Abstract
Selection has been a central issue in biology in eukaryotes as well as prokaryotes. Inference of selection in recombining bacterial species, compared with clonal ones, has been a challenge. It is not known how codons under diversifying selection are distributed along the chromosome or among functional categories or how frequently such codons are subject to mutual homologous recombination. Here, we explored these questions by analysing genes present in >90% among 29 genomes of Helicobacter pylori, one of the bacterial species with the highest mutation and recombination rates. By a method for recombining sequences, we identified codons under diversifying selection (dN/dS > 1), which were widely distributed and accounted for ∼0.2% of all the codons of the genome. The codons were enriched in genes of host interaction/cell surface and genome maintenance (DNA replication, recombination, repair, and restriction modification system). The encoded amino acid residues were sometimes found adjacent to critical catalytic/binding residues in protein structures. Furthermore, by estimating the intensity of homologous recombination at a single nucleotide level, we found that these codons appear to be more frequently subject to recombination. We expect that the present study provides a new approach to population genomics of selection in recombining prokaryotes.
Keywords: selection, dN/dS, recombination, population genomics, bacteria
1. Introduction
Selection is one of the most important processes behind evolution of organisms. Inference of selection is made possible by statistical methods that utilize DNA sequence data of multiple individuals.1–4 A common way is to align protein-coding DNA sequences at the codon level to distinguish between non-synonymous (amino acid changing) and synonymous (amino acid maintaining) codon changes. Based on the distinction, the selective pressure at the protein-coding level can be measured by the ratio of non-synonymous substitution rate to synonymous substitution rate: dN/dS = ɷ. dN/dS > 1 is a signal of selection that favours recurrent non-synonymous substitutions (amino acid changes) in the protein or diversifying selection. It has been widely applied to sequences of various species including highly recombining bacteria.5–7
Inference of diversifying selection (here, we define it as selection for amino acid changes that maintains diversity of amino acid residues at a codon) by dN/dS in recombining bacterial genomes is, however, methodologically challenging. A reason is that estimation of dN/dS usually requires a phylogenetic tree that assumes no recombination between sequences. Recombination changes phylogeny and affects estimation of dN/dS, and ignoring recombination causes false positives.8,9 A common way to deal with it is to exclude recombining sequences in advance.7,10 Otherwise, to analyse recombining sequences, there has been only one popular method called ‘omegaMap’, which can infer diversifying selection at the codon level by estimating dN/dS after averaging over all possible phylogenetic trees.11 The method is thus applicable to highly recombining sequences although it is computationally intensive.
Due to this methodological difficulty, an unanswered question is how codons under diversifying selection (with dN/dS > 1) in a highly recombining bacterial species are distributed in a genome physically and functionally. Furthermore, a related and more general unanswered question is whether codons under diversifying selection are more frequently subject to mutual homologous recombination between lineages compared with the other regions in a bacterial genome.
In this study, we explore these two questions in Helicobacter pylori, one of the bacterial species with the highest mutation and recombination rates.12–14 Helicobacter pylori infects the human stomach during childhood and evolves inside each host by mutation and mutual homologous recombination primarily through natural transformation. A previous search for codons under diversifying selection in this organism focused on some parts of the genome.6 We extend this to the entire dataset of all one-to-one orthologous genes and genes present in >90% among 29 genomes of H. pylori. We then infer intensity of homologous recombination at each polymorphic nucleotide site by a recently developed method for the bacterial core genome,15 and explore a potential relationship between diversifying selection and intensity of homologous recombination.
Our genome-wide survey revealed that the codons under diversifying selection were widely distributed and accounted for ∼0.2% in the genome. These codons were enriched in gene categories of host interaction/cell surface and genome maintenance, and appeared to be more frequently subject to mutual homologous recombination.
2. Materials and methods
2.1. Helicobacter pylori genome sequences of global strains and alignment of genes
We used complete genome sequences of 29 H. pylori strains from various regions in the world, which were analysed in our preceding study.16 They were annotated and classified into phylogeographic groups: hpAfrica1, J99, Gambia94; hpEurope, 26695, HPAG1, Lithuania75, P12, G27, B38, B8; hpAsia2, India 7, Santal49 (SNT49); hspAmerind, Puno120, Puno135, Sat464, Shi470, Cuz20, v225d; hspEastAsia, 35A, F57, F30, F16, 83, OK310, 51, 52, F32, OK113. There were also two hybrid strains: SJM180 and PeCan4. We then prepared sequence alignment dataset of each gene (orthologous group). An entire dataset of orthologous genes was prepared based on clustering by DomClust17 and RECOG (http://mbgd.genome.ad.jp/RECOG/).
For each gene (orthologous group), we aligned the nucleotide sequences by PRANK with the ‘codon’ option, which uses a codon substitution matrix.18 PRANK uses evolutionary information in determining where to place gaps. This improves the quality of alignment,19 which has been shown to affect detection of selection considerably.20,21
2.2. Search for codons under diversifying selection
We used the omegaMap11 to infer dN/dS and diversifying selection at the codon level in recombining sequences of all one-to-one orthologous genes and genes present in >90% among 29 genomes of H. pylori. The omegaMap uses a population genetics approximation to the coalescent with recombination, and reversible-jump Markov chain Monte Carlo (MCMC) to perform Bayesian inference on variation of dN/dS ratio along a DNA sequence.11 It does not assume a specific phylogenetic tree, but instead considers averaging over all possible trees based on the Hidden Markov model. Two independent MCMC chains were run for 500,000 iterations with a thinning (sampling) interval of 10 and a burn-in of 10% of the iterations. The average length of a block of dN/dS was set to be 10 codons. We extracted codons carrying non-synonymous polymorphism that satisfied both estimated dN/dS > 1 and a posterior probability of diversifying selection >0.999. We use the stringent criterion because we put priority on decreasing false positives although it in turn increases false negatives. Among 5,374 codons that satisfied these criteria and were located in the reference genome, we excluded those in hypothetical genes with >7% nucleotide diversity (3,629 codons, ∼68%) listed in Supplementary Table S1 because their entire coding region seemed to have decayed. We assessed the convergence of the MCMC chains by the Gelman–Rubin convergence diagnostic (PSRF)22 that is calculated from within-chain and between-chain variance of two independent runs; PSRF <1.2 is usually taken as indicating convergence.23,24 We confirmed that about 76% of the 1,745 codons after the filtering above showed the convergence. We did not include the remaining 24% into codons under diversifying selection. The convergence diagnostic (PSRF) was calculated by the R package CODA25 and EDISON.26
2.3. Functional categorization of each gene
We classified genes based on the functional categories in Microbial Genome Database for Comparative Analysis (MBGD).27,28 We defined five major functional categories for this study as follows: ‘basic cellular functions’ defined as ‘Energy metabolism’, ‘Transport and binding proteins’, ‘Translation’, and ‘Cellular processes’ major categories (except for ‘Cell killing’ and ‘Motility’ subcategories) in MBGD; ‘host interaction/cell surface’ including outer membrane and lipopolysaccharide-related proteins, ‘Cell envelope’, major category, ‘Cell killing’, ‘Motility’, ‘Membranes, lipoproteins, and porins’, ‘Murein sacculus and peptidoglycan’, ‘Surface polysaccharides, lipopolysaccharides and antigens’, and ‘Surface structures’ subcategories; Restriction modification (‘RM’) including the RM system; ‘DNA replication, recombination, and repair’ including ‘DNA replication, restriction, modification, recombination, and repair’ except for the RM system; ‘hypothetical’ as ‘Hypothetical’ and ‘Hypothetical (no functional assignment)’ categories; ‘others’ as all the other genes.
2.4. Protein 3D structure prediction and modelling
For genes carrying a codon under diversifying selection, in order to investigate possible structure–function implication of the residues under diversifying selection (dN/dS > 1), we mapped them together with functionally important residues (e.g. active sites and ligand-binding sites) on the structural models of their protein products. Protein fold recognition (identification of templates with experimentally determined structure) was carried out using the GeneSilico MetaServer.29 Alignments between the sequences of H. pylori proteins and the structures of the best templates identified by different FR methods were used to carry out comparative modelling using the ‘FRankenstein's Monster’ approach,30,31 with the use of MODELLER32 and Swiss-Model33 methods. HP0279, HP0462, HP0790, and HP1553 structures were modelled using crystal structures with the following Protein Data Bank34 codes: 1psw, 1yf2, 1yf2, and 3u4q, respectively.
2.5. Calculation of nucleotide diversity along the genome
We calculated nucleotide diversity of each gene by VariScan version 2.0.35 We also calculated nucleotide diversity at a polymorphic site by sliding windows implemented in VariScan version 2.0.35 We used the sliding windows with step size 1 bp and window size 15 or 30 bp. We checked that the two different sizes of the window showed consistent results. We wrote results that used the windows size of 15 bp, which is almost the same as a previous estimate of median size of chunks of DNA sharing the same ancestry (donor of the most recent recombination) in the H. pylori genome.16
2.6. Estimation of intensity of homologous recombination per nucleotide
We used a recently developed method to infer relative intensity of homologous recombination at a single nucleotide resolution in the bacterial genome.15 As its input, we prepared genome-wide haplotype data by combining single nucleotide polymorphisms (SNPs) without missing data on each orthologous gene while preserving information of SNP positions. The method focuses on genetic elements that have high rates of movement by homologous recombination in a species, and calculates values of the statistic Di that is highly correlated with the number of recombination events or DNA imports at a nucleotide i. The statistic captures deviation from genome average at a nucleotide due to recombination, and the value reflects how frequently DNA is transferred between individuals. It can be calculated for polymorphic nucleotides that do not have missing data.
We calculated a normalized version of Di as a statistic Hi (representing recombination hotness), so that its mean and S.D. become 0 and 1, respectively.36
3. Results
3.1. Genes and codons under diversifying selection
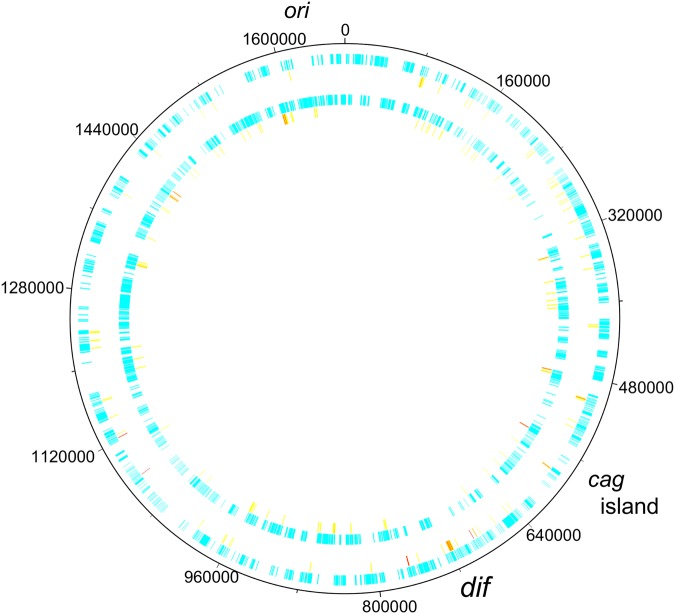
In all one-to-one orthologous genes and genes present in >90% among 29 global strains of H. pylori, we searched for codons under diversifying selection (with dN/dS > 1) by using the omegaMap method developed for recombining sequences.11 As a result, we found codons under diversifying selection in 134 loci. They accounted for ∼11% of the genes. These genes were widely distributed across the genome (Fig. 1, sorted by their positions in Supplementary Table S2). The codons accounted for ∼0.2% in the genome. There was no relationship between distribution of the codons and direction of replication (i.e. ori to dif, or from dif to ori, in which there was no significant difference by the χ2 test). The inferred dN/dS value and probability of diversifying selection are listed for each of the codons in Supplementary Table S3.
Figure 1.
Distribution of codons under diversifying selection in the H. pylori genome. The reference genome of strain 26695 is shown with all the genes. The two set of circles distinguish the plus (outer) and minus (inner) strand, respectively. On each strand, the codons under diversifying selection that allowed estimation of intensity of homologous recombination (that is, no gap in 29 genomes' alignment) are shown in dark (red), while the others are in light (yellow). This figure is available in black and white in print and in colour at DNA Research online.
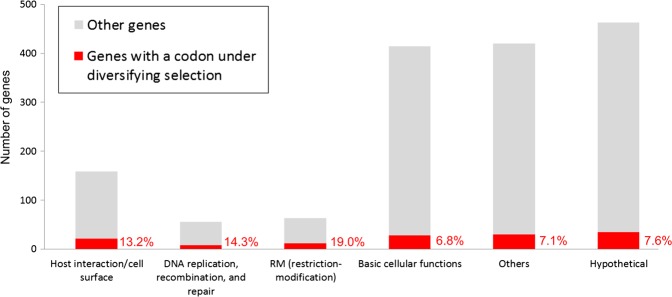
We then analysed some of these genes and codons for biological significance. These codons were accounted for >13% in each of the following three categories (Fig. 2): ‘host interaction/cell surface’, ‘DNA replication, recombination, and repair’, and ‘RM (restriction modification)’, as detailed below. The proportion in ‘host interaction/cell surface’ is significantly higher than other categories (P= 0.01, χ2 test). It is also significantly higher across the other two groups (P= 0.0003, χ2 test), both of which are responsible for the maintenance of bacterial genome and usually classified into a group as in the MBGD database.
Figure 2.
The number and proportion of genes with a codon under diversifying selection in the functional categories. For the categories, see Section 2. This figure is available in black and white in print and in colour at DNA Research online.
3.2. Genes for host interaction/cell surface with a codon under diversifying selection
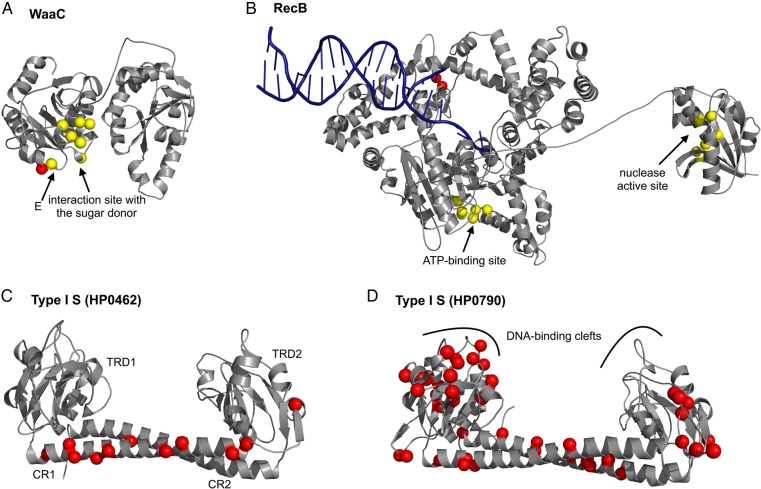
In this category, we examined genes for outer membrane proteins and for lipopolysaccharide synthesis in detail. Of the genes for lipopolysaccharide synthesis, we mapped the selected site of WaaC (HP0279) onto its structural model (Fig. 3A). WaaC catalyses the addition of the first heptose moiety to the inner core of a lipopolysaccharide. The residue (Y229) is located close to the critical glutamic acid residue (‘E’ in Fig. 3A) that forms hydrogen bonds with the pentose ring of the ADP moiety of the sugar donor.37
Figure 3.
Amino acid residues under diversifying selection mapped on structural models. Residues under diversifying selection (dN/dS > 1) in H. pylori proteins are shown as red spheres, and those interacting with any ligand in homologous proteins are shown as yellow spheres. (A) WaaC (HP0279). Red sphere, Y229. (B) RecB (HP1553). Red spheres, L680 and R681. (C) HsdS (Type I S subunit) (HP0462). Red spheres, 153, 158, 159, 163, 166, 177, 180, 188, 190, 198, and 346. (D) HsdS (Type I S subunit) (HP0790). The 44 positions shown in red correspond to the codons under diversifying selection.
Among other genes in this category, amiA (HP0772) produces N-acetylmuramoyl-l-alanine amidase, which is required for modification of the cell wall peptidoglycan, an essential step for morphological transition into the coccoid form.38 tlpA (HP0099) produces a chemotaxis receptor protein for sensing arginine and bicarbonate.39
3.3. Genes for DNA replication, recombination, repair, and RM system with a codon under diversifying selection
Helicase/nuclease/recombinase encoded by addA (=adnA=pcrA) (HP1553) is functionally related to the RecBCD enzyme of Escherichia coli, which degrades non-self DNA of invading genetic elements but repairs self DNA marked by a genome identification sequence called chi.40,41 Large divergence in this gene was previously reported between East Asian and European H. pylori.42 We mapped the residue under diversifying selection onto the structural model of the RecB subunit of RecBCD enzyme (Fig. 3B). The residues (680 and 681, red spheres) are located in the 2A motor domain responsible for ATP-dependent translocation of the protein along DNA in the vicinity of the ATP-binding site.43 We also found a codon with diversifying selection in other components of DNA replication/repair/recombination machinery (gyrA, ruvA, and rnhA).
RM systems consist of a modification enzyme activity that transfers a methyl group to a specific DNA sequence and a restriction enzyme activity that cleaves DNA lacking such methylation. They are responsible for distinction of self DNA from non-self DNA by epigenetic methylation of a specific DNA sequence. They destroy viral DNA or other invading DNA genome of a different epigenome. Their genes behave as mobile genetic elements.44 They frequently change their recognition sequences and their expression, and it is to be expected that codons under diversifying selection were found in such genes. Comparative analyses predicted that H. pylori contains an unusually large number of diverse RM systems with a high degree of heterogeneity.45 However, further biochemical analyses showed that only some of them retain enzymatic activity.46,47
Among the RM systems, type I RM systems consist of R, M, and S (specificity) subunits, encoded by three host specificity determinant (hsd) genes. The S–M complex exhibits the modification activity, while the S–M–R complex exhibits the restriction activity. The restriction enzyme cleaves random DNA between distant target sites. In HP0464, the nine residues under diversifying selection correspond to the C-terminal unclassified domain of predicted alpha-helical structure of R subunit from E. coli pR124.48 The specificity (S) subunit is essential in determining the recognition sequence through its two target recognition domains (TRDs). The TRDs are highly variable even between members of the same family49 and can be ‘swapped’ between related systems to generate novel DNA specificities.50 In addition, the S subunit contains two conserved regions (CRs) in the middle and at the C terminus, that form a long antiparallel a colied-coil.51 The coiled coil acts as a molecular ruler for the separation between two recognized DNA sequences.51 Codons of diversifying selection were found in two S proteins (HP0462 and HP0790) in addition to the R protein (HP0464). The genes, encoding HP0462 and HP0464, are located within the same genomic RM cluster, and possibly act in one complex. Tertiary structures of HP0462 and HP0790 were modelled using the structure of the S subunit from Methanococcus jannaschii51 (PDB: 1YF2) as a template. All but one codons (10/11) of diversifying selection in HP0462 correspond to the alpha-helical CRs (Fig. 3C), in agreement with the known mechanism that the orientation of DNA-binding sites is an essential feature defining the mode of DNA recognition by S proteins.52 In HP0790, the 44 residues under diversifying selection are distributed across each domain: 21 positions are located within TRD1, 4 within CR1, 8 within TRD2, 10 within CR2, and 1 on the C-terminal beta-strand (Fig. 3D). Interestingly, some of the residues mapping to TRD1 are located close to the predicted DNA-binding cleft (P38, K42, N43, E52, K89, Q107, and I141 in HP0790). In contrast to HP0462, sites under diversifying selection in HP0790 are scattered throughout the entire protein. We hypothesize that this is due to the fact that HP0790 is not co-expressed with other RM components, and thus might be subjected to stronger evolutionary pressure that influences not only positioning of DNA-binding clefts, but also their function.
Unlike the type I RM systems, type II R and M subunits act independently, and therefore must independently recognize the same target sequence. Therefore, they may not be able to rapidly alter their DNA sequence specificity: even a small change in a sequence recognized by the R subunit, without an equivalent change in the M subunit, specificity would result in host toxicity. Among them, HP0668 (putative R subunit) and mHP0669 (putative M subunit) have unusually a high number of codons with diversifying selection: 124 and 337, respectively. However, transcriptome annotations for H. pylori indicate that only mHP0669 is expressed.53 Therefore, despite the conservation of the residues from a potential methyltransferase active site in mHP0669, both genes might be in the process of pseudogenization.
3.4. Other genes with a codon under diversifying selection
We noted several genes related to virulence and host colonization in the lists. These include previously reported cagPAI genes (cagC and cagY in addition to the most famous cagA6), flagella-related genes, and poly E-rich protein. We earlier identified several such codons in cagA.54 They also included a transcription factor (rpoD), genes for chemotaxis, metabolism of amino acids, and nucleotides. We do not know whether they represent their adaptation to nutritional environments. The hemA (HP0239) product catalyses the first step in the biosynthesis of haem and copes with oxidative DNA damage.55 ackA encoding acetate kinase decayed in several strains in different geographical regions.42 Their non-synonymous mutations may have inactivated those genes to advantage.
3.5. Intensity of homologous recombination in nucleotides under diversifying selection
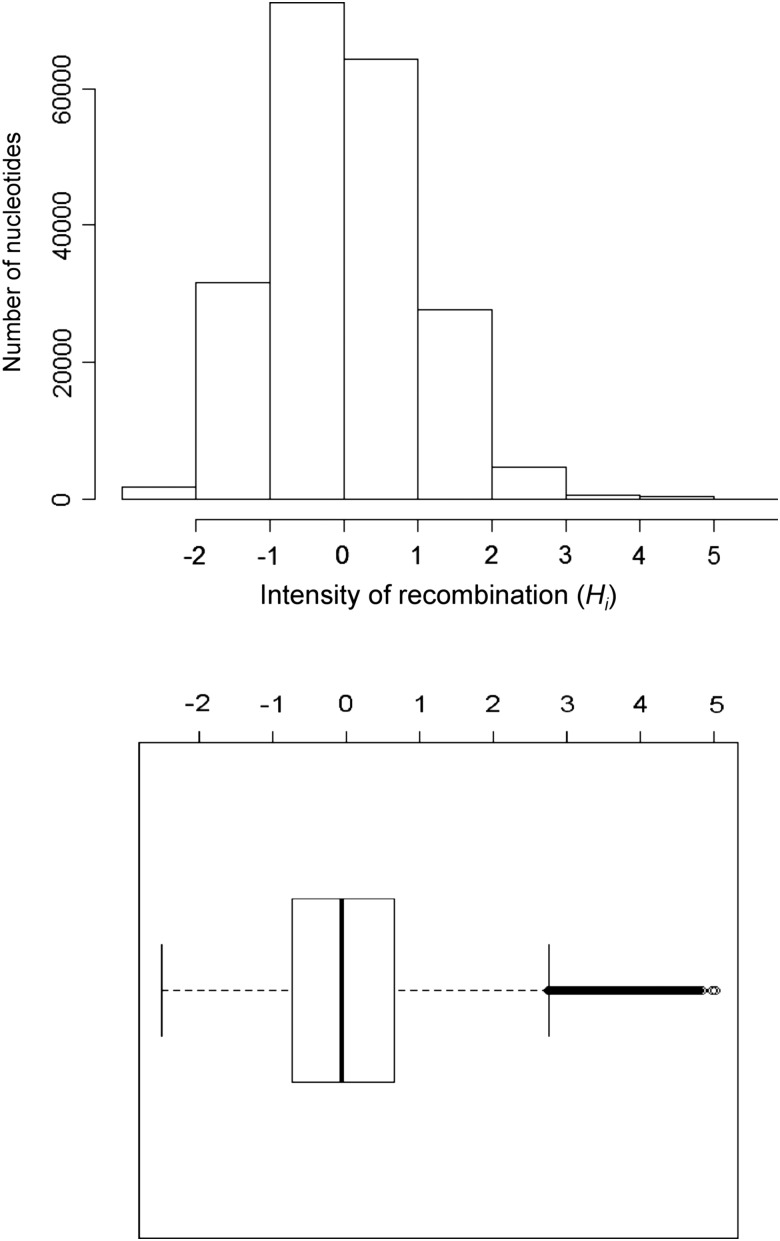
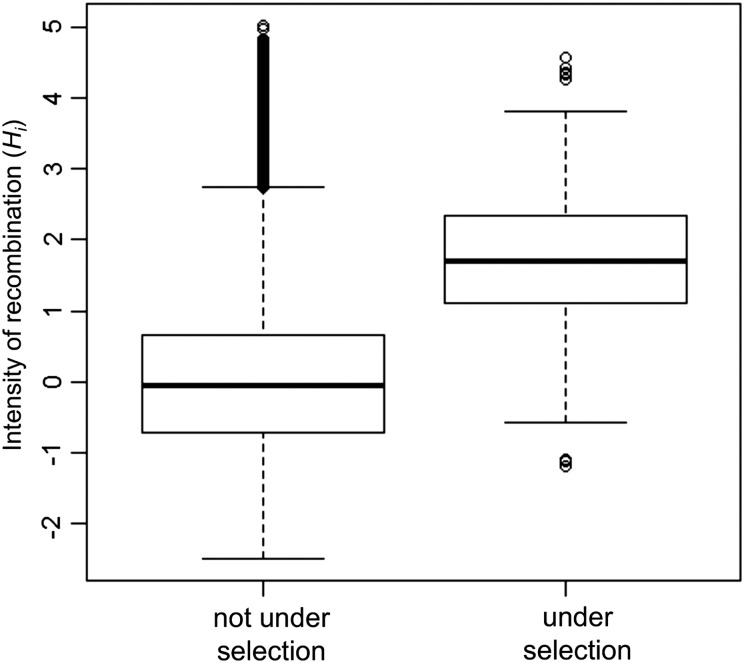
Next, we estimated the intensity of homologous recombination at each polymorphic nucleotide by a statistic Hi representing recombination hotness at nucleotide i. This is a normalized version of statistic Di, which well correlates with the number of homologous recombination events or DNA imports covering the nucleotide.15 The average and standard deviation of Hi is normalized to be 0 and 1 in a genome, and its high values indicate recombination hot nucleotides in the genome that are apparently frequently transferred among individual lineages as a result of homologous recombination. We found that distribution of the Hi values in the H. pylori genome was almost normal but had some outliers, which can be regarded as the recombination hot nucleotides (Fig. 4). Comparison of the intensity of homologous recombination between nucleotides under diversifying selection (red in Fig. 1) and others not under diversifying selection clearly showed elevation in the former (Fig. 5). The median value is 2.20 in the codons under diversifying selection and −0.05 in the other codons (P < 10−6, Wilcoxon's rank sum test).
Figure 4.
Distribution of estimated intensity of homologous recombination per nucleotide (Hi) in the H. pylori genome. In the box plot at the bottom, a bold vertical line indicates median, while the left and right of a box indicate 25th and 75th percentiles, respectively. The left whisker indicates the lowest value within the 25th percentile − 1.5 interquartile range, whereas right whisker indicates the highest value within the 75th percentile+1.5 interquartile range.
Figure 5.
Relation between diversifying selection and estimated intensity of homologous recombination. Intensity of homologous recombination in the nucleotides under diversifying selection compared with those not under diversifying selection.
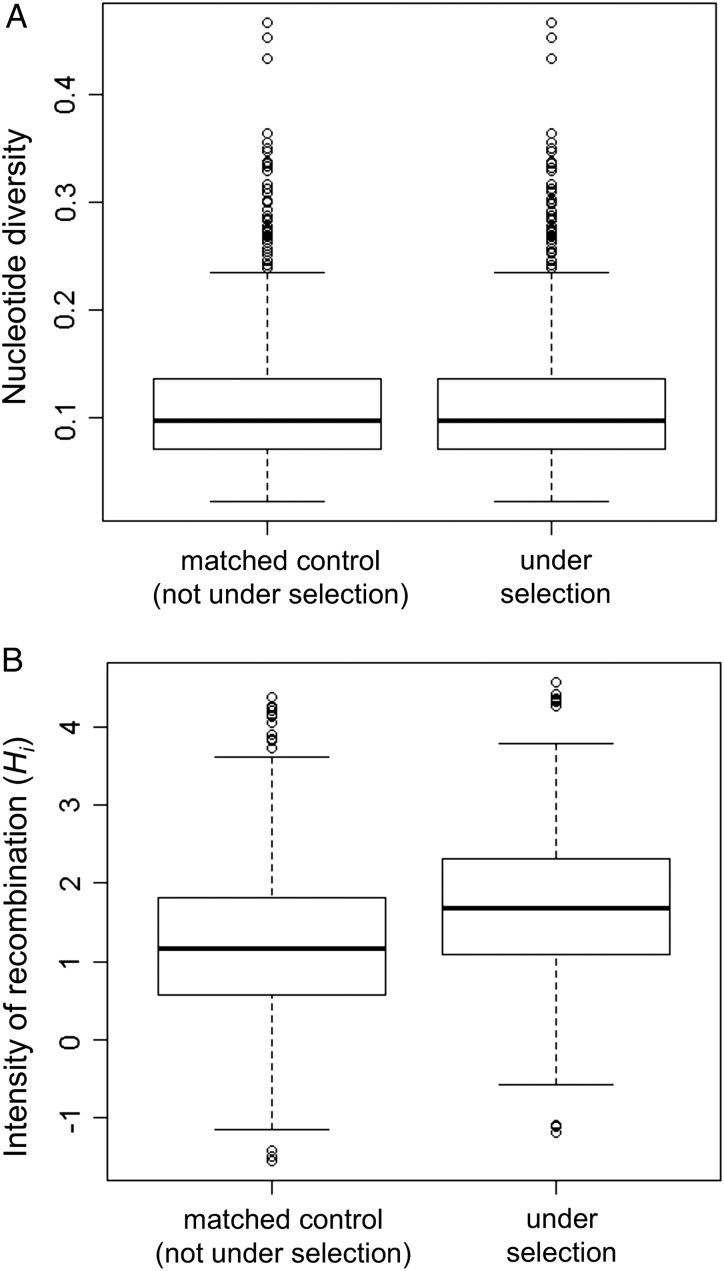
However, the estimated intensity of homologous recombination was positively correlated with nucleotide diversity in the genome (Supplementary Fig. S1) as reported previously.15 In addition, nucleotide diversity was higher in the codons under diversifying selection (Supplementary Fig. S2, P< 10−15, Wilcoxon rank sum test). Therefore, nucleotide diversity is a confounding factor, which is positively correlated with both diversifying selection and the intensity of homologous recombination estimated from the sequence data. We thus took a matching-based approach in which for each ‘case’ nucleotide that is under diversifying selection and allowed estimation of intensity of recombination (Hi), we chose a ‘control’ nucleotide that is not under diversifying selection and has the closest level of nucleotide diversity. We confirmed that there is almost no difference in nucleotide diversity between them (Fig. 6A), and examined difference in the intensity of recombination. As a result, we found that the intensity of recombination is significantly higher in the nucleotides under diversifying selection (Fig. 6B, P < 10−13).
Figure 6.
Comparison between matched control nucleotides and nucleotides under diversifying selection. (A) Nucleotide diversity. (B) Intensity of homologous recombination.
4. Discussion
In this study, we first conducted a genome-wide survey of genes with a codon under diversifying selection by utilizing the omegaMap method11 developed for recombining sequences. Although the method is computationally intensive, we conducted the genome-wide survey for the 29 genomes in various geographical regions by using super computers. It succeeded in identifying ∼0.2% of codons in the genome as under diversifying selection, reflecting high genetic diversity of the H. pylori genomes.
The genes carrying a codon under diversifying selection were statistically enriched in the ‘host interaction/cell surface’ category. Diversifying selection on outer membrane protein genes was reported for six strains of E. coli and Shigella flexneri.7 Diversifying selection on a gene (rfaC) involved in lipopolysaccharide biosynthesis was reported in the same study. They are likely to change bacterial interaction with the host.56
Diversifying selection in RM genes may reflect the evolutionary combat with viruses and other invading DNA elements consistent with the Red Queen model.57 Diversifying selection of the methyltransferases will cause changes in bacterial methylome, which would affect global gene expression and phenotype and might change interaction with their host. RM systems indeed frequently change their presence/absence, their sequence specificity, and their expression, which affect gene expression.44,58,59
Recently, a study explored signatures of selective sweep in each of several geographical subpopulations.60 Our study is complementary to that analysis, because we explored signatures of diversifying selection that are maintained across different strains of H. pylori. In addition, our analyses are codon level rather than gene level, and have higher resolution in identifying signatures of selection across a genome.
Next, based on the estimation of intensity of homologous recombination at the nucleotide level by the recently developed method,15 we revealed that the codons under diversifying selection could be more frequently transferred by recombination between individual genomes in this bacterial species. This sheds a new light on evolutionary roles of bacterial recombination.61 It would be consistent with the Red Queen-like selection regime62 in interaction with the host, in which recombination plays an advantageous role by bringing an unusual allele into a population and promoting diversification of the recombined region.
Some preceding studies reported association between diversifying selection and recombination in bacterial genomes.5,63,64 Our studies have three major differences from these works. First, they used the PAML for inference of diversifying selection, which is not applicable to recombining sequences. Second, they examined the association at gene level rather than codon level. Third, they did not mention and account for the confounding effect of nucleotide diversity. They listed genes with signs of recombination and those under diversifying selection, and reported the proportion of overlap between them.
To our knowledge, this is the first study that revealed distribution and proportion of the codons under diversifying selection across a genome of a highly recombining bacterial species. We also showed that the codons under diversifying selection could be more frequently subject to recombination. Our approach and the results provide a new direction of population genomic studies of selection in recombining prokaryotes.
5. Availability
The genome sequences of the H. pylori strains [J99, Gambia94, 26695, HPAG1, Lithuania75, P12, G27, B38, B8, India 7, Santal49 (SNT49), Puno120, Puno135, Sat464, Shi470, Cuz20, v225d, 35A, F57, F30, F16, 83, OK310, 51, 52, F32, OK113, SJM180, and PeCan4] are available in the public database.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by ‘Grant-in-Aid for Scientific Research’ from MEXT (15K21554 to K.Y. and 25291080 to I.K.), Ishibashi Foundation grant (to K.Y.), ‘Grant-in-Aid for Scientific Research on Innovative Areas’ from MEXT (24113506 and 26113704, to I.K.), Grant in Promotion of Basic Research Activities for Innovative Biosciences (grant no. 121205003001002100019) from Bio-oriented Technology Research Advance Institution to I.K., The Science and technology research promotion program for agriculture, forestry, fisheries and food industry (grant no. 26025A) from MAFF (Ministry of Agriculture, Forestry, and Fisheries) to I.K. and the statutory funds of the IIMCB (to J.M.B.) and the REGPOT Grant FishMed from the European Commission (316125 to Jacek Kuźnicki in IIMCB). S.D.-H. was supported by a fellowship for outstanding young scientists from the Polish Ministry of Science and Higher Education and by the Polish National Science Centre (grant 2011/03/D/NZ8/03011). Funding to pay the Open Access publication charges for this article was provided by Kakenhi (Correlative gene system: establishing next-generation genetics; 26113704) from MEXT (Ministry of Education, Culture, Sports, Science & Technology) in Japan.
Supplementary Material
Acknowledgements
We are grateful for Dr Kenji Kojima for comments. We thank Dr Hideki Innan for discussion. The computational calculations were done at the Institute of Medical Science (the University of Tokyo), the HPCI system provided by the Institute of Statistical Mathematics through the HPCI System Research Project (Project ID: hpci002244), and HPC Wales.
References
- 1.Sabeti P.C., Schaffner S.F., Fry B. et al. 2006, Positive natural selection in the human lineage, Science, 312, 1614–20. [DOI] [PubMed] [Google Scholar]
- 2.Karlsson E.K., Kwiatkowski D.P., Sabeti P.C.. 2014, Natural selection and infectious disease in human populations, Nat. Rev. Genet., 15, 379–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kosiol C., Anisimova M.. 2012, Selection on the protein-coding genome, In: Anisimova M. (ed.), Evolutionary Genomics. Humana Press, c/o Springer Science + Business Media: New York, pp. 113–40. [DOI] [PubMed] [Google Scholar]
- 4.Shapiro B.J., Alm E.J.. 2008, Comparing patterns of natural selection across species using selective signatures, PLoS Genet., 4, e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lefebure T., Stanhope M.J.. 2007, Evolution of the core and pan-genome of Streptococcus: positive selection, recombination, and genome composition, Genome Biol., 8, R71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olbermann P., Josenhans C., Moodley Y. et al. 2010, A global overview of the genetic and functional diversity in the Helicobacter pylori cag pathogenicity island, PLoS Genet., 6, e1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petersen L., Bollback J.P., Dimmic M., Hubisz M., Nielsen R.. 2007, Genes under positive selection in Escherichia coli, Genome Res., 17, 1336–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anisimova M., Nielsen R., Yang Z.. 2003, Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites, Genetics, 164, 1229–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shriner D., Nickle D.C., Jensen M.A., Mullins J.I.. 2003, Potential impact of recombination on sitewise approaches for detecting positive natural selection, Genet. Res., 81, 115–21. [DOI] [PubMed] [Google Scholar]
- 10.Su F., Ou H.Y., Tao F., Tang H., Xu P.. 2013, PSP: rapid identification of orthologous coding genes under positive selection across multiple closely related prokaryotic genomes, BMC Genomics, 14, 924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson D.J., McVean G.. 2006, Estimating diversifying selection and functional constraint in the presence of recombination, Genetics, 172, 1411–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Didelot X., Bowden R., Wilson D.J., Peto T.E., Crook D.W.. 2012, Transforming clinical microbiology with bacterial genome sequencing, Nat. Rev. Genet., 13, 601–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suerbaum S., Josenhans C.. 2007, Helicobacter pylori evolution and phenotypic diversification in a changing host, Nat. Rev. Microbiol., 5, 441–52. [DOI] [PubMed] [Google Scholar]
- 14.Kennemann L., Didelot X., Aebischer T. et al. 2011, Helicobacter pylori genome evolution during human infection, Proc. Natl. Acad. Sci. USA, 108, 5033–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yahara K., Didelot X., Ansari M.A., Sheppard S.K., Falush D.. 2014, Efficient inference of recombination hot regions in bacterial genomes, Mol. Biol. Evol., 31, 1593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yahara K., Furuta Y., Oshima K. et al. 2013, Chromosome painting in silico in a bacterial species reveals fine population structure, Mol. Biol. Evol., 30, 1454–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uchiyama I. 2006, Hierarchical clustering algorithm for comprehensive orthologous-domain classification in multiple genomes, Nucleic Acids Res., 34, 647–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kosiol C., Holmes I., Goldman N.. 2007, An empirical codon model for protein sequence evolution, Mol. Biol. Evol., 24, 1464–79. [DOI] [PubMed] [Google Scholar]
- 19.Loytynoja A., Goldman N.. 2008, Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis, Science, 320, 1632–5. [DOI] [PubMed] [Google Scholar]
- 20.Jordan G., Goldman N.. 2012, The effects of alignment error and alignment filtering on the sitewise detection of positive selection, Mol. Biol. Evol., 29, 1125–39. [DOI] [PubMed] [Google Scholar]
- 21.Fletcher W., Yang Z.. 2010, The effect of insertions, deletions, and alignment errors on the branch-site test of positive selection, Mol. Biol. Evol., 27, 2257–67. [DOI] [PubMed] [Google Scholar]
- 22.Gelman A., Rubin D.B.. 1992, Inference from iterative simulation using multiple sequences, Stat. Sci., 7, 457–511. [Google Scholar]
- 23.Congdon P.D. 2010, Applied Bayesian Hierarchical Methods. Chapman and Hall/CRC: Boca Raton, FL. [Google Scholar]
- 24.Brooks S.P., Gelman A.. 1998, General methods for monitoring convergence of iterative simulations, J. Comput. Graph. Stat., 7, 434–55. [Google Scholar]
- 25.Plummer M., Best N., Cowles K., Vines K.. 2006, CODA: convergence diagnosis and output analysis for MCMC, R News, 6, 7–11. [Google Scholar]
- 26.Dondelinger F., Lèbre S., Husmeier D.. 2013, Non-homogeneous dynamic Bayesian networks with Bayesian regularization for inferring gene regulatory networks with gradually time-varying structure, Mach. Learn., 90, 191–230. [Google Scholar]
- 27.Uchiyama I., Mihara M., Nishide H., Chiba H.. 2014, MBGD update 2015: microbial genome database for flexible ortholog analysis utilizing a diverse set of genomic data, Nucleic Acids Res., 43, D270–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uchiyama I. 2003, MBGD: microbial genome database for comparative analysis, Nucleic Acids Res., 31, 58–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurowski M.A., Bujnicki J.M.. 2003, GeneSilico protein structure prediction meta-server, Nucleic Acids Res., 31, 3305–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kosinski J., Cymerman I.A., Feder M., Kurowski M.A., Sasin J.M., Bujnicki J.M.. 2003, A ‘FRankenstein's monster’ approach to comparative modeling: merging the finest fragments of Fold-Recognition models and iterative model refinement aided by 3D structure evaluation, Proteins, 53(Suppl 6), 369–79. [DOI] [PubMed] [Google Scholar]
- 31.Kosinski J., Gajda M.J., Cymerman I.A. et al. 2005, FRankenstein becomes a cyborg: the automatic recombination and realignment of fold recognition models in CASP6, Proteins, 61(Suppl 7), 106–13. [DOI] [PubMed] [Google Scholar]
- 32.Sali A., Blundell T.L.. 1993, Comparative protein modelling by satisfaction of spatial restraints, J. Mol. Biol., 234, 779–815. [DOI] [PubMed] [Google Scholar]
- 33.Bordoli L., Kiefer F., Arnold K., Benkert P., Battey J., Schwede T.. 2009, Protein structure homology modeling using SWISS-MODEL workspace, Nat. Protoc., 4, 1–13. [DOI] [PubMed] [Google Scholar]
- 34.Berman H.M., Westbrook J., Feng Z. et al. 2000, The Protein Data Bank, Nucleic Acids Res., 28, 235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hutter S., Vilella A.J., Rozas J.. 2006, Genome-wide DNA polymorphism analyses using VariScan, BMC Bioinformatics, 7, 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yahara K., Didelot X., Jolley K.A. et al. in press, The landscape of realized homologous recombination in pathogenic bacteria, Mol. Biol. Evol., 33, 456–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grizot S., Salem M., Vongsouthi V. et al. 2006, Structure of the Escherichia coli heptosyltransferase WaaC: binary complexes with ADP and ADP-2-deoxy-2-fluoro heptose, J. Mol. Biol., 363, 383–94. [DOI] [PubMed] [Google Scholar]
- 38.Chaput C., Ecobichon C., Cayet N. et al. 2006, Role of AmiA in the morphological transition of Helicobacter pylori and in immune escape, PLoS Pathog., 2, e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cerda O., Rivas A., Toledo H.. 2003, Helicobacter pylori strain ATCC700392 encodes a methyl-accepting chemotaxis receptor protein (MCP) for arginine and sodium bicarbonate, FEMS Microbiol. Lett., 224, 175–81. [DOI] [PubMed] [Google Scholar]
- 40.Amundsen S.K., Fero J., Hansen L.M. et al. 2008, Helicobacter pylori AddAB helicase-nuclease and RecA promote recombination-related DNA repair and survival during stomach colonization, Mol. Microbiol., 69, 994–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Handa N., Yang L., Dillingham M.S., Kobayashi I., Wigley D.B., Kowalczykowski S.C.. 2012, Molecular determinants responsible for recognition of the single-stranded DNA regulatory sequence, chi, by RecBCD enzyme, Proc. Natl. Acad. Sci. USA, 109, 8901–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawai M., Furuta Y., Yahara K. et al. 2011, Evolution in an oncogenic bacterial species with extreme genome plasticity: Helicobacter pylori East Asian genomes, BMC Microbiol., 11, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singleton M.R., Dillingham M.S., Gaudier M., Kowalczykowski S.C., Wigley D.B.. 2004, Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks, Nature, 432, 187–93. [DOI] [PubMed] [Google Scholar]
- 44.Furuta Y., Kobayashi I.. 2013, Restriction-modification systems as mobile epigenetic elements, In: Roberts A., Mullany P. (eds.), Bacterial Integrative Mobile Genetic Elements, Landes Bioscience, Austin, Texas, pp. 85–103. [Google Scholar]
- 45.Nobusato A., Uchiyama I., Kobayashi I.. 2000, Diversity of restriction-modification gene homologues in Helicobacter pylori, Gene, 259, 89–98. [DOI] [PubMed] [Google Scholar]
- 46.Lin L.F., Posfai J., Roberts R.J., Kong H.. 2001, Comparative genomics of the restriction-modification systems in Helicobacter pylori, Proc. Natl. Acad. Sci. USA, 98, 2740–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kong H., Lin L.F., Porter N. et al. 2000, Functional analysis of putative restriction-modification system genes in the Helicobacter pylori J99 genome, Nucleic Acids Res., 28, 3216–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lapkouski M., Panjikar S., Janscak P. et al. 2009, Structure of the motor subunit of type I restriction-modification complex EcoR124I, Nat. Struct. Mol. Biol., 16, 94–5. [DOI] [PubMed] [Google Scholar]
- 49.Obarska A., Blundell A., Feder M. et al. 2006, Structural model for the multisubunit Type IC restriction-modification DNA methyltransferase M.EcoR124I in complex with DNA, Nucleic Acids Res., 34, 1992–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bickle T.A., Kruger D.H.. 1993, Biology of DNA restriction, Microbiol. Rev., 57, 434–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim J.S., DeGiovanni A., Jancarik J. et al. 2005, Crystal structure of DNA sequence specificity subunit of a type I restriction-modification enzyme and its functional implications, Proc. Natl. Acad. Sci. USA, 102, 3248–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Price C., Lingner J., Bickle T.A., Firman K., Glover S.W.. 1989, Basis for changes in DNA recognition by the EcoR124 and EcoR124/3 type I DNA restriction and modification enzymes, J. Mol. Biol., 205, 115–25. [DOI] [PubMed] [Google Scholar]
- 53.Sharma C.M., Hoffmann S., Darfeuille F. et al. 2010, The primary transcriptome of the major human pathogen Helicobacter pylori, Nature, 464, 250–5. [DOI] [PubMed] [Google Scholar]
- 54.Furuta Y., Yahara K., Hatakeyama M., Kobayashi I.. 2011, Evolution of cagA oncogene of Helicobacter pylori through recombination, PLoS One, 6, e23499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elgrably-Weiss M., Park S., Schlosser-Silverman E., Rosenshine I., Imlay J., Altuvia S.. 2002, A Salmonella enterica serovar typhimurium hemA mutant is highly susceptible to oxidative DNA damage, J. Bacteriol., 184, 3774–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kennemann L., Brenneke B., Andres S. et al. 2012, In vivo sequence variation in HopZ, a phase-variable outer membrane protein of Helicobacter pylori, Infect. Immun., 80, 4364–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kobayashi I. 2001, Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution, Nucleic Acids Res., 29, 3742–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Furuta Y., Namba-Fukuyo H., Shibata T.F. et al. 2014, Methylome diversification through changes in DNA methyltransferase sequence specificity, PLoS Genet., 10, e1004272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mruk I., Kobayashi I.. 2014, To be or not to be: regulation of restriction-modification systems and other toxin-antitoxin systems, Nucleic Acids Res., 42, 70–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Montano V., Didelot X., Foll M. et al. 2015, Worldwide population structure, long-term demography, and local adaptation of Helicobacter pylori, Genetics, 200, 947–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Michod R.E., Levin B.R.. 1988, The Evolution of Sex: An Examination of Current Ideas. Sinauer: Sunderland, MA. [Google Scholar]
- 62.Hamilton W.D. 1990, Sexual reproduction as an adaptation to resist parasites (a review), Proc. Natl. Acad. Sci. USA, 87, 3566–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Joseph S.J., Didelot X., Gandhi K., Dean D., Read T.D.. 2011, Interplay of recombination and selection in the genomes of Chlamydia trachomatis, Biol. Direct., 6, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Orsi R.H., Sun Q., Wiedmann M.. 2008, Genome-wide analyses reveal lineage specific contributions of positive selection and recombination to the evolution of Listeria monocytogenes, BMC Evol. Biol., 8, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.