Abstract
Epigenetic modifications, including DNA methylation, represent a potential mechanism for environmental impacts on human disease. Maternal smoking in pregnancy remains an important public health problem that impacts child health in a myriad of ways and has potential lifelong consequences. The mechanisms are largely unknown, but epigenetics most likely plays a role. We formed the Pregnancy And Childhood Epigenetics (PACE) consortium and meta-analyzed, across 13 cohorts (n = 6,685), the association between maternal smoking in pregnancy and newborn blood DNA methylation at over 450,000 CpG sites (CpGs) by using the Illumina 450K BeadChip. Over 6,000 CpGs were differentially methylated in relation to maternal smoking at genome-wide statistical significance (false discovery rate, 5%), including 2,965 CpGs corresponding to 2,017 genes not previously related to smoking and methylation in either newborns or adults. Several genes are relevant to diseases that can be caused by maternal smoking (e.g., orofacial clefts and asthma) or adult smoking (e.g., certain cancers). A number of differentially methylated CpGs were associated with gene expression. We observed enrichment in pathways and processes critical to development. In older children (5 cohorts, n = 3,187), 100% of CpGs gave at least nominal levels of significance, far more than expected by chance (p value < 2.2 × 10−16). Results were robust to different normalization methods used across studies and cell type adjustment. In this large scale meta-analysis of methylation data, we identified numerous loci involved in response to maternal smoking in pregnancy with persistence into later childhood and provide insights into mechanisms underlying effects of this important exposure.
Introduction
Despite years of advisories regarding health risks to the developing fetus from maternal smoking, many pregnant women still smoke, including 12.3% in the US.1 Maternal smoking during pregnancy is regarded as a cause of low birth weight, reduced pulmonary function (PLF [MIM: 608852]), orofacial clefts (OFC1 [MIM: 119530]), and sudden infant death syndrome (SIDS [MIM: 272120]) in exposed newborns.2 Other adverse birth outcomes3 have been associated with maternal smoking in pregnancy, along with common health problems in children, including asthma (ASRT [MIM: 600807]), otitis media (OMS [MIM: 166760]), and neurobehavioral disorders.2
The mechanisms for the adverse health effects of maternal smoking during pregnancy on offspring remain poorly understood.2 Recently, studies have examined the potential role of epigenetic modifications such as DNA methylation at specific CpG sites (CpGs). These include studies examining genome-wide DNA methylation in newborns in relation to maternal smoking in pregnancy with the Illumina Infinium HumanMethylation27 (27K) BeadChip4, 5, 6 or the newer platform with wider coverage, the HumanMethylation450 (450K) BeadChip.7, 8, 9, 10 A number of differentially methylated loci have been identified in offspring in relation to maternal smoking in pregnancy in individual studies (references in the Supplemental Note). One study examined the top CpGs with respect to timing of exposure and found that the signals reflect sustained, rather than short-term, exposure to maternal smoking during pregnancy,11 but this has not been evaluated genome-wide. A few studies suggest that some of these methylation signals persist into later childhood and adolescence, but data are limited.9, 12 The combination of genome-wide data across studies via meta-analysis to generate large sample sizes for the discovery of loci that would not have been identified from individual studies has been very successful in genetics, but this approach has rarely been used with methylation data.
To address the impact of maternal smoking during pregnancy on newborns with much greater power, we recruited 13 birth cohort studies with data on maternal smoking during pregnancy and DNA methylation in offspring from the 450K BeadChip into the Pregnancy and Childhood Epigenetics consortium (PACE). We meta-analyzed harmonized cohort-specific associations between maternal smoking during pregnancy and DNA methylation in the offspring. We examined both sustained maternal smoking and any smoking during pregnancy. We also examined persistence of DNA methylation patterns related to maternal smoking in newborns among older children, including adjustment for postnatal secondhand tobacco smoke exposure. For functional follow-up of findings, we evaluated the associations between methylation status in the newly identified CpGs and expression levels of nearby genes and performed pathway and functional network analyses. This study represents a large and comprehensive evaluation of the impact of maternal smoking during pregnancy on DNA methylation in offspring.
Material and Methods
Participating Cohorts
A total of 13 PACE cohorts participated in the meta-analysis of maternal smoking during pregnancy and 450K DNA methylation in newborns. These studies, listed in alphabetical order, are the Avon Longitudinal Study of Parents and Children (ALSPAC), the Center for Health Assessment of Mothers and Children of Salinas (CHAMACOS), the Children’s Health Study (CHS), the GECKO Drenthe cohort, the Generation R Study, Isle of Wight (IOW), Mechanisms of the Development of Allergy (MeDALL), three independent datasets from the Norwegian Mother and Child Cohort Study (MoBa1, MoBa2, and MoBa3), the Norway Facial Clefts Study (NFCS), the Newborn Epigenetics Study (NEST), and Project Viva. MeDALL represents a pooled analysis of four cohorts with coordinated methylation measurements: Infancia y Medio Ambiente (INMA), Etudes des Déterminants pré et postnatals précoces du développement et de la santé de l’Enfant (EDEN), Children’s Allergy Environment Stockholm Epidemiology study (BAMSE), and Prevention and Incidence of Asthma and Mite Allergy (PIAMA). Two of the MeDALL cohorts contributed to the newborn meta-analysis (INMA and EDEN). There were five studies with data on older children: ALSPAC, Genes-environments and Admixture in Latino Americans (GALA II), the Study to Explore Early Development (SEED), MeDALL (INMA, EDEN, BAMSE, and PIAMA), and an independent methylation dataset from BAMSE subjects. Ethical approval for study protocols was obtained for all participating cohorts. Further information on this as well as the study methods for each cohort are described in detail in the Supplemental Note.
For this paper, participating cohorts shared only results files from in-house analyses. No individual data were shared for this paper. Therefore, access to the individual cohort-level data for the purpose of reproducing results would require individual data transfer agreements to be negotiated with and approved by each of the contributing cohorts.
Harmonization of Maternal Smoking Variables
Cohorts assessed maternal smoking during pregnancy via questionnaires completed by the mothers. The MoBa study (MoBa1 and MoBa2) also used cotinine measurements from maternal blood samples taken during pregnancy as part of the definition of maternal smoking during pregnancy. More details on the cohort-specific smoking variables are in the Supplemental Note. In a previous publication from the MoBa1 study, significant associations between maternal smoking during pregnancy and DNA methylation in newborns were driven not by transient smoking that ended early in pregnancy but rather by sustained smoking during pregnancy.11 Thus, each cohort ran separate models to evaluate both sustained smoking and any smoking during pregnancy. The variable (yes/no) for sustained smoking during pregnancy was designed to capture women who smoked at least one cigarette per day through most of pregnancy. To cleanly contrast the effect of sustained smoking through pregnancy with that of never smoking during pregnancy, we excluded women who reported quitting smoking during pregnancy from the sustained smoking models. The variable (yes/no) for any maternal smoking during pregnancy was designed to capture any amount of smoking during pregnancy, at any time, even if a woman reported quitting. Because we did not exclude women who quit smoking during pregnancy from the models representing any smoking during pregnancy, the total sample sizes are slightly larger than those of the models representing sustained smoking during pregnancy. Genome-wide analyses use large sample statistics. We limited meta-analyses to cohorts with at least 15 subjects in both the exposed and unexposed groups. This excluded four cohorts (CHAMACOS, CHS, IOW, and Project Viva) from the sustained smoking models. However these cohorts did participate in the meta-analysis of any smoking during pregnancy.
Methylation Measurements and Quality Control
Each cohort independently conducted laboratory measurements and quality control. The samples for each cohort underwent bisulfite conversion via the EZ-96 DNA Methylation kit (Zymo Research). Samples were processed with the Illumina Infinium HumanMethylation450 (450K) BeadChip (Illumina) at Illumina or in cohort-specific laboratories.
Quality control of samples was performed by each cohort and failed samples were excluded on the basis of Illumina’s detection p value, low sample DNA concentration, sample call rate, CpG-specific percentage of missing values, bisulfite conversion efficiency, gender verification with multidimensional scaling plots, and other quality control metrics specific to cohorts. Cohorts could also use validated, published statistical methods for normalizing their methylation data on the untransformed methylation beta values (ranging from 0 to 1). Some cohorts also made independent probe exclusions. More details are provided in the Supplemental Note. For the meta-analysis, additional probe exclusions were made across all cohorts. Specifically, we excluded control probes (n = 65), probes that mapped to the X (n = 11,232) or Y (n = 416) chromosomes, probes with an underlying SNP mapping to the last five nucleotides of the probe sequence (N = 9,168) as previously described,7 and CpGs with an implausible (zero) value for the SE (n = 67). This left a total of 464,628 CpGs included in the meta-analysis.
Cohort-Specific Statistical Analyses
Each cohort ran independent statistical analyses according to a common pre-specified analysis plan. Robust linear regression was used in R13 to evaluate the association between maternal smoking during pregnancy and cord blood DNA methylation for each probe while accounting for potential heteroskedasticity and/or influential outliers. Each cohort ran the following covariate-adjusted statistical models: (1) the primary model, which used sustained maternal smoking during pregnancy as the exposure and the normalized betas as the outcome, (2) sustained maternal smoking during pregnancy as the exposure and raw betas (not normalized) as the outcome, (3) any maternal smoking during pregnancy as the exposure and normalized betas as the outcome, (4) any maternal smoking during pregnancy as the exposure and raw betas as the outcome, and (5) sustained maternal smoking during pregnancy as the exposure and normalized betas as the outcome, with additional adjustment for cell type proportion. All models were adjusted for maternal age, maternal education (or a surrogate socioeconomic metric), parity, and technical covariates such as batch or plate. Some cohorts used ComBat14 to account for batch effects and therefore did not include batch or plate as covariates in the models with normalized betas (see Supplemental Note). Additional correction for study design or sampling factors was done as needed in some cohorts. Because maternal smoking during pregnancy is not related to the child’s sex, it cannot be a confounder and thus was not included in models. We did not adjust for principal components (PCs) because not all cohorts had genome-wide genotype data and cohorts with genotype data had it only for a subset of subjects with methylation data. Furthermore, in one large cohort with PC data, models adjusted for PCs showed little variation in the results (correlation of betas = 0.991; correlation of log(p values) = 0.996) when compared to models without this adjustment, despite a reduction in sample size. The statistical models for cohorts with DNA methylation measured in older children were the same, with the additional adjustment for second-hand tobacco smoke exposure.
All cohorts independently estimated cell type proportion by using the reference-based Houseman method15 in the minfi package16 with the Reinius et al. dataset for reference.17 Cell type correction was applied by inclusion of the six estimated cell type proportions (CD8T, CD4T, NK cells, B cells, monocytes, granulocytes) as covariates in cohort-specific statistical models.
Meta-analysis
We performed inverse variance-weighted fixed-effects meta-analysis with METAL.18 We accounted for multiple testing by controlling the false discovery rate (FDR) at 5%, implementing the method by Benjamini and Hochberg.19 This method was applied to all instances of FDR correction described in this paper unless otherwise specified. CpGs with an FDR-corrected p value less than 0.05 were considered statistically significant. CpGs that were statistically significant based on the more stringent Bonferroni correction (uncorrected p value < 1.08 × 10−7 to account for 464,628 tests) were also noted.
To determine the robustness of our models and findings, we performed an additional analysis in which we removed the cohorts of non-European ancestry (Table S1). We compared the effect estimates, SEs, and the distribution of the p values for the model to the estimates for our primary model to evaluate the consistency of our findings.
Examination in Older Children of CpGs Associated with Smoking in Cord Blood
The FDR-significant CpGs identified in the primary model from the newborn meta-analyses were followed up with a lookup replication approach in the results from five older children cohorts, and FDR correction was applied to account for the number of CpGs tested.
Literature Review to Identify Genes Previously Associated with Smoking and Methylation
We performed a systematic literature review to determine which CpGs represented findings not previously related to smoking exposure and methylation in the literature. A query of NCBI’s PubMed database was performed with the search terms ((“DNA Methylation”[Mesh] OR methylation) AND (“Smoking”[Mesh] OR smoking)) in order to be broad enough to capture all past studies reporting such results. CpGs with previously reported associations with smoking, both from prenatal exposure or in adults, were considered. This search yielded 789 results when performed on March 1, 2015. All results were then reviewed by title and abstract to determine whether they met inclusion criteria. First, results were limited to those performed in healthy human populations. That is, participants could not exclusively have been drawn from disease cases and studies could not have been performed only in cell lines or animals. Case-control analyses that included healthy controls were accepted as meeting this criterion, and no limitation was applied concerning the tissue used for DNA extraction. Second, studies were required to have performed DNA methylation analysis agnostically on a large scale as opposed to targeted interrogation of candidate CpGs. This was operationalized by including only analyses that examined >1,000 sites simultaneously. The Illumina 450K, 27K, and GoldenGate arrays all met this criterion. Third, the exposure was restricted to tobacco cigarette smoking. Related exposures, such as to other forms of tobacco use or smoke exposure, were not included. Lastly, studies had to have reported their significant results publicly. Studies that failed to report p values or gene annotations were excluded.
Review of the existing literature on the effect of smoking on DNA methylation identified 25 publications meeting inclusion criteria. Of these, 16 studies reported results for adult smoking exposure,20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35 and nine provided results of association between maternal smoking during pregnancy on child DNA methylation.4, 7, 8, 9, 12, 36, 37, 38, 39 CpG level results (p values and gene annotations) for sites showing significant association between smoking exposure and DNA methylation were extracted and compiled for comparison with the results from the meta-analysis. Results were considered significant if they met the multiple testing criteria implemented within the publication. For studies failing to implement any multiple testing correction, a naive Bonferroni threshold for the number of tests performed in the individual study was used. Genes previously associated with either adult smoking or maternal smoking in pregnancy (Table S6) were excluded from our list of meta-analysis results.
CpG Annotation
The official gene name was noted for each CpG via Illumina’s genome coordinate.40 We enhanced the annotation provided by Illumina by using the UCSC Genome Browser (including data the RefSeq and Ensembl databases), as well as annotation data in Bioconductor. All of the annotations use the human February 2009 (GRCh37/hg19) assembly. We also used the program Snipper to annotate the nearest genes within 10 Mb of each CpG. We include this expanded Snipper gene annotation in our tables and the Discussion.
For selected genes, we used coMet41 to graphically display additional information about CpGs, including physical location, correlation, statistical significance, and functional annotation.
Enrichment Analysis
We evaluated whether the CpGs significantly associated with smoking (based on the FDR p value < 0.05) were enriched, relative to all CpGs analyzed, for several biologic annotations provided in the Illumina annotation file. We assessed enrichment by using the two-sided doubling mid p value of the hypergeometric test.24 We also evaluated enrichment in a subset of CpGs mapping to imprinted differentially methylated regions (DMRs) described by Court et al.42
Pathway Analyses
We linked the CpGs significantly associated with smoking (based on an FDR p value < 0.05) to genes on the basis of only the 450K BeadChip annotation file.43 Probes lacking an annotated Entrez Gene ID were filtered (n = 1,971), as were duplicate gene entries (n = 1,473). A total of 2,629 unique gene identifiers were used in gene ontology enrichment analysis with three different procedures as described below.
This resulted in 2,235 genes that mapped to gene ontologies of biological processes, and using topGO in R,44 we tested for gene enrichment over the background array (16,119 unique annotated Entrez Gene IDs) by using Fisher’s exact tests with a minimum of five genes per node. In addition, we used the DAVID bioinformatics resource45 to test for enrichment in gene ontology biological processes with a threshold of five, and we used the Benjamini-Hochberg procedure to control for false discoveries. Finally, we used QIAGEN’s Ingenuity Pathway Analysis (IPA) to identify relevant signaling and functional pathways.
Functional Network Analysis
To construct a functional association network, it was desirable to reduce the list of tested CpGs, so we prioritized the FDR-significant CpGs from the primary model in a stepwise manner. First, we only included those CpGs that were FDR significant in both the primary and cell-type-adjusted models (FDR p values < 0.05). Next, we sorted these CpGs according to their effect size (beta coefficient), and selected the top quartile (n = 980). The genes mapping to these prioritized CpGs were then used as input for the construction of a functional interaction network. We used the GeneMANIA algorithm, as well as its functional association data, including genetic interaction, physical interactions, co-expression, shared protein domains, and co-localization networks.46 We selected the “all available networks” option with a 500-gene output (accessed March 11, 2015). Functional enrichment analysis was then performed on all genes from the constructed interaction network against Gene Ontology (GO) terms to detect significantly enriched GO terms.47 FDR correction was applied to this analysis based on the q value; a threshold of q < 0.01 was used.
Methylation Transcription Analysis
To further explore the associations between methylation and gene expression, we performed methylation-expression analyses, evaluating the association between the methylation status of CpGs and differences in quantitative levels of gene expression. All identified CpGs that reached FDR-corrected significance and that we identified as not previously reported in the literature were tested for association with expression levels of genes within a region of 250 kb upstream or downstream of the CpG48 (total region 500 kb) to evaluate whether the CpG-methylation status influenced transcript levels of genes. We had two datasets available for this analysis. One dataset included mRNA gene expression (Illumina HumanHT-12 v.4) and 450K methylation data, both from whole-blood samples from 730 adults over 45 years of age in the Rotterdam Study, a population-based prospective cohort study in Rotterdam, the Netherlands.49 This gene expression dataset is available at the GEO public repository under the accession number GEO: GSE33828. The second dataset included mRNA gene expression (Affymetrix Human Transcriptome Array 2.0) and 450K methylation data on whole-blood samples from 107 children at 4 years of age from the INMA study in Spain. Study population details for the Rotterdam Study and INMA are in the Supplemental Note. In the Rotterdam dataset, 2,636 of the 2,965 CpGs examined mapped to a transcript within the 500 kb window. We created residuals for mRNA expression after regressing out the Houseman-estimated white-blood-cell proportions, the erythrocyte and platelet cell counts, fasting state, RNA quality score, plate number, age, and sex on the mRNA expression levels by using a linear mixed model. We then created residuals for DNA methylation, regressing out the Houseman-estimated white-blood-cell proportions, age, sex, batch effects on the dasen-normalized50 beta-values of the CpGs by using a linear mixed model. We used a linear regression model to evaluate the association between the residuals of the mRNA expression levels and the residuals of the dasen-normalized beta-values of the CpGs.
The INMA gene expression data were normalized with Expression Console Software from Affymetrix, and probes were clustered to the transcript level. Only transcripts within the 500 kb window of selected CpGs were considered in the analysis (n = 45,076 transcripts). To control for technical variation in the DNA methylation dataset, a PC analysis of 600 negative control probes was performed with 10,000 permutations, and the residuals of a linear regression model including the first five PCs were estimated. The effect of sex and Houseman cell-type-proportion estimates were adjusted for in a second-stage linear regression model. Two models were applied to control for technical and unwanted biological variation when estimating gene expression residuals. In the first one, sex and Houseman estimates were regressed out. In the second one, 14 surrogate variables estimated with the sva R package14 were adjusted for in a second model including sex and Houseman estimates. A linear regression model of residuals of gene expression versus residuals of methylation was performed. Multiple testing for both Rotterdam and INMA gene expression analyses was controlled with Benjamini-Hochberg FDR correction.
Examination of Polymorphic and Cross-Reactive Probes
The list of FDR-significant CpGs was matched to the list of polymorphic and cross-reactive CpGs provided by Chen et al.51 to identify potential problematic probes. We additionally performed the dip test52 for unimodality for each CpG to test for non-unimodal distributions in the MoBa1 cohort (n = 1,068). Also using the MoBa1 cohort, we visually inspected density plots for each of the probes that matched to the list of polymorphic probes from Chen et al. to assess departures from unimodality, including from small numbers of outlier values.
Results
Study Characteristics
A total of 13 cohorts participated in the meta-analysis of maternal smoking during pregnancy and 450K DNA methylation in newborns. Among these 6,685 newborns, 897 (13%) were exposed to sustained maternal smoking during pregnancy and 1,646 (25%) were exposed to any maternal smoking during pregnancy. We also included five cohorts of older children (n = 3,187, average age = 6.8 years); 266 children (8%) were exposed to sustained smoking during pregnancy and 404 (13%) were exposed to any maternal smoking during pregnancy. The cohort-specific summary statistics for maternal smoking are presented in Table 1 and covariates in Table S1. The majority of participants were of European ancestry (Table S1).
Table 1.
Smoking Variable Frequencies for the Cohorts Participating in Meta-analyses: Newborns and Older Children
| Studya | Study Population | Particpantsb | No. of Participants Exposed to Sustained Maternal Smoking during Pregnancy (%) | No. of. Participants Exposed to Any Maternal Smoking during Pregnancy (%) |
|---|---|---|---|---|
| ALSPAC | newborns | 860 | 87 (10.1) | 120 (14.0) |
| CHAMACOS | newborns | 378 | 7 (1.9)c | 24 (6.3) |
| CHS | newborns | 85 | NAc | 22 (25.9) |
| GECKO | newborns | 255 | 70 (27.5) | 129 (50.6) |
| Generation R | newborns | 883 | 129 (14.6) | 220 (24.9) |
| IOW | newborns | 90 | 9 (10.0)c | 23 (25.6) |
| MeDALL | newborns | 362 | 43 (11.9) | 63 (17.5) |
| MoBa1 | newborns | 1,063 | 156 (14.7) | 312 (29.4) |
| MoBa2 | newborns | 671 | 70 (10.4) | 173 (25.8) |
| MoBa3 | newborns | 252 | 28 (11.1) | 73 (29.0) |
| NEST | newborns | 413 | 69 (16.7) | 136 (32.9) |
| NFCS | newborns | 889 | 245 (27.6) | 325 (36.6) |
| Project Viva | newborns | 485 | 14 (2.9)c | 26 (5.4) |
| ALSPAC | older children | 840 | 89 (10.6) | 115 (13.7) |
| BAMSE | older children | 347 | 26 (7.5) | 43 (12.4) |
| GALA II | older children | 569 | 40 (7.0) | 76 (13.4) |
| MeDALL | older children | 851 | 86 (10.2) | 121 (14.3) |
| SEED | older children | 584 | 25 (4.3) | 49 (8.4) |
NA, not available.
Study names and additional information: The Avon Longitudinal Study of Parents and Children (ALSPAC), the Center for Health Assessment of Mothers and Children of Salinas (CHAMACOS), the Children’s Health Study (CHS), the GECKO Drenthe cohort, the Generation R Study, Isle of Wight (IOW), Mechanisms of the Development of Allergy (MeDALL), three independent datasets from the Norwegian Mother and Child Cohort Study (MoBa1, MoBa2, and MoBa3), the Norway Facial Clefts Study (NFCS), the Newborn Epigenetics Study (NEST), and Project Viva. MeDALL represents a pooled analysis of four cohorts with coordinated methylation measurements: Infancia y Medio Ambiente (INMA), Etudes des Déterminants pré et postnatals précoces du développement et de la santé de l’Enfant (EDEN), Children’s Allergy Environment Stockholm Epidemiology study (BAMSE), and Prevention and Incidence of Asthma and Mite Allergy (PIAMA). Two of the MeDALL cohorts contributed to the newborn meta-analysis (INMA and EDEN). Studies with data on older children: ALSPAC, Genes-environments and Admixture in Latino Americans (GALA II), the Study to Explore Early Development (SEED), MeDALL (INMA, EDEN, BAMSE, and PIAMA), and an independent methylation dataset from BAMSE subjects.
Number of participants with smoking data, 450K methylation, and covariates. Participants who quit smoking during pregnancy were not included in the sustained smoking models.
Cohorts in which the sustained smoking category had n < 15 or insufficient information to create the requested category, resulting in exclusion from the sustained smoking analysis models. All cohorts were included in the models evaluating the exposure of any smoking during pregnancy.
Meta-analysis
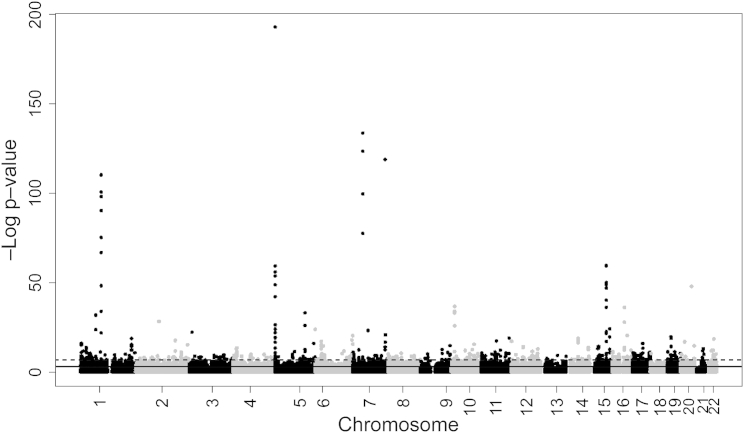
Our primary model evaluated the association between sustained maternal smoking during pregnancy and differential DNA methylation in newborns by using normalized methylation betas as the outcome, adjusting for covariates (Figure 1). The cohort-specific lambdas and number of CpGs included in each model are listed in Table S2. Among the 6,073 CpGs with FDR significance (Table S3), 568 also met the strict Bonferroni threshold for statistical significance (p value < 1.08 × 10−7, correcting for 464,628 independent tests). Results were quite robust to cell type adjustment (Table S3): all 568 Bonferroni-significant CpGs from the primary model remained FDR significant in the cell-type-adjusted model, and 78% were Bonferroni significant in both models. The log10(p values) for the primary model and cell-type-adjusted models were highly correlated (correlation coefficient = 0.92 across all CpGs, 0.98 for the FDR-significant CpGs in the primary model, Figure S1). Given the general similarity of the results before and after cell type adjustment and the fact that the available reference panel is from a small number (n = 6) of adult men,17 we regard the covariate-adjusted model as the primary model. The results for other models (the cell-type-adjusted model, the model representing any smoking during pregnancy, and the methylation model representing sustained smoking during pregnancy associated with older children) and the mean methylation values in newborns and older children are shown for all 6,073 CpGs in Table S3.
Figure 1.
Meta-analysis of the Association between Sustained Maternal Smoking during Pregnancy and DNA Methylation in Newborn Cord Blood
A total of 6,073 CpGs were considered statistically significant when using FDR correction (solid horizontal line); 568 were Bonferroni significant (dashed horizontal line).
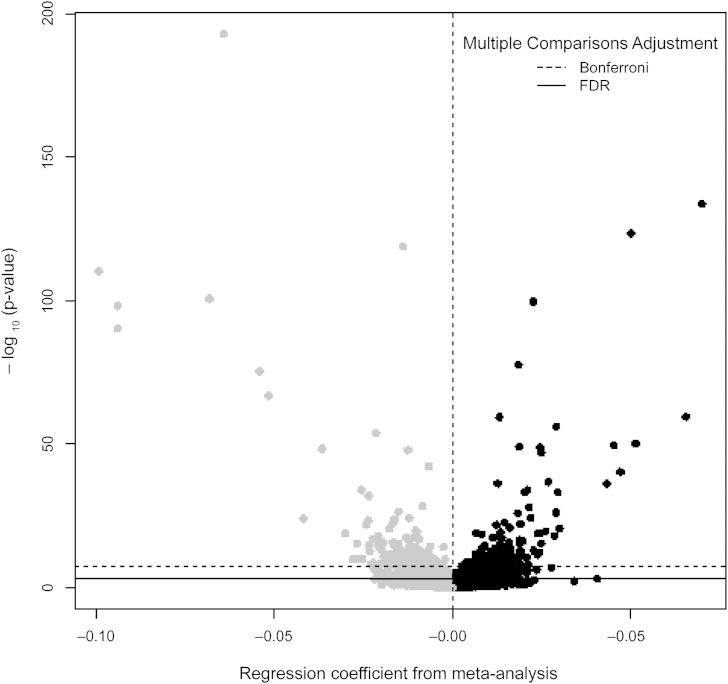
Among the 6,073 FDR-significant CpGs, smoking during pregnancy was associated approximately equally with increased methylation (52%) and decreased methylation (48%) (Figure 2). Out of the 3,932 CpGs that were also FDR-significant after cell type adjustment, there were 967 CpGs in or within 10 Mb of the 1,185 genes we identified in our systematic literature review (see Supplemental Note and Table S4) as previously reported to be differentially methylated in relation to smoking. This left 2,965 CpGs (corresponding to 2,017 annotated mapped or nearest genes) that had not previously been reported (Table S5; genes highlighted in discussion shown in Table 2). For comprehensive comparison with the previous literature, we also present our results for all CpGs that were either not FDR significant after cell type adjustment and/or that annotated to genes already described in the literature as related to smoking and methylation (n = 3,108 CpGs, Table S6). Our top finding among the 6,073 FDR-significant CpGs was for AHRR (MIM: 606517) cg05575921 (p value = 1.64 × 10−193), which is the top most statistically significant CpG in many other studies evaluating either personal smoking or maternal smoking during pregnancy.
Figure 2.
Volcano Plot Indicating the Direction of Effects for the Meta-analysis of the Association between Sustained Maternal Smoking during Pregnancy and DNA Methylation in Newborn Cord Blood
Table 2.
Meta-analysis Results from Newborns for Selected Loci Not Previously Reported with Genome-wide Statistically Significant Differential Methylation in Newborn DNA in Relation to Sustained Maternal Smoking in Pregnancy
| Chr. | Position | CpG | Mapped Genea | Nearest Gene (10 Mb)b | Gene Groupc | Regression Coefficient | SE | p Value | Direction of Effect across Cohortsd | Mean Betae |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 24648203 | cg06376426 | GRHL3 | GRHL3 | TSS1500; body | −0.004 | 0.001 | 1.84E-04 | −+−−−−−−− | 0.262 |
| 2 | 43685377 | cg20629315 | THADA | THADA | body | 0.003 | 0.001 | 4.62E-04 | −−+++++++ | 0.896 |
| 2 | 206628553 | cg22308949 | NRP2 | NRP2 | body | −0.016 | 0.002 | 7.83E-12 | −−−−−−−−− | 0.413 |
| 2 | 206628625 | cg05348875 | NRP2 | NRP2 | body | −0.026 | 0.004 | 1.13E-10 | −−−−−−−−− | 0.613 |
| 2 | 206628692 | cg14157435 | NRP2 | NRP2 | body | −0.028 | 0.004 | 1.61E-10 | −−−−−−−−− | 0.413 |
| 2 | 206692685 | cg14400541 | – | NRP2 | – | −0.008 | 0.002 | 5.19E-05 | +−−−−−−−− | 0.501 |
| 3 | 189348936 | cg05129081 | TP63 | TP63 | TSS1500 | 0.012 | 0.002 | 1.21E-07 | ++++++−++ | 0.539 |
| 3 | 189349021 | cg06720722 | TP63 | TP63 | TSS200 | 0.009 | 0.002 | 8.49E-06 | ++++++−++ | 0.798 |
| 4 | 10117479 | cg22821355 | WDR1 | WDR1 | body | −0.007 | 0.002 | 1.50E-04 | −−−−−−−−− | 0.382 |
| 4 | 81109888 | cg01789499 | PRDM8 | PRDM8 | 5′ UTR | −0.009 | 0.002 | 1.86E-04 | −−−−−−+−− | 0.852 |
| 4 | 81110205 | cg09595050 | PRDM8 | PRDM8 | 5′ UTR | −0.018 | 0.004 | 1.71E-06 | −−−−−−−−− | 0.739 |
| 4 | 81110459 | cg14197071 | PRDM8 | PRDM8 | 5′ UTR | −0.021 | 0.005 | 5.33E-06 | −−−−−−+−− | 0.723 |
| 4 | 81111177 | cg27111250 | PRDM8 | PRDM8 | 5′ UTR | −0.020 | 0.005 | 1.61E-05 | −−−−−−+−− | 0.708 |
| 4 | 81111393 | cg27639662 | PRDM8 | PRDM8 | 5′ UTR | −0.016 | 0.004 | 3.33E-05 | −−−−−−+−− | 0.702 |
| 4 | 81117647 | cg05452645 | PRDM8 | PRDM8 | TSS1500; 5′ UTR | −0.022 | 0.004 | 8.95E-09 | −−−−−−+−− | 0.520 |
| 4 | 81117665 | cg00138041 | PRDM8 | PRDM8 | TSS1500; 5′ UTR | −0.021 | 0.004 | 1.39E-06 | −−−−−−+−− | 0.556 |
| 4 | 81117853 | cg06373870 | PRDM8 | PRDM8 | TSS1500; 5′ UTR | −0.017 | 0.003 | 1.24E-08 | −−−−−−+−− | 0.422 |
| 4 | 81118188 | cg03463411 | PRDM8 | PRDM8 | TSS1500; 5′ UTR | −0.014 | 0.003 | 1.09E-06 | −−−−−−+−− | 0.372 |
| 4 | 81118343 | cg04235768 | PRDM8 | PRDM8 | TSS1500; 5′ UTR | −0.014 | 0.002 | 1.35E-09 | −−−−−−+−− | 0.159 |
| 4 | 81118588 | cg26299084 | PRDM8 | PRDM8 | 5′ UTR; TSS200 | −0.012 | 0.003 | 1.98E-06 | −−−−−−+−− | 0.247 |
| 4 | 81118794 | cg06307913 | PRDM8 | PRDM8 | 5′ UTR; 1st exon | −0.020 | 0.003 | 3.72E-09 | −−−−−−+−− | 0.424 |
| 4 | 81119178 | cg27242132 | PRDM8 | PRDM8 | 5′ UTR | −0.022 | 0.004 | 2.98E-09 | −−−−−−+−− | 0.240 |
| 4 | 81119198 | cg18073471 | PRDM8 | PRDM8 | 5′ UTR | −0.018 | 0.003 | 1.21E-08 | −−−−−−+−− | 0.178 |
| 4 | 81119249 | cg02458885 | PRDM8 | PRDM8 | 5′ UTR | −0.010 | 0.002 | 5.94E-06 | −−−−−−+−− | 0.189 |
| 4 | 81119299 | cg11388320 | PRDM8 | PRDM8 | 5′ UTR | −0.023 | 0.004 | 1.12E-08 | −−−−−−−−− | 0.324 |
| 4 | 81119473 | cg22902505 | PRDM8 | PRDM8 | 5′ UTR | −0.027 | 0.004 | 1.21E-10 | −−−−−−+−− | 0.433 |
| 4 | 81122726 | cg05522011 | PRDM8 | PRDM8 | body | −0.015 | 0.004 | 2.00E-04 | −−−−−−+−− | 0.799 |
| 5 | 78365647 | cg01856645 | DMGDH; BHMT2 | BHMT2 | TSS200; body | 0.008 | 0.002 | 3.35E-06 | ++++++−++ | 0.177 |
| 5 | 78365687 | cg06501366 | BHMT2; DMGDH | BHMT2 | body; TSS1500 | 0.018 | 0.003 | 1.11E-10 | +++++++++ | 0.408 |
| 5 | 78365691 | cg08328513 | BHMT2; DMGDH | BHMT2 | body; TSS1500 | 0.017 | 0.003 | 3.94E-09 | ++++++−++ | 0.265 |
| 5 | 78365710 | cg23911707 | BHMT2; DMGDH | BHMT2 | body; TSS1500 | 0.006 | 0.001 | 5.69E-06 | +++++++++ | 0.260 |
| 5 | 78365801 | cg03400060 | BHMT2; DMGDH | BHMT2 | body; TSS1500 | 0.012 | 0.002 | 2.96E-10 | +++++++++ | 0.392 |
| 5 | 78366076 | cg01902605 | BHMT2; DMGDH | BHMT2 | body; TSS1500 | 0.013 | 0.002 | 1.50E-09 | +++++++++ | 0.707 |
| 6 | 7673306 | cg25370658 | – | BMP6 | – | 0.004 | 0.001 | 2.63E-04 | +++−+−−++ | 0.806 |
| 6 | 7698374 | cg17951878 | – | BMP6 | – | 0.013 | 0.003 | 1.15E-06 | +++++++++ | 0.286 |
| 6 | 7731280 | cg23623251 | BMP6 | BMP6 | body | 0.006 | 0.002 | 3.25E-04 | +++++++++ | 0.783 |
| 6 | 10405499 | cg16199280 | TFAP2A | TFAP2A | body | 0.006 | 0.002 | 3.26E-04 | +−+++++++ | 0.342 |
| 6 | 55767865 | cg16728651 | – | BMP5 | – | −0.010 | 0.002 | 1.05E-06 | −−−−−−−−− | 0.729 |
| 6 | 152011415 | cg08161546 | ESR1 | ESR1 | TSS1500 | 0.008 | 0.002 | 3.50E-04 | +−+++++++ | 0.709 |
| 6 | 152124815 | cg08415493 | ESR1 | ESR1 | 5′ UTR | −0.003 | 0.001 | 1.74E-04 | −+−−−−−−− | 0.706 |
| 6 | 152126736 | cg20893956 | ESR1 | ESR1 | 5′ UTR; TSS200 | −0.009 | 0.002 | 4.13E-05 | −−+−−−+−− | 0.620 |
| 6 | 152126785 | cg07746998 | ESR1 | ESR1 | 5′ UTR; TSS200 | −0.006 | 0.002 | 1.18E-04 | −+−−−−−−− | 0.594 |
| 6 | 152126895 | cg21157690 | ESR1 | ESR1 | 5′ UTR; 1st exon | −0.008 | 0.002 | 5.70E-05 | −++−−−−−− | 0.747 |
| 6 | 152126938 | cg17264271 | ESR1 | ESR1 | 5′ UTR; 1st exon | −0.009 | 0.002 | 1.26E-06 | −++−−−−−− | 0.627 |
| 6 | 152130058 | cg04063345 | ESR1 | ESR1 | body | −0.013 | 0.004 | 1.22E-04 | −++−−−+−− | 0.507 |
| 6 | 152130207 | cg15626350 | ESR1 | ESR1 | body | −0.018 | 0.004 | 1.42E-06 | −+−−−−+−− | 0.444 |
| 6 | 152130332 | cg00601836 | ESR1 | ESR1 | body | −0.014 | 0.003 | 1.19E-06 | −++−−−−−− | 0.676 |
| 8 | 1403050 | cg16442298 | – | DLGAP2 | – | −0.004 | 0.001 | 3.28E-04 | −−−−−−−−− | 0.716 |
| 8 | 1404023 | cg03551508 | – | DLGAP2 | – | −0.007 | 0.002 | 2.74E-05 | −−−−−−−−− | 0.746 |
| 8 | 1427491 | cg00827210 | – | DLGAP2 | – | −0.007 | 0.001 | 4.83E-07 | −−−−−−+−− | 0.882 |
| 8 | 1442292 | cg13063207 | – | DLGAP2 | – | −0.006 | 0.001 | 1.85E-05 | −−−−−−−−− | 0.847 |
| 8 | 1458508 | cg24526596 | DLGAP2 | DLGAP2 | 5′ UTR | −0.005 | 0.001 | 2.52E-04 | −−−−−−+−− | 0.590 |
| 8 | 1462903 | cg25955692 | DLGAP2 | DLGAP2 | 5′ UTR | −0.005 | 0.001 | 1.67E-05 | ++−−−−−−− | 0.874 |
| 8 | 1468625 | cg00598912 | DLGAP2 | DLGAP2 | 5′ UTR | −0.003 | 0.001 | 1.39E-04 | −−−−−−−−− | 0.831 |
| 8 | 1494546 | cg23424125 | DLGAP2 | DLGAP2 | 5′ UTR | −0.010 | 0.003 | 3.23E-04 | −−−−−−+−− | 0.850 |
| 8 | 1501226 | cg03185622 | DLGAP2 | DLGAP2 | body | −0.005 | 0.001 | 5.33E-07 | +−−−−+−−− | 0.825 |
| 8 | 1526540 | cg15833940 | DLGAP2 | DLGAP2 | body | −0.013 | 0.003 | 6.70E-06 | −−−−−−−−− | 0.659 |
| 8 | 1534376 | cg02840179 | DLGAP2 | DLGAP2 | body | −0.004 | 0.001 | 1.05E-04 | −−−−−−−−− | 0.816 |
| 8 | 1615080 | cg02709139 | DLGAP2 | DLGAP2 | body | −0.007 | 0.002 | 1.32E-05 | −−−−−−+−− | 0.870 |
| 8 | 1616381 | cg04687241 | DLGAP2 | DLGAP2 | body | −0.008 | 0.002 | 6.92E-06 | −+−−−−−−− | 0.666 |
| 8 | 1618448 | cg06040034 | DLGAP2 | DLGAP2 | body | −0.013 | 0.003 | 2.42E-06 | −−−−−−−−− | 0.619 |
| 8 | 1649758 | cg02083412 | DLGAP2 | DLGAP2 | 3′ UTR | −0.004 | 0.001 | 3.15E-05 | −−−−−−−−+ | 0.129 |
| 8 | 1649868 | cg22763586 | DLGAP2 | DLGAP2 | 3′ UTR | −0.013 | 0.003 | 5.83E-07 | −−−−−−+−− | 0.450 |
| 8 | 1650172 | cg27351978 | DLGAP2 | DLGAP2 | 3′ UTR | −0.015 | 0.004 | 8.69E-05 | −−−−−−+−− | 0.566 |
| 8 | 1650309 | cg02690013 | DLGAP2 | DLGAP2 | 3′ UTR | −0.013 | 0.003 | 9.92E-06 | −−−−−−+−− | 0.599 |
| 14 | 54412780 | cg23104439 | – | BMP4 | – | 0.005 | 0.001 | 2.08E-04 | ++++++++− | 0.741 |
| 14 | 54418728 | cg05928290 | BMP4 | BMP4 | body | 0.024 | 0.003 | 1.48E-19 | +++++++++ | 0.759 |
| 14 | 54418804 | cg05923197 | BMP4 | BMP4 | body | 0.029 | 0.003 | 1.08E-18 | +++++++++ | 0.699 |
| 14 | 54418851 | cg09367901 | BMP4 | BMP4 | body | 0.019 | 0.002 | 3.98E-17 | +++++++++ | 0.827 |
| 14 | 54419614 | cg08046044 | BMP4 | BMP4 | 5′ UTR | 0.005 | 0.001 | 2.70E-09 | +++−+++++ | 0.077 |
| 14 | 54424149 | cg24526899 | BMP4 | BMP4 | TSS1500 | 0.007 | 0.002 | 5.14E-04 | ++++++++− | 0.441 |
| 17 | 76930245 | cg04999637 | – | TIMP2 | – | 0.005 | 0.001 | 6.91E-05 | +++++++++ | 0.583 |
Meta-analysis results of the association between sustained maternal smoking during pregnancy and DNA methylation in newborns, adjusted for covariates, using normalized methylation betas as the outcome. Selected not previously reported loci genome-wide significant after FDR correction. Results sorted by the chromosome (chr.) and position of the CpG sites listed. Selection limited to genes prioritized for discussion.
UCSC Genome Browser annotated gene.
Nearest gene (within 10 Mb) symbol, determined with the Snipper program.
UCSC gene region feature category. Regions for the gene and related isoforms are listed.
Direction of effect across cohorts included in the statistical model: maternal smoking during pregnancy associated with increased (+) or decreased (−) methylation in alphabetical order of cohorts.
Average of the mean methylation beta values across the newborn cohorts. For complete listing of CpGs differentially methylated in relation to sustained maternal smoking during pregnancy and for results from meta-analysis models unadjusted for covariates and adjusted for covariates and cell type, see Table S3.
We found our results to be robust to different analytic approaches. We present results from models using normalized betas as the outcome. When using raw betas as the outcome, we observed little difference in the results (Spearman’s correlation coefficient = 0.96 for regression coefficients; 0.98 for log10[p values] for our significant findings). Furthermore, exclusion of the one cohort with newborns of non-European ancestry (NEST) from the model representing sustained maternal smoking provided similar results (Spearman’s correlation coefficient = 0.99 for regression coefficients; 0.89 for log10[p values]).
Examination of Potentially Polymorphic and Cross-Reactive Probes
A total of 742 of the 6,073 FDR-significant CpGs overlapped with the list of 70,889 potentially polymorphic probes provided by a table of Chen et al.51 Only 137 of the 6,073 FDR-significant CpGs overlapped with the list of 29,233 cross-reactive probes annotated by Chen et al. Many of the probes flagged by Chen et al. are associated with very low-frequency SNPs and thus are likely to have minimal impact on results in most datasets. In visual inspection of the density plots of all 742 such probes, we flagged 19 CpGs as having a possible deviation from unimodality (listed in Table S7). However, results from the dip test52 applied to all 6,073 FDR significant CpGs identified only four CpGs as statistically significantly deviated from unimodality (FDR adjusted p < 0.05; cg11459648, cg17847044, cg15028160, and cg25849281).
Persistence in Older Children of DNA Methylation Related to Maternal Smoking during Pregnancy
Because of the smaller sample size and smaller proportion of children exposed to maternal smoking in the older children models, we had less statistical power than we did for the newborn models. When we compared the coefficients for newborns and older children for all 6,073 CpGs that were significantly associated with maternal smoking in newborns, 4,403 (73%) had a consistent direction of effect and all 6,073 (100%) gave nominal p values <0.05 for the older children models, which is higher than the 5% expected by chance alone (Kolmogorov p value < 2.2 × 10−16). Among these, 3,722 CpGs (61%) had a weaker effect size (attenuation) in the older children than in the newborns, but the attenuation overall was very small in magnitude and not significant (mean attenuation = −0.00039, SD = 0.0059). Compared to CpGs in newborns, of the 148 CpGs that met FDR significance at lookup replication level in the older children (Table S8), 100% were consistent in the direction of effect, and there was attenuation for 32%, again small in magnitude and not significant (mean attenuation = −0.00008, SD = 0.018).
Enrichment Analysis
For our 6,073 FDR-significant CpGs, we observed enrichment for localization to CpG island shores (35% versus 23% overall as compared to all CpGs on the array, p value = 2.8 × 10−100), enhancers (29% versus 22% overall, p value = 5.7 × 10−45), and DNase hypersensitivity sites (14% versus 12% overall, p value = 2.8 × 10−7). Conversely, we found relative depletion in CpG islands (18% versus 31% overall, p value = 9.1 × 10−116), FANTOM promoters (2.5% versus 6.7% overall, p value = 2.1 × 10−49), and promoter-associated regions (13% versus 19% overall, p value = 2.3 × 10−33). There was no statistically significant enrichment or depletion of sites mapping to imprinted DMRs (0.082% versus 0.16% overall, p value = 0.107).
Pathway Analysis
Our pathway analyses indicated that the FDR-significant CpG sites corresponded to genes enriched for several categories of biological processes, including anatomical development, phosphate-containing compound metabolism, nervous system development, and cell communication processes (Figure S2). Based on DAVID, eight biological processes were enriched, including GTPase signal transduction, neuronal differentiation, and protein kinase activity (Figure S3). The top statistically significantly enriched diseases and biofunctions identified through Ingenuity software included tumor adhesion, neuron development, and connective tissue differentiation (Figure S4).
Functional Network Analysis
Functional network analysis revealed 447 significantly enriched GO terms after FDR correction was applied (q value < 0.01 for this analysis, Table S9). The majority of the enriched terms, and particularly the most statistically significant ones, pointed toward biological processes related to cell, tissue, or organ development, proliferation, morphogenesis, differentiation, growth, and other biologically relevant processes. There were also several enriched processes related to embryonic morphogenesis or development.
Methylation Transcription Analysis
To assess transcriptional effects related to methylation differences, we investigated whether methylation status correlated with gene expression levels for our 2,965 CpGs associated with sustained maternal smoking in newborns that we identified through literature review as not having been previously reported. In the Rotterdam Study dataset of adults, out of the 2,636 (of the 2,965) CpGs that we were able to match to a gene transcript (+/− 250 kb), 254 unique CpGs (343 total CpG-gene transcript associations) were significantly associated with expression of a nearby gene in whole blood from adults (FDR p value < 0.05, Table S10). We observed strong associations for several CpGs annotated to the same gene and corresponding gene expression levels, most strikingly for IL32 (MIM: 606001), which had four CpGs associated with its expression, and HOXB2 (MIM: 142967), which had several CpGs related to its expression (lowest p value 2.38 × 10−72, Table S10). In the much smaller study of children at age four from INMA (n = 107), 35 CpGs were associated with gene expression (FDR p value < 0.05). The following six genes had CpGs with methylation that was statistically significantly related to gene expression in both the Rotterdam Study adults and INMA children: ENOSF1 (MIM: 607427), HOXB2 (MIM: 142967, IL32 (MIM: 606001), NLRP2 (MIM: 609364), PASK (MIM: 607505), and TDRD9. In both the adult and child datasets, for the majority of CpGs statistically significantly associated with expression, the direction was inverse (higher methylation, lower expression). This inverse relationship represented 68% (Clopper-Pearson 95% confidence interval = 62%–73%) of the adult associations and 79% (Clopper-Pearson 95% confidence interval = 62%–91%) of the associations for children.
Newborn DNA Methylation Related to Any Maternal Smoking during Pregnancy
In addition to the sustained smoking model, we meta-analyzed the effect of any maternal smoking during pregnancy on newborn methylation. As expected, based on previous literature,12 we found that despite the much larger number of women with any smoking during pregnancy, there were fewer statistically significant findings for this less specific exposure (4,653 FDR-significant CpGs, Table S3).
Discussion
We combined data across studies in a large-scale epigenome-wide meta-analysis to evaluate the association between maternal smoking during pregnancy and DNA methylation in offspring. We established the PACE consortium to study this association and used 13 birth cohort studies from the US and Europe that, with the same reproducible platform, measured CpG-specific DNA methylation across the epigenome in newborns. Combining these studies resulted in the discovery of 6,073 statistically significant CpGs; 3,932 remained statistically significant after adjustment for cell type proportion. Our results are remarkably robust to different modeling techniques. Our findings were very similar when using either the raw methylation betas or the normalized betas as the outcome. This is despite the variety of data processing methods used across the cohorts for normalization and corrections for technical variables such as batch (described in the Supplemental Note). This consistency is reassuring given the range of published methods available for researchers to apply to 450K DNA methylation data for quality control, normalization, and adjustment for technical variation. Furthermore, our main findings persisted after cell type adjustment (Table S3, Figure S1).
As predicted based on earlier evaluation of top findings for maternal smoking in the MoBa cohort,12 we had fewer statistically significant findings for any smoking during pregnancy than for sustained smoking during pregnancy (Figure S5). Nonetheless, with the large sample size of this meta-analysis, we still observed many statistically significant CpGs after FDR correction in the any smoking models, and the directions of effect and p values were similar to those from the sustained smoking models (Table S3, Figure S6). However, the stronger signal for sustained smoking suggests that this might be the more powerful variable for studying epigenetic effects and possible health outcomes from this exposure in offspring.
Our observation of a large number of genome-wide significant CpGs related to maternal smoking is not surprising given reports of multiple genome-wide significant loci identified in single studies, all with smaller sample sizes.7, 8, 10, 22, 37, 38 Reassuringly, among our myriad findings, the top hit in all newborn models was AHRR cg05575921 (p value < 1.64 × 10−193), which has been observed as differentially methylated in relation to smoking in many studies of adults and children.7, 8, 10, 28, 35, 53
Our enrichment testing of the genome-wide results is in line with previous findings showing that island shores, enhancers, and DNase I hypersensitive sites are more dynamic (susceptible to methylation changes) than promoter regions54 and imprinted loci.55 These regions might be more resistant to changes in DNA methylation in response to in utero exposure.55 Thus, it is not surprising that associations between maternal smoking and newborn methylation might be more likely to be found in island shore and enhancer regions as opposed to promoters or CpG islands.
To assess the underlying biology involved in the associated genomic regions, we applied pathway and functional analyses, as well as tests of enrichment. These results implicated numerous neurological pathways, pathways involved in embryogenesis, and various developmental pathways. These observations could provide insight into the etiology of childhood health outcomes related to maternal smoking during pregnancy.
We focus discussion on some specific genes among the associations that, according to our literature review, had not been previously reported (2,965 CpGs annotating to 2,017 mapped or nearest genes). For 27 of these genes, mutations or SNPs have been implicated in susceptibility to orofacial clefts (as identified with the Snipper program described in the Supplemental Note). This includes the following genes (each representing one FDR-significant CpG unless otherwise specified): BHMT2 ([MIM: 605932] six CpGs), GRHL3 (MIM: 608317), THADA (MIM: 611800), GAD67 (MIM: 605363), TP63 ([MIM: 603273] two CpGs), MSX1 (MIM: 142983), WDR1 (MIM: 604734), SPP1 (MIM: 166490), BMP6 (MIM: 112266), TFAP2A (MIM: 107580), COL11A2 ([MIM: 120290] three CpGs), PDGFRA (MIM: 173490), MN1 (MIM: 156100), MSX2 ([MIM: 123101] four CpGs), PVT1 (MIM: 165140), ZIC2 (MIM: 603073), HOXA2 ([MIM: 604685] ten CpGs), WNT3 (MIM: 165330), RUNX2 ([MIM: 600211] two CpGs), TERT (MIM: 187270), SPATA13 ([MIM: 613324] two CpGs), VAX1 (MIM: 604294), TIMP2 (MIM: 188825), NOG (MIM: 602991), BEST3 (MIM: 607337), MYH9 (MIM: 160775), and BMP4 ([MIM: 112262] six CpGs) (results in Table S5). Although this does not imply that the smoking-related CpGs are on the causal pathway, we note that the Surgeon General’s Report summarizes the evidence as sufficient to infer a causal relationship between maternal smoking during pregnancy and these birth defects.2 Many of these genes also have varied biological effects relevant to other aspects of development.
Among this group of genes previously related to orofacial clefts, bone morphogenetic protein 4 (BMP4) is especially interesting. Maternal smoking might interact with SNPs in this gene in relation to oral clefts.56 We identified six CpGs in BMP4 at genome-wide significance in newborns; two remained statistically significant in the older children. In addition to orofacial clefts, SNPs in BMP4 are related to tooth development and eruption, as well as to colorectal cancer in genome-wide association studies (GWASs).57 BMPs, including BMP4,58 also play an important role in lung development: reduced lung function among infants is an established consequence of maternal smoking during pregnancy.2 A plot showing greater detail on the CpGs in or near BMP4 is provided in Figure S7.
We observed six CpGs significantly related to maternal smoking in betaine-homocysteine methyltransferase (BHMT2; Figure S8). Genetic variants in this gene have been associated with orofacial clefts in candidate-gene studies59 and with selenium levels in GWASs.60, 61 Of note, in experimental studies, selenium has been shown to protect against orofacial clefts induced by exposure to teratogens.62 In the Cancer Genome Atlas, methylation of BHMT2 in lung adenocarcinoma (lung cancer [MIM: 211980]) was strongly correlated (3rd rank genome wide) with smoking history.63
The gene PRDM8 (PR domain containing 8 [MIM: 616639]) has the largest number of CpGs (18 of 61, based on Illumina annotation) significantly associated with maternal smoking during pregnancy. Maternal smoking during pregnancy was associated with decreased methylation throughout the gene. PRDM8 is one of several PRDMs belonging to the SET domain family of histone methyltransferases.64 PRDM genes either act as direct histone methyltransferases or recruit a suite of histone-modifying enzymes to target promoters.65 PRDM8 specifically methylates H3K9 of histones to repress transcriptional activity.66 PRDM8 expression is tightly regulated in a spatiotemporal manner during neural development;67 it regulates morphological transition in neocortical development68 and forms part of a repressor complex that directs, through regulation of cadherin-11, neural circuit assembly.69 Thus PRDM8 appears to play an important role in neurologic development.
DLGAP2 (discs large homolog-associated protein 2 [MIM: 605438]) is another gene with a large number of significant CpGs (14 of 192 tested) associated with maternal smoking in our study. DLGAP2, also known as SAPAP2, belongs to a gene family that encodes SAP90/PSD95-associated proteins (SAPAPs), and is known to be involved in the molecular organization of synapses and in neuronal cell signaling.70 DLGAP2 was first identified in studies of progressive epilepsy with mental retardation (EPMR [MIM: 610003])71 and has been associated with other CNS disorders such as schizophrenia (SCZD [MIM: 181500])72 and autism spectrum disorders (ASD [MIM: 209850]).73, 74 Differential methylation at this locus in a rat model appears to play a role in the development of post-traumatic stress disorder.75
The neuropilin-2 (NRP2 [MIM: 602070]) gene had three CpGs located in close proximity (among 48 tested) that were statistically significantly associated with maternal smoking during pregnancy. NRP2 is one of two transmembrane receptors for axonal guidance cues of the class 3 semaphorin (SEMA) family and is expressed in sympathetic neural crest cells and their progeny.76 It might also be required in vivo for sorting migrating cortical and striatal interneurons to their correct destination.77 NRP2 also functions as a receptor for some forms of vascular endothelial growth factor, thereby playing a crucial role in angiogenesis and lymphangiogenesis.78 Polymorphisms in NRP2 have been associated with several diseases, including autism77 and multiple cancers.79, 80, 81, 82, 83
Hypermethylation of ESR1 (estrogen receptor 1 [MIM:133430], a key nuclear transcription factor) on chromosome 6q25.1 is well-studied in relation to presence and prognosis of various malignancies such as breast cancer and hepatocellular carcinoma,84, 85 as well as asthma.86, 87 We found an inverse association between maternal smoking and methylation levels for seven out of the eight FDR-significant ESR1 CpGs. ESR1 hypomethylation has been reported in relation to induced microRNA expression (synthetic miR-29b oligonucleotides) in acute myeloid leukemia cells.88
To evaluate possible functional gene expression effects of methylation at the CpGs that we found to be significantly related to maternal smoking, we analyzed data from two studies—one of adults and another of children at age four years. Although on first pass, one might expect a higher proportion of the CpGs related to maternal smoking to also be related to gene expression, there are several factors that decrease the likelihood of seeing significant associations. Most importantly, the sample size for discovery of the methylation association with smoking was much larger than that of the datasets available to correlate gene expression and methylation (about 10-fold smaller for the adult gene expression dataset and about 60-fold smaller for the childhood dataset). In addition, gene expression in blood might be more transient than methylation, decreasing the ability to find significant associations with a single gene-expression measurement. Furthermore, constitutive gene expression is measured in this setting, whereas many genes are inducible and methylation might contribute to this process. Lastly, some in-utero-induced changes to methylation could have affected transcription during fetal development but not in postnatal life, and might have transcription-independent functional mechanisms. Nonetheless, we observed significant associations between methylation and gene expression at six genes in both the adults and the children. The majority of CpGs significantly associated with expression were in the commonly expected direction of methylation related to gene silencing. Notably, CpGs in IL32 (Figure S9), a proinflammatory cytokine involved in several diseases such as asthma89 and cancer,90 HOXB2, a transcription factor involved in development91 and several cancer forms,92 and PASK (PAS domain containing serine/threonine kinase), involved in glucose homeostasis,93 were significantly associated with expression in both datasets.
We analyzed the associations of CpGs with expression levels of genes within a region of 250 kb up- or downstream of the CpG. Consensus on the optimal physical distance for these analyses is lacking. However, in a recent study, associations between CpGs and SNPs were the strongest when within close proximity (500 kb) of the CpG.48 Despite the limitations with the expression datasets included in our study, we believe that the transcriptomics data provide functional support for our maternal smoking findings.
In older children, all of the CpGs significantly associated with maternal smoking in newborns gave at least nominal levels of significance (p value < 0.05). This skew of the distribution of p values toward small values was much more than expected by chance (Kolmogorov p value < 2.2 × 10−16), demonstrating a very high level of replication and persistence of findings at birth into later childhood. This is consistent with and substantially extends a few previous reports.9, 12 We had only very limited data with repeat measures in the same individuals so we could not meta-analyze change in methylation over time.
This inaugural paper from the PACE consortium represents a major effort to combine data from many studies in a large-scale meta-analysis of epigenome-wide association studies of maternal smoking in relation to methylation in newborns. We report at least an order of magnitude more genes differentially methylated in response to maternal smoking than have been identified in any previous study. This suggests that meta-analysis in epigenome-wide association studies produces similar success to that of genome-wide association SNP studies in the identification of biologically meaningful loci. The similarity in the results obtained when using the raw betas compared to those obtained when using normalized betas generated with various methods indicates that cohort-specific processing methods do not interfere with the ability to perform meta-analysis.
We identified nearly 3,000 CpGs corresponding to genes differentially methylated in offspring in relation to whether their mothers smoked during pregnancy. Some of these genes have been implicated in genetic studies of orofacial clefts or asthma, both conditions related to maternal smoking in pregnancy, and others in the pathogenesis of cancers that are associated with adult smoking, including lung, colorectal (CRC [MIM: 114500]), and liver (HCC [MIM: 114550]).2 We also find substantial persistence of effects of maternal smoking identified in newborns into later childhood. Our findings might implicate epigenetic mechanisms in the etiology of these exposure-disease relationships. This large-scale study also provides confirmation of previously reported loci, many of which have not been previously replicated. Pathway analysis highlights the involvement of identified genes in various developmental pathways, and functional effects at the transcriptomics level were observed for many of the identified CpGs. These findings could provide new insights into the mechanisms involved in the detrimental health outcomes that arise from this important in utero exposure.
Acknowledgments
For all studies, information on funding and acknowledgments can be found in the Supplemental Data.
Published: March 31, 2016
Footnotes
Supplemental Data include a Supplemental Note, nine figures, and ten tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.02.019.
Web Resources
The URLS for data presented herein are as follows:
A Catalog of Published Genome-Wide Association Studies, National Human Genome Research Institute, http://www.genome.gov/gwastudies
Bioconductor, http://www.bioconductor.org/
Infinium HumanMethylation450K v1.2 Product Files, http://support.illumina.com/downloads/infinium_humanmethylation450_product_files.html
Ingenuity, www.ingenuity.com
OMIM, http://www.omim.org/
The R Project for Statistical Computing, R v.3.0.2, http://www.r-project.org/
UCSC Human Genome Browser, http://hgdownload.soe.ucsc.edu/goldenPath/hg19/database/
Supplemental Data
References
- 1.Tong V.T., Dietz P.M., Morrow B., D’Angelo D.V., Farr S.L., Rockhill K.M., England L.J., Centers for Disease Control and Prevention (CDC) Trends in smoking before, during, and after pregnancy--Pregnancy Risk Assessment Monitoring System, United States, 40 sites, 2000-2010. MMWR Surveill. Summ. 2013;62:1–19. [PubMed] [Google Scholar]
- 2.US Department of Health and Human Services . US Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health; Atlanta, GA: 2014. The health consequences of smoking—50 years of progress: A report of the surgeon general. [Google Scholar]
- 3.Moritsugu K.P. The 2006 Report of the Surgeon General: the health consequences of involuntary exposure to tobacco smoke. Am. J. Prev. Med. 2007;32:542–543. doi: 10.1016/j.amepre.2007.02.026. [DOI] [PubMed] [Google Scholar]
- 4.Breton C.V., Siegmund K.D., Joubert B.R., Wang X., Qui W., Carey V., Nystad W., Håberg S.E., Ober C., Nicolae D., Asthma BRIDGE consortium Prenatal tobacco smoke exposure is associated with childhood DNA CpG methylation. PLoS ONE. 2014;9:e99716. doi: 10.1371/journal.pone.0099716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flom J.D., Ferris J.S., Liao Y., Tehranifar P., Richards C.B., Cho Y.H., Gonzalez K., Santella R.M., Terry M.B. Prenatal smoke exposure and genomic DNA methylation in a multiethnic birth cohort. Cancer Epidemiol. Biomarkers Prev. 2011;20:2518–2523. doi: 10.1158/1055-9965.EPI-11-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suter M., Ma J., Harris A., Patterson L., Brown K.A., Shope C., Showalter L., Abramovici A., Aagaard-Tillery K.M. Maternal tobacco use modestly alters correlated epigenome-wide placental DNA methylation and gene expression. Epigenetics. 2011;6:1284–1294. doi: 10.4161/epi.6.11.17819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joubert B.R., Håberg S.E., Nilsen R.M., Wang X., Vollset S.E., Murphy S.K., Huang Z., Hoyo C., Midttun Ø., Cupul-Uicab L.A. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ. Health Perspect. 2012;120:1425–1431. doi: 10.1289/ehp.1205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Markunas C.A., Xu Z., Harlid S., Wade P.A., Lie R.T., Taylor J.A., Wilcox A.J. Identification of DNA methylation changes in newborns related to maternal smoking during pregnancy. Environ. Health Perspect. 2014;122:1147–1153. doi: 10.1289/ehp.1307892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richmond R.C., Simpkin A.J., Woodward G., Gaunt T.R., Lyttleton O., McArdle W.L., Ring S.M., Smith A.D., Timpson N.J., Tilling K. Prenatal exposure to maternal smoking and offspring DNA methylation across the lifecourse: findings from the Avon Longitudinal Study of Parents and Children (ALSPAC) Hum. Mol. Genet. 2015;24:2201–2217. doi: 10.1093/hmg/ddu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Küpers L.K., Xu X., Jankipersadsing S.A., Vaez A., la Bastide-van Gemert S., Scholtens S., Nolte I.M., Richmond R.C., Relton C.L., Felix J.F. DNA methylation mediates the effect of maternal smoking during pregnancy on birthweight of the offspring. Int. J. Epidemiol. 2015;44:1224–1237. doi: 10.1093/ije/dyv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joubert B.R., Haberg S.E., Bell D.A., Nilsen R.M., Vollset S.E., Midttun O., Ueland P.M., Wu M.C., Nystad W., Peddada S.D. Maternal smoking and DNA methylation in newborns: in utero effect or epigenetic inheritance. Cancer Epidemiol. Biomarkers Prev. 2014;23:1007–1017. doi: 10.1158/1055-9965.EPI-13-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee K.W., Richmond R., Hu P., French L., Shin J., Bourdon C., Reischl E., Waldenberger M., Zeilinger S., Gaunt T. Prenatal exposure to maternal cigarette smoking and DNA methylation: epigenome-wide association in a discovery sample of adolescents and replication in an independent cohort at birth through 17 years of age. Environ. Health Perspect. 2015;123:193–199. doi: 10.1289/ehp.1408614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2013. R: A language and environment for statistical computing.http://www.R-project.org/ [Google Scholar]
- 14.Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houseman E.A., Accomando W.P., Koestler D.C., Christensen B.C., Marsit C.J., Nelson H.H., Wiencke J.K., Kelsey K.T. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D., Irizarry R.A. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reinius L.E., Acevedo N., Joerink M., Pershagen G., Dahlén S.E., Greco D., Söderhäll C., Scheynius A., Kere J. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS ONE. 2012;7:e41361. doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benjamini Y., Hochberg Y. Controlling for False Discovery Rate: a Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 20.Besingi W., Johansson A. Smoke-related DNA methylation changes in the etiology of human disease. Hum. Mol. Genet. 2014;23:2290–2297. doi: 10.1093/hmg/ddt621. [DOI] [PubMed] [Google Scholar]
- 21.Breitling L.P., Yang R., Korn B., Burwinkel B., Brenner H. Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am. J. Hum. Genet. 2011;88:450–457. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dogan M.V., Shields B., Cutrona C., Gao L., Gibbons F.X., Simons R., Monick M., Brody G.H., Tan K., Beach S.R., Philibert R.A. The effect of smoking on DNA methylation of peripheral blood mononuclear cells from African American women. BMC Genomics. 2014;15:151. doi: 10.1186/1471-2164-15-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elliott H.R., Tillin T., McArdle W.L., Ho K., Duggirala A., Frayling T.M., Davey Smith G., Hughes A.D., Chaturvedi N., Relton C.L. Differences in smoking associated DNA methylation patterns in South Asians and Europeans. Clin. Epigenetics. 2014;6:4. doi: 10.1186/1868-7083-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flanagan J.M., Brook M.N., Orr N., Tomczyk K., Coulson P., Fletcher O., Jones M.E., Schoemaker M.J., Ashworth A., Swerdlow A. Temporal stability and determinants of white blood cell DNA methylation in the breakthrough generations study. Cancer Epidemiol. Biomarkers Prev. 2015;24:221–229. doi: 10.1158/1055-9965.EPI-14-0767. [DOI] [PubMed] [Google Scholar]
- 25.Guida F., Sandanger T.M., Castagné R., Campanella G., Polidoro S., Palli D., Krogh V., Tumino R., Sacerdote C., Panico S. Dynamics of smoking-induced genome-wide methylation changes with time since smoking cessation. Hum. Mol. Genet. 2015;24:2349–2359. doi: 10.1093/hmg/ddu751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Philibert R.A., Beach S.R., Brody G.H. Demethylation of the aryl hydrocarbon receptor repressor as a biomarker for nascent smokers. Epigenetics. 2012;7:1331–1338. doi: 10.4161/epi.22520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Philibert R.A., Beach S.R., Lei M.K., Brody G.H. Changes in DNA methylation at the aryl hydrocarbon receptor repressor may be a new biomarker for smoking. Clin. Epigenetics. 2013;5:19. doi: 10.1186/1868-7083-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shenker N.S., Polidoro S., van Veldhoven K., Sacerdote C., Ricceri F., Birrell M.A., Belvisi M.G., Brown R., Vineis P., Flanagan J.M. Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum. Mol. Genet. 2013;22:843–851. doi: 10.1093/hmg/dds488. [DOI] [PubMed] [Google Scholar]
- 29.Siedlinski M., Klanderman B., Sandhaus R.A., Barker A.F., Brantly M.L., Eden E., McElvaney N.G., Rennard S.I., Stocks J.M., Stoller J.K. Association of cigarette smoking and CRP levels with DNA methylation in alpha-1 antitrypsin deficiency. Epigenetics. 2012;7:720–728. doi: 10.4161/epi.20319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Y.V., Smith A.K., Conneely K.N., Chang Q., Li W., Lazarus A., Smith J.A., Almli L.M., Binder E.B., Klengel T. Epigenomic association analysis identifies smoking-related DNA methylation sites in African Americans. Hum. Genet. 2013;132:1027–1037. doi: 10.1007/s00439-013-1311-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsaprouni L.G., Yang T.P., Bell J., Dick K.J., Kanoni S., Nisbet J., Vinuela A., Grundberg E., Nelson C.P., Meduri E. Cigarette smoking reduces DNA methylation levels at multiple genomic loci but the effect is partially reversible upon cessation. Epigenetics. 2014;9:1382–1396. doi: 10.4161/15592294.2014.969637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wan E.S., Qiu W., Baccarelli A., Carey V.J., Bacherman H., Rennard S.I., Agusti A., Anderson W., Lomas D.A., Demeo D.L. Cigarette smoking behaviors and time since quitting are associated with differential DNA methylation across the human genome. Hum. Mol. Genet. 2012;21:3073–3082. doi: 10.1093/hmg/dds135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wan E.S., Qiu W., Carey V.J., Morrow J., Bacherman H., Foreman M.G., Hokanson J.E., Bowler R.P., Crapo J.D., DeMeo D.L. Smoking-Associated Site-Specific Differential Methylation in Buccal Mucosa in the COPDGene Study. Am. J. Respir. Cell Mol. Biol. 2015;53:246–254. doi: 10.1165/rcmb.2014-0103OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaghlool S.B., Al-Shafai M., Al Muftah W.A., Kumar P., Falchi M., Suhre K. Association of DNA methylation with age, gender, and smoking in an Arab population. Clin. Epigenetics. 2015;7:6. doi: 10.1186/s13148-014-0040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeilinger S., Kühnel B., Klopp N., Baurecht H., Kleinschmidt A., Gieger C., Weidinger S., Lattka E., Adamski J., Peters A. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS ONE. 2013;8:e63812. doi: 10.1371/journal.pone.0063812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chhabra D., Sharma S., Kho A.T., Gaedigk R., Vyhlidal C.A., Leeder J.S., Morrow J., Carey V.J., Weiss S.T., Tantisira K.G. Fetal lung and placental methylation is associated with in utero nicotine exposure. Epigenetics. 2014;9:1473–1484. doi: 10.4161/15592294.2014.971593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harlid S., Xu Z., Panduri V., Sandler D.P., Taylor J.A. CpG sites associated with cigarette smoking: analysis of epigenome-wide data from the Sister Study. Environ. Health Perspect. 2014;122:673–678. doi: 10.1289/ehp.1307480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ivorra C., Fraga M.F., Bayón G.F., Fernández A.F., Garcia-Vicent C., Chaves F.J., Redon J., Lurbe E. DNA methylation patterns in newborns exposed to tobacco in utero. J. Transl. Med. 2015;13:25. doi: 10.1186/s12967-015-0384-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maccani J.Z., Koestler D.C., Houseman E.A., Marsit C.J., Kelsey K.T. Placental DNA methylation alterations associated with maternal tobacco smoking at the RUNX3 gene are also associated with gestational age. Epigenomics. 2013;5:619–630. doi: 10.2217/epi.13.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Infinium HumanMethylation450K v1.2 Product Files (2014). http://support.illumina.com/downloads/infinium_humanmethylation450_product_files.html.
- 41.Martin T.C., Yet I., Tsai P.C., Bell J.T. coMET: visualisation of regional epigenome-wide association scan results and DNA co-methylation patterns. BMC Bioinformatics. 2015;16:131. doi: 10.1186/s12859-015-0568-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Court F., Tayama C., Romanelli V., Martin-Trujillo A., Iglesias-Platas I., Okamura K., Sugahara N., Simón C., Moore H., Harness J.V. Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment. Genome Res. 2014;24:554–569. doi: 10.1101/gr.164913.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Triche, T. (2014). IlluminaHumanMethylation450k.db: Illumina Human Methylation 450k annotation data. R package version 2.0.8. http://www.bioconductor.org/packages/release/data/annotation/html/IlluminaHumanMethylation450k.db.html.
- 44.Alexa, A., and Rahnenfuhrer, J. (2010). topGO: Enrichment analysis for Gene Ontology. R package version 2.18.0. http://bioconductor.org/packages/release/bioc/html/topGO.html.
- 45.Huang W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 46.Warde-Farley D., Donaldson S.L., Comes O., Zuberi K., Badrawi R., Chao P., Franz M., Grouios C., Kazi F., Lopes C.T. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:214–220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaez A., Jansen R., Prins B.P., Hottenga J.J., de Geus E.J., Boomsma D.I., Penninx B.W., Nolte I.M., Snieder H., Alizadeh B.Z. In Silico Post Genome-Wide Association Studies Analysis of C-Reactive Protein Loci Suggests an Important Role for Interferons. Circ Cardiovasc Genet. 2015;8:487–497. doi: 10.1161/CIRCGENETICS.114.000714. [DOI] [PubMed] [Google Scholar]
- 48.Zhang H., Wang F., Kranzler H.R., Yang C., Xu H., Wang Z., Zhao H., Gelernter J. Identification of methylation quantitative trait loci (mQTLs) influencing promoter DNA methylation of alcohol dependence risk genes. Hum. Genet. 2014;133:1093–1104. doi: 10.1007/s00439-014-1452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hofman A., Darwish Murad S., van Duijn C.M., Franco O.H., Goedegebure A., Ikram M.A., Klaver C.C., Nijsten T.E., Peeters R.P., Stricker B.H. The Rotterdam Study: 2014 objectives and design update. Eur. J. Epidemiol. 2013;28:889–926. doi: 10.1007/s10654-013-9866-z. [DOI] [PubMed] [Google Scholar]
- 50.Pidsley R., Y Wong C.C., Volta M., Lunnon K., Mill J., Schalkwyk L.C. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14:293. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Y.A., Lemire M., Choufani S., Butcher D.T., Grafodatskaya D., Zanke B.W., Gallinger S., Hudson T.J., Weksberg R. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hartigan J.A., Hartigan P.M. The Dip Test of Unimodality. Ann. Stat. 1985;13:70–84. [Google Scholar]
- 53.Monick M.M., Beach S.R., Plume J., Sears R., Gerrard M., Brody G.H., Philibert R.A. Coordinated changes in AHRR methylation in lymphoblasts and pulmonary macrophages from smokers. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2012;159B:141–151. doi: 10.1002/ajmg.b.32021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ziller M.J., Gu H., Müller F., Donaghey J., Tsai L.T., Kohlbacher O., De Jager P.L., Rosen E.D., Bennett D.A., Bernstein B.E. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ivanova E., Chen J.H., Segonds-Pichon A., Ozanne S.E., Kelsey G. DNA methylation at differentially methylated regions of imprinted genes is resistant to developmental programming by maternal nutrition. Epigenetics. 2012;7:1200–1210. doi: 10.4161/epi.22141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Q., Wang H., Schwender H., Zhang T., Hetmanski J.B., Chou Y.H., Ye X., Yeow V., Chong S.S., Zhang B. Joint testing of genotypic and gene-environment interaction identified novel association for BMP4 with non-syndromic CL/P in an Asian population using data from an International Cleft Consortium. PLoS ONE. 2014;9:e109038. doi: 10.1371/journal.pone.0109038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burdett, T., Hall, P.N., Hasting, E., Hindorff, L.A., Junkins, H.A., Klemm, A.K., MacArthur, J., Manolio, T.A., Morales, J., Parkinson, H., et al. The NHGRI-EBI Catalog of published genome-wide association studies. www.ebi.ac.uk/gwas. Accessed June, 2015.
- 58.Arora R., Metzger R.J., Papaioannou V.E. Multiple roles and interactions of Tbx4 and Tbx5 in development of the respiratory system. PLoS Genet. 2012;8:e1002866. doi: 10.1371/journal.pgen.1002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mostowska A., Hozyasz K.K., Biedziak B., Misiak J., Jagodzinski P.P. Polymorphisms located in the region containing BHMT and BHMT2 genes as maternal protective factors for orofacial clefts. Eur. J. Oral Sci. 2010;118:325–332. doi: 10.1111/j.1600-0722.2010.00757.x. [DOI] [PubMed] [Google Scholar]
- 60.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hindorff, L.A., MacArthur, J., Morales, J., Junkins, H.A., Hall, P.N., Klemm, A.K., and Manolio, T.A. A Catalog of Published Genome-Wide Association Studies. http://www.genome.gov/gwastudies. Accessed June 26, 2015.
- 62.Ozolins T.R., Siksay D.L., Wells P.G. Modulation of embryonic glutathione peroxidase activity and phenytoin teratogenicity by dietary deprivation of selenium in CD-1 mice. J. Pharmacol. Exp. Ther. 1996;277:945–953. [PubMed] [Google Scholar]
- 63.Broad Institute TCGA Genome Data Analysis Center . Broad Institute of MIT and Harvard; 2015. Correlation between gene methylation status and clinical features. [Google Scholar]
- 64.Fog C.K., Galli G.G., Lund A.H. PRDM proteins: important players in differentiation and disease. BioEssays. 2012;34:50–60. doi: 10.1002/bies.201100107. [DOI] [PubMed] [Google Scholar]
- 65.Hohenauer T., Moore A.W. The Prdm family: expanding roles in stem cells and development. Development. 2012;139:2267–2282. doi: 10.1242/dev.070110. [DOI] [PubMed] [Google Scholar]
- 66.Eom G.H., Kim K., Kim S.M., Kee H.J., Kim J.Y., Jin H.M., Kim J.R., Kim J.H., Choe N., Kim K.B. Histone methyltransferase PRDM8 regulates mouse testis steroidogenesis. Biochem. Biophys. Res. Commun. 2009;388:131–136. doi: 10.1016/j.bbrc.2009.07.134. [DOI] [PubMed] [Google Scholar]
- 67.Komai T., Iwanari H., Mochizuki Y., Hamakubo T., Shinkai Y. Expression of the mouse PR domain protein Prdm8 in the developing central nervous system. Gene Expr. Patterns. 2009;9:503–514. doi: 10.1016/j.gep.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 68.Inoue M., Kuroda T., Honda A., Komabayashi-Suzuki M., Komai T., Shinkai Y., Mizutani K. Prdm8 regulates the morphological transition at multipolar phase during neocortical development. PLoS ONE. 2014;9:e86356. doi: 10.1371/journal.pone.0086356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ross S.E., McCord A.E., Jung C., Atan D., Mok S.I., Hemberg M., Kim T.K., Salogiannis J., Hu L., Cohen S. Bhlhb5 and Prdm8 form a repressor complex involved in neuronal circuit assembly. Neuron. 2012;73:292–303. doi: 10.1016/j.neuron.2011.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takeuchi M., Hata Y., Hirao K., Toyoda A., Irie M., Takai Y. SAPAPs. A family of PSD-95/SAP90-associated proteins localized at postsynaptic density. J. Biol. Chem. 1997;272:11943–11951. doi: 10.1074/jbc.272.18.11943. [DOI] [PubMed] [Google Scholar]
- 71.Ranta S., Zhang Y., Ross B., Takkunen E., Hirvasniemi A., de la Chapelle A., Gilliam T.C., Lehesjoki A.E. Positional cloning and characterisation of the human DLGAP2 gene and its exclusion in progressive epilepsy with mental retardation. Eur. J. Hum. Genet. 2000;8:381–384. doi: 10.1038/sj.ejhg.5200440. [DOI] [PubMed] [Google Scholar]
- 72.Li J.M., Lu C.L., Cheng M.C., Luu S.U., Hsu S.H., Hu T.M., Tsai H.Y., Chen C.H. Role of the DLGAP2 gene encoding the SAP90/PSD-95-associated protein 2 in schizophrenia. PLoS ONE. 2014;9:e85373. doi: 10.1371/journal.pone.0085373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pinto D., Pagnamenta A.T., Klei L., Anney R., Merico D., Regan R., Conroy J., Magalhaes T.R., Correia C., Abrahams B.S. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chien W.H., Gau S.S., Liao H.M., Chiu Y.N., Wu Y.Y., Huang Y.S., Tsai W.C., Tsai H.M., Chen C.H. Deep exon resequencing of DLGAP2 as a candidate gene of autism spectrum disorders. Mol. Autism. 2013;4:26. doi: 10.1186/2040-2392-4-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chertkow-Deutsher Y., Cohen H., Klein E., Ben-Shachar D. DNA methylation in vulnerability to post-traumatic stress in rats: evidence for the role of the post-synaptic density protein Dlgap2. Int. J. Neuropsychopharmacol. 2010;13:347–359. doi: 10.1017/S146114570999071X. [DOI] [PubMed] [Google Scholar]
- 76.Maden C.H., Gomes J., Schwarz Q., Davidson K., Tinker A., Ruhrberg C. NRP1 and NRP2 cooperate to regulate gangliogenesis, axon guidance and target innervation in the sympathetic nervous system. Dev. Biol. 2012;369:277–285. doi: 10.1016/j.ydbio.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu S., Yue W., Jia M., Ruan Y., Lu T., Gong X., Shuang M., Liu J., Yang X., Zhang D. Association of the neuropilin-2 (NRP2) gene polymorphisms with autism in Chinese Han population. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2007;144B:492–495. doi: 10.1002/ajmg.b.30495. [DOI] [PubMed] [Google Scholar]
- 78.Neufeld G., Kessler O., Herzog Y. The interaction of Neuropilin-1 and Neuropilin-2 with tyrosine-kinase receptors for VEGF. Adv. Exp. Med. Biol. 2002;515:81–90. doi: 10.1007/978-1-4615-0119-0_7. [DOI] [PubMed] [Google Scholar]
- 79.Nasarre P., Gemmill R.M., Potiron V.A., Roche J., Lu X., Barón A.E., Korch C., Garrett-Mayer E., Lagana A., Howe P.H., Drabkin H.A. Neuropilin-2 Is upregulated in lung cancer cells during TGF-β1-induced epithelial-mesenchymal transition. Cancer Res. 2013;73:7111–7121. doi: 10.1158/0008-5472.CAN-13-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jubb A.M., Sa S.M., Ratti N., Strickland L.A., Schmidt M., Callahan C.A., Koeppen H. Neuropilin-2 expression in cancer. Histopathology. 2012;61:340–349. doi: 10.1111/j.1365-2559.2012.04224.x. [DOI] [PubMed] [Google Scholar]
- 81.Yasuoka H., Kodama R., Hirokawa M., Takamura Y., Miyauchi A., Inagaki M., Sanke T., Nakamura Y. Neuropilin-2 expression in papillary thyroid carcinoma: correlation with VEGF-D expression, lymph node metastasis, and VEGF-D-induced aggressive cancer cell phenotype. J. Clin. Endocrinol. Metab. 2011;96:E1857–E1861. doi: 10.1210/jc.2011-1180. [DOI] [PubMed] [Google Scholar]
- 82.Samuel S., Gaur P., Fan F., Xia L., Gray M.J., Dallas N.A., Bose D., Rodriguez-Aguayo C., Lopez-Berestein G., Plowman G. Neuropilin-2 mediated β-catenin signaling and survival in human gastro-intestinal cancer cell lines. PLoS ONE. 2011;6:e23208. doi: 10.1371/journal.pone.0023208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yasuoka H., Kodama R., Tsujimoto M., Yoshidome K., Akamatsu H., Nakahara M., Inagaki M., Sanke T., Nakamura Y. Neuropilin-2 expression in breast cancer: correlation with lymph node metastasis, poor prognosis, and regulation of CXCR4 expression. BMC Cancer. 2009;9:220. doi: 10.1186/1471-2407-9-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gaudet M.M., Campan M., Figueroa J.D., Yang X.R., Lissowska J., Peplonska B., Brinton L.A., Rimm D.L., Laird P.W., Garcia-Closas M. DNA hypermethylation of ESR1 and PGR in breast cancer: pathologic and epidemiologic associations. Cancer Epidemiol Biomarkers Prev. 2009;18:3036–3043. doi: 10.1158/1055-9965.EPI-09-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dai B., Geng L., Yu Y., Sui C., Xie F., Shen W., Zheng T., Yang J. Methylation patterns of estrogen receptor α promoter correlate with estrogen receptor α expression and clinicopathological factors in hepatocellular carcinoma. Exp. Biol. Med. (Maywood) 2014;239:883–890. doi: 10.1177/1535370214536651. [DOI] [PubMed] [Google Scholar]
- 86.Dijkstra A., Howard T.D., Vonk J.M., Ampleford E.J., Lange L.A., Bleecker E.R., Meyers D.A., Postma D.S. Estrogen receptor 1 polymorphisms are associated with airway hyperresponsiveness and lung function decline, particularly in female subjects with asthma. J. Allergy Clin. Immunol. 2006;117:604–611. doi: 10.1016/j.jaci.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 87.Koppelman G.H., Sayers I. Evidence of a genetic contribution to lung function decline in asthma. J. Allergy Clin. Immunol. 2011;128:479–484. doi: 10.1016/j.jaci.2011.05.036. [DOI] [PubMed] [Google Scholar]
- 88.Garzon R., Liu S., Fabbri M., Liu Z., Heaphy C.E., Callegari E., Schwind S., Pang J., Yu J., Muthusamy N. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Meyer N., Christoph J., Makrinioti H., Indermitte P., Rhyner C., Soyka M., Eiwegger T., Chalubinski M., Wanke K., Fujita H. Inhibition of angiogenesis by IL-32: possible role in asthma. J. Allergy Clin. Immunol. 2012;129:964–973. doi: 10.1016/j.jaci.2011.12.1002. [DOI] [PubMed] [Google Scholar]
- 90.Joosten L.A., Heinhuis B., Netea M.G., Dinarello C.A. Novel insights into the biology of interleukin-32. Cell. Mol. Life Sci. 2013;70:3883–3892. doi: 10.1007/s00018-013-1301-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tümpel S., Maconochie M., Wiedemann L.M., Krumlauf R. Conservation and diversity in the cis-regulatory networks that integrate information controlling expression of Hoxa2 in hindbrain and cranial neural crest cells in vertebrates. Dev. Biol. 2002;246:45–56. doi: 10.1006/dbio.2002.0665. [DOI] [PubMed] [Google Scholar]
- 92.Boimel P.J., Cruz C., Segall J.E. A functional in vivo screen for regulators of tumor progression identifies HOXB2 as a regulator of tumor growth in breast cancer. Genomics. 2011;98:164–172. doi: 10.1016/j.ygeno.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.DeMille D., Grose J.H. PAS kinase: a nutrient sensing regulator of glucose homeostasis. IUBMB Life. 2013;65:921–929. doi: 10.1002/iub.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.