Abstract
Bivalve mitochondrial genomes show many uncommon features, like additional genes, high rates of gene rearrangement, high A-T content. Moreover, Doubly Uniparental Inheritance (DUI) is a distinctive inheritance mechanism allowing some bivalves to maintain and transmit two separate sex-linked mitochondrial genomes. Many bivalve mitochondrial features, such as gene extensions or additional ORFs, have been proposed to be related to DUI but, up to now, this topic is far from being understood. Several species are known to show this unusual organelle inheritance but, being widespread only among Unionidae and Mytilidae, DUI distribution is unclear. We sequenced and characterized the complete female- (F) and male-transmitted (M) mitochondrial genomes of Meretrix lamarckii, which, in fact, is the second species of the family Veneridae where DUI has been demonstrated so far. The two mitochondrial genomes are comparable in length and show roughly the same gene content and order, except for three additional tRNAs found in the M one. The two sex-linked genomes show an average nucleotide divergence of 16%. A 100-aminoacid insertion in M. lamarckii M-cox2 gene was found; moreover, additional ORFs have been found in both F and M Long Unassigned Regions of M. lamarckii. Even if no direct involvement in DUI process has been demonstrated so far, the finding of cox2 insertions and supernumerary ORFs in M. lamarckii both strengthens this hypothesis and widens the taxonomical distribution of such unusual features. Finally, the analysis of inter-sex genetic variability shows that DUI species form two separate clusters, namely Unionidae and Mytilidae+Veneridae; this dichotomy is probably due to different DUI regimes acting on separate taxa.
Introduction
Doubly Uniparental Inheritance (DUI) [1–4] is an interesting alternative to the common Strict Maternal Inheritance (SMI) for cytoplasmic organelles in Eukaryotes.
Species with DUI are characterized by the presence of two different sex-linked mitochondrial lineages, being transmitted independently by the two sexes. One lineage is called F (from Female-transmitted) and it is transmitted by females through eggs, while the other is called M (Male-transmitted) and it is transmitted by males through sperm.
After fertilization, the heteroplasmic zygote contains both F and M mitochondrial lineages. During embryonic development, females become essentially homoplasmic for F, whereas males remain heteroplasmic, the M lineage being localized in germ line and (often in traces) in soma, and the F one in soma only [5]. As a result, in adult DUI bivalves, somatic tissues of both sexes are dominated by the F-mtDNA lineage, while germ-line cells contain the sex-specific mtDNA lineage [6–8]. The two sex-linked mtDNAs are therefore inherited separately, and thus they evolve independently. This results in a high level of sequence divergence between the two genomes, comprised between 10% and 50% (see, f.i., [9, 8, 5, 10]).
Up to now, DUI has only been found in ten bivalve families: Arcticidae, Donacidae, Hyriidae, Margaritiferidae, Mactridae, Mytilidae, Nuculanidae, Solenidae, Unionidae and Veneridae ([11, 6, 12, 7, 9, 8, 13, 10, 14–15]; and reference therein). With the exception of mytilids and unionids, where several DUI species have been discovered, in other families only few DUI species have been found, thus affecting the possibility of comparisons between phylogenetically related species.
Until very recently, among the Veneridae, DUI has been detected in Venerupis philippinarum only [16]. Although it was suggested for the species Cyclina sinensis [12], this claim was based solely on three GenBank sequences that were recently questioned [10]. In a recent paper, we investigated the presence of DUI in seven species of the subclass Heterodonta and we found DUI only in the venerid clam Meretrix lamarckii [10]. M. lamarckii, also known as the Korean hard clam, is a medium size clam widespread around coasts of Pacific Ocean, including China, Korea, Japan and South-East Asia [17–18]. This economically important mollusk lives in sandy sediments on subtidal flats [19]. A complete mitochondrial genome of M. lamarckii has already been sequenced from somatic cells by [18]: this is most likely the F-type, as indeed confirmed by phylogenetic analysis [10]. In the present work, we sequenced the complete M and F mitochondrial genomes of M. lamarckii.
Materials and Methods
Samples Collection, DNA Extraction and Dilution
24 individuals of Meretrix lamarckii were commercially purchased at the Tsukiji Wholesale Fish Market (Tokyo, Japan) in June 2012. All specimens were screened alive by microscopic inspection of gonadal extract to confirm sexual maturity and to determine the sex. A standard phenol:chloroform protocol [20] was used to extract total nucleic acid from gametes; samples were re-suspended in TE 1× and are conserved in the Mozoo Lab at the Department of Biological, Geological and Environmental Sciences (Bologna, Italy).
Total nucleic acid was quantified using a Nanodrop spectrophotometer and further diluted to reach optimal Long-PCR concentration (125 ng/μL). Two individuals, whose specimen numbers are BES:TKJ:004 (female) and BES:TKJ:009 (male), were selected because of yield results for further sequencing.
PCR Amplifications, Electrophoresis and Sequencing
The two complete genomes were amplified in three large overlapping fragments using Long-PCR technique paired with primer-walking on a Gene Amp® PCR System 2720 (Applied Biosystem). To perform Long PCR amplification from 2,000 bp up to 10,000 bp we used the Herculase® II Fusion Enzyme kit (Stratagene). Reaction volume of 50 μL was composed as follows: 2 μL DNA template (125 ng/μL), 0.5 μL Herculase II Fusion DNA Polymerase, 10 μL 5× Herculase II reaction Buffer (containing Mg2+ 10 mM), 0.5 μL dNTPs mix (25 mM for each dNTP), 1.25 μL each Primer (10 μM) and 34.5 μL of sterilized distilled water.
Reaction conditions (S1 Table) were first set up according to manufacturer’s instruction and further modified whenever necessary. After the initial denaturation step (92°C for 2’), 30 cycles were used as follows: denaturation at 92°C for 20”, annealing at 48–56°C for 20–30” and extension at 68°C for 10’. The final extension step was carried out at 68°C for 8’. Primers were designed using the Primer3 online tool [21].
For amplification of fragments < 2,000 bp we used a standard PCR approach using the GoTaq® Flexi DNA Polymerase kit (Promega). The reaction volume was 30 μL composed of 10.85 μL of sterilized distilled water, 6 μL of 5× Green GoTaq® Flexi Buffer, 2.4 μL of dNTPs (2.5 mM for each dNTP), 3.6 μL of MgCl2 (25 mM), 1.5 μL of each primer (10 μM), 0.15 μL of Taq Polymerase (5u/μL) and 4 μL of appropriately diluted DNA template. Typically, the cycle (S1 Table) was composed by an initial denaturation step (95°C for 2’), then 35 cycles as follows: denaturation at 95°C for 1’, annealing at 48–56°C for 1’ and extension at 72°C for 1–2’ depending on amplicons length. The final extension was carried out at 72°C for 5’.
PCR results were visualized by electrophoresis onto a 1% agarose gel stained with ethidium bromide and then purified through a standard isopropanol protocol, Wizard® SV Gel and PCR Clean-Up System (Promega) and, in some cases, with an empirically modified PEG precipitation protocol [22]. Successfully purified products were sequenced with the Sanger method thanks to Macrogen Europe facility (Amsterdam, The Netherland).
Data analysis and genome annotation
MEGA 6.06 [23] was used to examine and edit electropherograms and further merge the entire mitochondrial genomes according to overlapping sequences. This software was also used to compute codon usage and nucleotide percentages.
Nucleotide trends and A-T skew at four-fold degenerate sites were used to identify the Control Region (CR) and the Origin of Replication (OR) of either strand. A dedicated R [24] script was written to (i) compute these statistics over a sliding window, (ii) plot results and (iii) test for significance of the linear correlation. Any size and step for the sliding window can be specified: for the present work, we used a 700-bp sliding window with a step of 300 bp. Autocorrelograms for each nucleotide are also produced in order to evaluate the amount of autocorrelation between sliding windows and therefore the validity of the linear model approach. The user is allowed to analyze several genomes at the same time and to set any gene as starting point for the four-fold degenerate sites analysis; the script is available as S1 Script along with test files and a detailed tutorial. A GitHub repository was created, which can be found at the URL https://github.com/mozoo/4F.git.
Protein Coding Genes (PCGs) were predicted using both MITOS [25] and NCBI's ORF Finder online tool [26], using invertebrate mitochondrial genetic code and alternative start codons. Potential PCGs were identified through homologous sequence similarity using BLAST [27–28]. F- and M-cox2 sequences were aligned using the software MUSCLE [29] and the alignment was graphically edited thanks to the TeXshade package [30]. Putative tRNA genes were detected using MITOS, tRNAScan-SE [31–32] and ARWEN v1.2 [33] online softwares, using default settings. rRNA sequences have been identified by comparisons with other rRNAs present in GenBank using BLAST. In both F and M genomes, the rRNAs sequences were then annotated assuming that the first base comes immediately after the last base of the previous gene and that the last base comes immediately before the first base of the following gene. Finally, the two whole-genome maps were created using GenomeVx online tool [34], conventionally setting cox1 as the starting point of the mtDNA. All putative secondary structures of rRNAs and non-coding regions were predicted using Mfold server [35] and then graphically edited through VARNA 3.7 [36].
Repeats were identified through the online software Tandem Repeat Finder [37]. Phobius [38], InterProScan [39], TMPred [40] and HMMTOP [41] online softwares were all used to predict additional Trans Membrane Helices (TMHs). Introns were searched using @TOME2 [42]. The analysis of additional putative ORFs was done using the software Glimmer3 [43]. The EMBOSS [44] package was used to extract all the possible ORFs from all available bivalve complete mitochondrial genomes (GenBank consulted in August, 2014). An alignment was computed for F_ORF141 and M_ORF138 using HHBlits [45] and the last UniProt release. The computed hidden Markov model was used as a query against the database of all bivalve mitochondrial ORFs, which was built using HHBlits and the Pdb70 database.
Phylogeny and evolutionary comparisons
A phylogenetic analysis was carried out using other complete mitochondrial genomes from the family Veneridae that were available at August, 2014. Three heterodonts, Coelomactra antiquata (Mactridae), Hiatella arctica (Hiatellidae), and Acanthocardia tuberculata (Cardiidae) were selected as outgroups; the complete dataset is available as S2 Table. The 13 PCGs and the 2 rRNAs were extracted and aligned with PSI-BLAST [46], MUSCLE, ProbconsRNA [47], RNAplfold [48], and MAFFT [49] through the T-Coffee algorithm [50–51], using the pipeline PSI-Coffee > Expresso > accurate for PCGs and the MR-Coffee mode for rRNAs. Alignments were masked using BMGE 1.1 [52]; the best partitioning scheme was selected using PartitionFinder 1.1.0 [53] under the Bayesian Information Criterion and a greedy approach. The final Maximum Likelihood (ML) tree search was carried out with RAxML 8.2.0 [54] performing 1,000 bootstrap replicates. The consensus tree was computed with PhyUtility 2.2 [55] and graphically edited with Dendroscope 3 [56].
Finally, we compared sex-linked divergence in different DUI systems; all the DUI species whose complete mitochondrial genomes were available in January, 2015 were selected for this analysis. M and F coding sequences of M. lamarckii and other DUI species were aligned gene by gene using the T-Coffee algorithm: as above, the accurate method was used for PCGs, while MR-Coffee was chosen for rRNAs and tRNAs. To account for multiple substitutions, nucleotide Jin-Nei [57] and aminoacid Kimura [58] corrected distances were then computed using the EMBOSS suite along with uncorrected p-distances. Principal Component Analysis was carried out through R on concatenated Jin-Nei and Kimura distances using the packages FactoMineR [59] for computations and ggplot2 [60] for graphics. We also computed average distances within Unionidae, within Amarsipobranchia sensu [61] (i.e., Pteriomorphia + Heterodonta; in this case, Mytilidae + Veneridae), and within the complete dataset. Nucleotide single-gene average values were ranked and rankings within Unionidae and Amarsipobranchia were compared through R using the Spearman ρ and the Kendall's τ.
Results
Overall genomic features
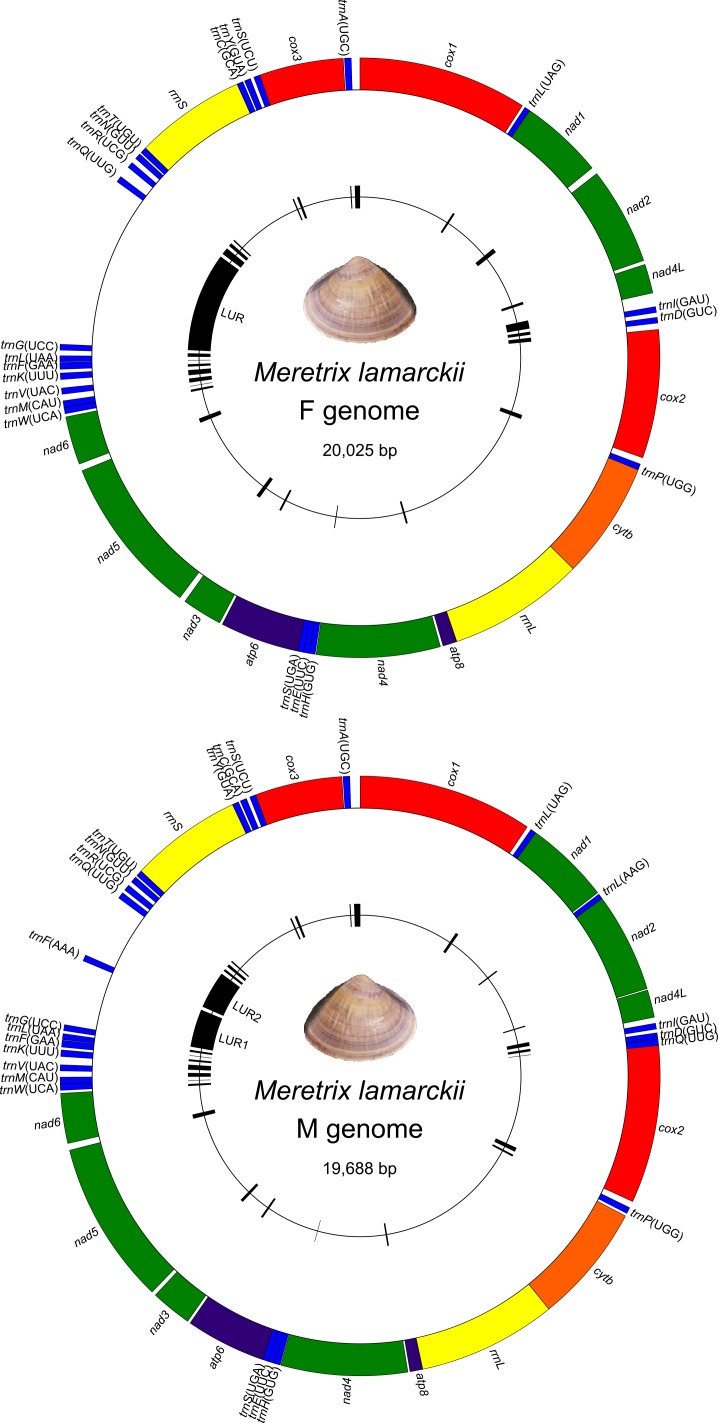
The Meretrix lamarckii complete F and M mitochondrial genomes are 20,025 bp and 19,688 bp long, respectively (Fig 1). Sequences are available in GenBank under the accession numbers KP244451 and KP244452, respectively. All genes are located on the same “+” strand and the two lineages share the same gene order. The only exceptions are three tRNAs, which were found only in M-mtDNA: trnL(AAG), an additional copy of trnQ(UUG), and trnF(AAA). Genome annotations are shown in Table 1.
Fig 1. Meretrix lamarckii sex-linked mitochondrial genomes.
Genomic map of F- (above) and M- (below) mtDNA of Meretrix lamarckii starting from cox1. Genes are all on "+" strand; genome lengths are shown in the middle of each map. Unassigned Regions (URs) are reported in black in the internal circle.
Table 1. Annotation of F and M mitochondrial genomes of Meretrix lamarckii.
| Name | Typea | Startb | End | Lenght (bp) | UNsc | Anticodon | Start Codon | Stop Codon | |
|---|---|---|---|---|---|---|---|---|---|
| cox1 | F | PCG | 1 | 1,824 | 1,824 | 32 | GTG | TAG | |
| M | PCG | 1 | 1,851 | 1,851 | 46 | GTG | TAA | ||
| trnL(NAG) | F | tRNA | 1,857 | 1,918 | 62 | 0 | TAG | ||
| M | tRNA | 1,898 | 1,959 | 62 | 0 | TAG | |||
| nad1 | F | PCG | 1,919 | 2,821 | 903 | 77 | ATC | TAA | |
| M | PCG | 1,960 | 2,862 | 903 | 19 | ATT | TAA | ||
| trnL(NAG) | M | tRNA | 2,882 | 2,939 | 58 | 0 | AAG | ||
| nad2 | F | PCG | 2,899 | 3,954 | 1,056 | 42 | GTG | TAA | |
| M | PCG | 2,940 | 3,995 | 1,056 | 15 | ATG | TAA | ||
| nad4L | F | PCG | 3,997 | 4,311 | 315 | 148 | GTG | TAG | |
| M | PCG | 4,011 | 4,304 | 294 | 63 | ATT | TAG | ||
| trnI | F | tRNA | 4,460 | 4,523 | 64 | 51 | GAT | ||
| M | tRNA | 4,368 | 4,430 | 63 | 38 | GAT | |||
| trnD | F | tRNA | 4,575 | 4,636 | 62 | 68 | GTC | ||
| M | tRNA | 4,469 | 4,533 | 65 | 4 | GTC | |||
| trnQ | M | tRNA | 4,538 | 4,604 | 67 | -2 | TTG | ||
| cox2 | F | PCG | 4,705 | 6,090 | 1,386 | 71 | ATG | TAG | |
| M | PCG | 4,603 | 6,276 | 1,674 | 70 | ATG | TAA | ||
| trnP | F | tRNA | 6,162 | 6,227 | 66 | -1 | TGG | ||
| M | tRNA | 6,347 | 6,414 | 68 | 29 | TGG | |||
| cytb | F | PCG | 6,227 | 7,492 | 1,266 | 0 | ATG | TAG | |
| M | PCG | 6,444 | 7,712 | 1,269 | 0 | TTG | TAG | ||
| rrnL | F | rRNA | 7,493 | 8,964 | 1,472 | 0 | |||
| M | rRNA | 7,713 | 9,185 | 1,473 | 0 | ||||
| atp8 | F | PCG | 8,965 | 9,117 | 153 | 28 | ATA | TAA | |
| M | PCG | 9,186 | 9,320 | 135 | 22 | ATG | TAG | ||
| nad4 | F | PCG | 9,146 | 10,480 | 1,335 | 9 | ATA | TAA | |
| M | PCG | 9,343 | 10,683 | 1,341 | 7 | GTG | TAA | ||
| trnH | F | tRNA | 10,490 | 10,551 | 62 | 0 | GTG | ||
| M | tRNA | 10,691 | 10,752 | 62 | 0 | GTG | |||
| trnE | F | tRNA | 10,552 | 10,617 | 66 | -3 | TTC | ||
| M | tRNA | 10,753 | 10,818 | 66 | -3 | TTC | |||
| trnS(NGA) | F | tRNA | 10,615 | 10,679 | 65 | 0 | TGA | ||
| M | tRNA | 10,816 | 10,880 | 65 | 0 | TGA | |||
| atp6 | F | PCG | 10,680 | 11,531 | 852 | 31 | ATG | TAG | |
| M | PCG | 10,881 | 11,732 | 852 | 31 | GTG | TAG | ||
| nad3 | F | PCG | 11,563 | 11,997 | 435 | 65 | ATG | TAG | |
| M | PCG | 11,764 | 12,198 | 435 | 46 | GTG | TAG | ||
| nad5 | F | PCG | 12,063 | 13,787 | 1,725 | 76 | GTG | TAG | |
| M | PCG | 12,245 | 13,981 | 1,737 | 72 | GTG | TAA | ||
| nad6 | F | PCG | 13,864 | 14,385 | 522 | 34 | ATA | TAA | |
| M | PCG | 14,054 | 14,584 | 531 | 31 | ATA | TAA | ||
| trnW | F | tRNA | 14,420 | 14,487 | 68 | 2 | TCA | ||
| M | tRNA | 14,616 | 14,682 | 67 | 2 | TCA | |||
| trnM | F | tRNA | 14,490 | 14,559 | 70 | 70 | CAT | ||
| M | tRNA | 14,685 | 14,754 | 70 | 64 | CAT | |||
| trnV | F | tRNA | 14,630 | 14,696 | 67 | 90 | TAC | ||
| M | tRNA | 14,819 | 14,883 | 65 | 86 | TAC | |||
| trnK | F | tRNA | 14,787 | 14,856 | 70 | 39 | TTT | ||
| M | tRNA | 14,970 | 15,038 | 69 | 30 | TTT | |||
| trnF | F | tRNA | 14,896 | 14,969 | 74 | 7 | GAA | ||
| M | tRNA | 15,069 | 15,140 | 72 | 6 | GAA | |||
| trnL(NAA) | F | tRNA | 14,977 | 15,040 | 64 | 59 | TAA | ||
| M | tRNA | 15,147 | 15,210 | 64 | 31 | TAA | |||
| trnG | F | tRNA | 15,100 | 15,163 | 64 | 1,855 | TCC | ||
| M | tRNA | 15,242 | 15,305 | 64 | 694 | TCC | |||
| trnF | M | tRNA | 16,000 | 16,066 | 67 | 690 | AAA | ||
| trnQ | F | tRNA | 17,019 | 17,086 | 68 | 125 | TTG | ||
| M | tRNA | 16,757 | 16,825 | 69 | 35 | TTG | |||
| trnR | F | tRNA | 17,212 | 17,277 | 66 | 57 | TCG | ||
| M | tRNA | 16,861 | 16,926 | 66 | 45 | TCG | |||
| trnN | F | tRNA | 17,335 | 17,400 | 66 | 19 | GTT | ||
| M | tRNA | 16,972 | 17,037 | 66 | 20 | GTT | |||
| trnT | F | tRNA | 17,420 | 17,487 | 68 | 0 | TGT | ||
| M | tRNA | 17,058 | 17,122 | 65 | 0 | TGT | |||
| rrnS | F | rRNA | 17,488 | 18,680 | 1,193 | 0 | |||
| M | rRNA | 17,123 | 18,310 | 1,188 | 0 | ||||
| trnC | F | tRNA | 18,681 | 18,747 | 67 | 19 | GCA | ||
| M | tRNA | 18,311 | 18,378 | 68 | 21 | GCA | |||
| trnY | F | tRNA | 18,767 | 18,834 | 68 | 38 | GTA | ||
| M | tRNA | 18,400 | 18,469 | 70 | 37 | GTA | |||
| trnS(NCT) | F | tRNA | 18,873 | 18,939 | 67 | 0 | TCT | ||
| M | tRNA | 18,507 | 18,573 | 67 | 0 | TCT | |||
| cox3 | F | PCG | 18,940 | 19,848 | 909 | 12 | ATG | TAA | |
| M | PCG | 18,574 | 19,497 | 924 | 11 | ATG | TAA | ||
| trnA | F | tRNA | 19,861 | 19,933 | 73 | 92 | TGC | ||
| M | tRNA | 19,509 | 19,579 | 71 | 109 | TGC |
a PCG, Protein Coding Gene.
b All genes are located on the same strand.
c Unassigned nucleotides after the gene (negative values for overlapping nucleotides).
Nucleotide composition is reported in Table 2. M. lamarckii A-T content reaches 66.01% in F-mtDNA and 67.17% in M-mtDNA. This value increases considering only the 3rd base nucleotide composition of PCG codons (73.31% for F-mtDNA and 75.08% for M-mtDNA).
Table 2. Nucleotide composition (%) of Meretrix lamarckii F and M genomes.
| Name | Sex | Length | T | C | A | G | A-T | T3a | C3a | A3a | G3a | A-T3a |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cox1 | F | 1,824 | 43.10 | 12.10 | 22.10 | 22.70 | 65.20 | 56.00 | 4.90 | 18.90 | 19.70 | 74.90 |
| M | 1,851 | 44.30 | 11.30 | 22.70 | 21.60 | 67.00 | 58.35 | 4.21 | 20.10 | 17.34 | 78.44 | |
| nad1 | F | 903 | 45.96 | 11.52 | 19.27 | 23.26 | 65.23 | 55.81 | 4.98 | 18.94 | 20.27 | 74.75 |
| M | 903 | 47.07 | 10.63 | 18.83 | 23.48 | 65.89 | 57.81 | 3.32 | 17.61 | 21.26 | 75.42 | |
| nad2 | F | 1,056 | 44.41 | 7.39 | 23.20 | 25.00 | 67.61 | 46.02 | 3.98 | 30.68 | 19.32 | 76.70 |
| M | 1,056 | 44.89 | 6.63 | 21.69 | 26.80 | 66.57 | 48.01 | 2.27 | 23.58 | 26.14 | 71.59 | |
| nad4L | F | 315 | 50.16 | 8.25 | 19.37 | 22.22 | 69.52 | 52.38 | 3.81 | 17.14 | 26.67 | 69.52 |
| M | 294 | 51.70 | 9.52 | 18.71 | 20.07 | 70.41 | 53.06 | 7.14 | 16.33 | 23.47 | 69.39 | |
| cox2 | F | 1,386 | 38.89 | 9.52 | 25.69 | 25.90 | 64.57 | 56.28 | 5.63 | 18.83 | 19.26 | 75.11 |
| M | 1,674 | 37.48 | 9.15 | 28.21 | 25.16 | 65.69 | 53.23 | 4.84 | 22.40 | 19.53 | 75.63 | |
| cytb | F | 1,266 | 43.68 | 13.03 | 21.88 | 21.41 | 65.56 | 54.27 | 7.11 | 21.33 | 17.30 | 75.59 |
| M | 1,269 | 44.52 | 13.00 | 20.72 | 21.75 | 65.25 | 54.85 | 6.62 | 20.80 | 17.73 | 75.65 | |
| atp8 | F | 153 | 45.10 | 12.42 | 22.22 | 20.26 | 67.32 | 43.14 | 3.92 | 27.45 | 25.49 | 70.59 |
| M | 135 | 50.37 | 9.63 | 19.26 | 20.74 | 69.63 | 51.11 | 0.00 | 17.78 | 31.11 | 68.89 | |
| nad4 | F | 1,335 | 45.39 | 10.49 | 19.78 | 24.34 | 65.17 | 51.24 | 5.62 | 21.80 | 21.35 | 73.03 |
| M | 1,341 | 44.74 | 10.07 | 22.22 | 22.97 | 66.96 | 49.66 | 5.37 | 25.28 | 19.69 | 74.94 | |
| atp6 | F | 852 | 43.78 | 10.92 | 23.12 | 22.18 | 66.90 | 45.07 | 6.34 | 25.00 | 23.59 | 70.07 |
| M | 852 | 44.13 | 10.21 | 23.47 | 22.18 | 67.61 | 46.83 | 3.87 | 29.93 | 19.37 | 76.76 | |
| nad3 | F | 435 | 43.45 | 8.97 | 25.75 | 21.84 | 69.20 | 45.52 | 4.83 | 29.66 | 20.00 | 75.17 |
| M | 435 | 42.99 | 7.13 | 25.52 | 24.37 | 68.51 | 44.14 | 2.76 | 24.83 | 28.28 | 68.97 | |
| nad5 | F | 1,725 | 46.09 | 10.96 | 17.74 | 25.22 | 63.83 | 49.39 | 8.70 | 16.00 | 25.91 | 65.39 |
| M | 1,737 | 47.67 | 8.52 | 19.80 | 24.01 | 67.47 | 54.06 | 3.63 | 20.03 | 22.28 | 74.09 | |
| nad6 | F | 522 | 47.51 | 10.34 | 18.20 | 23.95 | 65.71 | 56.32 | 6.32 | 16.67 | 20.69 | 72.99 |
| M | 531 | 48.96 | 8.10 | 18.27 | 24.67 | 67.23 | 58.76 | 4.52 | 12.99 | 23.73 | 71.75 | |
| cox3 | F | 909 | 44.66 | 11.88 | 19.47 | 23.98 | 64.14 | 62.05 | 4.62 | 15.51 | 17.82 | 77.56 |
| M | 924 | 46.10 | 9.63 | 19.59 | 24.68 | 65.69 | 62.34 | 1.95 | 15.91 | 19.81 | 78.25 | |
| rrnS | F | 1,193 | 36.46 | 10.73 | 28.50 | 24.31 | 64.96 | |||||
| M | 1,188 | 37.29 | 9.76 | 28.96 | 23.99 | 66.25 | ||||||
| rrnL | F | 1,472 | 39.27 | 9.71 | 29.82 | 21.20 | 69.09 | |||||
| M | 1,473 | 39.24 | 9.91 | 30.69 | 20.16 | 69.93 | ||||||
| All PCGsb | F | 12,681 | 44.21 | 10.79 | 21.30 | 23.70 | 65.51 | 52.78 | 5.82 | 20.53 | 20.87 | 73.31 |
| M | 13,002 | 44.67 | 9.75 | 22.05 | 23.52 | 66.73 | 53.88 | 4.15 | 21.20 | 20.77 | 75.08 | |
| All rRNAs | F | 2,665 | 38.01 | 10.17 | 29.23 | 22.59 | 67.24 | |||||
| M | 2,661 | 38.37 | 9.85 | 29.91 | 21.87 | 68.28 | ||||||
| All tRNAs | F | 1,464 | 37.70 | 10.93 | 29.71 | 21.65 | 67.42 | |||||
| M | 1,653 | 39.21 | 9.76 | 29.45 | 21.58 | 68.67 | ||||||
| All coding DNAc | F | 16,810 | 42.66 | 10.70 | 23.29 | 23.35 | 65.95 | |||||
| M | 17,316 | 43.18 | 9.77 | 23.97 | 23.08 | 67.15 | ||||||
| All URsd | F | 3,216 | 40.14 | 7.37 | 26.18 | 26.31 | 66.32 | |||||
| M | 2,374 | 41.29 | 6.59 | 26.20 | 25.91 | 67.50 | ||||||
| Complete genome | F | 20,025 | 42.26 | 10.17 | 23.75 | 23.83 | 66.01 | |||||
| M | 19,688 | 42.95 | 9.39 | 24.23 | 23.43 | 67.19 |
a Computed only on third codon positions.
b PCGs, Protein Coding Genes.
c PCGs + rRNAs + tRNAs
d URs, Untranslated Regions.
Both F-type and M-type mtDNAs contain a large numbers of Unassigned Regions (URs; 27 in F-mtDNA and 29 in M-mtDNA), which are detailed in S3 Table.
Protein Coding Genes (PCGs)
We found all 13 canonical protein coding genes, including the atp8 gene, reported as missing in several bivalve species [7, 62–63]. ATG start codon is used in cox2, cytb, atp6, nad3, and cox3 of the F genome and in nad2, cox2, atp8 and cox3 of the M one. Like most invertebrate mitochondrial genomes, the two M. lamarckii mtDNAs show alternative start codons: GTG, ATC, ATA, ATT and TTG (see Table 1). Observed stop codons are TAG and TAA, as expected. Overall, TAA is the most common stop codon, while TAG is used in F- cox1, nad4L, cox2, cytb, atp6, nad3, and nad5 and in M- nad4L, cytb, atp8, atp6, and nad3. Truncated TA-/T—stop codons ([8, 63]; and reference therein) were not found in M. lamarckii mtDNAs.
M. lamarckii F and M protein coding genes (PCGs) contain 4,227 codons and 4,333 codons, respectively (Table 3). In both F and M mtDNAs, the most used codons is UUU (410 and 434 hits, respectively). Less used codons are CGC and ACC (both 7 hits) in F-mtDNA and UGC (2 hits) in M-mtDNA. The most common aminoacid in both F- and M-mtDNA is leucine, while the rarest is glutamic acid.
Table 3. Meretrix lamarckii codon count (#) and usage (%).
| aa | codona | # | % | aa | codona | # | % | aa | codona | # | % | aa | codona | # | % | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phe(F) | UUUb | F | 410 | 9.70 | Ser(S) | UCU | F | 161 | 3.81 | Ala(A) | GCU | F | 134 | 3.17 | Asp(D) | GAU | F | 93 | 2.20 |
| M | 434 | 10.02 | M | 158 | 3.65 | M | 122 | 2.82 | M | 114 | 2.63 | ||||||||
| UUC | F | 42 | 0.99 | UCC | F | 9 | 0.21 | GCC | F | 9 | 0.21 | GAC | F | 19 | 0.45 | ||||
| M | 17 | 0.39 | M | 11 | 0.25 | M | 7 | 0.16 | M | 15 | 0.35 | ||||||||
| Leu(L) | UUA | F | 230 | 5.44 | UCA | F | 16 | 0.38 | GCA | F | 26 | 0.62 | Glu(E) | GAA | F | 46 | 1.09 | ||
| M | 244 | 5.63 | M | 21 | 0.48 | M | 26 | 0.60 | M | 67 | 1.55 | ||||||||
| UUG | F | 190 | 4.49 | UCG | F | 15 | 0.35 | GCG | F | 28 | 0.66 | GAG | F | 78 | 1.85 | ||||
| M | 193 | 4.45 | M | 18 | 0.42 | M | 21 | 0.48 | M | 70 | 1.62 | ||||||||
| CUUb | F | 82 | 1.94 | AGU | F | 88 | 2.08 | Tyr(Y) | UAU | F | 152 | 3.60 | Cys(C) | UGU | F | 84 | 1.99 | ||
| M | 91 | 2.10 | M | 103 | 2.38 | M | 171 | 3.95 | M | 99 | 2.28 | ||||||||
| CUC | F | 9 | 0.21 | AGC | F | 13 | 0.31 | UAC | F | 25 | 0.59 | UGC | F | 10 | 0.24 | ||||
| M | 3 | 0.07 | M | 11 | 0.25 | M | 18 | 0.42 | M | 2 | 0.05 | ||||||||
| CUA | F | 25 | 0.59 | AGA | F | 66 | 1.56 | STOP(*) | UAA | F | 6 | 0.14 | Trp(W) | UGA | F | 58 | 1.37 | ||
| M | 11 | 0.25 | M | 52 | 1.20 | M | 8 | 0.18 | M | 52 | 1.20 | ||||||||
| CUG | F | 17 | 0.40 | AGG | F | 51 | 1.21 | UAG | F | 7 | 0.17 | UGG | F | 66 | 1.56 | ||||
| M | 17 | 0.39 | M | 67 | 1.55 | M | 5 | 0.12 | M | 72 | 1.66 | ||||||||
| Ile (I) | AUU | F | 198 | 4.68 | Pro(P) | CCU | F | 89 | 2.11 | His(H) | CAU | F | 51 | 1.21 | Arg(R) | CGU | F | 40 | 0.95 |
| M | 235 | 5.42 | M | 95 | 2.19 | M | 51 | 1.18 | M | 39 | 0.90 | ||||||||
| AUC | F | 14 | 0.33 | CCC | F | 11 | 0.26 | CAC | F | 20 | 0.47 | CGC | F | 7 | 0.17 | ||||
| M | 9 | 0.21 | M | 8 | 0.18 | M | 16 | 0.37 | M | 4 | 0.09 | ||||||||
| Met(M) | AUA | F | 84 | 1.99 | CCA | F | 16 | 0.38 | Gln(Q) | CAAc | F | 27 | 0.64 | CGA | F | 23 | 0.54 | ||
| M | 114 | 2.63 | M | 18 | 0.42 | M | 25 | 0.58 | M | 15 | 0.35 | ||||||||
| AUG | F | 117 | 2.77 | CCG | F | 8 | 0.19 | CAG | F | 30 | 0.71 | CGG | F | 13 | 0.31 | ||||
| M | 90 | 2.08 | M | 11 | 0.25 | M | 29 | 0.67 | M | 14 | 0.32 | ||||||||
| Val(V) | GUU | F | 275 | 6.51 | Thr(T) | ACU | F | 87 | 2.06 | Asn(N) | AAU | F | 102 | 2.41 | Gly(G) | GGU | F | 185 | 4.38 |
| M | 266 | 6.14 | M | 90 | 2.08 | M | 102 | 2.35 | M | 165 | 3.81 | ||||||||
| GUC | F | 14 | 0.33 | ACC | F | 7 | 0.17 | AAC | F | 24 | 0.57 | GGC | F | 13 | 0.31 | ||||
| M | 11 | 0.25 | M | 6 | 0.14 | M | 21 | 0.48 | M | 21 | 0.48 | ||||||||
| GUA | F | 71 | 1.68 | ACA | F | 18 | 0.43 | Lys(K) | AAA | F | 85 | 2.01 | GGA | F | 71 | 1.68 | |||
| M | 95 | 2.19 | M | 13 | 0.30 | M | 100 | 2.31 | M | 57 | 1.32 | ||||||||
| GUG | F | 116 | 2.74 | ACG | F | 20 | 0.47 | AAG | F | 47 | 1.11 | GGG | F | 79 | 1.87 | ||||
| M | 120 | 2.77 | M | 16 | 0.37 | M | 53 | 1.22 | M | 104 | 2.40 |
a Codons that match a corresponding mtDNA-encoded tRNA are underlined.
b The corresponding tRNA is present only in the M genome.
c Two trnQ(UUG) tRNAs were detected in the M genome.
Post-transcriptional cleavage sites could be indicated by the presence of a tRNA between two PCGs [64]. In absence of a tRNA, the cleavage role can be played by intergenic non-coding sequences that form a stem-loop secondary structure ([8]; and reference therein). According to the previous statement, for each M. lamarckii unassigned region located between a pair of PCGs, the predicted hairpin was determined and reported in S1 Fig.
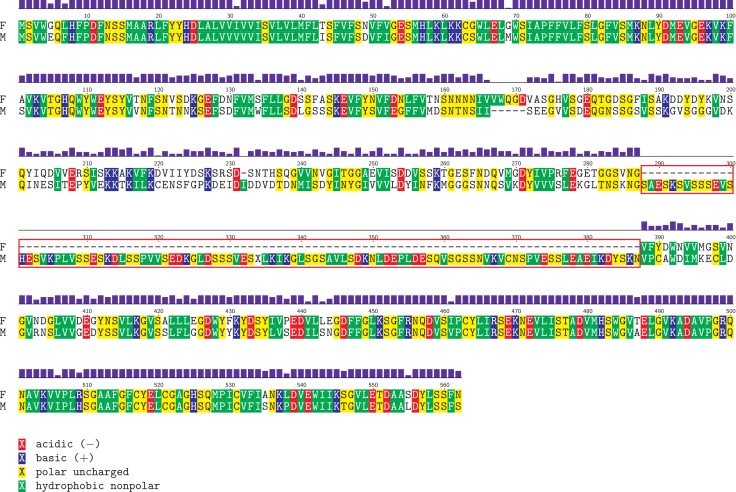
Finally, M-cox2 gene is significantly different from the F one. More specifically, it includes a 100-aminoacid long region in the middle of the gene, which is not present in F-cox2 (Fig 2).
Fig 2. Meretrix lamarckii cox2 gene alignment.
Aminoacid alignment between female-type (F) and male-type (M) Meretrix lamarckii sequences of the cox2 gene. Identical aminoacids are shaded following their hydropathy (see the legend below the figure for the meanings of the different colors); purple bars show aminoacid similarity. The 100-aminoacid insertion found in M-mtDNA is boxed in red. Sites are numbered above the sequences, conventionally starting at 1.
rRNAs and tRNAs
Standard rRNAs were found in both genomes: rrnS is located between trnT and trnC, while rrnL between cytb and atp8. The F and the M rRNAs predicted secondary structures are reported in S2 Fig.
F-mtDNA shows all 22 canonical tRNAs, with two serine-encoding tRNAs and two leucine-encoding tRNAs. They may differ from each other in terms of anticodon. Like many other metazoan taxa (see, f.i., [65, 8, 63]), both F- and M-trnS(UCU) present a shortened DHU arm.
In addition, the M genome presents three sex-specific tRNAs, totaling 25 tRNAs: supernumerary trnL(AAG) (between nad1 and nad2), trnQ(UUG) (between trnD and cox2), and trnF(AAA) (within M Long Unassigned Region). All secondary structures of tRNAs are reported in S3 and S4 Figs.
To better understand their origin, M supernumerary tRNAs were compared with the URs mapped in the same position of F-mtDNA. In all cases, a very similar sequence was found, albeit the canonical cloverleaf structure is essentially unrecoverable (S5 Fig).
Long Unassigned Region (LUR)
A Long Unassigned Region (LUR) is located between trnG and trnQ. F-LUR measures 1,855 bp, whereas M-LUR is apparently divided in two regions (LUR1 and LUR2 of 694 bp and 690 bp, respectively) by the putative supernumerary trnF(AAA) (see above).
A complex secondary structure was found in both M and F mtDNAs in the middle LUR sequence. This highly folded structure is comprised between bases 15,698 and 15,952 in F-LUR and between bases 15,838 and 16,512 in M-LUR. In the F-LUR two tandem-repeated motifs were also found, both with two tandem copies. The first motif is 15 bp long (positions 15,722–15,736 and 15,737–15,751) and the second one is 109 bp long (positions 16,767–16,875 and 16,880–16,988, at the end of F-LUR region) (S6 Fig). In M-mtDNA only the 15 bp-long motif was found (positions 15,845–15,858 and 15,860–15,873). A BLAST search of the Termination-Associated Sequence (TAS; [66–67]) element found a significant hit (S7 Fig) in the F-LUR (positions 16,340–16,354), but not in the M-LUR.
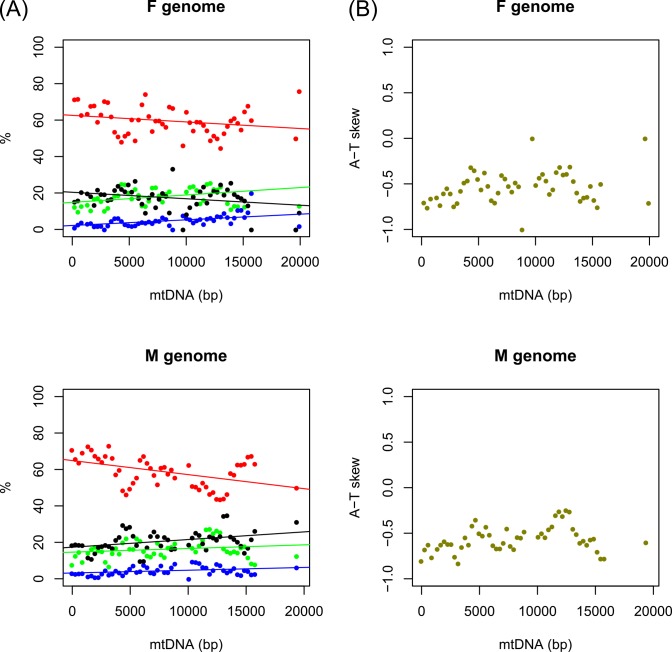
The first PCG downstream of the LUR is cox3; therefore, we set cox3 as the starting point of the sliding window computing nucleotide composition at four-fold degenerate sites. We found 1,911 degenerate sites in the F M. lamarckii genome (15.01% of PCG sites) and 1,907 in the M one (14.67%). In both cases, four-fold degenerate sites are highly T-rich (59.65% for F and 59.20% for M) and definitely weak, but significant trends were uncovered (Fig 3A): while a significant A trend was never found, we detected a significant increase in C in both sexes, even if with very low R2 values. A negative trend for G was found in F-mtDNA, while a positive one for G and T was found in M-mtDNA, again with very low R2 values. The autocorrelograms show a significant value of the autocorrelation function (acf) only at lag-1 for F genomes, or at lag-1 and lag-2 (or lag-5) for three M nucleotides (S8 Fig). Given the T-richness of four-fold degenerate sites, the A-T skew is always negative or equal to 0, but two peaks were found, corresponding to the LUR and to the atp6/nad3 boundary (Fig 3B).
Fig 3. Origins of Replication.
A, nucleotide composition at four-fold degenerate sites, using a sliding window of 700 bp with a step of 300 bp. The starting point is the first PCG after the LUR, i.e. cox3. Equations are as follows, for F/M, respectively. A (green): y = 0.0004x+14.84; R2 = 0.0681; p = 0.0616 / y = 0.0002x+14.62; R2 = 0.0409; p = 0.1546. C (blue): y = 0.0003x+2.15; R2 = 0.2263; p = 0.0004*** / y = 0.0002x+3.17; R2 = 0.1087; p = 0.0181*. G (black): y = −0.0004x+20.39; R2 = 0.0796; p = 0.0428* / y = 0.0004x+17.33; R2 = 0.1448; p = 0.0059**. T (red): y = −0.0004x+62.61; R2 = 0.0595; p = 0.0815 / y = −0.0008x+64.88; R2 = 0.2123; p = 0.0007***. B, A-T skew at four-fold degenerate sites, using a sliding window as for (A); the starting point is again cox3.
The structure of the F-LUR is comparable to the LUR of the published genome of M. lamarckii (GenBank Accession Number NC_016174), with some differences. The LUR of the available M. lamarckii mtDNA is found at positions 14,982–18,044; again, a highly folded region can be inferred (15,470–16,433). At the 5' side of the highly folded region there is a sequence very similar to that of the F-ORF (15,242–15,392); this sequence would be a putative ORF located on the reverse strand, were it not for a stop codon (TAA) right after the start one and for an insertion of a G, which triggers a frameshift mutation leading to the loss of the stop codon (S9 Fig). Conversely, at the 3' side of the highly folded region a 100-bp repeated motif was found; the repeat unit shows some similarities with the 109-bp motif of our F genome (S10 Fig). However, in the GenBank M. lamarckii mtDNA it is repeated 13 times; this higher number of repeats accounts for the great difference in length between the two LURs (1,855 against 3,063 bp).
Supernumerary Open Reading Frames (ORFs)
Several additional putative Open Reading Frames (ORFs) were found within the LUR of both F and M M. lamarckii mtDNAs. Among all these sequences, we found only one ORF in each genome (F_ORF141 and M_ORF138) that does not overlap with the highly folded structure revealed in the LUR (see above). To better understand whether these putative ORFs are expressed or not, the prediction software Glimmer3 was used. At first, the software was trained with M. lamarckii standard gene data. All mitochondrial PCGs were given a score comprised between 8.71 and 16.34 (for F) and between 10.47 and 18.08 (for M). According to these values, the two potential supernumerary ORFs should not be considered as expressed, because they showed extremely low scores (i.e., 2.34 for F_ORF141 and 3.36 for M_ORF138; see S4 Table).
The presence of F_ORF141 was also searched for in all available bivalve complete mitochondrial genomes using HHBlits. In all the other available Meretrix species, a homolog was found in the reverse strand, within the LUR. All homologous ORFs have a probability over 90%, while E-value and p-value were always lower than 0.05; this holds also for the F_ORF141/M_ORF138 comparison (S5 Table).
Phylogenetic analysis
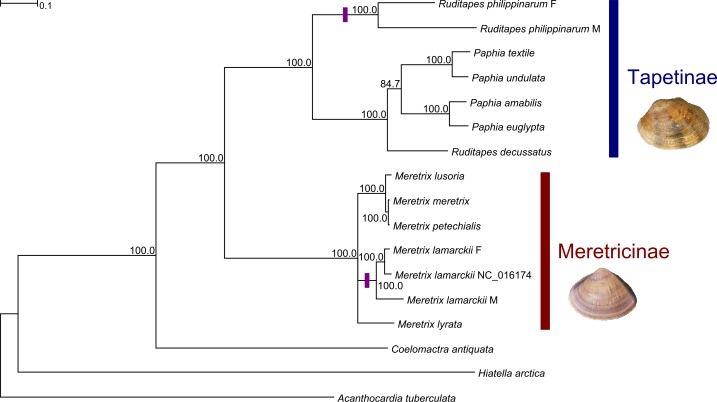
The complete dataset was composed by 5,035 aminoacids (PCGs) and 4,554 nucleotides (rRNAs). 3,841 aminoacids and 1,232 nucleotides were left for phylogenetic analysis (69.14% and 27.05%, respectively) after masking with BMGE; the most affected PCG was cox2 (only 29.82% aminoacids were selected), while the least affected was nad4 (85.57%); rrnL and rrnS were similarly affected by the masking phase (28.20% and 25.57%, respectively). PartitionFinderProtein suggested the partition of PCGs in two clusters, namely ATP synthase/NADH dehydrogenase subunits (atp6, atp8, nad1, nad2, nad3, nad4, nad4L, nad5, and nad6) and cytochrome c oxidase subunits/cytochrome b (cox1, cox2, cox3, and cytb); conversely, PartitionFinder suggested to keep together the two ribosomal genes (rrnL and rrnS). Best-fitting molecular evolution models were JTT [68], LG [69], and GTR [70], respectively. The consensus tree computed over 1,000 bootstrap replicates is highly supported (Fig 4), being the bootstrap proportion equal to 100% for all nodes, with one exception (the Paphia clade). Veneridae were recovered as monophyletic, being the mactrid Coelomactra antiquata the sister taxon. Tapetinae and Meretricinae are also monophyletic; within Tapetinae, the deepest split separates the DUI species R. philippinarum from R. decussatus + Paphia; within Meretricinae, only the genus Meretrix is represented in our tree and the available M. lamarckii mtDNA clusters with our F genome.
Fig 4. Phylogenetic analysis.
Maximum Likelihood phylogenetic analysis of the family Veneridae using complete mitochondrial genomes and Acanthocardia tuberculata (Cardiidae), Hiatella arctica (Hiatellidae), and Coelomactra antiquata (Mactridae) as outgroups. Shown is the consensus of ML trees obtained from 1,000 bootstrap replicates; number at the nodes are bootstrap proportions. Purple bars mark known DUI species.
Genetic variability
Nucleotide Jin-Nei distances and aminoacid Kimura distances were calculated between M. lamarckii F-mtDNA and M-mtDNA for each PCG, tRNA, rRNA, UR and for concatenations of these, up to the whole genome. Jin-Nei and Kimura distance values are reported in Table 4 and S6 Table (for single tRNAs).
Table 4. Bivalves nucleotide and aminoacid (boldface) distances.
| Reference | All coding DNAa | PCGsb | rRNAs | tRNAs | atp6 | atp8c | cox1 | cox2 | cox3 | cytb | nad1 | nad2 | nad3 | nad4 | nad4L | nad5 | nad6 | rrnL | rrnS | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Anodonta anatina | F | NC_022803 | 92.24 | 90.02 | 98.76 | 112.28 | 114.22 | 109.70 | 80.86 | 88.62 | 78.97 | 85.97 | 90.20 | 100.78 | 90.00 | 96.48 | 48.03 | 88.90 | 119.64 | 98.99 | 98.70 |
| M | KF030963 | 81.46 | 87.98 | 186.71 | 42.34 | 82.92 | 54.98 | 54.35 | 66.80 | 133.98 | 98.29 | 150.76 | 92.06 | 101.01 | 194.11 | ||||||
| Hyriopsis cumingii | F | NC_011763 | 10.09 | 11.59 | 5.80 | 4.85 | 10.70 | 4.32 | 7.04 | 14.78 | 9.96 | 12.80 | 13.82 | 16.56 | 5.59 | 10.23 | 5.47 | 15.95 | 7.32 | 5.17 | 6.76 |
| M | HM347668 | 1.98 | 0.43 | 4.20 | 0.20 | 2.71 | 1.17 | 1.58 | 2.39 | 3.18 | 1.72 | 1.03 | 2.96 | 3.57 | 0.00 | ||||||
| Hyriopsis schlegelii | F | NC_015110 | 7.52 | 8.18 | 3.51 | 7.17 | 6.94 | 11.77 | 8.58 | 26.10 | 4.57 | 10.03 | 7.32 | 6.79 | 5.46 | 5.79 | 3.26 | 6.26 | 7.32 | 4.53 | 1.93 |
| M | HQ641407 | 1.34 | 1.29 | 7.81 | 0.39 | 3.63 | 0.39 | 1.05 | 2.04 | 0.94 | 0.00 | 1.03 | 1.12 | 1.40 | 2.51 | ||||||
| Meretrix lamarckii | F | KP244451 | 49.65 | 55.03 | 37.86 | 27.41 | 61.65 | 43.82 | 45.83 | 59.95 | 45.08 | 51.38 | 41.22 | 52.56 | 51.58 | 58.08 | 40.76 | 70.81 | 62.34 | 37.47 | 38.34 |
| M | KP244452 | 19.57 | 13.55 | 9.71 | 12.95 | 43.17 | 15.42 | 17.81 | 4.82 | 13.75 | 18.90 | 9.93 | 16.90 | 33.45 | 28.73 | ||||||
| Musculista senhousia | F | GU001953 | 64.73 | 71.53 | 44.63 | 45.51 | 80.66 | 54.79 | 71.26 | 74.24 | 36.14 | 72.38 | 94.98 | 76.24 | 82.73 | 63.26 | 90.98 | 73.89 | 61.15 | 24.44 | |
| M | GU001954 | 23.19 | 27.21 | 5.52 | 43.66 | 17.40 | 6.02 | 25.84 | 39.22 | 23.54 | 21.05 | 29.18 | 34.45 | 31.22 | |||||||
| Mytilus californianus | F | GQ527172 | 86.82 | 90.97 | 73.94 | 76.00 | 97.13 | 61.28 | 93.29 | 78.24 | 79.71 | 75.76 | 89.64 | 106.51 | 124.15 | 127.89 | 105.55 | 135.27 | 77.64 | 69.09 | |
| M | GQ527173 | 39.82 | 35.96 | 11.87 | 35.05 | 21.50 | 26.53 | 35.36 | 72.89 | 46.92 | 43.94 | 49.80 | 79.85 | 79.85 | |||||||
| Mytilus edulis | F | NC_006161 | 62.25 | 71.85 | 39.72 | 32.36 | 73.33 | 60.52 | 64.70 | 68.87 | 62.87 | 72.10 | 79.52 | 82.80 | 80.57 | 88.82 | 75.66 | 82.04 | 43.20 | 35.14 | |
| M | AY823623 | 16.66 | 17.47 | 4.70 | 11.60 | 9.24 | 19.40 | 14.85 | 27.86 | 12.17 | 16.85 | 16.94 | 20.60 | 56.50 | |||||||
| Mytilus galloprovincialis | F | NC_006886 | 64.68 | 73.22 | 45.18 | 36.47 | 74.60 | 53.95 | 76.90 | 69.80 | 67.62 | 77.26 | 92.88 | 38.73 | 82.16 | 87.21 | 81.51 | 83.56 | 45.76 | 44.43 | |
| M | AY363687 | 15.20 | 14.86 | 4.50 | 10.66 | 11.91 | 15.28 | 16.07 | 25.51 | 4.44 | 22.44 | 16.42 | 22.26 | 25.29 | |||||||
| Mytilus trossulus | F | HM462080 | 65.35 | 75.63 | 36.44 | 36.82 | 67.12 | 99.83 | 55.70 | 70.59 | 75.06 | 66.15 | 71.69 | 78.44 | 89.01 | 85.48 | 97.43 | 91.61 | 90.81 | 37.64 | 34.79 |
| M | HM462081 | 19.12 | 13.84 | 67.99 | 5.86 | 12.11 | 12.61 | 13.61 | 14.24 | 27.31 | 19.65 | 22.44 | 17.58 | 46.48 | 23.50 | ||||||
| Pyganodon grandis | F | NC_013661 | 102.42 | 101.92 | 102.53 | 117.28 | 119.56 | 32.19 | 75.66 | 91.70 | 90.26 | 108.94 | 118.28 | 131.56 | 149.81 | 99.09 | 97.67 | 101.23 | 106.48 | 100.56 | 106.33 |
| M | FJ809755 | 85.18 | 88.68 | 111.47 | 43.24 | 78.68 | 56.89 | 57.66 | 100.86 | 132.61 | 118.81 | 138.89 | 97.42 | 116.94 | 143.55 | ||||||
| Quadrula quadrula | F | NC_013658 | 113.61 | 115.31 | 100.34 | 113.18 | 83.49 | 377.36 | 76.67 | 83.03 | 84.97 | 108.53 | 97.81 | 112.58 | 87.45 | 80.06 | 120.22 | 92.09 | 125.57 | 109.20 | 88.44 |
| M | FJ809751 | 84.23 | 88.77 | n/ad | 40.23 | 62.47 | 61.38 | 52.44 | 93.92 | 141.42 | 79.85 | 223.27 | 109.69 | 103.39 | 175.18 | ||||||
| Ruditapes philippinarum | F | NC_003354 | 75.33 | 81.28 | 59.10 | 64.07 | 86.49 | 63.97 | 101.96 | 77.73 | 74.37 | 65.14 | 99.00 | 80.88 | 44.73 | 50.78 | 84.46 | 85.83 | 62.20 | 54.80 | |
| M | AB065374 | 47.04 | 46.36 | 12.46 | 87.16 | 47.50 | 39.61 | 25.59 | 66.01 | 38.69 | 259.28 | 44.58 | 46.13 | 99.67 | |||||||
| Solenaia carinatus | F | NC_023250 | 96.23 | 96.22 | 98.70 | 107.45 | 104.10 | 100.16 | 94.19 | 84.21 | 78.87 | 115.07 | 101.08 | 120.46 | 101.90 | 88.66 | 122.68 | 91.81 | 98.03 | 100.43 | 96.16 |
| M | KC848655 | 79.16 | 86.13 | 140.88 | 40.45 | 75.17 | 55.77 | 58.77 | 73.80 | 135.10 | 89.38 | 156.15 | 81.05 | 101.35 | 141.96 | ||||||
| Utterbackia peninsularis | F | HM856636 | 101.63 | 100.76 | 102.52 | 122.15 | 106.38 | 100.69 | 86.15 | 99.73 | 90.59 | 91.34 | 105.68 | 121.58 | 102.85 | 99.22 | 149.86 | 95.98 | 144.58 | 103.13 | 102.13 |
| M | NC_015477 | 86.40 | 95.57 | 186.71 | 41.77 | 79.85 | 58.33 | 60.88 | 78.05 | 137.17 | 112.12 | 193.71 | 101.15 | 108.45 | 175.68 | ||||||
| Venustaconcha ellipsiformis | F | FJ809753 | 97.71 | 96.96 | 103.10 | 109.14 | 98.23 | 85.24 | 73.69 | 96.01 | 87.25 | 97.83 | 143.08 | 106.92 | 71.45 | 99.65 | 142.04 | 91.67 | 130.07 | 101.71 | 105.26 |
| M | NC_013659 | 83.34 | 81.05 | 137.48 | 38.50 | 68.98 | 50.66 | 52.83 | 96.40 | 137.10 | 90.95 | 150.13 | 103.84 | 113.59 | 231.95 | ||||||
| Unionoideae (N = 6) | 100.64 | 100.20 | 100.99 | 113.58 | 104.33 | 134.22 | 81.20 | 90.55 | 85.15 | 101.28 | 109.36 | 115.65 | 100.58 | 93.86 | 113.42 | 93.61 | 120.73 | 102.34 | 99.50 | ||
| 83.30 | 88.03 | 152.65 | 41.09 | 74.68 | 56.34 | 56.16 | 84.97 | 136.23 | 98.23 | 168.82 | 97.54 | 107.46 | 177.07 | ||||||||
| Amarsipobranchiaf (N = 7) | 66.97 | 74.22 | 48.12 | 45.52 | 77.28 | 71.83 | 56.58 | 76.95 | 69.86 | 62.61 | 67.94 | 83.86 | 75.11 | 79.70 | 79.45 | 85.80 | 87.68 | 52.15 | 43.00 | ||
| 25.80 | 24.18 | 38.85 | 8.27 | 34.77 | 19.37 | 19.75 | 19.54 | 38.94 | 23.47 | 56.56 | 27.34 | 40.46 | 49.25 | ||||||||
| Overallg (N = 15) | 72.68 | 76.03 | 63.48 | 67.48 | 78.97 | 96.51 | 59.93 | 74.86 | 67.63 | 71.25 | 76.85 | 86.95 | 76.02 | 75.81 | 83.03 | 78.96 | 90.18 | 65.92 | 60.45 | ||
| 45.58 | 46.61 | 94.77 | 20.33 | 46.52 | 31.68 | 31.85 | 43.40 | 72.94 | 50.36 | 94.06 | 52.05 | 62.19 | 93.98 |
a Complete genome excluding untranslated regions.
b Protein Coding Genes.
c atp8 gene is not annotated in some species. See text for details.
d The sequences are too divergent to compute the Kimura distance.
e Average values of Unionoidea. Hyriopsis cumingii and H. schlegelii were excluded because we are most probably comparing two female genomes; see text for details.
f Average values of Amarsipobranchia.
g Average values of all species.
Between the F- and the M-mtDNA the nucleotide Jin-Nei distance of the complete genome (coding + non-coding) is 53.13, corresponding to a 16.19% divergence. Jin-Nei nucleotide distances are 81.53 (25.81%) for non-coding regions and 49.65 (14.89%) for coding genes. PCG concatenation has an aminoacid Kimura score of 19.57 and, within that, the highest values belong to cox2 (43.17) and nad5 (33.45), while lowest values are associated with nad1 (4.82) and atp8 (9.71). The average Jin-Nei distance between rRNAs is 37.86; the average Jin-Nei distance between tRNAs is 27.41.
We also compared the two M. lamarckii mitochondrial genomes obtained in this paper (F and M) to the already sequenced M. lamarckii mtDNA present in literature (GenBank Accession Number NC_016174). The uncorrected distance (p-distance) between this genome and our F genome is 9.81% for all the coding DNA, being 10.67% for all PCGs, 8.17% for all rRNAs and 5.33% for all tRNAs. On the other side, the divergence between this genome and our M genome scored is 14.65% for all coding DNA, scoring 15.76% for all PCGs, 11.43% for all rRNAs and 10.72% for all tRNAs. In both cases, the most divergent gene is cox2 (15.09% and 21.72%, respectively), whereas the less divergent is atp8 (4.17% and 9.17%, respectively). The aminoacid Kimura distance was also computed to account for synonymous substitutions (S7 Table): again, the NC_016174 sequence is always more similar to the F-mtDNA than to the M-mtDNA and the most divergent gene is cox2 (19.28 and 39.86, respectively), while the less divergent genes are atp8 (0.00 and 8.13) and nad1 (0.67 and 4.12). Generally speaking, with the exception of cox1 and cox2, the Kimura distance from M-mtDNA is always one order of magnitude higher than from F-mtDNA.
The F vs M distances were also computed for all known DUI species whose complete mitochondrial genomes have been published (see Table 4). M. lamarckii has lower divergence values, when compared to other DUI families such as Unionidae or Mytilidae. The Unionidae show, by far, the highest values, with divergence scores of coding DNA ranging from 92.24 of Anodonta anatina to 113.61 of Quadrula quadrula; Hyriopsis species show abnormally low distance values, not even comparable with average Unionidae family values. Unionidae are followed by Mytilidae with an all-coding Jin-Nei distance comprised between 62.25 of Mytilus edulis and 86.82 of M. californianus. Within the Veneridae, Venerupis philippinarum has a higher divergence value (75.33) with respect to M. lamarckii (49.65). The most divergent PCGs are atp8, nad4, and nad6 with average Kimura distances of 94.77, 94.06, and 93.98, respectively; the most conserved is cox1 (20.33).
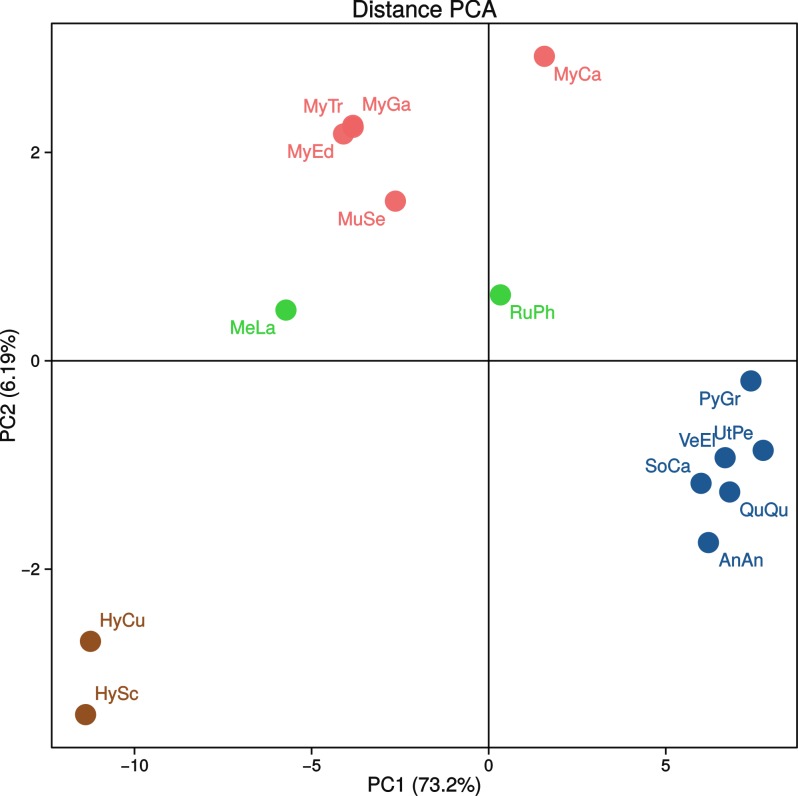
The resulting PCA plot (Fig 5) uses the first two components to explain the 73.20% + 6.19% = 79.39% of distance variability (using Jin-Nei and Kimura distances together). Datasets are roughly arranged by overall divergence levels along the first principal component (Hyriopsis spp. < Mytilidae+Veneridae < Unionidae); the second principal component further separates venerids and mytilids from unionids. Finally, it is impossible to reject the null hypothesis that nucleotide distance rankings among single PCGs in Unionidae and Mytilidae + Veneridae are unrelated, using both the Spearman ρ (p = 0.2392) and the Kendall's τ (p = 0.2044). For example, cytb and nad1 are highly divergent for Unionidae, but they are among the least variable for Amarsipobranchia, while the opposite is true for nad4 and nad5.
Fig 5. PCA plot.
Principal Component Analysis (PCA) based on both Jin-Nei and Kimura distances reported in Table 4. Colors refer to different families: blue, Unionidae with the exception of Hyriopsis spp. (brown; see text for details); Indian red, Mytilidae; green, Veneridae. AnAn, Anodonta anatina; HyCu, Hyriopsis cumingii; HySc, Hyriopsis schlegelii; MeLa, Meretrix lamarckii; MuSe, Musculista senhousia; MyCa, Mytilus californianus; MyEd, Mytilus edulis; MyGa, Mytilus galloprovincialis; MyTr, Mytilus trossulus; PyGr, Pyganodon grandis; QuQu; Quadrula quadrula; RuPh, Ruditapes philippinarum; SoCa, Solenaia carinatus; UtPe; Utterbackia peninsularis; VeEl, Venustaconcha ellipsiformis.
Discussion
Comparison with the previously published Meretrix lamarckii mitogenome
The two mitochondrial genomes of Meretrix lamarckii (F and M) sequenced here are slightly shorter than the one previously reported in GenBank: 20,025 (F) and 19,688 (M) bp against 21,209 bp [18]. This genome was extracted from a somatic tissue (the foot muscle; [18]) and, indeed, it was previously attributed to the female type by Plazzi and colleagues [10]. The phylogenetic analysis of the present work further corroborates this hypothesis (Fig 4). It shares the same gene content and gene order, with the exception of trnL(AAG), trnQ(UUG) and trnF(AAA), which have been found only in M-mtDNA. This, again, strengthens the idea that the previously published genome is from the female lineage.
However, comparison between either sex and the published F genome showed surprisingly high divergence values. These were generally one order of magnitude higher when comparing it with our M genome, and, as in the case of our F/M comparison, divergence ranking is similar: f.i., highest values are obtained from cox2, while lowest scores are obtained from atp8. Significant divergence values are still observed at the aminoacid level for both sexes; the distances between our M-mtDNA and the published F genome are comparable to those computed between the two sexes in the present study (Table 4 and S6 Table).
It is possible to find similarities in the LUR structure between the two F genomes, a highly folded region being the divide between a supernumerary ORF and a region with tandem repeats of about 100 bp in length. However, in the published F genome it was not possible to find a functional ORF (S9 Fig). This may be due to sequencing errors; if the available sequence is confirmed, it is hard to say whether an ORF was originally present in the species and was subsequently pseudogenized in some populations or a novel ORF appeared in some others. Given the widespread presence of supernumerary mitochondrial ORFs in bivalves [71, 62, 72, 63, 5, 73–74], we largely favor the first hypothesis. On the other side, it is possible to align the 3' repeated motifs (S10 Fig). The great variation in length between the two LURs is due to the different number of repeats: 2 in our F genome, 13 in the published one. This difference, in turn, accounts for the aforementioned difference in length between the two genomes.
Interestingly, intra-specific variability in the number of repeats in a mitochondrial LUR has been reported elsewhere [75–76] for (DUI) bivalves. The two M. lamarckii specimens sampled for this research come from the Tokyo area (Japan), whereas the F genome available in GenBank comes from Zhejiang (China) [18]. Recall that the Chinese specimen was only tentatively identified as M. lamarckii due to its similar morphology, despite showing some differences in color, shell shape and thickness [18], we cannot completely rule out the hypothesis that these specimens belong to different species; however, it is not unconceivable that the differences found here simply reflect the high degree of taxonomic distinctness between Japanese and Chinese clams belonging to very distant populations.
Nucleotide composition and codon usage
A-T content in M. lamarckii F and M genomes is slightly higher with respect to the average A-T content in bivalves, but it is comparable with those of other DUI organisms such as V. philippinarum and M. senhousia [63]. The coding strand (+) is G-T rich: this is expected [77–78] and in good agreement with [63], where it was stated that a higher G-T percentage is related with mtDNAs characterized by most (if not all) genes located on the same strand.
Codons usage reflects the general nucleotide composition of the two genomes, with a high presence of T in most used codons. In almost all cases, except for trnL(TAA), trnK(TTT), and trnM(CAT) in F-mtDNA and for trnL(TAA), trnF(AAA) and trnK(TTT) in M-mtDNA, within four-fold or two-fold degenerate codon families the most used codons do not have a complementary anticodon in mitochondrially-encoded tRNAs (Table 3). Moreover, they differ for only one base (the third one) with respect to the synonymous codon for which a complementary tRNA exists in the mitochondrion. The codon usage table demonstrates the presence of high degrees of third-base wobbling in M. lamarckii, as previously seen in other bivalves [8, 63]: a tRNA can have a non-standard base at the first anticodon position pairing with more than one base and allowing to bind codons that are not perfectly complementary.
PCGs and the cox2 insertion
With the exception of the supernumerary ORF, all genes are located on the same strand in both F and M mtDNAs of M. lamarckii. This is commonly found in all Amarsipobranchia, while unionids [71, 9, 63] and Solemya velum [63] encode genes on either strand. This finding reinforces the hypothesis that the one of M. lamarckii is a derived state, which evolved once in the common ancestor of Pteriomorphia and Heterodonta ([9]; and reference therein).
The atp8 gene was declared as missing in several bivalve species [8], especially in the genus Mytilus [79], even if it was recently found in some bivalves like Solemya velum [63], Musculista senhousia [8], Venerupis philippinarum [80], and presently in M. lamarckii. In addition, recent studies [9, 81] found this gene in species in which it was not previously annotated.
The use of Jin-Nei corrected distance to evaluate nucleotide divergence unveiled that atp8 is not less conserved than other mitochondrial genes (see Table 4). As suggested elsewhere [5, 82], there is a strong possibility that the absence off this gene is simply due to past annotation difficulties or inaccuracy. Up to now, the presence/absence of ATPase subunit 8 does not appear linked with DUI [9], but rather, if confirmed, to phylogeny [63].
It was suggested to be the commonest situation in metazoans that the two ATPase subunits, atp8 and atp6, are adjacent and overlapping [79]. This especially holds if the co-translation of these genes from a bicistronic transcript (as is the case in mammals; [83]) is confirmed as a widespread rule. In fact, this association is present in the Unionidae [71] and it was recently found in S. velum [63]; however, a disjointed location of atp8 and atp6 has already been highlighted for some heterodont bivalves, like Hiatella arctica [80] and Macoma balthica [82]. Similarly, in M. lamarckii atp8 and atp6 are not neighboring in either F- and M-mtDNA: again, the contiguity of these genes may be an example of an ancestral state that was subsequently lost in derived bivalves.
M M. lamarckii genome presents an insertion of 100 codons in cox2 gene, which is totally absent in the F counterpart (Fig 2). It is not the first time that a M-cox2 gene is longer than F-cox2; generally, however, these extensions map to the 3’ end of the gene. In fact, the M-cox2 3’ tail is present in all three subfamilies of Unionidae [5]. This extension (Mcox2e) has been found only in M mtDNAs and varies in length between 177 and 192 bp [84–85]. Mcox2e has been found in poly-adenylated transcripts of cox2 obtained from male gonads, and also proved to be translated and localized in both inner and outer mitochondrial membranes [84–86]. The structural analysis of unionid Mcox2e sequences reveals the presence of the two canonical N-terminal trans-membrane helices (TMHs). In addition to that, several additional TMHs were found in Mcox2e [87]. For the above mentioned reasons, a proposed hypothesis was that such extension may be a mitochondrial tag implicated in male mitochondria survival to elimination and differential segregation during development [87].
Outside from the unionid family, the pattern of cox2 variations among DUI M and F lineages is unclear and not easy to unravel. In mytilids, no extensions were found in M genomes of Mytilus [5], but a duplicated cox2 gene (cox2b) is found in M M. senhousia [8], with the duplicated gene being longer than the original one at 3’. A putative TMH of 41 residues was found in the cox2b tail [8], allowing the authors to hypothesize a correlation between the unionid Mcox2e and the cox2b tail of M. senhousia.
V. philippinarum is with M. lamarckii the only known DUI species of the family Veneridae: a duplication of the cox2 gene, similar to that of M. senhousia (i.e. longer at 3’), was found, but, contrastingly, it is located in the F genome [8]. However, additional TMHs (either in insertions or tails) are not detectable in V. philippinarum cox2, nor in M. lamarckii (S11 Fig). Moreover, @TOME analysis did not find any intron, and the coding frame is apparently kept (see Fig 2).
Concluding, it was impossible to properly assign a function in silico to this region, and further analyses are therefore mandatory in this regard. Non-canonical features in cox2 gene are often coupled with DUI, but a general rule is still not evident and each DUI system seems to follow its own evolutionary pathway. However, despite the relationship between cox2 variations and DUI phenomenon has not been demonstrated yet, the finding of a new M-cox2 gene insertion (albeit differently located in the gene) in another DUI bivalve is an interesting clue.
Supernumerary tRNAs in M-mtDNA
As mentioned above, M and F genomes basically share the same gene arrangement, the only difference being three tRNAs in M-mtDNA. As a consequence of the high variability of their mitochondrial genomes, additional tRNA copies are common in bivalves [88–90]. In fact, when aligning the M additional tRNAs with the region mapped in the same position in the F-mtDNA, high levels of sequence similarity were always detected (see S5 Fig). Therefore, we may hypothesize that the duplication of trnL, trnQ, and trnF took place before the separation of the two sex-linked lineages, and that, afterwards, the F copies became pseudogenes or remain functional tRNAs that the in silico methods are not able to retrieve. Anyway, it has also to be noted that the anticodon region of the F counterpart of M-trnQ(TTG) would be complementary to the stop codon TAA.
The presence of a tRNA in the middle of M-LUR (trnF(AAA)) is intriguing and deserves further investigation: possibly, the cloverleaf structure of a tRNA was co-opted as part of the signaling structure of the putative control region (see below) and, thus, would not correspond to a functional tRNA. However, it is noteworthy that the anticodon of the middle-LUR tRNA is AAA, which is complementary to TTT, the most used codon in both genomes (see Table 3).
The presence of a functional tRNA in the middle of a control region, where it may work also as a signaling sequence, would make of the trnF(AAA) gene of M-mtDNA of M. lamarckii a good example of an evolutionary spandrel [91] and/or a case of molecular exaptation: this region, being a tRNA, necessarily had a complex secondary structure, and this became useful in the wider context of the control region as well (or vice versa, even if the presence of a degenerated tRNA in the F-mtDNA makes us to prefer the first hypothesis). The presence of a tRNA-like structure was already signaled by [67] in the Mytilus spp. LUR, but in the case of M. lamarckii it seems that the tRNA maintained its functionality.
Other expected non-canonical tRNA structures are found in our genomes: f.i., in both F- and M-mtDNA two trnS were found and the DHU arm was not recovered in trnS(UCU). However, as mentioned by [8], this unusual tRNA has been found in several other animal groups and it evolved early in Metazoans group [92]. In vitro analysis further confirmed its functionality [93].
Control Region (CR) and the Origin of Replication (OR)
Several parameters have been proposed to identify the mtDNA control region (CR). The most used are the presence of repetitive elements, palindromes, length, high A-T content and secondary structures with T-rich loops [67, 94, 71]. M. lamarckii Long Unassigned Region (LUR) is the longest UR in both F and M mtDNAs, although the M one is apparently split into two parts by a phenylalanine supernumerary tRNA (as mentioned above). A-T content is roughly the same found in the entire genomes, 64.2% for F and 65.8% for M, even if several poly-T have been found during (and heavily hampered) sequencing.
The short (15-bp long) repeat is essentially a stretch of G and A and may simply reflect the general G-T-/A-T-richness of both genomes in a region where less selective constraints are working; however, this repeat is conserved in both F and M genomes and it is known that similar G-rich sequences are present in Mytilus and human control regions, being related with replication and/or transcription [67].
The 109-bp long repeats are located near to the 3' end of the F-LUR sequence, and, due to their proximity with the putative origin of replication (see below), they may play a functional role in F mtDNA duplication (but recall that they are not detectable in M). Both M- and F-LUR present a central region which appears to be heavily folded (S6 Fig): again, this secondary structure may play some role for the replication/transcription process to begin [67, 71].
The nucleotide composition at fourfold degenerate sites is related with single-strand state duration during mtDNA replication. As detailed in ([95–98, 71]; and reference therein), the more the heavy (H) strand remains unpaired, the more the spontaneous hydrolytic deamination of C to U and A to hX (hypoxanthine) takes place. Such an increase of T and hX in the H strand leads to a corresponding increase in the percentages of A and C in the complementary lagging (L) strand where the H strand remains for longer time in the single-stranded condition, i.e. near to the OR. Moreover, single-stranded-guanine may spontaneously oxidize to 8-hydroxyguanine, which basepairs with adenine: thus, in this case, G decreases and T increases on the H strand. In a nutshell, T will only tend to accumulate near to the origin of replication of the H strand, while the opposite is true for A and C; finally, G may behave in either way [95, 98]. This asymmetrical composition can leave a neutral signature in fourfold degenerate sites, being them under no or weak selection.
The 700-bp sliding window analysis on these sites is in agreement with this model (Fig 3): with the exception of A (and of T in F-mtDNA), all correlations are significant, even if R2 values are very low (<25%). Setting cox3 as the starting point of the pattern, T in M-mtDNA tends to decrease, C tends to increase, and G decreases in the F-mtDNA and increases in the M one, which would be expected if the OR is located upstream to cox3. This also point to the conclusion that the "+" strand is in fact the H strand (as predictable, being all genes located on it).
The A-T skew at four-fold degenerate sites is known to be correlated with the position of the ORs as well: extreme (i.e., closer to ±1) values are associated with PCGs located near to the OR of the H strand, while balanced (i.e., closer to 0) values are associated with PCGs located near to the OR of the L strand [95, 97, 99]. Given the overall high T-richness of these genomes, the A-T skew at four-fold degenerate sites is always negative: however, lowest values (i.e., closer to −1) are associated with the LUR and with the cox3 gene (Fig 3B), while highest values (i.e., closer to 0), are associated with the nad2/nad4L and atp6/nad3 regions.
Therefore, we have further evidence that the LUR contains the OR of the H strand; moreover, it is tempting to conclude that either the nad2/nad4L or the atp6/nad3 region is the OR of the L strand. Both regions are neighbored by a two- or three-tRNA cassette, and it has been shown that an array of tRNAs on a strand may act as OR in the opposite one through alternative secondary structure [100]. If the OR of the L strand were located in the atp6/nad3 region, that shows A-T skews closer to 0 (Fig 3B) and is near to three tRNAs in either sex (Fig 1), this would leave the OR of the L strand quite distant from the OR of the H strand, a situation very similar (if not more extreme) to that of Mytilus [98] and unionids [71]. However, it is possible that more complex patterns are shadowed by the presence of all genes on the same strand and by the high T-richness of both genomes (recall that the A-T skew for the third codon position of PCGs is −0.44 for both genomes; Table 2).
As a conclusion, we gathered seven pieces of evidence that the F-LUR and the M-LUR are the control regions of M. lamarckii mtDNAs; as detailed above, most of these features are shared with other DUI species, namely Mytilus spp. [67, 98] and Unionidae [71]. First of all, we have (i) a complex secondary structure, that, if the supernumerary ORFs are expressed (see below), would involve the complete LUR. Within that, we found, approximately from 5' to 3': (ii) the presence of G-rich elements; (iii) the presence of a tRNA (only in M); (iv) a sequence with some homology to the human TAS element (only in F); (v) the 109-bp long repeats (only in F). Finally, downstream from the LUR, we detected (vi-vii) the two above-mentioned nucleotide composition trends. Being these features unique to this region, we propose the LUR to act as the CR and to contain the OR of the H strand.
Supernumerary ORFs
Many DUI species, like M. senhousia and Mytilus spp., present supernumerary ORFs with no known homologies with other proteins (i.e., ORFans; [101]), which are located in the LUR. M. lamarckii is no exception. For such ORFs a correlation with the DUI phenomenon has been suggested [71–72, 5], even if the opposite was also proposed, interpreting the RNA transcripts as degradation intermediates [102]. Supernumerary ORFs were also found in the basal species S. velum, leading to the hypothesis that they constitute a plesiomorphy among bivalves [63]. Although some of these ORFs have uncontrovertibly proved to be translated [71–73], it is uncertain whether the M. lamarckii ones are even transcribed. This issue can be assessed only by looking at expression data, but, currently, without an available transcriptome/proteome of M. lamarckii, we cannot confirm nor disprove the functionality of either ORF.
However, a precise homology between F and M ORFans was detected, which would not be expected if these sequences did not share a common ancestor; furthermore, an ORFan with high homology to F_ORF141 has been found in all species belonging to genus Meretrix. Interestingly, this would make of this supernumerary ORF the only gene located on the reverse strand in all the Meretrix genomes (S5 Table).
Sex-linked mtDNA diversification and evolution in DUI bivalves
The two entire M. lamarckii genomes diverge by a 16% on average (see also [10]), hence the divergence between F and M mtDNAs is somewhat lower than other DUI species. On the other hand, the most diversified genomes belong to unionids (around 35%), followed by mytilids (around 25%). The other venerid, V. philippinarum, shows levels of divergence comparable to those of mytilids (26%).
In this work, nucleotide Jin-Nei and aminoacid Kimura distance values of all DUI species (whose complete mitochondrial genomes are available in GenBank) were calculated between M- and F-type mitogenomes to estimate divergences and give an idea of the rate of independent evolution between the two sex-linked genomes. We strongly advocate the use of corrected distance methods, like the Jin-Nei and Kimura formulae, over the uncorrected p-distance, because of the high divergence between sex-linked mtDNAs in many DUI species and the overall high variability of the molluscan mitochondrial genome, where significant level of saturation and multiple-hits events are quite common (see, f.i., [61]).
Both Hyriopsis cumingii and H. schlegelii Jin-Nei distance values are surprisingly low, in contrast with all other sequenced unionids (and, in general, DUI) mtDNAs. However, there is a chance that there was an error in assigning the paternal route of transmission to genomes retrieved from males. Actually, as reported in GenBank, H. cumingii M genome (GenBank Accession Number HM347668) was indeed extracted from mantle tissue, whereas the source of the F (GenBank Accession Number NC_011763) is not reported. Conversely, H. schlegelii F and M genomes (GenBank Accession Numbers NC_015110 and HQ641407, respectively) were extracted by gonad tissue–not from gametes–which is known to contain somatic cells carrying the F genome. Furthermore, the study of cox2 gene reveals that M mtDNAs of both species do not have additional putative TMHs typical of all other unionids M-cox2 (see above; S11 Fig). These evidences point to the fact that both genomes do belong to F-type and their minimal divergence is due to normal intraspecific variability.
More interestingly, in the PCA of distance scores (Fig 5), DUI species clustering follows the taxonomic arrangement of bivalves. Two large assemblages are visible: unionid species, one side, and Amarsipobranchia (i.e. Veneridae + Mytilidae), the other. In fact, the divergence of the two mtDNAs is higher in unionids (Table 4). This may point to the conclusion that DUI is somehow different in these two lineages, leading to distinct patterns of sequence evolution. This is not a new observation, since differences in many respects of DUI were repeatedly evidenced between Unionidae and Mytilidae + Veneridae (see, e.g., [5, 14]; and reference therein). In particular, the main difference is that unionids have established M- and F-mtDNA lineages earlier than species radiation, thus leading to a higher divergence between sex-linked lineages and, thus, to a very strict "gender-joining" phylogenetic pattern [84–85, 5].
Conversely, in Amarsipobranchia, two tentative, perhaps overlapping explanations were given to account for the observed "species-joining" pattern, with the only exception of the fairly recent Mytilus edulis species complex ([5]; see also Fig 4 for Veneridae): (i) multiple role reversal events, as well as reversions to SMI, may have blurred the phylogenetic and diversification pattern [103–104, 5], and/or (ii) DUI and the establishment of the two sex-linked mitogenomes may have happened many times in different lineages. This was a conceivable hypothesis given the model described in [105, 5], where a relatively simple switch, called factor Z, is proposed to trigger DUI/SMI swaps.
However, it is also worth pointing out that genetic divergence behaves differently in single-genes pairwise comparisons, and this is not expected if we consider the observed variability as a function of the DUI onset time only. For example, unionids show high distance values for cytb and nad1, which are among the most conserved within Amarsipobranchia, and vice versa for nad4 and nad5 (Table 4). Currently, it is not possible to speculate on the reasons of such divergence patterns, and more comparative and structural analyses have to be done.
Conclusions
The present phylogenetic reconstruction (Fig 4) corroborates previous evolutionary trees of venerids [106, 10] and, above all, indicates future research lines: the detection of DUI in other genera of the family Veneridae and/or in other species of the genus Meretrix would add consistency in the single DUI origin hypothesis (at least for Heterodonta; [14]), while the direct observation of SMI in those groups would probably lead to a re-evaluation of the parsimony approach to the origin of DUI proposed in [14]. Furthermore, investigating the distribution of DUI within the genus Meretrix would open the field for comparisons with the Mytilus species complex, which is the only known case of a gender-joining pattern among Amarsipobranchia.
The great genome variability shown by bivalves at the mitochondrial level may somehow veil mtDNA similarities between distantly related DUI species, so that comparisons between taxonomically closer DUI species are needed to further characterize and understand the DUI mechanism and the related molecular machinery. This opportunity was unavailable for venerids so far. Therefore, the sequencing and characterization of M. lamarckii mtDNAs presented here makes this species a useful experimental counterpart of V. philippinarum, which in turn has been thoughtfully characterized in recent years (see, f.i., [107–111, 74]).
Supporting Information
Inferred stem-loop secondary structures of all Unassigned Regions (URs) comprised between two neighboring protein coding genes (PCGs). The label of each structure is obtained by concatenating "UNs" (Unassigned Nucleotides) and the two PCG names.
(PDF)
A, F-rrnL; B, F-rrnS; C, M-rrnL; D, M-rrnS.
(PDF)
All aminoacids are reported with their one-letter code; anticodons are highlighted in yellow.
(PDF)
All aminoacids are reported with their one-letter code; anticodons are highlighted in yellow.
(PDF)
The M anticodons are highlighted, while stretches of nucleotides involved in tRNA stem-loop structures are underlined. Only the relevant part of the corresponding F-UR is shown.
(PDF)
F (A) and M (B) M. lamarckii Long Unassigned Region (LUR) inferred secondary structures. The 109-bp tandem repeat that was detected in F-LUR is detailed in the upper-right insert; yellow lines, first repeat; purple lines, second repeat.
(PDF)
Numbers refer to the positions on the mitochondrial genomes. The TAS element was taken from [66] and located on the revised Cambridge Reference Sequence (GenBank Accession Number NC_012920).
(PDF)
Autocorrelograms for nucleotide trends shown in Fig 3: the autocorrelation function (acf) is plotted for lags from 0 to 17. Page 1, female mitochondrial genome; page 2, male mitochondrial genome; dashed lines, large-lag 95% standard errors.
(PDF)
Alignment between the unassigned ORF found in the LUR of the female mitochondrial genome (MeLaF) and the corresponding region of the published Meretrix lamarckii mitochondrial genome (MeLaNC_016174); numbers refer to positions on the GenBank sequences. Regions of mutations pseudogenizing the putative lost ORF are shaded in black in the published sequence; asterisks mark identical nucleotides.
(PDF)
MUSCLE alignment between the 109-bp repeated motif of the female LUR (MeLaF) and the 100-bp repeated motif of the published LUR (MeLaFNC_016174). Asterisks mark identical nucleotides.
(PDF)
Phobius predictions of cox2 residue locations for all DUI species used for this work. The cox2 length is reported in the x axis; the y axis refers to the posterior probability of a given position to be part of a TMH (gray), cytoplasmic (green), non-cytoplasmic (blue), or part of a signal peptide (red).
(PDF)
The script is called 4F; example files and a tutorial are also provided. The same script can be downloaded at the GitHub repository https://github.com/mozoo/4F.git.
(GZ)
Primers are listed by pairs, showing for each pair the forward (F) and the reverse (R) primers in the column "Strand". In the column "Sex" it is specified if a given pair was used for the female genome (F), for the male genome (M), or for both (both). For amplicons > 2,000 bp the Herculase enzyme was used (see text for details). Where two annealing temperatures are listed, the first one refers to the female genome and the second one to the male genome.
(PDF)
Sequences in boldface were obtained for this study. Taxonomy is taken from GenBank.
(PDF)
Meretrix lamarckii F (A) and M (B) Unassigned Regions (URs).
(PDF)
The canonical 13 PCGs (bold) and all other Open Reading Frames (ORFs) are reported along with their start base, stop base, frame, and Glimmer score. F_ORF141 and M_ORF138 are shown in bold as well.
(PDF)
Homologies of F_ORF141 with ORFs in other Meretrix mitochondrial genomes; the first entry is F_ORF141 itself.
(PDF)
(PDF)
The Kimura aminoacid distance is listed for each PCG. F, female genome; M, male genome; NC_016174, published Meretrix lamarckii mitochondrial genome. The F-atp8 gene has 11 aminoacids at the 5' end that are lacking in the published atp8 gene; as the remaining part of the peptide sequence is identical, the pairwise deletion led to a Kimura distance of 0.
(PDF)
Acknowledgments
Thanks are due to Prof. Norihiro Okada and Dr Hidenori Nishihara of the Tokyo Institute of Technology for having supported one of us (FP) in sampling animals and inspecting specimens. We would also like to thank Fabrizio Ghiselli, Davide Guerra, Mariangela Iannello, Liliana Milani and Emanuele Procopio for their kind help in different steps of the work. Finally, this work was greatly improved thanks to two anonymous reviewers.
Data Availability
Sequences are available in GenBank under the accession numbers KP244451 and KP244452. All other relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors have no support or funding to report.
References
- 1.Skibinski DOF, Gallagher C, Beynon CM. Mitochondrial DNA inheritance. Nature. 1994a;368: 817–818. [DOI] [PubMed] [Google Scholar]
- 2.Skibinski DOF, Gallagher C, Beynon CM. Sex limited mitochondrial DNA transmission in the marine mussel Mytilus edulis. Genetics. 1994b;138: 801–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zouros E, Oberhauser Ball A, Saavedra C, Freeman KR. An unusual type of mitochondrial DNA inheritance in the blue mussel Mytilus. Proc Natl Acad Sci USA. 1994a;91: 7463–7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zouros E, Oberhauser Ball A, Saavedra C, Freeman KR. Mitochondrial DNA inheritance—Reply. Nature. 1994b;368: 818. [DOI] [PubMed] [Google Scholar]
- 5.Zouros E. Biparental Inheritance Through Uniparental Transmission: The Doubly Uniparental Inheritance (DUI) of Mitochondrial DNA. Evol Bio. 2013;40: 1–31. [Google Scholar]
- 6.Breton S, Doucet-Beaupré HD, Stewert DT, Hoeh WR, Blier PU. The unusual system of doubly uniparental inheritance of mtDNA: isn't one enough? Trends Genet. 2007;23: 465–474. [DOI] [PubMed] [Google Scholar]
- 7.Passamonti M, Ghiselli F. Doubly uniparental inheritance: two mitochondrial genomes, one precious model for organelle DNA inheritance and evolution. DNA Cell Biol. 2009;28: 1–10. [DOI] [PubMed] [Google Scholar]
- 8.Passamonti M, Ricci A, Milani L and Ghiselli F. Mitochondrial genomes and Doubly Uniparental Inheritance: new insights from Musculista senhousia sex linked mitochondrial DNAs (Bivalvia Mytilidae). BMC Genomics. 2011;12: 442 10.1186/1471-2164-12-442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doucet-Beaupré H, Breton S, Chapman EG, Blier PU, Bogan AE, Stewart DT, et al. Mitochondrial phylogenomics of the Bivalvia (Mollusca): searching for the origin and mitogenomic correlates of doubly uniparental inheritance of mtDNA. BMC Evol Biol. 2010;10: 50 10.1186/1471-2148-10-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plazzi F, Cassano A, Passamonti M. The quest for Doubly Uniparental Inheritance in heterodont bivalves and its detection in Meretrix lamarckii (Veneridae: Meretricinae). J Zoolog Syst Evol Res. 2014;53: 87–94. [Google Scholar]
- 11.Hoeh WR, Stewart DT, Guttman SI. High fidelity of mitochondrial genome transmission under the doubly uniparental mode of inheritance in freshwater mussels (Bivalvia: Unionoidea). Evolution. 2002;56: 2252–2261. [DOI] [PubMed] [Google Scholar]
- 12.Theologidis I, Fodelianakis S, Gaspar MB, Zouros E. Doubly Uniparental Inheritance (DUI) of mitochondrial DNA in Donax trunculus (Bivalvia: Donacidae) and the problem of its sporadic detection in Bivalvia. Evolution. 2008;62: 959–970. 10.1111/j.1558-5646.2008.00329.x [DOI] [PubMed] [Google Scholar]
- 13.Boyle EE, Etter RJ. Heteroplasmy in a deep-sea protobranch bivalve suggests an ancient origin of doubly uniparental inheritance of mitochondria in Bivalvia. Mar Biol. 2013;160: 413–422. [Google Scholar]
- 14.Plazzi F. The detection of sex-linked heteroplasmy in Pseudocardium sachalinense (Bivalvia: Mactridae) and its implications for the distribution of doubly uniparental inheritance of mitochondrial DNA. J Zoolog Syst Evol Res. 2015;53: 205–210. [Google Scholar]
- 15.Dégletagne C, Abele D, Held C. A distinct mitochondrial genome with DUI-like inheritance in the ocean quahog Arctica islandica. Mol Biol Evol. 2016;33: 375–383. 10.1093/molbev/msv224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Passamonti M, Scali V. Gender-associated mitochondrial DNA heteroplasmy in the venerid clam Tapes philippinarum (Mollusca Bivalvia). Curr Genet. 2001;39: 117–124. [DOI] [PubMed] [Google Scholar]
- 17.Zhuang Q. Fauna Sinica: Invertebrata: Mollusca: Bivalvia: Veneridae. Beijing: Science Press; 2001. p. 229–236. [Google Scholar]
- 18.Wang H, Zhang S, Xiao G, Liu B. Complete mtDNA of the Meretrix lamarckii (Bivalvia: Veneridae) and molecular identification of suspected M. lamarckii based on the whole mitochondrial genome. Mar Genom. 2011;4: 263–271. [DOI] [PubMed] [Google Scholar]
- 19.Qi Z. Seashells of China. Beijing: China Ocean Press; 2004. p. 306. [Google Scholar]
- 20.Sambrook J, Russell DW. Purification of nucleic acids by extraction with phenol:chloroform. CSH Protoc. 2006; 10.1101/pdb.prot4455 [DOI] [PubMed] [Google Scholar]
- 21.Rozen S, Skaletsky HJ. Primer3 on the WWW for generals users and for biologist programmers. Methods in Mol Biol. 2000;132: 365–386. [DOI] [PubMed] [Google Scholar]
- 22.Lis JT, Schleif R. Size Fractionation of Double-Stranded DNA by Precipitation with Polyethylene-Glycol. Nucleic Acids Res. 1975;2: 383–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.R Development Core Team. R: A language and environment for statistical computing Vienna: R Foundation for Statistical Computing; 2013. Available: http://www.R-project.org/. [Google Scholar]
- 25.Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, et al. MITOS: Improved de novo Metazoan Mitochondrial Genome Annotation. Mol Phylogenet Evol. 2013;69: 313–319. 10.1016/j.ympev.2012.08.023 [DOI] [PubMed] [Google Scholar]
- 26.Tatusov T, Tatusov R. ORF Finder. Available: http://www.ncbi.nlm.nih.gov/projects/gorf/.
- 27.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215: 403–410. [DOI] [PubMed] [Google Scholar]
- 28.Ye J, McGinnis S, Madden TL. BLAST: improvements for better sequence analysis. Nucleic Acids Res. 2006;34: W6–W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beitz E. TeXshade: shading and labeling multiple sequence alignments using LaTeX 2ε. Bioinformatics 2000;16: 135–139. [DOI] [PubMed] [Google Scholar]
- 31.Lowe TM, Eddy SR. tRNAscan-SE: A Program for Improved Detection of Transfer RNA Genes in Genomics Sequence. Nucleic Acids Res. 1997;25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schattner P, Brooks AN, Lowe TM. tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33: W686–W689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laslett D, Canbäck B. ARWEN, a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 2008;24: 172–175. [DOI] [PubMed] [Google Scholar]
- 34.Conant GC, Wolfe KH: GenomeVx: simple web-based creation of editable circular chromosome maps. Bioinformatics. 2008;24: 861–862. 10.1093/bioinformatics/btm598 [DOI] [PubMed] [Google Scholar]
- 35.Zucker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Darty K, Denise A, Ponty J. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009;25: 1974–1975. 10.1093/bioinformatics/btp250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acid Res. 1999;27: 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Käll L, Krogh A, Sonnhammer ELL. A Combined Transmembrane Topology and Signal Peptide Prediction Method. J Mol Biol. 2004;338: 1027–1036. [DOI] [PubMed] [Google Scholar]
- 39.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30: 1236–1240. 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hofmann K, Stoffel W. TMbase—A database of membrane spanning proteins segments Biol Chem. 1993;347: 166. [Google Scholar]
- 41.Tusnády GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17: 849–850. [DOI] [PubMed] [Google Scholar]
- 42.Pons JL, Labesse G. @TOME-2: a new pipeline for comparative modeling of protein-ligand complexes. Nucleic Acids Res. 2009;37: W485–W491. 10.1093/nar/gkp368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delcher A, Harmon D, Kasif S, White O, Salzberg S. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27: 4636–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16: 276–277. [DOI] [PubMed] [Google Scholar]
- 45.Remmert M, Biegert A, Hauser A, Söding J. HHblits: Lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods. 2012;9: 173–175. [DOI] [PubMed] [Google Scholar]
- 46.Altschul S, Madden T, Schaffer A, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res. 1997;25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Do CB, Mahabhashyam MSP, Brudno M, Batzoglou S. PROBCONS: Probabilistic Consistency-based Multiple Sequence Alignment. Genome Research. 2005;15: 330–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lorenz R, Bernhart SH, Höner zu Siederdissen C, Tafer H, Flamm C, Stadler PF, et al. ViennaRNA Package 2.0. Algorithm Mol Biol. 2011;6: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katoh K, Standley DM. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol Biol Evol. 2013;30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302: 205–217. [DOI] [PubMed] [Google Scholar]
- 51.Di Tommaso P, Moretti S, Xenarios I, Orobitg M, Montanyola A, Chang JM, et al. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011;39: W13–W17. 10.1093/nar/gkr245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): selection of phylogenetic informative regions from multiple sequence alignments. BMC Evolutionary Biology. 2010;10: 210 10.1186/1471-2148-10-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lanfear R, Calcott B, Ho SYW, Guindon S. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution. 2012;29: 1695–1701. 10.1093/molbev/mss020 [DOI] [PubMed] [Google Scholar]
- 54.Stamatakis A. RAxML Version 8: A tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics. 2014;30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith SA, Dunn C. Phyutility: a phyloinformatics utility for trees, alignments, and molecular data. Bioinformatics. 2008;24: 715–716. 10.1093/bioinformatics/btm619 [DOI] [PubMed] [Google Scholar]
- 56.Huson DH, Scornavacca C. Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks. Syst Biol. 2012;61: 1061–1067. 10.1093/sysbio/sys062 [DOI] [PubMed] [Google Scholar]
- 57.Jin L, Nei M. Limitations of the evolutionary parsimony method of phylogenetic analysis. Mol Biol Evol. 1990;7: 82–102. [DOI] [PubMed] [Google Scholar]
- 58.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge: Cambridge University Press; 1983. [Google Scholar]
- 59.Lê S, Josse J, Husson F: FactoMineR: An R Package for Multivariate Analysis. J Stat Softw. 2008;25: 1–18. [Google Scholar]
- 60.Wickham H. ggplot2 Elegant Graphics for Data Analysis. New York: SpringerScience+Business Media; 2010. [Google Scholar]
- 61.Plazzi F, Ceregato A, Taviani M, Passamonti M. A molecular phylogeny of bivalve mollusks: ancient radiations and divergences as revealed by mitochondrial genes. PLoS ONE. 2011;6: e27174 10.1371/journal.pone.0027147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Breton S, Stewart DT, Hoeh WR. Characterization of a mitochondrial ORF from the gender-associated mtDNA of Mytilus spp. (Bivalvia: Mytilidae): Identification of the missing ATPase 8 gene. Mar Genom. 2010;3: 11–18. [DOI] [PubMed] [Google Scholar]
- 63.Plazzi F, Ribani A, Passamonti M. The complete mitochondrial genome of Solemya velum (Mollusca: Bivalvia) and its relationships with Conchifera. BMC Genomics. 2013;14: 409 10.1186/1471-2164-14-409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290: 470–474. [DOI] [PubMed] [Google Scholar]
- 65.Hoffmann RJ, Boore JL, Brown WM. A novel mitochondrial genome organization for the blue mussel, Mytilus edulis. Genetics. 1992;131: 397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Doda JN, Wright CT, Clayton DA. Elongation of displacement-loop strands in human and mouse mitochondrial DNA is arrested near specific template sequences. Proc Natl Acad Sci USA. 1981;78: 6116–6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cao L, Kenchington E, Zouros E, Rodakis GC. Evidence that the large noncoding sequence is the main control region of maternally and paternally transmitted mitochondrial genomes of the marine mussel (Mytilus spp.). Genetics 2004;167: 835–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kosiol C, Goldman N. Different versions of the Dayhoff rate matrix. Mol Biol Evol. 2005;22: 193–199. [DOI] [PubMed] [Google Scholar]
- 69.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25: 1307–1320. 10.1093/molbev/msn067 [DOI] [PubMed] [Google Scholar]
- 70.Tavaré S. Some probabilistic and statistical problems in the analysis of DNA sequences. Lectures on Mathematics in the Life Sciences. 1986;17: 57–86. [Google Scholar]
- 71.Breton S, Doucet-Beaupré H, Stewart DT, Piontkivska H, Karmakar M, Bogan AE, et al. Comparative mitochondrial genomics of freshwater mussels (Bivalvia: Unionida) with Doubly Uniparental Inheritance of mtDNA: gender-specific Open Reading Frames and putative origins of replication. Genetics. 2009;183: 1575–1589. 10.1534/genetics.109.110700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Breton S, Stewart DT, Shepardson S, Trdan RJ, Bogan AE, Chapman EG, et al. Novel protein genes in animal mtDNA: a new sex determination system in freshwater mussels (Bivalvia: Unionoida)? Mol Biol Evol. 2011;28: 1645–1659. 10.1093/molbev/msq345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Milani L, Ghiselli F, Maurizii MG, Nuzhdin S, Passamonti M. Paternally transmitted mitochondria express a new gene of potential viral origin. Genome Biol Evol. 2014;6: 391–405. 10.1093/gbe/evu021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Milani L, Ghiselli F, Pecci A, Maurizii MG, Passamonti M. The expression of a novel mitochondrially-encoded gene in gonadic precursors may drive paternal inheritance of mitochondria. PLoS One. 2015;10: e0137468 10.1371/journal.pone.0137468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guerra D, Ghiselli F, Passamonti M. The largest unassigned regions of the male- and female-transmitted mitochondrial DNAs in Musculista senhousia (Bivalvia Mytilidae). Gene. 2014;536: 316–325. 10.1016/j.gene.2013.12.005 [DOI] [PubMed] [Google Scholar]
- 76.Ghiselli F, Milani L, Guerra D, Chang PL, Breton S, Nuzhdin SV, et al. Structure, transcription and variability of metazoan mitochondrial genome. Perspectives from an unusual mitochondrial inheritance sistem. Genome Biol Evol. 2013;5: 1535–1554. 10.1093/gbe/evt112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Francino MP, Ochman H. Strand asymmetries in DNA evolution. Trends Genet. 1997;13: 240–245. [DOI] [PubMed] [Google Scholar]
- 78.Fonseca MM, Harris DJ, Posada D. The Inversion of the Control Region in Three Mitogenomes Provides Further Evidence for an Asymmetric Model of Vertebrate mtDNA Replication. PLoS ONE. 2014;9: e106654 10.1371/journal.pone.0106654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27: 1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dreyer H, Steiner G. The complete sequences and gene organization of the mitochondrial genomes of the heterodont bivalves Acanthocardia tubercolata and Hiatella arctica and the first record for a putative Atpase subunit 8 gene in marine bivalves. Front Zool. 2006;3: 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Smietanka B, Burzynski A, Wenne R. Comparative genomics of marine mussels (Mytilus spp.) gender associated mtDNA: Rapidly evolving atp8. J Mol Evol. 2010;71: 385–400. 10.1007/s00239-010-9393-4 [DOI] [PubMed] [Google Scholar]
- 82.Saunier A, Garcia P, Becquet V, Marsaud N, Escudié F, Pante E. Mitochondrial genomes of the Baltic clam Macoma balthica (Bivalvia: Tellinidae): setting the stage for studying mito-nuclear incompatibilities. BMC Evol Biol. 2014;14: 259 10.1186/s12862-014-0259-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fearnley IM, Walker JE. Two overlapping genes in bovine mitochondrial DNA encode membrane components of ATP synthase. EMBO J. 1986;5: 2003–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Curole JP, Kocher TD. Ancient sex-specific extension of the cytochrome c oxidase II gene in bivalves and the fidelity of doubly-uniparental inheritance. Mol Biol Evol. 2002;19: 1323–1328. [DOI] [PubMed] [Google Scholar]
- 85.Curole JP, Kocher TD. Evolution of a unique mitotype-specific protein-coding extension of the cytochrome c oxidase II gene in freshwater mussels (Bivalvia: Unionida). J Mol Evol. 2005;61: 381–389. [DOI] [PubMed] [Google Scholar]
- 86.Chakrabarti R, Walker JM, Chapman EG, Shepardson SP, Trdan RJ, Curole JP, et al. Reproductive function for a C-terminus extended, male-transmitted cytochrome c oxidase subunit II protein expressed in both spermatozoa and eggs. FEBS Lett. 2007;581: 5213–5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chapman EG, Piontkivska H, Walker JM, Stewart DT, Curole JP, Hoeh WR. Extreme primary and secondary protein structure variability in the chimeric male-transmitted cytochrome c oxidase subunit II protein in freshwater mussels: Evidence for an elevated amino acid substitution rate in the face of domain-specific purifying selection. BMC Evol Biol. 2008;8: 165 10.1186/1471-2148-8-165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Milbury CA, Gaffney PM. Complete mitochondrial DNA sequence of the eastern oyster Crassostrea virginica. Mar Biotechnol. 2005;7: 697–712. [DOI] [PubMed] [Google Scholar]
- 89.Xu K, Kanno M, Yu H, Li Q, Kijima A. Complete mitochondrial DNA sequence and phylogenetic analysis of Zhikong scallop Chlamys farreri (Bivalvia: Pectinidae). Mol Biol Rep. 2011;38: 3067–3074. 10.1007/s11033-010-9974-8 [DOI] [PubMed] [Google Scholar]
- 90.Sun S, Kong L, Yu H, Li Q. The complete mitochondrial DNA of Tegillarca granosa and comparative mitogenomic analyses of three Arcidae species. Gene. 2015;557: 61–70. 10.1016/j.gene.2014.12.011 [DOI] [PubMed] [Google Scholar]
- 91.Gould SJ, Lewontin RC. The Spandrels of San Marco and the Panglossian Paradigm: A Critique to the Adaptationist Programme. Proc R Soc B-Biol Sci. 1979;205: 581–598. [DOI] [PubMed] [Google Scholar]
- 92.Garey JR, Wolstenholme DR. Platyhelminth mitochondrial DNA: Evidence for early evolutionary origin of a tRNAserAGN that contains a dihydrouridine arm replacement loop, and of serine-specifying AGA and AGG codons. J Mol Evol. 1989;28: 374–387. [DOI] [PubMed] [Google Scholar]
- 93.Hanada T, Suzuki T, Yokogawa T, Takemoto-Hori C, Sprinzl M, Watanabe K. Translation ability of mitochondrial tRNAsSer with unusual secondary structures in an in vitro translation system of bovine mitochondria. Genes Cells. 2001;6: 1019–1030. [DOI] [PubMed] [Google Scholar]
- 94.Arunkumar KP, Nagaraju J. Unusually long palyndromes are abundant in mitochondrial control regions of insects and nematodes. PLoS ONE. 2006;1: e110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Reyes A, Gissi C, Pesole G, Saccone C. Asymmetrical directional mutation pressure in the mitochondrial genome of mammals. Mol Biol Evol. 1998;15: 957–966. [DOI] [PubMed] [Google Scholar]
- 96.Saccone C, Gissi C, Reyes A, Larizza A, Sbisà E, Pesole G. Mitochondrial DNA in metazoa: degree of freedom in a frozen event. Gene 2002;286: 3–12. [DOI] [PubMed] [Google Scholar]
- 97.Faith JJ, Pollock DD. Likelihood analysis of asymmetrical mutation bias gradients in vertebrate mitochondrial genomes. Genetics 2003;165: 735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rodakis GC, Cao L, Mizi A, Kenchington EL, Zouros E. Nucleotide content gradients in maternally and paternally inherited mitochondrial genomes of the mussel Mytilus. J Mol Evol. 2007;65: 124–136. [DOI] [PubMed] [Google Scholar]
- 99.Fonseca MM, Posada D, Harris JD. Inverted replication of vertebrate mitochondria. Mol Biol Evol. 2008;25: 805–808. 10.1093/molbev/msn050 [DOI] [PubMed] [Google Scholar]
- 100.Seligmann H, Krishnan NM, Rao BJ. Possible multiple origins of replication in primate mitochondria: alternative role of tRNA sequences. J Theor Biol. 2006;241: 321–332. [DOI] [PubMed] [Google Scholar]
- 101.Fischer D, Eisenberg D. Finding families for genomic ORFans. Bioinformatics. 1999;15: 759–762. [DOI] [PubMed] [Google Scholar]
- 102.Kyriakou E, Chatzoglou E, Rodakis GC, Zouros E. Does the ORF in the control region of Mytilus mtDNA code for a protein product? Gene 2014;546: 448–450. 10.1016/j.gene.2014.06.031 [DOI] [PubMed] [Google Scholar]
- 103.Hoeh WR, Stewart DT, Saavedra C, Sutherland BW, Zouros E. Phylogenetic evidence for role-reversals of gender-associated mitochondrial DNA genomes in Mytilus (Bivalvia: Mytilidae). Mol Biol Evol. 1997;14: 959–967. [DOI] [PubMed] [Google Scholar]
- 104.Quesada H, Wenne R, Skibinski DOF. Interspecies transfer of female mitochondrial DNA is coupled with role-reversals and departure from neutrality in the mussel Mytilus trossulus. Mol Biol Evol. 1999;16: 655–665. [DOI] [PubMed] [Google Scholar]
- 105.Diz AP, Dudley E, Cogswell A, MacDonald BW, Kenchington ELR, Zouros E, et al. Proteomic Analysis of Eggs from Mytilus edulis Females Differing in Mitochondrial DNA Transmission Mode. Mol Cell Proteomics. 2013;12: 3068–3080. 10.1074/mcp.M113.031401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kappner I, Bieler R. Phylogeny of venus clams (Bivalvia: Venerinae) as inferred from nuclear and mitochondrial gene sequences. Mol Phylogenet Evol. 2006;40: 317–331. [DOI] [PubMed] [Google Scholar]
- 107.Ghiselli F, Milani L, Passamonti M. Strict sex-specific mtDNA segregation in the germ line of the DUI species Venerupis philippinarum (Bivalvia: Veneridae). Mol Biol Evol. 2011;28: 949–961. 10.1093/molbev/msq271 [DOI] [PubMed] [Google Scholar]
- 108.Ghiselli F, Milani L, Chang PL, Hedgecock D, Davis JP, Nuzhdin SV, et al. De novo assembly of Ruditapes philippinarum trascriptome provides new insights on expression bias, mitochondrial Doubly Uniparental Inheritance and sex-determination. Mol Biol Evol. 2012;29: 771–786. 10.1093/molbev/msr248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Milani L, Ghiselli F, Passamonti M. Sex-linked mitochondrial behavior during early embryo development in Ruditapes philippinarum (Bivalvia Veneridae) a species with the Doubly Uniparental Inhetitance (DUI) of mitochondria. J Exp Zool Part B. 2012;318: 182–189. [DOI] [PubMed] [Google Scholar]
- 110.Milani L, Ghiselli F, Nuzhdin S, Passamonti M. Nuclear genes with sex bias in Ruditapes philippinarum (Bivalvia Veneridae): mitochondrial inheritance and sex determination in DUI species. J Exp Zool Part B. 2013;320B: 442–454. [DOI] [PubMed] [Google Scholar]
- 111.Milani L, Ghiselli F, Iannello M, Passamonti M. Evidence for somatic transcription of male-transmitted mitochondrial genome in the DUI species Ruditapes philippinarum (Bivalvia: Veneridae). Curr Genet. 2014;60: 163–173. 10.1007/s00294-014-0420-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Inferred stem-loop secondary structures of all Unassigned Regions (URs) comprised between two neighboring protein coding genes (PCGs). The label of each structure is obtained by concatenating "UNs" (Unassigned Nucleotides) and the two PCG names.
(PDF)
A, F-rrnL; B, F-rrnS; C, M-rrnL; D, M-rrnS.
(PDF)
All aminoacids are reported with their one-letter code; anticodons are highlighted in yellow.
(PDF)
All aminoacids are reported with their one-letter code; anticodons are highlighted in yellow.
(PDF)
The M anticodons are highlighted, while stretches of nucleotides involved in tRNA stem-loop structures are underlined. Only the relevant part of the corresponding F-UR is shown.
(PDF)
F (A) and M (B) M. lamarckii Long Unassigned Region (LUR) inferred secondary structures. The 109-bp tandem repeat that was detected in F-LUR is detailed in the upper-right insert; yellow lines, first repeat; purple lines, second repeat.
(PDF)
Numbers refer to the positions on the mitochondrial genomes. The TAS element was taken from [66] and located on the revised Cambridge Reference Sequence (GenBank Accession Number NC_012920).
(PDF)
Autocorrelograms for nucleotide trends shown in Fig 3: the autocorrelation function (acf) is plotted for lags from 0 to 17. Page 1, female mitochondrial genome; page 2, male mitochondrial genome; dashed lines, large-lag 95% standard errors.
(PDF)
Alignment between the unassigned ORF found in the LUR of the female mitochondrial genome (MeLaF) and the corresponding region of the published Meretrix lamarckii mitochondrial genome (MeLaNC_016174); numbers refer to positions on the GenBank sequences. Regions of mutations pseudogenizing the putative lost ORF are shaded in black in the published sequence; asterisks mark identical nucleotides.
(PDF)
MUSCLE alignment between the 109-bp repeated motif of the female LUR (MeLaF) and the 100-bp repeated motif of the published LUR (MeLaFNC_016174). Asterisks mark identical nucleotides.
(PDF)
Phobius predictions of cox2 residue locations for all DUI species used for this work. The cox2 length is reported in the x axis; the y axis refers to the posterior probability of a given position to be part of a TMH (gray), cytoplasmic (green), non-cytoplasmic (blue), or part of a signal peptide (red).
(PDF)
The script is called 4F; example files and a tutorial are also provided. The same script can be downloaded at the GitHub repository https://github.com/mozoo/4F.git.
(GZ)
Primers are listed by pairs, showing for each pair the forward (F) and the reverse (R) primers in the column "Strand". In the column "Sex" it is specified if a given pair was used for the female genome (F), for the male genome (M), or for both (both). For amplicons > 2,000 bp the Herculase enzyme was used (see text for details). Where two annealing temperatures are listed, the first one refers to the female genome and the second one to the male genome.
(PDF)
Sequences in boldface were obtained for this study. Taxonomy is taken from GenBank.
(PDF)
Meretrix lamarckii F (A) and M (B) Unassigned Regions (URs).
(PDF)
The canonical 13 PCGs (bold) and all other Open Reading Frames (ORFs) are reported along with their start base, stop base, frame, and Glimmer score. F_ORF141 and M_ORF138 are shown in bold as well.
(PDF)
Homologies of F_ORF141 with ORFs in other Meretrix mitochondrial genomes; the first entry is F_ORF141 itself.
(PDF)
(PDF)
The Kimura aminoacid distance is listed for each PCG. F, female genome; M, male genome; NC_016174, published Meretrix lamarckii mitochondrial genome. The F-atp8 gene has 11 aminoacids at the 5' end that are lacking in the published atp8 gene; as the remaining part of the peptide sequence is identical, the pairwise deletion led to a Kimura distance of 0.
(PDF)
Data Availability Statement
Sequences are available in GenBank under the accession numbers KP244451 and KP244452. All other relevant data are within the paper and its Supporting Information files.