Significance
Gastric cancer is the third leading cause of cancer mortality, with an overall 5-y survival rate of <25%. Although sequencing and comprehensive molecular profiling of human gastric cancer have identified a wide spectrum of genes that drive gastric cancer development, we are still far from understanding the complete pathophysiology of this disease. Here, we used Sleeping Beauty transposon mutagenesis to identify genes and evolutionary forces driving gastric cancer development. This study identified pathways driving gastric cancer development, including WNT, TGF-β, PI3K signaling, ubiquitin-mediated proteolysis, adherens junctions, RNA degradation, and chromatin modification, and discovered many previously unidentified genes, such as the LDL receptor-related protein 1B (LRP1B) tumor suppressor gene, with potential clinical importance in human gastric cancer.
Keywords: SB mutagenesis, Smad4-cooperating genes, gastric cancer
Abstract
Mutations in SMAD4 predispose to the development of gastrointestinal cancer, which is the third leading cause of cancer-related deaths. To identify genes driving gastric cancer (GC) development, we performed a Sleeping Beauty (SB) transposon mutagenesis screen in the stomach of Smad4+/− mutant mice. This screen identified 59 candidate GC trunk drivers and a much larger number of candidate GC progression genes. Strikingly, 22 SB-identified trunk drivers are known or candidate cancer genes, whereas four SB-identified trunk drivers, including PTEN, SMAD4, RNF43, and NF1, are known human GC trunk drivers. Similar to human GC, pathway analyses identified WNT, TGF-β, and PI3K-PTEN signaling, ubiquitin-mediated proteolysis, adherens junctions, and RNA degradation in addition to genes involved in chromatin modification and organization as highly deregulated pathways in GC. Comparative oncogenomic filtering of the complete list of SB-identified genes showed that they are highly enriched for genes mutated in human GC and identified many candidate human GC genes. Finally, by comparing our complete list of SB-identified genes against the list of mutated genes identified in five large-scale human GC sequencing studies, we identified LDL receptor-related protein 1B (LRP1B) as a previously unidentified human candidate GC tumor suppressor gene. In LRP1B, 129 mutations were found in 462 human GC samples sequenced, and LRP1B is one of the top 10 most deleted genes identified in a panel of 3,312 human cancers. SB mutagenesis has, thus, helped to catalog the cooperative molecular mechanisms driving SMAD4-induced GC growth and discover genes with potential clinical importance in human GC.
Gastric cancer (GC) is the third leading cause of cancer mortality worldwide, with an overall 5-y survival rate that is <25% (1). The majority of GCs are associated with infectious agents, including the bacterium Helicobacter pylori (2) and EBV. A small minority of GCs are associated with germ-line mutations, including mutations in E-cadherin (CDH1) (3) or mismatch repair genes (4) (Lynch syndrome). Understanding the molecular basis of GC could, therefore, offer new insights into its pathogenesis, help to identify new biomarkers and treatment strategies, and be central to developing individualized treatment strategies for treating GC.
Whole-genome/exome sequencing and comprehensive molecular profiling of GC have greatly improved our understanding of the molecular aspects and pathogenesis of GC (5–9). These studies have identified a wide spectrum of key genetic influences, including chromosomal instability, microsatellite instability, and changes in microRNA expression, somatic gene mutations, and functional SNPs. Despite these efforts, however, it is clear that we are still far from understanding the complete pathophysiology of GC.
Animal models have greatly enriched our understanding of the molecular mechanisms of numerous types of cancers. To generate animal models that mimic human GC, researchers have infected mice with H. pylori and then, treated them with carcinogens. They have also used genetic engineering to develop a variety of transgenic and KO mouse models of GC (10). Smad4 KO mice are one GC model that has been of particular interest to us (11, 12). Heterozygous Smad4 KO mice develop polyps in the pyloric region of the stomach at 50–100 wk of age and the small intestine in older animals at a lower frequency (11, 12). Similarly, juvenile polyposis patients carrying heterozygous germ-line mutations in SMAD4 are predisposed to developing hamartomatous polyps and gastrointestinal cancer. These studies and others showing that ∼8% of human GCs have mutations in SMAD4 (9) show that SMAD4 is an important GC tumor suppressor gene (TSG).
In previous studies, we performed a Sleeping Beauty (SB) transposon mutagenesis screen in the intestine of Smad4+/− mutant mice (13). In the study reported here, we performed a similar screen, but this time we focused on the stomach. SB transposon mutagenesis screens performed in mice that carry germ-line mutations in genes that promote cancer have been proven to be invaluable for identifying the genes and evolutionary forces that drive cancer development (13). This screen has helped to catalog the cooperative molecular mechanisms that drive human SMAD4-induced GC and discover many genes with potential clinical importance in human GC.
Results
SB Mutagenesis Accelerates Gastric Tumor Development in Smad4KO/+ Mice.
To identify genes that cooperate with Smad4 loss to promote GC development, we performed an SB transposon mutagenesis screen in Smad4KO/+ mutant mice (subsequently referred to as Smad4KO mice) (11) that also carried 11 tandem copies of the mutagenic transposon T2/Onc3 (14) and a conditional transposase allele, LSL-SB11 (15). T2/Onc3 carries an internal CMV promoter and downstream splice donor site, which is used to deregulate the expression of oncogenes, and transcriptional stop cassettes in both orientations, which are used to inactivate TSGs. A β-actin-Cre transgene, which is expressed in most body tissues, including the stomach (Fig. S1A), was used to activate transposase expression. We used this transgene, because at the time that these experiments were initiated, a Cre transgene that was specifically active in gastric epithelial and stem cells had not been identified. In total, we generated 35 T2/Onc3:LSL-SB11:β-actin-Cre:Smad4KO/+ mice (hereafter referred to as Smad4KO:SB mice) and aged them until they showed signs of tumor development.
Fig. S1.
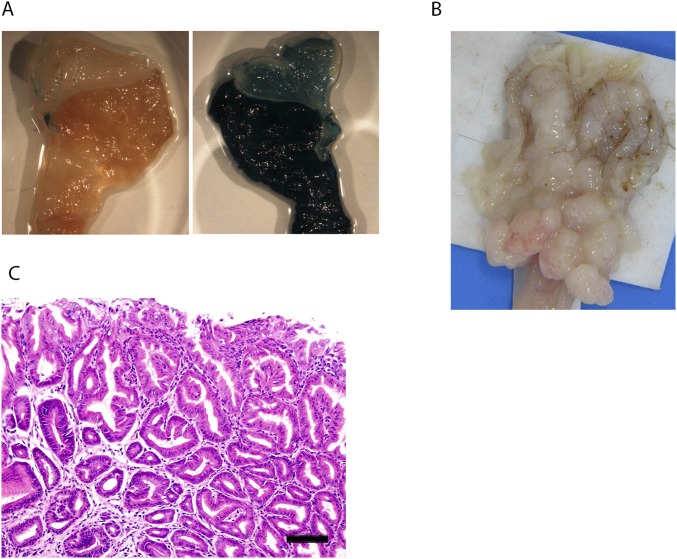
Analysis of β-actin-Cre:R26R3, Smad4KO:SB, and Smad4KO mice. (A) Detection of β-actin-Cre activity in the stomach using R26R3 LacZ reporter mice (55). X-Gal staining was performed using the stomach of (Left) a WT mouse or (Right) a β-actin-Cre:R26R3 mouse. (B) Macroscopic appearance of gastric tumors that developed in an Smad4KO:SB mouse. (C) H&E staining of a tumor from an Smad4KO mouse. (Scale bar: 100 μm.)
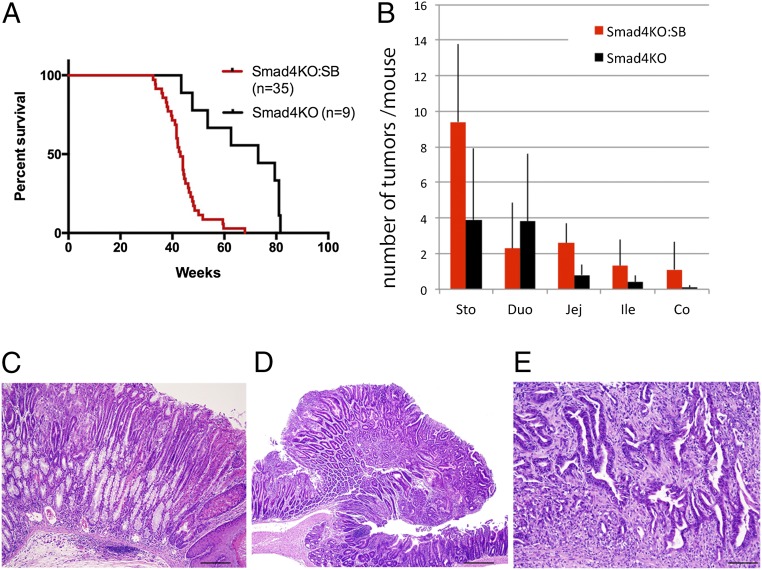
The median survival of Smad4KO:SB mice was 43.1 wk, which was significantly reduced compared with that of Smad4KO mice (67.8 wk; P < 0.01) (Fig. 1A). As expected, because the Cre transgene was expressed throughout the body, these mice developed multiple different types of tumors. In this study, we focused exclusively on the gastric tumors. Smad4KO:SB mice developed more gastric tumors (9.4 ± 4.4 tumors per mouse) than Smad4KO mice (3.9 ± 4.0 tumors per mouse) (Fig. 1B). They also developed more tumors in the jejunum, ileum, and colon but did not develop more tumors in the duodenum (Fig. 1B). All gastric tumors developing in Smad4KO:SB mice were located in the glandular stomach region (Fig. S1B), similar to those in Smad4KO mice.
Fig. 1.
SB mutagenesis accelerates GC development in Smad4+/− mutant mice. (A) Smad4KO:SB mice develop tumors earlier than Smad4KO mice (P < 0.01). (B) Smad4KO:SB mice also develop more tumors per mouse in the stomach, jejunum, ileum, and colon than Smad4KO mice, but they do not develop more tumors per mouse in the duodenum. (C–E) Histopathology of gastric lesions identified in Smad4KO:SB. C shows gastric intestinal metaplasia and inflammation. D shows a gastric adenoma, and E illustrates a gastric adenocarcinoma. Co, colon; Duo, duodenum; Ile, ileum; Jej, jejunum; Sto, stomach. (Scale bars: C, 100 μm; D, 0.5 mm; E, 200 μm.)
Histopathological analysis showed that gastric tumors in Smad4KO:SB (Fig. 1 C and D) and Smad4KO (Fig. S1C) mice were mostly benign tumors (adenomas). There were, however, some cases of adenocarcinoma observed exclusively in Smad4KO:SB mice (Fig. 1E), suggesting that SB mutagenesis can also accelerate tumor histopathology. Inflammation and parietal cell atrophy, which are hallmarks of human GC, were also observed in Smad4KO:SB mice (Fig. 1C). These data show that tumors that are developed in Smad4KO:SB mice are histopathologically similar to human gastric tumors.
Identification of Common Transposon Insertion Sites in Smad4KO:SB Gastric Tumors.
To identify genes that cooperate with Smad4 loss to promote GC development, we PCR-amplified and sequenced the transposon insertion sites from 66 gastric tumors isolated from 35 Smad4KO:SB mice (13) (Table S1). Using the Wellcome Trust Sanger Institute’s SB transposon informatics pipeline (16), we identified 163,078 mapped reads corresponding to 49,610 nonredundant transposon insertion sites in these 66 tumors. Comparison of the insertion site sequences in tumors from the same animal showed that they all had independent origins (Fig. S2). To identify common transposon insertion sites (CISs), we analyzed the data using two different statistical methods. CISs are genomic regions that contain more transposon insertions than predicted by chance, and thus, they are likely to mark the location of candidate cancer genes (CCGs). The “locuscentric” Gaussian Kernel Convolution (GKC) method detects significantly mutated genomic regions within kernel widths of 15, 30, 50, 75, 120, and 240 kb (17), whereas the “genecentric” common transposon insertion site (gCIS) method calculates the observed and expected numbers of insertions within each RefSeq gene and then, looks for RefSeq genes that have more insertions than predicted by chance (18). gCIS identified 702 CIS genes that have human orthologs (13) (Table S2), whereas GKC identified 736 CIS genes with human orthologs (Table S3) and also, six microRNAs (Table S4). The function of CIS genes was predicted by the location of SB insertions (Table S3). Most of the genes identified by SB were TSGs, which is consistent with previous studies showing that, in SB-induced solid tumors, >90% of the CCGs seem to function as TSGs. We also identified nine pairs of genes that were significantly comutated in the same tumor (Table S5).
Fig. S2.
Hamming distance dendrogram showing the biological independence of transposon insertions in different SB-induced gastric tumors from the same mouse.
Concordance between the genes identified by these two methods was far greater than expected by chance (P < 0.001). We then generated a nonredundant list of 941 CIS genes by merging the GKC and gCIS gene lists to define a GC discovery gene set for additional oncogenomic analysis (Table S6). The large number of CIS genes identified in only 66 tumors likely reflects high tumor polyclonality resulting from branching tumor evolution and high rates of mutagenesis induced by SB.
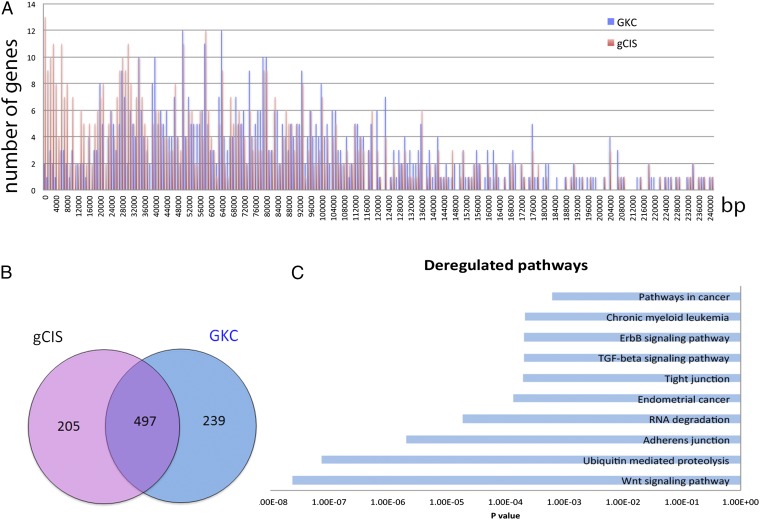
Comparison of genes identified by these two methods showed that gCIS could identify genes smaller than 15,000 more efficiently than GKC (Fig. 2A); 497 CIS genes were identified by both methods, whereas 239 CISs genes were unique to GKC, and 205 CIS genes were unique to gCIS (Fig. 2B and Table S6).
Fig. 2.
Identification of CIS genes in Smad4KO:SB gastric tumors. (A) Chart showing the distribution of CIS genes according to their corresponding size. The blue bars show the numbers of CIS genes and their corresponding sizes identified by GKC, whereas the red bars show the numbers of CIS genes and their corresponding sizes identified by gCIS. Note that gCIS identified more small genes than GKC. (B) Venn diagram showing the number of CIS genes identified by both gCIS and GKC (497) and the numbers of CIS genes that were unique to GKC (239) or gCIS (205). (C) Deregulated pathways identified by the DAVID pathway analysis of SB-identified GC CIS genes.
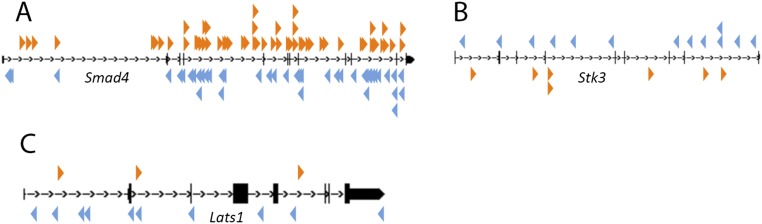
The most highly mutated gene identified in this screen was Smad4, which was mutated in 71.2% of the tumors (Fig. 3A and Table S3). This preference to the mutation for Smad4 likely reflects the strong selective pressure to inactivate the WT Smad4 allele present in tumor cells. Consistent with this prediction, the transposons were inserted throughout the coding region of Smad4 in both transcriptional orientations (Fig. S3A) as expected if these were inactivating insertions. These results are also consistent with other studies performed in Smad4KO/+ mice, which showed that Smad4 haploinsufficiency is sufficient for gastric tumor initiation and that loss of the WT allele only occurs in later stages of tumor progression (12).
Fig. 3.
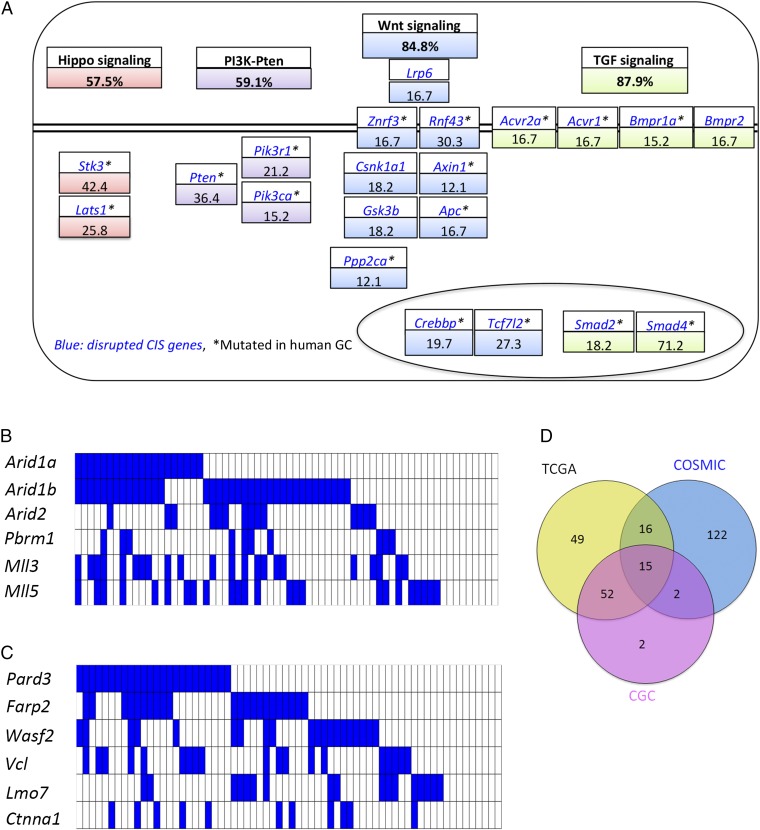
Genes and pathways that are significantly mutated in Smad4KO:SB stomach tumors. A, Upper shows that percentages of tumors that have at least one gene mutated in a given pathway. A, Lower shows the percentages of genes within each pathway that are mutated in tumors. The double black line denotes the plasma membrane. The oval denotes the nucleus. Heat maps showing the landscape of SB insertions in (B) six SWI/SNF (switch/sucrose nonfermentable) complex genes and (C) six adherens junction genes that are extensively mutated in Smad4KO:SB gastric tumors. (D) Venn diagram showing the number of CIS genes commonly mutated in the TCGA, the COSMIC, or the Cancer Gene Census (CGC) databases.
Fig. S3.
SB transposon insertion patterns in three candidate GC TSGs. SB insertion patterns in the (A) Smad4, (B) Stk3, and (C) Lats1 genomic loci. Orange arrowheads indicate transposon insertions in the same orientation as the gene, and blue arrowheads indicate transposon insertions in the inverse orientation as the gene. All insertion patterns are what would be expected for a TSG.
Interestingly, activating insertions in Rspo1 or Rspo2 were not identified in gastric tumors from Smad4KO:SB mice. In the case of Apc, only 16.7% of gastric tumors had insertional mutations in Apc (10 of 66 tumors). The frequency for mutations of Rspo1, Rspo2, and Apc is different from what we reported for intestinal tumors induced in Smad4KO:SB mice using Vil1-CreERT2 (13), where 21% and 55% of these tumors had activating insertions in Rspo1 and Rspo2, respectively. R spondins are secreted proteins that potentiate Wnt signaling by binding to Lgr5 homologs (19). These results suggest that gastric tumors have other preferred methods for fine-tuning their Wnt signaling pathway and that cell type matters in selecting the different components of Wnt pathway in tumorigenesis.
Arid1a and Arid1b were also highly mutated in Smad4KO:SB GCs (Fig. 3B). These genes were mutated in 30.3% and 56.1% of tumors, respectively. Arid1a and Arid1b are members of the SWI/SNF (switch/sucrose nonfermentable) chromatin-remodeling complex. In human GC, inactivating mutations in ARID1A have been identified in 83% of cancers with microsatellite instability, 73% of cancers associated with EBV infection, and 11% of cancers that are microsatellite stable and not infected with EBV. These results highlight the importance of this gene in human GC (5).
Identification of Trunk Driver Genes.
Previous studies have suggested that most of the CCGs identified by SB mutagenesis function during late stages of tumor progression in the branches and leaves of the cancer evolutionary tree (13). To identify the subset of SB-identified CCGs that function early in gastric tumor development, we selected the transposon insertion sites represented by the highest number of sequencing reads (SB insertions in three or more tumors with nine or more sequencing reads), arguing that these insertions would be present in the largest number of tumor cells. This analysis identified 59 genes that we subsequently refer to as “trunk driver” genes (Table S7). Strikingly, 22 of these SB-identified trunk drivers are known or candidate human cancer genes. These 22 CIS genes include PTEN, SMAD4, RNF43, and NF1, which are known human GC trunk drivers. A role for other SB-identified trunk drivers in human GC has been suggested, but the role in GC is less firmly established. For example, expression of USP10, the SB-identified trunk driver with the fifth highest average read count, is inversely correlated with gastric wall invasion and nodal metastasis in GC patients (20). SB mutagenesis, thus, provides strong evidence for a role for USP10 in human GC.
Other SB-identified trunk drivers with suggested roles in human GC include USP47, a ubiquitin-specific protease and regulator of Wnt signaling with expression that is up-regulated in GC cells through down-regulation of miR-204–5p (21). Down-regulation of miR-204–5p promotes GC cell proliferation, suggesting that miR-204–5p is a GC TSG that functions, in part, through its inhibition of USP47. Another SB-identified trunk driver CRK is an adapter protein that binds to several tyrosine-phosphorylated proteins. Like USP47, CRK expression is up-regulated in GC through down-regulation of miR-126. Overexpression of miR-126 has been shown to inhibit GC cell invasion in part by targeting CRK (22). ROCK1 is a Rho-associated kinase and a target of miR-135a (23) with expression that is increased in 33.9% of GC patients because of decreased miR-135a expression. Patients with increased ROCK1 expression show a significantly more advanced stage of GC and higher rates of lymph node metastases. ROCK1 expression is also increased in GC patients with lymph node metastasis. The SB insertion patterns in Crk, USP47, and ROCK1 are, however, more consistent with what would be expected for a TSG. In future studies, it will be important to validate these CCGs one by one to confirm whether they are oncogenes or TSGs.
SF3B1 is an RNA splicing factor that is mutated in a large number of myelodysplastic syndromes. Recent studies have shown that SF3B1 is also mutated in solid tumors, including GC (24). FBXW7 is an F-box protein that mediates the ubiquitin-dependent proteolysis of several oncoproteins, including cyclin E1, Myc, Jun, and Notch1. FBXW7 is inactivated in a diverse range of human cancers, including bile duct (cholangiocarcinomas; 35%), blood (T-cell acute lymphocytic leukemia; 31%), endometrium (9%), and stomach (6%) cancers (25).
Finally, several SB-identified trunk drivers are known or candidate human cancer genes, but there is not yet any published evidence for a role for these genes in human GC. Our results provide suggestive evidence for a role for these genes in human GC. STK3 (MST2) is a serine-threonine kinase and a component of Hippo signaling. Combined deficiency of Mst1/2 in the intestine results in intestinal adenoma formation in mice (26), consistent with Mst2 being a TSG. SUV420H1 is an H4-K20 histone methyltransferase. Loss of H4-K20 trimethylation is associated with multiple cancers, including breast cancer, where genetic alterations in SUV420H1 have been identified (27). MKLN1 is a gene that functions in cell adhesion and cytoskeletal organization, and it is frequently amplified in glioblastoma (28). PARD3 is a gene that modulates Hippo pathway signaling in response to cell–cell contact and cell polarity signals (29). Loss of PARD3 promotes breast tumorigenesis and metastasis (30), whereas inactivating mutations in PARD3 have been identified in various cancers. USP9X is a deubiquitinase that suppresses pancreatic ductal adenocarcinoma, and it is mutated in over 50% of mouse pancreatic ductal tumors induced on a KrasG12D background (31). ARID1B is a gene that is mutated in a number of human cancers, including neuroblastoma, hepatocelluar carcinoma, breast, colorectal, and pancreatic cancer.
DAVID Pathway Analysis.
To identify signaling pathways and cellular processes driving gastric tumor development, we analyzed 941 CCGs identified in Smad4KO:SB gastric tumors using the DAVID functional annotation platform (32). The most highly deregulated signaling pathway was Wnt signaling (P = 2.33 × 10−8) (Fig. 2C and Table S8). Nearly 85% of gastric tumors had a mutation in at least one Wnt pathway gene (Fig. 3A). In addition, ubiquitin-mediated proteolysis, adherens junctions, and RNA degradation were also highly deregulated in Smad4KO:SB gastric tumors (Fig. 2C). Our result is consistent with driver pathway analysis in human GC, which showed that the top enriched pathways/cellular processes are Wnt signaling, adherens junctions, and focal adhesions (7). The role of Wnt signaling in GC is thought to help keep gastric progenitor cells in an undifferentiated state (33). Collectively, these results show that activation of Wnt signaling is one of the most important molecular events driving GC development.
Smad4KO:SB gastric tumors have mutations in Lpr6 (Fig. 3A), which were not observed in Smad4KO:SB intestinal tumors (13). Other Wnt signaling genes were mutated in both tumor types, but their mutation frequencies varied between the two tumor types. For example, 27% and 12% of intestinal tumors had mutations in Znrf3 and Rnf43, respectively (13), whereas 16.7% and 30.3% of gastric tumors had mutations in these genes, respectively (Fig. 3A). ZNRF3 and its homolog RNF43 are ubiquitin ligases and negative feedback regulators of Wnt signaling. ZNRF3 inhibits Wnt signaling by promoting the turnover of the Wnt coreceptors Frizzled and LDL receptor-related protein 6 (LRP6). Interestingly, R spondins interact with the extracellular domain of ZNRF3 and induce an association between ZNRF3 and LGR4, which results in the membrane clearance of ZNRF3. R spondins, therefore, enhance Wnt signaling by inhibiting ZNFR3 (34). Thus, a significant number of these two cancers has mutations that function upstream in Wnt signaling, but the distribution and nature of the Wnt pathway genes mutated in these two cancer types are different.
It is interesting to note that Wnt signaling in SB-induced intestinal tumors produced on a WT background is predominantly activated through loss-of-function mutations in Apc, which functions downstream in Wnt signaling (16). Thus, Wnt signaling seems to be activated at two different levels in intestinal tumors depending on whether they have a preexisting Smad4 mutation. These results show how much the presence of another mutation matters. If another mutation occurs first, it can profoundly affect the nature of the mutations that occur second. Our results presented here also show how much tissue type matters. These results have clear therapeutic implications for those who might use pathway drugs to treat cancer.
TGF-β signaling is another important pathway for GC (Fig. 2C). TGF-β signaling regulates diverse processes, such as cell proliferation, differentiation, motility, adhesion, organization, and programmed cell death (35). Smad4 is a TSG and one of the most important transcriptional cofactors regulating the TGF-β pathway. Loss of heterozygosity for SMAD4 occurs in late-stage human GC (36) and confers resistance against TGF-β signaling. In our SB-induced gastric tumors, >70% of tumors carried insertional mutations in Smad4, which is likely to induce biallelic inactivation of Smad4 and confer an additional growth advantage to tumor cells. An additional five genes were identified by SB mutagenesis that also function in TGF-β signaling, including Acvr2a, Acvr1, Bmpr1a, Bmpr2, and Smad2 (Fig. 3A). Two of these genes, Smad2 and Bmpr2, were not identified in Smad4KO:SB intestinal tumors (13), showing once again how much cell type matters. Multiple mutations in genes in the same pathway are a common event in human tumors as well as SB-induced tumors (16). For example, SB mutagenesis on an Apc heterozygous mutant background identified 183 CIS genes that are candidate Wnt signaling genes. March et al. (16) hypothesized that multiple mutations in the Wnt pathway in the same tumor fine-tune Wnt signaling during tumor progression. Similar selective pressure could be occurring here in the case of the TGF-β pathway.
The Hippo pathway genes Stk3 (Mst2) and Lats1 were mutated in 42.4% and 25.8% of the tumors, respectively (Fig. 3A). Lats1 and Stk3 (Mst2) are known tumor suppressors that negatively regulate Hippo signaling (26, 37, 38), consistent with the inactivating pattern of SB insertions observed in these genes (Fig. S3 B and C). These results suggest that Hippo signaling is also an important contributor to GC.
Finally, PI3K-Pten signaling also seems to be of critical importance for SB-induced GCs (Fig. 3A). The most highly mutated gene was Pten, which was mutated in 36.4% of the tumors. PTEN is also frequently mutated in human GC (9), showing the importance of PI3K-Pten signaling in GC.
Importance of Chromatin-Modifying and Adherence Junction Genes in GC.
Gene ontology classification showed that SB-identified CCGs are also highly enriched in genes involved in “chromatin modification” and “chromatin organization” (P = 8.99 × 10−15 and P = 1.3 × 10−13, respectively) (Table S9). These genes include Arid1a, Arid1b, Arid2, Pbrm1, Mll3, and Mll5 (Fig. 3B). Three of these genes, ARID1A, ARID1B, and ARID2, are components of the SWI/SNF complex and also highly mutated in human GC (6). These results show the importance of chromatin-modifying genes in GC.
Recently, an in silico 3D model of tumor evolution has been developed (39) that also reveals the dynamics of mutational spread. This model argues that cell migration might be the key to tumor shape, spread, and drug resistance. This study opens up the possibility of treatments that target genes related to cell motility and adhesion rather than the conventional targets governing cell division, death, and differentiation. Previously, it was thought that cell migration was mostly involved in the invasion of tissues by tumors or in metastasis. If this model is correct, it would argue that cell migration is also critically important for the initial stages of tumor growth. Therefore, genes identified by SB in early-stage tumors that are predicted to be involved in cell migration/invasion could represent excellent drug targets for future cancer treatment.
Cell migration is regulated by several genes involved in cell–cell and cell–ECM adhesion, actin remodeling and EGF signaling (40). In SB-induced tumors, DAVID pathway analysis showed that SB-identified CIS genes are enriched in genes that regulate adherens junctions (P < 0.001), tight junctions (P < 0.001), focal adhesions (P = 0.0059), ErbB signaling (P < 0.001), and the actin cytoskeleton (P = 0.015) (Fig. 2C and Table S8), suggesting that deregulation of cell migration is also important in SB-induced GC. A key molecule involved in adherens junctions is PARD3, which is frequently lost during metastasis in human breast cancers. Down-regulation of Pard3 in mammary cancer cells induces an invasive phenotype in vitro and promotes metastasis in vivo by disrupting actin dynamics at cell–cell junctions (41). Interestingly, 86.8% of SB-induced gastric tumors carry mutations in genes involved in adherens junctions (Fig. 3C), suggesting that disruption of adherens junction could also have a significant impact on cell migration in mouse GCs.
Comparative Oncogenomic Filtering.
Next, we asked whether the CCGs identified by SB in mouse GC are significantly enriched among the known genes mutated in human GC. In our initial analysis, we asked whether our CCGs are enriched in the genes listed in version 66 of the Catalogue of Somatic Mutations in Cancer (COSMIC) for genes mutated in ≥4% of human GCs. Of 814 COSMIC genes that met this criterion, 155 were CIS genes (Fig. 3D and Table 1), which is highly significant (P = 1.19 × 10−45). Next, using the Cancer Genome Atlas (TCGA; downloaded in May of 2015), we identified 1,384 mutated genes from a collection of 443 human GC samples and found that 132 are CIS genes (Fig. 3D and Table 1), which is also highly significant (P = 6.96 × 10−11). Finally, we compared our CIS genes with 513 human genes listed in the Cancer Gene Census database (42), which have mutations causally implicated in cancer, and identified 71 overlapping genes (Fig. 3D and Table 1) (P = 1.30 × 10−13). These results show that CCGs identified by SB in mouse GC are highly relevant for human GC.
Table 1.
CIS genes identified by SB in mouse GC are highly relevant for human GC
| Name of the dataset | No. of genes in the dataset | No. of overlapping CIS genes | P value |
| Human GC somatic mutations in ≥4% samples in the COSMIC database | 814 | 155 | 1.19E−45 |
| Human GC somatic mutations in the TCGA database | 1,384 | 132 | 6.96E−11 |
| Cancer Gene Census in the COSMIC database | 513 | 71 | 1.30E−13 |
The most interesting of these overlapping genes are 15 SB-identified CCGs that were present in all three databases (Fig. 3D, Table 2, and Table S10). Five of these genes, including Arid1a, Fbxw7, Pten, Pik3ca, and Kras, are known human GC driver genes. Among the other 10 genes, APC mutations are common in human GC, suggesting that APC is also a human GC driver gene. EXT is an endoplasmic reticulum-resident type II transmembrane glycoprotein with expression that alters the synthesis and displays of cell surface heparan sulfate glycosaminoglycans. Mutation or epigenetic silencing of EXT1 leads to abrogation of heparan sulfate biosynthesis, which is important for tumor onset and progression (43). AKAP9 is a scaffolding protein that assembles several protein kinases and phosphatases on centrosomes and the Golgi apparatus. AKAP9 is a breast (44) and colorectal cancer susceptibility gene (45). Inactivation of another gene, ATRX, has been shown to activate the alternative lengthening of telomeres pathway, which uses homologous recombination to maintain telomere length and sustain the limitless replicability of cancer cells. ATRX mutations have been identified in many human cancers but so far, have not been identified in GC. SETBP1 is known to recruit the NuRD complex to repress gene expression. Mutations in SETBP1 are often seen in atypical chronic myeloid leukemia and related diseases. SETBP1 has also been identified as a likely breast cancer susceptibility gene in genome-wide association studies (46).
Table 2.
A list of 15 CIS genes identified by SB in mouse GC that were present in three human somatic cancer databases
| Gene symbol | TCGA (Sto) | COSMIC (Sto) | CGC | Human ortholog | Gene name | Chromosome | P value | Framework |
| Arid1a | ✓ | ✓ | ✓ | ARID1A | AT-rich interactive domain 1A (SWI-like) | chr4 | 0 | GKC |
| Fbxw7 | ✓ | ✓ | ✓ | FBXW7 | F-box and WD-40 domain protein 7 | chr3 | 0 | GKC |
| Pten | ✓ | ✓ | ✓ | PTEN | Phosphatase and tensin homolog | chr19 | 0 | GKC |
| Rnf43 | ✓ | ✓ | ✓ | RNF43 | Ring finger protein 43 | chr11 | 0 | GKC |
| Apc | ✓ | ✓ | ✓ | APC | Adenomatosis polyposis coli | chr18 | 1.0E−06 | GKC |
| Pik3ca | ✓ | ✓ | ✓ | PIK3CA | PI3K, catalytic, α-polypeptide | chr3 | 8.0E−06 | GKC |
| Ext1 | ✓ | ✓ | ✓ | EXT1 | Exostoses (multiple) 1 | chr15 | 8.0E−06 | GKC |
| Akap9 | ✓ | ✓ | ✓ | AKAP9 | A kinase (PRKA) anchor protein 9 | chr5 | 8.0E−06 | GKC |
| Setbp1 | ✓ | ✓ | ✓ | SETBP1 | SET binding protein 1 | chr18 | 1.0E−05 | GKC |
| Nsd1 | ✓ | ✓ | ✓ | NSD1 | Nuclear receptor-binding SET-domain protein 1 | chr13 | 5.0E−05 | GKC |
| Atrx | ✓ | ✓ | ✓ | ATRX | α-Thalassemia/mental retardation syndrome X-linked | chrX | 7.0E−05 | GKC |
| Crebbp | ✓ | ✓ | ✓ | CREBBP | CREB binding protein | chr16 | 1.0E−09 | gCIS |
| Kdm5a | ✓ | ✓ | ✓ | KDM5A | Lysine (K)-specific demethylase 5A | chr6 | 6.0E−07 | gCIS |
| Kras | ✓ | ✓ | ✓ | KRAS | Kirsten rat sarcoma viral oncogene homolog | chr6 | 7.0E−08 | gCIS |
| Mkl1 | ✓ | ✓ | ✓ | MKL1 | Megakaryoblastic leukemia 1 | chr15 | 5.0E−08 | gCIS |
CGC, Cancer Gene Census; Sto, stomach.
Finally, we compared our list of SB-identified CCGs with the combined list of mutated genes identified in five large-scale human GC sequencing studies (5–9). Genes found in common are listed in Table S11. The CCG that is most highly mutated in these five sequencing studies is LRP1B; 129 mutations in LRP1B were identified from 462 GC samples. The LRP1B was originally isolated based on its homozygous deletion in human lung cancer cell lines, and it is one of the top 10 most significantly deleted genes in a panel of 3,312 human cancers (47). Inactivation of LRP1B leads to changes in the tumor microenvironment that confer cancer cells with an increased growth and invasive capacity (48). LRP1B is hypermethylated in 61% of human GC, and this hypermethylation correlates with low RNA expression (49). Restoration of LRP1B expression in GC cell lines reduces cell proliferation, colony formation, and tumorigenicity in nude mice. These results combined with our SB mutagenesis results strongly indicate that LRP1B is a human GC TSG that is epigenetically silenced in a majority of GC. Reactivation of LRP1B in GC might, therefore, be clinically beneficial in the treatment of GC.
Discussion
In the studies presented here, we describe a transposon-based screen in mice aimed at identifying genes that cooperate with mutant Smad4 in the generation of gastric tumors. To our knowledge, this study is the first transposon screen performed for gastric tumors. Transposons, by their nature, are well-suited to capture the full complement of the genetic complexity present in tumor cells, because they can identify genes that are somatically mutated in human cancer in addition to cancer genes that are deregulated by transcriptional or epigenetic means. In addition, transposon-induced mutations that function in the branches and leaves of the cancer evolutionary tree can be identified in these screens, because only the transposon insertion sites and not the rest of mouse genome are PCR-amplified and sequenced. Therefore, transposon insertions in cancer genes that are present in only a small number of cells in the tumor can be identified by SB. Therefore, SB makes it possible to characterize the genetic complexity of tumors on a much more in-depth scale than is currently possible in human tumors using whole-genome/exome sequencing.
A real strength of our model is that it seems to be identifying similar genetic changes as those observed in human GC, which is best shown by the highly significant number of GC genes identified by SB that are also mutated in human GC as well as other forms of cancer. One noted difference was the paucity of p53 mutations identified in our screen. In human GC, p53 is one of the most frequently mutated genes. The primary biological importance of p53 loss in cancer cells is to induce genomic instability, which leads to an increase in the number of mutations. In SB-induced tumors, SB is a very powerful mutagen, abrogating the need for mutations induced by p53 loss.
Many of the genes identified by SB also seem to function in signaling pathways known to be important in human GC. One of the most important of these pathways is Wnt signaling, which is similar to what occurs in gastrointestinal tract cancer in mouse and humans, where Wnt signaling is also critical for tumor formation. What we have learned, however, is that the mechanism of Wnt pathway activation is not always the same and seems to be effected by the presence of preexisting mutations in other genes in the cancer cell in addition to the tissue type. In human and mouse colorectal cancers, Wnt activation primarily occurs through loss-of-function mutations in APC (50). However, in mouse intestinal epithelial cells that have a preexisting mutation in Smad4, the preferred route of Wnt activation is through gain-of-function mutations in Rspo1 and Rspo2 (13). Similarly, ∼10% of human colorectal tumors with no APC mutations tend to carry chromosomal translocation involving RSPO2 and RSPO3, and also show decreased expression of SMAD4 (51).
Another study showing how much a preexisting mutation can affect subsequent tumor evolution has recently been reported (52). These researchers used exome sequencing to study the evolution of thymic lymphomas in p53 null mice. They found that all thymic lymphomas had a deletion in Pten that occurred before rearrangement of the TCRβ locus (52). This event was followed by amplification or overexpression of cyclin Ds and Cdk6. The initial inherited loss of p53 functions, therefore, seemed to delineate an order of genetic alterations selected for during the evolution of these thymic lymphomas.
In light of these studies, it was surprising that activating insertions in Rspo1 and Rspo2 were not observed in gastric tumors from Smad4KO:SB mice. Instead, mutations were identified in other upstream Wnt signaling components, such as Lrp6. These results show how much tissue type also matters for tumor evolution. These results have clear therapeutic implication for those who might want to use pathway-targeted drugs to treat cancer.
By performing SB screens in the intestines of mice that carried one of four different mutations in genes known to be important in human colorectal cancer development (i.e., APC, KRAS, SMAD4, and TP53), it was possible to identify 111 genes that are highly likely to be involved in human colorectal cancer by looking for genes that were commonly mutated in all four screens (13). A similar approach used here to identify genes that are highly likely to be involved in human GC was to look for SB-identified CIS genes that are known to be mutated in human GC that are also listed in the TCGA, which contains a collection of 513 genes that have mutations causally implicated in human cancer. The 15 CIS genes identified in this analysis are highly likely to be involved in human GC as evidenced by the fact that 5 of these genes are known drivers of human GC.
Materials and Methods
Mice.
The T2/Onc3 (12740) transposon allele and the conditional SB transposase (lox-STOP-lox-SB11) line were described previously (14). The β-actin-Cre line was purchased from The Jackson Laboratory (019099), whereas Smad4+/− mice were described in a previous report (11). All animal experiments were performed following institutional animal care and use committee protocols. Briefly, animals were sent for necropsy when they showed signs of sickness. Mouse survival was analyzed using GraphPad Prism6. Animals were surgically opened, and tumor counts were performed for each gastrointestinal tract. All gastric tumors (>2 mm) were collected and snap frozen. Tumors (>5 mm) were fixed in 10% (wt/vol) formalin and processed for H&E slides. All tumors were examined by an expert rodent pathologist (J.M.W.) who graded and staged the tumors.
Cloning of the Transposon Insertion Sites.
Tumors were lysed, and genomic DNA was purified using the Gentra Puregene Cell Kit (158767; QIAGEN); 300 ng to 1 μg DNA was digested with NalIII or BfaI at 37 °C overnight. The NlaIII or BfaI linkers were ligated by T4 ligase (NEB) at 4 °C overnight: BfaI linker(−): 5′-Phos-TAGTCCCTTAAGCGGAG-C3spacer-3′; BfaI linker(+): 5′-GTAATACGACTCACTATAGGGCTCCGCTTAAGGGAC-3′; NlaIII linker(−): 5′-Phos-GTCCCTTAAGCGGAG-C3spacer-3′; and NlaIII linker(+): 5′-GTAATACGACTCACTATAGGGCTCCGCTTAAGGGACCATG-3′. DNA was purified by column filtration (28706; QIAGEN). The NlaIII-digested DNA was then digested with XhoI, and BfaI-digested DNA was digested with BamHI to eliminate unmobilized transposons. DNA was purified again by column filtration. The primary PCR was done using linker primer GTAATACGACTCACTATAGGGC and IRDR(R1): GCTTGTGGAAGGCTACTCGAAATGTTTGACCC (for NlaIII-digested DNA) or IRDR(L1): CTGGAATTTTCCAAGCTGTTTAAAGGCACAGTCAAC (for BfaI-digested DNA). For secondary PCR, FusBT-LilL-SB nested linker primer CCTATCCCCTGTGTGCCTTGGCAGTCTCAGAGGGCTCCGCTTAAGGGAC and barcoded primers (SIGMA) were used for both NlaIII-digested DNA and BfaI-digested DNA. PCR products were sequenced using the 454 GS-Titanium (Roche) platform as previously reported (16).
CIS Detection by GKC and gCIS Frameworks.
The T2/Onc3 transposon concatamer used in this study is located on chromosome 9. Therefore, all insertions on chromosome 9 were removed from data because of the problems caused by local hopping. All informative sequence reads were mapped to the B6 mouse genome. To detect CISs, a GKC method was applied, and kernel widths of 15,000, 30,000, 50,000, 75,000, 120,000, and 240,000 nt were used as previously reported (18). When CISs were detected by several kernel widths, CISs were merged, and the smallest P value was reported. When a CIS contained more than one gene, all genes within the CIS were called. Human orthologs were determined using BioMart (www.biomart.org/martservice_9.html).
Comparison with Human Mutation Datasets.
The COSMIC, version 66 database was downloaded, and somatic mutations identified for GC were subsequently used for data comparisons; 513 genes in the Cancer Gene Census were also downloaded from the COSMIC website (cancer.sanger.ac.uk/census/). For the TCGA dataset, the list of mutated genes in 443 stomach adenocarcinomas was downloaded from cBioPortal (www.cbioportal.org/study.do?cancer_study_id=stad_tcga#summary) and used for subsequent comparisons. This information contained the size of the gene and the number of mutations. For the other four GC genome sequencing studies, corresponding supplementary files were downloaded and used for comparisons (5–8) (Table S11). Bio-Venn (53) was used to identify the overlap between CIS genes and those mutated in human GC.
Pathway Analysis.
The DAVID online gene annotation tool was used for pathway analysis (54).
Supplementary Material
Acknowledgments
We thank Keith Rogers, Susan Rogers, and the Institute of Molecular and Cell Biology histopathology core for performing the histopathological analyses; Makoto M. Taketo for supplying the Smad4+/− mutant mice; and Pamela Goh, Pearlyn Cheok, Nicole Lim, Dorothy Chen, and Cherylin Wee for animal technical assistance. The Biomedical Research Council, Agency for Science, Technology and Research, Singapore and the Cancer Prevention Research Institute of Texas (CPRIT) supported this research. H.T is supported by Grants-in-Aid for Scientific Research. N.A.J. and N.G.C. are both CPRIT Scholars in Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1603223113/-/DCSupplemental.
References
- 1.Jemal A, et al. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Uemura N, et al. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345(11):784–789. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 3.Richards FM, et al. Germline E-cadherin gene (CDH1) mutations predispose to familial gastric cancer and colorectal cancer. Hum Mol Genet. 1999;8(4):607–610. doi: 10.1093/hmg/8.4.607. [DOI] [PubMed] [Google Scholar]
- 4.Keller G, et al. Analysis for microsatellite instability and mutations of the DNA mismatch repair gene hMLH1 in familial gastric cancer. Int J Cancer. 1996;68(5):571–576. doi: 10.1002/(SICI)1097-0215(19961127)68:5<571::AID-IJC3>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 5.Wang K, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011;43(12):1219–1223. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- 6.Zang ZJ, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44(5):570–574. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 7.Wang K, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46(6):573–582. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 8.Kakiuchi M, et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet. 2014;46(6):583–587. doi: 10.1038/ng.2984. [DOI] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayakawa Y, et al. Mouse models of gastric cancer. Cancers (Basel) 2013;5(1):92–130. doi: 10.3390/cancers5010092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takaku K, et al. Gastric and duodenal polyps in Smad4 (Dpc4) knockout mice. Cancer Res. 1999;59(24):6113–6117. [PubMed] [Google Scholar]
- 12.Xu X, et al. Haploid loss of the tumor suppressor Smad4/Dpc4 initiates gastric polyposis and cancer in mice. Oncogene. 2000;19(15):1868–1874. doi: 10.1038/sj.onc.1203504. [DOI] [PubMed] [Google Scholar]
- 13.Takeda H, et al. Transposon mutagenesis identifies genes and evolutionary forces driving gastrointestinal tract tumor progression. Nat Genet. 2015;47(2):142–150. doi: 10.1038/ng.3175. [DOI] [PubMed] [Google Scholar]
- 14.Dupuy AJ, et al. A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer Res. 2009;69(20):8150–8156. doi: 10.1158/0008-5472.CAN-09-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Starr TK, et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323(5922):1747–1750. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.March HN, et al. Insertional mutagenesis identifies multiple networks of cooperating genes driving intestinal tumorigenesis. Nat Genet. 2011;43(12):1202–1209. doi: 10.1038/ng.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uren AG, et al. Large-scale mutagenesis in p19(ARF)- and p53-deficient mice identifies cancer genes and their collaborative networks. Cell. 2008;133(4):727–741. doi: 10.1016/j.cell.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mann KM, et al. Australian Pancreatic Cancer Genome Initiative Sleeping Beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proc Natl Acad Sci USA. 2012;109(16):5934–5941. doi: 10.1073/pnas.1202490109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Lau W, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476(7360):293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- 20.Zeng Z, et al. Prognostic significance of USP10 as a tumor-associated marker in gastric carcinoma. Tumour Biol. 2014;35(4):3845–3853. doi: 10.1007/s13277-013-1509-1. [DOI] [PubMed] [Google Scholar]
- 21.Shi J, et al. Deubiquitinase USP47/UBP64E Regulates β-Catenin Ubiquitination and Degradation and Plays a Positive Role in Wnt Signaling. Mol Cell Biol. 2015;35(19):3301–3311. doi: 10.1128/MCB.00373-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, Wang F, Qi Y. MiR-126 inhibits the invasion of gastric cancer cell in part by targeting Crk. Eur Rev Med Pharmacol Sci. 2014;18(14):2031–2037. [PubMed] [Google Scholar]
- 23.Shin JY, et al. MicroRNA 135a suppresses lymph node metastasis through down-regulation of ROCK1 in early gastric cancer. PLoS One. 2014;9(1):e85205. doi: 10.1371/journal.pone.0085205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Je EM, Yoo NJ, Kim YJ, Kim MS, Lee SH. Mutational analysis of splicing machinery genes SF3B1, U2AF1 and SRSF2 in myelodysplasia and other common tumors. Int J Cancer. 2013;133(1):260–265. doi: 10.1002/ijc.28011. [DOI] [PubMed] [Google Scholar]
- 25.Akhoondi S, et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007;67(19):9006–9012. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- 26.Zhou D, et al. Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proc Natl Acad Sci USA. 2011;108(49):E1312–E1320. doi: 10.1073/pnas.1110428108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu LL, et al. FoxD3-regulated microRNA-137 suppresses tumour growth and metastasis in human hepatocellular carcinoma by targeting AKT2. Oncotarget. 2014;5(13):5113–5124. doi: 10.18632/oncotarget.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nord H, et al. Characterization of novel and complex genomic aberrations in glioblastoma using a 32K BAC array. Neuro-oncol. 2009;11(6):803–818. doi: 10.1215/15228517-2009-013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lv XB, et al. PARD3 induces TAZ activation and cell growth by promoting LATS1 and PP1 interaction. EMBO Rep. 2015;16(8):975–985. doi: 10.15252/embr.201439951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCaffrey LM, Montalbano J, Mihai C, Macara IG. Loss of the Par3 polarity protein promotes breast tumorigenesis and metastasis. Cancer Cell. 2012;22(5):601–614. doi: 10.1016/j.ccr.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pérez-Mancera PA, et al. Australian Pancreatic Cancer Genome Initiative The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature. 2012;486(7402):266–270. doi: 10.1038/nature11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bongers G, et al. The cytomegalovirus-encoded chemokine receptor US28 promotes intestinal neoplasia in transgenic mice. J Clin Invest. 2010;120(11):3969–3978. doi: 10.1172/JCI42563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oshima H, et al. Carcinogenesis in mouse stomach by simultaneous activation of the Wnt signaling and prostaglandin E2 pathway. Gastroenterology. 2006;131(4):1086–1095. doi: 10.1053/j.gastro.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 34.Hao HX, et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature. 2012;485(7397):195–200. doi: 10.1038/nature11019. [DOI] [PubMed] [Google Scholar]
- 35.Padua D, Massagué J. Roles of TGFbeta in metastasis. Cell Res. 2009;19(1):89–102. doi: 10.1038/cr.2008.316. [DOI] [PubMed] [Google Scholar]
- 36.Wang LH, et al. Inactivation of SMAD4 tumor suppressor gene during gastric carcinoma progression. Clin Cancer Res. 2007;13(1):102–110. doi: 10.1158/1078-0432.CCR-06-1467. [DOI] [PubMed] [Google Scholar]
- 37.Yu T, Bachman J, Lai ZC. Mutation analysis of large tumor suppressor genes LATS1 and LATS2 supports a tumor suppressor role in human cancer. Protein Cell. 2015;6(1):6–11. doi: 10.1007/s13238-014-0122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou D, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16(5):425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waclaw B, et al. A spatial model predicts that dispersal and cell turnover limit intratumour heterogeneity. Nature. 2015;525(7568):261–264. doi: 10.1038/nature14971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simpson KJ, et al. Identification of genes that regulate epithelial cell migration using an siRNA screening approach. Nat Cell Biol. 2008;10(9):1027–1038. doi: 10.1038/ncb1762. [DOI] [PubMed] [Google Scholar]
- 41.Xue B, Krishnamurthy K, Allred DC, Muthuswamy SK. Loss of Par3 promotes breast cancer metastasis by compromising cell-cell cohesion. Nat Cell Biol. 2013;15(2):189–200. doi: 10.1038/ncb2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Futreal PA, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4(3):177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ropero S, et al. Epigenetic loss of the familial tumor-suppressor gene exostosin-1 (EXT1) disrupts heparan sulfate synthesis in cancer cells. Hum Mol Genet. 2004;13(22):2753–2765. doi: 10.1093/hmg/ddh298. [DOI] [PubMed] [Google Scholar]
- 44.Milne RL, et al. GENICA Network kConFab Investigators Australian Ovarian Cancer Study Group TNBCC Common non-synonymous SNPs associated with breast cancer susceptibility: Findings from the Breast Cancer Association Consortium. Hum Mol Genet. 2014;23(22):6096–6111. doi: 10.1093/hmg/ddu311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Webb EL, et al. Search for low penetrance alleles for colorectal cancer through a scan of 1467 non-synonymous SNPs in 2575 cases and 2707 controls with validation by kin-cohort analysis of 14 704 first-degree relatives. Hum Mol Genet. 2006;15(21):3263–3271. doi: 10.1093/hmg/ddl401. [DOI] [PubMed] [Google Scholar]
- 46.Michailidou K, et al. BOCS kConFab Investigators AOCS Group NBCS GENICA Network Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat Genet. 2015;47(4):373–380. doi: 10.1038/ng.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prazeres H, et al. Chromosomal, epigenetic and microRNA-mediated inactivation of LRP1B, a modulator of the extracellular environment of thyroid cancer cells. Oncogene. 2011;30(11):1302–1317. doi: 10.1038/onc.2010.512. [DOI] [PubMed] [Google Scholar]
- 49.Lu YJ, et al. Aberrant methylation impairs low density lipoprotein receptor-related protein 1B tumor suppressor function in gastric cancer. Genes Chromosomes Cancer. 2010;49(5):412–424. doi: 10.1002/gcc.20752. [DOI] [PubMed] [Google Scholar]
- 50.Pinto D, Clevers H. Wnt, stem cells and cancer in the intestine. Biol Cell. 2005;97(3):185–196. doi: 10.1042/BC20040094. [DOI] [PubMed] [Google Scholar]
- 51.Seshagiri S, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488(7413):660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dudgeon C, et al. The evolution of thymic lymphomas in p53 knockout mice. Genes Dev. 2014;28(23):2613–2620. doi: 10.1101/gad.252148.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hulsen T, de Vlieg J, Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang W, et al. DAVID gene ID conversion tool. Bioinformation. 2008;2(10):428–430. doi: 10.6026/97320630002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21(1):70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.