Abstract
Congenital hemangioma is a rare vascular tumor that forms in utero. Postnatally, the tumor either involutes quickly (i.e., rapidly involuting congenital hemangioma [RICH]) or partially regresses and stabilizes (i.e., non-involuting congenital hemangioma [NICH]). We hypothesized that congenital hemangiomas arise due to somatic mutation and performed massively parallel mRNA sequencing on affected tissue from eight participants. We identified mutually exclusive, mosaic missense mutations that alter glutamine at amino acid 209 (Glu209) in GNAQ or GNA11 in all tested samples, at variant allele frequencies (VAF) ranging from 3% to 33%. We verified the presence of the mutations in genomic DNA using a combination of molecular inversion probe sequencing (MIP-seq) and digital droplet PCR (ddPCR). The Glu209 GNAQ and GNA11 missense variants we identified are common in uveal melanoma and have been shown to constitutively activate MAPK and/or YAP signaling. When we screened additional archival formalin-fixed paraffin-embedded (FFPE) congenital cutaneous and hepatic hemangiomas, 4/8 had GNAQ or GNA11 Glu209 variants. The same GNAQ or GNA11 mutation is found in both NICH and RICH, so other factors must account for these tumors’ different postnatal behaviors.
Main Text
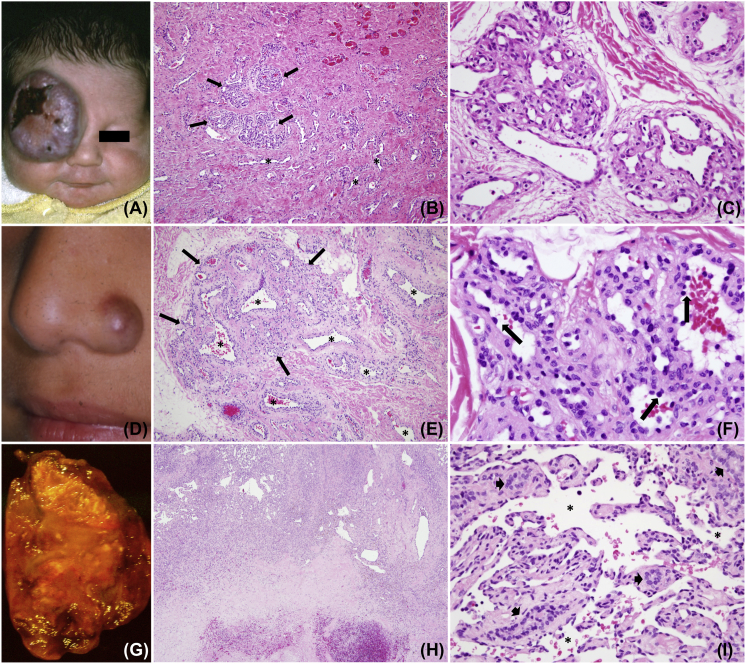
Congenital hemangiomas are rare vascular tumors that are present at birth. They are different from common infantile hemangiomas that rapidly enlarge postnatally and immunostain for the cell surface marker GLUT1.1 In contrast, congenital hemangiomas are GLUT1 negative and display one of two clinical patterns: “rapidly involuting congenital hemangioma” (RICH)2, 3 or “non-involuting congenital hemangioma” (NICH).4 RICH can be detected prenatally (as early as 12 weeks gestation) and presents in a newborn as a raised, gray-violaceous solitary cutaneous tumor with fine telangiectasias, ectatic veins, and a pale halo (Figure 1). RICH demonstrates fast-flow and can be associated with congestive cardiac failure and transient low-grade thrombocytopenia. If there are not overwhelming complications from heart failure or hemorrhagic ulceration, the tumor rapidly regresses by 6–14 months, leaving a patch of subcutaneous atrophy, dilated veins, and persistent fast-flow. RICH is also well documented to occur in the liver (Figure 1), where it spontaneously regresses just as in skin.5 The second category of congenital hemangioma (NICH) presents as a well-circumscribed, plaque-like tumor with a purple-pink hue, pale rim, coarse telangiectasia, and fast-flow (Figure 1). It remains unchanged throughout childhood; however, there are uncommon examples of growth and expansion in adolescence.6 In some cases RICH can cease regressing and transform into NICH.7, 8 There are histopathological similarities between RICH and NICH (Figure 1) and between congenital hemangioma and placental chorangioma, suggesting that chorangioma might be the placental counterpart of cutaneous and intrahepatic congenital hemangioma.9
Figure 1.
Representative Gross and Histological Images of RICH, NICH, and Solitary Congenital Hepatic Hemangioma from Three Participants
Shown are RICH (A–C), NICH (D–F), and solitary congenital hepatic hemangioma (G–I).
(A) Supraorbital purple tumor with central depression and ulceration in a 1-week-old infant (participant 8 with RICH).
(B) Hematoxylin and eosin (H&E) staining of the affected tissue imaged at 100× reveals small lobules (arrows) surrounded by abundant dense fibrous tissue where lobules have involuted and only draining channels remain (asterisks).
(C) H&E staining at 400× reveals lobular capillaries containing enlarged lumens and minimally prominent endothelium.
(D) Raised erythematous nodule at the left nasal ala in a 5-year-old child (participant 4 with NICH).
(E) H&E staining at 100× reveals a large lobule (arrows) with prominent intra- and peri-lobular channels (asterisks).
(F) H&E staining at 600× shows a hobnailed endothelium (arrows) lining lobular channels surrounded by fibrous tissue.
(G) Excised 5 cm hepatic mass with pallor, extensive infarction, focal calcification, and hemorrhage in a 3-month-old infant (participant 16 with hepatic RICH).
(H) H&E staining at 20× reveals a highly vascular rim superiorly and an acellular infarcted and hemorrhagic zone inferiorly.
(I) H&E staining at 400× shows a minimally prominent endothelium lining widened hepatic sinusoids (asterisks) with cords containing a few bile ducts (arrowheads) but devoid of hepatocytes.
We hypothesized that somatic mutations initiate the formation of congenital hemangiomas and searched for these mutations by performing massively parallel sequencing on fresh frozen specimens.
The Committee on Clinical Investigation of Boston Children’s Hospital approved this study. Congenital hemangioma samples were collected during a clinically indicated procedure, and all participants provided written consent. Tissues were immediately flash-frozen or placed in RNAlater (Thermo Fisher Scientific) and stored at −80°C until further processing. The original intent was to perform RNA-seq looking for coding sequence mutations or genetic alterations that might produce chimeric or miss-spliced transcripts and to perform whole-exome sequencing (WES) looking for coding sequence mutations in genes that were not highly expressed in affected tissue. RNA-seq libraries were prepared via standard Illumina TruSeq protocols and sequenced on one flowcell of an Illumina HiSeq 2500 system. We did not perform WES because we were able to successfully identify mutations with RNA-seq.
We generated 32–47 million 100-bp paired-end reads per RNA-seq library. Following the Genome Analysis Toolkit (GATK) best practices workflow,10 we mapped each set of reads to the reference human genome (GRCh37) with STAR aligner,11 removed PCR duplicates with Picard, realigned the reads around small insertions and deletions, and recalibrated the base quality scores with GATK.12 We then compiled single-nucleotide variant lists with Samtools13 and identified positions with a minimum of 20× read depth, 3× variant read depth, and 10% variant allele frequency using VarScan.14 We removed variants with greater than 40% variant allele frequency, assuming that these changes most likely represent germline variants rather than somatic changes. We further filtered the variants by removing those previously reported in the Exome Variant Server (ESP6500), 1000 Genomes Project, Exome Aggregation Consortium (v.0.3), and dbSNP as a non-clinical entry (build 138). We annotated the remaining variants with ANNOVAR.15
A total of 43 genes exhibited protein sequence altering variants in ≥3 of 8 specimens (Figure S1). We narrowed this list further by comparing these variants to those found in RNA-seq data from unrelated vascular lesions (arteriovenous malformation [n = 3] and common infantile hemangioma [n = 1]). A total of 12 genes contained a variant found only in the congenital hemangioma samples (Table 1). The variants found in the other RNA-seq datasets were assumed to be associated with RNA editing or with errors in sequencing, mapping, or annotation. Close inspection of variants in the 12 remaining genes for potential mismapping, sequencing errors, and lack of evolutionary conservation left only variants in GNAQ (MIM: 600998) as likely true-positive somatic mutations (Tables 2 and S1). GNAQ encodes guanine nucleotide binding protein G(q) alpha, a subunit within a complex that hydrolyzes the intracellular messenger GTP to GDP. GNAQ shares 90% protein sequence similarity with GNA11 (MIM: 139313). Somatic missense mutations that alter codon 209 in GNAQ (GenBank: NP_002063.2) and GNA11 (GenBank: NP_002058.2) have been reported in >80% of uveal melanomas.16, 17 A different somatic mutation in GNAQ (GRCh37; GenBank: NM_002072.4, NP_002063.2; c.548G>A [p.Arg183Gln]) occurs in isolated capillary malformations and in individuals with Sturge-Weber syndrome (MIM: 185300).18 Stringent filtering revealed evidence for a GNAQ missense mutation in three of eight samples (Figure S1). When we reanalyzed with less stringent filtering, we found that six of eight samples had a somatic GNAQ mutation and the remaining two samples had a somatic GNA11 mutation (Table 2).
Table 1.
Variant Filtering Strategy Employed in the Analysis of RNA-Seq Data
| Participant | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | Average |
|---|---|---|---|---|---|---|---|---|---|
| Min 20× read depth, 3× var read depth, 10% var allele freq | 56,331 | 49,047 | 40,440 | 38,348 | 37,672 | 32,473 | 39,380 | 26,900 | 40,074 |
| Known variants filtered | 24,333 | 18,454 | 12,671 | 12,893 | 11,756 | 10,206 | 11,236 | 9,570 | 13,890 |
| Coding sequence variants | 279 | 208 | 234 | 253 | 241 | 276 | 261 | 348 | 263 |
| Nonsynonymous variants | 172 | 120 | 144 | 138 | 134 | 155 | 164 | 221 | 156 |
| Variants without strand bias | 133 | 83 | 94 | 105 | 99 | 114 | 110 | 167 | 113 |
| Variants with <40% allele freq | 93 | 62 | 57 | 92 | 70 | 92 | 87 | 139 | 87 |
| Genes with variants in ≥3 samples, not in negative controls | ATN1, B3GNT2, GNAQ, IL13RA1, NBPF11, PLEKHO1, SMC4, SSC5D, TBX3, TCF7L2, TCHP, POLR2J2/UPK3BL | 12 | |||||||
Table 2.
Participants Screened for GNAQ and GNA11 Variants
| Participant | Phenotype | Age | Sex | Location | Source | Variant | RNA | DNA (MIP-seq) | DNA (ddPCR) | Control DNA (ddPCR) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | NICH | 8 years | male | chest | frozen tissue | GNAQ pGlu209Leu (c.626A>T) | 2/6 (33%) | – | 101/1,853 (5%) | 0/1,499 (0%) |
| 2 | NICH | 9 years | male | neck | frozen tissue | GNAQ pGlu209Pro (c.626A>C) | 1/10 (10%) | – | 392/6,061 (6%) | 0/2,215 (0%) |
| 3 | NICH | 14 years | female | temple | frozen tissue | GNA11 pGlu209Leu (c.626A>T) | 15/124 (12%) | 13/211 (6%) | 84/1,042 (8%) | 0/2,498 (0%) |
| 4 | NICH | 5 years | male | nose | frozen tissue | GNAQ pGlu209Leu (c.626A>T) | 3/15 (20%) | – | 235/3,233 (7%) | 0/2,359 (0%) |
| 5 | NICH | 7 years | male | nose | frozen tissue | GNAQ pGlu209Leu (c.626A>T) | 7/51 (14%) | – | 61/759 (8%) | – |
| 6 | NICH | 2.5 years | male | neck | frozen tissue | GNA11 pGlu209Leu (c.626A>T) | 3/103 (3%) | – | 15/633 (2%) | – |
| 7 | RICH | 3 years | male | lower extremity | frozen tissue | GNAQ pGlu209His (c.627A>C) | 7/30 (23%) | – | – | – |
| 8 | RICH | 1 week | male | orbital area | frozen tissue | GNAQ pGlu209Leu (c.626A>T) | 6/31 (19%) | – | – | – |
| 9 | NICH | 12 years | male | lower extremity | FFPE tissue | GNAQ pGlu209Pro (c.626A>C) | – | 1139/19,657 (6%) | 125/1,089 (11%) | – |
| 10 | NICH | 14 months | male | lower extremity | FFPE tissue | NA | – | 3/1,383 (< 1%) | 0/640 (0%) | – |
| 11 | RICH | 3 months | male | lower extremity | FFPE tissue | GNAQ pGlu209Leu (c.626A>T) | – | - | 26/717 (4%) | – |
| 12 | NICH | 3 years | male | ear | FFPE tissue | NA | – | 1/400 (< 1%) | - | – |
| 13 | RICH | 2 weeks | female | upper extremity | FFPE tissue | NA | – | 2/772 (< 1%) | 0/516 (0%) | – |
| 14 | NICH | 2 years | male | lower extremity | FFPE tissue | NA | – | 2/179 (1%) | 0/152 (0%) | – |
| 15 | RICH | 2 weeks | male | lower extremity | FFPE tissue | GNA11 pGlu209Leu (c.626A>T) | – | – | 27/2,576 (1%) | – |
| 16 | RICH | 4 months | male | liver | FFPE tissue | GNA11 pGlu209Leu (c.626A>T) | – | – | 52/537 (10%) | – |
| 17 | CHOR | neonatal | female | placenta | FFPE tissue | NA | – | 0/850 (0%) | – | – |
| 18 | CHOR | neonatal | female | placenta | FFPE tissue | NA | – | 3/1,855 (< 1%) | – | – |
| 19 | CHOR | neonatal | female | placenta | FFPE tissue | NA | – | 3/1,875 (< 1%) | – | – |
| 20 | CHOR | neonatal | female | placenta | FFPE tissue | NA | – | 7/2,233 (< 1%) | – | – |
RNA and DNA (MIP-seq) columns indicate read depth, and DNA (ddPCR) column indicates number of droplets. The rate of variant/total alleles is also depicted in the aforementioned columns. For samples that were not associated with a mutation (indicated with NA), available MIP-seq and ddPCR results with the lowest power (in terms of read depth and number of droplets) are depicted. Control DNA samples were retrieved from blood or saliva. Abbreviations are as follows: NICH, non-involuting congenital hemangioma; RICH, rapidly involuting congential hemangioma; CHOR, chorangioma. Dash (–) indicates that the assay was not performed.
We confirmed that the mutations were real by testing DNA from six of eight samples via an orthologous method, digital droplet PCR (ddPCR), and/or molecular inversion probe sequencing (MIP-seq) (Table 2 and Figure 2). We performed ddPCR (Table S2) as previously described19 and considered variants present at frequencies ≥1% to represent true positives. We also verified that the mutations we identified are somatic by testing control DNA (extracted from blood or saliva) of four participants via ddPCR (Table 2). For MIP-seq, we enriched the genomic DNA samples for the protein-coding sequences of GNAQ and GNA11 by hybridization to probes (Table S3) containing an 8-nucleotide barcode that uniquely identifies individual MIPs. MIP capture and sequencing were performed as previously described.20 Raw reads were mapped to the reference human genome sequence (GRCh37) with BWA; PCR duplicates were removed with FastqMcf and Picard. The minimum read depths for GNAQ c.626A>T and GNA11 c.626A>T (GRCh37, GenBank: NM_002067.4) were 2,032× and 179×, respectively. We considered true positive somatic variants to have allele frequencies >2%. Using a combination of the aforementioned ddPCR and MIP-seq assays, we also analyzed eight archival formalin-fixed paraffin-embedded (FFPE) congenital hemangiomas and four chorangioma samples. Four of the hemangiomas contained a likely somatic GNAQ or GNA11 mutation, whereas no such mutations were found in the chorangiomas (Table 2).
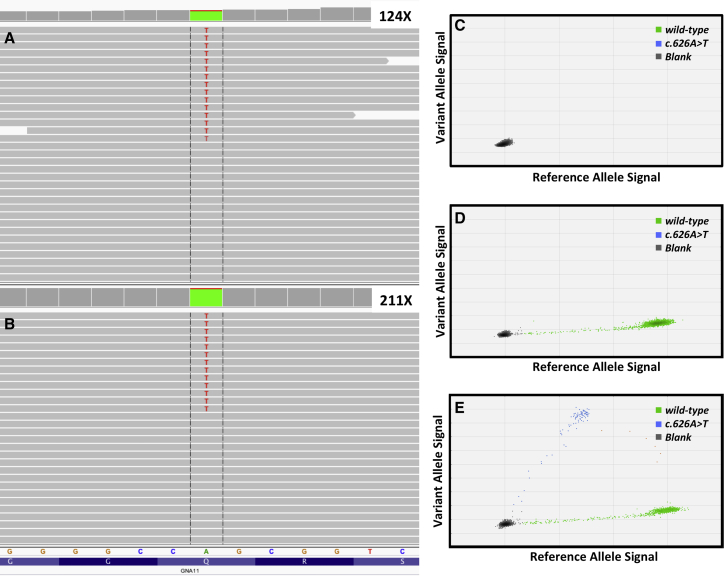
Figure 2.
Detection of the GNA11 c.626A>T (p.Glu209Leu) Allele in Congenital Hemangioma Specimen from Participant 3 with NICH
(A) IGV screenshot indicating 12% variant allele frequency in GNA11 transcripts measured by RNA-seq. 124× indicates the depth of coverage at position 626.
(B) IGV screenshot indicating 6% variant allele frequency in GNA11 measured by MIP-seq. 211× indicates the depth of coverage at position 626.
(C) Water-negative control for the ddPCR assay shows no flourescence for GNA11 wild-type or variant alleles.
(D) ddPCR results of human DNA from unaffected control tissue indicates the presence of only the GNA11 wild-type allele.
(E) ddPCR results of congenital hemangioma indicates the presence of wild-type and variant (8%) alleles.
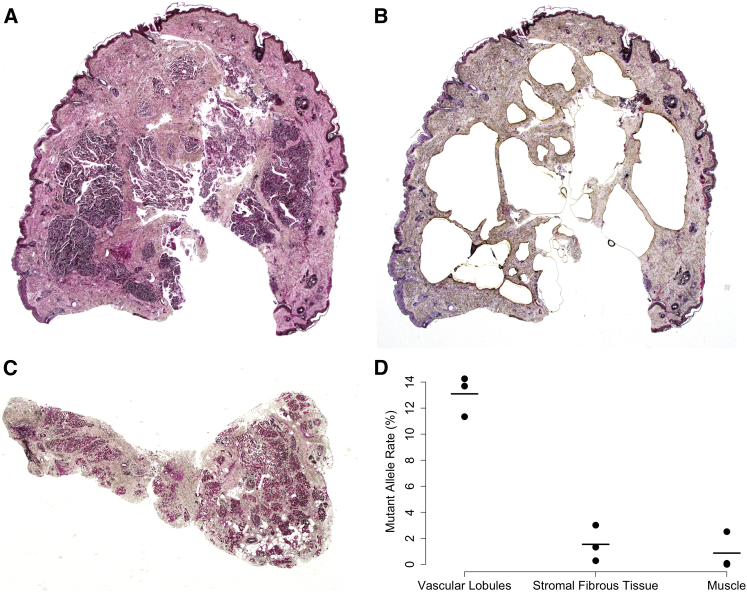
Our findings expand the spectrum of genes and alleles that are somatically altered in congenital vascular anomalies (Figure S2).18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29 Of interest is that the same mutation (i.e., GNAQ c.626A>T and GNA11 c.626A>T) occurs in RICH and NICH, implying that other genetic, epigenetic, and/or environmental factors probably influence these lesions’ postnatal behavior. Because GNAQ and GNA11 mutations are common in highly metastatic uveal melanomas,16, 17 identifying factors that determine the postnatal behavior of RICH and NICH might suggest new strategies for treating uveal melanoma. Thus far, we observed mutations affecting Glu209 in GNAQ or GNA11 only in congenital hemangioma, whereas we and other investigators18, 30, 31 have identified mutations affecting Arg183 only in capillary malformations. Our data indicate that, with respect to congenital hemangiomas, GNAQ and GNA11 have functional overlap. Consistent with functional overlap in nonmalignant conditions, somatic mutations affecting either codon 183 or 209 in GNAQ and GNA11 have also been found in phakomatosis pigmentovascularis and extensive dermal melanocytosis.32 However, a role for GNA11 that does not overlap with that of GNAQ can be inferred from recent studies of inherited hypocalcemia and hypercalcemia caused by mutations in GNA11 but not GNAQ.33, 34, 35, 36 In vitro studies indicated that missense mutations altering Arg183 and Glu209 both activate GTP-dependent signaling, but might do so via different pathways and with different efficiency.18, 37, 38 Therefore, it is critical to understand the cell-type-specific, context-specific, and allele-specific effects of somatic GNAQ or GNA11 mutations during vasculogenesis. For example, the GNAQ c.548G>A (p.Arg183Gln) mutation in sporadic capillary malformations was shown to be enriched in endothelial cells that line blood vessels,30 whereas in lymphatic malformations activating PIK3CA (MIM: 171834), mutations were present in the lymphatic endothelial cells.39, 40, 41 To determine which features within a congenital hemangioma sample are enriched for mutant cells, we performed laser capture microdissection on a FFPE tissue section from participant 4. We observed a mutant allele frequency of 13% in the vascular-enriched lobules within the lesion, whereas the remaining tissues exhibit less than 2% mutant allele frequency (Figure 3).
Figure 3.
Mutant Allele Frequency in Capillary-Enriched Lobules Is Higher than That in the Surrounding Stromal Tissue within Congenital Hemangioma
(A) Pre-laser capture image of NICH showing multiple dermal vascular lobules from participant 4 (staining dark blue in hematoxylin and eosin-stained section, ×20 magnification).
(B) Post-laser capture image of the section depicted in (A) showing absence of virtually all vascular lobules (hematoxylin and eosin-stained section, ×20 magnification).
(C) Pre-laser capture of skeletal muscle, fibrous tissue, and fat uninvolved tissue by the NICH shown in (A) and (B) (hematoxylin and eosin-stained section, ×20 magnification) that was resected en bloc.
(D) Graph depicting the GNAQ c.626A>T allele frequencies in DNA extracted from the capillary enriched lobules (A), the residual tissue (B), and the adjacent muscle tissue (C) measured in triplicate by ddPCR (dots). Solid lines indicate the average of the three measurements.
We were unable to identify strongly suggestive GNAQ or GNA11 mutations in 4/16 congenital hemangiomas, leading us to conclude that the frequency of mutated cells in some specimens was either below our threshold of detection or that congenital hemangiomas exhibit further locus heterogeneity. Although our sample size for chorangiomas is small, our data indicate that chorangioma is probably not the placental counterpart of congenital hemangiomas associated with mutations altering Glu209 in GNAQ or GNA11.
In contrast to an earlier study in which we detected somatic mutations in PIK3CA with WES but not with RNA-seq,23 RNA-seq was useful in this study. For WES, the sensitivity of detecting somatic mutations is dependent on depth of coverage and frequency of mutant cells (Figure S3). Interestingly, when we compared the relative frequencies of the GNAQ and GNA11 mutant allele at the RNA and DNA levels in the affected tissue specimens for which we had paired RNA and DNA, the mutant allele frequency was always higher in the RNA (Table 2). This latter finding is compatible with mutation-containing cells within the affected tissue expressing GNAQ or GNA11 at a greater level relative to other cells. Thus, in addition to the advantages of using RNA-seq to find mutations that produce abnormal splicing or chimeric transcripts, RNA-seq can be advantageous for detecting somatic coding sequence mutations.
Acknowledgments
We are grateful for the contributions of the participants to this study. This work was supported by NIH grants R01-AR064231 (to M.L.W.), R01-HL096384 (to J.B.), and R21-HD081004 (to A.K.G.).
Published: April 7, 2016
Footnotes
Supplemental Data include three figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.03.009.
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes, http://browser.1000genomes.org
Burrows-Wheeler Aligner, http://bio-bwa.sourceforge.net/
dbSNP, build 138, http://www.ncbi.nlm.nih.gov/projects/SNP/
ExAC Browser, http://exac.broadinstitute.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
samtools, https://github.com/samtools/
STAR Aligner, https://github.com/alexdobin/STAR/releases
VarScan, http://varscan.sourceforge.net/
Supplemental Data
References
- 1.North P.E., Waner M., Mizeracki A., Mihm M.C., Jr. GLUT1: a newly discovered immunohistochemical marker for juvenile hemangiomas. Hum. Pathol. 2000;31:11–22. doi: 10.1016/s0046-8177(00)80192-6. [DOI] [PubMed] [Google Scholar]
- 2.Berenguer B., Mulliken J.B., Enjolras O., Boon L.M., Wassef M., Josset P., Burrows P.E., Perez-Atayde A.R., Kozakewich H.P. Rapidly involuting congenital hemangioma: clinical and histopathologic features. Pediatr. Dev. Pathol. 2003;6:495–510. doi: 10.1007/s10024-003-2134-6. [DOI] [PubMed] [Google Scholar]
- 3.Boon L.M., Enjolras O., Mulliken J.B. Congenital hemangioma: evidence of accelerated involution. J. Pediatr. 1996;128:329–335. doi: 10.1016/s0022-3476(96)70276-7. [DOI] [PubMed] [Google Scholar]
- 4.Enjolras O., Mulliken J.B., Boon L.M., Wassef M., Kozakewich H.P., Burrows P.E. Noninvoluting congenital hemangioma: a rare cutaneous vascular anomaly. Plast. Reconstr. Surg. 2001;107:1647–1654. doi: 10.1097/00006534-200106000-00002. [DOI] [PubMed] [Google Scholar]
- 5.Fishman S.J., Burrows P.E. Treatment of visceral vascular tumors. In: Mulliken J.B., Burrows P.E., Fishman S.J., editors. Mulliken and Young’s Vascular Anomalies: Hemangiomas and Malformations. Oxford University Press; 2013. pp. 242–245. [Google Scholar]
- 6.Mulliken J.B. Diagnosis and natural history of hemangiomas. In: Mulliken J.B., Burrows P.E., Fishman S.J., editors. Mulliken and Young’s Vascular Anomalies: Hemangiomas and Malformations. Oxford University Press; 2013. pp. 69–110. [Google Scholar]
- 7.Mulliken J.B., Enjolras O. Congenital hemangiomas and infantile hemangioma: missing links. J. Am. Acad. Dermatol. 2004;50:875–882. doi: 10.1016/j.jaad.2003.10.670. [DOI] [PubMed] [Google Scholar]
- 8.Nasseri E., Piram M., McCuaig C.C., Kokta V., Dubois J., Powell J. Partially involuting congenital hemangiomas: a report of 8 cases and review of the literature. J. Am. Acad. Dermatol. 2014;70:75–79. doi: 10.1016/j.jaad.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 9.Mulliken J.B., Bischoff J., Kozakewich H.P. Multifocal rapidly involuting congenital hemangioma: a link to chorangioma. Am. J. Med. Genet. A. 2007;143A:3038–3046. doi: 10.1002/ajmg.a.31964. [DOI] [PubMed] [Google Scholar]
- 10.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013;11 doi: 10.1002/0471250953.bi1110s43. 1, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koboldt D.C., Zhang Q., Larson D.E., Shen D., McLellan M.D., Lin L., Miller C.A., Mardis E.R., Ding L., Wilson R.K. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Raamsdonk C.D., Bezrookove V., Green G., Bauer J., Gaugler L., O’Brien J.M., Simpson E.M., Barsh G.S., Bastian B.C. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Raamsdonk C.D., Griewank K.G., Crosby M.B., Garrido M.C., Vemula S., Wiesner T., Obenauf A.C., Wackernagel W., Green G., Bouvier N. Mutations in GNA11 in uveal melanoma. N. Engl. J. Med. 2010;363:2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shirley M.D., Tang H., Gallione C.J., Baugher J.D., Frelin L.P., Cohen B., North P.E., Marchuk D.A., Comi A.M., Pevsner J. Sturge-Weber syndrome and port-wine stains caused by somatic mutation in GNAQ. N. Engl. J. Med. 2013;368:1971–1979. doi: 10.1056/NEJMoa1213507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Couto J.A., Vivero M.P., Kozakewich H.P., Taghinia A.H., Mulliken J.B., Warman M.L., Greene A.K. A somatic MAP3K3 mutation is associated with verrucous venous malformation. Am. J. Hum. Genet. 2015;96:480–486. doi: 10.1016/j.ajhg.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luks V.L., Kamitaki N., Vivero M.P., Uller W., Rab R., Bovée J.V., Rialon K.L., Guevara C.J., Alomari A.I., Greene A.K. Lymphatic and other vascular malformative/overgrowth disorders are caused by somatic mutations in PIK3CA. J. Pediatr. 2015;166 doi: 10.1016/j.jpeds.2014.12.069. 1048–54.e1, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eerola I., Boon L.M., Mulliken J.B., Burrows P.E., Dompmartin A., Watanabe S., Vanwijck R., Vikkula M. Capillary malformation-arteriovenous malformation, a new clinical and genetic disorder caused by RASA1 mutations. Am. J. Hum. Genet. 2003;73:1240–1249. doi: 10.1086/379793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groesser L., Peterhof E., Evert M., Landthaler M., Berneburg M., Hafner C. BRAF and RAS mutations in sporadic and secondary pyogenic granuloma. J. Invest. Dermatol. 2016;136:481–486. doi: 10.1038/JID.2015.376. [DOI] [PubMed] [Google Scholar]
- 23.Kurek K.C., Luks V.L., Ayturk U.M., Alomari A.I., Fishman S.J., Spencer S.A., Mulliken J.B., Bowen M.E., Yamamoto G.L., Kozakewich H.P., Warman M.L. Somatic mosaic activating mutations in PIK3CA cause CLOVES syndrome. Am. J. Hum. Genet. 2012;90:1108–1115. doi: 10.1016/j.ajhg.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liaw D., Marsh D.J., Li J., Dahia P.L., Wang S.I., Zheng Z., Bose S., Call K.M., Tsou H.C., Peacocke M. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat. Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 25.Lim Y.H., Douglas S.R., Ko C.J., Antaya R.J., McNiff J.M., Zhou J., Choate K.A., Narayan D. Somatic activating RAS mutations cause vascular tumors including pyogenic granuloma. J. Invest. Dermatol. 2015;135:1698–1700. doi: 10.1038/jid.2015.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Limaye N., Kangas J., Mendola A., Godfraind C., Schlögel M.J., Helaers R., Eklund L., Boon L.M., Vikkula M. Somatic activating PIK3CA mutations cause venous malformation. Am. J. Hum. Genet. 2015;97:914–921. doi: 10.1016/j.ajhg.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindhurst M.J., Sapp J.C., Teer J.K., Johnston J.J., Finn E.M., Peters K., Turner J., Cannons J.L., Bick D., Blakemore L. A mosaic activating mutation in AKT1 associated with the Proteus syndrome. N. Engl. J. Med. 2011;365:611–619. doi: 10.1056/NEJMoa1104017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vikkula M., Boon L.M., Carraway K.L., 3rd, Calvert J.T., Diamonti A.J., Goumnerov B., Pasyk K.A., Marchuk D.A., Warman M.L., Cantley L.C. Vascular dysmorphogenesis caused by an activating mutation in the receptor tyrosine kinase TIE2. Cell. 1996;87:1181–1190. doi: 10.1016/s0092-8674(00)81814-0. [DOI] [PubMed] [Google Scholar]
- 29.Rivière J.B., Mirzaa G.M., O’Roak B.J., Beddaoui M., Alcantara D., Conway R.L., St-Onge J., Schwartzentruber J.A., Gripp K.W., Nikkel S.M., Finding of Rare Disease Genes (FORGE) Canada Consortium De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat. Genet. 2012;44:934–940. doi: 10.1038/ng.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Couto J.A., Huang L., Vivero M.P., Kamitaki N., Maclellan R.A., Mulliken J.B., Bischoff J., Warman M.L., Greene A.K. Endothelial cells from capillary malformations are enriched for somatic GNAQ mutations. Plast. Reconstr. Surg. 2016;137:77e–82e. doi: 10.1097/PRS.0000000000001868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakashima M., Miyajima M., Sugano H., Iimura Y., Kato M., Tsurusaki Y., Miyake N., Saitsu H., Arai H., Matsumoto N. The somatic GNAQ mutation c.548G>A (p.R183Q) is consistently found in Sturge-Weber syndrome. J. Hum. Genet. 2014;59:691–693. doi: 10.1038/jhg.2014.95. [DOI] [PubMed] [Google Scholar]
- 32.Thomas A.C., Zeng Z., Rivière J.B., O’Shaughnessy R., Al-Olabi L., St-Onge J., Atherton D.J., Aubert H., Bagazgoitia L., Barbarot S. Mosaic activating mutations in GNA11 and GNAQ are associated with phakomatosis pigmentovascularis and extensive dermal melanocytosis. J. Invest. Dermatol. 2016 doi: 10.1016/j.jid.2015.11.027. Published online January 14, 2016. S0022-202X(16)00332-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piret S.E., Gorvin C.M., Pagnamenta A.T., Howles S.A., Cranston T., Rust N., Nesbit M.A., Glaser B., Taylor J.C., Buchs A.E. Identification of a G-protein subunit-α11 gain-of-function mutation, Val340Met, in a family with autosomal dominant hypocalcemia type 2 (ADH2) J. Bone Miner. Res. 2016 doi: 10.1002/jbmr.2797. Published online January 28, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorvin C.M., Cranston T., Hannan F.M., Rust N., Qureshi A., Nesbit M.A., Thakker R.V. G-protein subunit-α11 loss-of-function mutation, Thr54Met, causing familial hypocalciuric hypercalcemia type 2 (FHH2) J. Bone Miner. Res. 2016 doi: 10.1002/jbmr.2778. Published online January 5, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nesbit M.A., Hannan F.M., Howles S.A., Babinsky V.N., Head R.A., Cranston T., Rust N., Hobbs M.R., Heath H., 3rd, Thakker R.V. Mutations affecting G-protein subunit α11 in hypercalcemia and hypocalcemia. N. Engl. J. Med. 2013;368:2476–2486. doi: 10.1056/NEJMoa1300253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mannstadt M., Harris M., Bravenboer B., Chitturi S., Dreijerink K.M., Lambright D.G., Lim E.T., Daly M.J., Gabriel S., Jüppner H. Germline mutations affecting Gα11 in hypoparathyroidism. N. Engl. J. Med. 2013;368:2532–2534. doi: 10.1056/NEJMc1300278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng X., Degese M.S., Iglesias-Bartolome R., Vaque J.P., Molinolo A.A., Rodrigues M., Zaidi M.R., Ksander B.R., Merlino G., Sodhi A. Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated rho GTPase signaling circuitry. Cancer Cell. 2014;25:831–845. doi: 10.1016/j.ccr.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu F.X., Luo J., Mo J.S., Liu G., Kim Y.C., Meng Z., Zhao L., Peyman G., Ouyang H., Jiang W. Mutant Gq/11 promote uveal melanoma tumorigenesis by activating YAP. Cancer Cell. 2014;25:822–830. doi: 10.1016/j.ccr.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Couto J.A., Vivero M.P., Upton J., Padwa B.L., Warman M.L., Mulliken J.B., Greene A.K. Facial infiltrating lipomatosis contains somatic PIK3CA mutations in multiple tissues. Plast. Reconstr. Surg. 2015;136(4, Suppl):72–73. [Google Scholar]
- 40.Boscolo E., Coma S., Luks V.L., Greene A.K., Klagsbrun M., Warman M.L., Bischoff J. AKT hyper-phosphorylation associated with PI3K mutations in lymphatic endothelial cells from a patient with lymphatic malformation. Angiogenesis. 2015;18:151–162. doi: 10.1007/s10456-014-9453-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Osborn A.J., Dickie P., Neilson D.E., Glaser K., Lynch K.A., Gupta A., Dickie B.H. Activating PIK3CA alleles and lymphangiogenic phenotype of lymphatic endothelial cells isolated from lymphatic malformations. Hum. Mol. Genet. 2015;24:926–938. doi: 10.1093/hmg/ddu505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.