Significance
Pluripotency factors, such as NANOG, play a critical role in the maintenance and specification of cancer stem cells, which are required for primary tumor formation and metastasis. In this study, we report that exposure of breast cancer cells to hypoxia (i.e., reduced O2 availability), which is a critical feature of the tumor microenvironment, induces N6-methyladenosine (m6A) demethylation and stabilization of NANOG mRNA, thereby promoting the breast cancer stem cell (BCSC) phenotype. We show that inhibiting the expression of AlkB homolog 5 (ALKBH5), which demethylates m6A, or the hypoxia-inducible factors (HIFs) HIF-1α and HIF-2α, which activate ALKBH5 gene transcription in hypoxic breast cancer cells, is an effective strategy to decrease NANOG expression and target BCSCs in vivo.
Keywords: hypoxia-inducible factors, metastasis, pluripotency factors, self-renewal, tumorigenesis
Abstract
N6-methyladenosine (m6A) modification of mRNA plays a role in regulating embryonic stem cell pluripotency. However, the physiological signals that determine the balance between methylation and demethylation have not been described, nor have studies addressed the role of m6A in cancer stem cells. We report that exposure of breast cancer cells to hypoxia stimulated hypoxia-inducible factor (HIF)-1α- and HIF-2α–dependent expression of AlkB homolog 5 (ALKBH5), an m6A demethylase, which demethylated NANOG mRNA, which encodes a pluripotency factor, at an m6A residue in the 3′-UTR. Increased NANOG mRNA and protein expression, and the breast cancer stem cell (BCSC) phenotype, were induced by hypoxia in an HIF- and ALKBH5-dependent manner. Insertion of the NANOG 3′-UTR into a luciferase reporter gene led to regulation of luciferase activity by O2, HIFs, and ALKBH5, which was lost upon mutation of the methylated residue. ALKBH5 overexpression decreased NANOG mRNA methylation, increased NANOG levels, and increased the percentage of BCSCs, phenocopying the effect of hypoxia. Knockdown of ALKBH5 expression in MDA-MB-231 human breast cancer cells significantly reduced their capacity for tumor initiation as a result of reduced numbers of BCSCs. Thus, HIF-dependent ALKBH5 expression mediates enrichment of BCSCs in the hypoxic tumor microenvironment.
Breast cancer has the highest incidence of all cancers affecting women (1). It is also the leading cause of cancer-related death for women worldwide (2). Although progress has been made in treating early-stage breast cancer, there is no effective strategy to prevent or treat metastasis, which is the major cause of breast cancer-related mortality (3). Within breast cancers, a small subpopulation of cells, which are designated as tumor-initiating cells or breast cancer stem cells (BCSCs), have the unique property of generating daughter BCSCs, which are capable of infinite proliferation through self-renewal, and transient amplifying cells, which are capable of a limited number of cell divisions and give rise to differentiated breast cancer cells (4, 5). Only BCSCs are capable of forming a secondary (recurrent or metastatic) tumor (5, 6). As in the case of ES cells (ESCs) (7), the BCSC phenotype is specified by the expression of core pluripotency factors, including Kruppel-like factor 4 (KLF4), Octamer-binding transcription factor 4 (OCT4), SRY-box 2 (SOX2), and NANOG (8–12). Delineation of the molecular mechanisms that regulate the BCSC phenotype is needed to better understand metastasis and design more effective therapies.
Intratumoral hypoxia is a common finding in advanced cancers. The median partial pressure of oxygen (pO2) within primary breast cancers is 10 mm Hg [1.4% (vol/vol) O2], compared with 65 mm Hg [9.3% (vol/vol) O2] in normal breast tissue (13). Hypoxia-inducible factors (HIFs) HIF-1 and HIF-2 are heterodimeric transcriptional activators, consisting of an O2-regulated HIF-1α or HIF-2α subunit and a constitutively expressed HIF-1β subunit (14). In hypoxic breast cancer cells, HIFs activate the transcription of target genes that play important roles in tumor growth, angiogenesis, metabolic reprogramming, motility, invasion, metastasis, and resistance to chemotherapy (15, 16). Recent studies have demonstrated that HIFs are required for the specification and/or maintenance of BCSCs in response to hypoxia (17–21) or chemotherapy (22, 23), leading to direct or indirect transcriptional regulation of genes encoding the pluripotency factors NANOG, SOX2, and KLF4. In addition, HIF-1α is required for hypoxia-induced epithelial-mesenchymal transition (24), which is also linked to the BCSC phenotype (25). In contrast to WT cells, the capacity of mouse HIF-1α–KO (26) or human HIF-1α–knockdown (21) breast cancer cells to initiate tumorigenesis is markedly impaired.
Recent studies have indicated that the expression of pluripotency factors is also regulated by changes in mRNA stability. N6-methyladenosine (m6A) is the most prevalent modification of mRNA in mammalian cells, consisting of 0.1–0.4% of all adenosine residues, particularly at the beginning of the 3′-UTR near the translation termination codon, usually embedded within the consensus sequence 5′-RRACU-3′ (R = A or G; methylated adenosine residue is underscored) (27–29). This reversible modification is catalyzed by a methyltransferase complex consisting of the proteins methyltransferase-like 3 (METTL3), METTL14, and Wilms tumor 1 associated protein (WTAP), the activity of which is opposed by two m6A demethylases, fat mass and obesity-associated protein (FTO) and AlkB homolog 5 (ALKBH5) (27, 30). In human cells, thousands of mRNAs are subject to m6A modification (28, 29), which has been implicated in mRNA splicing, nuclear export, and translation (31) as well as microRNA processing (32). However, m6A modification has been most strongly linked to increased mRNA degradation (33). NANOG, SOX2, and KLF4 mRNA were reported to contain m6A modification in ESCs (29). METTL3 KO led to decreased m6A modification of NANOG mRNA, increased NANOG mRNA stability, and decreased ESC differentiation (29, 34). These studies indicated that m6A methylation of mRNA plays a critical role in regulating ESC pluripotency. However, the physiological signals that regulate the balance between methylation and demethylation have not been described, nor have studies addressed the role of m6A in cancer stem cells.
ALKBH5 is a dioxygenase that uses α-ketoglutarate and O2 as substrates in the m6A demethylation reaction (35). Even before its identification as an RNA demethylase, hypoxic induction of ALKBH5 expression was reported (36), but, at that time, the functional significance of this observation was not known. In the study reported here, we tested the hypothesis that exposure of breast cancer cells to hypoxia is sufficient to stimulate HIF-dependent ALKBH5 expression, leading to decreased m6A methylation of NANOG mRNA, increased NANOG mRNA stability, increased NANOG protein levels, and BCSC enrichment, thereby establishing hypoxia as a major physiological stimulus in the tumor microenvironment that directly connects regulation of m6A RNA methylation to the specification and maintenance of BCSCs.
Results
HIF-Dependent ALKBH5 Expression Promotes m6A RNA Demethylation in Hypoxic Breast Cancer Cells.
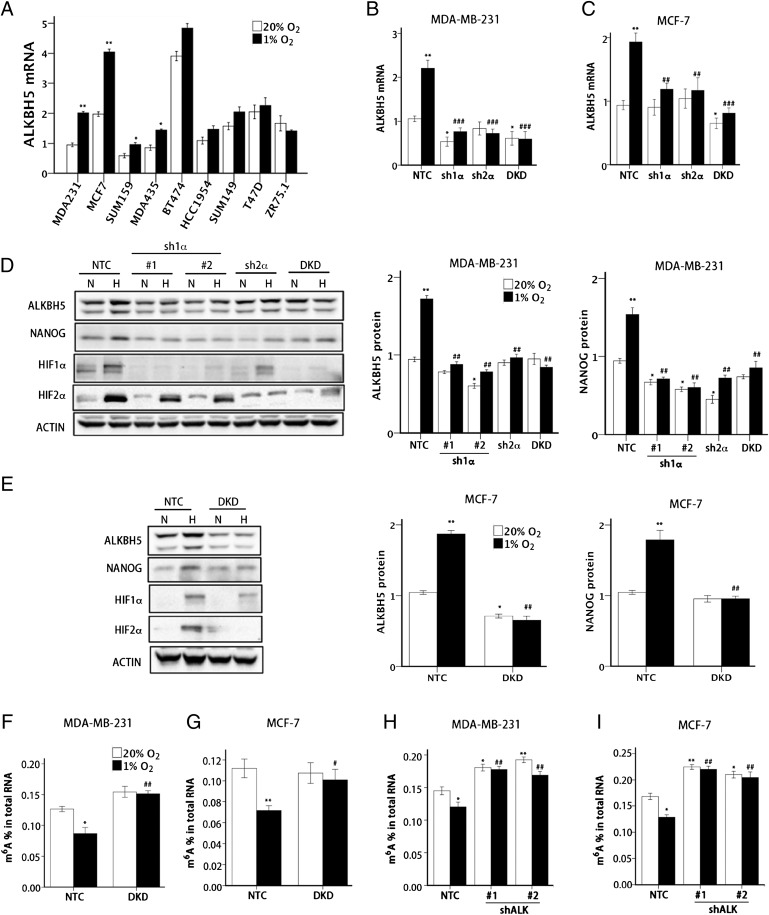
Breast cancers are divided into three groups based on expression of the estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). We first investigated whether ALKBH5 expression is induced by hypoxia in nine human breast cancer cells that are ER+ (ZR75.1), ER+PR+ (MCF-7 and T47D), ER+PR+HER2+ (BT-474), HER2+ (HCC-1954), or triple-negative (MDA-MB-231, MDA-MB-435, SUM-149, and SUM-159) (37). These nine breast cancer cell lines were exposed to 20% or 1% O2 for 24 h, and RNA was isolated for reverse transcription and quantitative real-time PCR (RT-qPCR). ALKBH5 mRNA levels were significantly increased under hypoxic conditions by greater than 1.6-fold in SUM-159 and MDA-MB-435 cells and greater than twofold in MDA-MB-231 and MCF-7 cells (Fig. 1A).
Fig. 1.
Hypoxia-induced and HIF-dependent ALKBH5 expression mediates a reduction in total RNA m6A levels. (A) Nine human breast cancer cell lines were exposed to 20% or 1% O2 for 24 h, and ALKBH5 mRNA levels were determined by RT-qPCR relative to 18S rRNA and normalized to the mean value for MDA-MB-231 cells (MDA231) at 20% O2 (mean ± SEM; n = 3; *P < 0.05 and **P < 0.01 vs. same cell line at 20% O2). (B and C) NTC, sh1α, sh2α, and DKD subclones of MDA-MB-231 (B) and MCF-7 (C) were exposed to 20% or 1% O2 for 24 h, and RT-qPCR was performed to determine ALKBH5 mRNA levels relative to 18S rRNA. The results were normalized to NTC at 20% O2 (mean ± SEM; n = 3; *P < 0.05 and **P < 0.01 vs. NTC at 20% O2; ##P < 0.01 and ###P < 0.001 vs. NTC at 1% O2). (D and E) MDA-MB-231 (D) and MCF-7 (E) subclones were exposed to 20% or 1% O2 for 48 h, whole cell lysates were prepared, and immunoblot assays were performed to analyze HIF-1α, HIF-2α, ALKBH5, and NANOG protein expression. Actin was also analyzed as a loading control. The intensity of ALKBH5 and NANOG bands were determined, and the data were normalized to NTC at 20% O2 (mean ± SEM; n = 3; *P < 0.05 and **P < 0.01 vs. NTC at 20% O2; ##P < 0.01 vs. NTC at 1% O2). (F and G) MDA-MB-231 (F) and MCF-7 (G) NTC and DKD subclones were exposed to 20% or 1% O2 for 24 h. Total RNA was extracted and m6A levels were determined as a percentage of all adenosine residues in RNA (mean ± SEM; n = 3; *P < 0.05 and **P < 0.01 vs. NTC at 20%; #P < 0.05 and ##P < 0.01 vs. NTC at 1% O2). (H and I) MDA-MB-231 (H) and MCF-7 (I) NTC and ALKBH5 knockdown (i.e., shALK) subclones were exposed to 20% or 1% O2 for 24 h. Total RNA was extracted, and m6A content was determined as a percentage of all adenosine residues (mean ± SEM; n = 3; *P < 0.05 and **P < 0.01 vs. NTC at 20% O2; ##P < 0.01 vs. NTC at 1% O2).
To determine whether hypoxia-induced ALKBH5 expression was dependent on HIF-1α, HIF-2α, or both, we analyzed MDA-MB-231 and MCF-7 subclones that were stably transfected with lentiviral vectors encoding shRNA to inhibit the expression of HIF-1α (sh1α), HIF-2α (sh2α), or both HIF-1α and HIF-2α [double knockdown (DKD)], as well as a subclone expressing a nontargeting control shRNA (NTC) (21, 38). In contrast to the NTC subclone, hypoxia-induced ALKBH5 mRNA expression was abrogated in sh1α, sh2α, and DKD subclones of MDA-MB-231 (Fig. 1B) and MCF-7 (Fig. 1C). Immunoblot assays revealed that ALKBH5 protein levels were increased by hypoxia in NTC subclones of MDA-MB-231 (Fig. 1D) and MCF-7 (Fig. 1E), but not in HIF-knockdown subclones. Thus, hypoxia induces ALKBH5 expression in a subset of ER+ and ER− breast cancer cell lines in an HIF-dependent manner.
Next, we investigated whether increased ALKBH5 expression in hypoxic cells leads to changes in levels of m6A RNA modification in total cellular RNA pools. We hypothesized that m6A levels in RNA represent the net effect of methylation mediated by the METTL3/METTL14 complex and demethylation mediated by ALKBH5 and FTO. Thus, increased ALKBH5 expression should lead to decreased m6A levels in RNA. Exposure of NTC subclones of MDA-MB-231 (Fig. 1F) and MCF-7 (Fig. 1G) to 1% O2 for 24 h led to a significant decrease (>30%) in the percentage of m6A residues among total adenosine residues in RNA. In contrast, there was no significant change in total m6A levels in RNA when the DKD subclones were exposed to hypoxia.
To demonstrate that decreased m6A modification of RNA in hypoxic cells was caused by increased ALKBH5 expression, we generated subclones of MDA-MB-231 and MCF-7 that were stably transfected with a lentiviral vector encoding either of two different shRNAs targeting ALKBH5 (shALK1#1 and shALK#2). Compared with the NTC subclone, knockdown of ALKBH5 in MDA-MB-231 (Fig. 1H) and MCF-7 (Fig. 1I) cells significantly increased m6A levels in total RNA isolated from cells incubated at 20% O2, and ALKBH5 knockdown blocked hypoxia-induced RNA demethylation. Taken together, the data in Fig. 1 indicate that HIF-dependent ALKBH5 expression mediates m6A demethylation in total cellular RNA pools of hypoxic breast cancer cells.
Hypoxia Induces HIF- and ALKBH5-Dependent NANOG Expression.
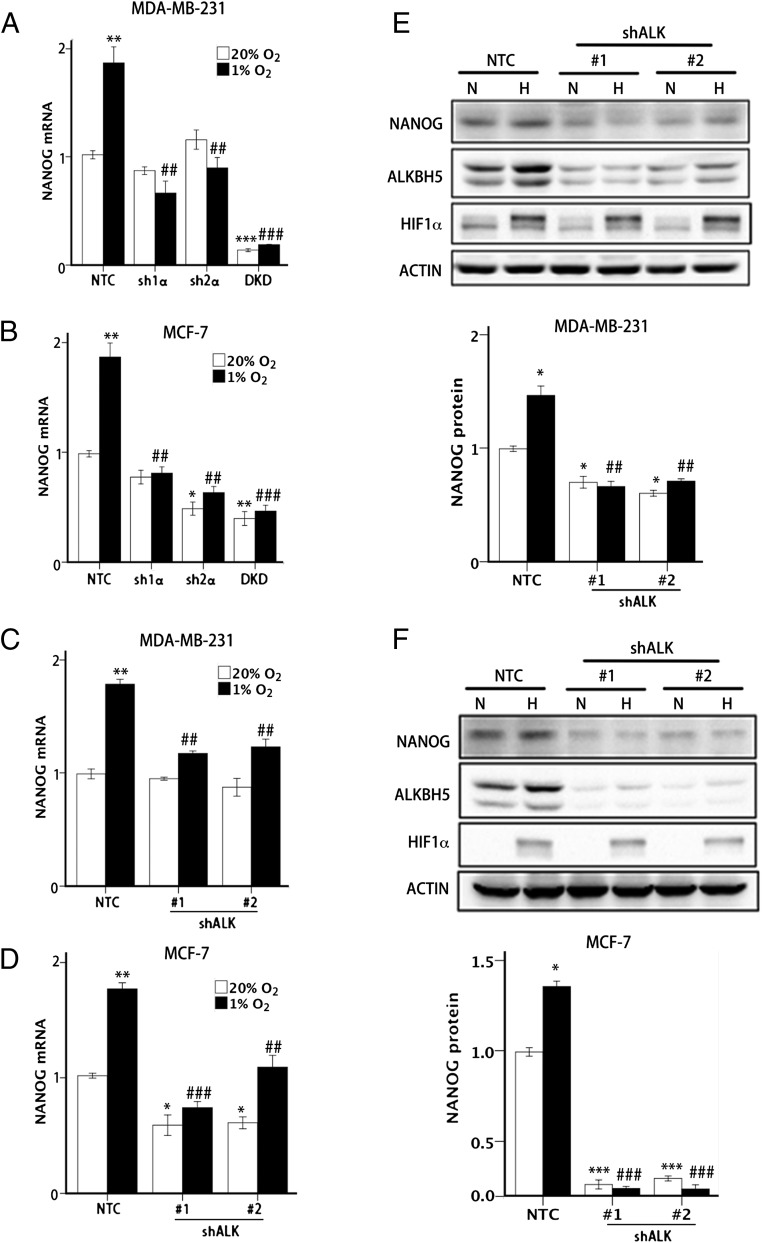
We next analyzed the effect of hypoxia on expression of the core pluripotency factor NANOG in human breast cancer cells. In NTC subclones of MDA-MB-231 (Fig. 2A) and MCF-7 (Fig. 2B), hypoxia significantly increased NANOG mRNA levels, whereas the hypoxic induction was abrogated in the sh1α, sh2α, and DKD subclones. Hypoxic induction of NANOG protein expression was also abrogated in the HIF knockdown subclones (Fig. 1 D and E).
Fig. 2.
HIF and ALKBH5 are required for hypoxia-induced NANOG expression. (A and B) MDA-MB-231 (A) and MCF-7 (B) NTC and HIF knockdown subclones were exposed to 20% or 1% O2 for 24 h, and NANOG mRNA levels were determined by RT-qPCR. The results were normalized to NTC at 20% O2 (mean ± SEM; n = 3; *P < 0.05, **P < 0.01, and ***P < 0.001 vs. NTC at 20% O2; ##P < 0.01 and ###P < 0.001 vs. NTC at 1% O2). (C and D) MDA-MB-231 (C) and MCF-7 (D) NTC and shALK knockdown subclones were exposed to 20% or 1% O2 for 24 h, and NANOG mRNA levels were determined by RT-qPCR. The results were normalized to NTC at 20% O2 (mean ± SEM; n = 3; *P < 0.05 and **P < 0.01 vs. NTC at 20%; ##P < 0.01 and ###P < 0.001 vs. NTC at 1% O2). (E and F) MDA-MB-231 (E) and MCF-7 (F) NTC and shALK subclones were exposed to 20% or 1% O2 for 48 h, and immunoblot assays were performed to analyze HIF-1α, ALKBH5, and NANOG protein expression. Actin was analyzed as a loading control. NANOG band intensity was quantified and normalized to NTC at 20% O2 (mean ± SEM; n = 3; *P < 0.05 and ***P < 0.001 vs. NTC at 20% O2; ##P < 0.01 and ###P < 0.001 vs. NTC at 1% O2).
ALKBH5 knockdown by either of two different shRNAs also significantly impaired the hypoxic induction of NANOG mRNA expression in MDA-MB-231 (Fig. 2C) and MCF-7 (Fig. 2D) cells. Finally, immunoblot assays demonstrated that ALKBH5 knockdown led to decreased NANOG protein expression in MDA-MB-231 (Fig. 2E) and MCF-7 (Fig. 2F) cells. Decreased levels of NANOG mRNA and protein were also observed in ALKBH5-knockdown subclones at 20% O2, indicating that there is significant ALKBH5 activity under nonhypoxic conditions, which is further increased by hypoxia.
ALKBH5 Enhances NANOG mRNA Stability by Catalyzing m6A Demethylation.
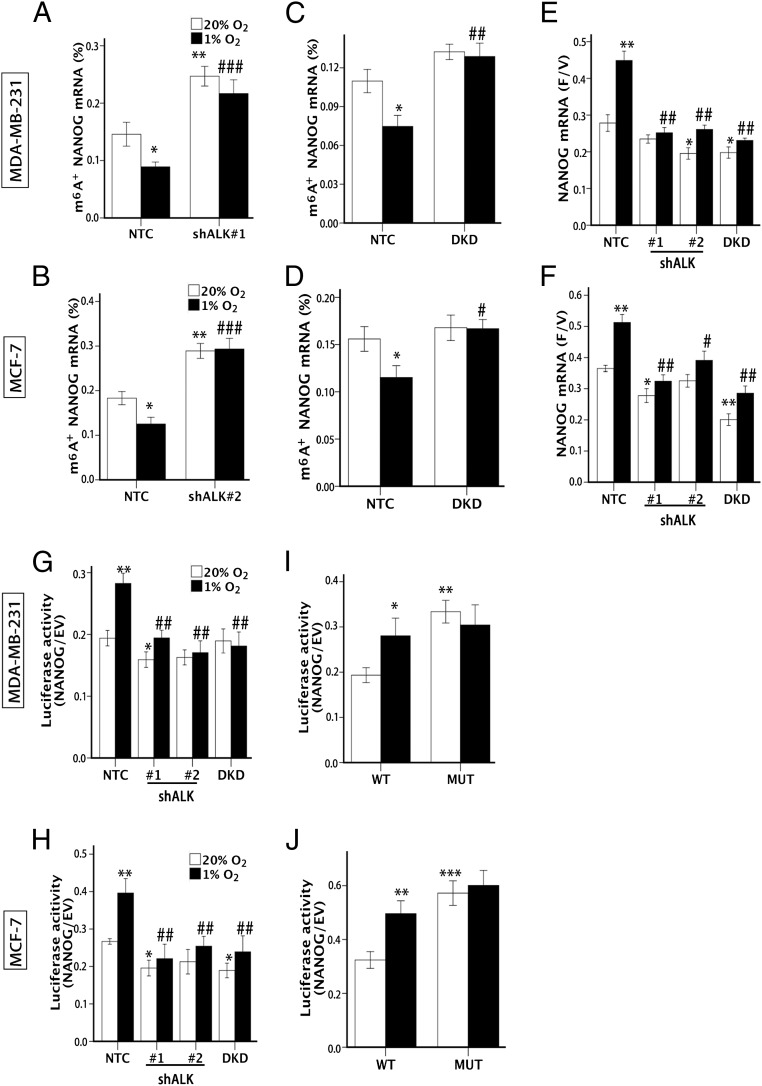
To investigate the mechanism by which ALKBH5 regulates NANOG expression, we first explored whether hypoxia-induced ALKBH5 targeted NANOG mRNA for m6A demethylation. Total cellular RNA was immunoprecipitated with an antibody that recognizes m6A, and the immunoprecipitated RNA was subjected to RT-qPCR to amplify the beginning of the NANOG 3′-UTR, which contains a match to the m6A consensus sequence, 5′-RRACU-3′. Hypoxic exposure led to decreased levels of m6A+ NANOG mRNA in NTC subclones of MDA-MB-231 (Fig. 3A) and MCF-7 (Fig. 3B) cells. In contrast, expression of either of two different shRNAs targeting ALKBH5 led to increased levels of m6A+ NANOG mRNA under normoxic and hypoxic conditions. HIF DKD also increased m6A+ NANOG mRNA levels, although to a lesser degree, and blocked the hypoxia-induced decrease in m6A+ NANOG mRNA levels that was observed in NTC subclones of MDA-MB-231 (Fig. 3C) and MCF-7 (Fig. 3D). These data indicate that, under hypoxic conditions, NANOG mRNA is subject to m6A demethylation that is HIF- and ALKBH5-dependent.
Fig. 3.
ALKBH5 demethylates and stabilizes NANOG mRNA. (A and B) MDA-MB-231 (A) and MCF-7 (B) NTC and shALK subclones were exposed to 20% or 1% O2 for 48 h. m6A immunoprecipitation and RT-qPCR were performed to determine the percentage of NANOG mRNA with methylation (m6A+; mean ± SEM; n = 3; *P < 0.05 and **P < 0.01 vs. NTC at 20% O2; ###P < 0.001 vs. NTC at 1% O2). (C and D) MDA-MB-231 (C) and MCF-7 (D) NTC and DKD subclones were exposed to 20% or 1% O2 for 48 h, and the percentage of m6A+ NANOG mRNA was determined (mean ± SEM; n = 3; *P < 0.05 vs. NTC at 20% O2; #P < 0.05 and ##P < 0.01 vs. NTC at 1% O2). (E and F) MDA-MB-231 (E) and MCF-7 (F) subclones were exposed to 20% or 1% O2 for 24 h and treated with vehicle or flavopiridol for 3 h (E) or 6 h (F), and NANOG mRNA levels were measured by RT-qPCR and the F/V ratio of NANOG mRNA was determined (mean ± SEM; n = 3; *P < 0.05 and **P < 0.01 vs. NTC at 20% O2; #P < 0.05 and ##P < 0.01 vs. NTC at 1% O2). (G and H) Luciferase assays were performed by transfecting MDA-MB-231 (G) and MCF-7 (H) subclones with a reporter plasmid containing luciferase coding sequences followed by the NANOG-3′UTR or EV containing luciferase only. The ratio of luciferase activity in cells transfected with NANOG-3′UTR relative to EV was determined (mean ± SEM; n = 3; *P < 0.05 and **P < 0.01, vs. NTC at 20% O2; ##P < 0.01 vs. NTC at 1% O2). (I and J) Luciferase assays were performed in MDA-MB-231 (I) or MCF-7 (J) cells transfected with WT or mutant (MUT) luciferase-NANOG-3′UTR reporter (mean ± SEM; n = 3; *P < 0.05, **P < 0.01, and ***P < 0.001 vs. NTC at 20% O2).
Hypoxia induced decreased m6A modification of NANOG mRNA and increased total NANOG mRNA levels, which is consistent with increased degradation of m6A+ mRNA (29, 33). To measure NANOG mRNA stability, flavopiridol was used to inhibit global mRNA transcription as previously described (29), and the ratio of NANOG mRNA in flavopiridol-treated cells relative to vehicle-treated cells (F/V ratio) was calculated. Exposure of NTC subclones of MDA-MB-231 (Fig. 3E) and MCF-7 (Fig. 3F) to hypoxia increased the NANOG mRNA F/V ratio, indicating decreased degradation of NANOG mRNA. However, this effect of hypoxia was lost in ALKBH5 and HIF knockdown subclones, indicating that hypoxia enhances NANOG mRNA stability through HIF-dependent ALKBH5 expression.
Sequence analysis revealed three matches to the 5′-RRACU-3′ (methylated adenosine residue is underscored) m6A consensus sequence within the NANOG 3′-UTR. Reporter plasmids, which contained or lacked the entire NANOG 3′-UTR immediately downstream of luciferase coding sequences, were transiently transfected into MDA-MB-231 (Fig. 3G) and MCF-7 (Fig. 3H) subclones, followed by incubation at 20% or 1% O2 for 24 h and measurement of luciferase activity. In NTC subclones exposed to 20% O2, luciferase activity was reduced four- to fivefold by insertion of the NANOG 3′-UTR into the reporter, but hypoxia significantly increased luciferase activity, whereas this effect was lost in the shALK and DKD subclones (Fig. 3 G and H).
Next, the adenine residue embedded within the consensus sequence located closest to the translation termination codon in the NANOG 3′-UTR was mutated (5′-AAACU-3′ to 5′-AAUCU-3′ in mRNA), and luciferase assays were performed comparing WT and mutant reporters. Mutation of the adenosine residue resulted in constitutively increased luciferase activity in parental MDA-MB-231 (Fig. 3I) and MCF-7 (Fig. 3J) cells, suggesting that the mutation prevented methylation and thereby increased the stability of the luciferase-NANOG 3′-UTR fusion mRNA. These results suggest that the mutated adenosine residue was a target for methylation, which was consistent with the identification of this site by others using m6A immunoprecipitation and RNA sequencing in ESCs (29). Taken together, the results presented in Fig. 3 indicate that hypoxia-induced and HIF-dependent ALKBH5 expression increases NANOG mRNA demethylation, stabilization, and accumulation.
ALKBH5 Deficiency Impairs Hypoxia-Induced BCSC Enrichment.
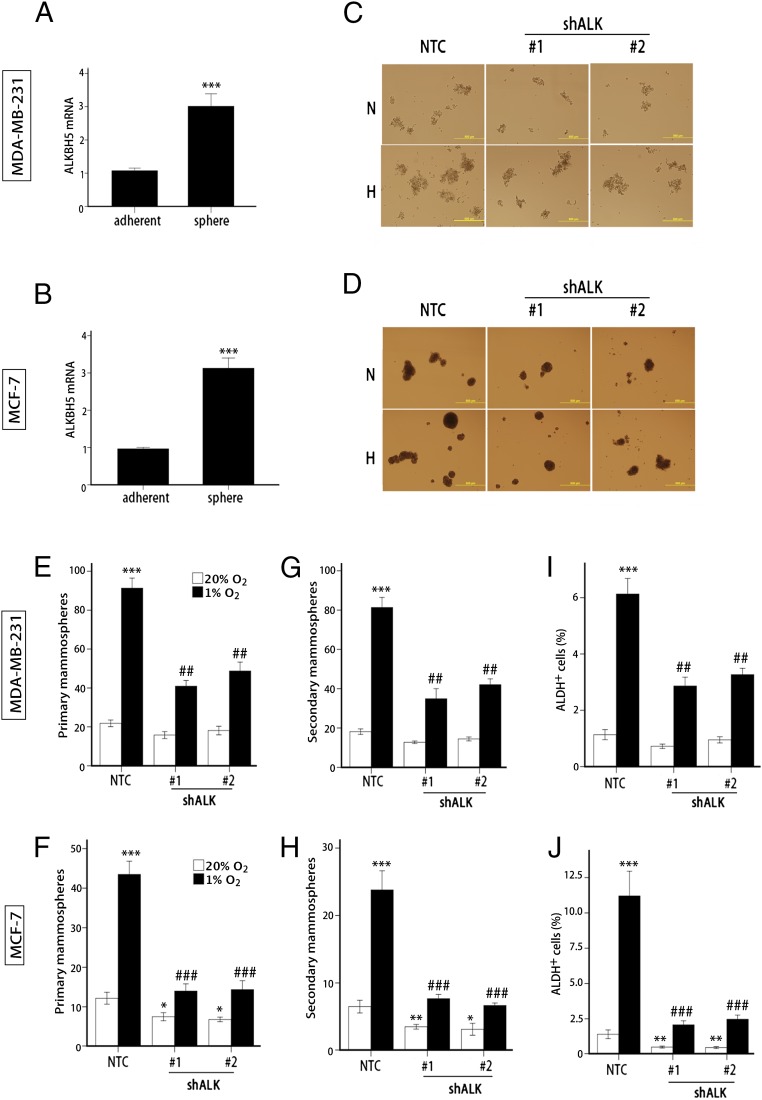
We next investigated whether ALKBH5 plays a role in regulation of the BCSC phenotype. Two properties that identify populations of breast cancer cells that are enriched in BCSCs are the ability to generate clusters of daughter cells when cultured on ultra-low adherence plates (mammosphere assay) (39) and increased aldehyde dehydrogenase (ALDH) 1 activity, which can be quantified by flow cytometry using a fluorogenic substrate (40). Analysis of MDA-MB-231 (Fig. 4A) and MCF-7 (Fig. 4B) cells grown as adherent monolayers or as nonadherent mammospheres revealed significantly increased ALKBH5 mRNA expression in mammosphere cultures, which are enriched for BCSCs.
Fig. 4.
ALKBH5 is required for hypoxia-induced BCSC enrichment. (A and B) ALKBH5 mRNA levels were determined by RT-qPCR in MDA-MB-231 (A) and MCF-7 (B) parental cells, which were cultured at 20% O2 as adherent monolayers or as mammospheres (mean ± SEM; n = 3; ***P < 0.001 vs. adherent cells). (C–H) Monolayer cultures of MDA-MB-231 (C, E, and G) and MCF-7 (D, F, and H) subclones were exposed to 20% or 1% O2 for 72 h and transferred to ultra-low attachment plates. The number of primary (E and F) and secondary (G and H) mammospheres per 1,000 cells initially seeded was determined (mean ± SEM; n = 3; *P < 0.05, **P < 0.01, and ***P < 0.001 vs. NTC at 20% O2; ##P < 0.01 and ###P < 0.001 vs. NTC at 1% O2). (I and J) MDA-MB-231 (I) and MCF-7 (J) subclones were exposed to 20% or 1% O2 for 72 h, and the percentage of cells expressing ALDH (ALDH+) was determined (mean ± SEM; n = 3; **P < 0.01 and ***P < 0.001 vs. NTC at 20% O2; ##P < 0.01 and ###P < 0.001 vs. NTC at 1% O2).
To determine the effect of ALKBH5 depletion on the specification/maintenance of BCSCs, we exposed NTC and shALK subclones to 20% or 1% O2 for 72 h, then transferred the cells to ultra-low adherence plates, incubated them at 20% O2 for 1 wk, and counted the number of primary mammospheres that had formed (Fig. 4 C–F). We then dissociated the primary mammospheres, replated the cells on ultra-low adherence plates, incubated them at 20% O2 for 1 wk, and counted the number of secondary mammospheres (Fig. 4 G and H). Hypoxic exposure of NTC subclones for 72 h dramatically increased primary and secondary mammosphere formation at 1 and 2 wk after the hypoxic exposure was terminated, as previously reported (21). In contrast, ALKBH5 knockdown significantly impaired hypoxia-induced mammosphere formation and, in MCF-7 cells, significantly decreased mammosphere formation even when cells were not exposed to hypoxia (Fig. 4 F and H). Analysis of BCSCs by the ALDH assay gave remarkably similar results: ALKBH5 knockdown significantly impaired the hypoxia-induced enrichment of ALDH+ cells in MDA-MB-231 (Fig. 4I) and MCF-7 (Fig. 4J) cells and decreased the percentage of ALDH+ cells even when MCF-7 cells were not exposed to hypoxia. Thus, mammosphere and ALDH assays indicate that ALKBH5 expression is necessary for hypoxia-induced BCSC enrichment.
Overexpression of ALKBH5 Is Sufficient to Enhance NANOG Expression and BCSC Enrichment.
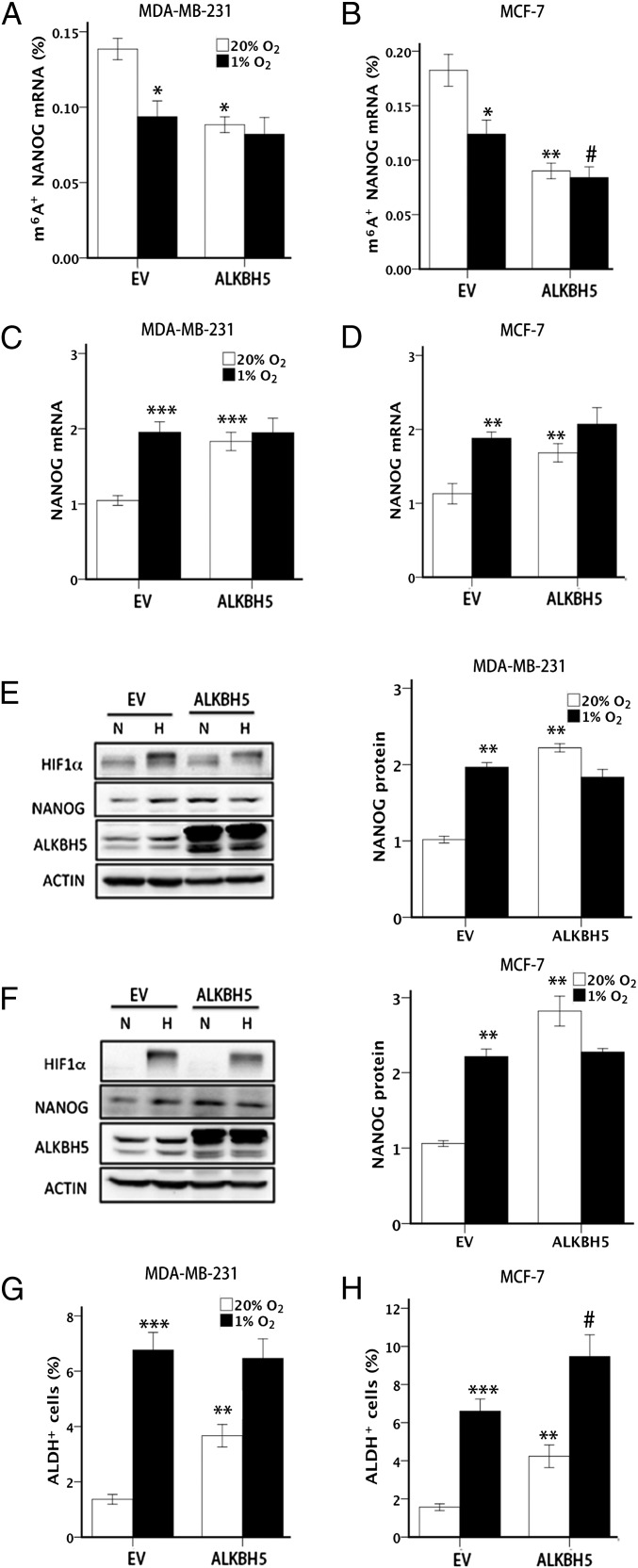
To determine whether ALKBH5 induction is sufficient to decrease the methylation of NANOG mRNA and thereby increase the BCSC population, we stably transfected MDA-MB-231 and MCF-7 cells with a lentiviral vector encoding human ALKBH5 or empty vector (EV), and exposed the cells to 20% or 1% O2. Under nonhypoxic conditions, ALKBH5 overexpression led to significantly decreased m6A modification of NANOG mRNA (Fig. 5 A and B), increased levels of NANOG mRNA (Fig. 5 C and D) and protein (Fig. 5 E and F), and increased percentage of ALDH+ cells (Fig. 5 G and H). In MCF-7 cells, ALKBH5 overexpression also potentiated the decrease in m6A+ NANOG mRNA levels (Fig. 5B) and increase in ALDH+ cells (Fig. 5H) that was observed in response to hypoxia. These data indicate that, under nonhypoxic conditions, overexpression of ALKBH5 is sufficient to demethylate NANOG mRNA, increase NANOG expression, and enhance BCSC enrichment, thereby phenocopying the effects of hypoxia.
Fig. 5.
ALKBH5 overexpression promotes BCSC enrichment. (A and B) MDA-MB-231 (A) and MCF-7 (B) subclones transfected with EV or vector encoding ALKBH5 were exposed to 20% or 1% O2 for 48 h, and m6A immunoprecipitation coupled with RT-qPCR was performed to determine the percentage of input NANOG mRNA with m6A methylation (m6A+; mean ± SEM; n = 3; *P < 0.05 and **P < 0.01 vs. EV at 20% O2; #P < 0.05 vs. EV at 1% O2). (C and D) MDA-MB-231 (C) and MCF-7 (D) subclones were exposed to 20% or 1% O2 for 24 h and NANOG mRNA levels were quantified and normalized to EV at 20% O2 (mean ± SEM; n = 3; **P < 0.01 and ***P < 0.001 vs. EV at 20% O2). (E and F) MDA-MB-231 (E) and MCF-7 (F) subclones were exposed to 20% or 1% O2 for 48 h, and immunoblot assays were performed. NANOG protein expression was quantified and normalized to EV at 20% O2 (mean ± SEM; n = 3; **P < 0.01 vs. EV at 20% O2). (G and H) MDA-MB-231 (G) and MCF-7 (H) subclones were exposed to 20% or 1% O2 for 72 h, and the percentage of ALDH+ cells was determined (mean ± SEM; n = 3; **P < 0.01 and ***P < 0.001 vs. EV at 20% O2; #P < 0.05 vs. EV at 1% O2).
Knockdown of ALKBH5 Impairs Tumor Formation and Decreases BCSCs in Vivo.
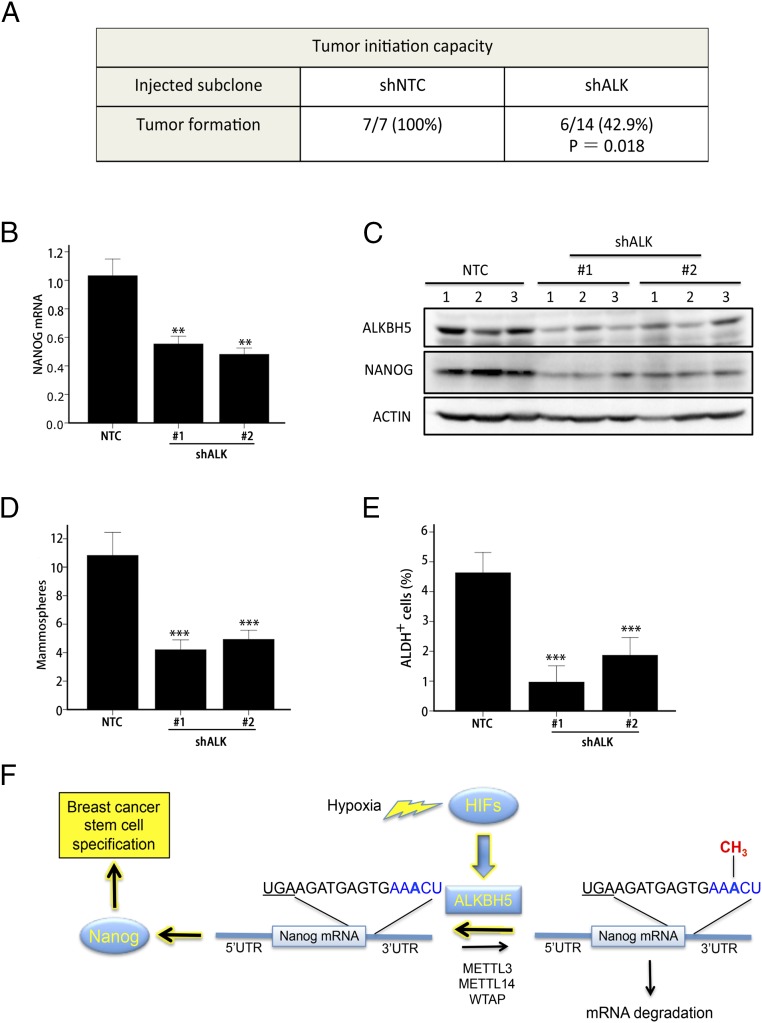
To analyze the effect of ALKBH5 deficiency on the tumor-initiating potential of breast cancer cells, we injected a limiting number of cells (1 × 103) from MDA-MB-231 subclones into the mammary fat pad of female nonobese diabetic/SCID/IL2Rγ-null (NSG) immunodeficient mice. Ten weeks after injection, 100% of the mice (n = 7) that were injected with NTC cells developed palpable tumors, compared with only 43% (n = 6 of 14) of the mice injected with ALKBH5-deficient cells (Fig. 6A). To further study the effect of ALKBH5 deficiency on the BCSC population in vivo, the tumors that formed after injection of NTC or shALK cells were harvested for analysis of NANOG expression and BCSCs. NANOG mRNA (Fig. 6B) and protein (Fig. 6C) levels were significantly decreased in tumors formed by ALKBH5-knockdown subclones compared with the NTC subclone. Both mammosphere (Fig. 6D) and ALDH (Fig. 6E) assays revealed a significant decrease in the BCSC population in tumors derived from injection of shALK compared with NTC cells. These results indicate that loss of ALKBH5 expression significantly impairs tumor formation and decreases the BCSC population in breast tumors.
Fig. 6.
Knockdown of ALKBH5 impairs tumor formation and decreases the BCSC population in orthotopic breast tumors. (A) A total of 1,000 MDA-MB-231 NTC or shALK subclone cells were injected into the mammary fat pad of female NSG immunodeficient mice. The number of mice that developed palpable tumors after 10 wk is shown. The NTC and shALK groups were compared by using Fisher exact test (n = 7 for NTC group; n = 14 for shALK group; n = 7 mice each were injected with shALK#1 or shALK#2 cells). (B) Tumors were harvested and a portion of the tissue was used for isolation of total RNA. RT-qPCR assays of NANOG mRNA were performed and normalized to the mean value for NTC (mean ± SEM; n = 3; **P < 0.01 vs. NTC). (C) Immunoblot assays were performed to analyze ALKBH5, NANOG, and Actin protein levels in lysates prepared from NTC and shALK tumor samples. (D) Single cell suspensions were prepared from NTC and shALK tumor samples and seeded on ultra-low attachment plates, and mammospheres were counted 7 d later (mean ± SEM; n = 3; ***P < 0.001 vs. NTC). (E) The percentage of ALDH+ cells in NTC and shALK tumor samples was determined (mean ± SEM; n = 3; ***P < 0.001 vs. NTC). (F) Hypoxia induces HIF-dependent expression of ALKBH5, which mediates demethylation of an adenosine residue in the 3′-UTR of NANOG mRNA, leading to increased NANOG mRNA stability and protein expression, and increased specification of BCSCs. The translation termination codon is underscored, m6A consensus sequence match is in blue, and methyl group of m6A residue is in red.
Discussion
The results presented here demonstrate that an essential physiological stimulus, i.e., hypoxia, regulates m6A modification of RNA in human cancer cells and that hypoxia induces the BCSC phenotype in part by increased demethylation of an mRNA encoding a core pluripotency factor. Although we focused on the regulation of NANOG mRNA stability, it is likely that the stability of other mRNAs is increased in breast cancer cells as a result of HIF-dependent and ALKBH5-mediated m6A demethylation. It is remarkable that exposure of MCF-7 or MDA-MB-231 breast cancer cells to 1% O2, which is comparable to the median pO2 in advanced breast cancers, decreased the m6A content of the total cellular RNA pool by as much as one third within only 24 h (Fig. 1 F–I). In addition, our studies have demonstrated a critical role for ALKBH5 in mediating NANOG expression and BCSC specification and/or maintenance within the hypoxic microenvironment of human breast cancer orthotopic tumors. ALKBH5 expression was induced by hypoxia in ER+ (MCF-7) and ER− (MDA-MB-231, SUM-159, and MDA-MB-435) cells, but not in other ER+ (T47D, ZR75.1, and BT-474) and ER− (HCC-1954 and SUM-149) cell lines. Further studies are required to determine whether FTO, the other known m6A demethylase, is induced by hypoxia in these latter cell lines.
We have previously shown that shRNA-mediated inhibition of NANOG expression in MDA-MB-231 cells results in a profound depletion of BCSCs (23). High NANOG mRNA (23) or protein (41) expression in primary human breast cancers is significantly associated with increased patient mortality. Other groups have implicated HIFs in the direct transcriptional activation of NANOG and other genes encoding pluripotency factors, both in human ESCs (42) and human cancer cells (17). We demonstrated that expression of ALKBH5 was necessary (Figs. 1–4) and sufficient (Fig. 5) to mediate a posttranscriptional induction of NANOG expression and BCSC enrichment. Taken together with prior studies, these results suggest that HIFs may coordinately regulate NANOG expression at the transcriptional and posttranscriptional levels in hypoxic breast cancer cells. Other HIF-regulated genes may also encode ALKBH5-regulated mRNAs, as dual regulation by HIFs and ALKBH5 provides a means to rapidly accumulate a target mRNA by coordinately increasing its production and decreasing its destruction.
A single multiprotein complex (consisting of METTL3, METTL14, and WTAP) mediates m6A modification of RNA, whereas two different proteins (FTO and ALKBH5) mediate demethylation, and it is not known whether FTO and ALKBH5 have distinct or redundant functions. Knockdown of ALKBH5 expression in hypoxic MCF-7 and MDA-MB-231 cells increased the m6A content of the total cellular RNA pool by as much as one third (Fig. 1 H and I), indicating that ALKBH5 plays a major role in regulating RNA methylation in human breast cancer cells.
Finally, it should be noted that, in addition to activating transcription of the RNA demethylase ALKBH5, HIFs also regulate the expression of two other O2-dependent dioxygenases that play key roles in breast cancer metastasis and stem cell maintenance (43, 44): JMJD2C, which mediates demethylation of histones (45), and TET1, which mediates hydroxymethylation of DNA (46). It is interesting that, in the absence of HIFs, the activity of these dioxygenases should decline under hypoxic conditions as a result of reduced substrate (i.e., O2) availability. However, HIF activity does not merely compensate for reduced O2 levels but significantly increases ALKBH5, JMJD2C, and TET1 activity in hypoxic breast cancer cells, leading to transcriptional and posttranscriptional changes in gene expression that promote the specification and/or maintenance of BCSCs. In addition to O2, these dioxygenases use α-ketoglutarate as a substrate. Further studies are warranted to investigate whether competitive antagonists of α-ketoglutarate that inhibit ALKBH5 (and possibly other dioxygenases) might be useful as anticancer agents that target BCSCs.
Materials and Methods
Cell Culture.
MCF-7, MDA-MB-231, MDA-MB-435, and T47D cells were cultured in DMEM. BT-474, HCC-1954, and ZR75.1 cells were cultured in RPMI-1640 medium. SUM-149 and SUM-159 cells were cultured in DMEM/F12 (50:50) media supplemented with hydrocortisone and insulin. All culture media were supplemented with 10% (vol/vol) FBS and 1% (vol/vol) penicillin/streptomycin. Cells were maintained in a 5% CO2 and 95% air incubator [20% (vol/vol) O2]. For hypoxia exposure, cells were placed in a modular incubator chamber (Billups-Rothenberg), which was flushed for 2 min at 2 psi with a gas mixture containing 1% O2, 5% CO2, and 94% N2.
RT-qPCR.
Total RNA was extracted using TRIzol (Invitrogen) according to the manufacturer’s instructions. cDNA synthesis was performed using the High Capacity RNA-to-cDNA Kit (Applied Biosystems). qPCR was performed by using the following primers: ALKBH5, 5′-CGG CGA AGG CTA CAC TTA CG-3′ and 5′-CCA CCA GCT TTT GGA TCA CCA-3′; NANOG, 5′-TTT GTG GGC CTG AAG AAA ACT-3′ and 5′-AGG GCT GTC CTG AAT AAG CAG-3′; and 18S rRNA, 5′-CGG CGA CGA CCC ATT CGA AC-3′ and 5′-GAA TCG AAC CCT GAT TCC CCG TC-3′. For each sample, the ratio of NANOG mRNA:18S rRNA was determined.
Immunoblot Assay.
Whole cell lysates were prepared in modified RIPA lysis buffer (50 mM Tris⋅HCl, pH 7.5, 1 mM β-mercaptoethanol, 150 mM NaCl, 1 mM Na3VO4, 1 mM NaF, 1 mM EDTA, 0.25% sodium deoxycholate and 1% Igepal CA-630). Blots were probed with HIF-1α (BD Biosciences), HIF-2α (Novus Biologicals), ALKBH5 (Novus Biologicals), NANOG (Novus Biologicals), and Actin (Santa Cruz) antibodies. HRP-conjugated anti-rabbit and anti-mouse secondary antibodies were used and the chemiluminescent signal was detected by using ECL Plus (GE Healthcare). Densitometric analysis was performed by using ImageJ (NIH).
Lentiviral Vectors and Transduction.
Lentiviral vectors encoding shRNA targeting HIF-1α and HIF-2α were described previously (38). pLKO.1-puro lentiviral vectors encoding shRNA targeting ALKBH5 mRNA (clone ID, NM_017758.2–1625s1c1 and NM_017758.2–1176s1c1) were purchased from Sigma-Aldrich. pLenti-GIII-CMV lentiviral vector containing cDNA encoding human ALKBH5 was purchased from Applied Biological Materials (cat. no. LV073201). All lentiviral vectors were transfected into 293T cells for packaging. Viral supernatant was collected after 48 h and used for transfection as described before (38). Puromycin (0.5 μg/mL) was added to the medium of cells transduced with lentivirus for selection.
Measurement of Total m6A and m6A+ NANOG mRNA Levels.
Total m6A content was measured in 200-ng aliquots of total RNA extracted from MDA-MB-231 and MCF-7 subclones using an m6A RNA methylation quantification kit (cat. no. P-9005; Epigentek) according to the manufacturer’s instructions. To measure m6A+ NANOG mRNA levels, m6A immunoprecipitation was performed as described before (47). A 1-µg aliquot of m6A antibody was conjugated to protein A-agarose slurry (Millipore) overnight at 4 °C. A 100-μg aliquot of total RNA was incubated with the antibody in immunoprecipitation buffer (50 mM Tris⋅HCl, 750 mM NaCl, and 0.5% Igepal CA-630) supplemented with RNase inhibitor for 3 h at 4 °C, RNA was eluted from the beads by incubation in 300 μL of elution buffer (5 mM Tris⋅HCl, 1 mM EDTA, and 0.05% SDS) with 4.2 μL of proteinase K for 1.5 h at 50 °C, and m6A+ RNA was purified by phenol/chloroform extraction and analyzed by RT-qPCR. The nucleotide sequences of the primers used for detection for m6A+ NANOG mRNA were as follows: 5′-ATG CAA CCT GAA GAC GTG TG-3′ and 5′-GAG ATT GAC TGG ATG GGC AT-3′.
Measurement of NANOG mRNA Stability.
Cells were exposed to 20% or 1% O2 for 24 h and then treated with vehicle or flavopiridol (Sigma-Aldrich) at a concentration of 0.8 μΜ for 3 h (MDA-MB-231) or 3.2 μΜ for 6 h (MCF-7), followed by RNA extraction and RT-qPCR as described earlier.
Luciferase Assay.
The luciferase coding sequence NANOG 3′-UTR reporter (cat. no. S807466) and luciferase coding sequence-only reporter (cat. no. S890005) were purchased from SwitchGear Genomics. Cells were seeded in a 96-well plate and transfected with luciferase-NANOG 3′-UTR reporter or luciferase-only reporter, and exposed to 20% or 1% O2 for 24 h. Luciferase activity was measured using a commercial kit (SwitchGear Genomics) and is presented as the activity ratio of luciferase-NANOG 3′-UTR to luciferase only. Adenine-to-thymine mutation of the luciferase-NANOG 3′-UTR reporter plasmid was performed by using the Q5 Site-Directed Mutagenesis Kit (New England Biolabs) according to the manufacturer’s instructions.
Mammosphere Assay.
Cultured cells were trypsinized, whereas tumor tissue was minced and digested with 1 mg/mL of type 1 collagenase (Sigma) at 37 °C for 30 min to prepare single-cell suspensions, which were seeded in six-well ultra-low attachment plates (Corning) at a density of 5,000 cells per milliliter in Complete MammoCult Medium (Stem Cell Technologies). After 7 d, the cells were photographed under an Olympus TH4-100 microscope, and primary mammospheres (diameter ≥70 μm) were counted. For secondary mammosphere formation, primary mammospheres were trypsinized, plated at a density of 5,000 cells per milliliter, incubated for 7 d, and counted.
ALDH Assay.
Cells were exposed to 20% or 1% O2 for 72 h and harvested for Aldefluor assay (Stem Cell Technologies). Single-cell suspensions (5 × 105 cells in 1 mL of assay buffer) were prepared as described earlier and incubated with BODIPY-aminoacetaldehyde (1 μM) for 45 min, followed by flow cytometry analysis.
Orthotopic Transplantation.
Protocols were approved by the Johns Hopkins University Animal Care and Use Committee and were in accordance with the NIH Guide for the Care and Use of Laboratory Animals. A total of 1,000 breast cancer cells were injected into the mammary fat pad of 6–8-wk-old female NSG mice in a 1:1 suspension of Matrigel (BD Biosciences) in PBS solution. At 10 wk after injection, mice were examined for the presence of tumors, which were harvested for analysis.
Statistical Analysis.
Data are presented as mean ± SEM. Differences between two groups or multiple groups were analyzed by Student t test and ANOVA, respectively. Fisher exact test was used for analyzing tumor initiation capacity. P values <0.05 were considered significant.
Acknowledgments
We thank Karen Padgett (Novus Biologicals) for providing antibodies against ALKBH5, HIF-2α, and NANOG. This work was supported in part by American Cancer Society Grant 122437-RP-12-090-01-COUN, the Cindy Rosencrans Fund for Triple Negative Breast Cancer, and the China Scholarship Council (C.Z.). G.L.S. is an American Cancer Society Research Professor and the C. Michael Armstrong Professor at the Johns Hopkins University School of Medicine.
Footnotes
The authors declare no conflict of interest.
References
- 1.DeSantis C, Siegel R, Bandi P, Jemal A. Breast cancer statistics, 2011. CA Cancer J Clin. 2011;61(6):409–418. doi: 10.3322/caac.20134. [DOI] [PubMed] [Google Scholar]
- 2.DeSantis CE, et al. International variation in female breast cancer incidence and mortality rates. Cancer Epidemiol Biomarkers Prev. 2015;24(10):1495–1506. doi: 10.1158/1055-9965.EPI-15-0535. [DOI] [PubMed] [Google Scholar]
- 3.O’Shaughnessy J. Extending survival with chemotherapy in metastatic breast cancer. Oncologist. 2005;10(suppl 3):20–29. doi: 10.1634/theoncologist.10-90003-20. [DOI] [PubMed] [Google Scholar]
- 4.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charafe-Jauffret E, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69(4):1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oskarsson T, Batlle E, Massagué J. Metastatic stem cells: Sources, niches, and vital pathways. Cell Stem Cell. 2014;14(3):306–321. doi: 10.1016/j.stem.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ben-Porath I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40(5):499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu T, et al. Octamer 4 small interfering RNA results in cancer stem cell-like cell apoptosis. Cancer Res. 2008;68(16):6533–6540. doi: 10.1158/0008-5472.CAN-07-6642. [DOI] [PubMed] [Google Scholar]
- 10.Yu F, et al. Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene. 2011;30(18):2161–2172. doi: 10.1038/onc.2010.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leis O, et al. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene. 2012;31(11):1354–1365. doi: 10.1038/onc.2011.338. [DOI] [PubMed] [Google Scholar]
- 12.IV Santaliz-Ruiz LE, Xie X, Old M, Teknos TN, Pan Q. Emerging role of nanog in tumorigenesis and cancer stem cells. Int J Cancer. 2014;135(12):2741–2748. doi: 10.1002/ijc.28690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaupel P, Höckel M, Mayer A. Detection and characterization of tumor hypoxia using pO2 histography. Antioxid Redox Signal. 2007;9(8):1221–1235. doi: 10.1089/ars.2007.1628. [DOI] [PubMed] [Google Scholar]
- 14.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148(3):399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Semenza GL. The hypoxic tumor microenvironment: A driving force for breast cancer progression. Biochim Biophys Acta. 2016;1863(3):382–391. doi: 10.1016/j.bbamcr.2015.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Regan Anderson TM, et al. Breast tumor kinase (Brk/PTK6) is a mediator of hypoxia-associated breast cancer progression. Cancer Res. 2013;73(18):5810–5820. doi: 10.1158/0008-5472.CAN-13-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathieu J, et al. HIF induces human embryonic stem cell markers in cancer cells. Cancer Res. 2011;71(13):4640–4652. doi: 10.1158/0008-5472.CAN-10-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conley SJ, et al. Antiangiogenic agents increase breast cancer stem cells via the generation of tumor hypoxia. Proc Natl Acad Sci USA. 2012;109(8):2784–2789. doi: 10.1073/pnas.1018866109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iriondo O, et al. Distinct breast cancer stem/progenitor cell populations require either HIF1α or loss of PHD3 to expand under hypoxic conditions. Oncotarget. 2015;6(31):31721–31739. doi: 10.18632/oncotarget.5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, et al. HIF-1 regulates CD47 expression in breast cancer cells to promote evasion of phagocytosis and maintenance of cancer stem cells. Proc Natl Acad Sci USA. 2015;112(45):E6215–E6223. doi: 10.1073/pnas.1520032112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiang L, et al. Hypoxia-inducible factor 1 mediates TAZ expression and nuclear localization to induce the breast cancer stem cell phenotype. Oncotarget. 2014;5(24):12509–12527. doi: 10.18632/oncotarget.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samanta D, Gilkes DM, Chaturvedi P, Xiang L, Semenza GL. Hypoxia-inducible factors are required for chemotherapy resistance of breast cancer stem cells. Proc Natl Acad Sci USA. 2014;111(50):E5429–E5438. doi: 10.1073/pnas.1421438111. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Lu H, et al. Chemotherapy triggers HIF-1-dependent glutathione synthesis and copper chelation that induces the breast cancer stem cell phenotype. Proc Natl Acad Sci USA. 2015;112(33):E4600–E4609. doi: 10.1073/pnas.1513433112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoo YG, Christensen J, Gu J, Huang LE. HIF-1α mediates tumor hypoxia to confer a perpetual mesenchymal phenotype for malignant progression. Sci Signal. 2011;4(178):pt4. doi: 10.1126/scisignal.2002072. [DOI] [PubMed] [Google Scholar]
- 25.Scheel C, et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145(6):926–940. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwab LP, et al. Hypoxia-inducible factor 1α promotes primary tumor growth and tumor-initiating cell activity in breast cancer. Breast Cancer Res. 2012;14(1):R6. doi: 10.1186/bcr3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m⁶A RNA methylation. Nat Rev Genet. 2014;15(5):293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 28.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 29.Batista PJ, et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15(6):707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee M, Kim B, Kim VN. Emerging roles of RNA modification: m6A and U-tail. Cell. 2014;158(5):980–987. doi: 10.1016/j.cell.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 31.Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol. 2014;15(5):313–326. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519(7544):482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geula S, et al. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347(6225):1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 35.Zheng G, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thalhammer A, et al. Human AlkB homologue 5 is a nuclear 2-oxoglutarate dependent oxygenase and a direct target of hypoxia-inducible factor 1α (HIF-1α) PLoS One. 2011;6(1):e16210. doi: 10.1371/journal.pone.0016210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H, et al. HIF-1-dependent expression of angiopoietin-like 4 and L1CAM mediates vascular metastasis of hypoxic breast cancer cells to the lungs. Oncogene. 2012;31(14):1757–1770. doi: 10.1038/onc.2011.365. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Dontu G, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17(10):1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ginestier C, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagata T, et al. Prognostic significance of NANOG and KLF4 for breast cancer. Breast Cancer. 2014;21(1):96–101. doi: 10.1007/s12282-012-0357-y. [DOI] [PubMed] [Google Scholar]
- 42.Forristal CE, Wright KL, Hanley NA, Oreffo ROC, Houghton FD. Hypoxia inducible factors regulate pluripotency and proliferation in human embryonic stem cells cultured at reduced oxygen tensions. Reproduction. 2010;139(1):85–97. doi: 10.1530/REP-09-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo W, Chang R, Zhong J, Pandey A, Semenza GL. Histone demethylase JMJD2C is a coactivator for hypoxia-inducible factor 1 that is required for breast cancer progression. Proc Natl Acad Sci USA. 2012;109(49):E3367–E3376. doi: 10.1073/pnas.1217394109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu MZ, et al. Hypoxia drives breast tumor malignancy through a TET-TNFα-p38-MAPK signaling axis. Cancer Res. 2015;75(18):3912–3924. doi: 10.1158/0008-5472.CAN-14-3208. [DOI] [PubMed] [Google Scholar]
- 45.Pollard PJ, et al. Regulation of Jumonji-domain-containing histone demethylases by hypoxia-inducible factor (HIF)-1α. Biochem J. 2008;416(3):387–394. doi: 10.1042/BJ20081238. [DOI] [PubMed] [Google Scholar]
- 46.Mariani CJ, et al. TET1-mediated hydroxymethylation facilitates hypoxic gene induction in neuroblastoma. Cell Reports. 2014;7(5):1343–1352. doi: 10.1016/j.celrep.2014.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, Amariglio N, Rechavi G. Transcriptome-wide mapping of N6-methyladenosine by m6A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc. 2013;8(1):176–189. doi: 10.1038/nprot.2012.148. [DOI] [PubMed] [Google Scholar]