Abstract
As sequencing technologies progress, the amount of data produced grows exponentially, shifting the bottleneck of discovery towards the data analysis phase. In particular, currently available mapping solutions for RNA-seq leave room for improvement in terms of sensitivity and performance, hindering an efficient analysis of transcriptomes by massive sequencing. Here, we present an innovative approach that combines re-engineering, optimization and parallelization. This solution results in a significant increase of mapping sensitivity over a wide range of read lengths and substantial shorter runtimes when compared with current RNA-seq mapping methods available.
Keywords: RNA-seq, mapping, Burrows-Wheeler Transform, high-performance computing
1. Introduction
The last generations of high-throughput sequencers produce data at an unprecedented scale with associated sequencing costs in a continuous decrease. In particular, RNA sequencing (RNA-seq) technology,1 which provides a comprehensive profile of a transcriptome, is increasingly replacing conventional expression microarrays.2 Primary data processing in RNA-seq (as well as in other massive sequencing experiments, including genome resequencing) involves mapping reads onto a reference genome. This step constitutes a computationally expensive process in which, in addition, sensitivity is a serious concern.3 A variety of programs, many of them implementing the Burrows-Wheeler Transform (BWT) algorithm that is based on an indexing method which enormously speed up the searching process, have been developed.4–8
Recently proposed algorithms for DNA read mapping map ∼98–99% of DNA reads (a gain of only 2% with respect to the 96% obtained by previously available mappers at a modest speedup).9–11 Thus, the current software provides a reasonably quick and sensitive framework for read mapping in DNA resequencing experiments. However, the scenario in RNA-seq is far from these standards. Mapping sequencing reads in the context of transcripts is a much more complex problem than simply mapping reads onto the genomic sequence, as in the case of genome resequencing experiments. Eukaryotic transcriptomes are complex, with an average of more than nine transcripts per gene12 with exons of 250 bp on the average that span over several hundreds of kilobases, and more new transcript isoforms are continuously being discovered.13 It has been reported that widely used mappers, such as TopHat,14 can hardly reach 70% of 100 bp reads correctly mapped in a realistic scenario with sequencing errors, while the best sensitivity (82%) is attained by GSNAP.15 Although GSNAP seems to display a lower sensitivity in an ideal error-free scenario.16 According to this last report, other RNA-seq mappers like RUM,17 STAR,18 or MapSplice19 align sequencing reads with even lower accuracy. However, more recent reports20 support a better performance of STAR18 or MapSplice19 with respect to the other methods, in agreement with earlier benchmarks.17 Moreover, as read length increases (the natural trend of the continuous upgrades of sequencing instruments), the sensitivity of the methods drops down, causing serious problems in terms of sensitivity and runtime.16 Mapping is actually problematic with most BWT-based mappers, because they often allow only a small number of mismatches (insufficient for the current read lengths even in scenarios of low variability). New aligners, such TopHat2,14 which uses the new Bowtie2,10 have significantly accelerated the mapping step (with a small trade-off in sensitivity), although the strategy for junction detection remains unchanged. While this study was under review, a new mapper was published, HISAT,21 that combines the BWT and the Ferragina-Manzini index in its indexing scheme. Similarly, this speedup that shows this program seems to be attained at the exchange of a trade-off in sensitivity.
Despite the interest in the use of RNA-seq for transcriptome analysis and the advancements in algorithms either for quantifying reads22,23 or for transcript discovery,24 the basic problem of mapping reads in the context of transcripts is far from being solved.
Here, we propose an innovative solution for high-quality mapping of both short and long reads, based on a combination of mapping with BWT and local alignment with Smith-Waterman (SW), that drastically increases mapping accuracy (95 versus 60–85% by current mappers, in the most common scenarios) and substantially reduces runtimes (by about 15× when compared with TopHat2, 8× with MapSplice and 2× with STAR). In addition, the proposed strategy has also demonstrated to be quite robust against indels and mismatches. This proposal provides a simple, fast and elegant solution that maps almost all the reads, even those containing a high number of mismatches or indels. This solution also saves a substantial amount of time in the mapping step which, consequently, critically contributes to the acceleration of the current pipelines of sequencing data processing. This strategy, implemented in a program that makes use of different high-performance computing (HPC) technologies, HPG Aligner, shows an excellent performance with both, short and long reads, with runtimes presenting only a linear dependence with the number of reads.
2. Methods
Most modern mappers rely on the BWT indexing method to speed up the searching process. However, speedup by indexing is achieved at the exchange of sensitivity, which decreases as read length increases, since most common BWT implementations only allow a limited number of mismatches. On the other side of the spectrum, the well-known BLAST mapper25 uses SW as a local alignment algorithm, which presents a high sensitivity, being able to map reads with many mismatches or indels. However, its performance in terms of runtimes makes BLAST unsuitable for mapping the huge number of reads produced in next-generation sequencing (NGS) experiments.
Here, we have brought together the best of both algorithms: the performance of BWT and the sensitivity of SW, by implementing a combined strategy that allows an ultrafast and high sensitivity read mapping, even in the presence of high proportions of mutations and indels. The algorithm proposed also deals very efficiently with the problem of introns, no matter that only a small fragment of the end of the read is contained in the next exon. This is achieved by using a highly efficient data structure, which we called ‘metaexon’, that learns from high-quality mapped reads and helps to map low-quality or more complex reads. This metaexon structure stores information of the genomic coordinates and the number of aligned reads with low memory requirements (Supplementary Fig. S1). By doing so, the proportion of hard and soft clipping is drastically reduced.
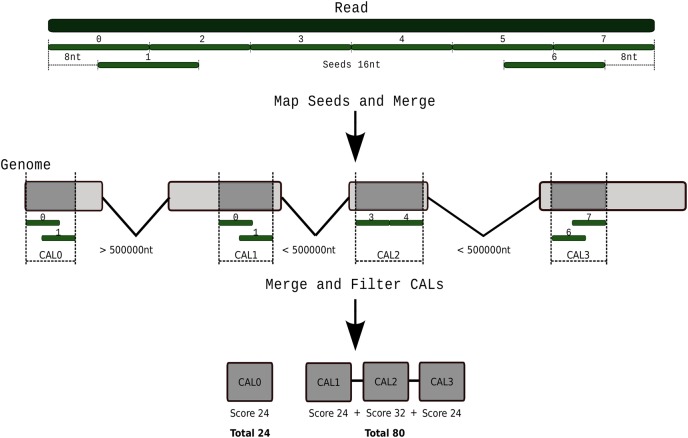
The aligning process is completed in two main steps: first, our BWT implementation, based on a previous version for exact alignment,26 is used to map reads containing up to two mismatches (BWT index is built with a default factor of 8). The resulting high-quality alignment information is stored in the metaexon data structure. Secondly, reads containing more than two mismatches, indels or introns are mapped following a multi-stage process: (i) contiguous seeds are generated covering the whole read and also two more reads overlapping the beginning and end of the read are generated (Fig. 1). These overlapping seeds will help to align multi-exon covering reads. A seed size of 16 nucleotides (nt) represents a good trade-off between performance and sensitivity. These seeds are mapped using BWT, with no mismatches allowed. (ii) Seeds mapping at distances closer than a read length and with the same strand orientation are brought together to generate candidate alignment locations (labelled as candidate alignment location in Fig. 1); (iii) candidate alignment locations are clustered together to define transcripts providing they are closer than 500,000 nt and in the same strand orientation. Note that several clusters can be obtained from the candidate alignment locations; (iv) for each cluster, the gaps produced by either not aligned seeds or exon–intron boundaries are aligned using a HPC implementation of SW. This implementation uses Streaming SIMD Extensions (SSE) instructions, which allow mismatches and indels and present a high sensitivity to determine intron boundaries. For aligning seeds mapping at the ends of the exons, the metaexon data structure is queried to obtain candidate exon junctions. Using this simple strategy, HPG Aligner can provide a high-quality alignment for spliced reads; (v) finally, a score is obtained and the alignments are reported.
Figure 1.
Schema of the implementation of the mapping process. (Top) Contiguous seeds of size (16 bp) are taken covering the whole read. Also, two more overlapping seeds near the ends of the read are taken for anchoring the ends to the exons. (Middle) Seeds are mapped without allowing any mismatch. Seeds mapped closer than read size and, in the same strand orientation, constitute a candidate alignment location (CAL). (Bottom) CALs closer than 500,000 bp and, in the same strand orientation, are clustered to form candidate exons and transcripts. These are evaluated and scores based on SW are assigned to them. This figure is available in black and white in print and in colour at DNA Research online.
Due to the HPC implementation, we have devised a simple but powerful strategy in which the original high sensitivity of the algorithms is maintained without sacrificing performance. HPG Aligner can map reads containing many mismatches and indels, which can cover one or more introns. The algorithms have been implemented in the most convenient accelerator hardware. Thus, BWT has been implemented in two ways: in a multi-thread library that exploits the modern multicore CPUs and also using Nvidia CUDA library for GPUs. Both implementations can be executed simultaneously to obtain higher performance. On the other hand, SW benefits from the parallelism of SIMD registers present in each core of modern CPUs and has been implemented using SSE4 instructions.
The performance of HPG aligner was greatly improved with the implementation of some specific strategies to overcome the bottlenecks detected in the alignment process. Most significant implementation improvements include: (i) the use of a custom SW implementation based on multisequence vectors;27 during the alignment, SW executions are buffered to execute them in parallel. (ii) Grouping seeds that map close in the genome (conceptually similar to clustering) was an expensive operation. This was easily solved by storing all the seeds from different chromosomes independently and sorted. (iii) BWT is a very fast algorithm and a significant fraction of the FASTQ file could be aligned with less than two mismatches. Therefore, reads are aligned with BWT before seeding. When the alignment fails (because there are more than two mismatches), the aligned part of the read is still stored as a seed (i.e. if after BWT there are 40 bp exact alignment, then this is considered a seed.). (iv) The metaexon data structure (see above and Supplementary Fig. S1) reduces the computing time for some of the more difficult alignments.
To improve the overall performance of the implementation, several advanced tools for high-performance profiling were used during the development (in particular, Paraver http://www.bsc.es/computer-sciences/performance-tools/paraver and Extrae http://www.bsc.es/computer-sciences/extrae). These tools revealed that some steps of the internal pipeline were not taking fully advantage of the hardware resources. A dynamic runtime engine was developed and built inside HPG Aligner.28 Each of the steps of the pipeline were considered as a task that needs some resource to be executed. Requested resources were assigned by the engine runtime in a dynamic and efficient way. In this way, the computing resources (such as cores) are assigned to the different tasks in an optimal way to avoid some cores to be waiting for other tasks to finish. This is a more complex implementation, which can usually be found in HPC applications. This allowed us to increase significantly the performance.
By leveraging the HPC implementation, the software runs many times faster than any other currently available solution and, in addition, it presents much higher sensitivity. Moreover, this implementation has proved to be technically much more efficient than current state-of-the art implementations since (i) the requirement of memory is much lower and it is kept below 10 GB, (ii) scalability in a multicore CPUs (up to 24 cores) is maintained and (iii) no large secondary indices or ‘temp’ files are generated during the execution, so no extra hard disk space is required.
2.1. Paired and unpaired RNA-seq simulated datasets comparison
We have used two popular programs for the simulations of paired and unpaired reads from the human transcriptome: BEERS (http://www.cbil.upenn.edu/BEERS/), a simulator specifically devised to produce mRNA populations17 and also the popular dwgsim 0.1.8 from SAMtools (http://sourceforge.net/apps/mediawiki/dnaa/index.php?title=Whole_Genome_Simulation).
Using BEERS, we generated 10 million reads for 10,000 genes in a duplicate of all the simulations, that is, FastQ files of 50, 75, 100, 150, 250 and 400 bp long, with different error ratios (0.001, 0.01 and 0.02 corresponding to 0.1, 1 and 2% mismatches, respectively). In all the configurations, the fraction of mutations that are indels was set to 10%.
The program dwgsim was run in ‘Illumina’ mode with option ‘-c 0’ for the six different length sizes tested. To study sensitivity against mutations and N's present in reads, three quality configurations were tested. First, a high-quality dataset containing 0.1% of mutations and a maximum of 2 N's per read was generated with options ‘-r 0.001 -n 2’. For the second and third datasets, higher proportion of mutations and N's were allowed to increase up to 1 and 2% of mutations, respectively, and 3 N's per read with options ‘-r 0.01 -n 3’ and ‘-r 0.02 -n 3’. In both configurations, the fraction of mutations that are indels was set to 10% with option ‘-R 0.1’.
The coordinates for cDNA sequences were taken from Ensembl 68 built upon GRCh37. For all combinations of length sizes and quality configurations, single-end and paired-end datasets were generated. For paired-end datasets, the inner distance between reads was set to 400 for reads smaller than 250 nt and a distance of 250 for the datasets of 250 and 400 length sizes.
Human assembly GRCh37.p8 was taken from Ensembl 68. HPG Aligner was run in ‘rna’ mode. The default command line parameters for the programs can be found in Supplementary data.
Benchmarks were performed in a high-end machine with two hexa-core Intel Xeon E5645 2.40 GHz CPUs and 48 GB of memory. All executions were done using the 12 cores available and memory use was monitored. HPG Aligner showed a memory peak of 10 GB.
All the FastQ file generated in the simulations and the BAM files generated in the benchmarking are available at: http://bioinfo.cipf.es/publications/supplementary_material.
2.2. Real RNA-seq datasets comparison
To test short- and long-read alignment with real data, two different datasets from Sequence Read Archive (SRA) were used, and the first dataset tested was SRR364003 (http://www.ncbi.nlm.nih.gov/sra?term=SRR364003) sequenced with Illumina HiSeq 2000 platform and contained 81.6 million single and paired-end short reads of 100 nt length; the second dataset was built by combining the two runs of SRX025091 (http://www.ncbi.nlm.nih.gov/sra?term=SRX025091) sequenced using the Roche 454 GS FLX platform; in total, a dataset of 1.26 million of single-end reads of about sequences, 600 nt long, was obtained.
The programs were run using default command line options in a single and paired-end mode (see above). Evaluation was performed by counting as correctly mapped the number of reads mapping within known transcript positions. The known transcripts are taken from Ensembl.29 Benchmarks were performed in a high-end machine with two hexa-core Intel Xeon E5645 2.40 GHz CPUs and 48 GB of memory.
3. Results
3.1. Performance of the method: speed and sensitivity
We have compared the proposed aligner with the most extensively used RNA-seq mapper, TopHat214 (version 2.0.9), STAR18 (version 2.3.0), MapSplice19 (version 2.1.3) and HISAT21 in simulated datasets with 10 million reads of lengths of 50, 75, 100, 150, 250 and 400 nt, in three scenarios of variability (0.1, 1 and 2% of mismatches, 10% of these being indels). Simulations were initially carried out with the BEERS program (see the Methods section).
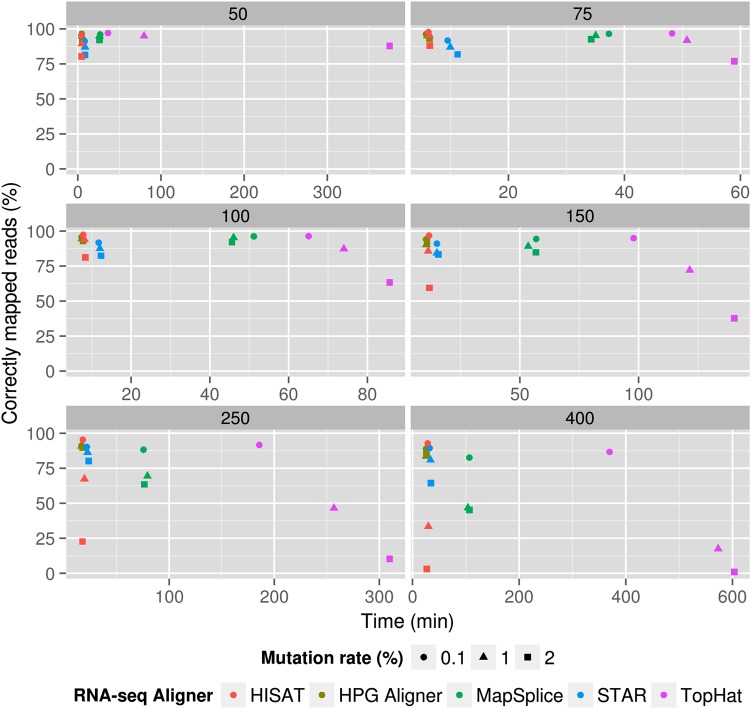
Figure 2 shows the sensitivity of the proposed aligner, measured as the number of reads generated by BEERS correctly mapped (that is, mapping correctly the start and end and covering splice sites in between). HPG Aligner outperforms TopHat2 with Bowtie 2, HISAT (except in unrealistic scenarios of no error with read lengths of 150 and 250), STAR and MapSplice, even when mapping short reads (see details in Table 1) and, as read length grows and the error rate increases, the difference in sensitivity increases even more. In current conventional lengths (100 bp) in an ideal scenario of low error, performances are quite similar (95–96%). However, if error is a bit higher (1%), HPG Aligner and MapSplice 2 maintain the sensitivity (94.50 and 95.34%, respectively). At this read length, TopHat2 and STAR sensitivities drop to a lower, but still reasonable value of ∼87%. For longer reads, the difference in sensitivity between HPG Aligner and STAR with respect to TopHat2 and MapSplice 2 grows considerably in scenarios of moderate error (see Table 1 reads 250 or 400 long at errors 1 or 2%). TopHat2 presents a peculiar behaviour: it keeps an extraordinarily low ratio of incorrect read mapping (below 2%) at the exchange of increasing the number of unmapped reads, which arrives to 27.13% for read lengths of 150 bp and 1% error and 53.29% for 250 bp (same error). It is remarkable that HPG Aligner, STAR and MapSplice keep the number of unmapped read below 5% across all the range of read lengths. Regarding runtimes, HPG Aligner runs much faster, ranging from 7× to almost than 10× for read lengths of 50–150, respectively, for TopHat2, 7× in the case of MapSplice and 2× in the case of STAR. HISAT runtimes are similar, although a bit slower, except in the case of very short reads (not produces anymore by modern sequencers), in which it is slightly faster than HPG Aligner.
Figure 2.
Representation of mapping sensitivity (percentage of reads correctly mapped) versus runtimes (in min) for simulated datasets of different single read lengths (from 50 bp, upper left, to 400 bp, lower right) containing 10 million single-end reads in two scenarios of variability (low variability represented by circles and high variability represented by triangles). The colour code corresponds to the different programs. In all the cases, HPG Aligner sensibilities and runtimes were better than the ones shown by the rest of programs (Supplementary Table S1). These differences increase as read length and error increase.
Table 1.
Benchmarking for the simulated dataset containing 10 million single-end reads simulated with the BEERS program
| RL | MR (%) | HPG Aligner 1 |
HISAT |
STAR 2 |
TopHat 2 + Bowtie 2 |
MapSplice 2 |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IMR | RNM | CMR | T | IMR | RNM | CMR | T | IMR | RNM | CMR | T | IMR | RNM | CMR | T | IMR | RNM | CMR | T | ||
| 0.1 | 3.08 | 0.42 | 96.5 | 4.8 | 4.42 | 3.22 | 92.36 | 4.45 | 7.32 | 1.46 | 91.22 | 8.28 | 2.05 | 0.93 | 97.02 | 36.35 | 2.82 | 1.16 | 96.02 | 26.93 | |
| 50 | 1 | 4.26 | 0.84 | 94.9 | 4.9 | 9.79 | 8.61 | 81.60 | 4.80 | 11.23 | 1.72 | 87.05 | 8.73 | 2.06 | 3.04 | 94.90 | 79.65 | 2.95 | 2.03 | 95.02 | 26.13 |
| 2 | 5.82 | 2.26 | 91.92 | 5.2 | 16.29 | 17.18 | 66.53 | 4.58 | 15.44 | 3.16 | 81.40 | 8.85 | 2.00 | 10.18 | 87.82 | 374.9 | 3.21 | 4.81 | 91.98 | 26.08 | |
| 0.1 | 3.51 | 0.24 | 96.25 | 5.8 | 2.30 | 1.13 | 96.57 | 6.21 | 7.81 | 0.46 | 91.73 | 9.52 | 0.28 | 2.98 | 96.74 | 48.22 | 3.31 | 0.24 | 96.45 | 37.32 | |
| 75 | 1 | 4.17 | 0.94 | 94.89 | 6.0 | 4.64 | 3.48 | 91.88 | 6.40 | 11.82 | 1.18 | 87.00 | 9.97 | 1.24 | 6.90 | 91.86 | 50.78 | 3.59 | 1.16 | 95.25 | 35.05 |
| 2 | 5.64 | 1.27 | 93.09 | 6.4 | 10.59 | 10.46 | 78.95 | 6.46 | 16.62 | 1.55 | 81.83 | 11.25 | 0.94 | 22.18 | 76.88 | 58.93 | 5.13 | 2.20 | 92.67 | 34.28 | |
| 0.1 | 4.07 | 0.24 | 95.69 | 7.4 | 2.57 | 1.24 | 96.19 | 7.86 | 7.59 | 0.64 | 91.77 | 11.77 | 1.69 | 1.94 | 96.37 | 65.08 | 3.54 | 0.26 | 96.20 | 51.17 | |
| 100 | 1 | 4.95 | 0.55 | 94.50 | 7.5 | 5.70 | 4.51 | 89.79 | 8.15 | 11.90 | 0.62 | 87.48 | 12.08 | 0.99 | 11.70 | 87.31 | 74.02 | 3.98 | 0.68 | 95.34 | 46.03 |
| 2 | 6.07 | 0.93 | 93.00 | 7.8 | 15.64 | 17.19 | 67.17 | 8.40 | 16.48 | 1.14 | 82.38 | 12.35 | 0.60 | 36.14 | 63.26 | 85.65 | 6.45 | 1.50 | 92.05 | 45.6 | |
| 0.1 | 5.58 | 0.31 | 94.11 | 10.5 | 3.15 | 1.56 | 95.29 | 11.75 | 8.40 | 0.57 | 91.03 | 15.07 | 1.49 | 3.55 | 94.96 | 97.9 | 5.45 | 0.21 | 94.34 | 56.88 | |
| 150 | 1 | 7.77 | 1.95 | 90.28 | 10.6 | 12.49 | 13.03 | 74.48 | 11.41 | 12.88 | 2.73 | 84.39 | 15.08 | 0.82 | 27.13 | 72.05 | 121.55 | 8.23 | 2.67 | 89.10 | 53.53 |
| 2 | 7.06 | 0.78 | 92.16 | 10.9 | 24.42 | 39.76 | 35.82 | 11.91 | 15.97 | 0.73 | 83.30 | 15.73 | 0.45 | 61.84 | 37.71 | 140.28 | 13.65 | 1.57 | 84.78 | 56.73 | |
| 0.1 | 8.21 | 0.66 | 91.13 | 16.55 | 4.49 | 2.47 | 93.04 | 17.80 | 9.13 | 0.62 | 90.25 | 21.58 | 1.62 | 6.70 | 91.68 | 185.78 | 11.51 | 0.23 | 88.26 | 75.55 | |
| 250 | 1 | 8.43 | 0.81 | 90.76 | 16.5 | 22.28 | 31.6 | 46.12 | 19.28 | 12.99 | 0.71 | 86.30 | 22.3 | 0.28 | 53.29 | 46.43 | 256.83 | 29.85 | 0.75 | 69.40 | 79.28 |
| 2 | 8.72 | 1.62 | 89.66 | 17.45 | 17.75 | 77.04 | 5.21 | 17.46 | 18.35 | 1.43 | 80.22 | 23.35 | 0.07 | 89.62 | 10.31 | 309.85 | 35.27 | 1.24 | 63.49 | 76.37 | |
| 0.1 | 10.02 | 1.08 | 88.90 | 25.45 | 6.88 | 5.04 | 88.08 | 28.5 | 9.98 | 0.56 | 89.46 | 32.12 | 0.99 | 12.32 | 86.69 | 369.58 | 17.33 | 0.03 | 82.64 | 106.67 | |
| 400 | 1 | 12.94 | 3.35 | 83.71 | 25.6 | 22.52 | 66.17 | 11.31 | 29.7 | 14.82 | 4.26 | 80.92 | 33.83 | 0.09 | 82.40 | 17.51 | 573.18 | 51.64 | 1.65 | 46.71 | 103.98 |
| 2 | 11.78 | 2.84 | 85.38 | 26.0 | 3.08 | 96.82 | 0.10 | 27.10 | 32.75 | 2.75 | 64.5 | 24.5 | 0.01 | 99.02 | 0.97 | 603.28 | 54.34 | 0.51 | 45.15 | 106.53 | |
First column, RL, indicated read length in bp. Second column represents the mutation rate (MR). For each program, the table contains the following columns with the percentages of: IMR: incorrectly mapped reads; RNM: reads not mapped; CMR: correctly mapped reads covering the corresponding splice junctions. The last column, T, represents runtimes for producing a BAM file in min.
We repeated the simulations in the same conditions with the dwgsim simulator (see the Methods section). The results obtained in the transcriptomics simulated scenario provided by dwgsim are similar although the percentages of correct mapping decrease for all the programs across the wide range of lengths (Supplementary Table S1). In particular, the sensitivity of TopHat2 notably decreases in the low variability (0.1%). As noise and read length increase, the sensitivity falls down again. One of the reasons for this general decrease in the sensitivity can be the fact that dwgsim has an extra parameter that accounts for the error of the sequencing technology, which introduces an extra amount of mismatches per read. As expected, the results obtained in the case of paired reads were quite similar (Supplementary Table S2).
Actually, sequencing technologies are progressively increasing the read length (currently, HiSeq 2500 or HiSeq 1500 in rapid run mode produces read lengths of 150 bp, 454 GS FLX+ Lifesciences average read length is of 700–800 bp and Pacific Biosciences is over 3 Kb). Therefore, a desirable property for aligners is robustness against increasingly large sequence lengths.
When the five aligners are tested against a real dataset of 20 million reads, 100 bp long, from an Illumina HiSeq (SRA accession SRR364003), HPG Aligner was able to map at known transcript positions almost 80% of the reads, whereas HISAT and MapSplice reached ∼76%, STAR 72% and TopHat2 only reached 63%. In terms of runtimes, HPG Aligner, HISAT and STAR are >10 times faster than TopHat2 and MapSplice (∼10 versus over 100 min; Table 2). When a dataset with longer reads, obtained with 454 GS FLX+ Lifesciences (average 600 bp), was analysed (SRA accession SRP003173), the number of reads mapped by HPG Aligner reduces to ∼50%, while STAR only reached 30%, HISAT and TopHat almost none and MapSplice did not get any result after 3 days running (Table 2).
Table 2.
Real RNA-seq datasets taken from sequence read archive (SRA)
| SRA Study | Run accession | Run description | HPG Aligner |
HISAT |
STAR |
TopHat2 + Bowtie2 |
MapSplice |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RNM | CMR | Time | RNM | CMR | Time | RNM | CMR | Time | RNM | CMR | Time | RNM | CMR | Time | |||
| SRP009262 | SRR364003_1 | Illumina HiSeq 81 M 100 nt (single-end) | 4.53 | 79.72 | 11.0 | 15.16 | 76.10 | 10.43 | 5.50 | 75.00 | 13.9 | 25.73 | 63.38 | 115.9 | 4.63 | 76.83 | 139.9 |
| SRR364003_1 SRR364003_2 |
Illumina HiSeq 81 M 100 nt (paired-end) | 4.90 | 78.20 | 21.6 | 15.00 | 77.8 | 21.46 | 8.00 | 72.02 | 22.7 | 29.26 | 59.9 | 230.6 | 5.04 | 77.5 | 288.1 | |
| SRP003173 | SRR063344 SRR063345 |
Roche 454 GS FLX 1.26 M ∼600 nt (single-end) | 11.40 | 48.04 | 6.9 | 99.87 | 0.10 | 3.45 | 41.6 | 30.25 | 6.6 | 99.97 | 0.02 | 520.6 | NA | NA | NA |
SRA SRP009262 study was sequenced with Illumina HiSeq and contains 81.6 M reads (20 M reads are used for the benchmarking) of 100 nt length. Aligners were run in single and paired-end mode. SRA SRP003173 study was sequenced with Roche 454 GS FLX and contains 1.26 M reads (1,265,460) of ∼600 nt.
For each program, the table contains the following columns with the percentages of: RNM: reads not mapped; CMR: correctly mapped reads (mapped reads that cover the corresponding to known transcript positions). The last column, T, represents runtimes for producing a BAM file in min.
Apart from read length, another important consideration is the continuous increase in the number of reads. Table 3 summarizes the different behaviours of the four aligners tested. HPG Aligner and HISAT clearly render much better runtimes than the rest of programs.
Table 3.
Scalability with the number of reads
| Million reads | HPG Aligner | HISAT | STAR | TopHat2+Bowtie2 | MapSplice |
|---|---|---|---|---|---|
| 2 | 0.9 | 0.95 | 4.5 | 38.7 | 11.8 |
| 5 | 1.9 | 1.7 | 5.6 | 40.5 | 18.65 |
| 10 | 3.8 | 4.2 | 7.75 | 75.2 | 36.4 |
| 20 | 7.6 | 7.28 | 13.5 | 112.2 | 68.5 |
Runtimes for producing a BAM file (in min) of the different aligners in datasets of growing size.
We have also assessed the performance of the method in specifically mapping reads containing splice junctions. Two sets containing 5 million reads 100 bp long, spliced and non-spliced, respectively, were constructed. Runtimes obtained were very similar (220.61 s non-spliced versus 222.45 s spliced). We repeated the simulation with longer reads (250 bp) obtaining a similar result (485.56 s non-spliced versus 488.74 s spliced).
3.2. Other technical advantages
In addition, the implementation presented here has additional advantages. The program can directly read gzipped, FASTQ files saving in this way both disc space and the time required for the decompression. It is also able to directly generate BAM formats, saving the SAM to BAM conversion step. From a technical point of view, the use of memory is highly efficient and can be managed by the user. For example, HPG Aligner only needs 9 GB of RAM in the analysis of the simulations shown in Fig. 2, whereas TopHat2 and STAR require 35 GB. HPC implementation guarantees that software will have a good runtime performance in most modern and future CPUs and GPUs. HPG Aligner scales quite efficiently with the number of cores (threads), which is a desirable property in a scenario in which the deep of sequencing (number of reads sequenced) reveals as a crucial factor to properly measure differential expression.22 Source code and development process have been opened to the community and released in GitHub; biologist and computational researchers are encouraged to use and contribute to HPG Aligner.
3.3. Program availability
HPG Aligner is free and open source. Documentation and software are available at: https://github.com/opencb/hpg-aligner/wiki.
4. Discussion
Transcriptomic studies are nowadays essential for understanding biological mechanisms.30 For most than one decade, expression microarrays have been the dominant technology for transcriptomic analysis. However, in a quick technological transition, RNA-seq is becoming today the mainstream technology. The continuous upgrades of NGS technologies result in an increase of both throughput and read lengths. This trend requires of algorithms able to efficiently and accurately process an increasing number of reads of increasing length. Current algorithms present serious limitations in both aspects, which preclude an optimal use of RNA-seq technologies.
Mapping sequencing reads in the context of transcripts is a problem of much higher complexity than simply mapping reads onto the genomic sequence, as in the case of genome resequencing experiments. Eukaryotic transcriptomes are complex, with an average of more than nine transcripts per gene12 with exons of 250 bp on the average that span over several hundreds of kilobases, and more new transcript isoforms are continuously being discovered.13 Particularly, splices near the ends of reads can be especially difficult to align, given that a minimum amount of sequence is needed to confidently identify exon boundaries.20 Accurate mapping of both short and long reads as well as a precise detection of potential splice junctions are crucial for accurate transcript isoform definition, which is essential for a proper RNA-seq data analysis.
In addition, new strategies to speed up runtimes are needed. Brute-force scaling-up strategies that rely only in the use of more powerful computers are expensive, inefficient and unsustainable. Contrarily, algorithmic solutions that take advantage of the intrinsic parallelism of NGS data and exploit the different possibilities of parallelization in the hardware used (multicore CPUs, SIMD and GPUs) offer a promising solution to the problem of the ever-growing genomic data. Actually, given the trend of increasing the number of cores in the new CPUs, scalability becomes a desirable property for any algorithm that aims to cope with the future growing sequencing datasets.
Recent comparative analysis reported that STAR18 or MapSplice19 performed better than other methods. Here, we have shown how the proposed approach is faster and more accurate than these ones over a wide range of read lengths and errors. Summarizing, here we present a solution for RNA-seq short-read mapping, which is optimal in terms of speed and accuracy alignment. This solution is robust to growing read lengths, it is highly sensitive to indels, and from a technological point of view, it is quite effective in memory management and efficiently scalable to many cores or processors. All these properties make of HPG Aligner, a particularly suitable solution for the analysis of current and future sequencing datasets.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work is supported by grants from the Spanish Ministry of Economy and Competitiveness (BIO2014-57291-R) and co-funded with European Regional Development Funds (ERDF), AECID (D/016099/08) and from the Conselleria d'Educacio of the Valencian Community (PROMETEOII/2014/025). This work has been carried out in the context of the HPC4G initiative (http://www.hpc4g.org) and the Bull-CIPF Chair for Computational Genomics. Funding to pay the Open Access publication charges for this article was provided by grant BIO2014-57291-R from the Spanish Ministry of Economy and Competitiveness (MINECO), co-funded with European Regional Development Funds (ERDF).
Supplementary Material
References
- 1.Garber M., Grabherr M.G., Guttman M., Trapnell C.. 2011, Computational methods for transcriptome annotation and quantification using RNA-seq, Nat. Methods, 8, 469–77. [DOI] [PubMed] [Google Scholar]
- 2.Marioni J.C., Mason C.E., Mane S.M., Stephens M., Gilad Y.. 2008, RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays, Genome Res., 18, 1509–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li H., Homer N.. 2010, A survey of sequence alignment algorithms for next-generation sequencing, Brief Bioinform., 11, 473–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Langmead B., Trapnell C., Pop M., Salzberg S.L.. 2009, Ultrafast and memory-efficient alignment of short DNA sequences to the human genome, Genome Biol., 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li R., Yu C., Li Y. et al. 2009, SOAP2: an improved ultrafast tool for short read alignment, Bioinformatics, 25, 1966–7. [DOI] [PubMed] [Google Scholar]
- 6.Homer N., Merriman B., Nelson S.F.. 2009, BFAST: an alignment tool for large scale genome resequencing, PLoS ONE, 4, e7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H., Durbin R.. 2009, Fast and accurate short read alignment with Burrows-Wheeler transform, Bioinformatics, 25, 1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H., Durbin R.. 2010, Fast and accurate long-read alignment with Burrows-Wheeler transform, Bioinformatics, 26, 589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marco-Sola S., Sammeth M., Guigo R., Ribeca P.. 2012, The GEM mapper: fast, accurate and versatile alignment by filtration, Nat. Methods, 9, 1185–8. [DOI] [PubMed] [Google Scholar]
- 10.Langmead B., Salzberg S.L.. 2012, Fast gapped-read alignment with Bowtie 2, Nat. Methods, 9, 357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tarraga J., Arnau V., Martinez H. et al. 2014, Acceleration of short and long DNA read mapping without loss of accuracy using suffix array, Bioinformatics, 30, 3396–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Birney E., Stamatoyannopoulos J.A., Dutta A. et al. 2007, Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project, Nature, 447, 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ameur A., Wetterbom A., Feuk L., Gyllensten U.. 2010, Global and unbiased detection of splice junctions from RNA-seq data, Genome Biol., 11, R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trapnell C., Pachter L., Salzberg S.L.. 2009, TopHat: discovering splice junctions with RNA-Seq, Bioinformatics, 25, 1105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu T.D., Nacu S.. 2010, Fast and SNP-tolerant detection of complex variants and splicing in short reads, Bioinformatics, 26, 873–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.. 2013, TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions, Genome Biol., 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grant G.R., Farkas M.H., Pizarro A.D. et al. 2011, Comparative analysis of RNA-Seq alignment algorithms and the RNA-Seq unified mapper (RUM), Bioinformatics, 27, 2518–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobin A., Davis C.A., Schlesinger F. et al. 2013, STAR: ultrafast universal RNA-seq aligner, Bioinformatics, 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang K., Singh D., Zeng Z. et al. 2010, MapSplice: accurate mapping of RNA-seq reads for splice junction discovery, Nucleic Acids Res., 38, e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engstrom P.G., Steijger T., Sipos B. et al. 2013, Systematic evaluation of spliced alignment programs for RNA-seq data, Nat. Methods, 10, 1185–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim D., Langmead B., Salzberg S.L.. 2015, HISAT: a fast spliced aligner with low memory requirements, Nat. Methods, 12, 357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tarazona S., Garcia-Alcalde F., Dopazo J., Ferrer A., Conesa A.. 2011, Differential expression in RNA-seq: a matter of depth, Genome Res., 21, 2213–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trapnell C., Roberts A., Goff L. et al. 2012, Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks, Nat. Protoc., 7, 562–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trapnell C., Williams B.A., Pertea G. et al. 2010, Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation, Nat. Biotechnol., 28, 511–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. 1990, Basic local alignment search tool, J. Mol. Biol., 215, 403–10. [DOI] [PubMed] [Google Scholar]
- 26.Salavert Torres J., Blanquer Espert I., Domínguez A. et al. 2012, Using GPUs for the exact alignment of short-read genetic sequences by means of the Burrows-Wheeler transform, IEEE/ACM Trans. Comput. Biol. Bioinform., 9, 1245–56. [DOI] [PubMed] [Google Scholar]
- 27.Alpern B., Carter L., Gatlin K.. 1995, Microparallelism and high-performance protein matching. In: Proceedings of the ACM/IEEE Supercomputing Conference, San Diego, California, December 8, 1995, p. 24. [Google Scholar]
- 28.Martinez H., Tarraga J., Medina I. et al. 2013, EuroMPI ‘13 Proceedings of the 20th European MPI Users’ Group Meeting. In: A Dynamic Pipeline for RNA Sequencing on Multicore Processors, Madrid, Spain, September 15–18, 2013, pp. 235–40. [Google Scholar]
- 29.Flicek P., Ahmed I., Amode M.R. et al. 2013, Ensembl 2013, Nucleic Acids Res., 41, D48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soon W.W., Hariharan M., Snyder M.P.. 2013, High-throughput sequencing for biology and medicine, Mol. Syst. Biol., 9, 640. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.