Significance
There is a growing awareness that receptor kinases (RKs) regulate various aspects of plant development in response to perception of specific ligands, but the highly redundant nature of this family's members often makes it difficult to identify their ligands by a conventional mutant screening approach. To sidestep gene redundancy problems, here we used a combination of a custom-made RK expression library and exhaustive photoaffinity labeling and identified three RKs that directly interact with root meristem growth factor (RGF), a peptide hormone regulating root meristem development. The receptor triple mutant was insensitive to externally applied RGF peptide and displayed a short root phenotype accompanied by a considerable decrease in meristematic cell number, representing a previously unidentified ligand–receptor system in plants.
Keywords: peptide hormone, receptor, plant, stem cell, sulfated peptide
Abstract
A peptide hormone, root meristem growth factor (RGF), regulates root meristem development through the PLETHORA (PLT) stem cell transcription factor pathway, but it remains to be uncovered how extracellular RGF signals are transduced to the nucleus. Here we identified, using a combination of a custom-made receptor kinase (RK) expression library and exhaustive photoaffinity labeling, three leucine-rich repeat RKs (LRR-RKs) that directly interact with RGF peptides in Arabidopsis. These three LRR-RKs, which we named RGFR1, RGFR2, and RGFR3, are expressed in root tissues including the proximal meristem, the elongation zone, and the differentiation zone. The triple rgfr mutant was insensitive to externally applied RGF peptide and displayed a short root phenotype accompanied by a considerable decrease in meristematic cell number. In addition, PLT1 and PLT2 protein gradients, observed as a gradual gradient decreasing toward the elongation zone from the stem cell area in wild type, steeply declined at the root tip in the triple mutant. Because RGF peptides have been shown to create a diffusion-based concentration gradient extending from the stem cell area, our results strongly suggest that RGFRs mediate the transformation of an RGF peptide gradient into a PLT protein gradient in the proximal meristem, thereby acting as key regulators of root meristem development.
Receptor kinases (RKs) constitute the largest family of cell surface transmembrane receptors in plants, representing 610 members in Arabidopsis (1). RKs have been shown to play a role in a wide variety of signal transduction pathways related to plant growth, development, environmental adaptation, and defense responses (2). In particular, there is a growing awareness that RKs regulate primary and secondary meristem development in response to binding of specific peptide ligands. These include CLAVATA1 (CLV1) and BARELY ANY MERISTEM1 (BAM1), which directly bind CLV3 peptide, involved in control of the stem cell population in the shoot apical meristem (3–5); TDIF RECEPTOR (TDR)/PHLOEM INTERCALATED WITH XYLEM (PXY), which interacts with TRACHEARY ELEMENT DIFFERENTIATION INHIBITORY FACTOR (TDIF)/CLAVATA3 ESR-related 41/44 (CLE41/44) peptides regulating vascular stem cell proliferation and differentiation (6); and CLV1 and ARABIDOPSIS CRINKLY4 (ACR4), which mediate CLE40 peptide signaling involved in stem cell differentiation in the distal meristem of the root cap (7). These findings highlight the importance of the RK-mediated peptide signaling pathway in controlling meristem development in plants (8), but it is still unknown whether RKs play roles in the regulation of proximal meristem activity in the root apical meristem (RAM).
An involvement of peptide signaling in proximal meristem development was initially suggested by phenotypic analysis of a loss-of-function mutant for tyrosylprotein sulfotransferase (TPST), which catalyzes posttranslational tyrosine sulfation of secreted peptides (9, 10). An Arabidopsis mutant deficient in TPST (tpst-1) shows a short-root phenotype characterized by a considerable decrease in proximal meristem cell number accompanied by reduced expression of stem cell transcription factors PLETHORA1 (PLT1) and PLT2. A subsequent search for sulfated peptides that rescue these meristem defects identified a 13-amino acid sulfated peptide, root meristem growth factor (RGF) (11). The RGF family of peptides comprises 11 genes in Arabidopsis, more than half of which are specifically expressed in quiescent center cells, columella stem cells, and the innermost layer of central columella cells in the root tip. Chemically synthesized RGF peptides restore meristem size by increasing meristematic cell number in the proximal meristem, accompanied by recovery of stem cell function in tpst-1 roots (11). Conversely, rgf1-1 rgf2-1 rgf3-1 triple mutation causes a short-root phenotype characterized by a decrease in meristematic cell number. RGF was later found to cause an irregular wavy growth pattern in roots when overexpressed and, although controversial (12), has been proposed to be involved in the gravitropic response (13).
RGF signaling targets PLT transcription factors, which define patterning of the root proximal meristem (11). PLT proteins display shootward gradient distributions that are highest in the stem cell area, with high levels of PLT maintaining stem cells, intermediate levels facilitating transit-amplifying cell divisions, and low levels allowing cellular differentiation (14). Because PLT1 and PLT2 protein expression and gradient dimensions are considerably reduced in tpst-1 roots, but recover after the application of RGF peptide, RGF is proposed to be a key factor regulating proximal meristem activity through the PLT pathway (11).
Given that accumulating evidence indicates the critical importance of RGF signaling in proximal meristem development, RKs that can transduce extracellular RGF signals to nuclear PLT transcription factors should exist in the proximal meristem region. To this end, the present study aimed to identify membrane-localized RKs involved in RGF perception in Arabidopsis. Functional redundancy of this family, however, often makes it difficult to clearly discern the role of individual members by a genetic approach. We thus used an exhaustive binding assay strategy taking advantage of a custom-made RK expression library, which allowed rapid identification of receptors without being hampered by their low abundance or gene redundancy. This approach enabled us to identify three leucine-rich repeat RKs (LRR-RKs) that redundantly act as specific receptors for RGF in the proximal meristem.
Results
Establishment of RK Expression Library.
From the functional point of view, RKs can be divided into two groups; namely, ligand-binding receptors, which directly interact with ligands, and coreceptors, which form heteromers with ligand-binding receptors to modulate signaling. Cumulative evidence suggests the extracellular domain of ligand-binding receptors is quite large and mostly exceeds 400 amino acid residues (Dataset S1). In contrast, the size of typical coreceptors is so small that fewer than 300 amino acid residues have been found in the extracellular domain. In addition, genes encoding RKs that directly interact with small oligopeptide ligands contain no introns within the gene regions corresponding to the extracellular domain, with the only exception being the ERECTA family (15). Receptors for small ligands in plants might have evolved through gradual structural changes via point mutations, rather than the addition or swapping of domains. According to this empirical rule, we expected that RGF receptors should exist among the Arabidopsis RKs that have a large extracellular domain with no introns.
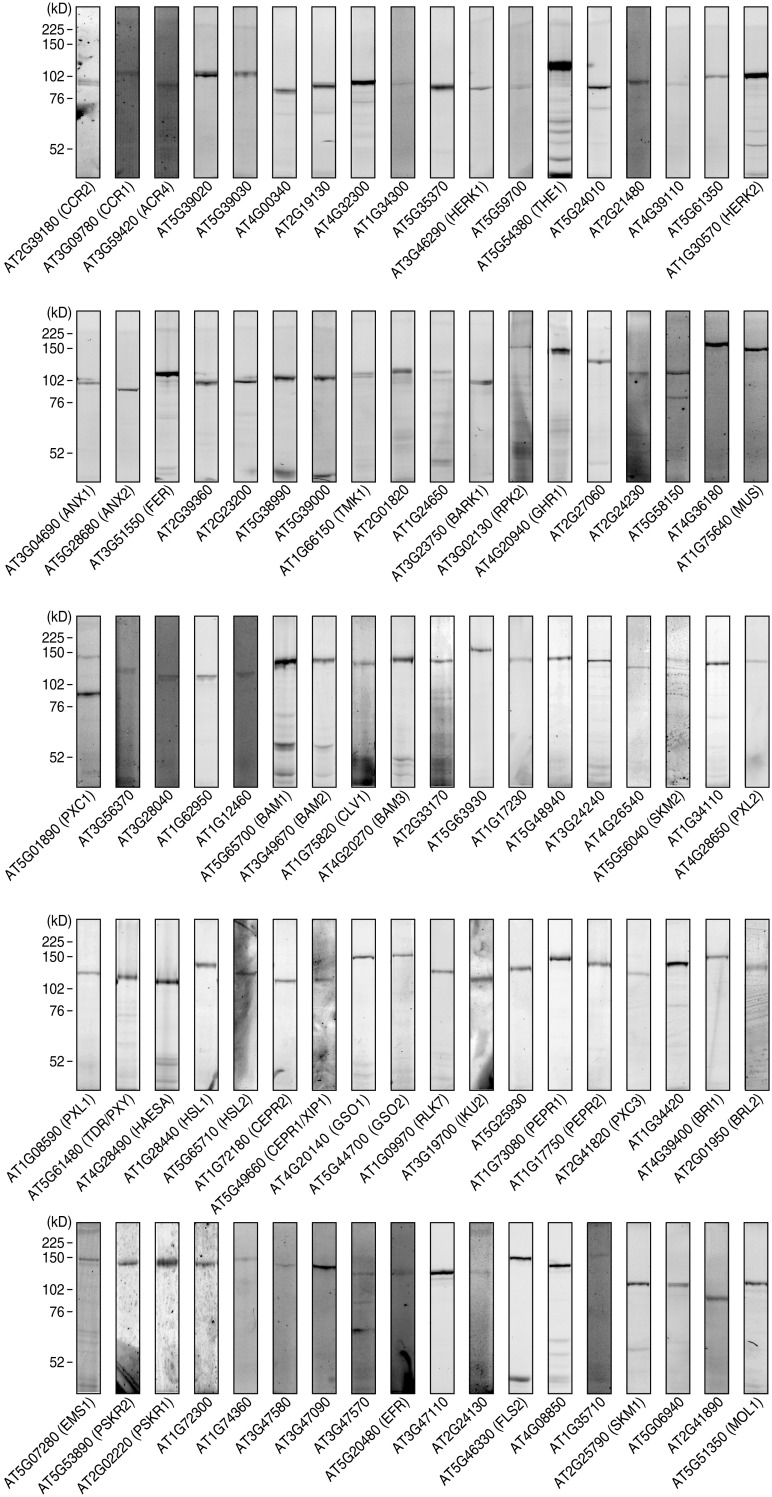
On the basis of this assumption, we analyzed the extracellular domain size of all the Arabidopsis RKs and found that 194 members carry an extracellular domain larger than 400 amino acid residues, including the N-terminal signal sequence. We excluded subfamilies to which members containing more than two introns belong, resulting in 95 members selected as primary candidates (Dataset S2). We individually overexpressed these RKs in tobacco BY-2 cells and established overexpression lines of 90 RKs (Fig. S1). No protein expression was detected for the remaining five RKs.
Fig. S1.
RK expression library covering 90 Arabidopsis RKs. Individual RK-HaloTag fusions expressed in tobacco BY-2 cells were labeled with the HaloTag TMR fluorescent dye, analyzed by SDS/PAGE, and detected by fluoroimaging.
Identification of Three RGF Receptors by Exhaustive Photoaffinity Labeling.
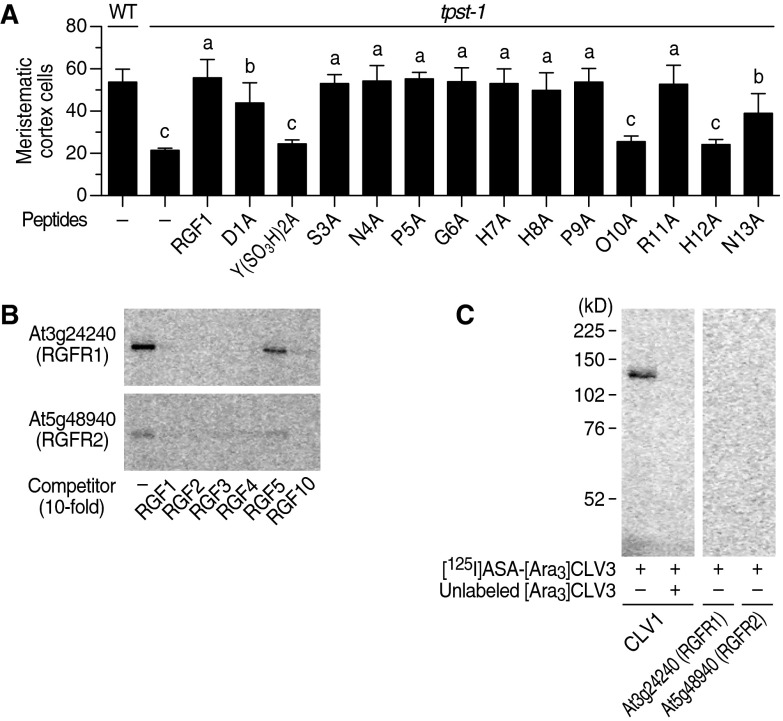
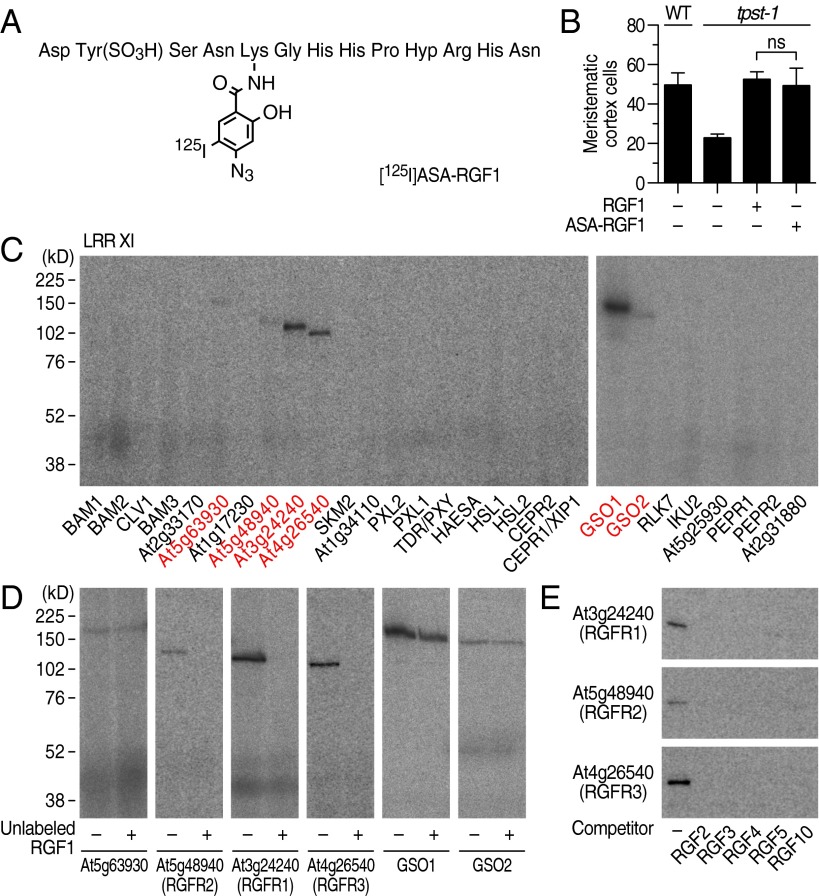
On the basis of the results from the alanine scanning experiments of RGF1 (Fig. S2A), we chemically synthesized a cross-linkable RGF1 derivative in which photoactivatable 4-azidosalicylic acid (ASA) is incorporated into the fifth residue from the N terminus. This ASA-RGF1 was radioiodinated to give [125I]ASA-RGF1 (Fig. 1A). ASA-RGF1 retained biological activity comparable to that of unmodified RGF1 in stimulating meristematic cell division in the tpst-1 mutant (Fig. 1B).
Fig. S2.
Identification of RKs that specifically interact with RGF by photoaffinity labeling. (A) Alanine-scanning experiments with RGF1. Data represent the number of meristematic cortex cells of tpst-1 seedlings grown in the presence of 100 nM alanine-scanned RGF1 analogs at 7 DAG (n = 3–6). Significant differences are indicated with different letters (P < 0.05, one-way ANOVA). (B) Competitive displacement of [125I]ASA-RGF1 binding to At3g24240 and At5g48940 by 10-fold excess unlabeled RGF family peptides. (C) Photoaffinity labeling using [125I]ASA-[Ara3]CLV3 against membrane fractions derived from At3g24240 and At5g48940 expression lines.
Fig. 1.
Identification of three RKs that specifically interact with RGF by photoaffinity labeling. (A) Structure of [125I]ASA-RGF1. (B) Biological activity of ASA-RGF1 determined by measuring meristematic cell number in the tpst-1 mutant at 100 nM. Data represent mean values ± SD (n = 10–16; ns, not significant; paired t test). (C) Exhaustive photoaffinity labeling using [125I]ASA-RGF1 against membrane fractions derived from individual RK expression lines. (D) Competitive displacement of [125I]ASA-RGF1 binding by 300-fold excess unlabeled RGF1. (E) Competitive displacement of [125I]ASA-RGF1 binding by 300-fold excess of other unlabeled RGF members.
To identify RKs that directly interact with RGF1, we performed an exhaustive binding assay, using [125I]ASA-RGF1 against membrane fractions derived from the 88 individual RK expression lines. Our binding assay used covalent photoaffinity labeling followed by immunoprecipitation of the target RKs to exclude false-positive bands arising from abundant proteases that potentially interact with peptide ligands. After SDS/PAGE and autoradiography, we detected binding of [125I]ASA-RGF1 to six LRR-RKs, all of which belong to LRR-RK subfamily XI (Fig. 1C). Competitive displacement of bound [125I]ASA-RGF1 by 300-fold excess unlabeled RGF1 confirmed that At3g24240, At5g48940, and At4g26540 specifically interact with RGF1 (Fig. 1D). These three LRR-RKs also interacted with RGF2, RGF3, RGF4, RGF5 [also known as GLV10 (16)], and RGF10 (At3g60650) peptides in a competitive manner with [125I]ASA-RGF1, indicating that they recognize all the RAM-expressing RGF family peptides (Fig. 1E). Notably, RGF2, RGF3, RGF4, and RGF10 showed complete competition for binding of RGF1 to At3g24240 and At5g48940, even at low excess (10-fold) concentrations, indicating that these four RGF family peptides interact with affinities comparable to that of RGF1 (Fig. S2B). Functionally distinct peptide ligand CLV3 (3, 4), as a negative control, showed no binding to these three LRR-RKs, confirming ligand binding specificity of these LRR-RKs (Fig. S2C). In contrast, binding of [125I]ASA-RGF1 to GSO1, GSO2, and At5g63930 was not competitively antagonized by excess unlabeled RGF1, indicating that the observed binding is nonspecific (Fig. 1D). We concluded that At3g24240, At5g48940, and At4g26540 are possible receptors for RGF and named them RGFR1, RGFR2, and RGFR3, respectively.
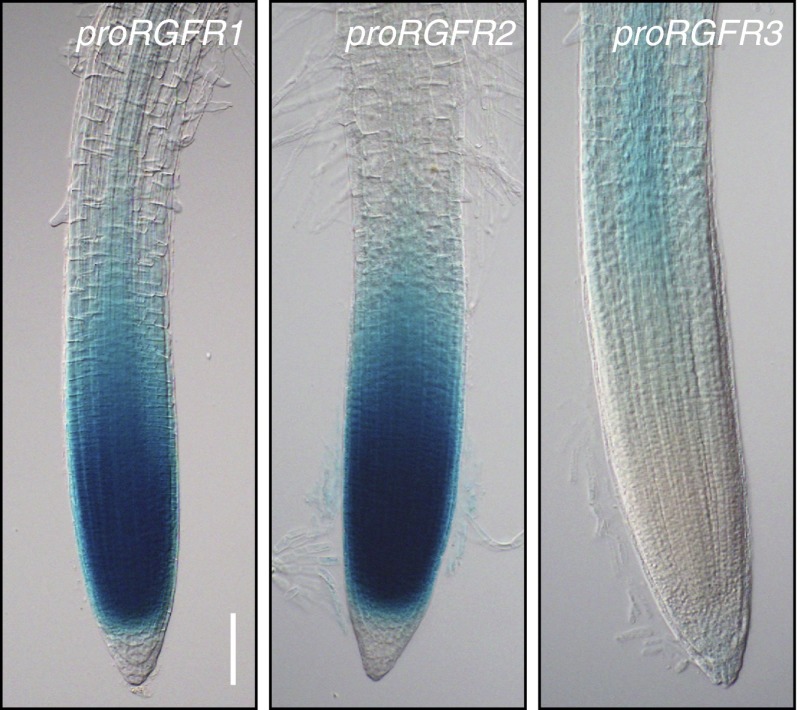
Promoter:GUS expression analysis in Arabidopsis plants revealed that promoter activity of RGFR1 and RGFR2 is predominantly observed in the proximal meristem, including the elongation zone, and gradually decreases toward the differentiation zone (Fig. 2). RGFR2 (At5g48940) is also known as ROOT CLAVATA-HOMOLOG1 (RCH1), the promoter of which has been used to express transgenes in root meristem (17). In contrast, RGFR3 promoter activity is detected in the more basal region of the elongation zone and the differentiation zone. Public microarray data also support these tissue-specific RGFR expression patterns (18).
Fig. 2.
Promoter activities of the RGFR1, RGFR2, and RGFR3 genes. GUS activity is evident in the proximal meristem and the elongation zone of proRGFR1:GUS plants and proRGFR2:GUS plants, and in the elongation zone and differentiation zone of proRGFR3:GUS plants 5 d after germination (DAG). (Scale bar, 100 μm.)
Phenotypic Analysis of rgfr Mutants.
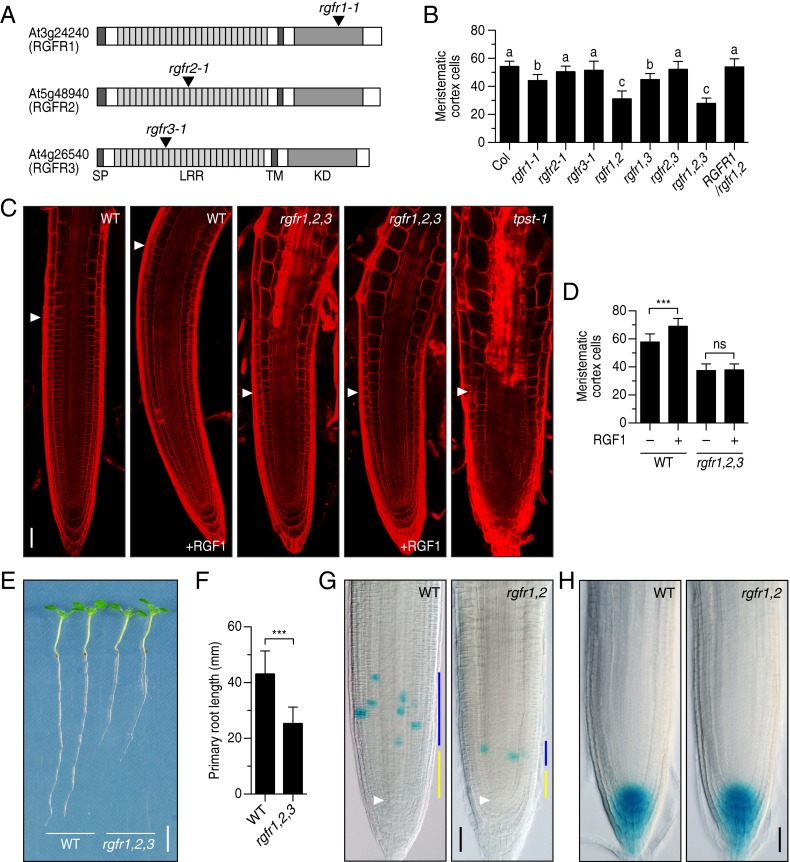
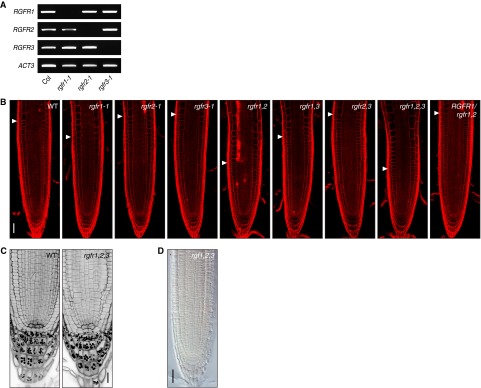
To analyze the effect of loss-of-function mutations of RGFRs on root meristem development, we identified T-DNA insertion mutants for each of the three RGFR genes (rgfr1-1, rgfr2-1, and rgfr3-1) (Fig. 3A). RT-PCR confirmed that a full-length transcript was absent in each mutant (Fig. S3A). The rgfr1-1 single mutant showed a slight decrease in meristematic cell number in the RAM, whereas the rgfr2-1 and rgfr3-1 mutants were indistinguishable from the wild type (Fig. 3B and Fig. S3B). We further generated all combinations of double and triple knockout mutant plants to uncover possible redundant roles of RGFRs. We observed a considerable decrease in meristematic cell number in the rgfr1-1 rgfr2-1 rgfr3-1 triple mutant to a level similar to that of the tpst-1 mutant, which is deficient in biosynthesis of all sulfated peptides, including RGFs (Fig. 3 B and C and Fig. S3B) (9, 10). A substantial decrease in meristematic cell number was also found in the rgfr1-1 rgfr2-1 double mutant, comparable to that in the rgfr1-1 rgfr2-1 rgfr3-1 triple mutant, indicating a major role of RGFR1 and RGFR2 in regulating proximal meristem activity (Fig. 3B and Fig. S3B). Complementation of the rgfr1-1 rgfr2-1 double mutant with RGFR1 genomic DNA reversed the double mutant phenotype, confirming that the observed morphological changes arise from the absence of functional RGFR proteins and validating the assumption of functional redundancy between RGFR1 and RGFR2 (Fig. 3B and Fig. S3B).
Fig. 3.
Phenotypic analysis of T-DNA insertion mutants of the RGFR genes. (A) Schematic representation of T-DNA insertion sites in rgfr1-1, rgfr2-1, and rgfr3-1. (B) Number of meristematic cortex cells in root meristem of wild-type, rgfr mutants, and complemented line at 5 DAG. Significant differences are indicated with different letters (n = 16–23; one-way ANOVA, see also Fig. S3B). (C) Confocal images of root meristem of wild-type and rgfr1-1 rgfr2-1 rgfr3-1 triple mutant treated with 100 nM RGF1 at 5 DAG. The root meristem of the tpst-1 mutant is also shown as a reference. (D) Quantitative analysis of the number of meristematic cortex cells in Fig. 3C (n = 16–35; ***P < 0.001; paired t test). (E) Root length of wild-type and rgfr1-1 rgfr2-1 rgfr3-1 triple mutant seedlings at 7 DAG. (F) Quantitative comparison of root length in Fig. 3E (n = 47–50; ***P < 0.001; paired t test). (G) CYCB1;1-GUS expression in wild type and rgfr1-1 rgfr2-1 root meristem at 5 DAG. Blue bar represents transit amplifying zone; yellow bar represents slow-dividing immature cell population zone that is thought to be a stem cell niche. White arrowheads indicate the quiescent center. (H) DR5:GUS expression in wild type and rgfr1-1 rgfr2-1 root meristems at 5 DAG. (Scale bar, C, G, and H, 50 μm; E, 5 mm.)
Fig. S3.
Root phenotypes of rgfr mutants. (A) Absence of full-length RGFR transcripts in each mutant, as demonstrated by RT-PCR. (B) Confocal images of root meristem of wild-type, rgfr mutants, and complemented line at 5 DAG. Arrowheads indicate the root meristem boundary. See also Fig. 3B. (C) Confocal images of distal meristem of wild-type and rgfr1-1 rgfr2-1 rgfr3-1 seedlings at 5 DAG, stained with the mPS-PI method. (D) Whole-mount in situ hybridization with PLT2 sense probe in root meristem of rgf1-1 rgf2-1 rgf3-1 ligand mutant. No signal was detected with a sense probe. (Scale bar, B and D, 50 μm; C, 20 μm.)
To examine whether the rgfr1-1 rgfr2-1 rgfr3-1 triple mutant is defective in RGF responses, wild-type and mutant seedlings were cultured in a liquid medium containing 100 nM RGF1. Under the conditions in which wild-type roots respond to RGF1, the rgfr1-1 rgfr2-1 rgfr3-1 triple mutant showed no detectable changes in meristematic cell number in the RAM (Fig. 3 C and D). Enhanced expansion and premature elongation of cells in the elongation zone, reminiscent of the phenotype conferred by tpst-1 mutation, was also not alleviated by RGF1 treatment in the rgfr1-1 rgfr2-1 rgfr3-1 triple mutant. Accordingly, the rgfr1-1 rgfr2-1 rgfr3-1 triple mutant developed shorter roots than wild type because of the decrease in meristematic activity (Fig. 3 E and F).
The cell cycle marker CYCB1;1-GUS in the rgfr1-1 rgfr2-1 double mutant indicated a reduction in cell division rate of the transit-amplifying zone and a decrease in the adjacent slow-dividing immature cell population zone that is thought to be a stem cell niche (Fig. 3G) (19). Importantly, the auxin distribution in rgfr1-1 rgfr2-1 root tips visualized by the DR5:GUS marker was comparable to that of wild type, indicating that this root meristem defect is not associated with changes in auxin distribution (Fig. 3H).
In contrast to the evident role of the RGFRs in the proximal meristem, the rgfr1-1 rgfr2-1 rgfr3-1 triple mutant displayed no obvious phenotype in the distal meristem (Fig. S3C). In addition, both public microarray data and our promoter activity analysis indicated no or low expression of these three RGFR genes in the distal meristem (Fig. 2) (18). Externally applied RGF has been shown to promote distal stem cell activity as well as proximal meristem activity in the tpst-1 mutant (11). This might be interpreted as external RGF indirectly affecting distal stem cell activity as a result of recovery of proximal meristem function in tpst-1. Taken together, we concluded that RGFR1, RGFR2, and RGFR3 act as specific receptors for RGF peptides in the proximal meristem.
Loss of RGFRs Abolished PLT Protein Gradient in Proximal Meristem.
PLT1 and PLT2 are AP2 class transcription factors that play a crucial role in root meristem development by mediating root stem cell niche patterning (14). PLT proteins show gradient expression patterns that extend from the stem cell area into the elongation zone across the region of rapidly dividing transit-amplifying cells. High PLT levels maintain stem cell identity, intermediate levels promote cell division of stem cell daughters, and low levels allow cell differentiation at the elongation zone (14). Because exogenous RGF application has been shown to cause enlargement of PLT1 and PLT2 expression domains (11), we investigated whether rgfr1-1 rgfr2-1 rgfr3-1 triple mutation affects the PLT gradient in the RAM.
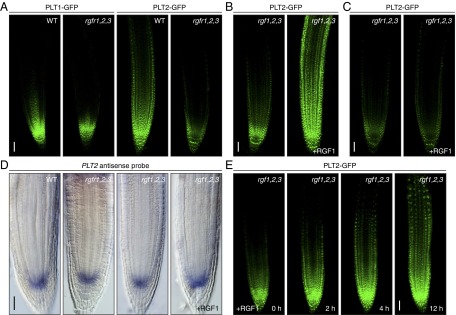
In wild-type plants, PLT1-GFP and PLT2-GFP exhibited a gradual shootward gradient from the root stem cell area (Fig. 4A). In contrast, gradients of PLT1-GFP and PLT2-GFP steeply declined at the root tip in the rgfr1-1 rgfr2-1 rgfr3-1 background, with weak expression being retained in the stem cell area. In particular, the PLT2-GFP expression domain was strikingly diminished in the triple mutant compared with wild type (Fig. 4A). A similar reduction in the PLT2-GFP expression domain was also observed in the ligand triple mutant rgf1-1 rgf2-1 rgf3-1, albeit milder than in the receptor triple mutant, indicating that RGFs and RGFRs are required for maintaining a proper PLT gradient in the proximal meristem (Fig. 4B). Exogenous application of RGF1 to the ligand mutant rgf1-1 rgf2-1 rgf3-1 caused drastic shootward enlargement of the PLT2-GFP expression domain to an even greater extent than the natural PLT2 gradient in wild type (Fig. 4B). This result can be interpreted as the intrinsic RGF peptide level defining the magnitude and the slope of the PLT gradient. In contrast, the receptor mutant rgfr1-1 rgfr2-1 rgfr3-1 was substantially less sensitive to RGF1 with respect to recovery of PLT2-GFP expression (Fig. 4C). Collectively, these results indicate that the three RGFRs act as receptors for RGFs in the root meristem to define the PLT gradient.
Fig. 4.
RGFRs are required for maintaining proper gradients of PLT transcription factors in proximal meristem. (A) Confocal image of root meristem of wild-type and rgfr1-1 rgfr2-1 rgfr3-1 receptor mutant seedlings expressing PLT1-GFP and PLT2-GFP at 5 DAG. (B) Root meristem of rgf1-1 rgf2-1 rgf3-1 ligand mutant seedlings expressing PLT2-GFP treated with or without 100 nM RGF1 for 24 h. (C) Root meristem of rgfr1-1 rgfr2-1 rgfr3-1 receptor mutant seedlings expressing PLT2-GFP treated with or without 100 nM RGF1 for 24 h. (D) Whole-mount in situ hybridization with PLT2 antisense probe in root meristem of wild type, rgfr1-1 rgfr2-1 rgfr3-1 receptor mutant, rgf1-1 rgf2-1 rgf3-1 ligand mutant, and the ligand mutant treated with 100 nM RGF1 for 24 h. (E) Changes in expression patterns of PLT2-GFP proteins in rgf1-1 rgf2-1 rgf3-1 ligand mutant after RGF1 treatment. (Scale bar, A–E, 50 μm.)
PLTs Are Very Proximal Downstream Targets of RGFRs.
In contrast to the drastic changes in PLT2 protein expression patterns in the mutants, in situ hybridization showed that localization of PLT2 transcripts was comparable among wild type, the rgfr1-1 rgfr2-1 rgfr3-1 receptor mutant, and the rgf1-1 rgf2-1 rgf3-1 ligand mutant, even after RGF1 treatment (Fig. 4D and Fig. S3D). In all lines, PLT2 transcripts were detected only in the stem cell region, irrespective of the PLT2 protein distribution. These expression profiles suggest that RGF defines PLT expression patterns at the protein level, possibly through stabilization of PLT proteins.
We also analyzed the time course of changes in expression patterns of PLT2-GFP proteins in the rgf1-1 rgf2-1 rgf3-1 ligand mutant after RGF1 treatment (Fig. 4E). Notably, the RGF response of the PLT2-GFP proteins was very rapid, and enlargement of PLT2-GFP expression domain was readily detected within 2–4 h after treatment. The PLT2-GFP expression domain further enlarged shootward to an even greater extent than in wild type after 12 h. These RGF response dynamics of PLT2-GFP proteins strongly suggest that PLTs are direct or very proximal downstream targets of RGFRs.
Discussion
We identified three LRR-RKs (RGFR1, RGFR2, and RGFR3) that specifically interact with RGF peptide by exhaustive photoaffinity labeling against a custom-made LRR-RK expression library. Multiple loss-of-function mutations in these LRR-RKs led to impaired RGF response and caused a short root phenotype characterized by a considerable decrease in meristematic cell number in the root proximal meristem. Competitive binding analysis revealed that the majority of the RAM-expressing RGF family peptides interact with RGFR1 and RGFR2 at affinities comparable that of RGF1, which indicates the potentially redundant action of these RGF family peptides. Competitive photoaffinity labeling, however, does not always precisely indicate the relative binding affinities of each peptide, especially when competitor peptides bind receptors at higher affinity than photoaffinity ligands. It has been recently reported that RGF1 and RGF2 play disparate roles in root development under phosphate depletion (20). One interpretation is that environmental stresses may affect expression levels and patterns of the three RGFR subtypes, which may have slightly different ligand recognition spectra.
We also found that RGFRs are required for maintaining proper gradients of PLT1 and PLT2 transcription factors, which regulate stem cell specification and transit-amplifying cell proliferation. In particular, PLT2 protein expression and gradient dimensions are considerably reduced both in the ligand mutant rgf1-1 rgf2-1 rgf3-1 and the receptor mutant rgfr1-1 rgfr2-1 rgfr3-1. Importantly, exogenous application of RGF1 caused drastic enlargement of the PLT2-GFP expression domain in the ligand mutant, but not in the receptor mutant, to even greater levels than in wild type. Moreover, the RGF response of the PLT2-GFP proteins was very rapid, readily detectable within 2–4 h after treatment, suggesting that PLTs are direct or very proximal downstream targets of RGFRs. Because RGF peptides secreted from the stem cell region create a diffusion-based concentration gradient extending shootward from the stem cell area (11), our results strongly suggest that RGFRs mediate the transformation of an RGF peptide gradient into a PLT protein gradient in the proximal meristem, thereby acting as key regulators of root meristem development.
From the biochemical point of view, the slope of the gradient of RGF is defined exclusively by the half-life of RGF degradation and the diffusion coefficient of the peptides. Degradation mechanisms of the peptide hormones are not well understood, but major proteolytic activity in the apoplast is attributable to nonspecific proteases that degrade a broad spectrum of peptides at a certain rate (21). The diffusion coefficient is a physical parameter that is solely proportional to the molecular weight, and independent of environmental fluctuations. Thus, even though transcription of RGF genes is influenced by environmental changes, the gradient slope of RGF itself would remain largely stable. Under this system, a particular RGF concentration zone may shift spatially, depending on the RGF gene expression levels, but is always maintained somewhere within the gradient, which enables buffering of short-term environmental fluctuations. PLT gene expression is affected by auxin (14), whereas the PLT protein gradient has been shown not to be a readout of the auxin distribution but, rather, is suggested to be regulated at the level of protein stability (11, 19). Therefore, it is tempting to propose that a simple diffusion-based RGF peptide gradient defines the PLT protein gradient by modulating the stability of these proteins to ensure robust root growth and development in fluctuating natural environments.
Materials and Methods
Expression of Arabidopsis Receptor Kinases in Tobacco BY-2 Cells.
To overexpress Arabidopsis receptor kinases in tobacco BY-2 cells, we amplified the genomic fragment of each receptor kinase gene that corresponds from the Met1 to the end of the juxtamembrane domain and the entire ORF of the HaloTag vector (Promega) by PCR. These two fragments were cloned in translational fusion by three-component ligation into the BamHI/SacI-digested binary vector pBI121, using an In-Fusion HD Cloning Kit (Clontech). Transformation of tobacco BY-2 cells and preparation of microsomal fractions were as described previously (4). Expressed HaloTag-fused receptors were specifically labeled using HaloTag TMR ligand (Promega), separated by SDS/PAGE, and visualized using a Typhoon 9400 fluorescent image analyzer (GE Healthcare) with a 523-nm excitation filter and a 580-nm emission filter.
Preparation of [125I]ASA-RGF1 and Photoaffinity Labeling.
The Fmoc-protected RGF1 analog Fmoc-[Lys5]RGF1 was synthesized by Fmoc chemistry, using a peptide synthesizer (Applied Biosystems Model 431A). This peptide was reacted with 4-azidosalicylic acid succinimidyl ester (Pierce), followed by deprotection with piperidine to yield [(4-azidosalicyl)-Lys5]RGF1 (ASA-RGF1). ASA-RGF1 was further radioiodinated by the chloramine T method, as previously described (4). The labeled peptide was purified by reverse-phase HPLC to yield analytically pure [125I]ASA-RGF1 with specific radioactivity of 80 Ci/mmol. Photoaffinity labeling, immunoprecipitation, and SDS/PAGE were performed according to a previous report (22), using 30 nM [125I]ASA-RGF1. Control binding experiments using [125I]ASA-[Ara3]CLV3 were performed as previously described (3).
Plant Materials and Growth Conditions.
Arabidopsis ecotype Columbia (Col) was used. T-DNA–tagged rgfr mutants were identified in the SALK T-DNA collections (rgfr1-1, SALK_040393; rgfr2-1, SALK_096206; rgfr3-1, SALK_053167) (23). Marker line CYCB1;1:GUS was a gift from P. Doerner, University of Edinburgh, Edinburgh (24), and the DR5:GUS marker was from T. Guilfoyle, University of Missouri, Columbia, MO (25). For PLT1/PLT2 expression analysis in the rgfr1 rgfr2 rgfr3 triple mutant, rgfr mutant lines were directly transformed with the PLT1-GFP and PLT2-GFP translational fusions (11), using the floral dipping method. For complementation analysis, a 5.8-kb genomic DNA fragment containing the entire ORF along with 2.5 kb upstream of the ATG start codon was amplified from Col genomic DNA. The PCR product was cloned into the pBI101-Hm vector, and the resulting construct was introduced into rgfr1-1 rgfr2-1 double-mutant plants. For the analysis of root meristem and PLT1/PLT2-GFP expression, surface-sterilized Arabidopsis seeds were directly sown into 1.0 mL B5 medium containing 1.0% sucrose in 24-well microplates and vernalized for 2 d at 4 °C, then incubated without shaking under continuous light at 22 °C for 5–8 d. For the analysis of root length, plants were vertically grown on the same medium solidified with 1.5% agar. All plants were grown under continuous light at 22 °C. Peptides were added to the medium at 100 nM.
Promoter Analysis of RGFR1, RGFR2, and RGFR3.
The 2.5-kb promoter regions upstream of RGFR1, RGFR2, and RGFR3 (including the initiation codon) were amplified and cloned by translational fusion in-frame with the β-glucuronidase (GUS) coding sequence in the binary vector pBI101 and transformed into Arabidopsis wild type. For GUS staining, β-glucuronidase activity was visualized by X-Gluc as substrate, using a conventional protocol.
Microscopy and Histochemistry.
For confocal root imaging, cell outlines were stained with 50 μg/mL propidium iodide for 2 min and observed under a confocal laser-scanning microscope (Olympus FV300) with helium-neon laser excitation at 543 nm. GFP images were collected after argon laser excitation at 488 nm. The number of root meristematic cells was obtained by counting cortex cells showing no signs of rapid elongation. For visualization of starch granules and cell walls in root tips, mPS-PI staining was performed according to a previous report (26). Whole-mount in situ hybridization experiments were performed as previously described (11).
Supplementary Material
Acknowledgments
We thank Mari Ogawa-Ohnishi for technical assistance with whole-mount in situ hybridization. This research was supported by a Grant-in-Aid for Scientific Research (S) (25221105 to Y.M.), a Grant-in-Aid for Scientific Research on Innovative Areas (15H05957 to Y.M. and 26113520 to H.S.), and a Grant-in-Aid for Young Scientists (B) (25840111 to H.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1522639113/-/DCSupplemental.
References
- 1.Shiu SH, Bleecker AB. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA. 2001;98(19):10763–10768. doi: 10.1073/pnas.181141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gish LA, Clark SE. The RLK/Pelle family of kinases. Plant J. 2011;66(1):117–127. doi: 10.1111/j.1365-313X.2011.04518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shinohara H, Matsubayashi Y. Reevaluation of the CLV3-receptor interaction in the shoot apical meristem: Dissection of the CLV3 signaling pathway from a direct ligand-binding point of view. Plant J. 2015;82(2):328–336. doi: 10.1111/tpj.12817. [DOI] [PubMed] [Google Scholar]
- 4.Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science. 2008;319(5861):294. doi: 10.1126/science.1150083. [DOI] [PubMed] [Google Scholar]
- 5.Guo Y, Han L, Hymes M, Denver R, Clark SE. CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification. Plant J. 2010;63(6):889–900. doi: 10.1111/j.1365-313X.2010.04295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirakawa Y, et al. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc Natl Acad Sci USA. 2008;105(39):15208–15213. doi: 10.1073/pnas.0808444105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stahl Y, et al. Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr Biol. 2013;23(5):362–371. doi: 10.1016/j.cub.2013.01.045. [DOI] [PubMed] [Google Scholar]
- 8.Matsubayashi Y. Posttranslationally modified small-peptide signals in plants. Annu Rev Plant Biol. 2014;65:385–413. doi: 10.1146/annurev-arplant-050312-120122. [DOI] [PubMed] [Google Scholar]
- 9.Zhou W, et al. Arabidopsis Tyrosylprotein sulfotransferase acts in the auxin/PLETHORA pathway in regulating postembryonic maintenance of the root stem cell niche. Plant Cell. 2010;22(11):3692–3709. doi: 10.1105/tpc.110.075721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komori R, Amano Y, Ogawa-Ohnishi M, Matsubayashi Y. Identification of tyrosylprotein sulfotransferase in Arabidopsis. Proc Natl Acad Sci USA. 2009;106(35):15067–15072. doi: 10.1073/pnas.0902801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuzaki Y, Ogawa-Ohnishi M, Mori A, Matsubayashi Y. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science. 2010;329(5995):1065–1067. doi: 10.1126/science.1191132. [DOI] [PubMed] [Google Scholar]
- 12.Meng L, Buchanan BB, Feldman LJ, Luan S. CLE-like (CLEL) peptides control the pattern of root growth and lateral root development in Arabidopsis. Proc Natl Acad Sci USA. 2012;109(5):1760–1765. doi: 10.1073/pnas.1119864109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitford R, et al. GOLVEN secretory peptides regulate auxin carrier turnover during plant gravitropic responses. Dev Cell. 2012;22(3):678–685. doi: 10.1016/j.devcel.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 14.Galinha C, et al. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature. 2007;449(7165):1053–1057. doi: 10.1038/nature06206. [DOI] [PubMed] [Google Scholar]
- 15.Lee JS, et al. Competitive binding of antagonistic peptides fine-tunes stomatal patterning. Nature. 2015;522(7557):439–443. doi: 10.1038/nature14561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez A, et al. Transcriptional and functional classification of the GOLVEN/ROOT GROWTH FACTOR/CLE-like signaling peptides reveals their role in lateral root and hair formation. Plant Physiol. 2013;161(2):954–970. doi: 10.1104/pp.112.206029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casamitjana-Martínez E, et al. Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr Biol. 2003;13(16):1435–1441. doi: 10.1016/s0960-9822(03)00533-5. [DOI] [PubMed] [Google Scholar]
- 18.Schmid M, et al. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37(5):501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 19.Mähönen AP, et al. PLETHORA gradient formation mechanism separates auxin responses. Nature. 2014;515(7525):125–129. doi: 10.1038/nature13663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cederholm HM, Benfey PN. Distinct sensitivities to phosphate deprivation suggest that RGF peptides play disparate roles in Arabidopsis thaliana root development. New Phytol. 2015;207(3):683–691. doi: 10.1111/nph.13405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delannoy M, et al. Identification of peptidases in Nicotiana tabacum leaf intercellular fluid. Proteomics. 2008;8(11):2285–2298. doi: 10.1002/pmic.200700507. [DOI] [PubMed] [Google Scholar]
- 22.Shinohara H, Moriyama Y, Ohyama K, Matsubayashi Y. Biochemical mapping of a ligand-binding domain within Arabidopsis BAM1 reveals diversified ligand recognition mechanisms of plant LRR-RKs. Plant J. 2012;70(5):845–854. doi: 10.1111/j.1365-313X.2012.04934.x. [DOI] [PubMed] [Google Scholar]
- 23.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301(5633):653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 24.Colón-Carmona A, You R, Haimovitch-Gal T, Doerner P. Technical advance: Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 1999;20(4):503–508. doi: 10.1046/j.1365-313x.1999.00620.x. [DOI] [PubMed] [Google Scholar]
- 25.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9(11):1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Truernit E, et al. High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of Phloem development and structure in Arabidopsis. Plant Cell. 2008;20(6):1494–1503. doi: 10.1105/tpc.107.056069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.