Significance
The majority of pharmaceuticals are inspired by natural product scaffolds that are functionalized by tailoring enzymes, such as halogenases. The degree of halogenation is an important determinant of natural product bioactivity, yet little is known regarding the molecular basis for the exquisite control exhibited by tailoring halogenases. Known pyrrole halogenases commonly perform up to two halogenations on the pyrrole. Our study of tetrabromopyrrole biosynthesis revealed a uniquely adapted halogenase–thioesterase enzyme pair that catalyzes an unprecedented series of halogenations on a pyrrole. Structural comparison of the pyrrole tetrahalogenase to a pyrrole dihalogenase revealed key residues involved in controlling the degree of halogenation. Our findings provide fundamental insights that might be applied in the rational design of biocatalysts toward directed biosynthesis of new chemicals.
Keywords: natural products, biosynthesis, halogenation, enzymology
Abstract
Halogenated pyrroles (halopyrroles) are common chemical moieties found in bioactive bacterial natural products. The halopyrrole moieties of mono- and dihalopyrrole-containing compounds arise from a conserved mechanism in which a proline-derived pyrrolyl group bound to a carrier protein is first halogenated and then elaborated by peptidic or polyketide extensions. This paradigm is broken during the marine pseudoalteromonad bacterial biosynthesis of the coral larval settlement cue tetrabromopyrrole (1), which arises from the substitution of the proline-derived carboxylate by a bromine atom. To understand the molecular basis for decarboxylative bromination in the biosynthesis of 1, we sequenced two Pseudoalteromonas genomes and identified a conserved four-gene locus encoding the enzymes involved in its complete biosynthesis. Through total in vitro reconstitution of the biosynthesis of 1 using purified enzymes and biochemical interrogation of individual biochemical steps, we show that all four bromine atoms in 1 are installed by the action of a single flavin-dependent halogenase: Bmp2. Tetrabromination of the pyrrole induces a thioesterase-mediated offloading reaction from the carrier protein and activates the biosynthetic intermediate for decarboxylation. Insights into the tetrabrominating activity of Bmp2 were obtained from the high-resolution crystal structure of the halogenase contrasted against structurally homologous halogenase Mpy16 that forms only a dihalogenated pyrrole in marinopyrrole biosynthesis. Structure-guided mutagenesis of the proposed substrate-binding pocket of Bmp2 led to a reduction in the degree of halogenation catalyzed. Our study provides a biogenetic basis for the biosynthesis of 1 and sets a firm foundation for querying the biosynthetic potential for the production of 1 in marine (meta)genomes.
Marine bacteria of the genus Pseudoalteromonas produce numerous small molecule natural products with varied roles in marine chemical ecology (1–4). Recently, tetrabromopyrrole (1, Fig. 1) was established as a chemical cue produced by Pseudoalteromonas that induces larval settlement and metamorphosis in reef-building Caribbean coral species (3). Additionally, the chemical structure of 1 is notable for its degree of halogenation (one-to-one carbon-to-halogen ratio), unique among naturally occurring aromatic organohalogens (5).
Fig. 1.
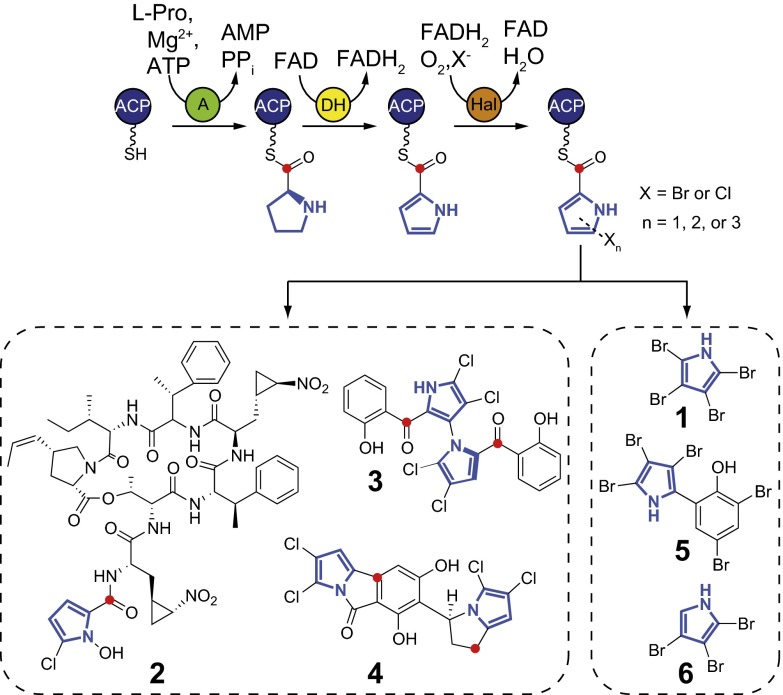
Structures and biosynthesis of halopyrrole natural products. Halopyrrole biosynthesis starts from the loading of l-proline onto an ACP phosphopantetheine thiol by an adenylation (A) enzyme, followed by a 4e− oxidation of the prolyl ring to a pyrrole (in blue) catalyzed by a dehydrogenase (DH) enzyme. Subsequent halogenation by a flavin-dependent halogenase (Hal) then installs one, two, or three halogens on the pyrrole ring. At this stage, the thiotemplated halopyrrole can proceed down assembly line biosynthetic routes to afford natural products, such as hormaomycin (2), marinopyrrole A (3), and chlorizidine A (4) (8–10). However, the biosyntheses of 1 and 5–6 do not entail modular assembly line extension of the l-proline–derived halopyrrole, but rather involve the loss of the prolyl alpha-carboxylate carbon atom (in red) via an unexplained mechanism.
Halopyrrole-containing microbial natural products are biosynthesized starting from l-proline. A highly conserved halopyrrole biosynthetic route involves oxidation of an acyl carrier protein (ACP)-loaded prolyl side chain and subsequent halogenation by a flavin-dependent halogenase (Fig. 1) (6). At this stage, two biosynthetic routes are possible. In the first route, the ACP-loaded halopyrrole is extended using modular assembly lines to yield nonribosomally synthesized peptide, or polyketide natural products (e.g., 2–4, Fig. 1) (7–10). During the biosynthesis of 2–4, and other related natural products, the halopyrrole carboxylic acid is transthioesterified to downstream ACPs during modular elongation reactions. Molecules arising from these modular biosynthetic pipelines are characterized by the preservation of the prolyl alpha-carboxyl carbon atom as a carbonyl, or embedded in the final natural product skeleton in varying oxidation states (in red, Fig. 1). In a recently discovered alternate route, the prolyl alpha-carboxyl carbon atom is lost during the biosynthesis of 1 and the cytotoxic marine bacterial natural product pentabromopseudilin (5, Fig. 1) (4, 11). Uniquely, 5 is biosynthesized via the coupling of 2,3,4-tribromopyrrole (6, Fig. 1) to 2,4-dibromophenol rather than via a modular assembly line. The molecular mechanism for the pyrrole offloading from the ACP and elimination of prolyl alpha-carboxyl carbon en route to the biosyntheses of 1 and 5–6 has not been determined. The structure of 1 poses an additional biosynthetic challenge due to the presence of a bromine atom in place of an acyl side chain. Although all previously described pyrrole halogenases catalyze one, two, or three halogen additions upon the ACP-loaded pyrrole ring, a halogenase capable of four halogenations on the pyrrole ring, as implied by the structure of 1, has not been characterized. These open biosynthetic questions, together with the ecological significance of 1 in coral larval settlement, motivated us to explore the genetic and molecular logic for its biosynthesis in marine bacteria.
Herein, we establish the biosynthesis of 1 by the total in vitro enzymatic reconstitution of its biosynthetic machinery. We show that a single flavin-dependent brominase installs an unprecedented four halogens on an ACP-bound pyrrole required for the progression of thioesterase (TE)-mediated offloading and decarboxylation reactions. We also exploit an opportunity to structurally characterize and investigate via site-directed mutagenesis the molecular basis for a differential number of halogen additions catalyzed by flavin-dependent halogenases on aromatic substrates. Our structural comparison of a highly homologous pyrrole dihalogenase to the pyrrole tetrahalogenase reveals subtle variations in biocatalyst design that lead to divergent molecular outcomes.
Results
Genetic Basis for the Biosynthesis of 1.
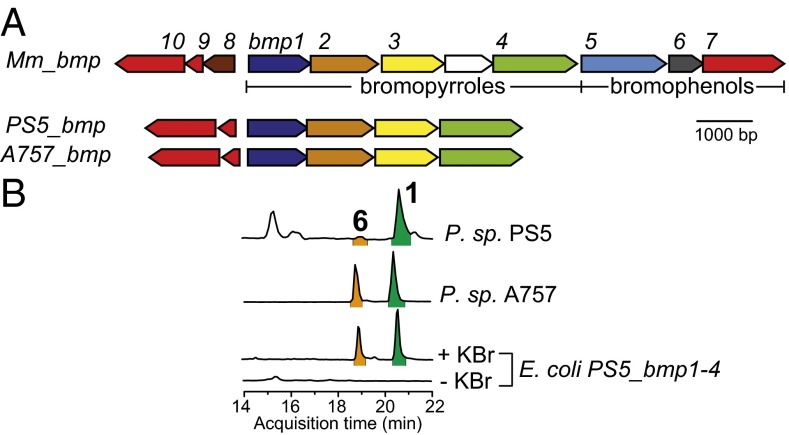
We recently reported that 1 produced by the marine bacterium Pseudoalteromonas sp. PS5 induces settlement of larvae associated with several Caribbean coral species (3). To investigate the genetic basis for the biosynthesis of 1, we sequenced and assembled a 5.08-Mbp draft genome for P. sp. PS5. Querying the draft genome for the presence of the halopyrrole biosynthetic genes, we identified a gene locus in P. sp. PS5 with high homology to a subset of bromopyrrole biosynthetic genes present in the previously reported bmp gene locus from marine bacteria of the genera Pseudoalteromonas and Marinomonas (11) (Fig. 2A and SI Appendix, Table S1). The organization of the bmp homologs identified in the genome of P. sp. PS5 (PS5_bmp) is identical to that of the bmp gene cluster found in Marinomonas mediterranea MMB-1 bmp (Mm_bmp) and others (11). Notably, the PS5_bmp gene locus maintains the bromopyrrole biosynthetic module Mm_bmp1–4 and the bromopyrrole/phenol coupling cytochrome P450 (CYP450) accessory genes Mm_bmp9–10, but lacks the genes associated with bromophenol biosynthesis (Mm_bmp5–6) and the CYP450-Mm_bmp7 (Fig. 2A). In addition, we also sequenced and assembled a 5.13-Mbp draft genome for Pseudoalteromonas sp. A757 that also produces 1 (12). Querying the genome of P. sp. PS5 as before revealed a gene cluster with high homology to PS5_bmp (A757_bmp), providing a second example of a stand-alone bromopyrrole biosynthetic pathway from the genus Pseudoalteromonas (Fig. 2A and SI Appendix, Table S1).
Fig. 2.
A genetic basis for production of 1. (A) Mm_bmp, PS5_bmp, and A757_bmp gene clusters with bromopyrrole and bromophenol biosynthetic genes indicated below the Mm_bmp gene cluster; a putative permease (uncolored) is inserted between Mm_bmp3 and Mm_bmp4. Mm_Bmp1–4 are colored per their catalytic roles shown in Fig. 1. Note that bmp1 encodes a di-domain protein with an ACP domain at the N terminus followed by a TE domain (11). (B) LC/MS extracted ion chromatograms (EICs) for [M-H]1- ions corresponding to 1 and 6 in organic extracts of P. sp. PS5, P. sp. A757, and E. coli expressing PS5_bmp1–4 grown in media with (+) or without (−) bromide.
To evaluate the role of PS5_bmp1–4 genes in the production of 1, we cloned and heterologously expressed PS5_bmp1–4 in Escherichia coli. Only in the presence of bromide in the culture media, we observed robust heterologous production of 1 along with a minor amount of 6, consistent with the product profile observed in organic extracts of cultures of P. sp. PS5 and P. sp. A757 (Fig. 2B). Although these results demonstrate that bmp1–4 genes are necessary for the production of 1, the molecular detail underlying the offloading of the l-proline–derived pyrrole moiety from the ACP, its timing relative to the bromination events, and the chemical logic for the loss of l-proline derived alpha-carboxyl was not discernible at this stage. Therefore, we next examined the biosynthetic pathway by total in vitro enzymatic reconstitution of the production of 1 using purified enzyme catalysts.
Decarboxylative Bromination of Thiotemplated Pyrrole.
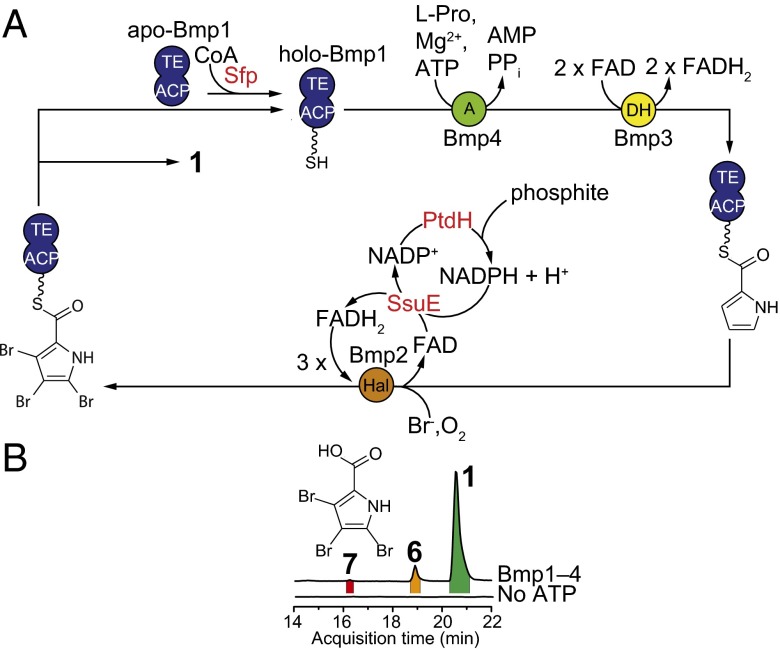
To examine the individual roles of Bmp1–4, we performed the total in vitro reconstitution of the biosynthesis of 1 (Fig. 3A). Due to the high sequence similarity of PS5_Bmp1–4 to Mm_Bmp1–4 (SI Appendix, Table S1), and production of 1 by both P. sp PS5 and M. mediterranea MMB-1 (11), we used recombinant Mm_Bmp1–4 proteins in our in vitro investigations. Bmp2 and Bmp4 were individually purified as N-His6–tagged proteins whereas N-His6–tagged apo-Bmp1 was purified in complex with untagged Bmp3 and converted to its pantetheine-loaded holo-form by the promiscuous Bacillus subtilis phosphopantetheinyl transferase Sfp (11, 13) (Fig. 3A). Incubation of l-proline and bromide with purified Bmp1–4 enzymes, along with flavin and nicotinamide cofactors (FAD and NADP+) and cofactor regeneration enzymes (PtdH and SsuE), led to the formation of 1 as the major product (Fig. 3 and SI Appendix, Fig. S1). The production of 1 was abolished in the absence of ATP, which is required for the loading of l-proline onto the ACP domain of Bmp1 by Bmp4 (Fig. 3B). By comparison with authentic synthetic standards, we also confirmed the production of 6 and 3,4,5-tribromo-pyrrole-2-carboxylic acid (7) as minor products of the reaction (Fig. 3B).
Fig. 3.
Enzymatic synthesis of 1. (A) Scheme for the total in vitro enzymatic synthesis of 1 starting from l-proline. Accessory enzymes not derived from the Mm_bmp gene cluster are indicated in red. FADH2 required as a cofactor for Bmp2 was regenerated in situ by E. coli flavin reductase SsuE (32), which in turn oxidizes NADPH to NADP+. NADP+ was recycled to NADPH in situ using NAD(P)+ reductase/phosphite dehydrogenase-PtdH (33). (B) EICs for [M-H]1- ions corresponding to 1, 6, and 7 for organic extracts of reactions described in A. Note that the production of 1, 6, and 7 was abolished in the absence of ATP.
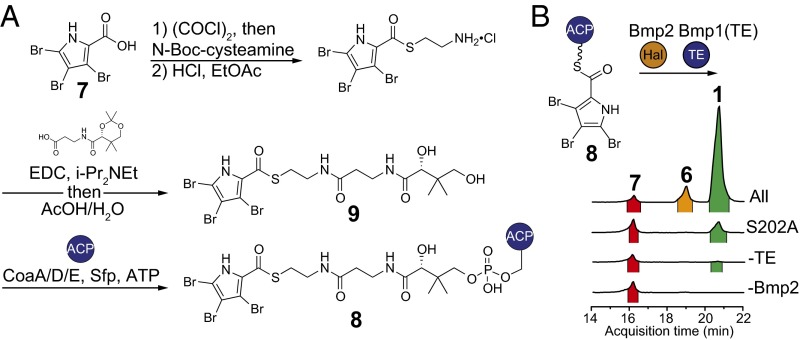
Having established that Bmp1–4 enzymes are sufficient for the production of 1, we next queried the timing and mechanism of the terminal bromination, offloading, and decarboxylation reactions. We previously established that Mm_Bmp2 converts pyrrolyl-S-Bmp1(ACP) to tribromopyrrolyl-S-Bmp1(ACP) (Fig. 3A), where the Bmp1(ACP) domain comprises residues 1–77 of Bmp1 (11). Therefore, we rationalized that 3,4,5-tribromopyrrolyl-S-Bmp1(ACP) (8, Fig. 4A) might be an intermediate en route to 1. To test this hypothesis, we synthesized 8 following a recently described chemoenzymatic method (14). Briefly, synthetic 7, generated by the bromination of pyrrole-2-carboxylic acid, was sequentially ligated to cysteamine and then to pantothenic acid to afford 3,4,5-tribromopyrrolyl-S-pantetheine 9 (Fig. 4A and SI Appendix for synthesis protocols and product characterization data). Compound 9 was then extended by the E. coli CoA biosynthetic enzymes CoaA, CoaD, and CoaE to generate 3,4,5-tribromopyrrolyl-S-CoA, which was used as the substrate for the in situ transfer of the 3,4,5-tribromopyrrolyl-S-phosphopantetheine moiety to the serine side chain hydroxyl of apo-Bmp1(ACP) by Sfp to yield 8 (Fig. 4A and SI Appendix, Fig. S2). We then expressed and purified the Bmp1 thioesterase domain, Bmp1(TE), whose activity we previously confirmed using a model esterase substrate, p-nitrophenylacetate (11). Incubation of 8 with Bmp2, Bmp1(TE), bromide, NADP+, FAD, and cofactor regeneration components led to formation of 1 as the major product, together with minor production of 6 and 7 (Fig. 4B and SI Appendix, Fig. S1). Exclusion of Bmp1(TE) from the reaction led to trace production of 1 (Fig. 4B). Additionally, we mapped the active site serine residue of Bmp1(TE) to Ser202 and confirmed that its mutation to alanine resulted in loss of esterase activity for the p-nitrophenylacetate substrate (SI Appendix, Fig. S3). Substitution of Bmp1(TE) with catalytically inactive Bmp1(TE)S202A mutant enzyme led to significantly reduced production of 1 (Fig. 4B and SI Appendix, Fig. S4). Finally, exclusion of Bmp2 from the reaction completely abolished the production of 1 (Fig. 4B and SI Appendix, Fig. S4). Similar levels of 7 were observed across all reactions, suggesting that it is likely an “off-pathway” product resulting from the hydrolysis of 8 (Fig. 4B). In support of 7 as a hydrolytic shunt product, no conversion of 7 to 1 or 6, or to any new products, was observed upon its incubation with Bmp2, Bmp1(TE), and bromide (SI Appendix, Fig. S5). These results provide two important findings. First, both catalysts, Bmp1(TE) and the halogenase Bmp2, are required to convert the intermediate 8 to 1. Second, two enzymatic activities [that is, hydrolysis via thioesterase Bmp1(TE) and bromination via Bmp2] lead to three chemical events: (i) offloading of the pyrrole from Bmp1(ACP), (ii) the fourth bromination on the 2′-position of the pyrrole, and (iii) decarboxylative loss of the l-proline–derived alpha-carboxylate carbon atom.
Fig. 4.
Preparation of 8 and in vitro conversion to 1 by Bmp2 and Bmp1(TE). (A) Scheme showing generation of 8 by one-pot in vitro extension of synthetically prepared 9 using E. coli CoA biosynthetic enzymes, and loading onto holo-Bmp1(ACP) by Sfp. (B) Scheme for conversion of 8 to 1, 6, and 7 by Bmp2 and Bmp1(TE). Omitted for clarity are the cofactor regeneration systems for Bmp2, as shown in Fig. 3. Only trace, or no production, of 1 was observed when either Bmp2 or Bmp1(TE) was omitted from the in vitro enzymatic reaction, or when the catalytically inactive Bmp1(TE)S202A enzyme was used.
We next evaluated the timing of formation of 6 relative to the formation of 1. We thus incubated 6 with Bmp2, with and without Bmp1(TE), and cofactor-regenerating enzymes. We observed trace conversion of 6 to 1 only after prolonged reaction times, consistent with our hypothesis that 6 is a nonphysiological substrate for Bmp2 (SI Appendix, Fig. S5). Therefore, we propose that 6 might be a nonenzymatic reductive degradation byproduct of 1. Indeed, incubation of synthetically prepared 1 with NADPH led to the conversion to 6 at levels comparable with total in vitro reconstitution reactions (SI Appendix, Fig. S6). Together with our previous results, we propose that 7 and 6 are both off-pathway products arising, respectively, at stages preceding and after formation of 1 (Fig. 5).
Fig. 5.
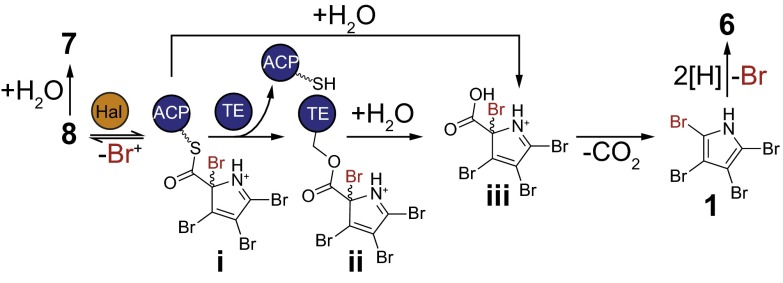
Scheme for bromination-dependent pyrrole offloading and decarboxylation. Proposed reaction mechanisms for terminal bromination, offloading, and decarboxylation reactions with 8; inferred intermediates are indicated by bold Roman numerals.
In sum, our data support the Bmp2-catalyzed bromination of 8 leading to transient intermediate i as shown in Fig. 5, which would undergo transesterification to generate a Bmp1(TE)S202 side chain-bound oxoester ii, thus offloading the pyrrole from Bmp1(ACP). Hydrolytic offloading from Bmp1(TE) would generate the labile alpha-bromo acid iii. Driven by the rearomatization of the pyrrole ring, iii would spontaneously decarboxylate to 1. Direct mass spectrometric evidence for i proved elusive, suggesting that i rapidly converts back to 8 via spontaneous debromination in the absence of Bmp1(TE). Therefore, the role of the Bmp1(TE) could be to trap the tetrabrominated pyrrole species by means of an irreversible transesterification step whereas the fourth bromination by Bmp2 activates the substrate for decarboxylation subsequent to hydrolytic offloading from Bmp1(TE).
Nonenzymatic hydrolysis of i may also lead to release of iii to afford 1, consistent with our observation of low levels of 1 from reactions excluding Bmp1(TE) and with inactive Bmp1(TE)S202A. The observation of similar levels of 7 in all reactions further supports direct hydrolytic bypass of TE-mediated offloading. Furthermore, the fourth bromination of 8 by Bmp2, followed by transesterification and decarboxylation of i, neatly differentiates the biosynthesis of 1 from nonribosomal peptide synthetase- and polyketide synthase-derived pathways involving halopyrrole units in which the l-proline–derived alpha-carboxylate carbon atom is preserved in the mature natural product (Fig. 1).
Structural Basis for Unique Tetrahalogenating Activity of Bmp2.
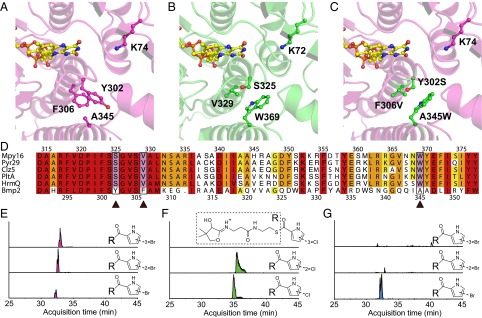
To realize the scheme shown in Fig. 5, the flavin-dependent brominase Bmp2 would need to halogenate all four positions of the pyrrole ring. This mechanistic requirement for the production of 1 is in stark contrast to all previously characterized pyrrole halogenases that halogenate the pyrrole ring only once or twice. To understand the structural basis for the tetrahalogenating activity of Bmp2, we determined the high-resolution crystal structure of FAD-bound holo-Bmp2 at a limiting resolution of 1.87 Å (Fig. 6A) and compared it with the FAD-bound holo-structure of dihalogenase Mpy16 participating in the biosynthesis of 3 that we determined at 1.95 Å resolution (Fig. 6B and SI Appendix, Table S2) (8). Bmp2 and Myp16 structures are highly homologous with all secondary structural elements in the vicinity of the active site strictly conserved between the two enzymes (SI Appendix, Fig. S7). Furthermore, the FAD cofactor isoalloxazine rings and the postulated active site lysine residues (15) (K74 for Bmp2, K72 for Mpy16) (Fig. 6 A and B) are superimposable. Although we could not determine a substrate pyrrolyl-S-ACP cocrystal structure for either Bmp2 or Mpy16, the position of the pyrrole binding sites for Bmp2 and Myp16 was inferred by a structural alignment of Bmp2 and Mpy16 with the crystal structures of substrate-bound forms of the flavin-dependent tryptophan-7-chlorinases PrnA (16) and RebH (17). Fittingly, the postulated pyrrole-binding site in Bmp2 and Mpy16 (Fig. 6 A and B) is in close proximity to the side chain of the catalytic lysine residue. An examination of the amino acids lining this putative pyrrole binding site in Bmp2 reveals a lack of conservation of three key sites (Y302, F306, and A345) that are otherwise strictly conserved among all pyrrolyl-S-ACP halogenases that selectively catalyze one [HrmQ (10)] or two [PltA (7), Mpy16 (8), Clz5 (9), and Pyr29 (18)] halogenations on the pyrrole ring (Fig. 6D). We thus mutated these three residues in Bmp2 to the corresponding residues in Mpy16 (Fig. 6B) to generate Bmp2-Y302S/F306V/A345W triple mutant enzyme (henceforth referred to as Bmp2-TM). We next investigated the effect of the triple mutation on the in vivo heterologous production of 1 in E. coli. Coexpression of Mm_bmp1, Mm_bmp3, and Mm_bmp4, with Mm_bmp2-TM in E. coli, led to a complete loss in the in vivo production of 1 in contrast to robust production with wild-type (WT) Mm_bmp2 (SI Appendix, Fig. S9).
Fig. 6.
Structural basis for tetrahalogenation activity of Bmp2. Comparison of the active sites of (A) holo-Bmp2, (B) holo-Mpy16, and (C) holo-Bmp2-TM. Note that side chains of Bmp2-TM residues S302, V306, and W345 in C structurally map to the corresponding side chains of Mpy16 residues S325, V329, and W369 shown in B. (D) Primary sequence alignments of tetrahalogenase Bmp2 with mono- and dihalogenating pyrrolyl-S-ACP halogenases demonstrate that the amino acids that were mutated on the basis of structural comparison with form Bmp2-TM (indicated by ▲) are conserved in all pyrrolyl-S-ACP halogenases but Bmp2 (see SI Appendix, Fig. S20 for alignment of Bmp2 homologs from different bacterial species). EICs for acyl-(cyclo)pantetheine MS2 product ions demonstrate that (E) WT Bmp2 generates mono-, di- and tribromopyrrolyl-S-Bmp1(ACP) products whereas (F) Mpy16 generates only mono- and dichloropyrrolyl-S-Mpy15, and (G) Bmp2-TM generates only monobromopyrrolyl-S-Bmp1(ACP) product.
To explore the molecular basis for the in vivo loss in production of 1 by the Bmp2-TM enzyme, we compared the in vitro enzymatic activities of Bmp2, Mpy16, and Bmp2-TM enzymes. We chemoenzymatically synthesized pyrrolyl-S-Bmp1(ACP) and pyrrolyl-S-Mpy15, the requisite substrates for Bmp2 and Mpy16, respectively (8, 11, 14). Using a previously reported mass spectrometry-based proteomic assay that relies on detection of acyl-(cyclo)pantetheine MS2 product ions (14), we verified that WT Bmp2 enzyme catalyzed three brominations on the pyrrole ring (Fig. 6E) and that Mpy16 catalyzed no more than two chlorinations on the pyrrole ring acylated to Mpy15 (Fig. 6F) (14). Note that, although Bmp2 tetrabrominates the pyrrole ring as established by our previous findings, the assay used here detects only up to tribromination of pyrrolyl-S-Bmp1(ACP). In contrast to the WT Bmp2, Bmp2-TM could catalyze only a single bromination on the pyrrole ring (Fig. 6G and SI Appendix, Fig. S8), thereby precluding the synthesis of 1.
Lastly, to eliminate the possibility that the reduced degree of halogenation catalyzed by Bmp2-TM was due to disruption of the enzyme active site by the three mutations, we determined the crystal structure of holo-Bmp2-TM at a resolution of 1.98 Å (Fig. 6C). Structural comparison of Bmp2-TM to Bmp2 and Mpy16 confirmed that the mutation of the three Bmp2 active site residues did not alter the positioning of the FAD isoalloxazine ring or the catalytic lysine side chain.
Discussion
In addition to characterizing the biosynthesis of a microbially produced coral settlement cue, our study reveals previously unidentified enzymology stemming from a biosynthetic motif ubiquitous among halopyrrole-containing natural products. Although our previous work had established tribromination of pyrrolyl-S-Bmp1(ACP) by Bmp2, the enzymatic activities responsible for the addition of the fourth bromine atom in 1 and offloading and decarboxylation of the pyrrolyl moiety from the ACP were unknown (11). Our current work demonstrates that a single halogenase, Bmp2, catalyzes the unprecedented tetrabromination of the ACP-bound pyrrole, which subsequently undergoes Bmp1(TE)-mediated offloading from the ACP and spontaneous decarboxylation. In light of the elucidation of the molecular and genetic details for the construction of 1, we posit that 1 is an intermediate en route to the production of 5, reconciling a previous report implicating an l-proline–derived symmetrical pyrrolic intermediate in the biosynthetic scheme for the production of 5 (19). Previously, based on primary sequence homology, we postulated that the enzyme Bmp8 (Fig. 2A) participates in the decarboxylation of 7 to produce 6 (11), a hypothetical route that is likely not operative in light of the biochemical data presented above. Therefore, the physiological transformations for the production of 6 en route to 5 remain to be elucidated. Furthermore, the previously reported production of 1 during the CYP450-Bmp7–mediated biradical homodimerization of 6 is likely an off-pathway route, with Bmp1–4 being the primary players in the production of 1. Successive biochemical studies promise to address these open questions to characterize all steps in the biosynthesis of 5 and assign physiological roles for each of the Bmp enzymes.
The complete in vitro reconstitution of the production of 1 using purified enzyme catalysts suggests a mechanism for the acyl-phosphopantetheine thioester to be sequestered and thereby inaccessible to Bmp1(TE) until the terminal fourth bromination. Sequestration of a pyrrole tethered to a type II peptidyl carrier protein (PCP) was recently demonstrated for a highly homologous PCP-PltL [57% amino acid similarity to Mm_Bmp1(ACP)] participating in the biosynthesis of the dichloropyrrole-containing natural product pyoluteorin (7, 20). By analogy, in the biosynthesis of 1, it is possible that the terminal halogenation on the pyrrole partially liberates the sequestered substrate from the ACP, allowing access to the thioesterase. Therefore, the terminal fourth bromination by Bmp2 serves the dual role of triggering the release of the sequestered pyrrole moiety by presumably making the thioester susceptible to hydrolysis, in addition to activating the substrate for elimination of the l-proline–derived alpha-carboxyl. Activating halogenation reactions in natural product biosynthetic pathways has been demonstrated in the biosynthesis of NRPS-PKS hybrid curacin A, in which a cryptic halogenation catalyzed by α-ketoglutarate–dependent halogenase-CurA promotes cyclopropane ring formation (21), and in the biosynthesis of meroterpenoid merochlorins A and B, in which a dearomatization/terpene cyclization reaction cascade is initiated by a chlorination catalyzed by vanadium-dependent haloperoxidase-Mcl24 (22). Most analogously, paralleling the decarboxylative bromination strategy used by Bmp2, flavin-dependent brominase-Bmp5 catalyzes a bromination reaction that drives decarboxylation in the conversion of free p-hydroxybenzoic acid to 2,4-dibromophenol, the bromophenol building block of 5 (11).
The tetrabromination activity of Bmp2 is unprecedented among the flavin-dependent halogenases described to date that selectively add a specific number of halogen atoms to an aromatic substrate. In this study, we embraced the opportunity to interrogate the structural basis for this halogenation control in flavin-dependent pyrrolyl-S-ACP halogenases. Although the previously reported 2.1-Å crystal structure of putative tyrosyl-S-ACP flavin-dependent halogenase CndH highlighted the general differences in the architectures of flavin-dependent halogenases acting on free versus ACP-bound substrates (23), our study sheds mechanistic insight into the tuning of the enzyme active site in the context of confirmed biochemistry. Comparison of the putative substrate-binding cavities of the highly conserved structures of Bmp2 and Mpy16 resulted in a catalyst, Bmp2-TM, exhibiting an altered halogenation profile. Notably, the active site of Bmp2-TM exhibited no perturbation of the bound FAD cofactor with respect to WT Bmp2, nor did it lead to any apparent change in halogen binding properties of the enzyme, as demonstrated by conservation of its specificity for bromide. Already, for flavin-dependent tryptophan halogenases, it has been demonstrated that amino acid side chains that constitute the substrate binding site control the regiochemical outcomes for halogen additions (17, 24, 25). Our findings extend this observation to flavin-dependent halogenases that catalyze halogenation of aromatic substrates acylated to ACPs in demonstrating that side chains of residues lining the putative halogenase active site play a role in controlling substrate access, and potentially in specifying the positions on the pyrrole ring that are accessible to halogenation. Unfortunately, in the case of Mpy16, efforts to alter the putative substrate-binding cavity to resemble that of Bmp2 resulted in insolubility of the mutant Mpy16 enzymes. Nonetheless, our results demonstrate that the active sites of otherwise highly homologous flavin-dependent halogenases are uniquely evolved to afford distinct product profiles. Biological activities of natural products are influenced by the number of halogens decorating their organic scaffolds (26, 27), and it would seem that Nature has taken note through the evolution of highly specialized halogenation catalysts.
In light of the existing literature and the findings of this study, several open questions remain regarding enzymatic halogenation (28). Of note is the question of the binding mode of both the ACP and the aromatic substrate by flavin-dependent halogenases that require acyl-S-ACP substrates. In addition, the primary question across all classes of halogenating enzymes is the structural determinant for halide specificity among these catalysts. Although Bmp2 and Mpy16 demonstrate different halide specificities, the amino acid side chains and the positioning of the FAD isoalloxazine ring relative to these side chains are remarkably conserved. Furthermore, the conservation of halide specificity in Bmp2-TM with respect to the WT enzyme raises the possibility that halide specificity in flavin-dependent halogenases is dictated not only by steric factors, such as halide ion radii, but also by the magnitude of the enthalpic penalty associated with desolvation of the halide ion in the halogenase active site before 2e− oxidation to generate the electrophilic halonium. Furthermore, the contribution of the redox potential of the flavin isoalloxazine ring necessary to oxidize the halide, and the stability of the activated lysine-amine intermediate generated en route to the transfer of the halonium to the aromatic substrate should be considered (15). As such, the currently proposed halide binding site for flavin-dependent halogenases is itself debatable (29), thus underscoring the challenges associated with teasing apart the role of the enzymatic halogenase catalyst in each of the requisite mechanistic steps. Discovery and characterization of additional flavin-dependent halogenases promises to provide opportunities to answer these mechanistic questions, ultimately leading to engineerable toolkits to tailor the biosynthesis of halogenated natural products (30).
Materials and Methods
Detailed materials and methods are provided in SI Appendix, Materials and Methods.
Database Deposition Information.
The sequences for P. sp. PS5- and P. sp. A757-derived bmp gene clusters have been deposited in GenBank under accession numbers KR011923 and KT808878, respectively. Structures for Mpy16, Bmp2, and Bmp2-TM are deposited in the Protein Data Bank (www.rcsb.org) under accession numbers 5BUK, 5BVA, and 5BUL, respectively.
Note.
During the review and publication of this manuscript, the crystal structure of the flavin-dependent halogenase PltA was reported in the literature (31).
Supplementary Material
Acknowledgments
We thank our colleague B. M. Duggan at University of California, San Diego for assistance in acquiring NMR data. This work was jointly supported by US National Science Foundation Grant OCE-1313747 (to B.S.M.) and US National Institutes of Health (NIH) Grants P01-ES021921 and R01-AI47818 (to B.S.M.) and R21-AI119311 (to K.E.W. and T.J.M.), the Mote Protect Our Reef Grant Program (POR-2012-3), the Dart Foundation, the Smithsonian Competitive Grants Program for Science (V.J.P.), the Howard Hughes Medical Institute (J.P.N.), NIH Marine Biotechnology Training Grant Predoctoral Fellowship T32-GM067550 (to A.E.), a Helen Hay Whitney Foundation Postdoctoral Fellowship (to V.A.), and a Swiss National Science Foundation Postdoctoral Fellowship (to S.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.A.F. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the Genbank database [accession nos. KR011923 (Pseudoalteromonas sp. PS5-derived bmp gene clusters) and KT808878 (Pseudoalteromonas sp. A757-derived bmp gene clusters)]. The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 5BUK (Mpy16), 5BVA (Bmp2), and 5BUL (Bmp2-TM)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1519695113/-/DCSupplemental.
References
- 1.Holmström C, Kjelleberg S. Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extracellular agents. FEMS Microbiol Ecol. 1999;30(4):285–293. doi: 10.1111/j.1574-6941.1999.tb00656.x. [DOI] [PubMed] [Google Scholar]
- 2.Tebben J, et al. Induction of larval metamorphosis of the coral Acropora millepora by tetrabromopyrrole isolated from a Pseudoalteromonas bacterium. PLoS One. 2011;6(4):e19082. doi: 10.1371/journal.pone.0019082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sneed JM, Sharp KH, Ritchie KB, Paul VJ. The chemical cue tetrabromopyrrole from a biofilm bacterium induces settlement of multiple Caribbean corals. Proc Biol Sci. 2014;281(1786):20133086. doi: 10.1098/rspb.2013.3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neu AK, Månsson M, Gram L, Prol-García MJ. Toxicity of bioactive and probiotic marine bacteria and their secondary metabolites in Artemia sp. and Caenorhabditis elegans as eukaryotic model organisms. Appl Environ Microbiol. 2014;80(1):146–153. doi: 10.1128/AEM.02717-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gribble GW. Naturally Occurring Organohalogen Compounds: A Comprehensive Update. Springer; Vienna: 2010. [Google Scholar]
- 6.Walsh CT, Garneau-Tsodikova S, Howard-Jones AR. Biological formation of pyrroles: Nature’s logic and enzymatic machinery. Nat Prod Rep. 2006;23(4):517–531. doi: 10.1039/b605245m. [DOI] [PubMed] [Google Scholar]
- 7.Dorrestein PC, Yeh E, Garneau-Tsodikova S, Kelleher NL, Walsh CT. Dichlorination of a pyrrolyl-S-carrier protein by FADH2-dependent halogenase PltA during pyoluteorin biosynthesis. Proc Natl Acad Sci USA. 2005;102(39):13843–13848. doi: 10.1073/pnas.0506964102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamanaka K, Ryan KS, Gulder TA, Hughes CC, Moore BS. Flavoenzyme-catalyzed atropo-selective N,C-bipyrrole homocoupling in marinopyrrole biosynthesis. J Am Chem Soc. 2012;134(30):12434–12437. doi: 10.1021/ja305670f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mantovani SM, Moore BS. Flavin-linked oxidase catalyzes pyrrolizine formation of dichloropyrrole-containing polyketide extender unit in chlorizidine A. J Am Chem Soc. 2013;135(48):18032–18035. doi: 10.1021/ja409520v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Höfer I, et al. Insights into the biosynthesis of hormaomycin, an exceptionally complex bacterial signaling metabolite. Chem Biol. 2011;18(3):381–391. doi: 10.1016/j.chembiol.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 11.Agarwal V, et al. Biosynthesis of polybrominated aromatic organic compounds by marine bacteria. Nat Chem Biol. 2014;10(8):640–647. doi: 10.1038/nchembio.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whalen KE, Poulson-Ellestad KL, Deering RW, Rowley DC, Mincer TJ. Enhancement of antibiotic activity against multidrug-resistant bacteria by the efflux pump inhibitor 3,4-dibromopyrrole-2,5-dione isolated from a Pseudoalteromonas sp. J Nat Prod. 2015;78(3):402–412. doi: 10.1021/np500775e. [DOI] [PubMed] [Google Scholar]
- 13.Quadri LE, et al. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry. 1998;37(6):1585–1595. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- 14.Agarwal V, et al. Chemoenzymatic synthesis of acyl coenzyme A substrates enables in situ labeling of small molecules and proteins. Org Lett. 2015;17(18):4452–4455. doi: 10.1021/acs.orglett.5b02113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeh E, Blasiak LC, Koglin A, Drennan CL, Walsh CT. Chlorination by a long-lived intermediate in the mechanism of flavin-dependent halogenases. Biochemistry. 2007;46(5):1284–1292. doi: 10.1021/bi0621213. [DOI] [PubMed] [Google Scholar]
- 16.Dong C, et al. Tryptophan 7-halogenase (PrnA) structure suggests a mechanism for regioselective chlorination. Science. 2005;309(5744):2216–2219. doi: 10.1126/science.1116510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu X, et al. Structural insights into regioselectivity in the enzymatic chlorination of tryptophan. J Mol Biol. 2009;391(1):74–85. doi: 10.1016/j.jmb.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, Parry RJ. Cloning and characterization of the pyrrolomycin biosynthetic gene clusters from Actinosporangium vitaminophilum ATCC 31673 and Streptomyces sp. strain UC 11065. Antimicrob Agents Chemother. 2007;51(3):946–957. doi: 10.1128/AAC.01214-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peschke JD, Hanefeld U, Laatsch H. Biosynthesis of the marine antibiotic pentabromopseudilin. 2. The pyrrole ring. Biosci Biotechnol Biochem. 2005;69(3):628–630. doi: 10.1271/bbb.69.628. [DOI] [PubMed] [Google Scholar]
- 20.Jaremko MJ, Lee DJ, Opella SJ, Burkart MD. Structure and substrate sequestration in the pyoluteorin type II peptidyl carrier protein PltL. J Am Chem Soc. 2015;137(36):11546–11549. doi: 10.1021/jacs.5b04525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu L, et al. Metamorphic enzyme assembly in polyketide diversification. Nature. 2009;459(7247):731–735. doi: 10.1038/nature07870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diethelm S, Teufel R, Kaysser L, Moore BS. A multitasking vanadium-dependent chloroperoxidase as an inspiration for the chemical synthesis of the merochlorins. Angew Chem Int Ed Engl. 2014;53(41):11023–11026. doi: 10.1002/anie.201405696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buedenbender S, Rachid S, Müller R, Schulz GE. Structure and action of the myxobacterial chondrochloren halogenase CndH: A new variant of FAD-dependent halogenases. J Mol Biol. 2009;385(2):520–530. doi: 10.1016/j.jmb.2008.10.057. [DOI] [PubMed] [Google Scholar]
- 24.Lang A, et al. Changing the regioselectivity of the tryptophan 7-halogenase PrnA by site-directed mutagenesis. Angew Chem Int Ed Engl. 2011;50(13):2951–2953. doi: 10.1002/anie.201007896. [DOI] [PubMed] [Google Scholar]
- 25.Shepherd SA, et al. Extending the biocatalytic scope of regiocomplementary flavin-dependent halogenase enzymes. Chem Sci (Camb) 2015;6(6):3454–3460. doi: 10.1039/c5sc00913h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eustáquio AS, et al. Clorobiocin biosynthesis in Streptomyces: Identification of the halogenase and generation of structural analogs. Chem Biol. 2003;10(3):279–288. doi: 10.1016/s1074-5521(03)00051-6. [DOI] [PubMed] [Google Scholar]
- 27.Harris CM, Kannan R, Kopecka H, Harris TM. The role of the chlorine substituents in the antibiotic vancomycin: Preparation and characterization of monodechlorovancomycin and didechlorovancomycin. J Am Chem Soc. 1985;107(23):6652–6658. [Google Scholar]
- 28.Neumann CS, Fujimori DG, Walsh CT. Halogenation strategies in natural product biosynthesis. Chem Biol. 2008;15(2):99–109. doi: 10.1016/j.chembiol.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 29.Blasiak LC, Drennan CL. Structural perspective on enzymatic halogenation. Acc Chem Res. 2009;42(1):147–155. doi: 10.1021/ar800088r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teufel R, Agarwal V, Moore BS. Unusual flavoenzyme catalysis in marine bacteria. Curr Opin Chem Biol. 2016;31:31–39. doi: 10.1016/j.cbpa.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pang AH, Garneau-Tsodikova S, Tsodikov OV. Crystal structure of halogenase PltA from the pyoluteorin biosynthetic pathway. J Struct Biol. 2015;192(3):349–357. doi: 10.1016/j.jsb.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 32.Eichhorn E, van der Ploeg JR, Leisinger T. Characterization of a two-component alkanesulfonate monooxygenase from Escherichia coli. J Biol Chem. 1999;274(38):26639–26646. doi: 10.1074/jbc.274.38.26639. [DOI] [PubMed] [Google Scholar]
- 33.Costas AMG, White AK, Metcalf WW. Purification and characterization of a novel phosphorus-oxidizing enzyme from Pseudomonas stutzeri WM88. J Biol Chem. 2001;276(20):17429–17436. doi: 10.1074/jbc.M011764200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.