Abstract
Most agricultural traits are controlled by quantitative trait loci (QTLs); however, there are few studies on QTL mapping of horticultural traits in pepper (Capsicum spp.) due to the lack of high-density molecular maps and the sequence information. In this study, an ultra-high-density map and 120 recombinant inbred lines (RILs) derived from a cross between C. annuum ‘Perennial’ and C. annuum ‘Dempsey’ were used for QTL mapping of horticultural traits. Parental lines and RILs were resequenced at 18× and 1× coverage, respectively. Using a sliding window approach, an ultra-high-density bin map containing 2,578 bins was constructed. The total map length of the map was 1,372 cM, and the average interval between bins was 0.53 cM. A total of 86 significant QTLs controlling 17 horticultural traits were detected. Among these, 32 QTLs controlling 13 traits were major QTLs. Our research shows that the construction of bin maps using low-coverage sequence is a powerful method for QTL mapping, and that the short intervals between bins are helpful for fine-mapping of QTLs. Furthermore, bin maps can be used to improve the quality of reference genomes by elucidating the genetic order of unordered regions and anchoring unassigned scaffolds to linkage groups.
Keywords: pepper, NGS, bin map, QTL, morphological trait
1. Introduction
Pepper (Capsicum spp.) is one of the first domesticated vegetables1 and shows enormous variation in plant architecture, flower-, leaf-, and fruit-related traits.2–4 Most yield and fruit quality traits important in pepper breeding are regulated by multiple genes or so-called quantitative trait loci (QTLs). QTL analyses of morphological traits in pepper have been performed using intraspecific and interspecific populations.5–8 Most QTL analyses for morphological traits have concentrated on fruit-associated traits. For instance, fs3.1 and fs10.1 control fruit shape (ratio of fruit length to fruit width).5,9 Major QTLs controlling fruit weight are present in chromosomes 2 and 4, and in comparison with the tomato genetic map, some of these QTLs were found to be conserved between the two crops.7,8 QTL analyses for plant height, leaf area, stem length, and other traits have also been performed,5,7,10 but no major QTLs were detected and the genes controlling QTLs have not been characterized.
For QTL mapping, restriction fragment length polymorphism, amplified fragment length polymorphism, random amplified polymorphic DNA, and microsatellite markers have been widely used.5,7,11,12 However, for accurate detection and characterization of QTLs, high-density genetic maps are required. Recently, a pepper GeneChip containing 30,815 unigenes derived from EST sequences was used for genotyping a recombinant inbred line (RIL) population.10,13 Using a high-density EST-based genetic map, 96 QTLs for 38 traits were mapped.10 However, the genes controlling the QTL regions were not discovered in this analysis.
Next-generation sequencing (NGS) methods are commonly used for whole-genome sequencing.14,15 Once a reference genome is available, SNP markers can be easily developed by aligning resequencing data to the reference genome. Although the time and cost involved in sequencing has been greatly reduced, resequencing large populations or organisms with a large genome remains challenging. Alternatively, low-coverage sequencing is a cost-efficient way to genotype for QTL analyses or association studies using a large number of samples.14 However, due to the low coverage of sequencing reads, this approach misses a large number of genotypes, and new computational tools have been needed to address the resulting reduction in mapping accuracy. Imputation of missing data using a reference genome can increase the power of SNP calling in genetic studies.15,16 Generally, imputation tools predict the genotype of missing data based on the haplotype ratio at that site. This method can increase the accuracy of mapping, and enables usage of most of the sequencing data. An alternative method is the sliding window approach, which has been used in cereal crops such as rice, sorghum, and maize to construct bin maps and for QTL analysis.17–20 Instead of using all SNPs as independent markers, the sliding window approach merges consecutive SNPs into one bin, which can greatly reduce errors caused by inaccurate SNP calling.
The first assembled reference genomes of cultivated hot pepper (Capsicum annuum) and wild pepper (C. chinense and C. annuum var. glabriusculum) were released in 2014.21,22 The availability of reference genome information facilitates the identification of horticulturally important genes, the development of useful markers, and improvement of cultivars. In this study, we identified QTLs controlling morphological traits using RILs derived from a cross between C. annuum ‘Perennial’ and C. annuum ‘Dempsey’. To increase the efficiency of QTL analysis, we improved the ultra-high-density genetic map for the reference genome using a sliding window approach. Horticultural traits were evaluated in four environments to detect major QTLs, and the ultra-high-density bin map increased the speed of QTL mapping. Furthermore, we improved the reference genome of pepper by correcting the genetic order of pseudomolecules and anchoring unassigned scaffolds into linkage groups.
2. Materials and methods
2.1. Plant materials
A total of 120 F7–F10 RILs obtained from a cross between pungent Capsicum annuum ‘Perennial’ and non-pungent C. annuum ‘Dempsey’ were used. RILs were grown for 3 years (2011, 2012, and 2014) to reduce environmental effects in the detection of QTLs. Plants were grown under greenhouse conditions in two locations, Anseong (2011 and 2012a) and Suwon, Korea (2012b and 2014). In 2011 and 2012a, plants were grown in the ground, whereas plants were planted in pots in 2012b and 2014. Seeds were disinfected using 2% sodium chlorate and 10% trisodium phosphate. Five plants per line were planted and fruits were harvested separately.
2.2. Phenotype evaluation
A total of 18 horticultural traits were evaluated for ‘Perennial’, ‘Dempsey’, and 120 RILs based on the definitions of RDA-Genebank, Korea. Stem colour, flower size, fruit position, calyx shape, and immature fruit colour were graded according to the definitions for efficient evaluations (Table 1). The other 14 traits were evaluated in three plants per line using the relevant units and average values were calculated. Traits related to plant architecture, leaf, and flower were evaluated 9 weeks after transplanting in 2011, and 10 weeks after transplanting in 2012a, 2012b, and 2014. Fruit-related traits were evaluated after harvest. For precise measurement of the fruit length and width, the image of hemisected fruits was scanned (Epson V30) and analysed using Tomato Analyser 3.0.23 Pearson correlation coefficients were calculated to analyse the relationship between traits in each environment.
Table 1.
Morphological traits evaluated in RILs
| Phenotype | Description |
|---|---|
| Plant architecture | |
| Plant height (cm) | From soil to head of the plant |
| Plant width (cm) | Wide part of the plant |
| Main stem length (cm) | From soil to the first branch |
| Stem thickness (cm) | Thickness of basal stem |
| Lateral branch number | Basal lateral branch number before the first branch |
| Internode length (cm) | Length of internode between the third and fourth node |
| Stem colour | 1: Green, 2: Green with purple, 3: Purple |
| Leaf | |
| Leaf length (cm) | Length of completely grown leaf |
| Leaf width (cm) | Width of completely grown leaf |
| Flower | |
| Flower size | 1: Small, 2: Intermediate, 3: Big |
| Stamen number | Most frequent number of stamen |
| Fruit | |
| Fruit length (cm) | Average length of fruit |
| Fruit diameter (cm) | Average width of fruit |
| Fruit shape | Ratio between fruit length and fruit diameter |
| Fruit weight (g) | Average weight of fresh fruit |
| Fruit position | 1: Erect, 2: Intermediate, 3: Pendant |
| Calyx shape | 1: Cup-shaped, 2: Intermediate, 3: Saucer-shaped |
| Immature fruit colour | 1: Light green, 2: Green, 3: Dark-green |
2.3. Genetic mapping and bin map construction
Sequencing of ‘Perennial’, ‘Dempsey’, and 120 RILs was as described by Kim et al.24 Parental lines were sequenced to 18× coverage and RILs were sequenced to 1× coverage. All sequence data are available under the Sequence Read Archive of NCBI (PRJNA298503). To redo SNP calling, sequencing reads were aligned to C. annuum ‘CM334’ chromosome v.1.55 using BWA version 0.7.12 with default parameters, and SNPs were called using GATK UnifiedGenotyper. High-quality SNPs with minimum sequencing depth 3 for each RIL, quality score 30, and called in at least 12 RILs were selected for further analysis.
A slightly modified sliding window approach17 was used to investigate recombination breakpoints and construct a bin map of RILs. The genotype of each window was called with a window size of 5 Mb and step size of 0.5 Mb. The ratio of SNPs with ‘Perennial’ and ‘Dempsey’ genotypes was calculated. When >70% of SNPs had one parental genotype, the window was called as homozygous; otherwise, the window was called as heterozygous. Recombination breakpoints of each RIL were determined by the physical locations where the genotype of window changes. On the basis of the recombination breakpoint position, a physical bin map was constructed as described by Huang et al.17 Briefly, when 120 RILs did not have the recombination breakpoints in 100 kb interval, these regions were combined to one bin. While there are more than two recombination breakpoints in 100 kb interval, they were determined as one breakpoint. To prevent detection of excessive crossovers, further processing was performed. When short heterozygous regions were located in the middle of consecutive ‘Perennial’ or ‘Dempsey’ genotype regions in the raw bin map and these heterozygous regions were shorter than window size, the genotype was manually corrected to match with the adjacent genotype. Bins were used as markers to construct an ultra-high-density genetic map using the Carthagene program21 with default threshold. The Kosambi mapping function was used to calculate the distance (cM) of each bin. Twelve linkage groups were drawn with the MapChart 2.2 program.22 The physical map and genetic map were compared using Marker Browser developed by the Phyzen Genomics Institute (Seoul, Korea).
2.4. Anchoring of unassigned scaffolds
Unmapped reads of ‘Perennial’, ‘Dempsey’, and 120 RILs were aligned to ‘CM334’ unassigned scaffolds v.1.55 (http://peppergenome.snu.ac.kr/, accessed 7 December 2015). The alignment and SNP calling pipeline was the same as that used for bin map construction. To assign the unassigned scaffolds, each scaffold was considered as one bin marker. The genotype of the scaffold in each RIL was determined based on the ratio between the SNPs and the ‘Perennial’ and ‘Dempsey’ genotypes. Genotyped scaffolds were mapped on the bin map using the Carthagene program21 with an logarithm of odds (LOD) threshold of 5 and a distance threshold of 20 cM.
2.5. QTL analysis
All phenotyping data were used for QTL analysis. Composite interval mapping was performed with Windows QTL Cartographer 2.5.25 Values for each trait in three to four locations were analysed separately to detect QTLs. The LOD threshold was determined by applying 500 permutation tests with 5% probability for each trait. The proportion of phenotypic variation explained by each QTL was estimated using R2 (%) value. The 95% confidence interval was used as the location of QTL, and QTLs detected in more than two different environments were considered as significant QTLs.
3. Results
3.1. Variation of morphological traits in RILs
The maternal parent ‘Perennial’ produces tall plants, with thin and purple-striped stems, and small leaves and flowers. ‘Perennial’ fruits are small, erect, and pungent. The paternal parent ‘Dempsey’ has relatively short plants, thick and green stems, and large leaves and flowers. ‘Dempsey’ produces large bell-type fruits that are pendent and non-pungent. We evaluated a total of 18 horticultural traits at least three different environments (Table 1), and all traits segregated in RILs generated from these two parents (Table 2 and Supplementary Fig. S1). Plant height, plant width, and internode length were greatly affected by environmental conditions. The plant height of both parents and RILs was highest in environment 2012a, and lowest in environment 2011. Plant width and internode length of ‘Dempsey’ and RILs were widest and longest in environment 2012a. Immature fruit colour was segregated only in RILs grown in 2012b and 2014. Both parents had green immature fruit, whereas the RILs showed 1 : 1 segregation of light green and green colour.
Table 2.
Phenotypic variation of ‘Perennial’, ‘Dempsey’, and RILs grown in four different environments
| Trait | 2011 |
2012a |
2012b |
2014 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | D | RIL | P | D | RIL | P | D | RIL | P | D | RIL | |
| Plant architecture | ||||||||||||
| Plant height | 132 | 53.5 | 139 ± 24.9a | 208 ± 2.5 | 128 ± 6.0 | 165 ± 26.6 | 157 ± 5.1 | 89 ± 1.2 | 130 ± 40.5 | 161 ± 3.3 | 60 ± 1.2 | 126 ± 26.9 |
| Plant width | 46 | 30 | 81 ± 12.4 | 64 ± 6.0 | 86 ± 2.5 | 86 ± 17.8 | 68 ± 5.0 | 63 ± 1.2 | 72 ± 9.8 | 68 ± 5.2 | 57 ± 6.2 | 65 ± 9.8 |
| Main stem length | –b | – | – | 41 ± 4.5 | 20 ± 1.0 | 25 ± 7.8 | 33 ± 1.2 | 21 ± 2.1 | 23 ± 6.8 | 38 ± 4.4 | 23 ± 0.6 | 25 ± 7.0 |
| Stem thickness | – | – | – | 2 ± 0.2 | 3 ± 0.4 | 2 ± 0.3 | 1 ± 0.0 | 2 ± 0.0 | 1 ± 0.2 | 1 ± 0.0 | 1 ± 0.0 | 1 ± 0.1 |
| Lateral branch number | – | – | – | 22 ± 1.5 | 12 ± 1.0 | 13 ± 2.3 | 20 ± 0.8 | 7 ± 0.5 | 13 ± 2.9 | 19 ± 2.1 | 14 ± 0.8 | 13 ± 3.0 |
| Internode length | – | – | – | 9 ± 2.3 | 10 ± 2.3 | 9 ± 2.4 | 16 ± 0.5 | 4 ± 1.1 | 6 ± 2.4 | 10 ± 2.1 | 6 ± 0.5 | 8 ± 2.3 |
| Stem colour | 2 | 1 | 1 ± 0.4 | – | – | – | 2 | 1 | 2 ± 0.8 | 2 | 1 | 2 ± 0.8 |
| Leaf | ||||||||||||
| Leaf length | 12.3 | 12.8 | 8 ± 1.6 | 12 ± 0.5 | 18 ± 1.0 | 11 ± 1.2 | 8 ± 0.1 | 14 ± 0.8 | 11 ± 1.5 | 12 ± 0.2 | 16 ± 0.6 | 13 ± 1.6 |
| Leaf width | 4.7 | 8.4 | 4 ± 1.0 | 7 ± 0.3 | 10 ± 0.0 | 7 ± 0.9 | 5 ± 0.3 | 8 ± 0.4 | 6 ± 1.0 | 6 ± 0.1 | 9 ± 0.7 | 7 ± 1.0 |
| Flower | ||||||||||||
| Flower size | 2 | 3 | 2 ± 0.4 | – | – | – | 1 | 3 | 2 ± 0.5 | 1 | 3 | 2 ± 0.5 |
| Stamen number | 6 | 6 | 5 ± 0.5 | – | – | – | 5 | 6 | 5 ± 0.5 | 5 | 6 | 5 ± 0.5 |
| Fruit | ||||||||||||
| Fruit length | 1.8 | 5.8 | 7 ± 1.6 | – | – | 6 ± 1.9 | 3 ± 0.3 | 8 ± 0.3 | 6 ± 1.7 | 3 ± 0.1 | 8 ± 0.4 | 6 ± 1.8 |
| Fruit diameter | 1.8 | 5.3 | 2 ± 0.8 | – | – | 2 ± 0.8 | 1 ± 0.0 | 8 ± 0.1 | 2 ± 0.6 | 1 ± 0.0 | 8 ± 0.1 | 2 ± 0.6 |
| Fruit shape | 1 | 1.1 | 3 ± 1.2 | – | – | 3 ± 1.4 | 4 | 1 | 3 ± 1.5 | 4 | 1 | 3 ± 1.4 |
| Fruit weight | 1.4 | 37.5 | 11 ± 7.2 | 1 | 90 | 10 ± 6.4 | 1 ± 0.0 | 115 ± 23.2 | 7 ± 4.0 | 1 ± 0.0 | 116 ± 9.6 | 6 ± 3.5 |
| Fruit position | 1 | 3 | 2 ± 0.9 | 1 | 3 | 2 ± 0.9 | 1 | 3 | 2 ± 0.9 | 1 | 3 | 2 ± 0.9 |
| Calyx shape | 1 | 3 | 2 ± 0.6 | 1 | 3 | 2 ± 0.6 | 1 | 3 | 2 ± 0.8 | 1 | 3 | 2 ± 0.8 |
| Immature fruit colour | 2 | 2 | 2 ± 0.0 | 2 | 2 | 2 ± 0.0 | 2 | 2 | 1 ± 0.5 | 2 | 2 | 1 ± 0.5 |
P, Perennial; D, Dempsey.
aMean ± standard deviation.
bPhenotypes were not evaluated.
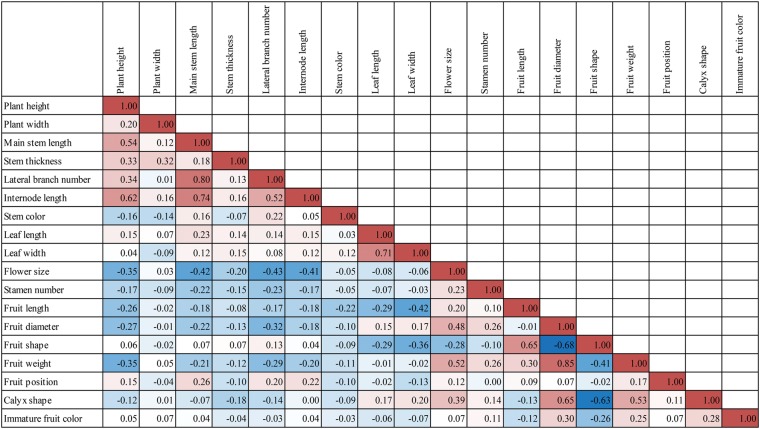
Positive and negative correlations were detected between evaluated traits (Fig. 1). In particular, fruit-related traits were highly correlated with each other compared with other traits. Fruit weight, fruit diameter, and calyx shape were positively correlated. As the fruit shape can be represented by fruit length divided by fruit diameter, it has positive and negative correlation with fruit length and fruit diameter, respectively. Flower size was positively correlated with fruit diameter and fruit weight, and negatively with main stem length. Phenotypic correlation in the parents and RILs indicated that one gene or closely linked genes control multiple fruit-related traits.
Figure 1.
Correlation between morphological traits evaluated in the RIL population. Average phenotypic values from four environments were used for Pearson correlation test. Red and blue blocks show positive and negative correlation, respectively. This figure is available in black and white in print and in colour at DNA Research online.
3.2. Construction of bin map using low-coverage sequence
‘Perennial’ and ‘Dempsey’ were sequenced with 18× coverage, whereas RILs were sequenced with 1× coverage. RILs had large amounts of missing data in SNP regions and the reliability of individual SNPs was very low due to the low coverage. Therefore, to construct an accurate ultra-high-density genetic map efficiently, we implemented a modified sliding window approach.17 Consecutive SNPs were joined in one bin, and the genotype of each bin was determined based on the ratio between SNPs from the two parents. Bins instead of individual SNPs were used as markers to increase mapping efficiency. A total of 1,431,214 SNPs were detected between ‘Perennial’ and ‘Dempsey’ and used to construct a bin map (Table 3 and Supplementary Table S1).
Table 3.
Comparison of physical length and genetic distance in the bin map
| Chr. | Number of SNPs | Number of bins | Physical length of bin (Mb) |
Genetic distance of bin (cM) |
||
|---|---|---|---|---|---|---|
| Mean | Total | Mean | Total | |||
| 1 | 82,966 | 370 | 0.74 | 272.6 | 0.56 | 208.5 |
| 2 | 80,141 | 195 | 0.88 | 171.1 | 0.55 | 107.5 |
| 3 | 87,793 | 261 | 0.99 | 257.9 | 0.45 | 118.5 |
| 4 | 54,657 | 216 | 1.03 | 222.5 | 0.54 | 116.5 |
| 5 | 82,413 | 190 | 1.23 | 233.4 | 0.53 | 100.6 |
| 6 | 107,015 | 220 | 1.08 | 236.9 | 0.47 | 102.6 |
| 7 | 84,339 | 175 | 1.33 | 231.9 | 0.53 | 92.5 |
| 8 | 24,383 | 217 | 0.67 | 144.8 | 0.71 | 153.7 |
| 9 | 275,842 | 161 | 1.57 | 252.7 | 0.54 | 86.2 |
| 10 | 230,360 | 154 | 1.52 | 233.6 | 0.67 | 103.9 |
| 11 | 252,765 | 196 | 1.33 | 259.7 | 0.44 | 86.8 |
| 12 | 68,540 | 223 | 1.06 | 235.7 | 0.43 | 94.9 |
| Total | 1,431,214 | 2,578 | 1.07 | 2,752.8 | 0.53 | 1,372.2 |
To set appropriate parameters for the sliding window approach, window length from 1 to 6 Mb and step size from 0.2 to 0.5 Mb for all chromosomes were tested. Window length and step sizes of 5 and 0.5 Mb, and 4 and 0.4 Mb showed a reasonable number of recombination breakpoints per chromosome, whereas the other values showed too large or low number (data not shown). A window length of 5 Mb and a step size of 0.5 Mb gave the best match of the recombination breakpoints determined manually in five randomly selected RILs. Therefore, this parameter was used to construct a bin map.
Using a sliding window approach, 3,983 recombination breakpoints were identified from 120 RILs, with an average of 33.2 per RIL (Supplementary Fig. S2A). For the RILs, the mean number of crossovers per chromosome per line was 2.8. All SNPs were grouped into 2,578 bins (Supplementary Table S2 and Fig. S3). The average length of bins was 1.07 Mb and ranged from 100 kb to 141 Mb (Table 3). The total genetic distance of the bin map was 1,372.2 cM, and the mean distance of the bins was 0.53 cM. On average, 564 SNPs were joined into each bin, and 54% of the bins contained <100 SNPs (Supplementary Fig. S2B). The proportion of heterozygous bins in the bin map was calculated to measure the heterozygosity of RILs (Supplementary Table S3). The average heterozygosity of the 120 RILs was 2.6%, whereas three RILs exhibited 31.8, 23.0, and 17.7% heterozygosity. Excluding these three RILs, the average heterozygosity of RILs in different generations was near 2.0%.
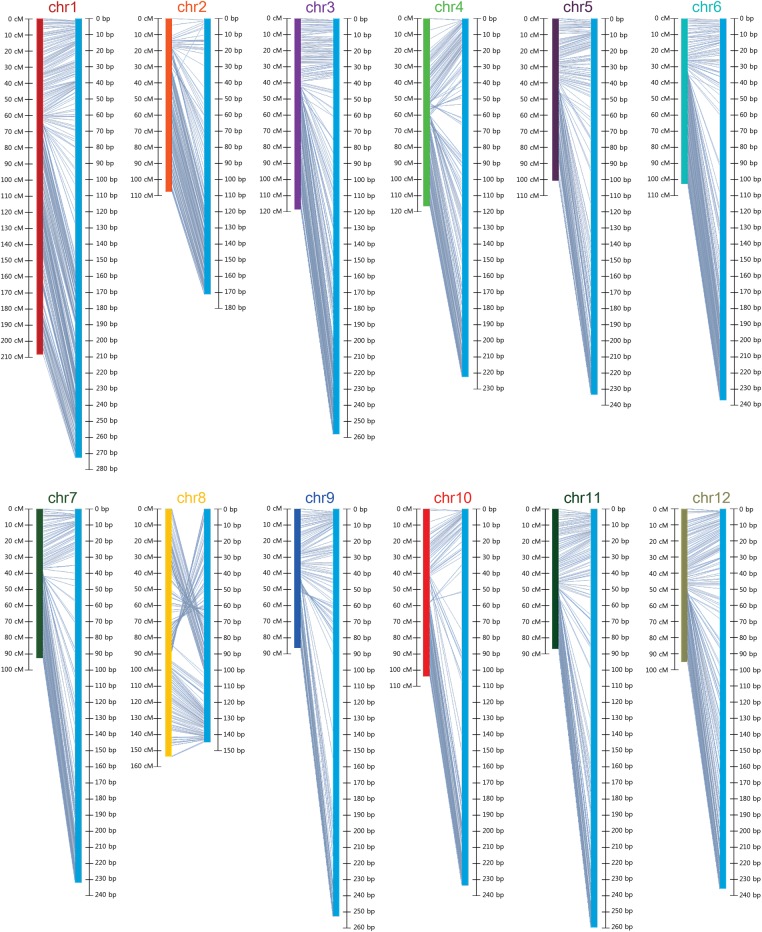
The locations of bins in the genetic map were compared with those of the physical map based on the ‘CM334’ reference genome (Fig. 2). Inconsistencies of bin order were detected in all chromosomes and most of the inconsistencies were located near the putative centromeric regions. For example, bins located 20–40 and 60–70 Mb on chromosome 4 were inverted in location on the genetic map. Similar patterns were detected at 95–120 Mb on chromosome 3, 80–100 Mb on chromosome 7, 40–50 Mb on chromosome 9, and 110–130 Mb on chromosome 11. Extensive inconsistencies were detected on the upper region of chromosome 8.
Figure 2.
Comparison of physical map and genetic map constructed with bins. Left and right map shows genetic and physical map, respectively. Markers are linked by grey lines between two maps. This figure is available in black and white in print and in colour at DNA Research online.
To confirm that the bin map was constructed properly, we mapped the Pun1 gene to the bin map and compared the results with the physical location of the Pun1 gene in ‘CM334’ reference genome. Pun1 is the gene controlling pungency, and the recessive allele pun1 confers non-pungency of pepper.26 Owing to the pungent parent ‘Perennial’ and non-pungent parent ‘Dempsey’, pungency in the RILs segregated to a 1 : 1 ratio (data not shown). The Pun1 gene was mapped between PD2-bin139 and PD2-bin141 on chromosome 2 of the bin map. The physical location of PD2-bin139 and PD2-bin141 was 149.8 and 151.0 Mb, respectively. On the ‘CM334’ reference genome, the Pun1 gene is located at 152.6 Mb, 1.6 Mb from the linked bin. Thus, although there was a slight difference from the physical location, the Pun1 gene was mapped quite precisely.
3.3. QTL analysis
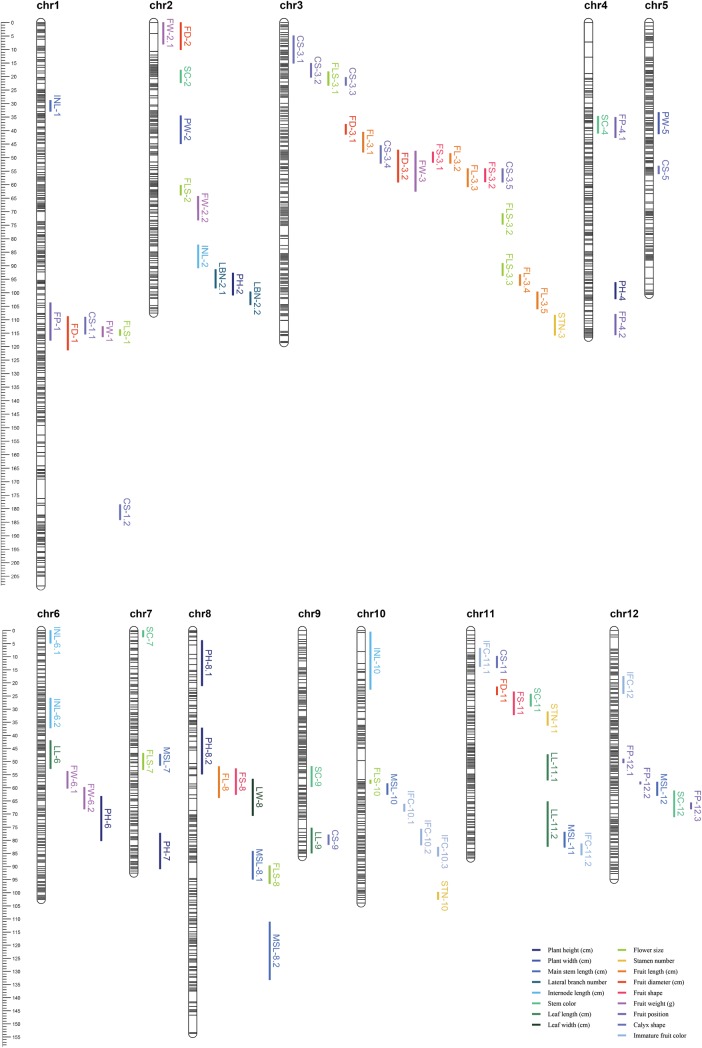
In four environments, a total of 434 QTLs were detected, and those QTLs detected in more than two environments were selected as significant. QTLs controlling the same trait and commonly detected in different environments with overlapped 95% confidence intervals were consolidated to one QTL. A total of 86 significant QTLs controlling 17 traits were selected (Table 4 and Fig. 3). Consolidated QTLs were named based on abbreviation of the trait name and the chromosome number. For each morphological trait, 2–10 QTLs were detected and these QTLs were distributed throughout the 12 pepper chromosomes. The phenotypic variation explained by each QTL ranged from 5.6 to 63.9%. Using a threshold R2 of 10%, 32 major QTLs were identified from 13 traits. LBN-2.1 and LBN-2.2, for lateral branch number, were each detected in three environments, and their R2 value ranges were 16–40 and 10–23, respectively. These large variations in R2 value indicate that lateral branch number is highly affected by the environment. The INL-2, for internode length, and FL-3.4, for fruit length, were also each detected in three environments, but their variations of R2 were smaller compared with those of LBN-2.1 and LBN-2.2. FS-3.1, for fruit shape, FP-1, and FP-12.2, for fruit position, were identified in all environments. In particular, FP-12.2 could explain >40% of phenotypic variation, which suggests the possible existence of a major gene controlling fruit position.
Table 4.
QTLs controlling morphological traits detected in more than two environments
| Trait | Year | QTL | Chr. | Location (cM) | LOD | R2 (%) | Direction | Additive effect |
|---|---|---|---|---|---|---|---|---|
| Plant height | 2012a, 2014 | PH-2 | 2 | 92.7–100.9 | 3.7–4.0 | 11.1–11.8 | + | 9.2–10.0 |
| 2011, 2014 | PH-4 | 4 | 96.2–102.3 | 2.3–2.6 | 7.8–10.0 | − | 8.3–10.4 | |
| 2011, 2012a, 2012b, 2014 | PH-6 | 6 | 63.2–80.2 | 2.1–3.4 | 6.4–11.6 | + | 7.8–12.8 | |
| 2011, 2012b, 2014 | PH-7 | 7 | 77.3–90.9 | 2.6–3.4 | 8.3–11.2 | + | 9.5–13.8 | |
| 2012b, 2014 | PH-8.1 | 8 | 3.8–21.1 | 2.6–2.8 | 5.6–8.3 | + | 9.3–11.7 | |
| 2011, 2012b, 2014 | PH-8.2 | 8 | 37.2–54.8 | 2.4–2.9 | 5.9–8.6 | − | 10.7–13.8 | |
| Plant width | 2012b, 2014 | PW-2 | 2 | 34.5–44.9 | 2.2–2.4 | 7.1–8.0 | − | 3.0–4.4 |
| 2012b, 2014 | PW-5 | 5 | 33.3–41.2 | 2.4–3.6 | 8.6–12.5 | + | 3.5–6.0 | |
| Main stem length | 2012b, 2014 | MSL-7 | 7 | 47.2–51.5 | 2.4–5.2 | 8.8–18.6 | + | 2.6–4.0 |
| 2012b, 2014 | MSL-8.1 | 8 | 84.2–94.9 | 4.6–6.3 | 14.0–18.3 | + | 3.3–4.5 | |
| 2012a, 2014 | MSL-8.2 | 8 | 111.1–133.2 | 2.2–4.3 | 7.4–13.4 | − | 2.6–3.0 | |
| 2012b, 2014 | MSL-10 | 10 | 58.4–62.6 | 2.6–3.3 | 8.7–12.3 | − | 5.9–7.5 | |
| 2012a, 2014 | MSL-11 | 11 | 76.9–82.7 | 2.6–2.7 | 9.8–10.4 | − | 2.8–3.0 | |
| 2012a, 2014 | MSL-12 | 12 | 57.8–63.0 | 1.9–2.6 | 6.5–9.9 | − | 2.6–3.0 | |
| Lateral branch number | 2012a, 2012b, 2014 | LBN-2.1 | 2 | 91.4–98.3 | 4.6–12.5 | 16.0–40.0 | + | 0.9–1.9 |
| 2012a, 2012b, 2014 | LBN-2.2 | 2 | 99.7–104.5 | 2.8–6.4 | 10.1–23.1 | + | 0.7–1.4 | |
| Internode length | 2012a, 2014 | INL-1 | 1 | 28.8–32.9 | 2.6–2.9 | 9.2–11.0 | + | 0.8–0.9 |
| 2012a, 2012b, 2014 | INL-2 | 2 | 82.3–90.8 | 3.2–4.8 | 11.0–16.9 | + | 0.8–1.0 | |
| 2012a, 2012b | INL-6.1 | 6 | 0.0–4.9 | 2.4–3.6 | 7.5–11.9 | + | 0.7–0.8 | |
| 2012a, 2014 | INL-6.2 | 6 | 25.8–37.2 | 2.2–3.1 | 7.4–10.2 | + | 0.7–0.8 | |
| 2012a, 2012b | INL-10 | 10 | 0.6–22.5 | 2.1–2.4 | 7.1–7.4 | + | 0.7–0.8 | |
| Stem colour | 2012b, 2014 | SC-2 | 2 | 17.6–22.3 | 3.0 | 11.4 | − | 0.5 |
| 2012b, 2014 | SC-4 | 4 | 34.7–41.0 | 2.6 | 9.7 | + | 0.3 | |
| 2012b, 2014 | SC-7 | 7 | 0.0–2.5 | 3.2 | 10.4 | − | 0.3 | |
| 2011, 2012b, 2014 | SC-9 | 9 | 51.8–59.6 | 2.4–3.4 | 9.3–10.3 | + | 0.1–0.6 | |
| 2012b, 2014 | SC-11 | 11 | 24.3–28.9 | 3.3 | 11.6 | + | 0.3 | |
| 2012b, 2014 | SC-12 | 12 | 61.1–71.1 | 2.8 | 11.1 | + | 0.5 | |
| Leaf length | 2012b, 2014 | LL-6 | 6 | 42.0–52.7 | 2.3–4.5 | 8.7–16.6 | + | 0.6–0.7 |
| 2012b, 2014 | LL-9 | 9 | 75.3–84.9 | 2.2–6.2 | 7.7–20.8 | − | 0.6–0.8 | |
| 2012a, 2014 | LL-11.1 | 11 | 47.3–57.1 | 2.3–4.2 | 7.9–16.1 | + | 0.4–1.0 | |
| 2012a, 2012b, 2014 | LL-11.2 | 11 | 65.3–82.4 | 2.4–3.6 | 8.0–12.3 | − | 0.4–0.9 | |
| Leaf width | 2012a, 2014 | LW-8 | 8 | 56.7–70.6 | 2.3–5.8 | 9.8–18.2 | − | 0.4–0.5 |
| Flower size | 2012b, 2014 | FLS-1 | 1 | 113.7–115.9 | 5.8 | 19.6 | − | 0.3 |
| 2012b, 2014 | FLS-2 | 2 | 60.2–63.9 | 2.8 | 9.0 | − | 0.2 | |
| 2012b, 2014 | FLS-3.1 | 3 | 18.2–23.3 | 3.7 | 11.3 | − | 0.2 | |
| 2012b, 2014 | FLS-3.2 | 3 | 70.7–74.7 | 2.9 | 9.0 | − | 0.2 | |
| 2012b, 2014 | FLS-3.3 | 3 | 89.1–93.7 | 2.6 | 7.8 | + | 0.2 | |
| 2012b, 2014 | FLS-7 | 7 | 46.8–53.1 | 2.4 | 8.8 | − | 0.1 | |
| 2012b, 2014 | FLS-8 | 8 | 89.7–96.6 | 2.7 | 12.2 | − | 0.2 | |
| 2012b, 2014 | FLS-10 | 10 | 57.0–58.4 | 2.4 | 8.7 | + | 0.5 | |
| Stamen number | 2012b, 2014 | STN-3 | 3 | 108.3–115.8 | 2.6 | 9.8 | − | 0.2 |
| 2012b, 2014 | STN-10 | 10 | 99.8–102.6 | 2.8 | 10.3 | − | 0.2 | |
| 2012b, 2014 | STN-11 | 11 | 31.0–36.2 | 2.3 | 8.5 | − | 0.2 | |
| Fruit length | 2011, 2012a, 2012b | FL-3.1 | 3 | 40.6–48.0 | 2.7–8.6 | 9.3–23.6 | + | 0.6–0.8 |
| 2011, 2012b | FL-3.2 | 3 | 48.5–52.1 | 4.4–10.0 | 14.4–26.7 | + | 0.9 | |
| 2012a, 2012b, 2014 | FL-3.3 | 3 | 54.1–60.8 | 2.5–6.3 | 8.5–21.3 | + | 0.6–0.9 | |
| 2011, 2012a, 2014 | FL-3.4 | 3 | 93.3–97.3 | 3.2–4.9 | 10.4–12.0 | − | 0.7–0.8 | |
| 2011, 2012a | FL-3.5 | 3 | 99.7–106.0 | 2.9–3.4 | 8.7–10.0 | − | 0.6–0.7 | |
| 2011, 2012a, 2012b, 2014 | FL-8 | 8 | 51.9–63.8 | 2.4–6.5 | 9.1–20.8 | + | 0.7–0.9 | |
| Fruit diameter | 2011, 2012a, 2012b, 2014 | FD-1 | 1 | 108.8–121.3 | 2.2–5.6 | 8.4–20.0 | − | 0.2–0.3 |
| 2012b, 2014 | FD-2 | 2 | 0.0–10.1 | 3.0–7.1 | 8.7–16.0 | + | 0.2–0.4 | |
| 2011, 2012a, 2012b, 2014 | FD-3.1 | 3 | 37.7–41.4 | 2.7–6.0 | 8.0–17.5 | − | 0.2–0.3 | |
| 2011, 2012a, 2012b, 2014 | FD-3.2 | 3 | 47.2–59.1 | 2.7–7.4 | 8.3–20.5 | − | 0.2–0.4 | |
| 2011, 2012b, 2014 | FD-11 | 11 | 21.5–24.6 | 2.1–2.8 | 7.7–10.1 | − | 0.2–0.3 | |
| Fruit shape | 2011, 2012a, 2012b, 2014 | FS-3.1 | 3 | 47.9–51.8 | 8.8–13.6 | 24.1–37.0 | + | 0.7–0.9 |
| 2011, 2012a, 2014 | FS-3.2 | 3 | 54.1–59.0 | 8.4–11.7 | 22.2–35.3 | + | 0.7–0.9 | |
| 2011, 2012a, 2012b, 2014 | FS-8 | 8 | 52.7–62.6 | 2.3–3.9 | 7.4–13.6 | + | 0.5–0.6 | |
| 2011, 2012b, 2014 | FS-11 | 11 | 23.4–32.2 | 2.5–2.8 | 9.6–10.2 | + | 0.5–0.6 | |
| Fruit weight | 2011, 2014 | FW-1 | 1 | 112.5–116.4 | 3.1–4.3 | 10.9–15.8 | − | 1.4–2.4 |
| 2012a, 2012b, 2014 | FW-2.1 | 2 | 0.0–8.0 | 2.9–3.0 | 9.8–11.4 | + | 1.4–2.9 | |
| 2011, 2014 | FW-2.2 | 2 | 64.3–73.1 | 2.2–3.5 | 6.6–12.1 | − | 1.2–2.0 | |
| 2011, 2012a | FW-3 | 3 | 47.6–62.5 | 3.4–3.7 | 9.8–12.7 | − | 2.3 | |
| 2012b, 2014 | FW-6.1 | 6 | 53.7–60.2 | 3.0 | 11.3–11.7 | − | 1.2–1.4 | |
| 2012a, 2014 | FW-6.2 | 6 | 59.8–68.1 | 2.2–2.3 | 6.8–8.5 | − | 1.0–1.7 | |
| Fruit position | 2011, 2012a, 2012b, 2014 | FP-1 | 1 | 103.7–117.7 | 3.1–3.8 | 10.2–14.3 | − | 0.3–0.4 |
| 2012b, 2014 | FP-4.1 | 4 | 35.1–42.6 | 3.3 | 11.0 | − | 0.3 | |
| 2012b, 2014 | FP-4.2 | 4 | 108.0–115.7 | 2.2 | 7.5 | − | 0.3 | |
| 2012b, 2014 | FP-12.1 | 12 | 49.0–50.5 | 20.0 | 53.6 | − | 0.7 | |
| 2011, 2012a, 2012b, 2014 | FP-12.2 | 12 | 57.7–58.6 | 12.7–26.9 | 40.2–63.9 | − | 0.6–0.8 | |
| 2011, 2012a | FP-12.3 | 12 | 65.7–68.1 | 4.4–17.1 | 11.6–47.0 | − | 0.5–0.7 | |
| Calyx shape | 2012b, 2014 | CS-1.1 | 1 | 109.0–115.4 | 3.7 | 12.9 | − | 0.3 |
| 2011, 2012a | CS-1.2 | 1 | 178.5–184.1 | 3.3–3.5 | 11.4–11.8 | − | 0.3 | |
| 2012b, 2014 | CS-3.1 | 3 | 4.9–15.1 | 2.2 | 7.8 | − | 0.2 | |
| 2011, 2012a | CS-3.2 | 3 | 15.1–20.3 | 2.6–3.5 | 8.0–10.2 | − | 0.2 | |
| 2012b, 2014 | CS-3.3 | 3 | 20.3–23.3 | 2.4 | 8.4 | − | 0.2 | |
| 2011, 2012a | CS-3.4 | 3 | 45.5–52.1 | 2.8–3.3 | 8.8–9.4 | − | 0.2–0.3 | |
| 2012b, 2014 | CS-3.5 | 3 | 54.1–59.1 | 3.3 | 11.1 | − | 0.3 | |
| 2011, 2012a | CS-5 | 5 | 53.1–56.0 | 2.8–3.1 | 10.8–11.3 | − | 0.3 | |
| 2012b, 2014 | CS-9 | 9 | 77.9–81.7 | 2.9 | 10.3 | − | 0.3 | |
| 2012b, 2014 | CS-11 | 11 | 9.8–14.2 | 2.6 | 9.5 | + | 0.3 | |
| Immature fruit colour | 2012b, 2014 | IFC-10.1 | 10 | 66.2–69.0 | 3.5 | 9.2 | − | 0.2 |
| 2012b, 2014 | IFC-10.2 | 10 | 75.7–81.8 | 14.3 | 40.2 | − | 0.3 | |
| 2012b, 2014 | IFC-10.3 | 10 | 82.6–86.1 | 5.8 | 19.3 | − | 0.2 | |
| 2012b, 2014 | IFC-11.1 | 11 | 6.8–13.8 | 2.3 | 8.2 | + | 0.2 | |
| 2012b, 2014 | IFC-11.2 | 11 | 81.3–85.6 | 2.6 | 8.8 | − | 0.2 | |
| 2012b, 2014 | IFC-12 | 12 | 17.5–24.1 | 3.2 | 11.3 | − | 0.2 |
Figure 3.
Chromosomal distribution of significant QTLs. Coloured bars show the location of QTLs, and the names of QTLs are listed in Table 4. This figure is available in black and white in print and in colour at DNA Research online.
On chromosome 3, 19 QTLs were detected and 15 of them controlled fruit-related traits (Fig. 3). Notably, 10 QTLs were located between 37.7 and 59.1 cM and all controlled fruit-related traits. FS-3.1, FS-3.2, FL-3.1, FL-3.2, and FL-3.3 had positive additive effects, meaning that RILs with the maternal genotype had higher values for fruit shape and fruit length (Supplementary Fig. S4A and B). The two traits controlled by these QTLs were positively correlated with each other. CS-3.4, CS-3.5, FD-3.1, FD-3.2, and FW-3 showed negative additive effects, and the traits showed positive correlation (Supplementary Fig. S4C and D). Co-localization of correlated QTLs was detected on chromosomes 1, 2, 3, 7, 8, 10, and 11. Most of these co-localized QTLs controlled fruit-related traits, whereas negatively correlated flower size and main stem length also showed co-localization of QTLs on chromosomes 7, 8, and 10.
3.4. Anchoring of unassigned scaffolds
Sequencing reads that did not map to ‘CM334’ chromosomes were realigned to the unassigned scaffolds. The longest scaffold was 3.4 Mb, and we used the assumption that there were no recombinant breakpoints within a scaffold. Therefore, scaffolds with more than 30 SNPs were used as markers. The ratio between the SNPs with ‘Perennial’ and ‘Dempsey’ genotypes was calculated to genotype the scaffolds of each RIL. Genotyped scaffolds were mapped to the bin map constructed with the sliding window approach. The number of unassigned scaffolds was 36,487, of which 263 scaffolds were anchored to the bin map. We removed 59 scaffolds that mapped to more than two chromosomes, and 204 scaffolds could ultimately be assigned using the bin map (Table 5 and Supplementary Table S4). As we used bins instead of individual SNPs, the precise position of each scaffold was not determined here.
Table 5.
Distribution of scaffolds assigned using the bin map
| Chromosome | Number of assigned scaffolds | Average length of scaffold (kb) |
|---|---|---|
| 1 | 39 | 624 |
| 2 | 12 | 512 |
| 3 | 24 | 511 |
| 4 | – | – |
| 5 | 14 | 572 |
| 6 | 26 | 457 |
| 7 | – | – |
| 8 | 8 | 1,228 |
| 9 | 27 | 552 |
| 10 | 10 | 247 |
| 11 | 24 | 393 |
| 12 | 20 | 601 |
| Total | 204 | 545 |
4. Discussion
In this work, we constructed an ultra-high-density bin map for pepper via a sliding window approach and used it to detect QTLs controlling morphological traits. Our bin map helps improve the accuracy of the pepper reference genome in two ways. First, the bins can elucidate the order of unordered scaffolds in the chromosomes. Comparison of the genetic and physical locations of bins revealed extended physical length of bins and inconsistencies for bins located near the middle of chromosomes (Fig. 2). These regions are likely centromeric or pericentromeric, as recombination rates tend to be low in the centromeric regions.27–29 In addition, unordered scaffolds are concentrated in putative centromeric regions of the reference genome.24 Thus, even though the lengths of bins in putative centromeric regions are longer than in other regions, they provide useful information for the reference genome by revealing the genetic order of bins. Second, our bin map refines the pepper reference genome by allowing us to anchor unassigned scaffolds to linkage groups. We were able to assign >200 scaffolds to chromosomes (Table 5). The total length of newly assigned scaffolds was 111 Mb, which is ∼3.2% of expected pepper genome size. This result shows that genetic maps constructed using the same population and sequencing data but with a different mapping approach can improve the reference genome by anchoring unassigned scaffolds.
The ultra-high-density bin map constructed in this study is the first SNP-based bin map in pepper, although the sliding window approach has been used to construct bin maps in cereal crops.17,19,20 To determine recombination breakpoints of 150 RILs in rice and 708 F2 plants in maize, 15 and 18 consecutive SNPs were used as one window in previous researches, respectively.17,20 To construct our bin map in pepper, we modified the sliding window approach. Instead of fixing the number of SNPs included in the window, we set the window length as 5 Mb. Non-uniform distributions of SNPs were detected by counting the number of SNPs in every 1.5 Mb (Supplementary Fig. S5A). When we set a window as 31 consecutive SNPs, recombination breakpoints were predicted to be excessively concentrated on regions where the density of SNPs was high (Supplementary Fig. S5B). Sequencing or genotyping error from low-coverage sequences can produce low-quality SNPs,30 and there will be high correlation between the number of SNPs and recombination breakpoints. The sliding window approach can reduce the noise created by individual SNPs, but the original (set SNP number per window) approach cannot reduce the effects of SNP density. In contrast, recombination breakpoints detected using a 5-Mb window were less affected by the density of SNPs (Supplementary Fig. S5B). Our constructed bin map contains 2,578 bins, with an average of 33 recombination breakpoints per line, similar to the bin map of rice.17 The number of recombination breakpoints was higher in rice and pepper RILs compared with that of maize F2 plants, confirming that the construction of bin maps using the sliding window approach can detect the differences in recombination frequency between RIL and F2 plants.
We confirmed the accuracy of our bin map by mapping the Pun1 gene controlling pungency of pepper. There was ∼1.6 Mb difference between the position in this study and the physical location of the Pun1 gene in the pepper genome. This may be caused by the misgenotyping of heterozygous Pun1/pun1 alleles as homozygous Pun1/Pun1 alleles. In this study, the Pun1 gene was genotyped using dominant molecular markers,31 but five RILs showed heterozygous bins near the Pun1 gene (Supplementary Table S2). Therefore, it is highly possible that the discrepancy was due to the misgenotyped RILs. In addition to the Pun1 gene, comparisons of the location of QTLs detected in this study with those in previous studies can verify the accuracy. Here, we detected FP-12.2 as a major QTL controlling fruit position and explaining >40% of the phenotypic variation. This QTL was located at 199.6 Mb on chromosome 12 in the ‘CM334’ reference genome. The up locus that is known to control fruit position is also located on chromosome 12 by previous research.32 The physical location of the up locus-linked marker is 197.1 Mb, and this marker is 4.3 cM apart from the up locus. Thus, mapping of the Pun1 gene and the up locus using our bin map demonstrate the accuracy of the bin map, and show that QTL mapping with an ultra-high-density bin map will appropriately map the genetic locus.
Fruit-related traits are of vital importance for pepper researchers and breeders. A number of QTLs controlling fruit weight, diameter, and shape have been identified in intraspecific and interspecific populations.5,7,10,33,34 In particular, QTLs controlling fruit weight were previously detected on chromosome 2.5,7,8 However, direct comparison of QTL locations has been difficult due to the lack of common markers. Therefore, we compared the physical locations of markers linked to QTLs using the ‘CM334’ reference genome sequence. fw2.1 detected in C. chinense and C. frutescens introgression lines were located between 156 and 158 Mb on chromosome 2.8 The QTL with the same name as fw2.1 was detected in a backcross population from a cross between C. annuum and C. frutescens and was also located ∼156 Mb of chromosome 2.7 FW-2.1 and FW-2.2, for fruit weight, detected in our research were located at 1.5 and 146 Mb, respectively. These findings show that FW-2.2 could be the same QTL as that previously reported, and that FW-2.1 is a newly detected QTL controlling fruit weight on chromosome 2. A slight difference in the physical location of FW-2.2 with fw2.1 could be caused by different statistical methods used for QTL analysis, or from the difference of the species used for the construction of genetic map. Using the same approach, we compared the location of QTLs controlling fruit shape on chromosome 3. The most tightly linked marker to the QTL, TG130, was located ∼194 Mb on chromosome 3.5,7,9 In the current work, we identified FS-3.1 and FS-3.2. Due to rearrangement of bins near FS-3.1, the physical location of FS-3.1 was spread in the ranges 60–75, 98–124, and 174–200 Mb on chromosome 3. FS-3.2 was located ∼206–211 Mb. It was not possible to determine whether the QTLs were the same or not, but the repeated detection of QTLs on chromosomes 2 and 3 for fruit weight and fruit shape demonstrates the significance and importance of those QTLs. Therefore, fine-mapping of FW-2.1, FW-2.2, FS-3.1, and FW-3.2 as well as comparative study with syntenic QTLs in tomato will likely be helpful to elucidate the underlying genetic factors.
In summary, in this study, we identified QTLs for not only fruit-related traits but also plant architecture- and leaf-related traits. In addition, we report QTLs controlling flower-related traits in pepper for the first time. The ultra-high-density bin map we constructed using a sliding window approach improves the pepper genetic map and increases the speed of QTL mapping. This bin map will help to revise the reference genome and as all bins were determined based on SNPs, the bins can be used directly to develop molecular markers linked to QTLs.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org online.
Funding
This work was carried out with the support of Cooperative Research Program for Agriculture Science and Technology Development (Plant Molecular Breeding Center No. PJ01120501 and the Agricultural Genome Center No. PJ01120401 and No. PJ01127501), Rural Development Administration, Republic of Korea. Funding to pay the Open Access publication charges for this article was provided by Seoul National University.
Supplementary Material
References
- 1.Paran I., van der Knaap E.. 2007, Genetic and molecular regulation of fruit and plant domestication traits in tomato and pepper, J. Exp. Bot., 58, 3841–52. [DOI] [PubMed] [Google Scholar]
- 2.Eshbaugh W.H. 1980, The taxonomy of the genus Capsicum (Solanaceae), Phytologia, 47, 153–66. [Google Scholar]
- 3.Sudré C.P., Gonçalves L.S.A., Rodrigues R., Júnior A.T.D.A., Riva-Souza E.M., Bento C.D.S.. 2010, Genetic variability in domesticated Capsicum spp. as assessed by morphological and agronomic data in mixed statistical analysis, Genet. Mol. Res., 9, 283–94. [DOI] [PubMed] [Google Scholar]
- 4.Thul S.T., Lal R.K., Shasany A.K. et al. 2009, Estimation of phenotypic divergence in a collection of Capsicum species for yield-related traits, Euphytica, 168, 189–96. [Google Scholar]
- 5.Ben-Chaim A., Paran I., Grube R.C., Jahn M., Wijk R.V., Peleman J.. 2001, QTL mapping of fruit-related traits in pepper (Capsicum annuum), Theor. Appl. Genet., 102, 1016–28. [Google Scholar]
- 6.Chaim A.B., Borovsky Y., Jong W.D., Paran I.. 2003, Linkage of the A locus for the presence of anthocyanin and fs10.1, a major fruit-shape QTL in pepper, Theor. Appl. Genet., 106, 889–94. [DOI] [PubMed] [Google Scholar]
- 7.Rao G.U., Chaim A.B., Borovsky Y., Paran I.. 2003, Mapping of yield-related QTLs in pepper in an interspecific cross of Capsicum annuum and C. frutescens, Theor. Appl. Genet., 106, 1457–66. [DOI] [PubMed] [Google Scholar]
- 8.Zygier S., Chaim A.B., Efrati A., Kaluzky G., Borovsky Y., Paran I.. 2005, QTLs mapping for fruit size and shape in chromosomes 2 and 4 in pepper and a comparison of the pepper QTL map with that of tomato, Theor. Appl. Genet., 111, 437–45. [DOI] [PubMed] [Google Scholar]
- 9.Ben-Chaim A., Borovsky Y., Rao G.U., Tanyolac B., Paran I.. 2003, fs3.1: a major fruit shape QTL conserved in Capsicum, Genome, 46, 1–9. [DOI] [PubMed] [Google Scholar]
- 10.Yarnes S.C., Ashrafi H., Reyes-Chin-Wo S., Hill T.A., Stoffel K.M., Van Deynze A.. 2013, Identification of QTLs for capsaicinoids, fruit quality, and plant architecture-related traits in an interspecific Capsicum RIL population, Genome, 56, 61–74. [DOI] [PubMed] [Google Scholar]
- 11.Kang B.C., Nahm S.H., Huh J.H. et al. 2001, An interspecific (Capsicum annuum× C. chinese) F2 linkage map in pepper using RFLP and AFLP markers, Theor. Appl. Genet., 102, 531–9. [Google Scholar]
- 12.Barchi L., Bonnet J., Boudet C. et al. 2007, A high-resolution, intraspecific linkage map of pepper (Capsicum annuum L.) and selection of reduced recombinant inbred line subsets for fast mapping, Genome, 50, 51–60. [DOI] [PubMed] [Google Scholar]
- 13.Hill T.A., Ashrafi H., Reyes-Chin-Wo S. et al. 2013, Characterization of Capsicum annuum genetic diversity and population structure based on parallel polymorphism discovery with a 30K unigene Pepper GeneChip, PLoS One, 8, e56200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nielsen R., Paul J.S., Albrechtsen A., Song Y.S.. 2011, Genotype and SNP calling from next-generation sequencing data, Nat. Rev. Genet., 12, 443–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y., Sidore C., Kang H.M., Boehnke M., Abecasis G.R.. 2011, Low-coverage sequencing: implications for design of complex trait association studies, Genome Res., 21, 940–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasaniuc B., Rohland N., McLaren P.J. et al. 2012, Extremely low-coverage sequencing and imputation increases power for genome-wide association studies, Nat. Genet., 44, 631–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang X., Feng Q., Qian Q. et al. 2009, High-throughput genotyping by whole-genome resequencing, Genome Res., 19, 1068–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L., Wang A., Huang X. et al. 2011, Mapping 49 quantitative trait loci at high resolution through sequencing-based genotyping of rice recombinant inbred lines, Theor. Appl. Genet., 122, 327–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou G., Zhai G., Feng Q. et al. 2012, Identification of QTLs for eight agronomically important traits using an ultra-high-density map based on SNPs generated from high-throughput sequencing in sorghum under contrasting photoperiods, J. Exp. Bot., 63, 5451–62. [DOI] [PubMed] [Google Scholar]
- 20.Chen Z., Wang B., Dong X. et al. 2014, An ultra-high density bin-map for rapid QTL mapping for tassel and ear architecture in a large F2 maize population, BMC Genomics, 15, 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Givry S., Bouchez M., Chabrier P., Milan D., Schiex T.. 2005, CARHTA GENE: multipopulation integrated genetic and radiation hybrid mapping, Bioinformatics, 21, 1703–4. [DOI] [PubMed] [Google Scholar]
- 22.Voorrips R.E. 2002, MapChart: software for the graphical presentation of linkage maps and QTLs, J Hered., 93, 77–8. [DOI] [PubMed] [Google Scholar]
- 23.Brewer M.T., Lang L., Fujimura K., Dujmovic N., Gray S., van der Knaap E.. 2006, Development of a controlled vocabulary and software application to analyze fruit shape variation in tomato and other plant species, Plant Physiol., 141, 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim S., Park M., Yeom S.I. et al. 2014, Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species, Nat. Genet., 46, 270–8. [DOI] [PubMed] [Google Scholar]
- 25.Wang S., Basten C.J., Zeng Z.B.. 2012, Windows QTL Cartographer 2.5. Department of Statistics. North Carolina State University; http://statgen.ncsu.edu/qtlcart/WQTLCart.htm. [Google Scholar]
- 26.Stewart C. Jr, Kang B.C., Liu K. et al. 2005, The Pun1 gene for pungency in pepper encodes a putative acyltransferase, Plant J., 42, 675–88. [DOI] [PubMed] [Google Scholar]
- 27.Puchta H., Hohn B.. 1996, From centiMorgans to base pairs: homologous recombination in plants, Trends Plant Sci., 1, 340–8. [Google Scholar]
- 28.Spindel J., Wright M., Chen C. et al. 2013, Bridging the genotyping gap: using genotyping by sequencing (GBS) to add high-density SNP markers and new value to traditional bi-parental mapping and breeding populations, Theor. Appl. Genet., 126, 2699–716. [DOI] [PubMed] [Google Scholar]
- 29.Si W., Yuan Y., Huang J. et al. 2015, Widely distributed hot and cold spots in meiotic recombination as shown by the sequencing of rice F2 plants, New Phytol., 206, 1491–502. [DOI] [PubMed] [Google Scholar]
- 30.Sims D., Sudbery I., Ilott N.E., Heger A., Ponting C.P.. 2014, Sequencing depth and coverage: key considerations in genomic analyses, Nat. Rev. Genet., 15, 121–32. [DOI] [PubMed] [Google Scholar]
- 31.Han K., Jeong H.-J., Sung J. et al. 2012, Biosynthesis of capsinoid is controlled by the Pun1 locus in pepper, Mol Breed, 31, 537–48. [Google Scholar]
- 32.Lee H.-R., Cho M.-C., Kim H.-J., Park S.-W., Kim B.-D.. 2008, Marker development for erect versus pendant-orientated fruit in Capsicum annuum L, Mol. Cells, 26, 548–53. [PubMed] [Google Scholar]
- 33.Barchi L., Lefebvre V., Sage-Palloix A.M., Lanteri S., Palloix A.. 2009, QTL analysis of plant development and fruit traits in pepper and performance of selective phenotyping, Theor. Appl. Genet., 118, 1157–71. [DOI] [PubMed] [Google Scholar]
- 34.Borovsky Y., Paran I.. 2011, Characterization of fs10.1, a major QTL controlling fruit elongation in Capsicum, Theor. Appl. Genet., 123, 657–65. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.