vulser, Bernadeta Walaszek, Robert Whelan, Steve Williams, Bing Xu, Juliana Yacubian, and Veronika Ziesch
References
- 1.Knutson B, Wimmer GE. Splitting the difference: How does the brain code reward episodes? Ann N Y Acad Sci. 2007;1104:54–69. doi: 10.1196/annals.1390.020. [DOI] [PubMed] [Google Scholar]
- 2.Nymberg C, et al. IMAGEN Consortium Neural mechanisms of attention-deficit/hyperactivity disorder symptoms are stratified by MAOA genotype. Biol Psychiatry. 2013;74(8):607–614. doi: 10.1016/j.biopsych.2013.03.027. [DOI] [PubMed] [Google Scholar]
- 3.Peters J, et al. IMAGEN Consortium Lower ventral striatal activation during reward anticipation in adolescent smokers. Am J Psychiatry. 2011;168(5):540–549. doi: 10.1176/appi.ajp.2010.10071024. [DOI] [PubMed] [Google Scholar]
- 4.Stacey D, et al. IMAGEN Consortium RASGRF2 regulates alcohol-induced reinforcement by influencing mesolimbic dopamine neuron activity and dopamine release. Proc Natl Acad Sci USA. 2012;109(51):21128–21133. doi: 10.1073/pnas.1211844110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plichta MM, et al. Simultaneous EEG and fMRI reveals a causally connected subcortical-cortical network during reward anticipation. J Neurosci. 2013;33(36):14526–14533. doi: 10.1523/JNEUROSCI.0631-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pleger B, Blankenburg F, Ruff CC, Driver J, Dolan RJ. Reward facilitates tactile judgments and modulates hemodynamic responses in human primary somatosensory cortex. J Neurosci. 2008;28(33):8161–8168. doi: 10.1523/JNEUROSCI.1093-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Serences JT. Value-based modulations in human visual cortex. Neuron. 2008;60(6):1169–1181. doi: 10.1016/j.neuron.2008.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salimpoor VN, et al. Interactions between the nucleus accumbens and auditory cortices predict music reward value. Science. 2013;340(6129):216–219. doi: 10.1126/science.1231059. [DOI] [PubMed] [Google Scholar]
- 9.Schultz W, Tremblay L, Hollerman JR. Reward processing in primate orbitofrontal cortex and basal ganglia. Cereb Cortex. 2000;10(3):272–284. doi: 10.1093/cercor/10.3.272. [DOI] [PubMed] [Google Scholar]
- 10.Girault JA. Integrating neurotransmission in striatal medium spiny neurons. Adv Exp Med Biol. 2012;970:407–429. doi: 10.1007/978-3-7091-0932-8_18. [DOI] [PubMed] [Google Scholar]
- 11.Ladepeche L, Yang L, Bouchet D, Groc L. Regulation of dopamine D1 receptor dynamics within the postsynaptic density of hippocampal glutamate synapses. PLoS One. 2013;8(9):e74512. doi: 10.1371/journal.pone.0074512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schumann G, et al. IMAGEN consortium The IMAGEN study: Reinforcement-related behaviour in normal brain function and psychopathology. Mol Psychiatry. 2010;15(12):1128–1139. doi: 10.1038/mp.2010.4. [DOI] [PubMed] [Google Scholar]
- 13.Mumford JA, et al. Detecting network modules in fMRI time series: A weighted network analysis approach. Neuroimage. 2010;52(4):1465–1476. doi: 10.1016/j.neuroimage.2010.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clatworthy PL, et al. Dopamine release in dissociable striatal subregions predicts the different effects of oral methylphenidate on reversal learning and spatial working memory. J Neurosci. 2009;29(15):4690–4696. doi: 10.1523/JNEUROSCI.3266-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gau SSF, Chiang HL. Association between early attention-deficit/hyperactivity symptoms and current verbal and visuo-spatial short-term memory. Res Dev Disabil. 2013;34(1):710–720. doi: 10.1016/j.ridd.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 16.Kirby KN, Maraković NN. Delay-discounting probabilistic rewards: Rates decrease as amounts increase. Psychon Bull Rev. 1996;3(1):100–104. doi: 10.3758/BF03210748. [DOI] [PubMed] [Google Scholar]
- 17.Cardinal RN, Pennicott DR, Sugathapala CL, Robbins TW, Everitt BJ. Impulsive choice induced in rats by lesions of the nucleus accumbens core. Science. 2001;292(5526):2499–2501. doi: 10.1126/science.1060818. [DOI] [PubMed] [Google Scholar]
- 18.Goodman R. Psychometric properties of the strengths and difficulties questionnaire. J Am Acad Child Adolesc Psychiatry. 2001;40(11):1337–1345. doi: 10.1097/00004583-200111000-00015. [DOI] [PubMed] [Google Scholar]
- 19.Morgan M, et al. The ESPAD study: Implications for prevention. Drugs Educ Prev Policy. 1999;6(2):243–256. [Google Scholar]
- 20.Plichta MM, Scheres A. Ventral-striatal responsiveness during reward anticipation in ADHD and its relation to trait impulsivity in the healthy population: A meta-analytic review of the fMRI literature. Neurosci Biobehav Rev. 2014;38:125–134. doi: 10.1016/j.neubiorev.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cloninger CR, Bayon C, Svrakic DM. Measurement of temperament and character in mood disorders: A model of fundamental states as personality types. J Affect Disord. 1998;51(1):21–32. doi: 10.1016/s0165-0327(98)00153-0. [DOI] [PubMed] [Google Scholar]
- 22.Li MX, Yeung JMY, Cherny SS, Sham PC. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum Genet. 2012;131(5):747–756. doi: 10.1007/s00439-011-1118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bloch B, Bernard V, Dumartin B. “In vivo” intraneuronal trafficking of G protein coupled receptors in the striatum: Regulation by dopaminergic and cholinergic environment. Biol Cell. 2003;95(7):477–488. doi: 10.1016/s0248-4900(03)00080-7. [DOI] [PubMed] [Google Scholar]
- 24.Pausova Z, et al. Genes, maternal smoking, and the offspring brain and body during adolescence: Design of the Saguenay Youth Study. Hum Brain Mapp. 2007;28(6):502–518. doi: 10.1002/hbm.20402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Golding J, Pembrey M, Jones R. ALSPAC Study Team ALSPAC--the Avon Longitudinal Study of Parents and Children. I. Study methodology. Paediatr Perinat Epidemiol. 2001;15(1):74–87. doi: 10.1046/j.1365-3016.2001.00325.x. [DOI] [PubMed] [Google Scholar]
- 26.Beyer A, et al. Comparative sequence and expression analyses of four mammalian VPS4 genes. Gene. 2003;305(1):47–59. doi: 10.1016/s0378-1119(02)01205-2. [DOI] [PubMed] [Google Scholar]
- 27.McBride WJ, et al. Gene expression in the ventral tegmental area of 5 pairs of rat lines selectively bred for high or low ethanol consumption. Pharmacol Biochem Behav. 2012;102(2):275–285. doi: 10.1016/j.pbb.2012.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riemensperger T, et al. Behavioral consequences of dopamine deficiency in the Drosophila central nervous system. Proc Natl Acad Sci USA. 2011;108(2):834–839. doi: 10.1073/pnas.1010930108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colantuoni C, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478(7370):519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korostynski M, Kaminska-Chowaniec D, Piechota M, Przewlocki R. Gene expression profiling in the striatum of inbred mouse strains with distinct opioid-related phenotypes. BMC Genomics. 2006;7:146. doi: 10.1186/1471-2164-7-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodgers S, et al. Externalizing disorders and substance use: Empirically derived subtypes in a population-based sample of adults. Soc Psychiatry Psychiatr Epidemiol. 2015;50(1):7–17. doi: 10.1007/s00127-014-0898-9. [DOI] [PubMed] [Google Scholar]
- 32.Chan YF, Dennis ML, Funk RR. Prevalence and comorbidity of major internalizing and externalizing problems among adolescents and adults presenting to substance abuse treatment. J Subst Abuse Treat. 2008;34(1):14–24. doi: 10.1016/j.jsat.2006.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poldrack RA. Can cognitive processes be inferred from neuroimaging data? Trends Cogn Sci. 2006;10(2):59–63. doi: 10.1016/j.tics.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 34.Hutzler F. Reverse inference is not a fallacy per se: Cognitive processes can be inferred from functional imaging data. Neuroimage. 2014;84:1061–1069. doi: 10.1016/j.neuroimage.2012.12.075. [DOI] [PubMed] [Google Scholar]
- 35.Stark R, et al. ADHD related behaviors are associated with brain activation in the reward system. Neuropsychologia. 2011;49(3):426–434. doi: 10.1016/j.neuropsychologia.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 36.Babst M, Davies BA, Katzmann DJ. Regulation of Vps4 during MVB sorting and cytokinesis. Traffic. 2011;12(10):1298–1305. doi: 10.1111/j.1600-0854.2011.01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. Escrt-III: An endosome-associated heterooligomeric protein complex required for mvb sorting. Dev Cell. 2002;3(2):271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- 38.Easton AC, et al. Rasgrf2 controls noradrenergic involvement in the acute and subchronic effects of alcohol in the brain. Psychopharmacology (Berl) 2014;231(21):4199–4209. doi: 10.1007/s00213-014-3562-x. [DOI] [PubMed] [Google Scholar]
- 39.Easton AC, et al. Rasgrf2 controls dopaminergic adaptations to alcohol in mice. Brain Res Bull. 2014;109:143–150. doi: 10.1016/j.brainresbull.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 40.Cuthbert BN, Insel TR. Toward the future of psychiatric diagnosis: The seven pillars of RDoC. BMC Med. 2013;11:126. doi: 10.1186/1741-7015-11-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schumann G, et al. Stratified medicine for mental disorders. Eur Neuropsychopharmacol. 2014;24(1):5–50. doi: 10.1016/j.euroneuro.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 42.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Isaac RE, Li C, Leedale AE, Shirras AD. Drosophila male sex peptide inhibits siesta sleep and promotes locomotor activity in the post-mated female. Proc Biol Sci. 2010;277(1678):65–70. doi: 10.1098/rspb.2009.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kong EC, et al. A pair of dopamine neurons target the D1-like dopamine receptor DopR in the central complex to promote ethanol-stimulated locomotion in Drosophila. PLoS One. 2010;5(4):e9954. doi: 10.1371/journal.pone.0009954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eguíluz VM, Chialvo DR, Cecchi GA, Baliki M, Apkarian AV. Scale-free brain functional networks. Phys Rev Lett. 2005;94(1):018102. doi: 10.1103/PhysRevLett.94.018102. [DOI] [PubMed] [Google Scholar]
- 46.Herjanic B, Reich W. Development of a structured psychiatric interview for children: Agreement between child and parent on individual symptoms. J Abnorm Child Psychol. 1997;25(1):21–31. doi: 10.1023/a:1025703323438. [DOI] [PubMed] [Google Scholar]
- 47.Stahre M, Naimi T, Brewer R, Holt J. Measuring average alcohol consumption: The impact of including binge drinks in quantity-frequency calculations. Addiction. 2006;101(12):1711–1718. doi: 10.1111/j.1360-0443.2006.01615.x. [DOI] [PubMed] [Google Scholar]
- 48.Lucas CP, et al. The DISC Predictive Scales (DPS): Efficiently screening for diagnoses. J Am Acad Child Adolesc Psychiatry. 2001;40(4):443–449. doi: 10.1097/00004583-200104000-00013. [DOI] [PubMed] [Google Scholar]
- 49.Faraone SV, et al. Molecular genetics of attention-deficit/hyperactivity disorder. Biol Psychiatry. 2005;57(11):1313–1323. doi: 10.1016/j.biopsych.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 50.Beaulieu JM, Gainetdinov RR. The physiology, signaling, and pharmacology of dopamine receptors. Pharmacol Rev. 2011;63(1):182–217. doi: 10.1124/pr.110.002642. [DOI] [PubMed] [Google Scholar]
- 51.Saunders C, Limbird LE. Localization and trafficking of alpha2-adrenergic receptor subtypes in cells and tissues. Pharmacol Ther. 1999;84(2):193–205. doi: 10.1016/s0163-7258(99)00032-7. [DOI] [PubMed] [Google Scholar]