Abstract
Bacteria with high nucleic acid (HNA) and low nucleic acid (LNA) content are commonly observed in aquatic environments. To date, limited knowledge is available on their temporal and spatial variations in freshwater environments. Here an investigation of HNA and LNA bacterial abundance and their flow cytometric characteristics was conducted in an exorheic river (Haihe River, Northern China) over a one year period covering September (autumn) 2011, December (winter) 2011, April (spring) 2012, and July (summer) 2012. The results showed that LNA and HNA bacteria contributed similarly to the total bacterial abundance on both the spatial and temporal scale. The variability of HNA on abundance, fluorescence intensity (FL1) and side scatter (SSC) were more sensitive to environmental factors than that of LNA bacteria. Meanwhile, the relative distance of SSC between HNA and LNA was more variable than that of FL1. Multivariate analysis further demonstrated that the influence of geographical distance (reflected by the salinity gradient along river to ocean) and temporal changes (as temperature variation due to seasonal succession) on the patterns of LNA and HNA were stronger than the effects of nutrient conditions. Furthermore, the results demonstrated that the distribution of LNA and HNA bacteria, including the abundance, FL1 and SSC, was controlled by different variables. The results suggested that LNA and HNA bacteria might play different ecological roles in the exorheic river.
Introduction
In natural aquatic environments, planktonic bacteria tend to cluster into two distinct subgroups, namely high nucleic acid content (HNA) bacteria and low nucleic acid content (LNA) bacteria, by flow cytometry (FCM) measurement in combination with nucleic acid staining [1]. This classification based on cellular size and fluorescence intensity of bacteria is widely observed in marine environments [2–6]. When first observed by FCM, LNA bacteria were regarded as inactive, dead or dying cells [7,8]. However, recent studies demonstrated that LNA bacteria were metabolically active [1,5,9]. It was found that LNA bacteria can survive and grow in oligotrophic environments due to their high affinity and binding-protein dependent uptake system [10]. Meanwhile, certain special cellular membrane constitutions could protect LNA bacteria from oxidation [11]. It was reported that LNA bacteria could adopt a dormancy strategy to overcome unfavorable environmental conditions [12].
In terms of ecological function, LNA bacteria were reported to play at least an equal niche role as HNA bacteria in the ecosystems [9,10,13–15]. Hence, information on the relation and variance between LNA and HNA bacteria are important to a better understanding of these two widespread groups of bacterioplankton in aquatic environments, and provide important foundation for further exploration of their environmental applications. The data on LNA and HNA bacterial abundance and activities have mainly come from marine environments [2,3]. It was found that the abundance and flow cytometric characteristics of LNA and HNA bacteria were strongly regulated by environmental variables, e.g. temperature, salinity, chlorophyll-α and nutrient conditions [5,6,16,17]. However, whether their variations on abundance and cytometric characteristics have similar pattern and how the variations respond to environmental factors are still unclear.
The distribution of LNA and HNA bacteria was found to change seasonally [18,19]. It was reported that although the growth rate of HNA bacteria in Lake Biwa generally exceeded that of LNA bacteria, LNA bacteria grew faster than and were grazed as fast as HNA bacteria in late August, when nutrients were severely limited [20]. Gomes and colleagues reported that HNA bacteria were apparently more responsive to the winter-spring phytoplankton bloom than LNA bacteria [21]. Nevertheless, to the best of our knowledge, information on the distribution and variations of LNA and HNA bacteria in freshwater environments is very limited, especially for the gradient along the river to the ocean.
The aims of the present study were 1) to analyze the temporal and spatial variations of the LNA and HNA bacterial abundance and cytometric characteristics along an exorheic river (the Haihe River); and 2) to estimate the effects of the driving factors on the distribution pattern of these two subgroups by multivariate analysis.
Materials and Methods
Ethics Statement
Water samples were taken from the Haihe River, where no specific permission is required. The Haihe River is a public river running through Tianjin, China. It is not privately owned or protected. The current study did not involve endangered or protected species. Water sampling procedures were reviewed and followed the Chinese Standard of Collection and Preservation of Water Samples (GB/T 5750.2–2006).
Sampling sites
The samples were collected at nine sampling stations along the Haihe river, which inflows into the Bohai Sea and is the largest river in Northern China (Fig 1) [22]. Water samples were collected from each station in four seasons: the autumn (September, 2011), winter (December, 2011), spring (April, 2012), and summer (July, 2012). At each sampling station, 2 L water samples were collected in clean and sterile bottles from a depth of about 0.5 m below the water surface. Samples were stored at 4°C during transportation and processed immediately within 24 hours after sampling.
Fig 1. Sampling sites in the Haihe River.
Black solid circles (●) indicate the sampling stations, and the black dash-dot curve (—·—) and blue solid curve (━) indicate the coastline and river, respectively.
Flow cytometry analysis
Flow cytometry analysis was performed as described in Ma et al. [22]. One milliliter water sample was stained with 10 μL/ml SYBR Green I (1:100 dilution in dimethyl sulfoxide as the working solution; Invitrogen, USA), and incubated in the dark for 15 min at room temperature before measurement. The FCM (CyFlow Space instrument, Partec, Germany) specific instrumental gain parameters settings were as follows: SSC = 369 (log3), FL1 = 380 (log4), FL3 = 750 (log4). Bacterial communities were gated through the two-parameter dot-plot of green fluorescence (FL1) and side scatter (SSC), then the LNA and HNA bacterial concentrations were counted separately and the respective geometrical means of FL1 and SSC of LNA and HNA were calculated. All samples were measured in triplicate. Water samples were diluted in Milli-Q water (cell-free) so that the bacterial concentration was always less than 2×105 cells/mL during the FCM measurement. The instrument detection limit was below 500 cells/s with an average standard deviation of 5%.
Water environmental parameters
The temperature, pH and salinity of sampling waters were measured using YSI EC300 Water Quality Sonde. Water physical and chemical properties e.g. total suspended solids (TSS), total nitrogen (TN), nitrate (NO3), total phosphorus (TP), total dissolved phosphorus (TDP), total organic carbon (TOC), and chlorophyll-α (Chl-a) were measured as described in our previous studies [22, 23].
Statistical Analysis
Analysis of variance (ANOVA) was conducted to test the significance of differences in measured or calculated parameters by using R statistical software (http://www.r-project.org/). A multivariate redundancy analysis (RDA) was performed by Canoco software (Canoco for Windows version 4.5) to further illustrate the changes of LNA and HNA bacteria in response to environmental factors [24]. The data were centered and standardized before redundancy analysis, and the Monte Carlo test was used to examine the significance of the RDA method. Meanwhile, a generalized linear model (GLM) and generalized additive model (GAM) were performed in a stepwise manner to predict the LNA and HNA bacterial abundance and flow cytometric characteristic response to environmental ordination axes in RDA. The axes were constrained by environmental variables, and the visualization formula constructed in terms of linear, quadratic or cubic degrees of GLM, then F statistics were used to test the significance in both GLM and GAM [25].
Results and Discussion
Temporal and spatial variations of LNA and HNA bacterial abundance and FCM characteristics
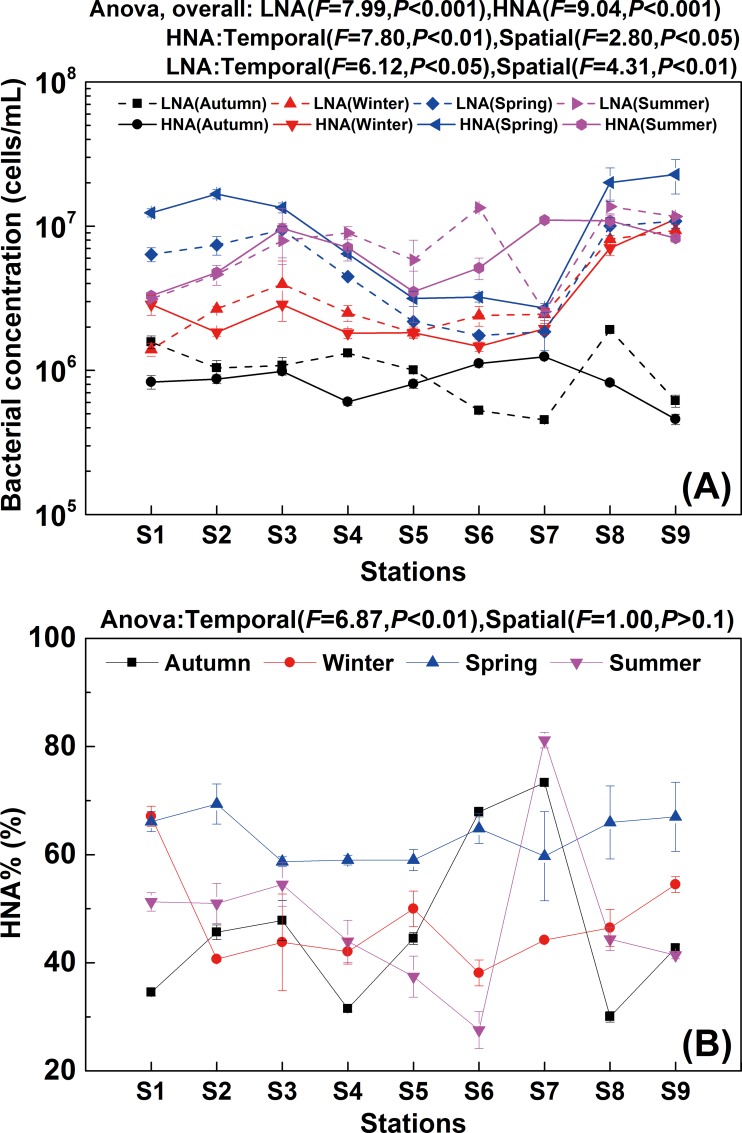
As shown in the flow cytogram, LNA and HNA bacteria in the Haihe River could be clearly discriminated on the basis of their side scatter (SSC) and fluorescence intensity (FL1) in all four seasons (Fig 2). It showed that the bimodal distribution phenomenon based on nucleic acid content and cell size was commonly present in the planktonic bacteria from river ecosystems to oceans irrespective of seasonal shifts. Both LNA and HNA bacterial concentrations in spring and summer were significantly higher than those in winter and autumn (P < 0.05) (Fig 3). Although there were similar trends in temporal distributions between HNA and LNA bacteria, the one-way ANOVA analysis of HNA and LNA bacterial abundance showed that the variability in HNA (F = 9.04, P < 0.001) was greater than the variability in LNA (F = 7.99, P < 0.001) in different seasons (Fig 3A). Clear spatial variations were observed for both HNA and LNA bacteria along the river to the Bohai Sea (Fig 3). Meanwhile, the results showed that LNA bacteria had an equal share with HNA bacteria in all seasons except spring. In contrast, HNA bacteria dominated the community in spring (64.8%) (Fig 3). This is consistent with previous reports on variations of LNA bacterial abundance in marine environments. For example, Calvo-Diaz and Moran investigated seasonal dynamics of picoplankton in the central Cantabrian Sea (southern Bay of Biscay), where it was observed that HNA bacteria dominated the community in winter and spring (64%), while the proportion of LNA and HNA bacteria was almost equal in autumn and summer [26]. The results suggested that both HNA and LNA bacteria could potentially dominate the community and were important components in the microbial community in both freshwater and marine ecosystems.
Fig 2. Flow cytogram example of LNA and HNA bacteria in the Haihe River.
Dashed lines indicate LNA bacteria, and solid lines indicate HNA bacteria.
Fig 3.
Variations of LNA and HNA bacterial concentration (A) and percentage of HNA bacteria (B) along the Haihe River. Error bars represent standard deviation of triplicate measurements.
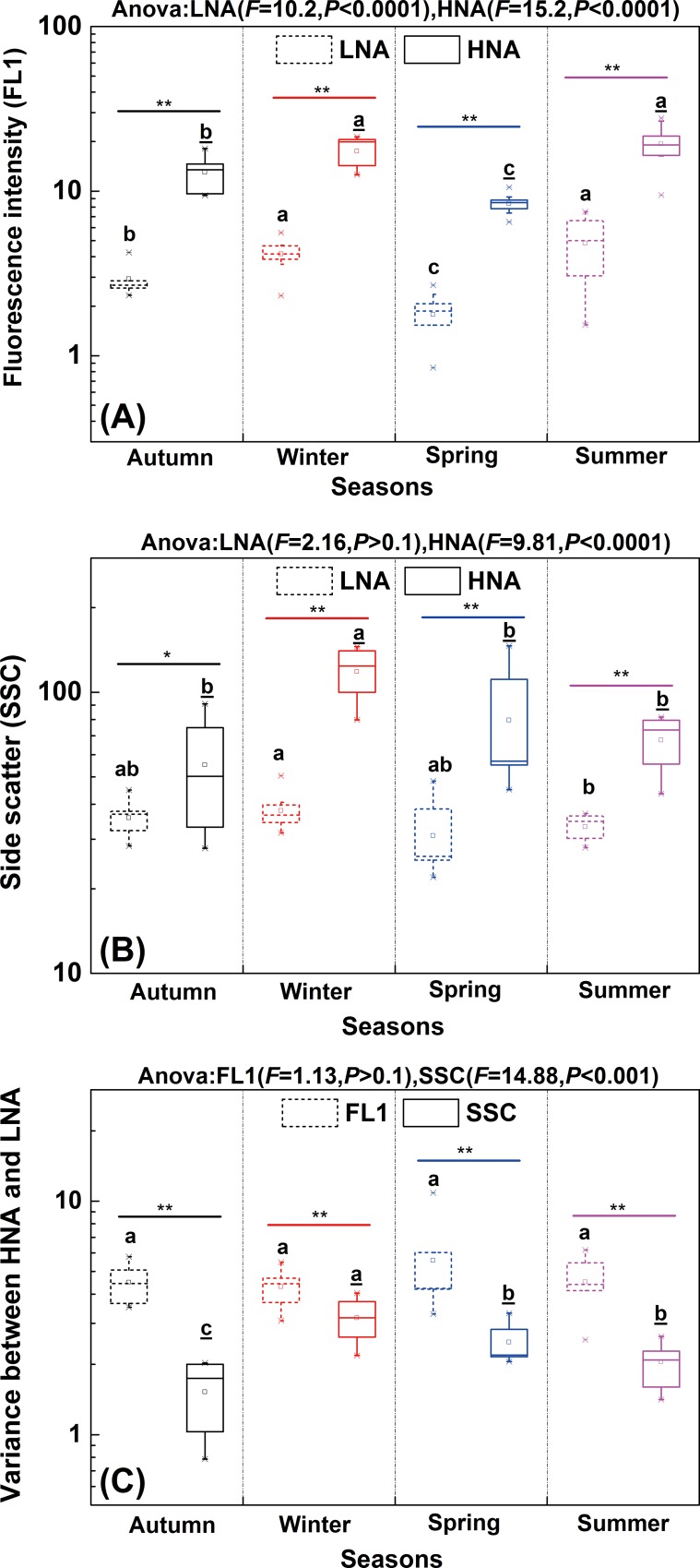
With respect to flow cytometric characteristics, both green fluorescence intensity (FL1) (Pearson’s R = 0.850, P < 0.01) and side scatter (SSC) (Pearson’s R = 0.600, P < 0.01) of HNA and LNA were significantly correlated along the river. In comparison, no significant correlation was found between the FL1 and SSC within each subgroup (HNA: Pearson’s R = 0.371, P < 0.05, LNA: Pearson’s P > 0.1). In terms of temporal variation, both FL1 and SSC of HNA as well as FL1 of LNA showed significant changes between seasons (Fig 4A and 4B). Meanwhile, similar to the abundance variation, the variabilities of FL1 (F = 15.2, P < 0.0001) and SSC (F = 9.81, P < 0.0001) in HNA were greater than that in LNA (FL1: F = 10.2, P < 0.0001; SSC: F = 2.16, P > 0.1). In addition, based on the differences in cytometric parameters (FL1 and SSC) between HNA and LNA, the derived variable (FL1HNA/FL1LNA and SSCHNA/SSCLNA) was used to figure out the relative distances between HNA and LNA in FCM. Such ratios could complementally represent variance within these two subgroups on FL1 and SSC, then further explain what and how different factors affect these variations [27]. The results showed that the relative distance of FL1 between HNA and LNA (median(min-max): 4.4(2.5–10.9)) was significantly higher than that of SSC (2.2(0.8–4.0)) (P < 0.01) in each season (Fig 4C). Furthermore, the relative distance of SSC between HNA and LNA showed significant temporal variation (F = 14.88, P < 0.001) (Fig 4C). The reason may lie in the seasonal variation of HNA and LNA bacterial activity which would result in the seasonal changes in cell size [28]. It was reported that the cell size of LNA bacteria became larger than that of HNA bacteria in the NW Mediterranean in March [21]. In contrast, no significant temporal variation was observed for the relative distance of FL1 (F = 1.13, P > 0.1) (Fig 4C). The different variability of FL1 and SSC suggested these two cytometric parameters would be related to different factors.
Fig 4.
Temporal changes of fluorescence intensity (FL1) (A), side scatter (SSC) (B) and variance between LNA and HNA of FL1 and SSC (C) in the Haihe River. The box represents the range from 25% to 75% percentiles, whisker lines represent the outlier percentiles and the middle line in the box shows the median value of all data points. X-marks and square dots represent the outlier and mean values, respectively. Letters with and without underline represent seasonal differences of HNA and LNA bacteria respectively. Different lowercase letters indicate significant difference of 0.05. The symbols "**" and "*" indicate significant differences of 0.01 and 0.05, respectively.
Partition of driving factors on LNA and HNA bacteria
Redundancy analysis (RDA) was conducted to estimate how environmental variables influenced LNA and HNA bacteria (Fig 5). The Monte Carlo test showed that RDA axes 1 and 2 were significant to elucidate the correlations between bacterial properties and environmental variables (F = 18.26, P = 0.004). RDA revealed that salinity was the main driving factor for LNA and HNA bacteria distribution patterns, accounting for 20% variance (F = 8.26, P < 0.01), followed by temperature (explain 11% variance, F = 5.51, P < 0.01), conductivity (explain 8% variance, F = 4.10, P < 0.05), total phosphorus (explain 6% variance, F = 3.64, P < 0.05) and total suspended solids (explain 6% variance, F = 3.27, P < 0.05) (Fig 5A). The salinity primarily controlled the variation of LNA and HNA in the Haihe River, which indicated the geographical gradient along freshwater river to oceans have considerable influence on the distribution of these two subgroups. While most samples were seasonally clustered together, samples from S8 and S9 were separated from the others in each season; station S8 is close to the estuary and S9 is located in the estuary of Bohai Sea (the salinity was higher than other stations). The changes in temperature reflect the seasonal dynamics and also remarkably affect the characteristics of LNA and HNA, which coincide with the significant temporal variability on the abundance and cytometric parameters (Figs 3 and 4). Overall, these two major factors shaped the variation of LNA and HNA (Fig 5B).
Fig 5. Redundancy analysis of LNA and HNA bacterial characteristic parameters with environmental factors in the Haihe River.
(A) Biplots between characteristics of LNA\HNA and environmental variables, (B) Samples ordination. Abbreviations: TSS, total suspended solids; TN, total nitrogen; NO3, nitrate; TP, total phosphorus; TDP, total dissolved phosphorus; TOC, total organic carbon; Chl-a, chlorophyll-α; FL1L and FL1H, FL1 of LNA and HNA bacteria, SSCL and SSCH, SSC of LNA and HNA bacteria; VFL1, FL1HNA/FL1LNA; VSSC, SSCHNA/SSCLNA; LNA and HNA, the LNA and HNA bacterial concentration, HNA%, the percentage of HNA in total bacterial concentration. Samples were connected with lines according to location along with the Haihe River in subgraph B.
It has been reported that HNA bacteria tend to grow in eutrophic and mesotrophic environments, while LNA bacteria reside in oligotrophic environments [29–31]. Our results showed that HNA and LNA bacterial abundance was significantly correlated to different environmental variables (Fig 5A). Specifically, LNA was negatively related to nitrate (NO3) and positively related to total suspended solids (TSS), while HNA was significantly related to total nitrogen (TN). A similar pattern between FL1 and SSC was observed, where the ordination of both cytometric parameters were divided in Fig 5A. The results revealed that the effects of geographical distance and temporal changes, which were characterized as salinity and temperature gradient respectively in RDA, were stronger than the effects of nutrient controls (e.g. TOC, TN and TP) on the LNA and HNA distribution (Fig 5A). The results are consistent with previous studies that seasonal changes in environmental variables have a more significant effect on microbial community patterns than trophic interactions [17,32]. Contemporary contingencies, e.g. local climate events, could drive the changes of biotic and abiotic factors on short time scales, then affects the microbial community dynamics [33–35].
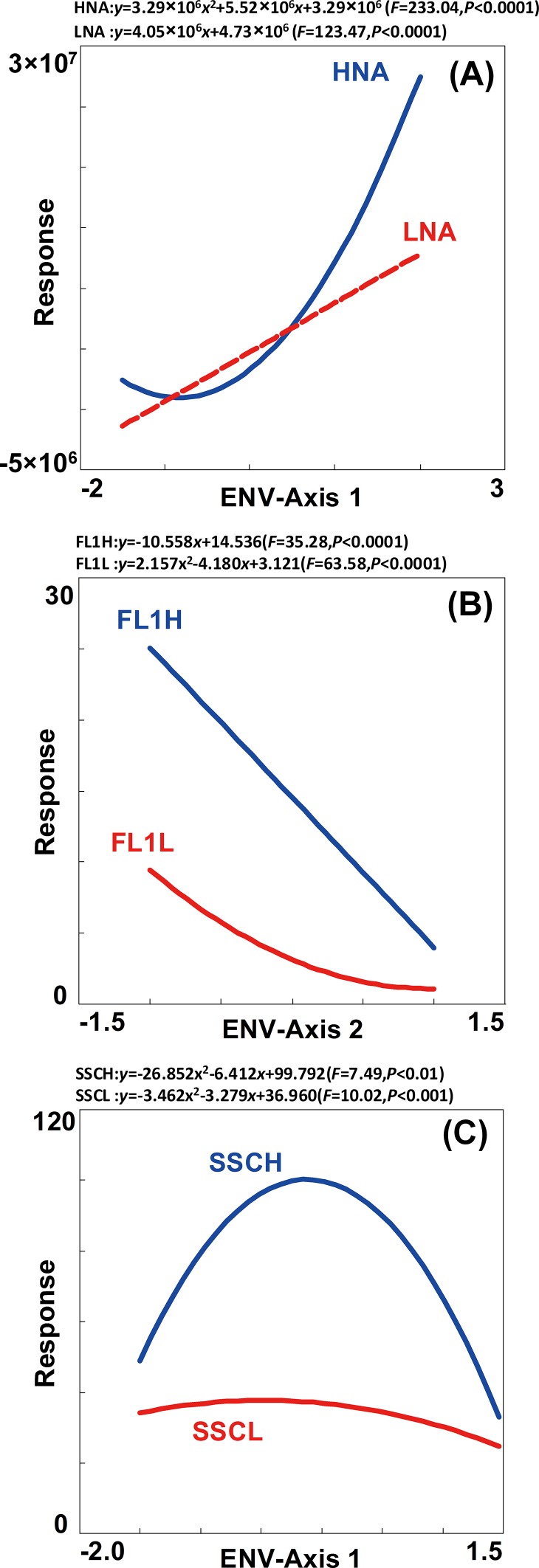
Furthermore, response analysis was performed to graphically compare the differentiation on the changes of LNA and HNA bacterial abundance and cytometric parameters (FL1 and SSC). The response of HNA showed more variation than that of LNA (Fig 6A), which indicated the changes in HNA abundance maybe more sensitive to environmental variation in comparison to LNA. Meanwhile, response analysis showed that the variations of FL1 and SSC significantly correlated to axis 2 (Fig 6B) and axis 1 (Fig 6C), respectively. Different variation was observed in SSC and FL1 within and between LNA and HNA. The results demonstrated that these two cytometric parameters represented dissimilar characteristics in bacterial cells and could be regulated differently [27]. The dissimilarity along gradients within an ecosystem suggested that LNA and HNA bacteria might play different ecological roles.
Fig 6.
Response of the abundance (A), FL1 (B) and SSC (C) of LNA and HNA bacteria to environmental ordination axis in RDA. GLM and GAM were selected to fit the response analysis in a stepwise manner, as well environmental explanatory ordination axis 1 and 2 in RDA analysis, where only explanatory axis be of significance was drawn in the analysis processes.
Conclusions
In summary, LNA and HNA bacteria make similar contributions to the total microbial abundance in an exorheic river on both spatial and temporal scale. The variability in HNA bacterial abundance and flow cytometric characteristics was greater than that of LNA bacteria. Meanwhile, the relative distance of SSC between HNA and LNA showed more variability than that of FL1. The present study demonstrated that the effects of geographical distance (salinity gradient along river to ocean) and temporal changes (temperature variation by seasonal succession) were stronger than the effects of nutrient conditions on the variations of LNA and HNA. Furthermore, the distribution of LNA and HNA bacteria, including the abundance and flow cytometric characteristics, were under the control of different environmental variables. The heterogeneity between LNA and HNA suggested those two subgroups may play different niche ecological roles in the microbial loop of aquatic ecosystems.
Data Availability
All relevant data are within the paper.
Funding Statement
The authors are grateful to the financial support from National Key Basic Research Program of China (2015CB459000) and National Natural Science Foundation, China (31322012). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wang YY, Hammes F, Boon N, Chami M, Egli T. Isolation and characterization of low nucleic acid (LNA)-content bacteria. ISME J. 2009;3(8):889–902. 10.1038/ismej.2009.46 . [DOI] [PubMed] [Google Scholar]
- 2.Vila-Costa M, Gasol JM, Sharma S, Moran MA. Community analysis of high- and low-nucleic acid-containing bacteria in NW Mediterranean coastal waters using 16S rDNA pyrosequencing. Environ Microbiol. 2012;14(6):1390–1402. 10.1111/j.1462-2920.2012.02720.x . [DOI] [PubMed] [Google Scholar]
- 3.Van Wambeke F, Catala P, Pujo-Pay M, Lebaron P. Vertical and longitudinal gradients in HNA-LNA cell abundances and cytometric characteristics in the Mediterranean Sea. Biogeosciences. 2011;8(7):1853–1863. 10.5194/bg-8-1853-2011 . [DOI] [Google Scholar]
- 4.Schattenhofer M, Wulf J, Kostadinov I, Gloeckner FO, Zubkov MV, Fuchs BM. Phylogenetic characterisation of picoplanktonic populations with high and low nucleic acid content in the North Atlantic Ocean. Syst Appl Microbiol. 2011;34(6):470–475. 10.1016/j.syapm.2011.01.008 . [DOI] [PubMed] [Google Scholar]
- 5.Santic D, Krstulovic N, Solic M, Kuspilic G. HNA and LNA bacteria in relation to the activity of heterotrophic bacteria. Acta Adriatica. 2012;53(1):25–40. . [Google Scholar]
- 6.Huete-Stauffer TM, Moran XAG. Dynamics of heterotrophic bacteria in temperate coastal waters: similar net growth but different controls in low and high nucleic acid cells. Aquat Microb Ecol. 2012;67(3):211–223. 10.3354/ame01590 . [DOI] [Google Scholar]
- 7.Lebaron P, Servais P, Baudoux AC, Bourrain M, Courties C, Parthuisot N. Variations of bacterial-specific activity with cell size and nucleic acid content assessed by flow cytometry. Aquat Microb Ecol. 2002;28(2):131–140. 10.3354/ame028131 . [DOI] [Google Scholar]
- 8.Lebaron P, Servais P, Agogué H, Courties C, Joux F. Does the high nucleic acid content of individual bacterial cells allow us to discriminate between active cells and inactive cells in aquatic systems? Appl Environ Microbiol. 2001;67(4):1775–1782. 10.1128/AEM.67.4.1775-1782.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Longnecker K, Sherr BF, Sherr EB. Activity and phylogenetic diversity of bacterial cells with high and low nucleic acid content and electron transport system activity in an upwelling ecosystem. Appl Environ Microbiol. 2005;71(12):7737–7749. 10.1128/aem.71.12.7737-7749.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salcher MM, Pernthaler J, Posch T. Seasonal bloom dynamics and ecophysiology of the freshwater sister clade of SAR11 bacteria 'that rule the waves' (LD12). ISME J. 2011;5(8):1242–1252. 10.1038/ismej.2011.8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramseier MK, von Gunten U, Freihofer P, Hammes F. Kinetics of membrane damage to high (HNA) and low (LNA) nucleic acid bacterial clusters in drinking water by ozone, chlorine, chlorine dioxide, monochloramine, ferrate(VI), and permanganate. Water Res. 2011;45(3):1490–1500. 10.1016/j.watres.2010.11.016 . [DOI] [PubMed] [Google Scholar]
- 12.Roesel S, Grossart H- P. Contrasting dynamics in activity and community composition of free-living and particle-associated bacteria in spring. Aquat Microb Ecol. 2012;66(2):169–181. 10.3354/ame01568 . [DOI] [Google Scholar]
- 13.Gozdereliler E, Boon N, Aamand J, De Roy K, Granitsiotis MS, Albrechtsen H- J, et al. Comparing metabolic functionalities, community structures, and dynamics of herbicide-degrading communities cultivated with different substrate concentrations. Appl Environ Microbiol. 2013;79(1):367–75. 10.1128/aem.02536-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams TJ, Ertan H, Ting L, Cavicchioli R. Carbon and nitrogen substrate utilization in the marine bacterium Sphingopyxis alaskensis strain RB2256. ISME J. 2009;3(9):1036–1052. 10.1038/ismej.2009.52 . [DOI] [PubMed] [Google Scholar]
- 15.Bouvier T, Maurice CF. A single-cell analysis of virioplankton adsorption, infection, and intracellular abundance in different bacterioplankton physiologic categories. Microb Ecol. 2011;62(3):669–678. 10.1007/s00248-011-9862-3 . [DOI] [PubMed] [Google Scholar]
- 16.Palesse S, Colombet J, Ram ASP, Sime-Ngando T. Linking host prokaryotic physiology to viral lifestyle dynamics in a temperate freshwater lake (Lake Pavin, France). Microb Ecol. 2014;68(4):740–750. 10.1007/s00248-014-0441-2. . [DOI] [PubMed] [Google Scholar]
- 17.Read DS, Gweon HS, Bowes MJ, Newbold LK, Field D, Bailey MJ, et al. Catchment-scale biogeography of riverine bacterioplankton. ISME J. 2015;9(2):516–526. 10.1038/ismej.2014.166 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santic D, Krstulovic N, Solic M, Ordulj M, Kuspilic G. Dynamics of prokaryotic picoplankton community in the central and southern Adriatic Sea (Croatia). Helgol Mar Res. 2013;67(3):471–481. 10.1007/s10152-012-0336-x . [DOI] [Google Scholar]
- 19.Santic D, Sestanovic S, Solic M, Krstulovic N, Kuspilic G, Ordulj M, et al. Dynamics of the picoplankton community from coastal waters to the open sea in the Central Adriatic. Mediterranean Marine Science. 2014;15(1):179–188. . [Google Scholar]
- 20.Nishimura Y, Kim C, Nagata T. Vertical and seasonal variations of bacterioplankton subgroups with different nucleic acid contents: possible regulation by phosphorus. Appl Environ Microbiol. 2005;71(10):5828–5836. 10.1128/aem.71.10.5828-5836.2005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomes A, Gasol JM, Estrada M, Franco-Vidal L, Diaz-Perez L, Ferrera I, et al. Heterotrophic bacterial responses to the winter-spring phytoplankton bloom in open waters of the NW Mediterranean. Deep-Sea Res Part I-Oceanogr Res Pap. 2015;96:59–68. 10.1016/j.dsr.2014.11.007 . [DOI] [Google Scholar]
- 22.Ma L, Sun R, Mao G, Yu H, Wang Y. Seasonal and spatial variability of virioplanktonic abundance in Haihe River, China. Biomed Research International. 2013;2013:526362 10.1155/2013/526362 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma L, Mao G, LIU J, Gao G, Zou C, Bartlam MG, et al. Spatial-temporal changes of bacterioplankton community along an exhorheic river. Front Microbiol. 2016;7:250 10.3389/fmicb.2016.00250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramette A. Multivariate analyses in microbial ecology. FEMS Microbiol Ecol. 2007;62(2):142–160. 10.1111/j.1574-6941.2007.00375.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J, Wang J, Gao G, Bartlam MG, Wang Y. Distribution and diversity of fungi in freshwater sediments on a river catchment scale. Front Microbiol. 2015;6:329 10.3389/fmicb.2015.00329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calvo-Diaz A, Moran XAG. Seasonal dynamics of picoplankton in shelf waters of the southern Bay of Biscay. Aquat Microb Ecol. 2006;42(2):159–174. 10.3354/ame042159 . [DOI] [Google Scholar]
- 27.Bouvier T, del Giorgio PA, Gasol JM. A comparative study of the cytometric characteristics of High and Low nucleic-acid bacterioplankton cells from different aquatic ecosystems. Environ Microbiol. 2007;9(8):2050–2066. 10.1111/j.1462-2920.2007.01321.x . [DOI] [PubMed] [Google Scholar]
- 28.Worden AZ, Nolan JK, Palenik B. Assessing the dynamics and ecology of marine picophytoplankton: The importance of the eukaryotic component. Limnol Oceanogr. 2004;49(1):168–179. . [Google Scholar]
- 29.Jochem FJ, Lavrentyev PJ, First MR. Growth and grazing rates of bacteria groups with different apparent DNA content in the Gulf of Mexico. Marine Biology. 2004;145(6):1213–1225. 10.1007/s00227-004-1406-7 . [DOI] [Google Scholar]
- 30.Solic M, Krstulovic N, Vilibic I, Bojanic N, Kuspilic G, Sestanovic S, et al. Variability in the bottom-up and top-down controls of bacteria on trophic and temporal scales in the middle Adriatic Sea. Aquat Microb Ecol. 2009;58(1):15–29. 10.3354/ame01342 [DOI] [Google Scholar]
- 31.Andrade L, Gonzalez AM, Rezende CE, Suzuki M, Valentin JL, Paranhos R. Distribution of HNA and LNA bacterial groups in the Southwest Atlantic Ocean. Braz J Microbiol. 2007;38(2):330–336. 10.1590/s1517-83822007000200028 . [DOI] [Google Scholar]
- 32.Gilbert JA, Steele JA, Caporaso JG, Steinbrueck L, Reeder J, Temperton B, et al. Defining seasonal marine microbial community dynamics. ISME J. 2012;6(2):298–308. 10.1038/ismej.2011.107 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carlson CA, Morris R, Parsons R, Treusch AH, Giovannoni SJ, Vergin K. Seasonal dynamics of SAR11 populations in the euphotic and mesopelagic zones of the northwestern Sargasso Sea. ISME J. 2009;3(3):283–295. 10.1038/ismej.2008.117 . [DOI] [PubMed] [Google Scholar]
- 34.Eiler A, Hayakawa DH, Church MJ, Karl DM, Rappe MS. Dynamics of the SAR11 bacterioplankton lineage in relation to environmental conditions in the oligotrophic North Pacific subtropical gyre. Environ Microbiol. 2009;11(9):2291–2300. 10.1111/j.1462-2920.2009.01954.x . [DOI] [PubMed] [Google Scholar]
- 35.Salter I, Galand PE, Fagervold SK, Lebaron P, Obernosterer I, Oliver MJ, et al. Seasonal dynamics of active SAR11 ecotypes in the oligotrophic Northwest Mediterranean Sea. ISME J. 2015;9(2):347–360. 10.1038/ismej.2014.129 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.