Significance
The discovery of natural anion channelrhodopsins (ACRs) opens possibilities for rapid and efficient inhibition of neuronal firing by hyperpolarization, but molecular mechanisms of these proteins’ function remain elusive. We studied spectral properties and photochemical conversions of purified wild-type and mutant ACRs to probe the coupling between chromophore photoexcitation and alteration of protein conformation leading to electrical conductance in the membrane. Our results show that not only the selectivity filter, but also the structural basis of channel gating in the cryptophyte ACRs differs from that in cation channelrhodopsins (CCRs) from chlorophyte algae used to depolarize the membrane. The knowledge of structure–function relationships in ACRs will benefit molecular engineering to improve their performance as tools for optogenetic research and therapies.
Keywords: photochemical conversions, Schiff base, channel gating, channelrhodopsins, optogenetics
Abstract
A recently discovered family of natural anion channelrhodopsins (ACRs) have the highest conductance among channelrhodopsins and exhibit exclusive anion selectivity, which make them efficient inhibitory tools for optogenetics. We report analysis of flash-induced absorption changes in purified wild-type and mutant ACRs, and of photocurrents they generate in HEK293 cells. Contrary to cation channelrhodopsins (CCRs), the ion conducting state of ACRs develops in an L-like intermediate that precedes the deprotonation of the retinylidene Schiff base (i.e., formation of an M intermediate). Channel closing involves two mechanisms leading to depletion of the conducting L-like state: (i) Fast closing is caused by a reversible L⇔M conversion. Glu-68 in Guillardia theta ACR1 plays an important role in this transition, likely serving as a counterion and proton acceptor at least at high and neutral pH. Incomplete suppression of M formation in the GtACR1_E68Q mutant indicates the existence of an alternative proton acceptor. (ii) Slow closing of the channel parallels irreversible depletion of the M-like and, hence, L-like state. Mutation of Cys-102 that strongly affected slow channel closing slowed the photocycle to the same extent. The L and M intermediates were in equilibrium in C102A as in the WT. In the position of Glu-123 in channelrhodopsin-2, ACRs contain a noncarboxylate residue, the mutation of which to Glu produced early Schiff base proton transfer and strongly inhibited channel activity. The data reveal fundamental differences between natural ACR and CCR conductance mechanisms and their underlying photochemistry, further confirming that these proteins form distinct families of rhodopsin channels.
The genomes of cryptophyte algae harbor nucleotide sequences that encode anion-conducting channelrhodopsins (ACRs) (1, 2). Although these proteins show distant sequence homology to cation-conducting channelrhodopsins (CCRs) from green (chlorophyte) algae, they completely lack permeability for protons and metal cations and, thus, represent a distinct structural and functional class among microbial rhodopsins. Using ACRs permits optogenetic inhibition of neuronal firing at much lower light intensities than other currently used silencers (1).
Analysis of photocurrents generated by ACR1 from Guillardia theta (GtACR1) in human embryonic kidney (HEK) cells under single-turnover conditions revealed that GtACR1 gating comprises two separate mechanisms with opposite dependencies on the membrane voltage and pH, and involving different amino acid residues (3). The first mechanism, characterized by fast closing of the channel, is regulated by Glu-68, a homolog of Glu-90 in channelrhodopsin-2 from the green alga Chlamydomonas reinhardtii (CrChR2). Replacement of Glu-90 with Arg in CrChR2 made the channel permeable for Cl− (4), whereas the same substitution of Glu-68 in GtACR1 did not influence its selectivity for anions, but inverted its gating, rendering the channel open in the dark and closed under illumination (3).
The second gating mechanism identified in GtACR1 is characterized by slow closing of the channel. It is not substantially affected by the E68Q mutation, but is controlled by Cys-102 (a homolog of Cys-128 in CrChR2), substitution of which with Ala dramatically extended the slow photocurrent decay (3). In CrChR2, mutation of either Cys-128 or Asp-156 that has been proposed to form an interhelical hydrogen bond (DC gate) (5) caused similar extension of the channel open state (6). In contrast, in GtACR1, mutation of Ser-130 that corresponds to Asp-156 in CrChR2 did not significantly alter the current kinetics unlike mutation of Cys-102 (3). Thus, both the selectivity filter and gating mechanisms of natural ACRs appear to be different from those of CCRs. In this manuscript, we use the term “ACRs” to denote only natural anion-conducting rhodopsins. There are significant differences in primary structure and mechanism between natural ACRs and CCRs and their chloride-conducting mutants.
To gain insight into coupling of ACR photochemical reactions and ionic conductance, we performed absorption spectroscopy and flash-photolysis experiments on purified pigments obtained from expression in the methylotrophic yeast Pichia pastoris, using wild-type and variants in which amino acid residues that have been shown to be important for channel gating (Glu-68, Asp-234, Cys-102, and Ser-97) were mutated.
The photochemical cycle of most microbial rhodopsins comprises a sequence of several spectral intermediates (K, L, M, N, O) that appear following all-trans to 13-cis photoisomerization of the retinal chromophore in the time domain between picoseconds and seconds, best studied in the haloarchaeal proton pump bacteriorhodopsin (BR) (for review, see ref. 7). The K state contains 13-cis retinal in a strained configuration, which is relaxed in the L state. Formation of the blue-shifted M intermediate reflects deprotonation of the retinylidene Schiff base. The M state is not formed in the photocycle of haloarchaeal Cl− pumps (halorhodopsins) (for review, see ref. 8) with which ACRs share a noncarboxylate residue in the position of Asp-85 that acts as a proton acceptor in BR. Reprotonation of the Schiff base gives rise to the N state in which the retinal is still 13-cis, and its reisomerization to a distorted all-trans configuration produces the O state, which then decays to the initial (unphotolyzed) state. In CCRs, the channel open state has been associated with the late M state and subsequent red-shifted (N/O) intermediates of the photocycle (for review, see refs. 9 and 10).
Here, our measurements of laser flash-induced absorbance changes identified at least four intermediates in the GtACR1 photocycle (K-like, L-like, M-like, and N/O-like). The rise and decay of the M intermediate in GtACR1 and GtACR2 are unprecedentedly slow compared with all previously studied microbial rhodopsins. Comparison of flash photolysis data with photocurrents generated under single-turnover conditions revealed that the GtACR1 channel opens ∼50-fold faster than the Schiff base deprotonates. Hence, in contrast to CCRs in which Schiff base deprotonation (M formation) precedes channel opening, we find that the L intermediate represents the conducting state in ACRs. The formation of M rather corresponds to fast channel closing, and its decay to slow channel closing. Furthermore, imposing early M formation on GtACR1 by engineering a proton acceptor adjacent to the protonated Schiff base nearly completely inhibits channel opening, indicating that the presence of a noncarboxylate residue in the position of the canonical proton acceptor (Asp-85 in BR) is critical for ACR function. Deionization of purified pigment or substitution of SO42− for Cl− in the buffer changed neither the position of the absorption maximum nor the photocycle, which argues against Cl− being a Schiff base counterion in GtACR1.
Results
Wild-Type GtACR1.
Absorption spectroscopy.
The absorption spectra of dark-adapted and light-adapted detergent-extracted wild-type GtACR1 at pH 7.4 are superimposable with the main peak at 515 nm (Fig. 1A). No significant light/dark spectral adaptation was found, indicating that both light and dark forms of the pigment contain retinal in the same configuration.
Fig. 1.
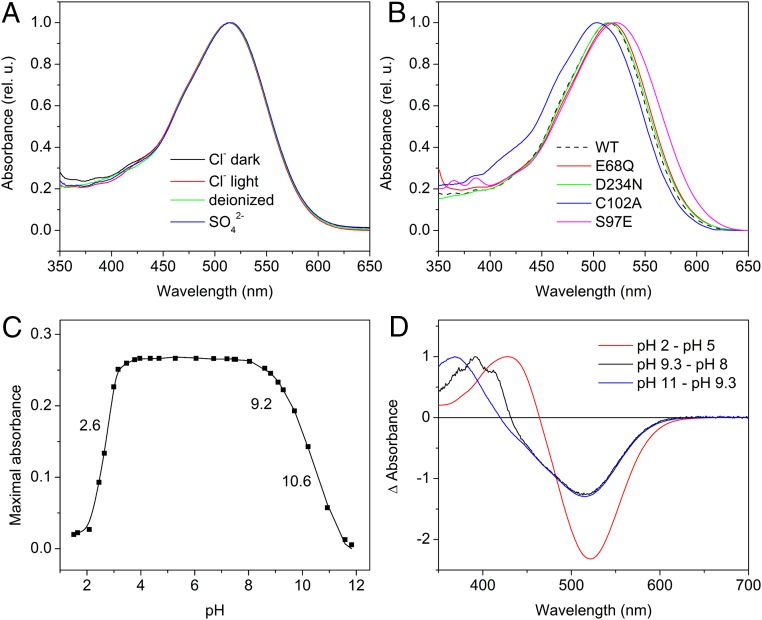
(A) Absorption spectra of GtACR1 detergent purified in buffer (pH 7.4) containing NaCl (black and red lines) or Na2SO4 (blue line), and a deionized sample (green line). The black and red lines show the spectra of the dark- and light-adapted samples, respectively. (B) Absorption spectra of the purified GtACR1_E68Q, D234N, C102A, and S97E mutants in NaCl buffer at pH 7.4. The WT spectrum from A is shown as a black dashed line for comparison. (C) The pH titration of the purified wild-type GtACR1 maximal absorbance of the ∼515-nm band. (D) The difference spectra obtained by subtraction of the absolute spectra as indicated.
A characteristic feature of all known ACRs is the presence of a noncarboxylate residue in the position of the proton acceptor Asp-85 in BR. It is also typical of halorhodopsins in which Cl− serves as a counterion to the protonated Schiff base (11). To test the possibility of Cl− acting as a Schiff base counterion in GtACR1, we made measurements in a deionized sample and a sample in which Cl− was replaced with SO42− in the buffer. In contrast to halorhodopsin (12), no spectral changes (Fig. 1A, green and blue lines, respectively) or substantial alteration of the photocycle kinetics (Fig. S1) were observed in GtACR1 upon such treatments, which argues against Cl− acting as a counterion in the latter protein.
Fig. S1.
Laser flash-induced absorption difference signals monitored at 580 nm (A) and 400 nm (B) in GtACR1 samples purified in Cl−-free (solid lines) or Cl−-containing (dashed lines) buffer. For comparison of the kinetics, the traces measured in the Cl−-containing buffer were scaled to yield the same peak amplitude as those in the Cl−-free buffer.
Detergent-extracted GtACR1 was stable within the pH range from 3.5 to 9 (Fig. 1C). At pH ≤ 3, irreversible absorption changes gave rise to a product with maximal absorption at ∼430 nm (Fig. 1D, red line), which is typical of partially unfolded microbial rhodopsins (13). Absorption changes in the alkaline region revealed two transitions. A transition with a pK ∼9 was reversible and resulted in the appearance of a characteristic structured spectrum of an M-like product that forms upon deprotonation of the Schiff base (Fig. 1D, black line). A further increase in pH led to irreversible conversion of the pigment to a product with maximal absorption at 370 nm, which likely indicated denaturing of the pigment (Fig. 1D, blue line).
Photochemical conversions.
The first laser-induced chemical conversion we observed was depletion of the unphotolyzed state and the appearance of a red-shifted K-like photoproduct with absorption maximum at 580 nm occurring within our time resolution of 20 ns (Fig. 2A, black line). Spectra of time-resolved transitions were obtained by global fit analysis and are shown by colored lines in the same graph. The time window for this fit does not include the fast component of the K-to-L transition, and, hence, its apparent amplitude reflects only a fraction of this transition. The K state decayed into a blue shifted L-like intermediate with a time constant (τ) of ∼1 µs, accompanied by a slower transition of a smaller amplitude with a τ value of ∼10–20 µs (Fig. 2B). The amplitudes of absorption changes at 580 nm and 450 nm were similar, which means that at least half of the pigment accumulates in the L state. The difference spectrum corresponding to the late K-to-L transition is shown in Fig. 2A, red line. The K intermediate did not convert fully, the first indication that the K and L intermediates were in equilibrium. The next major transition occurred with τ ∼20 ms (Fig. 2C) and resulted in formation of an M-like state with absorption maximum at 390–400 nm. The difference spectrum of this transition (Fig. 2A, green line) clearly indicated that the appearance of M occurred simultaneously with the depletion of both K and L intermediates, confirming the existence of a K⇔L equilibrium. A major component of M decay was characterized by τ ∼125 ms and resulted in formation of a species with a maximum at 510–520 nm, close to that of the initial (unphotolyzed) state (Fig. 2A, blue line).
Fig. 2.
(A) The spectra of transitions corresponding to the rise of K-like (unresolved), L-like (5 µs), and M-like (22 ms) intermediates and to M decay (125 ms) obtained by global fit analysis. The time window for the global fit did not include the fast component of the K-to-L transition, hence the apparent amplitude reflects only a small fraction of this transition. (B) The kinetics of K decay and L rise. (C) The kinetics of L decay and M rise. (D) The spectra of the slowest spectral transition obtained by global fit analysis and the long-lived intermediate(s) calculated as described in the text.
The slowest transition obtained by global fit analysis was in the range of seconds. Its difference spectrum exhibited a maximum at a wavelength longer than the peak of GtACR1 absorption (Fig. 2D, black symbols and line), which indicated the presence of a long-lived intermediate(s) with blue-shifted absorption. We derived its spectrum by subtraction of the difference spectrum from a properly scaled spectrum of unphotolyzed GtACR1. The resultant spectrum showed the maximum at ∼475 nm (Fig. 2D, red symbols and line), which resembled that of the N intermediate in the BR photocycle. Additional bands (shoulders) on the blue slope of the difference spectrum may either belong to this intermediate or reflect contributions of L and M intermediates.
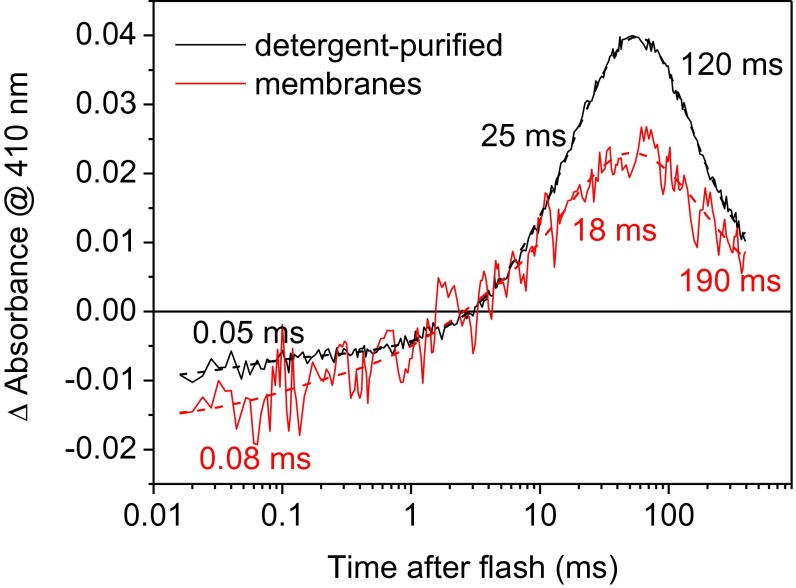
A slow formation of an M-like intermediate observed in detergent-purified GtACR1 is highly unusual for microbial rhodopsins. Photochemical conversions of some of them have been shown to become considerably slower upon purification in detergent, compared with those of the same pigments in biological membranes (e.g., ref. 14). However, in the case of GtACR1, the rate of M formation was almost the same in Pichia membranes and in detergent (Fig. S2). Therefore, the unusually slow M formation in GtACR1 could not be explained by the influence of detergent. Moreover, a similarly slow M formation was also observed in GtACR2, an orthologous protein from the same organism (Fig. S3), which indicated that the slow M formation is likely a common feature of the ACR family.
Fig. S2.
Laser flash-induced absorbance changes in detergent-purified GtACR1 (black lines) and GtACR1 in Pichia membranes (red lines). Solid lines are experimental data; dashed lines are three-exponential fits of the curves.
Fig. S3.
The formation of the M intermediate in GtACR2. The black line shows experimental data; the red line, a three-exponential fit.
Correlation of photochemical conversions with channel opening and closing.
Opening of the GtACR1 channel in response to a short laser flash (i.e., under single-turnover conditions) assessed by whole-cell patch clamp recording was >50-fold faster than the formation of the M intermediate (Fig. 3A). Therefore, we hypothesize that the conducting state of the channel occurs before Schiff base proton transfer during the lifetime of the L intermediate. The formation of the L spectral intermediate appears to be faster than the photocurrent rise, but the time course of fast electrogenic events in the cell membrane is distorted by the lower time resolution of patch-clamp recording (Fig. S4), and therefore precise temporal correlation between photocurrents and fast absorbance changes cannot be expected. Roughly, fast channel closing temporally correlated with formation of the M-like intermediate (Fig. 3A) at neutral and alkaline pH, where both processes accelerate (Fig. 3B), whereas slow channel closing correlated with M decay. Analysis of the photochemical conversions and photocurrents in the WT and the mutants further confirmed these correlations (see below).
Fig. 3.
(A) Laser flash-induced photocurrent generated by GtACR1 in a HEK293 cell (red line, left axis) and kinetics of the M intermediate in detergent-purified pigment monitored at 400 nm (thin black line) fitted with three exponentials (thick black dashed line). To correct for contribution of the formation and disappearance of the longer-wavelength (L-like) intermediate, the absorption changes at 410 nm were subtracted from those at 390 nm. (B) The pH dependencies of the rate of the M intermediate formation was monitored at 400 nm in purified pigments (solid lines, filled circles) and the rate of fast channel closing was measured by patch clamp in HEK293 cells (dashed lines, empty squares) in the WT (black symbols and lines) and the E68Q mutant (red symbols and lines). The data for M formation were pooled from five and two independent experiments in the WT and the mutant, respectively. The data for channel closing were obtained from ref. 3.
Fig. S4.
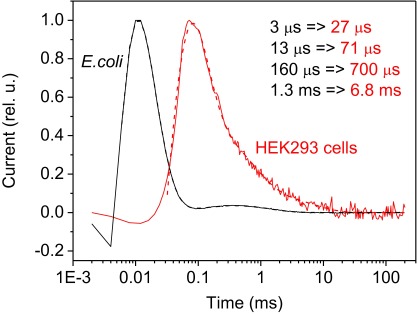
Testing the time resolution of whole-cell patch clamp recording using an electrogenic protein that can be functionally expressed in both E. coli and HEK293 cells (14). Photocurrents recorded by a suspension assay (24) from the AR-3_D96N mutant expressed in E. coli cells (black lines) and by patch clamp in HEK293 cells that express the same construct (red lines). Experimental data are shown in solid lines and four-exponential fit in dashed lines. All four kinetics components are considerably slower when recorded by patch clamp compared with the suspension assay.
The E68Q Mutant.
The pH titration of the rate of M formation and fast channel closing in the WT and E68Q mutant provided strong evidence for a tight relationship between these two processes. The E68Q mutation slowed both M formation and fast channel closing, and caused a shift of their pH dependencies to higher pH values (Fig. 3B). These results strongly indicate that generation of the M intermediate occurs concomitantly with fast channel closing in GtACR1. Different pH dependencies of the rate of M formation in the WT and the mutant indicate that the presence of Glu-68 in WT and its deprotonation in the initial state at high pH facilitates formation of M. These data and the predicted position of Glu-68 near the protonated Schiff base strongly suggest that Glu-68 serves as an acceptor of the Schiff base proton. However, formation of the M intermediate in the E68Q mutant, albeit at a lower amount, indicates that the presence of Glu-68 is not a strict prerequisite for Schiff base deprotonation.
The E68Q mutation also influenced the spectrum and pH dependence of transitions in the unphotolysed state of the pigment. At pH 7.4, the absorption spectrum of the E68Q mutant was shifted 3 nm to the red from that of the WT (Fig. 1B, red line). The appearance of an alkalization-induced M-like (deprotonated) state of the Schiff base in this mutant could be followed up to high pH (Fig. S5) and resembled that in the D85N mutant of BR (15, 16). This finding indicates a higher stability of the unphotolysed state in the mutant than in the WT, in which the M-like state could only be observed in a narrow pH range (Fig. 1 C and D) because of denaturation of pigment at higher pH. The pKa of the Schiff base deprotonation in the mutant was ∼10.3 (Fig. 4A).
Fig. S5.
A series of absorption spectra of the purified GtACR1_E68Q mutant at different pH values.
Fig. 4.
(A) The pH titration of the Schiff base in the purified GtACR1_E68Q mutant by measuring formation of an M-like state (the maximum at 390 nm at high pH in the dark). (Inset) The absorption spectrum of this state obtained by summation of the difference spectrum between pH 11 and pH 8 with the properly scaled spectrum of the unphotolysed state at pH 7.4. (B) The pH dependence of the maximal absorption wavelength for the E68Q mutant and WT. (C) Light-induced absorption changes of the E68Q mutant at 590 nm and 460 nm, associated with biphasic decay of K and formation of L. The incomplete K decay in submillisecond time ranges suggest establishment of equilibrium concentrations of K and L from a reversible K⇔L conversion. (D) Comparison of L-to-M transitions in WT and the mutant.
The dependence of the wavelength of maximal absorption on pH observed in the WT completely disappeared in the E68Q mutant (Fig. 4B, red line). This result indicates that Glu-68 serves not only as the proton acceptor, but also acts as the counterion of the Schiff base at high pH. If the pKa of Glu-68 decreases during the photocycle, this residue may play these roles also at neutral pH.
As in the WT, a fast biphasic reversible decay of K led to generation of an L-like intermediate in the mutant. The K⇔L transition in the photochemical reaction cycle of E68Q differed from that in the WT only quantitatively: Both its phases were several times slower than in the WT (Fig. 4C). However, the L-to-M transition in the E68Q mutant considerably differed from that in the WT. As mentioned above, the appearance of the M intermediate in the WT corresponded to the depletion of the L intermediate (reproduced for comparison in Fig. 4D, dashed lines). In contrast, in the E68Q mutant, the depletion of L was not observed, and the accumulation of M was fivefold smaller (Fig. 4D, solid lines). This result confirmed the association of M rise with fast channel closing in the WT that was absent in the mutant at neutral pH (3).
The D234N and D234N_E68Q Mutants.
The above results show that Glu-68 likely acts as a counterion and an acceptor of the Schiff base proton in GtACR1 at high pH. To assess the role of the second carboxyl residue near the Schiff base, Asp-234, and effects of removal of both carboxylic residues, we examined the D234N mutant and the E68Q_D234N double mutant. We were able to express and isolate the former for spectroscopic studies and conduct electrophysiological studies on the double mutant in HEK cells.
The absorption spectrum of the GtACR1_D234N mutant at pH 7.4 was indistinguishable from that of the WT (Fig. 1B, green line), indicating that this residue does not act as a counterion of the protonated Schiff base at this pH. The spectral maximum exhibited a pH-dependent transition with pKa 4.7 (Fig. 5A), indicating the appearance of a titratable group in this pH range in the D234N mutant. The correlation between the main component of M rise and fast channel closing, and of M decay and slow channel closing, remained in the mutants the same as in the WT. However, an additional fast component appeared in the M rise in the mutant (Fig. 5B), which could be interpreted as a transfer of the Schiff base proton to an acceptor. Indeed, the channel current generated by the D234N mutant upon excitation with short laser flashes (single-turnover conditions) was preceded by a fast positive current, the amplitude of which increased upon an increase in the bath pH (Fig. 5C). The sign, fast kinetics and the pH dependence of this current component, indicated that it reflected proton transfer from the protonated Schiff base to an outward acceptor. When Glu-68 was neutralized in addition to the D234N mutation, no fast positive current was observed even at pH 9.4 (Fig. 5D). This result suggests that Glu-68 is the residue that becomes titratable and serves as the Schiff base proton acceptor in the D234N mutant. Apparently, the D234N mutation causes a decrease in the pKa of Glu-68 because of a disruption of the coupling between these two residues in the WT predicted by our homology model (see below). This hypothesis is the most straightforward interpretation that does not exclude involvement of other interacting residues.
Fig. 5.
(A) The pH titration of the purified GtACR1_D234N mutant (red symbols and line) and WT (black symbols and line). Inset shows difference absorption spectrum between spectra at pH 3.5 and 6.5 for GtACR1_D234N. (B) The kinetics of the M intermediate in the D234N mutant (black line) and its multiexponential fit (red line). The numbers indicate τ values obtained by fitting. (C and D) Typical normalized laser flash-induced photocurrents generated by the GtACR1_D234N (C) and GtACR1_E68Q_D234N (D) mutants in HEK293 cells at varied bath pH. The absolute amplitude of channel currents in both mutants was the same within experimental error.
The C102A Mutant.
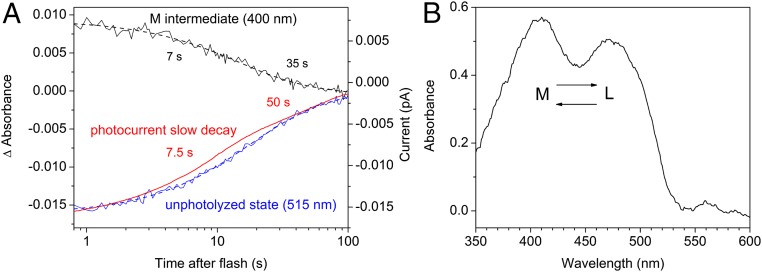
In the WT, channel is fully open well before M formation (Fig. 3A), which means that both fast and slow channel closing are due to the depletion of the conducting L state. The results obtained with the C102A mutant confirmed this conclusion. Cys-102 is the homolog of Cys-128 in CrChR2, and its replacement with Ala leads to a dramatic decrease in the slow channel closing in GtACR1 (3). We tested the influence of the C102A mutation on the photocycle of purified pigment. Both recovery of the unphotolyzed state and a decrease in absorption at 400 nm (close to the absorption peak of the M intermediate) were 100-fold slower in the mutant than in the WT, which matched perfectly the influence of this mutation on the slow phase of the photocurrent decay (Fig. 6A). This observation could be interpreted as the M intermediate being the open state in the gating mechanism characterized by slow channel closing. However, the spectrum of absorbance changes in the C102A mutant showed that the L and M intermediates were in equilibrium (Fig. 6B), and that therefore, L decay kinetically matches M decay. The simplest interpretation is that the L intermediate, rather than M, is the open state for both gating mechanisms also in the GtACR1_C102A mutant.
Fig. 6.
(A) The decay of the M intermediate (black lines) and the recovery of the unphotolyzed state (blue lines) in the purified GtACR1_C102A mutant, and the decay of laser flash-induced photocurrent generated by this mutant in a HEK293 cell. Solid lines are experimental data; dashed lines, two-exponential fit. (B) The absorption spectrum of the intermediates from which the unphotolyzed state recovers in the GtACR1_C102A mutant obtained by subtraction of the difference spectrum between spectra measured at 1 s and 100 s from the properly scaled absorption spectrum of dark-adapted pigment.
The S97E and S97A Mutants.
All so-far-known ACRs have a noncarboxylate residue in the position of Asp-85 in BR, in which most CCRs have a Glu residue. The corresponding residue in GtACR1 is Ser-97. When it was mutated to Glu, the pH titration of the wavelength position of the main absorption peak showed a large (35 nm) spectral transition with a pKa ∼8 (Fig. 7A, red symbols and line), as expected from its predicted position close to the Schiff base (3). The pH titration of the S97E mutant resembled that of the protonated Schiff base counterions in many other microbial retinal proteins, i.e., BR (17) and proteorhodopsins (18, 19). Introduction of this Glu results in appearance of a fast phase in M formation in the S97E mutant, which was absent in the WT. It is likely that the Glu facilitates M formation by acting as an acceptor of the Schiff base proton (Fig. 7B), the role of carboxylate residues at the corresponding position in most other microbial rhodopsins.
Fig. 7.
(A) The pH titration of the purified GtACR1_S97E mutant (red symbols and line). The data for the WT from Fig. 2A are shown for comparison as black symbols and lines. Inset shows the corresponding difference spectrum between pH 6 and pH 9.5 in the S97E mutant. (B) The kinetics of M formation in the S97E mutant monitored by absorbance changes at pH 8.
These findings also indirectly confirm that Cl− is unlikely to be the counterion in GtACR1. If it were, the introduction of another negatively charged group close to the Schiff base would not cause such a large red shift as was observed. However, a ∼10-nm difference between the absorption maxima of the GtACR1_S97E mutant and the WT at acidic pH suggests the presence of a negative charge, the displacement of which by the introduced bulky residue, neutral at this pH, led to the red shift of the spectrum in the mutant.
In contrast to the three other mutations that did not (E68Q) or only slightly (D234N and C102A) reduced the amplitude of photocurrents (3), the S97E mutation led to its dramatic (>30-fold) suppression (Fig. S6A). The peak current at −60 mV in response to the first laser flash in dark-adapted cells for the mutant was 47 ± 25 pA (mean ± SEM, n = 4 cells), compared with 1,444 ± 366 pA for the WT (mean ± SEM, n = 11 cells), whereas expression levels of the two constructs, as judged by the tag fluorescence, were comparable. This result confirms our hypothesis that the presence of a noncarboxylate residue in the position of Asp-85 in BR is critical for high ACR conductance.
Fig. S6.
(A) Typical laser flash-evoked photocurrents generated by GtACR1_S97E and wild-type GtACR1 in HEK293 cells at −60 mV at pH 7.4. (B) The decay of typical laser flash-evoked photocurrents generated by wild-type GtACR1, its S97A mutant, and wild-type PsuACR1. The traces were normalized at the peak value.
Whereas both G. theta ACRs contain a Ser residue in the position of Asp-85 in BR, an ACR from another cryptophyte, Proteomonas sulcata (PsuACR1), has Ala in this position. PsuACR1 exhibits an ∼eightfold more rapid channel closing than GtACR1 (2). To probe whether Ala in the position of Asp-85 determines fast channel closing in ACRs, we generated and tested the GtACR1_S97A mutant. Measurement of its laser flash-evoked photocurrents showed that GtACR1_S97A channel closed even slower than wild-type GtACR1 (Fig. S6B), which indicated that the fast closing of PsuACR1 was not controlled by the residue in question.
Discussion
Expression and purification of photoactive wild-type GtACR1 and its mutants in P. pastoris allowed us to characterize their photochemical conversions by flash photolysis and to measure the pH dependence of their absorption maxima. The most striking difference between the photocycle of ACRs and other microbial rhodopsins is an extremely slow appearance and decay of an M-like intermediate whose spectral shift to lower wavelengths is caused by the deprotonation of the retinylidene Schiff base. Opening of the GtACR1 channel occurs >50 times faster than M formation, which means that its open state is represented by the earlier L intermediate that appears on a submillisecond time scale. Fast channel closing corresponds to the depletion of the L state (and consequently generation of M), because of the reversible reaction between the L and M intermediates, whereas slow channel closing corresponds to the irreversible decay of M (and hence, L). A simplified scheme in Fig. 8 summarizes our model. Apparently, isomerization of the retinal chromophore from its all-trans to 13-cis configuration and accompanying conformational changes of the protein are sufficient to remove a barrier for Cl− conductance through the protein.
Fig. 8.
A simplified scheme of the GtACR1 photocycle and coupling of the photocycle intermediates with channel opening and closing.
The correspondence between the spectral intermediates and channel states in ACRs and CCRs appears to be quite different, which further confirms that these proteins represent two distinct families of microbial rhodopsins. In CCRs (or at least in the best studied CrChR2), deprotonation of the Schiff base occurs much earlier than channel opening, and, therefore, the conducting state of the channel was initially linked to the next red-shifted intermediate of the photocycle (P520) (20, 21). At least in the slow C128X mutants, photocurrents are quenched not only by green, but also by violet light (absorbed by an M-like intermediate, P-390), which led to the conclusion that CrChR2 opens already in the M state (6). Our observation of an extremely slow M formation in ACRs clearly shows that channel opening does not require deprotonation of the Schiff base. Furthermore, when early M formation occurs in the GtACR1_S97E mutant, in which a proton acceptor residue has been placed adjacent to the protonated Schiff base, the channel is nearly completely prevented from opening.
The scheme in Fig. 8 is a simplification that captures the main conclusions from our data but does not account for all our observations. Specifically, the two kinetically distinguished steps (∼1 µs and ∼20 µs) that we found in K decay and L formation may indicate the existence of either two K or two L species. We also cannot exclude a fast equilibrium between the L and N/O-like intermediates. However, this transition has small amplitude and is too slow to account for channel opening.
ACR sequences resemble those of haloarchaeal Cl− pumps and differ from most other microbial rhodopsins in the position corresponding to Asp-85 in BR, where they have a noncarboxylate residue. An exchange from Cl− to SO42− buffer caused a 24-nm red shift of the spectral maximum of purified halorhodopsin from Natronomonas pharaonis (12). In contrast, no spectral shift was detected upon the same substitution in GtACR1, which strongly argues against Cl− playing a role of the Schiff base counterion in the latter pigment. Nevertheless, the presence of a noncarboxylate residue in this position appears to be critically important for ACR function, because its replacement with Glu, found in this position in most CCRs, dramatically suppressed the photocurrent. It has, however, to be mentioned that application of a series of successive flashes led to a strong growth of photocurrents from the GtACR1_S97E mutant, which indicated the involvement of a two-photon process, investigation of which was beyond the present study.
Intriguing questions are the exact roles of carboxylic residues in the vicinity of the Schiff base, Glu-68 and Asp-234, in channel gating and Schiff base deprotonation. Neutralizing Glu-68 results in a four- to fivefold decrease in the accumulation of M and a corresponding disappearance of the fast channel closing at pH 7.4 (Fig. 4D and ref. 3). The pH dependence of the rates of both residual M formation and fast channel closing were shifted by this mutation to high (nonphysiological) pH values (Fig. 3B). Finally, the mutation fully eliminated the strong pH dependence of the maximum absorption wavelength of the unphotolysed state observed in the WT (Fig. 4B). These observations suggest that Glu-68 likely serves as a counterion and an acceptor of the proton from the Schiff base in the photocycle at least at neutral and high pH, a scenario supported by the features of the D234N mutant and D234N_E68Q double mutant (Fig. 5). However, residual M formation observed in the E68Q mutant indicates the existence of an alternative Schiff base proton acceptor in GtACR1. We cannot completely exclude that Glu-68 may only facilitate proton transfer to this acceptor.
An atomic-resolution structure of an ACR is not yet available. Accordingly, we built a homology model of GtACR1 by the Robetta server using the crystal structure of a chimeric CCR (C1C2), which the server identified as the best template (Fig. S7). Although predictions of this model have to be regarded with caution, several of them are consistent with our experimental data: (i) In contrast to the C1C2 structure, in which the homolog of Asp-234 is the closest protonatable residue to the Schiff base, in the GtACR1 model, Glu-68 is the closest, whereas Asp-234 is more than twice more distant (Table S1). The closer proximity of Glu-68 is in agreement with our results, indicating that Glu-68 may serve as a counterion and proton acceptor in GtACR1. (ii) pKa calculations using the PROPKA software (22) indicate that the only coupled carboxylate residues in the vicinity of the retinylidene Schiff base are Glu-68 and Asp-234 (Table S2), which agrees with the conclusion we draw from our experimental results. (iii) The predicted pKa of Glu-68 without coupling (∼4.5) is close to the experimental value obtained for the Glu-68 in the D234N mutant in which this coupling is disrupted.
Fig. S7.
A homology model of GtACR1 built by the Robetta server (robetta.bakerlab.org/submit.jsp) using PDB ID code 3ug9 (C1C2) as template and visualized by PyMOL 1.8.0.4. Shown are the side chains of the amino acid residues in the vicinity of the retinylidene Schiff base (RSB+). (A) Side view. (B) Top (from outside the cell) view.
Table S1.
The distances (in Å) between the indicated amino acid residues according to a homology model of GtACR1 and the crystal structure of the hybrid CCR C1C2
| Distance between residues | GtACR1 homology model | C1C2 crystal structure (3ug9) |
| RSB+⇔Glu-68/Glu-129 | 2.8 | 6.3 |
| RSB+⇔Asp-234/Asp-292 | 5.9 | 3.0 |
| RSB+⇔Ser-97/Glu-162 | 3.4 | 3.9 |
| Glu-68/Glu-129⇔Asp-234/Asp-292 | 4.7 | 5.8 |
Table S2.
Interactions of Glu-68 and Asp-234 residues in the homology model of GtACR1 (3) predicted by PROPKA software (22)
| Residue | Side chain H bond | Coulombic interaction | ||
| Strength | Partner | Strength | Partner | |
| Glu-68 | −0.72 | Thr-71 | −0.04 | Arg-94 |
| −0.83 | Ser-97 | — | — | |
| −0.85 | Lys-238 | −2.03 | Lys-238 | |
| Asp-234 | −0.84 | Tyr-72 | −0.39 | Arg-94 |
| −0.41 | Tyr-207 | 1.4 | Glu-68 | |
| — | — | −0.93 | Lys-238 | |
Our characterization of the GtACR1 photocycle contributes to understanding of the coupling between photochemistry of the chromophore and protein conformation changes leading to the appearance of ion conductance. Elucidating the conductance mechanism of ACRs is of particular interest because of the promise of these channels as optogenetic tools for research and potentially for gene therapy.
Materials and Methods
The opsin domain of GtACR1 in frame with an eight-His tag in the pPIC9K vector (Invitrogen) was expressed in P. pastoris SMD1168 (his4, pep4) and purified by using nickel-nitrilotriacetic acid agarose beads (Qiagen) in a buffer containing 20 mM Hepes, pH 7.4, 300 mM NaCl, 5% (vol/vol) glycerol, 0.02% dodecyl maltoside (DDM). Mutations were introduced by using a QuikChange XL site-directed mutagenesis kit (Agilent Technologies). Spectra of purified pigments in the UV-visible range were recorded on a Cary 4000 spectrophotometer (Varian). Light-induced absorption changes were measured with a laboratory-constructed crossbeam apparatus. Excitation flashes (532 nm, 6 ns, up to 40 mJ) were provided by a Surelite I Nd-YAG laser (Continuum). Signals were amplified by a low-noise current amplifier (model SR445A; Stanford Research Systems) and digitized with a GaGe Octopus digitizer board (model CS8327; DynamicSignals), maximal sampling rate 50 MHz. Data analysis was performed with pClamp 10.2 (Molecular Devices) and OriginPro 7 (OriginLab) software. Logarithmic filtration of the data was performed by using the GageCon program kindly provided by L. S. Brown, University of Guelph, Guelph, ON, Canada. Global fit of spectral transitions was performed by using the FITEXP program (23) kindly provided by A. K. Dioumaev, University of California, Irvine, CA. For expression in HEK293 cells, the constructs were transferred to the vector pcDNA3.1 (Life Technologies) in frame with an EYFP tag. Whole-cell patch clamp recording was carried out by using an Axopatch 200B amplifier (Molecular Devices). Laser excitation was provided by a Minilite Nd:YAG laser (532 nm, pulsewidth 6 ns, energy 12 mJ; Continuum). A detailed description of the acquisition, processing, and analysis of the data are provided in SI Materials and Methods.
SI Materials and Methods
Expression and Purification of GtACR1 from P. pastoris.
The opsin domain of human codon-adapted GtACR1 (amino acid residues 1–295) was fused in frame with a C-terminal eight-His tag and subcloned into the pPIC9K vector (Invitrogen) between EcoRI and NotI sites. Mutations were introduced by using a QuikChange XL site-directed mutagenesis kit (Agilent Technologies) and verified by DNA sequencing. The resultant plasmids were linearized with SalI and used for transformation of P. pastoris strain SMD1168 (his4, pep4) by electroporation. The transformants were first screened for their ability to grow on histidine-deficient medium, and second, for their geneticin resistance. Up to eight single colonies that grew on 4 mg/mL geneticin were screened by small-scale cultivation, and clones of the brightest pink color were selected for further use.
For protein purification, a starter culture was inoculated into buffered complex glycerol medium until A600 reached 4–8, after which the cells were harvested by centrifugation at 3,800 × g and transferred to buffered complex methanol medium supplemented with 5 μM all-trans retinal (Sigma Aldrich). Expression was induced by the addition of 0.5% methanol. After 24–30 h, the cells were harvested and disrupted in a bead beater (BioSpec Products) in buffer A [20 mM sodium phosphate, pH 7.4, 100 mM NaCl, 1 mM EDTA, 5% (vol/vol) glycerol]. Cell debris and unbroken cells were removed by centrifugation at 3,800 × g. Membrane fragments were collected by ultracentrifugation at 40,000 rpm in a Ti45 rotor, resuspended in buffer B (20 mM Hepes, pH 7.4, 300 mM NaCl, 5% glycerol), and solubilized by incubation with 1.5% DDM for 1.5 h or overnight at 4 °C. Nonsolubilized material was removed by ultracentrifugation at 50,000 rpm in a TLA-100 rotor. The supernatant was mixed with nickel-nitrilotriacetic acid agarose beads (Qiagen) and loaded on a column. After washing the column thoroughly with buffer C (20 mM Hepes, pH 7.4, 300 mM NaCl, 5% glycerol, 0.02% DDM) containing 20 mM and 40 mM imidazole, the protein was eluted with buffer C containing 300 mM imidazole. The pigment was concentrated, and imidazole was removed by repetitive washing with imidazole-free buffer C using YM-10 centrifugal filters (Amicon).
Absorption Spectroscopy and Flash Photolysis.
Absorption spectra of purified pigments in the UV-visible range were recorded on a Cary 4000 spectrophotometer (Varian). pH titration was carried out by the addition of small volumes of 1 M NaOH, 0.5 M Tris (pH 10), 0.5 M citric acid, or 1 M HCl.
Light-induced absorption changes were measured with a laboratory-constructed crossbeam apparatus. Excitation flashes (532 nm, 6 ns, up to 40 mJ) were provided by a Surelite I Nd-YAG laser (Continuum). Measuring light was from a 250-W incandescent tungsten lamp combined with a McPherson monochromator (model 272). A Hamamatsu Photonics photomultiplier tube (model R928) was protected from excitation laser flashes by a second monochromator of the same type and additionally with 12-nm bandwidth interference filters (Oriel Instruments). Signals were amplified by a low noise current amplifier (model SR445A; Stanford Research Systems) and digitized with a GaGe Octopus digitizer board (model CS8327, DynamicSignals), maximum sampling rate 50 MHz. The time interval between excitation flashes was 20 s, and up to 100 sweeps were averaged for each wavelength. Data analysis was performed with pClamp 10.2 (Molecular Devices) and OriginPro 7 (OriginLab) software. Logarithmic filtration of the data were performed by using the GageCon program provided by L. S. Brown, University of Guelph, Guelph, ON, Canada. Global fit of spectral transitions was performed by using the FITEXP program (23) provided by A. K. Dioumaev, University of California, Irvine, CA.
Photocurrent Measurements.
The opsin domain of the wild-type GtACR1 or its mutants was cloned into the mammalian expression vector pcDNA3.1 (Life Technologies) in frame with an EYFP (enhanced yellow fluorescent protein) tag. HEK293 (human embryonic kidney) cells were transfected by using the ScreenFectA transfection reagent (Waco Chemicals USA). All-trans retinal (Sigma) was added as a stock solution in ethanol at a final concentration of 5 µM. Measurements were performed 48–72 h after transfection with an Axopatch 200B amplifier (Molecular Devices). The signals were digitized with a Digidata 1440A by using pClamp 10 software (both from Molecular Devices) at the sampling rate of 4 µs per point. Patch pipettes with resistances of 2–5 MΩ were fabricated from borosilicate glass and filled with the following solution (in mM): KCl 126, MgCl2 2, CaCl2 0.5, EGTA 5, Hepes 25, pH 7.4. The bath solution contained (in mM): NaCl 150, CaCl2 1.8, MgCl2 1, glucose 5, Hepes, or Tris 10, pH was adjusted with N-methyl-d-glucamine to the desired value. A 4 M salt bridge was used in all experiments. All measurements were carried out at room temperature (25 °C). Laser excitation was provided by a Minilite Nd:YAG laser (532 nm, pulsewidth 6 ns, energy 12 mJ; Continuum). A laser artifact measured with a blocked optical path was digitally subtracted from the recorded traces. For further analysis, the signals were logarithmically averaged with a custom-created computer algorithm. Curve fitting was performed by using Origin 7 software (OriginLab).
Acknowledgments
We thank Dr. Sergei P. Balashov for helpful discussion and suggestions and critical reading of the manuscript, Dr. Leonid S. Brown for the GageCon software, and Dr. Andrei K. Dioumaev for the FITEXP program. This work was supported by NIH Grant R01GM027750 and Endowed Chair AU-0009 from the Robert A. Welch Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1525269113/-/DCSupplemental.
References
- 1.Govorunova EG, Sineshchekov OA, Janz R, Liu X, Spudich JL. NEUROSCIENCE. Natural light-gated anion channels: A family of microbial rhodopsins for advanced optogenetics. Science. 2015;349(6248):647–650. doi: 10.1126/science.aaa7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Govorunova EG, Sineshchekov OA, Spudich JL. Proteomonas sulcata ACR1: A fast anion channelrhodopsin. Photochem Photobiol. December 20, 2015 doi: 10.1111/php.12558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sineshchekov OA, Govorunova EG, Li H, Spudich JL. Gating mechanisms of a natural anion channelrhodopsin. Proc Natl Acad Sci USA. 2015;112(46):14236–14241. doi: 10.1073/pnas.1513602112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wietek J, et al. Conversion of channelrhodopsin into a light-gated chloride channel. Science. 2014;344(6182):409–412. doi: 10.1126/science.1249375. [DOI] [PubMed] [Google Scholar]
- 5.Nack M, et al. The DC gate in Channelrhodopsin-2: Crucial hydrogen bonding interaction between C128 and D156. Photochem Photobiol Sci. 2010;9(2):194–198. doi: 10.1039/b9pp00157c. [DOI] [PubMed] [Google Scholar]
- 6.Bamann C, Gueta R, Kleinlogel S, Nagel G, Bamberg E. Structural guidance of the photocycle of channelrhodopsin-2 by an interhelical hydrogen bond. Biochemistry. 2010;49(2):267–278. doi: 10.1021/bi901634p. [DOI] [PubMed] [Google Scholar]
- 7.Lanyi JK. Proton transfers in the bacteriorhodopsin photocycle. Biochim Biophys Acta. 2006;1757(8):1012–1018. doi: 10.1016/j.bbabio.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Essen LO. Halorhodopsin: Light-driven ion pumping made simple? Curr Opin Struct Biol. 2002;12(4):516–522. doi: 10.1016/s0959-440x(02)00356-1. [DOI] [PubMed] [Google Scholar]
- 9.Lórenz-Fonfría VA, Heberle J. Channelrhodopsin unchained: Structure and mechanism of a light-gated cation channel. Biochim Biophys Acta. 2014;1837(5):626–642. doi: 10.1016/j.bbabio.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 10.Schneider F, Grimm C, Hegemann P. Biophysics of channelrhodopsin. Annu Rev Biophys. 2015;44:167–186. doi: 10.1146/annurev-biophys-060414-034014. [DOI] [PubMed] [Google Scholar]
- 11.Kolbe M, Besir H, Essen LO, Oesterhelt D. Structure of the light-driven chloride pump halorhodopsin at 1.8 A resolution. Science. 2000;288(5470):1390–1396. doi: 10.1126/science.288.5470.1390. [DOI] [PubMed] [Google Scholar]
- 12.Scharf B, Engelhard M. Blue halorhodopsin from Natronobacterium pharaonis: Wavelength regulation by anions. Biochemistry. 1994;33(21):6387–6393. doi: 10.1021/bi00187a002. [DOI] [PubMed] [Google Scholar]
- 13.Sineshchekov OA, Sasaki J, Wang J, Spudich JL. Attractant and repellent signaling conformers of sensory rhodopsin-transducer complexes. Biochemistry. 2010;49(31):6696–6704. doi: 10.1021/bi100798w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sineshchekov OA, Govorunova EG, Wang J, Li H, Spudich JL. Intramolecular proton transfer in channelrhodopsins. Biophys J. 2013;104(4):807–817. doi: 10.1016/j.bpj.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turner GJ, et al. Bacteriorhodopsin D85N: Three spectroscopic species in equilibrium. Biochemistry. 1993;32(5):1332–1337. doi: 10.1021/bi00056a019. [DOI] [PubMed] [Google Scholar]
- 16.Brown LS, Bonet L, Needleman R, Lanyi JK. Estimated acid dissociation constants of the Schiff base, Asp-85, and Arg-82 during the bacteriorhodopsin photocycle. Biophys J. 1993;65(1):124–130. doi: 10.1016/S0006-3495(93)81064-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balashov SP, Imasheva ES, Govindjee R, Ebrey TG. Titration of aspartate-85 in bacteriorhodopsin: What it says about chromophore isomerization and proton release. Biophys J. 1996;70(1):473–481. doi: 10.1016/S0006-3495(96)79591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dioumaev AK, et al. Proton transfers in the photochemical reaction cycle of proteorhodopsin. Biochemistry. 2002;41(17):5348–5358. doi: 10.1021/bi025563x. [DOI] [PubMed] [Google Scholar]
- 19.Wang W-W, Sineshchekov OA, Spudich EN, Spudich JL. Spectroscopic and photochemical characterization of a deep ocean proteorhodopsin. J Biol Chem. 2003;278(36):33985–33991. doi: 10.1074/jbc.M305716200. [DOI] [PubMed] [Google Scholar]
- 20.Bamann C, Kirsch T, Nagel G, Bamberg E. Spectral characteristics of the photocycle of channelrhodopsin-2 and its implication for channel function. J Mol Biol. 2008;375(3):686–694. doi: 10.1016/j.jmb.2007.10.072. [DOI] [PubMed] [Google Scholar]
- 21.Ritter E, Stehfest K, Berndt A, Hegemann P, Bartl FJ. Monitoring light-induced structural changes of Channelrhodopsin-2 by UV-visible and Fourier transform infrared spectroscopy. J Biol Chem. 2008;283(50):35033–35041. doi: 10.1074/jbc.M806353200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olsson MHM, Søndergaard CR, Rostkowski M, Jensen JH. PROPKA3: Consistent treatment of internal and surface residues in empirical pK(a) predictions. J Chem Theory Comput. 2011;7(2):525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 23.Dioumaev AK. Evaluation of intrinsic chemical kinetics and transient product spectra from time-resolved spectroscopic data. Biophys Chem. 1997;67(1-3):1–25. doi: 10.1016/s0301-4622(96)02268-5. [DOI] [PubMed] [Google Scholar]
- 24.Sineshchekov OA, Spudich JL. Light-induced intramolecular charge movements in microbial rhodopsins in intact E. coli cells. Photochem Photobiol Sci. 2004;3(6):548–554. doi: 10.1039/b316207a. [DOI] [PubMed] [Google Scholar]