Significance
Bacterial biofilms are ubiquitous in the environment and serve beneficial roles in microbiota communities in the context of eukaryotic hosts and in industrial applications, yet biofilms of pathogenic bacteria can also cause devastating infections. Biofilm architectures are usually studied at a coarse morphological level, and consequently, little is known about the internal biofilm architecture and how it emerges. Here, we use an optical imaging technique to visualize every cell inside thousands of Vibrio cholerae biofilms to discover architectural transitions and the major phases of V. cholerae biofilm growth.
Keywords: biofilm, community, self-organization, emergent order, nematic order
Abstract
Many bacterial species colonize surfaces and form dense 3D structures, known as biofilms, which are highly tolerant to antibiotics and constitute one of the major forms of bacterial biomass on Earth. Bacterial biofilms display remarkable changes during their development from initial attachment to maturity, yet the cellular architecture that gives rise to collective biofilm morphology during growth is largely unknown. Here, we use high-resolution optical microscopy to image all individual cells in Vibrio cholerae biofilms at different stages of development, including colonies that range in size from 2 to 4,500 cells. From these data, we extracted the precise 3D cellular arrangements, cell shapes, sizes, and global morphological features during biofilm growth on submerged glass substrates under flow. We discovered several critical transitions of the internal and external biofilm architectures that separate the major phases of V. cholerae biofilm growth. Optical imaging of biofilms with single-cell resolution provides a new window into biofilm formation that will prove invaluable to understanding the mechanics underlying biofilm development.
Bacteria assemble into communities, termed biofilms, which are embedded in a secreted polymer matrix and often coat liquid–air or liquid–solid interfaces (1–4). Some biofilms are beneficial to human health, for example as part of the healthy gut and skin microbiota (5, 6) or in wastewater treatment systems (7). Other biofilms, however, cause serious problems in oral hygiene, chronic infections, and prosthetic contamination (8–10) and act as fouling agents in industrial flow systems (11). In all contexts, biofilms can be difficult to control due to their resilience against chemical and physical stresses (12, 13), including antibiotic treatment (14). Because of their ubiquity and their relevance to medicine and industry, the formation of biofilms has been studied intensively, with an emphasis on the genes, regulatory mechanisms, and transport properties that underlie transitions from planktonic growth to surface attachment (15–17), to proliferation and matrix secretion (2, 18), and finally to dispersal (19, 20). A basic understanding of several regulatory circuits and secreted matrix components governing biofilm formation has been developed (21–25). Nonetheless, the physical, biological, and chemical factors that interact to determine the biofilm architecture remain largely unknown.
Internal and global biofilm architectures are presumably consequences of emergent interactions between individual cell growth, physiological differentiation, secreted proteins, polymers and small molecules, and microenvironmental heterogeneity (21, 26–34). Attempts to dissect the individual and combined contributions of these factors to biofilm growth have increasingly relied on examination of bacterial communities in microfluidic devices that mimic central features of natural environments (11, 35, 36). Although sophisticated methods for fabricating biofilm microenvironments are available, a significant barrier to progress has been the lack of techniques capable of resolving all individual cells residing inside biofilms. Thus, the vast majority of studies to date have been limited to visualizing 3D biofilms as connected “clouds” of biomass, although some studies have used fixed samples to obtain cellular resolution (37–40). We therefore know little about the organizational principles that convert individual cell behavior into macroscopic growth and collective properties of biofilms.
Here, we develop and use experimental techniques to investigate at single-cell resolution the 3D architecture of biofilms containing thousands of cells. By using a customized spinning-disk confocal microscope that enables 3D imaging at high axial resolution with low-light doses and by combining this instrument with bespoke image analysis software, we were able to visualize and segment all individual cells in thousands of Vibrio cholerae biofilms grown on submerged glass surfaces under flow containing nutrients. From these data, we could construct ensemble averages of biofilm structure during every phase of growth. We discovered that the internal community architecture and global biofilm morphology undergo several distinct transitions, which manifest as changes in the relative arrangements of individual cells over the course of biofilm development. From these data, we identified four fundamental phases of biofilm growth, each characterized by its own unique architecture: 1D growth of 1–6 cells, 2D growth of 20–100 cells, 3D growth of 200–1,000 cells with low local order, and highly ordered growth of communities with more than 2,000 cells. These phases can be explained by transitions in the physical dimensionality of the particular biofilm combined with changes in local cell density. Of the three known V. cholerae matrix proteins RbmA, Bap1, and RbmC, only deletion of RbmA substantially perturbs cellular orientations and the overarching biofilm architecture. We thus provide, to our knowledge, the first steps toward resolving how the 3D biofilm architecture results from the interactions of the constituent cells.
Results and Discussion
Fluorescent proteins expressed from the V. cholerae chromosome do not provide a sufficient signal for live-cell imaging at single-cell resolution, even when the fluorescent proteins are expressed at levels just below those that inhibit growth. We therefore grew biofilms for different times in microfluidic flow channels and stained the biofilms in situ with a nucleic acid dye that we added to the medium immediately before imaging. This protocol makes it possible to investigate biofilm architecture at different growth stages. Using spinning disk confocal microscopy, we were able to resolve all individual cells inside biofilms. By applying our Matlab-based 3D image analysis software, we could localize and distinguish all cells in biofilms up to ∼30 µm in height, as shown in Fig. 1A (see Materials and Methods). From these gray-scale images, we obtained binary 3D images of each cell via image-processing techniques that are based on spatial band-pass filtering. A complete reconstruction of the positions and orientations of all cells in a biofilm with N = 4,543 cells is shown in Fig. 1B, where each cell is colored according to its distance from the substratum.
Fig. 1.
V. cholerae wild-type biofilm at single-cell resolution. (A) Planar cross-sections through the biofilm at heights z = 0.6 µm, 12.6 µm, and 24.6 µm. (B) All individual cells from A were automatically segmented into separate 3D objects, which are color-coded in panel B according to the heights of their centers of mass using the color scale indicated at the top right. Panels A and B show the identical biofilm, which was grown until time t = 24.5 h and contains N = 4,543 cells. Global morphological parameters for this particular biofilm are indicated by the red data points in Fig. 2.
Recent work has shown that V. cholerae biofilms grow clonally from single founder cells or founder cell clusters (41) into microcolonies. Here, we use the term “biofilm” to refer to these individual cell clusters on the glass substrate rather than the collection of all cell clusters on the surface. These biofilms strongly resist attachment and invasion by motile planktonic cells (42), although they permit increased surface attachment upon partial digestion of their extracellular matrix (43). Predominantly, clonal growth of biofilms also occurs in the infant rabbit model of V. cholerae infection (44), suggesting that clonal growth with strong segregation between different cell lineages is the dominant form of biofilm development for V. cholerae. Motivated by these findings, we used low seeding densities in our flow channels to ensure that we exclusively imaged biofilms that had arisen from single founder cells. To verify that biofilms originated from single founder cells in our experiments, we seeded our flow chambers with a mixture of three derivatives of wild-type V. cholerae, differing only in the fluorescent proteins expressed from the chromosome (mTFP, mKO, or mKate2). All biofilms were composed entirely of cells expressing the same fluorescent protein, as shown in Fig. S1, indicating that the biofilms were monoclonal and that attachment of planktonic cells to existing microcolonies was either rare or absent in our experiments.
Fig. S1.
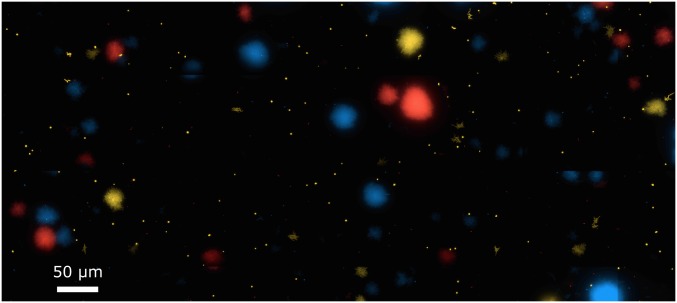
Biofilms investigated in this study are monoclonal and arise from founder cells. Biofilms grown for 30 h in the standard flow-channel assay are shown (see Materials and Methods). Strains are KDV103, KDV104, and KDV158 (see Table S1). These strains harbor genes encoding fluorescent proteins at the chromosomal lacZ site in wild-type V. cholerae N16961, under the control of the constitutive Ptac promoter. Fluorescent proteins are mTFP1 (blue), mKO (yellow), and mKate2 (red). The image demonstrates that biofilm colonies contain bacteria of a single color, indicating that cells of different lineages did not join biofilms that had already formed and that biofilms predominantly grow from a single founder cell or a cluster of clonal cells.
Biofilm Morphology and Internal Architecture During Growth.
Using the high-resolution microscopy technique noted above, we imaged a total of 2,429 V. cholerae biofilms of different sizes. This large number of 3D biofilm images allowed us to reconstruct ensemble averages to characterize morphological features, cell shapes, as well as relative and absolute cell arrangements throughout biofilm growth.
The resulting “growth laws” (45–47) for V. cholerae wild-type biofilms are summarized in Fig. 2. We found that the biofilm incubation period is a poor assessment parameter in ensemble averaged experiments, as shown in Fig. 2A, because dispersal and reattachment of cells to the glass substrate results in biofilms with a range of different ages and sizes at a given chamber incubation time. However, the global morphological features, such as biofilm volume (Fig. 2B), height (Fig. 2C), width (Fig. 2 D and E), and free surface area (Fig. S2), collapse tightly when measured as a function of the number of cells N in the biofilm. This finding implies that the cell number N is the key parameter that determines the biofilm architecture, and we therefore characterize our measurements in terms of N for the remainder of this work.
Fig. 2.
The growth laws and global morphology of V. cholerae wild-type biofilms. Each of the 2,429 data points in the subpanels represents a different biofilm. The red data point is the biofilm shown in Fig. 1. (A) Number of cells in biofilms as a function of incubation time. (B) The biofilm volume, calculated as the convex hull containing all of the cells inside each biofilm, shows a transition in slope around N = 102. The Inset shows the same data on a linear–linear scale. The green line depicts the running average of the data. (C) The peak height of the biofilm is the z-component of the center of the highest cell and reaches a maximum at ∼20 µm under the growth conditions tested. (D and E) Fitting an ellipse to the base of the biofilm in the XY plane yields a “width-A” and “width-B” for the biofilm, corresponding to the major and minor axes of the ellipse, respectively. For small N, ellipse fitting only provides qualitatively accurate results, whereas for large N, the ellipse fits are highly accurate. Panel D shows that the height of the biofilm remains lower than the width-A (a ratio height/width-A = 1 would correspond to a hemisphere), whereas the cross-section in the XY plane becomes nearly circular as the cell number in the biofilms increases.
Fig. S2.
Free surface and contact area of wild-type V. cholerae biofilms. (A) The free surface area of a biofilm is defined as the surface area of the biofilm colony (excluding the area that is in contact with the substratum). (B) The contact area of a biofilm colony is defined as the area that is occupied by the biofilm on the substratum. (C) The dimensionless ratio of the free surface area to the contact area. For a hemisphere, the ratio free surface/contact area = 2. (D) Free surface area as in panel A but on logarithmic axes. (E) Contact area as in panel B but on logarithmic axes. Each data point results from one biofilm.
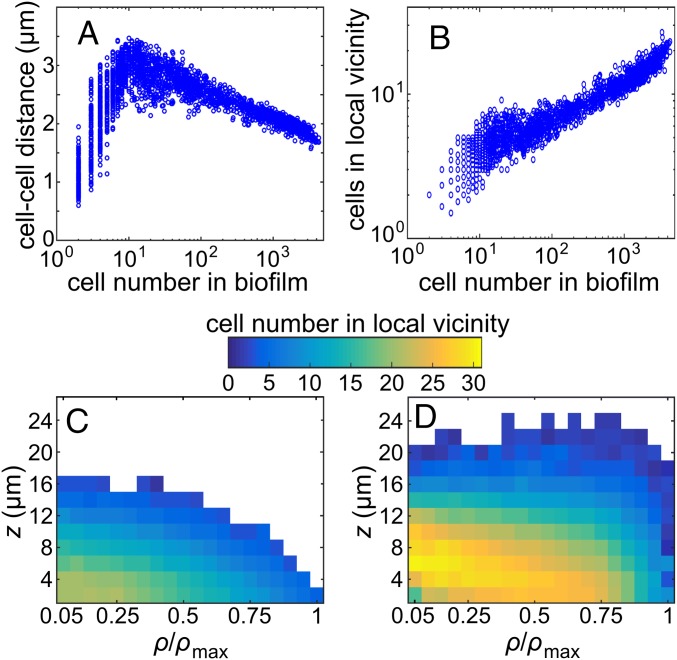
We find that the biofilm volume V increases more rapidly in biofilms with a low cell number than in biofilms with a large N (Fig. 2), indicating that an architectural or physiological transition occurs in the biofilm community at approximately N = 100 cells (Fig. 2B). The relationship between V and N can be captured by a simple power law V = c Nα, where c is a constant. For bacterial growth in well-mixed suspension cultures, a value of α = 1 would be expected due to a 1:1 correspondence between cell number and population biovolume. However, in the case of biofilms, we measure α = 2.5 for N < 100, whereas α = 0.85 for N > 100. The rapid accumulation of biovolume for N < 100 results from a significantly larger mean cell center–cell center distance in small biofilms, which reaches a maximum of ∼3 µm for biofilms with N = 10 (Fig. 3A). For biofilms with a large number of cells, the exponent α is less than unity, indicating that the biofilms compact with increasing cell number, which is reflected by reduced cell–cell distances (Fig. 3A), higher cell density (Fig. 3B), and smaller cell sizes (Fig. S3 A and B). All cell–cell distances and cell sizes were measured in 3D. The underlying mechanisms that drive the dramatic changes in biofilm expansion rate as a function of N are unknown. We speculate that the significant changes in cell–cell spacing between small and large biofilms are due to strong temporal variation in the extracellular matrix composition and/or production levels per cell.
Fig. 3.
Cell density depends on position inside biofilms and changes during V. cholerae biofilm growth. (A) The average distance to the center of mass of the nearest five neighboring cells from any individual cell center of mass in the biofilm, averaged over all cells in the entire biofilm, strongly depends on the total number of cells in the biofilm. (B) The cell number in the local vicinity, averaged over all cells in each biofilm and plotted as a function of the number of cells in the biofilm, shows a steady increase in cell density with biofilm size. The local vicinity in this figure is defined as a sphere of radius r = 3 µm around the center of each cell. (C and D) Spatially resolved average cell number in the local vicinity. To offset the effect that the cells in the bottom-most layers have fewer neighbors due to geometric constraints, the biofilm was mirrored at the z = 0 plane before calculating the cell number in the local vicinity in panels C and D. For panel C, 45 biofilms with N = 600–800 cells were averaged, using cylindrical coordinates. For panel D, 15 biofilms with N = 3,500–4,000 cells were averaged.
Fig. S3.
Cell sizes during biofilm growth and the heterogeneous distribution of cells inside biofilms. (A) Mean cell length for biofilms that contain different numbers of cells. (B) Mean aspect ratio of cells inside the biofilm for biofilms that contain different numbers of cells. For panels A and B, each data point results from one biofilm. (C) Spatially resolved average cell length in µm. For this graph, 15 biofilms with N = 3,500–4,000 cells were averaged using cylindrical coordinates. (D) The spatially resolved cell aspect ratio, averaged for the same set of biofilms shown in panel C.
Global biofilm shape also undergoes a clear transition between small and large cell number. Following initiation, biofilm height rapidly increases (Fig. 2C), saturating at ∼25 µm in our experimental conditions. Thus, the ratio of biofilm height to width and the ratio of their two cross-sectional widths (found by fitting an ellipse to the biofilm base) show that, initially, biofilms are highly asymmetric. However, for biofilms with N > 500 cells, the biofilm cross-section becomes nearly circular (Fig. 2E), and the height-to-width ratio approaches a steady state of 0.8 (Fig. 2D). In contrast to Bacillus subtilis biofilms (27), cell death does not seem to play a major role in determining the global architecture for V. cholerae biofilms (Fig. S4).
Fig. S4.
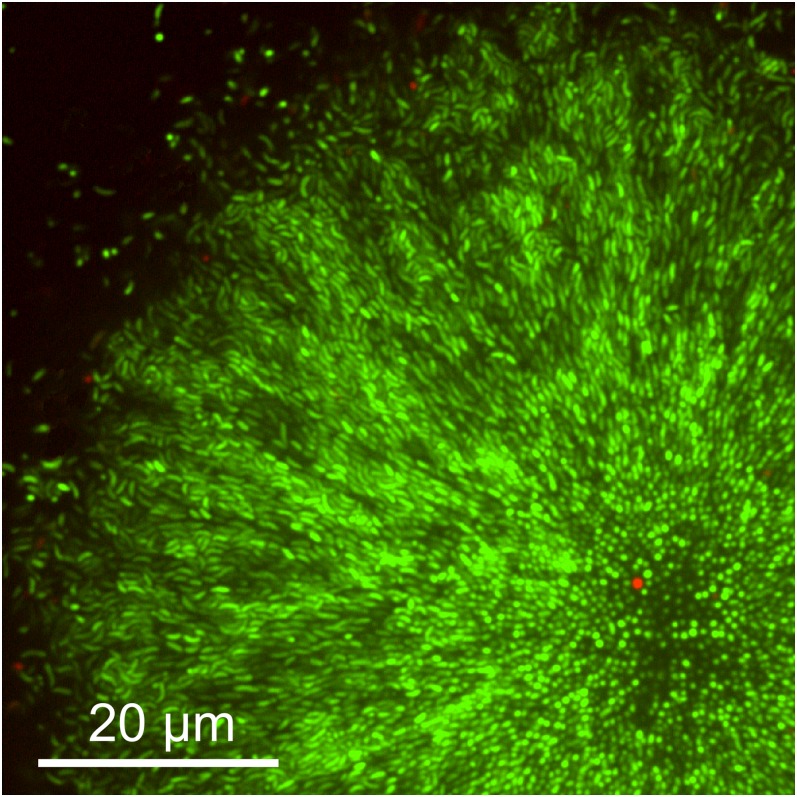
Imaging live and dead cells in V. cholerae biofilms. To assess whether dead cells might play a role in shaping biofilm architecture, we applied the LIVE/DEAD BacLight bacterial viability stain (Life Technologies) to V. cholerae biofilms. This procedure stains the nucleic acids of all cells green, and cells with a compromised membrane are stained red. Only a few cells in the biofilm appear to have compromised membranes.
The most striking features of the architectural transitions we observed in growing V. cholerae biofilms concerned the internal arrangement of the cells. To quantify such structural transitions, we defined the cellular local nematic order parameter S(r) = <3/2(ni · nj)2 – 1/2>, where ni is the orientation vector of a focal cell (see Fig. 4A), nj represents the orientation vectors of all other cells in the local vicinity, and <...> indicates an average, as described below. The local vicinity is defined as a 3D sphere of radius r around the center of the focal cell, as shown in Fig. 4A. The average local nematic order for each cell in its vicinity is subsequently averaged over all cells inside the biofilm to obtain a single order parameter S(r), which quantifies the degree of cellular alignment in the entire biofilm. The value S = 1 corresponds to perfect alignment of cells with their neighbors, whereas S = 0 corresponds to a random cellular arrangement. Fig. 4B shows that the average local order S(r) depends on the size of the local vicinity, with r = ∞ corresponding to the average global cellular order inside a biofilm (in practice, we set r = 50 µm to capture the global order). For a value r = 6 µm, which corresponds to a few cell lengths, the associated local order parameter S(6 µm) directly reflects the characteristic internal architectural transitions that biofilms undergo during growth (Fig. 4C): In very small biofilms, cells are nearly perfectly aligned. This high local order is rapidly lost during growth, and for N > 10 cells, the local order reaches S(6 µm) ∼ 0.2 and remains at this low, but not random, level for two orders of magnitude of population growth. The global cellular order drops to S(50 µm) ∼ 0 for N > 102, indicating globally heterogeneous cellular orientations. However, for very large biofilms (N > 2,000), the local order S(6 µm) increases sharply (see Fig. 4C), indicating that an architectural transition to high local cellular order occurs as biofilms grow to large population sizes. The characteristic internal cellular ordering for such large biofilms is shown in Fig. 4D: Globally the cells become oriented such that they appear to radiate from roughly the center of the biofilm’s basal plane. In the center of the biofilm, cells are aligned vertically relative to the substratum, transitioning gradually to fully parallel orientations relative to the base.
Fig. 4.
Transitions in local and global ordering of cells during wild-type V. cholerae biofilm growth. (A) Schematic drawing illustrating the definition of the local vicinity. The local vicinity is defined as a sphere of radius r around the center of each cell (denoted in blue; cells in the local vicinity are shown in pink). (B) The local nematic order parameter S(r) is color-coded and displayed for different radii r of the local vicinity, as a function of cell number in the biofilm. The red horizontal line in panel A indicates one value of r for which a more detailed graph is shown in panel C. (C) The biofilm architecture undergoes a transition in cell ordering as biofilms grow from N = 1–10 cells (a loss of order) and a further transition for biofilms for N > 2,000 cells (a gain in order). Solid lines are running averages of the red or black data points. (D) An XZ-slice through the center of a biofilm with N = 3,975 cells shows the internal architecture. Cells are colored according to their values of nz, the z-component of their orientation vector. (E) Biofilm local order parameter S from simulations for r = 6 µm. A “hedgehog”-type cellular arrangement corresponds to a von Mises–Fisher parameter κ >> 1, whereas complete disorder occurs for κ < 1. Our experimental data from panel C show that, for the simulated values of N, the local order parameter S ranges from 0.14–0.35, indicating that, for V. cholerae biofilms, κ = 3–10, which signifies an approximately hedgehog-type cellular arrangement.
Physical Aspects of Biofilm Internal Architecture.
V. cholerae cells can be coarsely approximated as growing and replicating rod-like objects with an aspect ratio of ∼2.5 (cell length/width; see Fig. S3B). Whereas in planktonic cultures V. cholerae has a characteristic curved cell shape, inside our biofilms, we observed predominantly straight, rod-shaped cell bodies (Fig. 1A and Fig. S5). In soft matter physics, it is well known that aligned nematic states of objects with aspect ratios > 1 form in dense suspensions due to steric and entropic constraints (48, 49). Nematic ordering is generally counteracted by orientational fluctuations. In molecular liquid crystals, these fluctuations are due to thermal rotational diffusion, which becomes large enough to disrupt the ordered phase around ∼50–250 °C, depending on pressure, concentration, and chemical composition of the liquid crystal molecules and solvent fluid (50–52). In suspensions of motile bacteria, cellular alignment and collective motion (53–56) are suppressed by orientational fluctuations due to variations in flagellar motor activity and cell–cell collisions that reorient cells with respect to one another (57). The factors that promote or inhibit nematic ordering in growing, nonmotile bacterial colonies remain largely unknown, yet recent experiments (58, 59) have suggested that for Escherichia coli growing in artificially confined 2D colonies, nematic alignment occurs at high area fractions due to a self-generated, growth-induced expansion flow. Our data for V. cholerae biofilms now allow us to quantify the emergence of orientational order in three dimensions, presumably as it occurs in natural settings.
Fig. S5.
Measuring the volume fraction of V. cholerae biofilms. To obtain an accurate visualization of the cell volume, we stained biofilms with the nucleic acid stain SYTO 9 and with the membrane stain FM4-64 (Life Technologies). (A) The nucleic acid stain labels the nucleoid, which fills most of the cytoplasm. (B) The membrane stain. (C) Merging panels A and B. (D) Binary image from our cell segmentation algorithm, obtained from panel A. (E) To obtain a binary image that more accurately describes the cell volume, we binarized both panels A and B to obtain the image shown in panel E. The volume fraction was calculated from images like panel E. The membrane stain revealed that 0.2 µm should be added to both the cell length and the cell width after extracting these values from images that are based on the nucleic acid stain.
We find that during growth and replication in biofilms, the average cell–cell distance between nearest neighbors first increases for cell numbers N ≲ 10, before this distance steadily decreases for all biofilms with N ≳ 10, implying that cell densities inside of biofilms steadily increase (Fig. 3A). The increase of cell density is confirmed by directly counting the number of cells within a local neighborhood of a reference cell (Fig. 3B), resulting in volume fractions of ϕ = 0.46 ± 0.05 in the densest regions of biofilms with N ∼ 4,000 cells (Fig. S5). At such high-volume fractions, systems of passive spherocylinders have been predicted to approach a phase in which isotropic regions and highly ordered crystalline domains coexist (60). In biofilms, both cell–cell adhesion and the extracellular matrix that fills the space between cells strongly inhibit rotational and translational diffusion of the bacterial cells, thereby effectively lowering the Brownian temperature. This reasoning suggests that the significantly increased cell density in large biofilms should promote local alignment. Our data indeed confirm that local orientational ordering increases at large cell numbers (Fig. 4C), with cells arranging in hedgehog-like configurations (Fig. 4D).
To quantify the orientational ordering, we compared the experimentally observed cell orientations with simulations of a basic stochastic biofilm model. In these simulations, we constructed model biofilms of different sizes N by sampling cell positions x uniformly in a 3D parabolic cap centered around the z axis. For a given number of cells N, the height and diameter of the paraboloid were matched to the corresponding experimentally determined values (Fig. 2). The individual cell orientations n were sampled from a spherical normal distribution f(n) = C(κ) exp(κR · n), where C(κ) is the normalization constant, R = x/|x| is the radial unit vector pointing from the coordinate origin to the cell position, and κ is a distribution parameter that determines the average deviation of the orientation vector n from the radial direction R. The exponential orientation distribution f(n), also known as the von Mises–Fisher distribution, defines the least biased cell orientation model as f(n) maximizes the Shannon entropy on a sphere for a given mean orientation R. The resulting stochastic biofilm model converges to a perfect radially symmetric hedgehog in the limit of a large von Mises–Fisher parameter κ >> 1, whereas κ = 0 yields a uniform distribution on a sphere, corresponding to completely disordered cell orientations. The numerically generated model biofilms were analyzed with the same methods and metrics as the experimental biofilms. This analysis shows that for biofilms with N = 200–4,000 cells, the experimentally obtained values of the local order parameter S are recovered for von Mises–Fisher parameters in the range κ = 3–10 (Fig. 4E). The fact that κ ∼ 10 is required to match the experimental data for N ∼ 4,000 confirms quantitatively that large biofilms exhibit approximately hedgehog-like orientational order.
The measured cellular aspect ratio of ∼2.5 (Fig. S3B) does not suffice to explain nematic ordering on purely thermodynamic grounds (48). We therefore hypothesize that cellular ordering inside biofilms is not a thermal process in which an isotropic phase of diffusing nematic objects is compressed until it transitions to orientational order. Instead, the biofilm matrix strongly suppresses rotational and translational diffusion so that the cellular ordering takes place at near-zero “effective temperature.” In addition to the absence of Brownian diffusion, there are two additional nonequilibrium effects that likely play important roles: First, the increase in cell number occurs due to cell growth by elongation and division, which is a directed process biased to favor local alignment, and second, the growth of cells inside an adhesive matrix leads to an increased effective pressure on the cells that favors nematic order. The observed ordering in our V. cholerae biofilms is consistent with recent experiments examining 2D confined colonies of E. coli, which have similar aspect ratios to V. cholerae (58, 59). Together, these findings suggest that a combination of local densification and radial expansion flows that occur as a consequence of cell division can trigger the 3D ordering transitions we observe in naturally growing biofilms.
Biofilm Architectures with Altered Matrix Production.
The secreted matrix is a defining feature of biofilms. To investigate its role in shaping global biofilm architecture, we analyzed mutants lacking individual biofilm matrix proteins (RbmA, Bap1, and RbmC), a mutant that is a matrix hyperproducer [carrying the vpvCW240R allele (61)], and mutants locked in either the low- or high-cell density quorum-sensing states (luxOD47E and ΔluxO, respectively). The proteins RbmA, Bap1, and RbmC are known to localize heterogeneously within biofilms, and their individual roles in biofilm structure have recently been examined (42, 43, 62–67). RbmA is implicated in binding mother and daughter cells to each other and to polysaccharide components of the matrix. Bap1 is involved in attaching cells to the surface, and RbmC forms envelopes surrounding clusters of cells. RbmA also plays an ecological role in restricting access to existing biofilms by planktonic cells (42). Mutants in quorum-sensing components were investigated because quorum sensing regulates matrix production in V. cholerae (23, 68). Specifically, a strain that mimics the quorum-sensing low-cell–density state (luxOD47E) is a matrix overproducer, whereas a strain that mimics the high-cell–density state (ΔluxO) displays decreased matrix production. In every strain we tested, the global morphological biofilm features were remarkably similar to those of the wild type, with respect to volume (Fig. S6), height (Fig. S7), and height/width ratio (Fig. S8), and depended on cell number N.
Fig. S6.
Biofilm volume for different V. cholerae strains. The vpvCW240R strain is a matrix hyperproducing strain. The ΔrbmA, ΔrbmC, and Δbap1 strains each lack a specific component of the extracellular matrix. The luxOD47E strain is locked in the quorum-sensing low-cell density state, whereas the ΔluxO strain is locked in the quorum-sensing high-cell density state. The dependence of the biofilm volume on the number of cells inside the biofilm is similar for all strains that were investigated. The Insets show log–log plots of the data from each panel and fits of the parameter α, defined in the main manuscript. Each data point in this figure results from one biofilm.
Fig. S7.
Peak height for biofilms made by different V. cholerae strains. Strains are as in Fig. S6. The dependence of the biofilm peak height for the ΔrbmA strain is different from all other strains investigated. Relative to all other strains examined, the ΔrbmA strain peak height increases more slowly with cell number. Each data point in every panel results from one biofilm.
Fig. S8.
Aspect ratios of biofilm morphologies for different V. cholerae strains. The height corresponds to the peak height shown in Fig. S7, the parameters width-A and width-B are defined in Fig. 2E. Strains are as in Fig. S6. Each data point in every panel results from one biofilm.
We also compared the internal structures of biofilms formed by the different mutants to that of the wild type. Fig. 5A shows the average of the z-component of the cellular orientations nz in biofilms with cell numbers in the range of N = 1,500–2,500, a size range for which the orientational order in wild-type biofilms is not particularly high (see Fig. 4C). The internal architectures of strains lacking the matrix protein RbmC or Bap1, the matrix hyperproducing strain, and the strains locked in low- or high-quorum-sensing states are similar to the wild-type architecture (Fig. 5A). The ΔrbmA strain, however, exhibits a significantly different internal structure, shown in detail in Fig. 5 B and C. In contrast to the wild type and the other strains analyzed here, the majority of cells in ΔrbmA biofilms are vertically oriented and consequently display a high orientational order at much smaller biofilm sizes. Even the surface-attached cells, which are predominantly horizontal in the wild type (Fig. 4D), are primarily vertical relative to the substratum in the rbmA mutant, revealing a new function for RbmA in surface attachment. Because the initial substrate-attached cells are vertically oriented, the consecutive layers of cells in ΔrbmA biofilm growth are also primarily vertical, as shown in Fig. 5C.
Fig. 5.
The internal biofilm architecture of the V. cholerae ΔrbmA mutant strongly deviates from that of wild-type V. cholerae. (A) The average z-component of each cell orientation, nz, is shown for V. cholerae wild type; the matrix mutants ΔrbmA, ΔrbmC, and Δbap1; the matrix hyperproducing mutant vpvCW240R; and the quorum-sensing mutants ΔluxO and luxOD47E. Error bars indicate the SD from at least three different biofilms with N = 1,500–2,500 cells for each strain. Detailed global and morphological comparisons between the biofilms of these mutants are provided in Figs. S6–S8. (B and C) The segmented cells in an XZ-slice through a biofilm with N = 114 cells (B) and N = 1,658 cells (C) are shown, in which individual cells are colored according to their orientation along the z-direction, nz, using the same color scale as in Fig. 4D and Fig. 6.
Phases of V. cholerae Biofilm Growth.
Based on our results, we propose that biofilm growth of V. cholerae on submerged glass surfaces in the presence of flow can be separated into four distinct phases, illustrated in Fig. 6. In phase I (N = 1–6 cells), the cells grow in a 1D line and are highly ordered. All cells in this growth phase are attached to the glass substratum (Fig. 6 A and B). This line of cells buckles into an in-plane 2D arrangement for N ≳ 6, presumably when cell–surface adhesion becomes stronger than bacterial pole–pole adhesion. In phase II (N = 20–100 cells), cells are mostly disordered locally and are growing in a 2D colony, with all cells still attached to the glass substratum (Fig. 6 C and D). This 2D colony buckles into a 3D colony, presumably when the cell–surface adhesion force becomes smaller than the force that cells exert when they push against each other due to cell growth (69). In phase III (N = 200–1,000 cells), the 3D colony is still locally disordered (Fig. 6 E and F) and at relatively low cell density (Fig. 3B). As the cell density inside the biofilm increases (Fig. 3 A and B), the biofilm undergoes an ordering transition to phase IV (N ≳ 2,000 cells), which is characterized by high local and long-range cellular alignment, with the architecture shown in Fig. 6 G and H. In phase IV, cells at large radial coordinate ρ and low z coordinates are located primarily horizontally, whereas cells in the center are primarily vertical (Fig. 6H). High cellular order and a radial cellular orientation inside biofilms likely provide a growth advantage compared with random cellular orientations, as radially oriented cells produce daughter cells at the outer edges of the 3D biofilm where the nutrient concentration is highest. Random cellular orientations, in contrast, will lead to nematic frustration of growth and fewer offspring directed to the high-nutrient regions.
Fig. 6.
The different phases of V. cholerae wild-type biofilm growth. (A and B) Biofilms in phase I consist of highly ordered N = 1∼6 cells growing along a line, and all cells are bound to the glass substratum. Panel A shows a representative image, and panel B shows the top and side views of the segmented cells in the community. (C and D) Biofilms in phase II consist of N = 20∼100 cells, which form a 2D community with low local order, in which all cells remain in contact with the glass substratum. (E and F) Biofilms in phase III consist of N = 200∼1,000 cells that form a disordered 3D community. Panel E shows the average magnitude of the z-component of the cellular orientation vector <nz>. For the heat map, 26 biofilms with N = 500–600 cells were averaged and the results plotted using cylindrical coordinates with radial coordinate ρ, normalized by the maximum cylindrical radius of the biofilm ρmax. Panel F shows all detected cells in an XZ-slice through a biofilm with N = 510 cells; individual cells are colored according to their value of nz, as indicated in the color bar. (G and H) Biofilms in phase IV are highly ordered 3D communities with N > 2,000 cells. Panel G shows <nz> averaged for 11 biofilms with N = 3,800–4,600 cells, using cylindrical coordinates. Panel H shows the cells in an XZ-slice through a biofilm with N = 3,975 cells; individual cells are colored according to their orientation along the z-direction, nz. Biofilms with cell numbers in between these four growth phases are undergoing transitions from 1D to 2D growth, from 2D to 3D disordered growth, or from 3D disordered to ordered growth, respectively.
Conclusion
By imaging and analyzing the position, size, and orientation of all individual cells inside V. cholerae biofilms, we were able to measure biofilm growth laws and we discovered architectural changes that occur during growth. The parameters that describe the internal and external biofilm architectures collapsed to a single curve, when measured as a function of cell number in the biofilm. Little variation was found between the biofilm structural parameters, when measured as a function of cell number for the wild type and mutants with varying degrees of matrix production levels, mutants defective in quorum-sensing regulation, and mutants deficient in production of specific matrix proteins. The experimentally observed global and internal biofilm architectures during and between the growth phases can, in part, be accounted for using mechanical arguments. However, mechanical effects alone are not sufficient to determine the entire biofilm architecture, as a mutant that does not produce the biofilm matrix protein RbmA exhibits a dramatically different biofilm architecture than the wild type and other mutant strains. Additional analyses to quantify the forces involved in V. cholerae surface adhesion (16, 70), cell–cell adhesion, and cell–matrix interaction strength are needed to develop accurate simulations of biofilm growth (71–74). Such simulations will be required to distinguish the effects of forces resulting from cell growth and division from those due to matrix production and adhesion in developing biofilms.
Materials and Methods
Strains and Media.
All V. cholerae strains used in this study are derivatives of the wild-type V. cholerae O1 biovar El Tor strain N16961. V. cholerae deletion mutations were engineered using pKAS32 harbored in E. coli S17-1λpir. All strains were grown in LB medium supplemented with appropriate antibiotics at 37 °C for strain construction. Biofilm experiments with V. cholerae were performed in M9 minimal medium, supplemented with 2 mM MgSO4 and 100 µM CaCl2, MEM vitamins, 0.5% glucose, and 15 mM triethanolamine (pH 7.1). A detailed strain list is provided in Table S1.
Table S1.
Bacterial strains and plasmids used in this study
| Strain/plasmid | Relevant features | Source |
| V. cholera | ||
| KDV101 | Wild-type strain N16961 (O1 El Tor, StrepR) | Meibom et al. (75) |
| KDV060 | ΔrbmA | This study |
| KDV062 | ΔrbmC | This study |
| KDV064 | Δbap1 | This study |
| KDV066 | ΔluxO | This study |
| KDV068 | luxOD47E | This study |
| KDV148 | vpvCW240R (matrix hyperproducer phenotype) | This study |
| KDV103 | lacZ:PtacmKO:lacZ | Drescher et al. (36) |
| KDV104 | lacZ:PtacmKate2:lacZ | Drescher et al. (36) |
| KDV158 | lacZ:PtacmTFP1:lacZ | This study |
| E. coli | ||
| S17-1λpir | Wild type | de Lorenzo and Timmis (76) |
| Plasmids | ||
| pNUT015 | pKAS32 suicide vector (origin pR6K, AmpR) | Skorupski and Taylor (77) |
| pNUT336 | pKAS32-based plasmid for making rbmA deletion (pCN007) | Nadell et al. (42) |
| pNUT337 | pKAS32-based plasmid for making rbmC deletion (pCN008) | Nadell et al. (42) |
| pNUT338 | pKAS32-based plasmid for making bap1 deletion | Nadell et al. (42) |
| pNUT029 | pKAS32-based plasmid for making luxO deletion | Bassler lab collection (BH111) |
| pNUT339 | pKAS32-based plasmid for making luxOD47E mutation | Bassler lab collection (BH032) |
| pNUT157 | pKAS32-based plasmid for making vpvCW240R mutation | Drescher et al. (36) |
Flow Chamber Biofilm Experiments.
V. cholerae biofilms were grown in microfluidic flow chambers (dimensions were 500 µm wide, 100 µm high, and 7 mm long) made from polydimethylsiloxane bonded to glass coverslips by an oxygen plasma, with four channels on a single coverslip. Microfluidic channels were designed and constructed according to standard soft lithography techniques. Channels were inoculated with V. cholerae strains that had been grown overnight at 37 °C in liquid LB medium under shaking conditions. Following inoculation of the channels, the cells were given 1 h to attach to the surface of the channel before a flow of 2 µL/min M9 medium (formulation as described above) was initiated to wash away nonadherent cells and to remove LB growth medium from the channels. The flow rate was then set to 0.1 µL/min, corresponding to a mean flow speed of 33 µm/s in the channel until the end of the experiment. Flow rates were controlled using a syringe pump. Immediately before imaging, the inflowing medium was exchanged from simple M9 to M9 containing 10 µM SYTO 9 nucleic acid stain (Life Technologies).
Microscopy and Image Processing.
Images were acquired with a Yokogawa CSU-X1 confocal spinning disk unit mounted on a Nikon Ti-E inverted microscope, using a 100× oil objective with N.A. 1.4, a 488-nm laser (Coherent Sapphire), and an Andor iXon EMCCD camera, cooled to –90 °C. To obtain sufficient magnification for automated cell segmentation, a 1.5× lens was placed between the CSU-X1 and the Nikon Ti-E side port. The distances between the spinning disk, the tube lens, and the camera sensor were fine-tuned to provide sufficient z-resolution for the single-cell segmentation. Point-scanning confocal microscopy with an adjustable pinhole size could also be used to obtain sufficient optical resolution. The magnification was 0.1 µm per pixel in the XY plane and 0.2 µm step size in the z-direction. To avoid residual movement of immersion oil due to changing the objective position during the z-scan, we introduced a 1-s lag time between changing the focal height and exposure. The exposure time was 100 ms per image. To segment the biofilms into individual cells from the 3D images, custom code was written in Matlab. Briefly, images were treated separately for each z-plane, band-pass filtered, thresholded, and after that, binary objects were connected between the different z-planes to obtain 3D volumes for each cell inside the biofilm. By manually verifying the segmentation of 10 biofilms of ∼200 and ∼500 cells, we determined that the algorithm correctly segments 95% of the cells, whereas the remaining 5% are clusters of >2 cells, of which 63% are clusters of two cells.
Acknowledgments
We thank members of the B.L.B., N.S.W., H.A.S., and K.D. laboratories for discussions and advice. This work was supported by the Howard Hughes Medical Institute (B.L.B.), National Institutes of Health Grant GM065859 (to B.L.B.), National Science Foundation Grants MCB-0948112 (to B.L.B.) and MCB-1119232 and MCB-1344191 (to B.L.B., H.A.S., and N.S.W.), the Max Planck Society (K.D.), the Human Frontier Science Program (K.D.), the Deutsche Forschungsgemeinschaft SPP1617 (K.D.), the Alfred P. Sloan Foundation (J.D.), and the Alexander von Humboldt Foundation (C.D.N.).
Footnotes
The authors declare no conflict of interest.
See Commentary on page 3711.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1601702113/-/DCSupplemental.
References
- 1.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: From the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2(2):95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 2.Teschler JK, et al. Living in the matrix: Assembly and control of Vibrio cholerae biofilms. Nat Rev Microbiol. 2015;13(5):255–268. doi: 10.1038/nrmicro3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Persat A, et al. The mechanical world of bacteria. Cell. 2015;161(5):988–997. doi: 10.1016/j.cell.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parsek MR, Greenberg EP. Sociomicrobiology: The connections between quorum sensing and biofilms. Trends Microbiol. 2005;13(1):27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Römling U, et al. Microbial biofilm formation: A need to act. J Intern Med. 2014;276(2):98–110. doi: 10.1111/joim.12242. [DOI] [PubMed] [Google Scholar]
- 6.Earle KA, et al. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe. 2015;18(4):478–488. doi: 10.1016/j.chom.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Escudié R, Cresson R, Delgenès JP, Bernet N. Control of start-up and operation of anaerobic biofilm reactors: An overview of 15 years of research. Water Res. 2011;45(1):1–10. doi: 10.1016/j.watres.2010.07.081. [DOI] [PubMed] [Google Scholar]
- 8.Høiby N, et al. ESCMID Study Group for Biofilms and Consulting External Expert Werner Zimmerli ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin Microbiol Infect. 2015;21(Suppl 1):S1–S25. doi: 10.1016/j.cmi.2014.10.024. [DOI] [PubMed] [Google Scholar]
- 9.Harmsen M, Yang L, Pamp SJ, Tolker-Nielsen T. An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunol Med Microbiol. 2010;59(3):253–268. doi: 10.1111/j.1574-695X.2010.00690.x. [DOI] [PubMed] [Google Scholar]
- 10.Marvig RL, Sommer LM, Molin S, Johansen HK. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat Genet. 2015;47(1):57–64. doi: 10.1038/ng.3148. [DOI] [PubMed] [Google Scholar]
- 11.Drescher K, Shen Y, Bassler BL, Stone HA. Biofilm streamers cause catastrophic disruption of flow with consequences for environmental and medical systems. Proc Natl Acad Sci USA. 2013;110(11):4345–4350. doi: 10.1073/pnas.1300321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fux CA, Costerton JW, Stewart PS, Stoodley P. Survival strategies of infectious biofilms. Trends Microbiol. 2005;13(1):34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 13.Donlan RM, Costerton JW. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15(2):167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen D, et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science. 2011;334(6058):982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao K, et al. Psl trails guide exploration and microcolony formation in Pseudomonas aeruginosa biofilms. Nature. 2013;497(7449):388–391. doi: 10.1038/nature12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maier B, Wong GCL. How bacteria use type IV pili machinery on surfaces. Trends Microbiol. 2015;23(12):775–788. doi: 10.1016/j.tim.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 17.Luo Y, et al. A hierarchical cascade of second messengers regulates Pseudomonas aeruginosa surface behaviors. MBio. 2015;6(1):e02456. doi: 10.1128/mBio.02456-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flemming H-C, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8(9):623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 19.McDougald D, Rice SA, Barraud N, Steinberg PD, Kjelleberg S. Should we stay or should we go: Mechanisms and ecological consequences for biofilm dispersal. Nat Rev Microbiol. 2012;10(1):39–50. doi: 10.1038/nrmicro2695. [DOI] [PubMed] [Google Scholar]
- 20.Kaplan JB. Biofilm dispersal: Mechanisms, clinical implications, and potential therapeutic uses. J Dent Res. 2010;89(3):205–218. doi: 10.1177/0022034509359403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seminara A, et al. Osmotic spreading of Bacillus subtilis biofilms driven by an extracellular matrix. Proc Natl Acad Sci USA. 2012;109(4):1116–1121. doi: 10.1073/pnas.1109261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hobley L, Harkins C, MacPhee CE, Stanley-Wall NR. Giving structure to the biofilm matrix: An overview of individual strategies and emerging common themes. FEMS Microbiol Rev. 2015;39(5):649–669. doi: 10.1093/femsre/fuv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50(1):101–104. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- 24.Chambers JR, Sauer K. Small RNAs and their role in biofilm formation. Trends Microbiol. 2013;21(1):39–49. doi: 10.1016/j.tim.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu J, et al. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 2002;99(5):3129–3134. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6(3):199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 27.Asally M, et al. Localized cell death focuses mechanical forces during 3D patterning in a biofilm. Proc Natl Acad Sci USA. 2012;109(46):18891–18896. doi: 10.1073/pnas.1212429109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prindle A, et al. Ion channels enable electrical communication in bacterial communities. Nature. 2015;527(7576):59–63. doi: 10.1038/nature15709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilking JN, et al. Liquid transport facilitated by channels in Bacillus subtilis biofilms. Proc Natl Acad Sci USA. 2013;110(3):848–852. doi: 10.1073/pnas.1216376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xavier JB, Picioreanu C, van Loosdrecht MCM. A framework for multidimensional modelling of activity and structure of multispecies biofilms. Environ Microbiol. 2005;7(8):1085–1103. doi: 10.1111/j.1462-2920.2005.00787.x. [DOI] [PubMed] [Google Scholar]
- 31.Nadell CD, Xavier JB, Foster KR. The sociobiology of biofilms. FEMS Microbiol Rev. 2009;33(1):206–224. doi: 10.1111/j.1574-6976.2008.00150.x. [DOI] [PubMed] [Google Scholar]
- 32.Lecuyer S, Stocker R, Rusconi R. Focus on the physics of biofilms. New J Phys. 2015;17(3):030401. [Google Scholar]
- 33.Oliveira NM, et al. Biofilm formation as a response to ecological competition. PLoS Biol. 2015;13(7):e1002191. doi: 10.1371/journal.pbio.1002191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghosh P, Mondal J, Ben-Jacob E, Levine H. Mechanically-driven phase separation in a growing bacterial colony. Proc Natl Acad Sci USA. 2015;112(17):E2166–E2173. doi: 10.1073/pnas.1504948112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rusconi R, Garren M, Stocker R. Microfluidics expanding the frontiers of microbial ecology. Annu Rev Biophys. 2014;43(1):65–91. doi: 10.1146/annurev-biophys-051013-022916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drescher K, Nadell CD, Stone HA, Wingreen NS, Bassler BL. Solutions to the public goods dilemma in bacterial biofilms. Curr Biol. 2014;24(1):50–55. doi: 10.1016/j.cub.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serra DO, Richter AM, Klauck G, Mika F, Hengge R. Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. MBio. 2013;4(2):e00103–e00113. doi: 10.1128/mBio.00103-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Serra DO, Richter AM, Hengge R. Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J Bacteriol. 2013;195(24):5540–5554. doi: 10.1128/JB.00946-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stewart EJ, Satorius AE, Younger JG, Solomon MJ. Role of environmental and antibiotic stress on Staphylococcus epidermidis biofilm microstructure. Langmuir. 2013;29(23):7017–7024. doi: 10.1021/la401322k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pavlovsky L, Sturtevant RA, Younger JG, Solomon MJ. Effects of temperature on the morphological, polymeric, and mechanical properties of Staphylococcus epidermidis bacterial biofilms. Langmuir. 2015;31(6):2036–2042. doi: 10.1021/la5044156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nadell CD, Bassler BL. A fitness trade-off between local competition and dispersal in Vibrio cholerae biofilms. Proc Natl Acad Sci USA. 2011;108(34):14181–14185. doi: 10.1073/pnas.1111147108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nadell CD, Drescher K, Wingreen NS, Bassler BL. Extracellular matrix structure governs invasion resistance in bacterial biofilms. ISME J. 2015;9(8):1700–1709. doi: 10.1038/ismej.2014.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith DR, et al. In situ proteolysis of the Vibrio cholerae matrix protein RbmA promotes biofilm recruitment. Proc Natl Acad Sci USA. 2015;112(33):10491–10496. doi: 10.1073/pnas.1512424112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Millet YA, et al. Insights into Vibrio cholerae intestinal colonization from monitoring fluorescently labeled bacteria. PLoS Pathog. 2014;10(10):e1004405. doi: 10.1371/journal.ppat.1004405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scott M, Hwa T. Bacterial growth laws and their applications. Curr Opin Biotechnol. 2011;22(4):559–565. doi: 10.1016/j.copbio.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maitra A, Dill KA. Bacterial growth laws reflect the evolutionary importance of energy efficiency. Proc Natl Acad Sci USA. 2015;112(2):406–411. doi: 10.1073/pnas.1421138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rozenfeld HD, et al. Laws of population growth. Proc Natl Acad Sci USA. 2008;105(48):18702–18707. doi: 10.1073/pnas.0807435105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Onsager L. The effects of shape on the interaction of colloidal particles. Ann N Y Acad Sci. 1949;51(4):627–659. [Google Scholar]
- 49.Fraden S, Maret G, Caspar DL, Meyer RB. Isotropic-nematic phase transition and angular correlations in isotropic suspensions of tobacco mosaic virus. Phys Rev Lett. 1989;63(19):2068–2071. doi: 10.1103/PhysRevLett.63.2068. [DOI] [PubMed] [Google Scholar]
- 50.Cladis P. New liquid-crystal phase diagram. Phys Rev Lett. 1975;35(1):48–51. [Google Scholar]
- 51.Keyes P, Weston H, Lin W, Daniels W. Liquid crystal phase diagrams: A study of seven thermotropic materials. J Chem Phys. 1975;63(11):5006–5010. [Google Scholar]
- 52.Dierking I. Chiral liquid crystals: Structures, phases, effects. Symmetry (Basel) 2014;6(2):444–472. [Google Scholar]
- 53.Zhang H-P, Be’er A, Florin E-L, Swinney HL. Collective motion and density fluctuations in bacterial colonies. Proc Natl Acad Sci USA. 2010;107(31):13626–13630. doi: 10.1073/pnas.1001651107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sokolov A, Aranson IS, Kessler JO, Goldstein RE. Concentration dependence of the collective dynamics of swimming bacteria. Phys Rev Lett. 2007;98(15):158102. doi: 10.1103/PhysRevLett.98.158102. [DOI] [PubMed] [Google Scholar]
- 55.Wensink HH, et al. Meso-scale turbulence in living fluids. Proc Natl Acad Sci USA. 2012;109(36):14308–14313. doi: 10.1073/pnas.1202032109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dunkel J, et al. Fluid dynamics of bacterial turbulence. Phys Rev Lett. 2013;110(22):228102. doi: 10.1103/PhysRevLett.110.228102. [DOI] [PubMed] [Google Scholar]
- 57.Drescher K, Dunkel J, Cisneros LH, Ganguly S, Goldstein RE. Fluid dynamics and noise in bacterial cell-cell and cell-surface scattering. Proc Natl Acad Sci USA. 2011;108(27):10940–10945. doi: 10.1073/pnas.1019079108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Volfson D, Cookson S, Hasty J, Tsimring LS. Biomechanical ordering of dense cell populations. Proc Natl Acad Sci USA. 2008;105(40):15346–15351. doi: 10.1073/pnas.0706805105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cho H, et al. Self-organization in high-density bacterial colonies: Efficient crowd control. PLoS Biol. 2007;5(11):e302. doi: 10.1371/journal.pbio.0050302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bolhuis P, Frenkel D. Tracing the phase boundaries of hard spherocylinders. J Chem Phys. 1997;106(2):666–687. [Google Scholar]
- 61.Beyhan S, Yildiz FH. Smooth to rugose phase variation in Vibrio cholerae can be mediated by a single nucleotide change that targets c-di-GMP signalling pathway. Mol Microbiol. 2007;63(4):995–1007. doi: 10.1111/j.1365-2958.2006.05568.x. [DOI] [PubMed] [Google Scholar]
- 62.Berk V, et al. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science. 2012;337(6091):236–239. doi: 10.1126/science.1222981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fong JCN, Karplus K, Schoolnik GK, Yildiz FH. Identification and characterization of RbmA, a novel protein required for the development of rugose colony morphology and biofilm structure in Vibrio cholerae. J Bacteriol. 2006;188(3):1049–1059. doi: 10.1128/JB.188.3.1049-1059.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fong JCN, Yildiz FH. The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae. J Bacteriol. 2007;189(6):2319–2330. doi: 10.1128/JB.01569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Absalon C, Van Dellen K, Watnick PI. A communal bacterial adhesin anchors biofilm and bystander cells to surfaces. PLoS Pathog. 2011;7(8):e1002210. doi: 10.1371/journal.ppat.1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Giglio KM, Fong JC, Yildiz FH, Sondermann H. Structural basis for biofilm formation via the Vibrio cholerae matrix protein RbmA. J Bacteriol. 2013;195(14):3277–3286. doi: 10.1128/JB.00374-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maestre-Reyna M, Wu W-J, Wang AHJ. Structural insights into RbmA, a biofilm scaffolding protein of V. cholerae. PLoS One. 2013;8(12):e82458. doi: 10.1371/journal.pone.0082458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krasteva PV, et al. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327(5967):866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grant MAA, Wacław B, Allen RJ, Cicuta P. The role of mechanical forces in the planar-to-bulk transition in growing Escherichia coli microcolonies. J R Soc Interface. 2014;11(97):20140400. doi: 10.1098/rsif.2014.0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thewes N, et al. Stochastic binding of Staphylococcus aureus to hydrophobic surfaces. Soft Matter. 2015;11(46):8913–8919. doi: 10.1039/c5sm00963d. [DOI] [PubMed] [Google Scholar]
- 71.Lardon LA, et al. iDynoMiCS: Next-generation individual-based modelling of biofilms. Environ Microbiol. 2011;13(9):2416–2434. doi: 10.1111/j.1462-2920.2011.02414.x. [DOI] [PubMed] [Google Scholar]
- 72.Borenstein DB, Meir Y, Shaevitz JW, Wingreen NS. Non-local interaction via diffusible resource prevents coexistence of cooperators and cheaters in a lattice model. PLoS One. 2013;8(5):e63304. doi: 10.1371/journal.pone.0063304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Estrela S, Brown SP. Metabolic and demographic feedbacks shape the emergent spatial structure and function of microbial communities. PLOS Comput Biol. 2013;9(12):e1003398. doi: 10.1371/journal.pcbi.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Melaugh G, et al. 2015 Shaping the growth behaviour of bacterial aggregates in biofilms. arXiv:1506.08168. [Google Scholar]
- 75.Meibom KL, et al. The Vibrio cholerae chitin utilization program. Proc Natl Acad Sci USA. 2004;101(8):2524–2529. doi: 10.1073/pnas.0308707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Lorenzo V, Timmis KN. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived mini transposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 77.Skorupski K, Taylor RK. Positive selection vectors for allelic exchange. Gene. 1996;169(1):47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]