Significance
Females have increased immune responsiveness than males, and they are more likely to develop autoimmune disorders. The mechanism underlying these observations is unclear, and hypotheses suggest an important role for the X chromosome. Here, we discover that the inactive X is predisposed to become partially reactivated in mammalian female lymphocytes, resulting in the overexpression of immunity-related genes. We also demonstrate that lymphocytes from systemic lupus erythematosus patients have different epigenetic characteristics on the inactive X that compromises transcriptional silencing. These findings are the first to our knowledge to link the unusual maintenance of X chromosome Inactivation (the female-specific mechanism for dosage compensation) in lymphocytes to the female bias observed with enhanced immunity and autoimmunity susceptibility.
Keywords: X chromosome inactivation, XIST RNA, epigenetics, female-biased autoimmunity
Abstract
Females have a greater immunological advantage than men, yet they are more prone to autoimmune disorders. The basis for this sex bias lies in the X chromosome, which contains many immunity-related genes. Female mammals use X chromosome inactivation (XCI) to generate a transcriptionally silent inactive X chromosome (Xi) enriched with heterochromatic modifications and XIST/Xist RNA, which equalizes gene expression between the sexes. Here, we examine the maintenance of XCI in lymphocytes from females in mice and humans. Strikingly, we find that mature naïve T and B cells have dispersed patterns of XIST/Xist RNA, and they lack the typical heterochromatic modifications of the Xi. In vitro activation of lymphocytes triggers the return of XIST/Xist RNA transcripts and some chromatin marks (H3K27me3, ubiquitin-H2A) to the Xi. Single-cell RNA FISH analysis of female T cells revealed that the X-linked immunity genes CD40LG and CXCR3 are biallelically expressed in some cells. Using knockout and knockdown approaches, we find that Xist RNA-binding proteins, YY1 and hnRNPU, are critical for recruitment of XIST/Xist RNA back to the Xi. Furthermore, we examined B cells from patients with systemic lupus erythematosus, an autoimmune disorder with a strong female bias, and observed different XIST RNA localization patterns, evidence of biallelic expression of immunity-related genes, and increased transcription of these genes. We propose that the Xi in female lymphocytes is predisposed to become partially reactivated and to overexpress immunity-related genes, providing the first mechanistic evidence to our knowledge for the enhanced immunity of females and their increased susceptibility for autoimmunity.
The X chromosome has the greatest density of immunity-related genes (1), and females, with two X chromosomes, have an immunological advantage over males (XY). Clinical studies have demonstrated that females have a more hyperresponsive immune system than males following immune challenges (2, 3). Females produce more serum IgM and antibodies (4, 5), which is immunologically advantageous, whereas males are more susceptible to bacterial and viral infections (5–7). This strong female-specific immune response is not always beneficial and can result in autoimmunity. Systemic lupus erythematosus (SLE) is an autoimmune disease where 85% of patients are women, yet the reason for this sex-based disparity is unknown (8, 9). The X chromosome is a critical factor for the breakdown of self-tolerance. Turner syndrome patients (XO) have a low risk of developing SLE (10), yet individuals suffering from Klinefelter’s syndrome (XXY) have 14-fold increased risk of developing SLE (11), suggesting that gene dosage from the X chromosome somehow influences SLE susceptibility.
Females select one X for chromosome-wide transcriptional silencing in a process called X chromosome inactivation (XCI), which equalizes the expression of X-linked genes between genders (12, 13). XCI first takes place during embryonic development, where one X is chosen at random for silencing. This process is initiated by the allele-specific expression of the long noncoding RNA XIST in humans (14) and Xist in mice (15). After XCI initiation, the inactive X (Xi) enters the maintenance phase where XIST/Xist RNA remains associated with the Xi after each cell division (16). The Xi becomes enriched with additional heterochromatic modifications (H3K27me3, macroH2A, H3K9me2/3, H4K20me1, ubiquitin-H2A) and DNA hypermethylation (17–21), which promote gene repression (13). Female mammals silence most X-linked genes with XCI, yet some genes escape silencing (22). Approximately 15% of human X-linked genes are biallelically expressed in hybrid fibroblasts (23), whereas 3% of the mouse Xi escapes silencing (24). The expression level of escapee genes from the Xi is usually lower than from the active X (Xa). Escape from XCI can also vary between individuals (which enhances phenotypic differences), among cells within a tissue (25), and also during development and aging. The number of genes exhibiting variable escape from XCI is small: In humans, 10–12% display variable escape (23, 26), and in mice approximately 18 genes escape (24).
Because XCI is a mechanism to equalize gene expression between the sexes, there should be equal levels of immunity-related proteins in female and male cells. However, some immunity-related X-linked genes exhibit sex-biased expression, and this variability may predispose females toward developing autoimmunity (27). Altered expression of X-linked genes is observed in female-biased autoimmune disorders and mouse models of autoimmunity (8), raising the provocative notion that reactivation of genes from the Xi leads to the overexpression of immunity-associated X-linked genes that contribute to disease. Consistently, female, but not male, SLE patient CD4+ T cells overexpress the X-linked genes CD40LG and CXCR3 and their promoter regions are demethylated, suggesting that these genes are not overexpressed from the Xa, but are instead expressed from a reactivated Xi (28, 29). Because increased dosage of immunity-related genes such as CD40LG and TLR7 are associated with SLE disease in human and mouse models, we investigated whether compromised maintenance of XCI in female lymphocytes from humans and mice could potentially contribute to predisposition toward autoimmunity. Notably, we demonstrate that fluctuations in the association of XIST/Xist RNA with the Xi may lead to partial reactivation, thereby increasing biallelic expression of autoimmune-associated genes.
Results
Female Lymphocytes Lack XIST RNA Transcripts on the Xi.
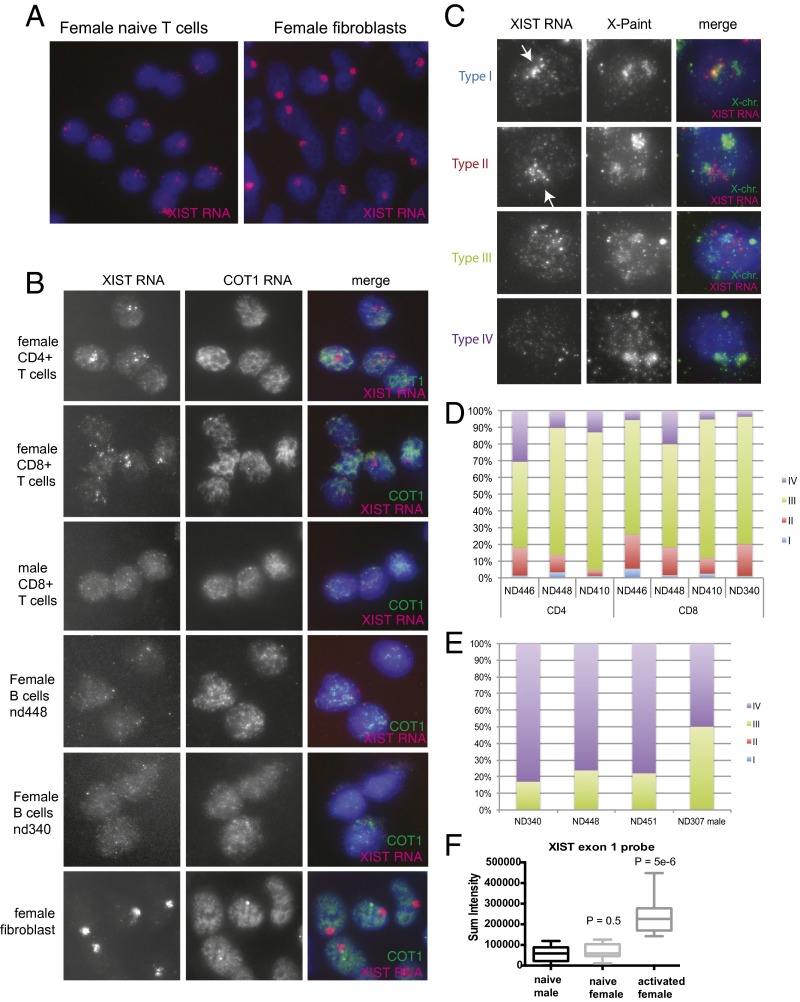
To determine whether noncanonical XCI could potentially contribute to the increased risk of SLE in females, we examined the epigenetic characteristics of the Xi chromosome in human female mature lymphocytes. First, we performed RNA fluorescence in situ hybridization (FISH) by using two types of XIST-specific probes (double-stranded exon 1 region and short oligos). XIST/Xist RNA is expressed from the Xi in somatic cells, and the transcripts remain tightly associated, forming a cloud-like structure that is visualized by RNA FISH (16, 30). Remarkably, bulk T cells from human females did not have the canonical XIST RNA cloud observed in other somatic cells (Fig. 1A). This result was consistent, regardless of T-cell purification method [fluorescence activated cell sorting (FACS) or negative selection with magnetic beads] or collection procedure (apheresis or traditional venipuncture). The majority of FACS sorted naïve CD4+ and CD8+ T cells and female naïve B cells also lacked XIST RNA clouds, resembling male T cells (Fig. 1B). Similar results were obtained by using single-molecule oligo probes for XIST (Fig. S1A, Left and B, Right). Mature naïve splenic T cells from female mice also lacked Xist RNA clouds (Fig. 2D, Top), in contrast with a previous study where Xist RNA signals were detected in 25–60% of CD4+ and CD8+ T cells (31).
Fig. 1.
Naïve human lymphocytes lack canonical XIST RNA clouds on the Xi. (A) RNA FISH analysis for XIST RNA and COT1 RNA, for naïve T cells and female fibroblast cell line (IMR-90). (B) XIST (red) and COT1 (green) RNA FISH for sorted mature naïve lymphocytes from human males and females. (C) Diversity of XIST RNA localization patterns (types I, II, III, IV) in naïve human lymphocytes. Sequential RNA FISH (for XIST RNA) followed by DNA FISH (to identify the two X chromosomes) at single-cell resolution. Arrows denote the inactive X chromosome. (D) Quantification of each type of XIST RNA localization pattern for naïve CD4+ and CD8+ T cells. (E) XIST RNA localization patterns for naïve B cells. (F) Quantification of total fluorescence for XIST RNA FISH using exon 1 probe for 12 nuclei for each cell type.
Fig. S1.
Quantification of XIST RNA transcripts in human T cells. (A) Representative field images of XIST RNA FISH using XIST oligo probes for human female T cells. (B) Quantification of total fluorescence and maximum intensities for naïve male and female T cells, and activated female T cells for RNA FISH performed using an exon 1 probe or XIST-specific oligos. Representative images of nuclei are shown, and below are distribution plots for 12 nuclei counted for each condition. P values were calculated comparing the naïve male cells to female cells (naïve or activated), and two-sided Welch t tests were used (which accounts for unequal variance).
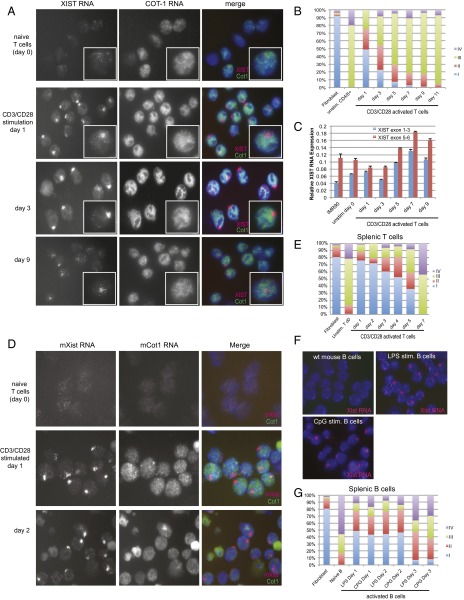
Fig. 2.
XIST/Xist RNA transcripts return to the Xi in activated T and B cells. (A) Representative XIST and COT1 RNA FISH images of human naïve T cells stimulated in vitro. Stimulation of five different individuals yielded similar results. (B) Quantification of each class of XIST RNA localization pattern (types I–IV) after stimulation for cells from the same individual. (C) qRT-PCR for XIST RNA in naïve and activated T cells using primer sets for both the 5′ and 3′ ends. (D) Xist RNA and Cot1 RNA FISH analyses of mouse naïve T and in vitro activated T cells. (E) Xist RNA localization in activated mouse T cells. (F) Xist RNA FISH using female mouse mature naïve B cells and activated B cells stimulated in vitro using lipopolysaccharaide (LPS) or CpG DNA (CpG). (G) Xist RNA localization patterns for activated mouse B cells.
We observed that female naïve lymphocytes have four distinct patterns of XIST RNA localization: Type I nuclei contain tightly clustered pinpoints localized with one X, similar to those in female fibroblasts; type II nuclei have diffuse pinpoints of XIST RNA that roughly encompass a nuclear area the size of one X; type III nuclei have dispersed pinpoints extending beyond the X chromosome territory; and type IV nuclei lack XIST RNA signal (Fig. 1C). We quantified the number of cells with each class of XIST RNA localization pattern and found that naïve T cells are mostly type III, followed by type II and IV patterns (Fig. 1D). Roughly 1–4% of human female naïve T cells had type I XIST RNA clouds. Next, we repeated RNA FISH by using single-molecule oligo probes specific for human XIST and found that naïve female T cells had greater fluorescence sum intensities (P = 0.05) and maximum intensities (P = 8.0e−6) compared with male T cells (Fig. S1B). We also quantified the numbers of each type of XIST RNA localization pattern for human female naïve B cells and observed a different distribution of XIST RNA localization pattern relative to T cells, irrespective of the collection method. Mature naïve B cells contain mostly type III and IV nuclei, and we never detected a type I XIST RNA cloud (Fig. 1E). In summary, naïve T and B cells are the first female somatic cells to our knowledge where the Xi is missing clustered XIST RNA transcripts.
XIST/Xist RNA Returns to the Xi upon Lymphocyte Activation.
Naïve lymphocytes are quiescent, and upon antigen recognition can become activated and reenter the cell cycle. We investigated the XIST RNA localization patterns in female T cells activated in vitro by using CD3/CD28 and performed a time course analysis of XIST RNA FISH following in vitro stimulation. We discovered that XIST RNA clouds return to the Xi 24 h after in vitro stimulation (Fig. 2A). The XIST clouds formed after 1–3 d of stimulation, and resembled clouds present in female fibroblasts (Fig. 1A), with the XIST RNA territory located in a COT-1 negative space within the nucleus (Fig. 2A). We observed similar RNA FISH results when using a double-stranded probe for XIST exon 1 or single-molecule oligo probes for XIST together with COT-1 (Fig. S1A). Activated female T cells exhibited greater nuclear fluorescence sum intensity and maximum intensity values compared with naïve female and male T cells, reflecting the presence of the XIST RNA transcript concentrated on the Xi (Fig. 1F and Fig. S1B). Because RNA FISH does not accurately reflect transcript absence, we used more sensitive quantification methods for XIST RNA (see below).
Because COT-1 RNA detects regions of nascent transcription within the nucleus (32–34), our observations of XIST clouds within COT-1 holes suggest that XIST localization promotes transcriptional silencing of the chromosome (Fig. 2A). Quiescent naïve T cells had a faint COT-1 signal, with speckled pinpoints distributed across the nucleus (Fig. 2A). Activated T cells had a greater overall COT-1 signal and defined nuclear distribution patterns for cells at days 1–3 following stimulation (Fig. 2A). Type I XIST RNA clouds decreased in activated T cells at day 5 following stimulation, with a concomitant increase in the number of cells with type III RNA pinpoints (Fig. S2 A and B). The percentages of type IV clouds, which lack XIST RNA signal, did not change after stimulation. In vitro stimulation resulted in a maximum of 30–50% of cells with type I XIST RNA clouds and 25–40% type II XIST pinpoints, and the distribution varied by individual (Fig. 2B and Fig. S2A). Type I cells disappeared by day 7, and COT-1 RNA patterns became diffuse by days 9–11 (Fig. S2B), consistent with increased apoptosis (visualized as nonspecific fluorescence and smaller DAPI nuclei).
Fig. S2.
Human XIST RNA FISH and FACS sorting of mouse T cells. (A) Quantification of each pattern of XIST RNA localization following in vitro stimulation (6 h, 1 d, 3 d, 5 d), using cells from the same female donor (ND407). (B) Representative XIST (red) and COT1 (green) RNA FISH images of human naïve T cells stimulated in vitro for 5, 7, or 11 d. Inset is an image of a nucleus. (C) FACS sorting of mature naïve T cells to quantify T-cell subsets, before in vitro stimulation, and used to profile Xist RNA expression in Fig. 2. Purified T cells are 93% pure (Left), of which 46% are CD4+ and 49% were CD8+. Approximately 10% of the CD4+ population is FoxP3+. Bulk T cells were also stained with Ki-67 (to determine proliferation), and 97% of cells were positive for this marker. (D) FACS sorting of activated T cells to determine the percentage of activated cells (39 h and 64 h after CD3/CD28 stimulation) determined by CD44 expression and CFSE labeling. At 39 h, 10–15% of the T cells have divided, but lack CD44 expression. At 64 h, 15–25% of the cells have divided and are CD44+.
We also examined mature naïve T and B cells isolated from female mouse spleens and found that, like human cells, these cells also lack canonical Xist RNA clouds (Fig. 2 D and F). Bulk female mouse T cells (typically 90% pure) contained similar amounts of helper T cells (CD4+) and cytotoxic T cells (CD8+), and roughly 10% of CD4+ cells were regulatory T cells (FoxP3+) (Fig. S2C). In vitro stimulation of mouse bulk T and B cells promoted the return of Xist RNA clouds to the Xi, with the greatest numbers of type I pattern observed 1 d after activation (Fig. 2 E and G). In vitro-activated mouse T cells had the most type I Xist RNA clouds (∼75%), even higher than human T cells where a maximum of 50% of cells contained the canonical XIST RNA cloud. We verified T-cell activation efficiency by using FACS to sort CD44+ cells (expressed on activated T cells) and the percentages of actively dividing cells using CFSE labeling (Fig. S2D). Type I Xist clouds persisted in activated mouse T cells for longer periods compared with activated human cells, yet by day 7, the cells reverted to type III and IV patterns and increased apoptotic cells (denoted by autofluorescence) (Fig. 2E and Fig. S3A). Next, we activated mature B cells in vitro two ways: either by adding lipopolysaccharide or CpG (35, 36). Type I Xist RNA clouds returned to the Xi following stimulation of B cells, similar to T-cell activation, irrespective of the method used for activation (Fig. 2 F and G). B cells stimulated for 1 d had the most type I Xist RNA clouds (45%), but never reached the levels in female mouse embryonic fibroblasts or T cells.
Fig. S3.
Xist RNA FISH analysis of activated mouse T cells and quantification of human XIST RNA expression during human T-cell stimulation. (A) Representative Xist (red) and COT1 (green) RNA FISH images of mouse T cells activated for 3, 4, and 5 d. (B) Human XIST RNA expression profiling in naïve CD4+ and CD8+ T cells and activated bulk T cells from the same donor (ND340). RNA samples were normalized for total RNA before cDNA synthesis. Two different female fibroblast cell lines are included as positive controls: fetal lung (IMR-90) and an adult fibroblast line (Coriell Institute; GM03956). Two different XIST intron-spanning primer sets were used (blue: exon 1 and exon 3; red: exon 5 and exon 6) and yielded similar results.
Because both human and mouse mature naïve T cells lack typical XIST/Xist RNA clouds, we investigated whether these cells express XIST/Xist. We used quantitative PCR (qPCR) to quantify the steady-state levels of human XIST RNA and mouse Xist RNA in naïve and activated lymphocytes. Remarkably, XIST/Xist RNA was abundant in naïve and activated T cells in human (Fig. 2C and Figs. S3B and S4A) and mouse (Fig. S4 C and D), and the levels were nearly equivalent to those found in female fibroblasts. We found that XIST/Xist was also highly expressed in naïve human (Fig. S4A) and mouse B cells (Fig. S4B), which had different XIST RNA localization patterns than naïve T cells. Because naïve T cells are quiescent and have reduced transcriptional activity, we repeated the quantitative RT-PCR (qRT-PCR) analyses normalizing for cell number. We also normalized XIST/Xist RNA expression by using a housekeeping gene (RPL13A), whose expression does not change with in vitro lymphocyte stimulation (37). We found that the levels of Xist RNA were similar for naïve and activated T cells, regardless of the method used for normalization (Fig. S4D). Similar to T cells, activated female mouse B cells also expressed equivalent levels of Xist RNA compared with naïve B cells (Fig. S4B). Northern blot analyses using equal amounts of RNA from mouse naïve and activated T cells support our qRT-PCR results that naïve female lymphocytes contain similar levels of Xist RNA transcripts as activated cells (Fig. S4E). We observed that Xist Northern blots of mouse T cells resembled Xist Northern blots of mouse embryonic stem cells and differentiating cells (38), with a multitude of high and low molecular weight bands of Xist transcripts. We found that female naïve T cells had a more intense signal of Xist RNA transcripts compared with activated T cells, but both samples had the same pattern of Xist RNA bands. We did not observe significant RNA degradation between naïve and activated female T-cell samples (Fig. S4E). Our results are in agreement with previous findings that XIST/Xist RNA transcription and transcript localization are independent processes (39, 40). We conclude that mammalian female naïve and activated lymphocytes have similar levels of XIST/Xist RNA transcripts to produce the canonical type I cloud, yet naïve lymphocytes are unable to properly localize these transcripts to the Xi.
Fig. S4.
XIST/Xist RNA quantification (using qPCR) in human and mouse lymphocytes. (A) Quantification of XIST RNA expression in female fibroblasts (fibro), and FACS-sorted CD4+, CD8+ T cells, and mature naïve B cells from human male and female donors. Two different XIST intron-spanning primer sets were used (blue: exon 1 and exon 3; red: exon 5 and exon 6) and yielded similar results. (B) Quantification of Xist RNA expression in splenic B cells after in vitro stimulation with LPS or CpG. Results from the same female mouse are shown; replicate experiments yielded similar results. (C) Quantification of Xist RNA transcripts in mature naïve T cells (mice) and 7 d after in vitro stimulation. (D) Comparison of different quantification methods to assess steady-state levels of mouse Xist RNA in naïve and activated (day 1) T cells. Equal amounts of total RNA (for cDNA synthesis) or total cell numbers were used. (E) Northern blot analysis of 20 μg of total RNA from female naïve and activated T cells hybridized using a probe for Xist (5′ end), and RNA Pol II was a loading control.
The Chromatin of the Xi Is More Euchromatic in Mammalian Lymphocytes.
Quiescent lymphocytes, which have a reduced transcriptional program, contain regions of facultative chromatin that become decondensed following activation (41). The mammalian Xi is enriched for histone H3K27me3, histone H2A ubiquitin (H2AUb), histone H4K20me1/3, histone H3K9me3, and the histone variant macroH2A (17, 20, 42–44). These marks form a focus that colocalizes with the Xi (and the XIST/Xist RNA cloud) when visualized cytologically in sequential RNA FISH and immunofluorescence (IF) experiments. Because naïve lymphocytes are quiescent and contain more heterochromatin than activated cells (45), they should theoretically exhibit enriched heterochromatic modifications on the Xi. However, because XIST RNA does not localize to the Xi in naïve lymphocytes, these modifications might also be missing. Therefore, we examined the localization of H2AUb, H3K27me3, macroH2A, or H4K20me1 with the Xi in mature naïve lymphocytes by using RNA FISH for detection of XIST/Xist RNA followed by IF for each modification, and finally DNA FISH to identify the two X chromosomes. Female fibroblasts were used as a positive control, and foci were observed overlapping XIST/Xist RNA signal for human and mouse cells. In human naïve T cells, we did not detect nuclear foci for H2AUb, H3K27me3, macroH2A, or H4K20me1 modifications that colocalized with an X chromosome (Fig. 3A). Mouse naïve T cells also lacked foci of H2AUb, H3K27me3, H3K9me3, and macroH2A that colocalized to the Xi (Fig. 3B). This observation is in agreement with a previous study showing that macroH2A is never detected over the Xi in female mouse mature lymphocytes (41). Although we detected abundant nuclear signal for all of these repressive modifications in both human and mouse T cells, we never observed a focus that overlapped with an X chromosome. We conclude that the mammalian Xi lacks enrichment of heterochromatic modifications in mature naïve lymphocytes.
Fig. 3.
The Xi has euchromatic features in mammalian lymphocytes. Sequential XIST RNA FISH, then immunofluorescence detection (and X-Paint to detect both X chromosomes) for naïve and activated T cells from humans (A) and mice (B). (C) Allele-specific expression (using RNA FISH) of CD40LG, CXCR3, and ATRX in activated human female T cells at single-cell resolution. White arrows indicate nascent transcripts from each X. (D) GSEA analysis comparing global gene expression differences with human female (seven samples) to male naïve B cells (two samples). Chromosome X map shows three regions on the X that have higher expression in female B cells. Heatmap lists the overexpressed genes in female naïve B cells from region chrX.q13 and fold-change (FC) values.
We repeated the serial RNA FISH/IF/DNA FISH analyses by using in vitro activated bulk T cells, and observed that some, but not all, heterochromatin marks returned to the Xi. Specifically, we found that H2AUb and H3K27me3 modifications formed foci in 70–80% of nuclei, and that these foci overlapped the type I XIST/Xist RNA cloud (Fig. 3 A and B). Remarkably, the return of these heterochromatic modifications to the Xi occurred in both human and mouse T cells. MacroH2A staining intensity was high throughout the nucleus of activated mouse and human T cells, yet focal enrichment on the Xi was observed for only 12–15% of nuclei with type I XIST/Xist patterns (Fig. 3 A and B). In activated human T cells, we observed the most pronounced macroH2A foci in type II nuclei with diffuse XIST RNA pinpoints (Fig. 3A). H4K20me1/3 modifications returned to the Xi in roughly 50% of human T cells (Fig. 3A). We conclude that the chromatin of the Xi in female T cells is different compared with fibroblasts, and that mature naïve cells have a more euchromatic Xi because it is missing H3K27me3, H4K20me, macroH2A, and H2AUb enrichment.
Female Lymphocytes Exhibit Partial X-Reactivation Resulting in Increased Expression of Immunity-Related X-Linked Genes.
We examined how this unusual chromatin affects expression of two X-linked genes, CD40LG and CXCR3, frequently overexpressed in the autoimmune disorder SLE. Our hypothesis was that the euchromatic features of the Xi would result in expression of genes from the Xi in naïve and stimulated human T cells. We used sequential RNA then DNA FISH to detect expression of three X-linked genes subject to XCI (CD40LG, CXCR3, and ATRX). To identify the location of the Xi, we used a probe for XIST RNA to identify the Xi (for RNA FISH). Lastly, we denatured the slides for DNA FISH, which detected the location of the two X chromosomes. We quantified the percentages of cells expressing these genes from just the Xa or both Xs by counting the number of cells containing one pinpoint (monoallelic) or two pinpoints (biallelic) that overlapped with an X chromosome. We examined mature naïve and in vitro stimulated T cells from healthy human female and male donors, which are a positive control for monoallelic expression. We found that human naïve T cells had dimmer (but still detectable) RNA FISH signals compared with day 3 stimulated cells (Fig. S5). Analysis of CD40LG, an X-linked gene overexpressed in CD4+ T cells from female SLE patients (23, 26, 28), revealed the presence of mostly monoallelic (96% with one pinpoint) and some biallelic expression (4% with two pinpoints), in activated female T cells (Fig. 3C). We were unable to detect a clear CD40LG nascent transcription signal in naïve female T cells, despite repeated efforts with cells from different female donors. Male T cells were exclusively monoallelic for CD40LG expression (Fig. S5A), as expected. Next, we repeated the analysis by using a probe specific for CXRC3, another X-linked gene subject to XCI that is frequently overexpressed in female SLE CD4+ T cells (23, 26, 28). Again, we detected some naïve (5%) and stimulated T cells (3%) with biallelic expression of CXCR3 (Fig. 3C and Fig. S5B). As a control, we examined an X-linked gene not involved in immune function, ATRX, which is known to undergo XCI in female cells (23, 26, 46). We found that ATRX is expressed in lymphocytes, and is predominantly monoallelic in naïve (99%) and activated (95%) female T cells, yet we also detected some biallelic-expressing cells (5%) present in activated T cells (Fig. 3C). Male cells only contained monoallelic-expressing cells for ATRX (100%) (Fig. S5A), similar to other X-linked genes examined. We conclude that the euchromatic nature of the Xi in female lymphocytes predisposes X-linked genes to become reactivated and biallelically expressed.
Fig. S5.
Allele-specific expression of CXCR3, CD40LG, and ATRX in naïve human male and female T cells using RNA FISH. (A) Representative RNA FISH images and counts of monoallelic and biallelic expression for each X-linked gene. Signals overlapping an X chromosome were counted. White arrow denotes nascent transcription from the X chromosome. XIST RNA (green) was used as a negative control. (B) Same as A, but for female naïve cells. Counts for female naïve T cells are shown in Fig. 3. (C) Comparison of differentially expressed genes from female and male fetal lung fibroblasts. The NES and FDR q values are shown for chrX q13 region, which had the greatest expression difference between samples. The heatmap lists the overexpressed genes in SLE samples from region chrX.q13 and FC values for the genes enriched at the leading edge for female samples.
Next, we investigated how the euchromatic-like features of the Xi correlate with X-linked gene expression chromosome-wide in lymphocytes. We tested the hypothesis that if there is partial reactivation from the Xi in female lymphocytes, we would expect that healthy female cells, with an Xa and Xi, would have higher expression of X-linked genes compared with healthy male cells containing one Xa. We used gene set enrichment analysis (GSEA) to query sex-specific differences in gene expression by using an unbiased approach, examining published microarray datasets from human male and female mature naïve B cells (GSE30153). We found that there are three regions on the X chromosome that are overexpressed in female B cells compared with male B cells: p11, p22, and q13 (Fig. 3D). Region chrX.q13, containing 21 genes, was the most significantly overexpressed genomic region in female B cells [normalized enrichment score (NES) = 2.25; false discovery rate (FDR) q = 0.003] compared with male cells (Fig. 3D). This region contains the XIST gene, exclusively expressed in females, and we observed the same sex-biased expression in B cells, serving as a positive control for our analysis (Fig. 3D). We identified a gene signature from chrX.q13 of overexpressed genes specific to female B cells, listed in Fig. 3D, that are enriched at the leading edge (red region of the x axis) of the enrichment score plot. Importantly, two of these genes, ITM2A and CXCR3, are immunity-related and CXCR3 is associated with autoimmunity (28, 47). The p arm of the X chromosome has more genes known to escape XCI compared with the q arm, explaining why genes from these two regions are expressed at higher levels in females compared with males. A number of genes known to escape XCI, including EIF1AX, EIF2S3, OFD1, and ZFX (23), are located within chrX.p22 and are enriched in female B cells. We found that the immunity-related genes IL3FA and FOXP3, located within regions chrX.p22 and chrX.p11, are overexpressed in female cells and are enriched at the leading edge of the enrichment score plots for female B cells. There is no evidence to date that IL3FA and FOXP3 escape XCI in humans (23, 26). To test for the significance of immunity-related genes in sex-specific cell comparisons, we repeated the unbiased GSEA by using human fetal lung fibroblasts. We found that chrX.q13 was the most differentially expressed region in lung fibroblasts (NES = 1.52; FDR q = 0.05) similar to GSEA results using human B cells (Fig. S5C). As expected, XIST was the most differentially expressed gene that distinguishes male and female lung fibroblasts (fold change = 127). However, the other significantly enriched genes (ATP7A, P2RY4, FOXO4, SLC16A2) are not immunity-related and are known to escape from XCI (23, 26). We conclude that human female B cells contain regions along the X that are expressed at higher levels compared with male cells, and these regions contain immunity-related genes. These results are consistent with the euchromatic nature of the Xi in mammalian lymphocytes that could facilitate higher expression in lymphocytes.
YY1 and hnRNPU Proteins Localize XIST/Xist RNA Transcripts to the Xi in Activated T and B Cells from Humans and Mice.
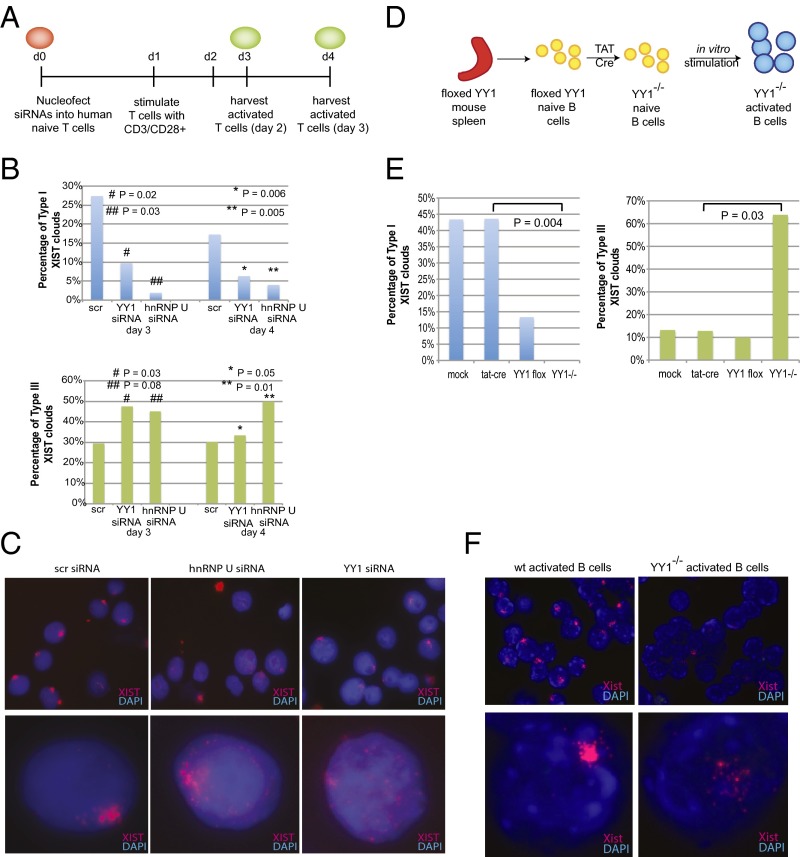
Naïve T and B cells express abundant levels of XIST/Xist RNA, but these transcripts do not associate with the Xi until these cells become activated. Therefore, we searched for candidate proteins known to bind XIST RNA that could function to recruit XIST RNA back to the Xi after lymphocyte stimulation. The nuclear scaffold protein SAF-A/hnRNPU is enriched on the Xi and is required for localizing Xist RNA to the Xi in embryonic stem cells during the initiation of XCI (48, 49). The transcription factor YY1 also tethers Xist RNA to the X during the initiation and maintenance stages of XCI, and activates Xist/XIST transcription (50, 51). Importantly, knockdown of hnRNPU or YY1 disrupts Xist RNA localization in post-XCI fibroblasts, resulting in a scattered pinpoint pattern similar to types II and III in lymphocytes. We found that YY1 and hnRNPU RNA and protein are expressed in naïve human female T cells, and that in vitro activation doubled transcriptional expression (Fig. S6A) and increased protein levels (Fig. S6B). Using siRNAs specific for YY1 or hnRNPU, we disrupted expression of these proteins in human mature naïve T cells, then stimulated the cells for 2–3 d. We verified knockdown efficiency for YY1 and hnRNPU in naïve and activated T cells by using qPCR and immunofluorescence (Fig. S6D). We found that hnRNPU or YY1 knockdown significantly reduced the number of canonical type I XIST RNA clouds from 25 to 10% (YY1 KD; P = 0.02) and 3% (hnRNPU KD; P = 0.03) (Fig. 4B). The percentage of activated human T cells with a type III XIST pattern increased from 30 to 47% (YY1 KD; P = 0.03) and 45% (hnRNPU KD; P = 0.08) (Fig. 4B), similar to what is observed when these proteins are disrupted in differentiating female mouse embryonic stem cells or embryonic fibroblasts (49, 50). Reduction of YY1 or hnRNPU in naïve or activated T cells did not affect XIST RNA transcript levels (Fig. S7B), further supporting a role for these proteins in localizing XIST RNA to the Xi rather than affecting XIST transcription.
Fig. S6.
YY1 and hnRNPU expression in female lymphocytes increases with in vitro stimulation. (A) Quantification of hnRNPU and YY1 expression in sorted human CD4+ (unstimulated CD4 T), CD8+ (unstimulated CD8 T) naïve T cells, and during in vitro stimulation of bulk T cells over time. Statistical significance was determined by using Student’s t test, comparing naïve to activated cells (days 3–9). (B) Immunofluorescence detection of hnRNPU and YY1 in naïve and activated human T-cell nuclei. The same exposure time was used for FITC and DAPI channels for imaging naïve and activated T cells. (C) Western blot analysis of YY1 protein levels in wild-type female mouse B cells, naïve and in vitro activated, and YY1 floxed B cells that were activated then infected with TAT-Cre (to delete YY1). Lamin B1 was used as a loading control. (D) Knockdown efficiency of YY1 and hnRNPU in activated human T cells, determined using qPCR and IF. Day 3 samples are naïve human T cells; day 4 samples are activated cells, which were stimulated for 3 d after knockdown. For IF, T cells were nucleofected with siRNAs on the first day, then stimulated for 3 d, and harvested. The experiment was repeated five times by using five different donor samples. IF images were captured by using equal exposure times.
Fig. 4.
YY1 and hnRNPU localize XIST/Xist RNA to the Xi in stimulated lymphocytes. (A) Experimental design for the knockdown experiments using human T cells. (B) Average percentages (for five experiments) quantifying type I and III XIST RNA patterns after YY1 or hnRNPU knockdown. Statistical significance calculated using Student’s t test. (C) Representative XIST RNA images for activated T cells treated with scrambled siRNA (scr), siRNAs against hnRNPU, and siRNAs against YY1. Nuclear distribution of XIST RNA transcripts for each condition are shown below. (D) Experimental design for YY1 deletion in mouse B cells. (E) Quantification of type I and type III Xist RNA patterns in wild-type and YY1−/− activated B cells. Statistical significance calculated for averages from two independent experiments using Student’s t test. (F) Representative Xist RNA images for wild-type and YY1−/− activated B cells.
Fig. S7.
Expression of X-linked genes in human B-cell lines (healthy female and female SLE patients) and sex-specific fetal lung fibroblasts. (A) Relative expression of XIST RNA in SLE patient B-cell lines (2411, 9383, 2190, 6109) and two age-matched healthy control cell lines (6141, 2747) determined using qPCR. (B) qRT-PCR analysis of XIST RNA levels after hnRNP U and YY1 knockdown in naïve and activated female T cells.
In complementary experiments, we asked whether YY1 deletion would affect Xist RNA localization to the Xi in activated B cells from mice. Similar to human T cells, YY1 levels increased upon activation in female mouse B cells (Fig. S6C). We isolated mature naïve B cells from mice containing loxP sites flanking exon 1 of YY1 (9, 52), then deleted YY1 by introducing TAT-Cre (53) and then stimulated with lipopolysaccharide treatment (Fig. 4D). This method eliminates 85–100% of YY1 expression (Fig. S6C) (9), depending on the efficiency of the TAT-Cre transfection into B cells, and is more effective at reducing YY1 levels than siRNAs. YY1 depletion significantly reduced type I RNA clouds from 43 to 0–5% (P = 0.004) in replicate experiments. As expected, type III Xist patterns increased with YY1 deletion from 11 to 65% (Fig. 4 E and F). In conclusion, the known Xist RNA binding proteins YY1 and hnRNPU also function in activated lymphocytes to recruit XIST RNA back to the Xi in activated T and B cells.
SLE Patient B Cells Have Altered Distributions of XIST RNA Localization Patterns and Biallelic Expression of Immunity-Related X-Linked Genes.
We next tested whether XIST RNA association to the Xi is further perturbed in SLE-derived lymphocytes. Using RNA FISH, we determined the percentage for each XIST RNA localization pattern (types I–IV) in immortalized B-cell lines derived from pediatric SLE patients and age-matched healthy controls (Fig. 5A). We did not observe a significant difference with the canonical type I XIST RNA clouds between SLE and control samples. However, SLE B cells had more type II patterns of XIST RNA localization relative to controls (P = 0.04). The most significant difference between SLE and control B cells was with the type III pattern, where 50% of the control cells were type III and SLE B cells had 10–30% type III cells (P = 0.006). We found that SLE B cells had more cells missing XIST RNA transcript accumulation within the nucleus (the type IV pattern) compared with normal B cells (P = 0.05). SLE B cells also had more type II patterns of XIST RNA localization relative to controls (P = 0.04). XIST RNA transcript levels were similar between normal and SLE B-cell lines (Fig. S7A), indicating that XIST RNA is not limiting in SLE cells. We also examined the levels of YY1 and hnRNPU transcripts and proteins in SLE B cells, and found similar expression for SLE and control lines (Fig. S8 B and C). Thus, XIST RNA exhibits abnormal localization patterns in SLE B cells, suggesting that X-silencing mechanisms may be different in SLE.
Fig. 5.
SLE patient B cells have different XIST RNA patterns and greater biallelic expression of immunity-related X-linked genes. (A) XIST (red) and COT1 (green) RNA FISH field images for immortalized B-cell lines from a pediatric SLE patient and a healthy age-matched control. Quantification of XIST RNA localization patterns for SLE B-cell lines and healthy controls. (B) RNA FISH analyses at single-cell resolution for allele-specific expression of CD40LG, CXCR3, and TRL7 in SLE patient and healthy control B-cell lines. (C) qRT-PCR analysis of CD40LG, CXCR3, and TRL7 in SLE and normal B-cell lines. P values were calculated by using Student’s t test. (D) GSEA comparing gene expression differences from the X (chrX.q13) in human female SLE naïve B cells (during inactive disease; 15 samples) to healthy female B cells (7 samples).
Fig. S8.
Gene expression differences between SLE patient B cells and healthy controls. (A) GSEA analysis comparing naïve B cells from female SLE patients and healthy controls. The heatmap lists the overexpressed genes in female SLE patients from the chrX.q13 region. FC values for the genes exhibiting greatest expression difference are shown. (B) qRT-PCR quantification of steady-state levels for YY1 and hnRNP U transcripts in SLE patient B-cell lines (same cell lines from Fig. 5). (C) Western blot analysis for YY1 and hnRNP U protein in B-cell lines from female SLE patients and healthy controls.
Next, we investigated whether these differences with XIST RNA localization were correlated with differences with allelic expression of X-linked immunity-related genes. We used RNA FISH to examine nascent transcription of three X-linked immunity-related genes typically overexpressed in lymphocytes from SLE patients: CD40LG, CXCR3, and TRL7. We determined the number of cells with either monoallelic (one pinpoint) or biallelic expression (two pinpoints), using immortalized B-cell lines generated from pediatric SLE patients or healthy female age-matched samples. We found that normal B-cell lines had mostly monoallelic expression of CD40LG (36–47% of cells), yet we also detected some biallelic-expressing B cells (4–20%; Fig. 5B), similar to our observations in activated T cells (Fig. 3C). CD40LG is normally overexpressed in SLE patient T cells, but is also ectopically expressed in SLE patient B cells (54). B-cell lines from three different pediatric SLE patients had slightly more biallelic-expressing SLE B cells for CD40LG (17–33%; P = 0.09) compared with healthy control lines (Fig. 5B). We found that SLE B-cell lines also had fewer cells with monoallelic expression of CD40LG (22–29%; P = 0.002) compared with healthy controls. Consistent with a trend toward more biallelic cells, SLE B cells also expressed more CD40LG transcripts compared with healthy controls (P = 0.033). In contrast to CD40LG, we saw no difference for monoallelic and biallelic expression of CXCR3 (Fig. 5B). We also examined the expression of TRL7, which is overexpressed in SLE patient B cells (55). We detected similar levels of biallelic expression of TLR7 in both normal (12–14%) and SLE B cells (9–23%), yet SLE lines had more TLR7 transcripts than normal lines (Fig. 5 B and C; P = 5.6e−6). In conclusion, biallelic expression of X-linked immunity-related genes was observed in both normal and SLE patient B cells, and the increased expression from the Xi could result from atypical XIST RNA localization patterns.
Lastly, we investigated whether altered distributions of XIST RNA patterns in SLE patients could reflect expression differences with X-linked genes. Because we found that a chrX.q13 gene signature distinguished male and female B cells (Fig. 3D), we examined whether this region is overexpressed in female SLE patients compared with healthy female controls. We used GSEA to query which regions of the X chromosome were overexpressed in female SLE B cells compared with healthy female B cells (GSE30153). As expected, we found that chrX.q13 had the greatest enrichment of overexpressed X-linked genes in SLE female B-cell samples (Fig. 5D). Six genes within this region, KIF4A, OGT, HMGN5, NONO, CXCR3, and ITM2A, were significantly enriched at the leading edge (red area of x axis) for the enrichment score plot (Fig. S8A). Importantly, both OGT and CXCR3 are overexpressed in female SLE patient lymphocytes (28), consistent with our results. Our findings suggest the intriguing hypothesis that altered XIST RNA localization to the Xi promotes higher expression of immunity-related X-linked genes from chrX.q13.
Discussion
Here, we have demonstrated that female lymphocytes in humans and mice do not maintain XCI with the same fidelity as other somatic cells. Specifically, whereas XIST/Xist RNA is thought to remain localized to the Xi in all somatic cells, mature naïve lymphocytes lack XIST/Xist RNA clouds. This study is the first, to our knowledge, to classify differences with XIST/Xist RNA localization patterns in lymphocytes. The molecular mechanisms responsible for these four types of XIST RNA patterns, and the significance of type II and III cells, are unknown at this time and warrant further investigation. Also unclear is how these XIST/Xist RNA patterns correlate with SLE disease severity, and whether other autoimmune disorders involving T and B cells also exhibit different localization. Moreover, we found that the Xi in naïve lymphocytes lacks enrichment of H3K27me3, H4K20me1/3, H2AUb, and macroH2A, all hallmarks of heterochromatin. The absence of these modifications is consistent with observed increased expression of immunity-related X-linked genes in human naïve B cells, and the presence of some T cells with biallelic expression of CXCR3. These euchromatic features of the lymphocyte Xi are in agreement with a recent study profiling the open regions of chromatin by using the Assay of Transposase Accessible Chromatin (ATAC) Seq comparing female and male CD4+ T cells (27). Qu et al. also found that females have more accessible elements on the X compared with male cells. Our study also observed that some activated T (Fig. 3C) and B cells (Fig. 5B) have biallelic expression of X-linked immunity-related genes. We conclude that this unusual maintenance of XCI in naïve and activated lymphocytes predisposes portions of the Xi to become reactivated and increase expression of X-linked genes, providing a potential mechanistic explanation for why female mammals and individuals with multiple X’s are more susceptible to autoimmunity.
The biological reason for XIST/Xist RNA localization changes in female lymphocytes is unknown. One possibility is that female mammals have evolved this mechanism to relax Xi silencing to increase the expression of immunity-related genes that may be beneficial for fighting infection. Lymphocyte gene expression from the Xi could be caused by escape from XCI or gene reactivation. Although 10% of human X-linked genes exhibit variable escape among individuals and in tissues (23, 26), it is unlikely that X-linked autoimmunity-related genes are escaping X silencing in lymphocytes. Escape from XCI has been profiled in human fibroblasts and peripheral blood mononuclear cells, and CD40LG and CXCR3 were not expressed from the Xi in any of these cell types (26). Our observations of T cells with biallelic expression suggests that the origin is likely due to gene-specific reactivation in a subpopulation of female lymphocytes, which results in increased expression. We speculate that immunity-related genes such as CD40LG and CXCR3 might be more susceptible to reactivation from the Xi in lymphocytes because of the euchromatic chromatin of the Xi. Another possibility is that immunity-related genes on chrX.q13, the region containing the XIST gene, can become reactivated and expressed from the Xi because XIST RNA is not localized to the Xi in naïve lymphocytes (Fig. 1). Support for this hypothesis is the observation that human female pluripotent stem cells lacking XIST RNA reactivate regions of the Xi and overexpress X-linked genes (56, 57).
Whereas the majority of naive human and mouse peripheral T cells lacked XIST/Xist RNA association with the Xi, a minority (∼1–5%) contained type I XIST RNA clouds. These few cells could either represent a specific subset of T cells where XIST remains localized to the Xi, or alternatively could represent circulating antigen-stimulated T cells. Our data contrasts with a previous study by Savarese et al. in which the absence of Xist RNA clouds and H3K27me3 was noted in immature lymphocytes, but Xist RNA clouds were observed in ∼50% in mature T and B cells (31). This discrepancy could be due to activation of an immune response in the animals from the Savarese et al. study before cell isolation. Savarese et al. also examined whether genes on the Xi were reactivated in immature lymphocytes by using F1 hybrid mice carrying an Xist deletion on the maternal allele, where reactivated genes would be expressed from the paternal allele. The authors examined four X-linked genes, Xist, G6pd, Pctk1, and Pgk1, and did not observe any expression from the paternal X by using allele-specific RT-PCR. However, this study did not examine the expression of X-linked immunity-related genes in lymphocytes from these F1 hybrid animals, which may be expressed at higher levels.
Recent work has identified new Xist RNA binding proteins that function in the initiation or maintenance of XCI in mice (58–60). YY1 and hnRNPU are known Xist RNA binding proteins that localize the RNA to the Xi during the initiation of XCI (YY1) and maintenance phase (hnRNPU) (49, 50). YY1 can also recruit a variety of proteins to DNA, most notably polycomb repressive complex 1 (PRC1) and PRC2 (61–63). Importantly, these complexes are involved in initiating XCI during early development (43, 64–66). Our studies have identified a previously unidentified localization function for these proteins: the return of XIST/Xist RNA to the Xi during the transition from quiescence to activation in female lymphocytes. The YY1 knockout and knockdown studies indicate that this protein is required for the early stages of Xist/XIST cloud formation during lymphocyte activation, suggesting that XIST RNA return to the Xi precedes PRC2 deposition of H3K27me3. Importantly, these observations provide a testable model system to investigate the mechanisms of Xist/XIST RNA cloud formation in both mouse and human lymphocytes. Naïve lymphocytes express low levels of YY1 and hnRNPU, which may explain why the XIST/Xist RNA transcript does not remain associated with the Xi in these cells. Activated lymphocytes contain greater amounts of these proteins, probably due to genome-wide transcriptional activation that occurs together with increased cell proliferation as naïve lymphocytes exit quiescence. Additional experiments are necessary to determine how the nuclear organization context (quiescent versus activated) and the concentration of Xist binding proteins influences Xist RNA recruitment back to the Xi. With the recent findings that Xist RNA physically interacts with 80–250 proteins (60), it is likely that additional proteins will be required to localize XIST/Xist back to the Xi.
The only human female cells previously shown to lack XIST RNA clouds are a predominant subset (class III) of female pluripotent stem cells (hPSCs) with an “eroded” Xi (56, 57, 67). Female hPSCs are epigenetically unstable for XCI, and these cells will irreversibly silence the XIST gene during routine culture. Class III hPSCs also lack cytologically visible enrichment of H3K27me3, H4K20me, macroH2A, and H2A-ub on the Xi (46, 68, 69), resulting in a partially reactivated Xi. These cells overexpress a variable set of X-linked genes (56, 57), indicating that loss of XIST RNA doesn’t always reactivate the same genes or regions in hPSCs. However, gene reactivation from the Xi in the absence of XIST/Xist RNA may be more uniform in human and mouse lymphocytes. Deletion of Xist in the hematopoietic cell lineage partially reactivates the Xi, resulting in increased expression of ∼86 X-linked genes in blood cells, some which are involved in hematopoiesis and cell cycle regulation (70). Interestingly, this list includes two immunity genes CXCR3 and TRL7 whose overexpression is associated with lupus (71–73), which suggests that immunity-related genes are somehow poised for reactivation in the blood lineage when Xist RNA is missing. Future work is required to determine the specificity of X-linked gene reactivation when Xist is deleted, and how partial reactivation of Xi changes depending on the cell type.
Materials and Methods
Mammalian Cell Isolation and Lines.
Human naïve lymphocytes (all deidentified) were collected by the University of Pennsylvania Pathology BioResouce Human Immunology Core facility. Immortalized B-cell lines [generated by infection with Epstein–Barr virus (EBV)] from five pediatric SLE patients and three healthy females (all deidentified) were derived by Hakon Hakonerson’s group at the Children’s Hospital of Philadelphia and approved by the Children's Hospital of Philadelphia Institutional Review Board (IRB). Patients and their families were recruited through the Children’s Hospital of Philadelphia clinic or CHOP outreach clinics. Written informed consent was obtained from the participants or their parents by using IRB-approved consent forms before enrollment in the project. In vitro activation of lymphocytes is described in SI Materials and Methods. Animal experiments were approved by the University of Pennsylvania Institutional Animal Care and Use Committee.
RNA FISH, DNA FISH, and Immunofluorescence.
RNA and DNA FISH were carried out as described (56, 67, 74). Human XIST probe (exon 1) and mouse Xist probe (Sx9) were labeled by nick translation with Cy3‐dUTP, and COT‐1 DNA was labeled with fluoroscein‐12‐dUTP. Three oligo probes (20 nt in length) specific for XIST were designed to recognize regions within exon 1, and labeled with one Cy3 molecule at the 5′ end (IDT). StarFISH X chromosome paints (Cambio) for DNA FISH were hybridized per manufacturer’s instructions.
SI Discussion
Our study also found that female SLE patient B cells exhibited different monoallelic and biallelic expression patterns for CD40LG, CXCR3, and TRL7. CD40LG is expressed by T cells, and B cells express low levels of this transcript, as observed in Fig. 5. However, we found that SLE patient B cells had greater percentages of biallelic expression of CD40LG and higher steady-state levels of this transcript compared with healthy controls. There are different mechanisms that could result in overexpression of CD40LG, CXCR3, and TRL7 in SLE lymphocytes. One possibility is partial X reactivation, where particular genes or regions on the X are not rigorously silenced and can become reactivated in lymphocytes. A second possibility is that Xi of female SLE patients can lose DNA methylation, causing X-linked genes to become expressed at higher levels. Recent work from Hewagama et al. found that the X-linked genes CD40LG, CXCR3, and OGT become overexpressed in healthy female (but not male) CD4+ T cells after treatment with the DNA methyltransferase inhibitor (28). Drugs known to induce lupus-like symptoms in mice, such as cainamide and hydralazine, inhibit DNA methylation in T cells (75). An alternative mechanism for increased expression of X-linked genes in female SLE patients is gene duplication of X-linked immunity related genes. For example, the BXSB lupus mouse model contains a translocated region of the X chromosome encompassing the TLR7 gene on the Y chromosome, resulting in overexpression of TLR7 in male animals that develop severe murine lupus (76). To our knowledge, X-linked gene duplication events in female SLE patients have not been examined.
Using methods that quantify cellular steady-state RNA levels (qPCR and Northern blot analysis), we observed that mammalian female naïve and activated lymphocytes have similar levels of Xist RNA but different nuclear localization patterns, visualized using RNA FISH. We can eliminate the possibility that our cDNA and oligo probes have reduced accessibility in female lymphocytes because we detect dispersed pinpoints of XIST/Xist RNA in some naïve cells and also activated cells (Figs. 1 and 2). Additional evidence that RNA accessibility does not account for the different XIST/Xist RNA localization patterns in female lymphocytes is that YY1 disruption generates the same diffuse pinpoint patterns of type III and type IV nuclei (Fig. 4 C and F). Previous work using locked nucleic acids (LNAs) and peptide nucleic acids (PNAs) indicates that Xist transcription and Xist RNA localization are not coupled, but independent from each other. Xist-specific LNAs and PNAs can displace Xist RNA away from the Xi, resulting in type III and IV clouds in RNA FISH analyses without changing steady-state levels of Xist (39, 40). These studies concluded that there are differences between Xist RNA expression and proper localization of Xist RNA transcripts to the inactive X. Our results in female lymphocytes are the first physiological example demonstrating that XIST/Xist transcription and its localization are distinct processes. The biological basis for the Xi lacking XIST/Xist RNA localization is not known at this time. We speculate that XIST/Xist binding proteins may be limiting in naïve cells (such as YY1 in Fig. S6), preventing accumulation on the Xi. Naïve lymphocytes are more heterochromatic compared with activated cells (41), which may inhibit the accessibility of XIST/Xist RNA binding proteins to keep the transcripts tethered to the Xi. Additional experimentation is required to determine how XIST/Xist RNA is prevented from accumulation on the Xi in naïve lymphocytes.
SI Materials and Methods
Lymphocyte Activation.
To stimulate human naïve T cells, we used Dynabeads Human T-Activator CD3/CD28 for T-Cell Expansion and Activation from Gibco (11161D); mouse cells were stimulated by using the Dynabeads Mouse T-activator CD3/CD28 kit (11456D). Beads were added 1:1 to the number of human naïve T cells, and then cultured by using T-cell medium (10% BSA, 1% Hepes, 1% penicillin streptomycin in DMEM16) for 1–10 d. Mouse T lymphocytes were isolated from pooled spleen and lymph nodes by using mouse CD3+ T-cell enrichment columns (R&D Systems). Cells were cultured in RPMI-1640 containing 10% FCS, 0.1% β-mercaptoethanol, 1% nonessential amino acids, 1% sodium pyruvate, and 100 U/mL IL-2. Mouse T cells were stimulated with 1 μg/mL plate-bound anti-CD3 and 1 μg/mL soluble anti-CD28, then cultured for 1–7 d. Mouse CD23+ splenic B cells were isolated from C57BL/6 female mice (8–12 mo old) by using negative selection via magnetic separation LS columns (Miltenyi Biotech). B cells were cultured in RPMI containing 10% FBS, 1% nonessential amino acids, 1% OPI medium supplement Hybri-Max, and 0.1% β-mercaptoethanol, and were activated with 5 μg/mL LPS.
siRNA Transfection and Cre-Mediated Deletion of YY1 and hnRNPU.
Human naïve T cells were transfected by using the Human T-Cell Nucleofector Kit from Lonza (VPA-1002), according to the manufacturer’s protocol. YY1 siRNA was from Ambion (AM16708). hnRNPU siRNA (L-013501-00-0005) and scramble control siRNA (D-001810-10-05) were from Dharmacon. Naïve human T cells were nucleofected with siRNAs immediately following purification (day 0), then stimulated 24 h later (day 1). Activated cells were harvested for RNA FISH and RNA 2 and 3 d after stimulation, corresponding to days 3 and 4 on the experimental timeline (Fig. 4A). For checking knockdown efficiency of YY1 and hnRNP U in naïve and stimulated cells, naïve T cells were harvested for RNA 48 h after nucleofection (day 2), and stimulated cells were harvested after 3 d of stimulation. To generate yy1Δ/Δ lymphocytes, splenic B from YY1-floxed female mice (49) were incubated with recombinant TAT-Cre protein (100 μg/10 × 106 cells) in OptiMEM before stimulation (9).
Quantitative RT-PCR and Northern Blots.
RNA was isolated from cells by using TRIzol reagent (Invitrogen), and cDNA was synthesized from the RNA by using qScript cDNA SuperMix (Quanta), according to the manufacturer's instructions. The cDNA samples were analyzed by using Power SYBR Green (Applied Biosystems) on the QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems), and PCR was performed by using these primers: hXISTex1 F: 5′-TTGCCCTACTAGCTCCTCGGAC, hXIST ex3 R: 5′-TTCTCCAGATAGCTGGCAACC, hXIST ex5 F: 5′- CTACAAGCAGTGCAGAGAGC, hXIST ex7 R: 5′- CTAAGACAAGAC ACAGACCAC; RPL13A F: 5′-GCCATCGTGGCTAAACAGGTA, R: 5′-GCCATCGTG GCTAAACAGGTA; CD40LG F: 5′-ACATACAACCAAACTTCTCCCCG, R: 5′-GCAAAA ATGTCTGACCCAATCA; CXCR3 F: 5′-ACATAGTTCATGCCACCCAGCTCT R: 5′-AAGATGAAGTCTGGGAGGGCGAAA; TLR7 F: 5′-CTAAAGACCCAGCTGTGACCAG, R: 5′-CCAGTCCCTTTCCTCGAGACAT. RPL13A was used as a housekeeping reference gene for calculating the relative expression level of target genes. The results are displayed as relative expression levels normalized to RPL13A (ΔΔCt). For experiments comparing mouse Xist RNA levels in naïve and activated T cells comparing different normalization methods (total RNA or cell number), cDNA synthesis was performed by using 1 μg of total RNA or 1 million cells each. The cDNA was diluted 1/3 and then used for Xist PCR amplification. The Ct values for Xist for each condition were normalized to RPL13A Ct (ΔΔCt) for relative expression. Northern blots were performed in four independent experiments, using 20 μg of total RNA for each cell type (female mouse naïve and activated T cells). A probe for the 5′ end of Xist (500 nt within exon 1) was used to probe for Xist RNA. An RNA Polymerase II probe was used as a loading control.
Sequential RNA/DNA FISH.
The RNA FISH was performed first, and 0.2-μm z‐section images were taken, and the x–y coordinates were marked. Slides were fixed again by using 4% PFA, then IF was performed by incubating the slide with 0.05% Trition X‐100, then blocking with 4% goat serum, and incubation with the primary antibody: H3K27me3 (Active Motif), macroH2A (Abcam), H4K20me1 (Abcam), H2AUb (Cell Signaling), YY1 (Santa Cruz). For DNA FISH (for X chromosome locations), the slides were denatured and images merged with RNA FISH signals and IF signals by using DAPI nuclear staining as a reference. For sequential IF and RNA FISH experiments, the percentage of nuclei containing a heterochromatin focus that colocalized with an X chromosome (naïve cells) or the XIST RNA cloud (for activated cells) was calculated by counting the number of nuclei with overlapping signals divided by the total number of cells. For allele-specific RNA/DNA FISH experiments, RNA pinpoints overlapping an X chromosome (DNA FISH) were counted. The different XIST RNA localization patterns were quantified for SLE B-cell lines (9383, 9248, 2190, 2411) and healthy controls (6141, 6868). For counting monoallelic versus biallelic expression at the single-cell level, nuclei with one pinpoint (monoallelic expression) or two pinpoints (biallelic expression) were counted. P values for comparison of normal and SLE samples were calculated by using Student’s t test. Oligo probes were used to detect CD40LG and CXCR3 in SLE/normal B-cell lines, and TRL-7 was detected by using a cDNA probe (red) together with an XIST probe (green). Normality of total fluorescence and maximum pixel intensities of XIST RNA exon 1 probe and oligo probes in male and female T cells was visualized by using probability plots and assessed by using the Shapiro–Wilk test (Matlab). Both total fluorescence and maximum intensity data were normally distributed (α < 0.05); however, some groups demonstrated unequal variance according to a two-sample F test for equality of variance (α > 0.05). To account for unequal variance, two-sided Welch t tests were used to compare total fluorescence and maximum pixel intensity between male and female samples. Significance was determined at α < 0.05.
GSEA Analyses.
GSEA was performed on Affymetrix Human Genome U133 Plus 2.0 Array datasets GSE30153 (77) (all samples) and GSE44723 (78) (samples GSM1089616, GSM1089617, GSM1089618, and GSM1089620). To identify enrichment of X-linked gene expression, GSEA was performed as described in ref. 79 by using C1 positional gene sets (MSigDB), and enrichment score plots are shown. Each gene (from the positional gene group) is plotted along the x axis, indicative of its contribution toward the enrichment score. Red indicates genes that are positively enriched at the leading edge of the enrichment score (positively correlated); genes in the blue region represent those that are negatively enriched for the enrichment score (negatively correlated). GSEA calculates the NES and FC values. Heatmaps were constructed in R studio by using heatmap.2 in gplots, and expression levels for all of the genes positively correlated (red region) are shown in heatmaps.
Acknowledgments
We thank J. Pearson, J. Wang, M. Bartolomei, K. Sarma, and L. King for reading this manuscript and critical discussions; the various laboratories across the University of Pennsylvania campus that shared mouse lymphocytes: S. Fuchs, A. Hu, C. Hunter, and J. Wherry; J. Riley and A. Medvec for instruction with human T-cell activation; D. Beiting for help with bioinformatic analysis; C. Berry for assistance with statistical analyses; all human blood donors recruited by the University of Pennsylvania; and J. Rinn and E. Hacisuleyman for assistance with hnRNPU siRNAs and knockdown. Human XIST oligo probes were designed by B. Del Rosario. The hnRNPU monoclonal antibody was a generous gift from G. Dreyfuss. EBV-immortialized patient B cells were collected by the Children’s Hospital of Philadelphia patient repository supervised by H. Hakonerson. This work was supported by the McCabe research foundation (M.C.A.), a grant with the Pennsylvania Department of Health (to M.C.A.), the Lupus Foundation (C.M.S. and M.C.A.), and NIH Grant T32 GM-07229 (to C.M.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1520113113/-/DCSupplemental.
References
- 1.Ross MT, et al. The DNA sequence of the human X chromosome. Nature. 2005;434(7031):325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Migeon BR. The role of X inactivation and cellular mosaicism in women’s health and sex-specific diseases. JAMA. 2006;295(12):1428–1433. doi: 10.1001/jama.295.12.1428. [DOI] [PubMed] [Google Scholar]
- 3.Spolarics Z. The X-files of inflammation: Cellular mosaicism of X-linked polymorphic genes and the female advantage in the host response to injury and infection. Shock. 2007;27(6):597–604. doi: 10.1097/SHK.0b013e31802e40bd. [DOI] [PubMed] [Google Scholar]
- 4.Butterworth M, McClellan B, Allansmith M. Influence of sex in immunoglobulin levels. Nature. 1967;214(5094):1224–1225. doi: 10.1038/2141224a0. [DOI] [PubMed] [Google Scholar]
- 5.Purtilo DT, Sullivan JL. Immunological bases for superior survival of females. Am J Dis Child. 1979;133(12):1251–1253. doi: 10.1001/archpedi.1979.02130120043008. [DOI] [PubMed] [Google Scholar]
- 6.Thompson DJ, Gezon HM, Rogers KD, Yee RB, Hatch TF. Excess risk of staphylococcal infection and disease in newborn males. Am J Epidemiol. 1966;84(2):314–328. doi: 10.1093/oxfordjournals.aje.a120645. [DOI] [PubMed] [Google Scholar]
- 7.Green MS. The male predominance in the incidence of infectious diseases in children: A postulated explanation for disparities in the literature. Int J Epidemiol. 1992;21(2):381–386. doi: 10.1093/ije/21.2.381. [DOI] [PubMed] [Google Scholar]
- 8.Libert C, Dejager L, Pinheiro I. The X chromosome in immune functions: When a chromosome makes the difference. Nat Rev Immunol. 2010;10(8):594–604. doi: 10.1038/nri2815. [DOI] [PubMed] [Google Scholar]
- 9.Selmi C, Brunetta E, Raimondo MG, Meroni PL. The X chromosome and the sex ratio of autoimmunity. Autoimmun Rev. 2012;11(6-7):A531–A537. doi: 10.1016/j.autrev.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 10.Cooney CM, et al. 46,X,del(X)(q13) Turner’s syndrome women with systemic lupus erythematosus in a pedigree multiplex for SLE. Genes Immun. 2009;10(5):478–481. doi: 10.1038/gene.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scofield RH, et al. Klinefelter’s syndrome (47,XXY) in male systemic lupus erythematosus patients: Support for the notion of a gene-dose effect from the X chromosome. Arthritis Rheum. 2008;58(8):2511–2517. doi: 10.1002/art.23701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 13.Payer B, Lee JT. X chromosome dosage compensation: How mammals keep the balance. Annu Rev Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- 14.Brown CJ, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349(6304):38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 15.Brockdorff N, et al. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. 1991;351(6324):329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- 16.Jonkers I, et al. Xist RNA is confined to the nuclear territory of the silenced X chromosome throughout the cell cycle. Mol Cell Biol. 2008;28(18):5583–5594. doi: 10.1128/MCB.02269-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plath K, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300(5616):131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 18.Mermoud JE, Popova B, Peters AH, Jenuwein T, Brockdorff N. Histone H3 lysine 9 methylation occurs rapidly at the onset of random X chromosome inactivation. Curr Biol. 2002;12(3):247–251. doi: 10.1016/s0960-9822(02)00660-7. [DOI] [PubMed] [Google Scholar]
- 19.Csankovszki G, Panning B, Bates B, Pehrson JR, Jaenisch R. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat Genet. 1999;22(4):323–324. doi: 10.1038/11887. [DOI] [PubMed] [Google Scholar]
- 20.Kohlmaier A, et al. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2004;2(7):E171. doi: 10.1371/journal.pbio.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith KP, Byron M, Clemson CM, Lawrence JB. Ubiquitinated proteins including uH2A on the human and mouse inactive X chromosome: Enrichment in gene rich bands. Chromosoma. 2004;113(6):324–335. doi: 10.1007/s00412-004-0325-1. [DOI] [PubMed] [Google Scholar]
- 22.Peeters SB, Cotton AM, Brown CJ. Variable escape from X-chromosome inactivation: Identifying factors that tip the scales towards expression. BioEssays. 2014;36(8):746–756. doi: 10.1002/bies.201400032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434(7031):400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 24.Yang F, Babak T, Shendure J, Disteche CM. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 2010;20(5):614–622. doi: 10.1101/gr.103200.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carrel L, Willard HF. Heterogeneous gene expression from the inactive X chromosome: An X-linked gene that escapes X inactivation in some human cell lines but is inactivated in others. Proc Natl Acad Sci USA. 1999;96(13):7364–7369. doi: 10.1073/pnas.96.13.7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cotton AM, et al. Chromosome-wide DNA methylation analysis predicts human tissue-specific X inactivation. Hum Genet. 2011;130(2):187–201. doi: 10.1007/s00439-011-1007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qu K, et al. Individuality and variation of personal regulomes in primary human T cells. Cell Syst. 2015;1(1):51–61. doi: 10.1016/j.cels.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hewagama A, et al. Michigan Lupus Cohort Overexpression of X-linked genes in T cells from women with lupus. J Autoimmun. 2013;41:60–71. doi: 10.1016/j.jaut.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu Q, et al. Demethylation of CD40LG on the inactive X in T cells from women with lupus. J Immunol. 2007;179(9):6352–6358. doi: 10.4049/jimmunol.179.9.6352. [DOI] [PubMed] [Google Scholar]
- 30.Brown CJ, et al. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71(3):527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 31.Savarese F, Flahndorfer K, Jaenisch R, Busslinger M, Wutz A. Hematopoietic precursor cells transiently reestablish permissiveness for X inactivation. Mol Cell Biol. 2006;26(19):7167–7177. doi: 10.1128/MCB.00810-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall LL, et al. An ectopic human XIST gene can induce chromosome inactivation in postdifferentiation human HT-1080 cells. Proc Natl Acad Sci USA. 2002;99(13):8677–8682. doi: 10.1073/pnas.132468999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clemson CM, Hall LL, Byron M, McNeil J, Lawrence JB. The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc Natl Acad Sci USA. 2006;103(20):7688–7693. doi: 10.1073/pnas.0601069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Namekawa SH, Payer B, Huynh KD, Jaenisch R, Lee JT. Two-step imprinted X inactivation: Repeat versus genic silencing in the mouse. Mol Cell Biol. 2010;30(13):3187–3205. doi: 10.1128/MCB.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andersson J, Sjöberg O, Möller G. Induction of immunoglobulin and antibody synthesis in vitro by lipopolysaccharides. Eur J Immunol. 1972;2(4):349–353. doi: 10.1002/eji.1830020410. [DOI] [PubMed] [Google Scholar]
- 36.Krieg AM, et al. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995;374(6522):546–549. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- 37.Ledderose C, Heyn J, Limbeck E, Kreth S. Selection of reliable reference genes for quantitative real-time PCR in human T cells and neutrophils. BMC Res Notes. 2011;4:427. doi: 10.1186/1756-0500-4-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5(4):695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- 39.Sarma K, Levasseur P, Aristarkhov A, Lee JT. Locked nucleic acids (LNAs) reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome. Proc Natl Acad Sci USA. 2010;107(51):22196–22201. doi: 10.1073/pnas.1009785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beletskii A, Hong YK, Pehrson J, Egholm M, Strauss WM. PNA interference mapping demonstrates functional domains in the noncoding RNA Xist. Proc Natl Acad Sci USA. 2001;98(16):9215–9220. doi: 10.1073/pnas.161173098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grigoryev SA, Nikitina T, Pehrson JR, Singh PB, Woodcock CL. Dynamic relocation of epigenetic chromatin markers reveals an active role of constitutive heterochromatin in the transition from proliferation to quiescence. J Cell Sci. 2004;117(Pt 25):6153–6162. doi: 10.1242/jcs.01537. [DOI] [PubMed] [Google Scholar]
- 42.Costanzi C, Stein P, Worrad DM, Schultz RM, Pehrson JR. Histone macroH2A1 is concentrated in the inactive X chromosome of female preimplantation mouse embryos. Development. 2000;127(11):2283–2289. doi: 10.1242/dev.127.11.2283. [DOI] [PubMed] [Google Scholar]
- 43.de Napoles M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7(5):663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 44.Changolkar LN, Pehrson JR. macroH2A1 histone variants are depleted on active genes but concentrated on the inactive X chromosome. Mol Cell Biol. 2006;26(12):4410–4420. doi: 10.1128/MCB.02258-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smetana K, Karhan J, Trneny M. Heterochromatin condensation in central and peripheral nuclear regions of maturing lymphocytes in the peripheral blood of patients suffering from B chronic lymphocytic leukemia - a cytochemical study. Neoplasma. 2011;58(6):476–481. doi: 10.4149/neo_2011_06_476. [DOI] [PubMed] [Google Scholar]
- 46.Tchieu J, et al. Female human iPSCs retain an inactive X chromosome. Cell Stem Cell. 2010;7(3):329–342. doi: 10.1016/j.stem.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Enghard P, et al. CXCR3+CD4+ T cells are enriched in inflamed kidneys and urine and provide a new biomarker for acute nephritis flares in systemic lupus erythematosus patients. Arthritis Rheum. 2009;60(1):199–206. doi: 10.1002/art.24136. [DOI] [PubMed] [Google Scholar]
- 48.Helbig R, Fackelmayer FO. Scaffold attachment factor A (SAF-A) is concentrated in inactive X chromosome territories through its RGG domain. Chromosoma. 2003;112(4):173–182. doi: 10.1007/s00412-003-0258-0. [DOI] [PubMed] [Google Scholar]
- 49.Hasegawa Y, et al. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell. 2010;19(3):469–476. doi: 10.1016/j.devcel.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 50.Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146(1):119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Makhlouf M, et al. A prominent and conserved role for YY1 in Xist transcriptional activation. Nat Commun. 2014;5:4878. doi: 10.1038/ncomms5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Affar B, et al. Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol Cell Biol. 2006;26(9):3565–3581. doi: 10.1128/MCB.26.9.3565-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Joshi SK, Hashimoto K, Koni PA. Induced DNA recombination by Cre recombinase protein transduction. Genesis. 2002;33(1):48–54. doi: 10.1002/gene.10089. [DOI] [PubMed] [Google Scholar]
- 54.Desai-Mehta A, Lu L, Ramsey-Goldman R, Datta SK. Hyperexpression of CD40 ligand by B and T cells in human lupus and its role in pathogenic autoantibody production. J Clin Invest. 1996;97(9):2063–2073. doi: 10.1172/JCI118643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.García-Ortiz H, et al. Association of TLR7 copy number variation with susceptibility to childhood-onset systemic lupus erythematosus in Mexican population. Ann Rheum Dis. 2010;69(10):1861–1865. doi: 10.1136/ard.2009.124313. [DOI] [PubMed] [Google Scholar]
- 56.Anguera MC, et al. Molecular signatures of human induced pluripotent stem cells highlight sex differences and cancer genes. Cell Stem Cell. 2012;11(1):75–90. doi: 10.1016/j.stem.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mekhoubad S, et al. Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell. 2012;10(5):595–609. doi: 10.1016/j.stem.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chu C, et al. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161(2):404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McHugh CA, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521(7551):232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Minajigi A, et al. Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science. 2015;349(6245):aab2276. doi: 10.1126/science.aab2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sawada S, Scarborough JD, Killeen N, Littman DR. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell. 1994;77(6):917–929. doi: 10.1016/0092-8674(94)90140-6. [DOI] [PubMed] [Google Scholar]
- 62.Wilkinson FH, Park K, Atchison ML. Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc Natl Acad Sci USA. 2006;103(51):19296–19301. doi: 10.1073/pnas.0603564103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Atchison L, Ghias A, Wilkinson F, Bonini N, Atchison ML. Transcription factor YY1 functions as a PcG protein in vivo. EMBO J. 2003;22(6):1347–1358. doi: 10.1093/emboj/cdg124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322(5902):750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kalantry S, et al. The Polycomb group protein Eed protects the inactive X-chromosome from differentiation-induced reactivation. Nat Cell Biol. 2006;8(2):195–202. doi: 10.1038/ncb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schoeftner S, et al. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 2006;25(13):3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Silva SS, Rowntree RK, Mekhoubad S, Lee JT. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc Natl Acad Sci USA. 2008;105(12):4820–4825. doi: 10.1073/pnas.0712136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen Y, et al. X-inactivation in female human embryonic stem cells is in a nonrandom pattern and prone to epigenetic alterations. Proc Natl Acad Sci USA. 2008;105(12):4709–4714. doi: 10.1073/pnas.0712018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hall LL, et al. X-inactivation reveals epigenetic anomalies in most hESC but identifies sublines that initiate as expected. J Cell Physiol. 2008;216(2):445–452. doi: 10.1002/jcp.21411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yildirim E, et al. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152(4):727–742. doi: 10.1016/j.cell.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hwang SH, et al. B cell TLR7 expression drives anti-RNA autoantibody production and exacerbates disease in systemic lupus erythematosus-prone mice. J Immunol. 2012;189(12):5786–5796. doi: 10.4049/jimmunol.1202195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pisitkun P, et al. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science. 2006;312(5780):1669–1672. doi: 10.1126/science.1124978. [DOI] [PubMed] [Google Scholar]
- 73.Moser K, et al. CXCR3 promotes the production of IgG1 autoantibodies but is not essential for the development of lupus nephritis in NZB/NZW mice. Arthritis Rheum. 2012;64(4):1237–1246. doi: 10.1002/art.33424. [DOI] [PubMed] [Google Scholar]
- 74.Erwin JA, Lee JT. 2010. Characterization of X-chromosome inactivation status in human pluripotent stem cells. Curr Protoc Stem Cell Biol, Chapter 1, Unit 1B 6.
- 75.Yung RL, Quddus J, Chrisp CE, Johnson KJ, Richardson BC. Mechanism of drug-induced lupus. I. Cloned Th2 cells modified with DNA methylation inhibitors in vitro cause autoimmunity in vivo. J Immunol. 1995;154:3025–3035. [PubMed] [Google Scholar]
- 76.Subramanian S. A Tlr7 translocation accelerates systemic autoimmunity in murine lupus. Proc Natl Acad Sci USA. 2006;103(26):9970–9975. doi: 10.1073/pnas.0603912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Garaud JC, et al. B cell signature during inactive systemic lupus is heterogeneous: Toward a biological dissection of lupus. PLoS One. 2011;6(8):e23900. doi: 10.1371/journal.pone.0023900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Peng R, et al. Bleomycin induces molecular changes directly relevant to idiopathic pulmonary fibrosis: A model for “active” disease. PLoS One. 2013;8(4):e59348. doi: 10.1371/journal.pone.0059348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]