Abstract
Abnormal protein aggregation is observed in an expanding number of neurodegenerative diseases. Here, we describe a mechanism for intracellular toxic protein aggregation induced by an unusual mutation event in families affected by axonal neuropathy. These families carry distinct frameshift variants in NEFH (neurofilament heavy), leading to a loss of the terminating codon and translation of the 3′ UTR into an extra 40 amino acids. In silico aggregation prediction suggested the terminal 20 residues of the altered NEFH to be amyloidogenic, which we confirmed experimentally by serial deletion analysis. The presence of this amyloidogenic motif fused to NEFH caused prominent and toxic protein aggregates in transfected cells and disrupted motor neurons in zebrafish. We identified a similar aggregation-inducing mechanism in NEFL (neurofilament light) and FUS (fused in sarcoma), in which mutations are known to cause aggregation in Charcot-Marie-Tooth disease and amyotrophic lateral sclerosis, respectively. In summary, we present a protein-aggregation-triggering mechanism that should be taken into consideration during the evaluation of stop-loss variants.
Introduction
Abnormal accumulations of protein aggregates are associated with a wide range of diseases mainly affecting the nervous system.1 The origins of these aggregates are diverse; however, they share similar structures and overlapping mechanisms of cellular toxicity in different diseases. Protein aggregates usually adopt high-ordered β sheet quaternary structures, forming insoluble fibrils termed amyloids.2 In Huntington disease (MIM: 143100), intranuclear inclusions and cytoplasmic aggregates are caused by polyglutamine expansion of the protein huntingtin.3 Parkinson disease (MIM: 168600) is characterized by the presence of inclusions known as Lewy bodies in the cytoplasm of neurons.4 In Alzheimer disease (MIM: 104300), aggregates can occur both extracellularly as neuritic plaques composed of Aβ peptide and intracellularly as neurofibrillary tangles of hyperphosphorylated TAU.5 In amyotrophic lateral sclerosis (ALS [MIM: 105400]), aggregation of ubiquitinated proteins, including FUS, TDP-43, and OPTN, occurs in degenerating motor neurons.6 Interestingly, these aggregated ubiquitinated proteins co-occur with perikaryal inclusions of neurofilament (NF).7 Abnormal NF aggregation and misassembly have also been reported in other motor neuron diseases such as giant axonal neuropathy (MIM: 256850)8 and Charcot-Marie-Tooth disease (CMT [MIM: 607736]),9, 10 the latter of which is the most common inherited motor neuron disease. However, the causes of NF accumulation appear to be heterogeneous and not fully understood.
NFs are a class of intermediate filaments exclusively expressed in neurons, and they represent major components of the cytoskeleton responsible for regulating axonal diameter and growth.11 NFs are composed of light (NEFL)-, medium (NEFM)-, and heavy (NEFH)-molecular-weight NF proteins that form heteropolymers in order to assemble into 10 nm filaments.12 Mutations in NF-encoding genes have been reported in multiple neurodegenerative diseases. In ALS, NEFH expansion and contraction of the KSP (Lys-Ser-Pro) repeat motif has been shown to be a risk factor, yet its clinical contribution to sporadic ALS remains to be determined.13, 14 Mutations in NEFL (MIM: 162280) have been associated with both axonal and demyelinated CMT (CMT2E [MIM: 607684] and CMT1F [MIM: 607734], respectively) and manifest with a wide spectrum of clinical phenotypes.10, 15, 16 The frequency of NEFL mutations in CMT is about 2%, and mutations can affect conserved protein domains, including the head (e.g., p.Pro8Arg [c.23C>G] [GenBank: NM_006158.4]), coil (e.g., p.Gln334Pro [c.1001A>C] [GenBank: NM_006158.4]), and tail (e.g., p.Glu527del [c.1579_1581delGAG] [GenBank: NM_006158.4]) domains.16 Most NEFL mutations have been identified as heterozygous, although homozygous mutations have also been reported.17 Most mutations lead to abnormal NF aggregates and disruption of NF axonal transport and assembly in transfected cells.10 HSBP1 (heat shock factor binding protein 1 [MIM: 602195]) mutations, which cause a subtype of CMT, also result in disruption and aggregation of NEFL, thus pointing to the importance of chaperone proteins in NF integrity.18, 19, 20
In the present study, we report previously unrecognized cryptic amyloidogenic elements (CAEs) encoded by the 3′ UTR of NEFH (MIM: 162230) and show that these lead to aggregation and neuronal degeneration in model systems and affected individuals. We demonstrate that frameshift variants in NEFH in CMT2-affected families result in stop loss and translation of a CAE encoded by the 3′ UTR. Expression of the mutant NEFH exhibited prominent abnormal protein aggregates, disruption of the NF network, and altered cell dynamics. Interestingly, we obtained the same aggregation-induced phenomenon by triggering the translation of the 3′ UTRs of NEFL and FUS (MIM: 137070). Our in vivo and in vitro results show that translation of CAEs encoded by the 3′ UTRs of NF-encoding genes causes axonopathy and could be of broader impact for neurodegenerative diseases.
Material and Methods
Families
The families were identified as part of our ongoing genetic studies in CMT. The families were ascertained in Austria and Great Britain. Participants were recruited, enrolled, and sampled according to the protocols of the institutional review board at the Universities of London and Vienna. A complete description of the study was provided to the subjects, and written informed consent was obtained. Whole blood was collected from all participants by venipuncture. Affection status was determined by consensus of physicians and clinical staff experienced in clinical CMT research and was based on medical records and in-person evaluation.
Plasmid Constructs and Immunocytochemistry
We created constructs encoding GFP-tagged wild-type protein (GFP-WT-NEFH) and frameshift mutant protein with 40 additional amino acids encoded by open reading frame 3 (ORF3) (GFP-FS-NEFH), as identified in family UK1 (p.Asp1004Glnfs∗58). The gene encoding human NEFH was synthesized by GenScript. Site-directed mutagenesis was used for generating the affected individual’s variant, c.3010_3011delGA (p.Asp1004Glnfs∗58) (GenBank: NM_021076.3). The GFP tag was introduced at the N terminus of the protein by cloning into pcDNA3.1/NT-GFP-Topo (Invitrogen). NEFL-Myc cloned into the pCMV6-Entry vector was obtained from OriGene Technologies. Neuro-2a cells were grown in complete DMEM media (GIBCO) to 75% confluence and transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. After 24 hr, cells were fixed with paraformaldehyde for 20 min, permeabilized with cold methanol for 5 min, and stained with anti-Myc antibody (Cell Signaling) and anti-tubulin (Invitrogen). Cells were mounted onto microscope slides and imaged with a confocal microscope, Zeiss LSM710, with a 60× objective lens.
Co-immunoprecipitation and Western Blot
Immunoprecipitation was performed with the Pierce Crosslink Magnetic IP/Co-IP Kit according to the manufacturer’s instructions. In brief, 500 μg of total cell lysates were incubated with 5 μg of either anti-GFP chromatin-immunoprecipitation-grade antibody (Abcam) or control rabbit immunoglobulin G (IgG, Santa Cruz) at 4°C overnight. The lysate-antibody mixture was incubated with protein A/G magnetic beads (Thermo Scientific) at room temperature for 1 hr on a rotator mixer. Beads were collected with a magnetic stand. Beads were washed twice with the manufacturer’s immunoprecipitation lysis and wash buffer. Samples were boiled with a lane marker containing reducing agent at 100°C for 5 min. Protein samples were analyzed by SDS-PAGE followed by western blot with appropriate antibodies. The following antibodies (diluted at 1:1,000) were used: mouse monoclonal anti-GFP (Santa Cruz), mouse monoclonal anti-Myc (Cell Signaling), and rabbit polyclonal anti-kinesin (Abcam).
IncuCyte Analysis
Neuro-2a cells transfected with GFP-WT-NEFH and GFP-FS-NEFH were plated in a 24-well plate, and 24 hr after transfection, cells were incubated in an IncuCyte live-cell imager system (Essen Instruments) for 2 days at 37°C and 5% CO2. Replicates of 12 wells were used for each group (wild-type and mutant). Time-lapse phase-contrast and GFP images were taken every 3 hr for 24 hr. A total of 36 images were acquired per well for each time point. The IncuCyte Zoom software was used for calculating cell size, confluence, and eccentricity (roundness).
Transmission Electron Microscopy and Analysis
Neuro-2a cells were cultured in 25 cm2 flasks (Corning) and transfected with plasmids as previously described. After 2 days, cells were fixed for 4 days in 2.5% glutaraldehyde in Millonig’s phosphate buffer, post-fixed in 1% OsO4 and uranyl acetate, dehydrated in an ethanol series, and embedded in Spurr’s resin. Ultrathin, 85 nm sections were made with a Leica microtome and were stained with cold lead citrate for 10 min. A total of 114 images were acquired with a Joel JEM-1400 transmission electron microscope with a digital Gatan camera. All images were processed and analyzed with Fiji (ImageJ and Photoshop CS5).
Zebrafish Studies
Experiments were carried out with wild-type or transgenic tg(Olig2:DsRed) fish in a mixed AB-TL background. Adults were kept on a 14/10 hr light/dark cycle at 28°C. Embryos were obtained from natural crosses after removal of a divider at first light. The mMESSAGE mMACHINE T7 Ultra Kit (Ambion) was used for synthesizing mRNAs from the GFP-WT-NEFH and GFP-FS-NEFH (pcDNA3.1/NT-GFP-Topo) plasmids after linearization with NotI, and 400 pg of RNA was microinjected into 1-cell-stage embryos. Embryos were reared in petri dishes in a 28°C incubator with the same light/dark cycle. Motor neuron outgrowth was assayed at 48 hr post-fertilization (hpf) in embryos obtained from tg(Olig2:DsRed) crosses. Live fish were anesthetized with tricaine methanesulfonate (Sigma), placed against a shelf of 1.5% agarose, and imaged with a Leica confocal microscope with a 20× lens. 1 μm z stacks were taken between segments 6 and 15, and the lengths of the first four caudal anterior primary axons were measured with the Simple Neurite Tracer in Fiji. Images in the figure were displayed with the menu option “Lookup Tables > Edges” for enhancing contrast. GFP fluorescence intensity was assayed in microinjected wild-type embryos at 24 hpf by similar methods. Maximum-intensity z stacks were compiled, and the trunk of the fish excluding the yolk was traced from the DIC image. Fluorescence intensity represents mean gray values 0–255, which were analyzed with Fiji. For western blots, 20–50 embryos were manually dechorionated at 24 hpf and were processed through a batch deyolking method.21 Deyolked and washed embryos were homogenized in 50 μl of RIPA with protease inhibitor in the Fisherbrand Disposable Pestle System, sonicated, and then frozen and thawed at −80. The supernatant was collected after centrifugation, and protein content was measured. NuPage sample buffer (Invitrogen) and reducing agent were added, samples were boiled, and approximately 12 μg of total fish proteins were loaded into SDS gel. For detection, anti-GFP tag antibody (Abcam) and anti-tubulin (Santa Cruz sc-9104) were used at a 1/1,000 concentration, and SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific) was used after incubation with the HRP-conjugated secondary antibodies. For RT-PCR validation, RNA was extracted from approximately 20 dechorionated embryos at 24 hpf in TRIzol (Invitrogen). The cDNA was generated with SuperScript III (Invitrogen) and random hexamers, and PCR was performed with the following GFP-NEFH transcript primers: 5′-TTTTACCAGACAACCATTACCTG-3′ and 5′-GGCTAGCGCGTAGTGGAG-3′ or control (slc25a46) 5′- GCCACTGGGTGACGACTC-3′ and 5′-GAAGCGGAAGAAGTCGTTTG-3′. All experiments were conducted in accordance with the guidelines of the University of Miami Institutional Animal Care and Use Committee.
Sanger and Exome Sequencing
We performed exome sequencing in 269 index individuals with autosomal-dominant CMT. 16 CMT-affected families were from the UK, and 48 families were from Austria. The remaining families were from several other countries from Europe and North and South America. The SureSelect Human All Exon 50 MB Kit (Agilent) was used for in-solution enrichment, and the HiSeq 2500 instrument (Illumina) was used to produce 100 bp paired-end sequence reads. The Burrows-Wheeler aligner, Picard, and the Genome Analysis Toolkit were used to align sequence reads and call variants. These data were imported into GENESIS (formerly GEM.app)22,23 for further analysis. Variants were filtered for those that segregated in an autosomal-dominant fashion and met the “strict” criteria, which required that variants be rare (minor allele frequency < 0.05% in the National Heart, Lung, and Blood Institute [NHLBI] Exome Sequencing Project [ESP] Exome Variant Server [ESP6500]), be present in fewer than three families within GEM.app (∼4,300 exomes), be conserved (GERP score > 2 or PhastCons score > 0.6), and have sufficient quality scores (genotype quality > 75). Mutations in known CMT-associated genes were absent in both Austrian and UK families. NEFH-variant calls identified were validated by conventional Sanger sequencing. In addition, we had access to 5,200 control samples from the GENESIS database. These samples broadly included 1,824 neuromuscular disorders (of which 470 were peripheral neuropathies), 286 cardiomyopathies, 188 dementia disorders, and 509 deafness disorders. For more details, refer to Gonzalez et al.22
Results
Identification of NEFH Frameshift Variants in CMT2-Affected Families
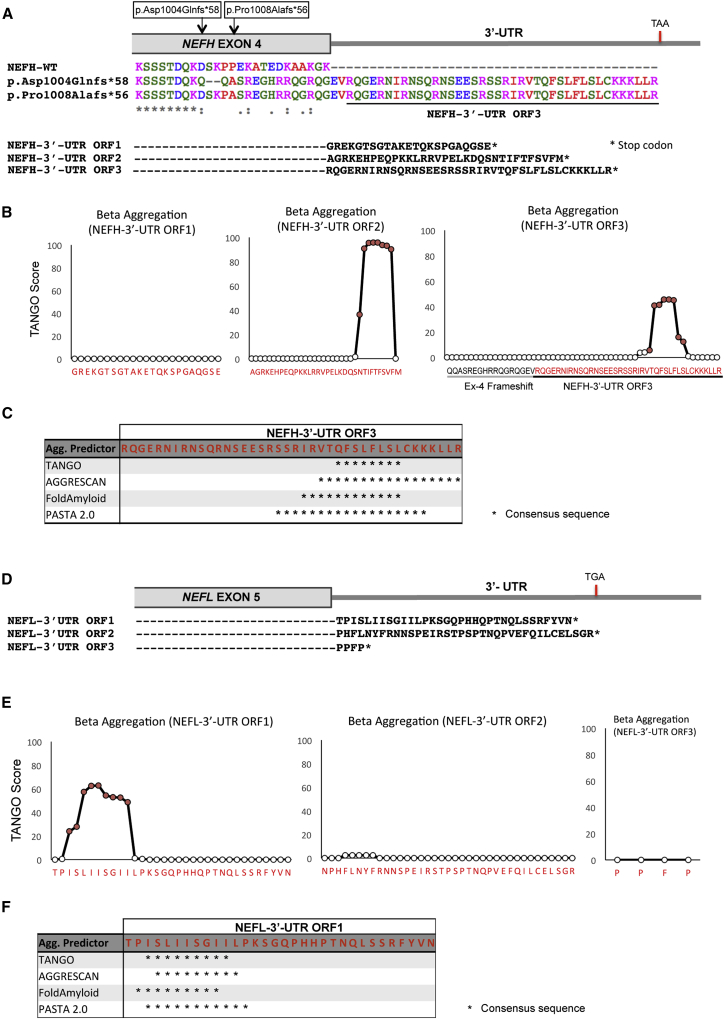
We performed whole-exome sequencing on three affected individuals belonging to different generations of a British family (UK1) diagnosed with autosomal-dominant CMT2 (Figure 1A). Exome data were analyzed with a strict filtering approach for segregation of non-synonymous heterozygous variants with the use of GENESIS software.22, 23 A heterozygous frameshift variant in NEFH was identified as a top candidate for the disease from a list containing six additional variants (Table S1). NEFH was selected because NF abnormalities have been previously reported in neurodegenerative diseases, including ALS.24 In addition, mutations in NEFL, another major NF component, also cause CMT.16 The variant co-segregated with the phenotype across three generations in this family (Figure 1A). This variant (c.3010_3011delGA [p.Asp1004Glnfs∗58] at chr22: 29,886,637) affects the last coding exon and shifts translation into an alternative ORF, resulting in continued translation of an additional 40 amino acids beyond the stop codon in the original ORF (Figure 2A). The mutant protein retains its major functional domains, including the head, rod, and tail domains (Figure 1C). We then screened an additional 322 CMT-affected families by whole-exome sequencing. We identified another CMT2-affected family (F2) with four individuals carrying a nearby heterozygous frameshift variant in NEFH (c.3017_3020dup [p.Pro1008Alafs∗56] at chr22: 29,886,645). This insertion interestingly also results in a stop-loss mutation and translation of the identical ORF, as observed in family UK1 (Figure 2A). Co-segregation of the distinct variants with the phenotype was confirmed by Sanger sequencing in both families (Figure 1B). We were unable to sequence DNA from the deceased parent in family F2, who was most likely a carrier of the pathogenic variant. In approximately 5,000 additional exomes22 with a wide range of clinical phenotypes (including other neuropathies) from our own collection, we did not observe these mutations and also did not observe other frameshift variants in the last exon of NEFH.
Figure 1.
NEFH Frameshift Variants in CMT2-Affected Families
Asterisks indicate probands.
(A) Pedigree and Sanger sequence traces of the CMT-affected family carrying the NEFH variant c.3010_3011delGA (p.Asp1004Glnfs∗58). Abbreviations are as follows: M, mutant c.3010_3011delGA allele; and +, wild-type allele.
(B) Pedigree and Sanger sequence traces of the CMT-affected family carrying the NEFH variant c.3017_3020dup (p.Pro1008Alafs∗56). Abbreviations are as follows: M, mutant c.3017_3020dup allele; and +, wild-type allele.
(C) Diagram shows NEFH domains and variants associated with diseases. Coding KSP deletions and insertions from reported ALS individuals are represented by triangles. CMT frameshift variants from families UK1 and F2 are indicated by arrows.
Figure 2.
Identification of CAEs Encoded by the NEFH and NEFL 3′ UTRs
(A) Clustal Omega multiple-sequence alignment of wild-type NEFH and frameshift variants harbored by the CMT-affected families. Translation of the 3′ UTR open reading frames (ORFs) is illustrated.
(B) TANGO score of NEHF 3′ UTR ORFs.
(C) Consensus sequence of positive residues (asterisks) for all aggregation predictors tested for NEFH 3′ UTR ORF3.
(D) NEFL 3′ UTR ORF sequences.
(E) TANGO score of NEFL 3′ UTR ORFs.
(F) Consensus sequence of positive residues (asterisks) for all aggregation predictors tested for NEFL 3′UTR ORF1.
Clinical Features of Family UK1, Harboring the p.Asp1004Glnfs∗58 NEFH Variant
The proband of UK1 (IV:1; Figure 1 and Table 1) had a normal birth and early development. He walked independently at the age of 18 months. Between the ages of 2 and 4 years, he started falling frequently and had a tendency to turn over on his ankles, and he was noted to have high arches and muscle wasting of the legs. Over the years, he noticed progressive lower-limb weakness, sensory loss in the feet, and occasional cramps. He started using support for walking in his early 20s and began using a wheelchair in his mid-20s. He had surgery for pyloric stenosis in his infancy; medical history was otherwise unremarkable.
Table 1.
Clinical Characteristics of Studied Individuals
|
Family UK1 |
Family F2 |
|||||
|---|---|---|---|---|---|---|
| IV:1 | III:2 | II:1 | I:1 | III:1 | ||
| Sex | male | male | female | male | male | |
| Current age | 26 years | 57 years | 84 years | died at 80 years | 56 years | |
| Age at first symptoms | 2–4 years | 15 years | early 20s | NA | 38 years | |
| First symptoms | frequent falls, tendency to turn over on ankles | shoes with ankle support, able to run until late 40s | unable to stand on tiptoes, flat-footed walk | NA | gait disturbance, problems with climbing stairs | |
| Age at exam | 24 years | 57 years | 79 years | NA | 49 years | |
| Muscle wastinga | upper limbs | + | + | + | NA | − |
| lower limbs | ++ | ++ | +++ | NA | ++ | |
| Muscle weaknessb | upper limbs | + | + | ++ | NA | normal |
| lower limbs | +++ | + | +++ | NA | ++ | |
| Pinprick sensationc | upper limbs | + | + | normal | NA | normal |
| lower limbs | ++ | ++ | ++ | NA | + | |
| Vibration sensec | upper limbs | normal | normal | ++ | NA | NA |
| lower limbs | ++ | ++ | +++ | NA | + | |
| Joint position sensec | upper limbs | normal | normal | normal | NA | normal |
| lower limbs | normal | normal | normal | NA | normal | |
| Reflexesd | upper limbs | + | + | − | NA | ↓ |
| lower limbs | − | − (ankle jerks only) | − | NA | ↓ or − | |
| Plantar responses (left/right)e | f/f | f/f | −/f | NA | f/f | |
| Walk with support | yes (20s) | yes (50s) | yes (50s) | yes (50s) | no | |
| Wheelchair bound | no | no | yes (70s) | yes (70s) | no | |
| Other features | increased tone in lower limbs, waddling and stiff gaitf | cramps in hands and lower limbs | hearing loss (70s) | hearing loss (50s) | none | |
| Creatine kinase | 1,288g | 442g | 150h | NA | 686 | |
| Brain and spinal-cord MRI | normal | normal | NA | NA | NA | |
NA, not available.
Muscle wasting: −, none; +, below wrist or ankle; ++, below elbow or knee; and +++, above elbow or knee.
Muscle weakness: +, >4 distal muscle groups (first dorsal interosseous, abductor pollicis brevis, ankle dorsiflexion, plantar flexion, or below); ++, <4 distal muscles; and +++, proximal weakness (knee flexion and extension, elbow flexion and extension, or above).
Sensory examination: +, reduced below wrist or ankle; ++, reduced below elbow or knee; and +++, reduced at or above elbow or knee.
Reflexes: −, absent; ↓, diminished; and +, present.
Plantar responses: −, mute; and f, flexor.
On subsequent assessments.
Reference range: 38–204 IU/L.
Reference range: 26–140 IU/L.
Neurological examination at age 16 revealed minimal distal wasting in the four limbs, mild weakness of the first dorsal interosseous and ankle dorsiflexion (Medical Research Council grade 4+), and reduced pinprick and vibration sense in the ankles. Subsequent examinations revealed proximal weakness and mildly increased tone in the lower limbs. At age 22, he walked with a slightly waddling and stiff gait and had a positive Gower’s maneuver. Examination at age 24 revealed wasting of the intrinsic hand muscles and below the knees and weakness of the first dorsal interosseous (4) and abductor pollicis brevis (4+), hip flexion (4+), knee flexion and extension (4+), ankle dorsiflexion (4), and ankle plantar flexion (2). Reflexes were present in the upper limbs and absent in the lower limbs. Plantar responses were flexor. Pinprick sensation was reduced in the fingers and to the proximal half of the calves. Vibration sense was reduced to the knees with a Rydel-Seiffer tuning fork. Proprioception was normal.
Nerve-conduction-velocity (NCV) studies (Table 1) were consistent with a motor and sensory axonal neuropathy predominantly affecting the lower limbs. Electromyography (EMG) revealed proximal and distal chronic neurogenic changes in a non-length-dependent pattern and additional myopathic features in proximal muscles. Central-motor-conduction times in the lower limbs were mildly prolonged bilaterally. Creatine kinase (CK) levels were raised at 721–1,288 IU/L (reference range: 38–204). Plasma lactate was elevated at 1.84 mmol/L (reference range: 0.5–1.65) in one out of three measurements. Analysis of cerebrospinal fluid was normal.
A muscle biopsy of the vastus lateralis showed increased variation in fiber size, a mild increase in connective tissue, frequent round and angular atrophic fibers of all types, nuclear bag fibers, group atrophy, and grouping of type I and IIA fibers. Split fibers and occasional regenerating fibers were also noted. Several fibers contained internal nuclei, and three contained rimmed vacuoles. Many fibers had a disturbed architecture, but no core-targetoid fibers were observed. There were no cytochrome-c-oxidase-negative or ragged-red fibers. Immunohistochemistry revealed a patchy increase in myotilin staining. Assays of respiratory-chain enzymes in muscle homogenate revealed reduced activity of all complexes (complex I: 0.077 [reference range: 0.104–0.268]; complex II + III: 0.032 [0.040–0.204]; and complex IV: 0.008 [0.014–0.034]). A repeated assay revealed complex I and IV activities at the lower limit of the reference range (complex I: 0.106; complex IV: 0.015). Sequence analysis failed to detect pathogenic variants in PMP22 (MIM: 601097), MPZ (MIM: 159400), GJB1 (MIM: 304040), MFN2 (MIM: 608507), GDAP1 (MIM: 606598), NEFL, BSCL2 (MIM: 606158), TRPV4 (MIM: 605427), HSPB1, and HSPB8 (MIM: 608014). 17p11.2 rearrangements were excluded.
The father of the proband (III:1; Figure 1A and Table 1) had used shoes with ankle support since the age of 15 years. In his 40s, he started noticing occasional tripping episodes and cramps but was able to run until age 48. He started using support for walking at age 53. Neurological examination at age 57 showed mild distal wasting in the upper and lower limbs and weakness of the first dorsal interosseous (4), ankle dorsiflexion (4), and ankle plantar flexion (4). Reflexes were present except at the ankles. Plantar responses were flexor. Pinprick sensation was reduced to the mid-palms and to just above the ankles. Vibration sense was reduced to the knees with a Rydel-Seiffer tuning fork. Proprioception was normal.
The grandmother (II:1; Figure 1A and Table 1) had had problems with standing on her toes and a flat-footed walk since her early 20s. In her 50s, she started using support for walking and developed problems with fine hand movements. She became wheelchair bound in her 70s and developed hearing loss in her mid-70s. Neurological examination at age 79 revealed muscle wasting of the intrinsic hand muscles and from the mid-thighs down. There was weakness of the first dorsal interosseous (3) and abductor pollicis brevis (3), hip flexion (4), knee extension (4), knee flexion (4+), ankle dorsiflexion (0), and ankle plantar flexion (4+). Reflexes were all absent. Pinprick was reduced to just below the knees. Vibration sense was reduced to the left elbow, right shoulder, and costal margins. Proprioception was normal.
Neurophysiological studies of individuals III:2 and II:1 (Table 2) revealed similar findings to those observed in the proband except for the absence of myopathic changes. NCV studies were consistent with a motor and sensory axonal neuropathy, which was severe in II:1. EMG revealed proximal and distal chronic neurogenic changes in a non-length-dependent pattern.
Table 2.
Electrophysiological Characteristics of Studied Individuals
|
Family UK1 |
Family F2 |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| IV:1 | III:2 | II:1 | III:1 | ||||||
| Age at examination | 24 years | 55 years | 72 years | 49 years | |||||
| Electromyography | |||||||||
| Deltoids and biceps | CN/M | CN | CN | NA | |||||
| FDIO and/or FDS | CN | CN | FP, PSW, CN | NA | |||||
| Vastus medialis | CN/M | CN | FP, PSW, CN | NA | |||||
| Tibialis anterior | CRD, CN | CN | FP, PSW, CN | FP, PSW, CN | |||||
| Nerve-Conduction Studies | L | R | L | R | L | R | L | R | |
| Radial nerve | sensory amp | NA | 20 μv | NA | 11 μv | NA | absent | NA | NA |
| sensory CV | NA | 56 m/s | NA | 60 m/s | NA | NA | NA | NA | |
| Median nerve | DML | NA | 4.1 ms | NA | 4.0 ms | NA | 4.8 ms | NA | 3.6 ms |
| motor Amp | NA | 10.7 mV | NA | 6.7 mV | NA | 1.7 mV | NA | 9.8 mV | |
| motor CV | NA | 46 m/s | NA | 54 m/s | NA | 38 m/s | NA | 59.2 m/s | |
| F wave latency | NA | 34.1 ms | NA | 33.9 ms | NA | 22.4 ms | NA | NA | |
| sensory amp | 3 μv | 6 μv | NA | 3 μv | NA | absent | NA | 9.5 μv | |
| sensory CV | 59 m/s | 56 m/s | NA | 51 m/s | NA | NA | NA | 50.9 | |
| Ulnar nerve | DML | 3.2 ms | 3.2 ms | NA | 2.8 ms | NA | 3.7 ms | NA | 2.6 ms |
| motor amp | 10.4 mV | 10.8 mV | NA | 12.1 mV | NA | 7.7 mV | NA | 20.6 mV | |
| motor CV | 54 m/s | 55 m/s | NA | 51 m/s | NA | 56 m/s | NA | 48.7 m/s | |
| F wave latency | NA | 31.0 ms | NA | 32.2 ms | NA | 34.2 ms | NA | NA | |
| sensory amp | NA | 3 μv | NA | 1 μv | NA | absent | NA | 10.1 μv | |
| sensory CV | NA | 52 m/s | NA | 52 m/s | NA | NA | NA | 52.1 m/s | |
| Peroneal nerve | DML | NA | 7.5 ms | NA | 6.0 ms | NA | NA | 4.4 ms | 4.0 ms |
| motor amp | NA | 1.9 mV | NA | 0.5 mV | NA | absent | 1.8 mV | 4.2 mV | |
| motor CV | NA | 35 m/sa | NA | 41 m/s | NA | NA | 32.3 m/s | 43.2 m/s | |
| F wave latency | NA | NA | NA | NA | NA | NA | NA | NA | |
| sensory amp | NA | absent | NA | absentb | NA | absent | NA | NA | |
| sensory CV | NA | NA | NA | NA | NA | NA | NA | NA | |
| Tibial nerve | DML | NA | 7.3 ms | NA | NA | NA | NA | 4.3 ms | 3.5 ms |
| motor amp | NA | 1.2 mV | NA | NA | NA | absent | 2.7 mV | 3.0 mV | |
| motor CV | NA | NA | NA | NA | NA | NA | 41.4 m/s | 40.8 m/s | |
| F wave latency | NA | 66.3 ms | NA | NA | NA | NA | NA | NA | |
| Sural nerve |
sensory amp | NA | absent | NA | absent | NA | absent | NA | 7.2 μV |
| sensory CV | NA | NA | NA | NA | NA | NA | NA | 49.7 m/s | |
| Central-Motor-Conduction Times | L | R | L | R | L | R | L | R | |
| Upper limbs | 8.1 msc | 6.0 msc | NA | NA | NA | NA | NA | NA | |
| Lower limbs | 19.5 msc | 22.2 msc | NA | NA | NA | NA | NA | NA | |
Abbreviations are as follows: NA, not assessed; CN, chronic neurogenic changes; CN/M, mixed chronic neurogenic and myopathic changes; CRD, complex repetitive discharges; CV, conduction velocity; DML, distal motor latency; FP, fibrillation potentials; FDIO, first dorsal interosseous; FDS, flexor digitorum superficialis; L, left; PSW, positive sharp waves; and R, right.
Cold extremity.
Absent in a previous study.
At age 22 years.
Our findings suggest an apparent anticipation pattern in this family, given that the proband had an earlier age of onset and more severe manifestations than did the previous generations. Increased severity of the disease over generations has been reported in other CMT-affected families;25, 26 however, the mechanism responsible for this phenomenon is unclear. It is possible that a modifier gene can affect the severity of the clinical phenotype; however, it is very challenging to identify a potential modifier gene from our exome data with such a small sample size.
Clinical Features of Family F2, Harboring the p.Pro1008Alafs∗56 NEFH Variant
The proband of family F2 (III:1; Figure 1B) was first brought to neurological attention at the age of 38 years, when he noticed weakness in the lower limbs. He initially complained of problems in climbing stairs, and running became impossible. There were no problems in the hands and no sensory disturbances. At examination, there was mild atrophy in the lower limbs, but this was more pronounced distally, as was muscle weakness. Tendon reflexes were reduced to absent. There was mild bilateral pes cavus. NCV studies revealed normal values in the upper limbs but bilateral slowing of the motor peroneal and tibial nerves and low amplitudes, pointing to an axonal neuropathy. NCV of the sural nerve was normal, but the amplitude was reduced. EMG of the tibialis anterior muscle revealed prominent chronic neurogenic disturbances with spontaneous activity. The disease was considerably progressive. Re-examination after 10 years revealed prominent proximal and distal weakness in the lower limbs. The individual walked with a very unsafe gait. Mild weakness and wasting then became evident in both distal and proximal muscles (Table 1). CK levels had been elevated from the beginning. Mutations in the following genes were excluded by direct sequencing: PMP22, MPZ, NEFL, LITAF (MIM: 603795), GDAP1, YARS (MIM: 603623), MFN2, HSPB1, HSPB8, DNM2 (MIM: 602378 [exons 13–16]), FBLN5 (MIM: 604580), and HINT1 (MIM: 601314).
Three siblings were reported to be similarly affected, and given the prominent proximal muscle involvement and elevated CK levels, a combination of neurogenic and myopathic disorders was suspected. Neurological evaluation of the parents was not performed during this study. One parent (II:1) had died early, whereas the other parent (II:2) was reported to be neurologically normal up to an advanced age.
Identification of CAEs Encoded by NEFH and NEFL 3′ UTRs
Because NFs have a considerable tendency to aggregate in neurodegenerative diseases, we decided to investigate the intrinsic aggregation propensity of the extension of amino acids present in NEFH in the two CMT2-affected families. We used the web-based aggregation prediction tool TANGO27 to analyze aggregation-prone segments on the basis of the physico-chemical principles of β sheet formation. According to this algorithm, a segment is predicted to aggregate when it contains at least five consecutive residues with a TANGO score above 5%.27 Analysis of the 3′ UTR-encoded extension of amino acids present in the NEFH mutant p.Asp1004Glnfs∗58 showed a hotspot for aggregation in a stretch of eight amino acids (QFSLFLSL), which had a combined score of 250 (Figure 2B). Aggregation propensity was also analyzed with other prediction tools, including AGGRESCAN,28 FoldAmyloid29 and PASTA 2.0,30 which use different algorithms to predict aggregation. All tested tools detected aggregation and an amyloidogenic region in an overlapping stretch of amino acids of the mutant extension (Figure 2C and Figure S2). We refer to this stretch of amino acids predicted to induce aggregation as a CAE. Prediction analysis of the other two ORFs of the NEFH 3′ UTR resulted in no aggregation for ORF1 and high aggregation scores for ORF2 (Figure 2B). This result suggests that any frameshift variants resulting in loss of the NEFH stop codon and translation of CAEs from the 3′ UTR would have the potential to cause protein aggregation.
Next, we investigated in silico the aggregation propensity of the NEFL 3′ UTR, given that NEFL aggregation has been reported for ALS6 and CMT.18 Interestingly, only ORF1 of the 3′ UTR, comprising the motif ISLIISGII, was positive for aggregation prediction with a high TANGO score of 443 (Figure 2E). Positive scores for ORF1 were also obtained with AGGRESCAN, FoldAmyloid, and PASTA2.0 (Figure 2F and Figure S2). Thus, our in silico results for NEFL suggest that only missense mutations affecting the stop codon and resulting in translation of the 3′ UTR ORF1 have the potential to cause aggregation. Importantly, because the first reading frame of the NEFL 3′ UTR encodes the CAE, we should also consider the possibility of leaky translational readthrough caused by reduced accuracy of translation termination. Therefore, NEFL aggregation would be a concern particularly for individuals treated with drugs that induce stop-codon readthrough, such as gentamicin G418.31
Prominent Aggregation in Cultured Neuro-2a Cells Expressing NF Genes with CAE-Encoding 3′ UTRs
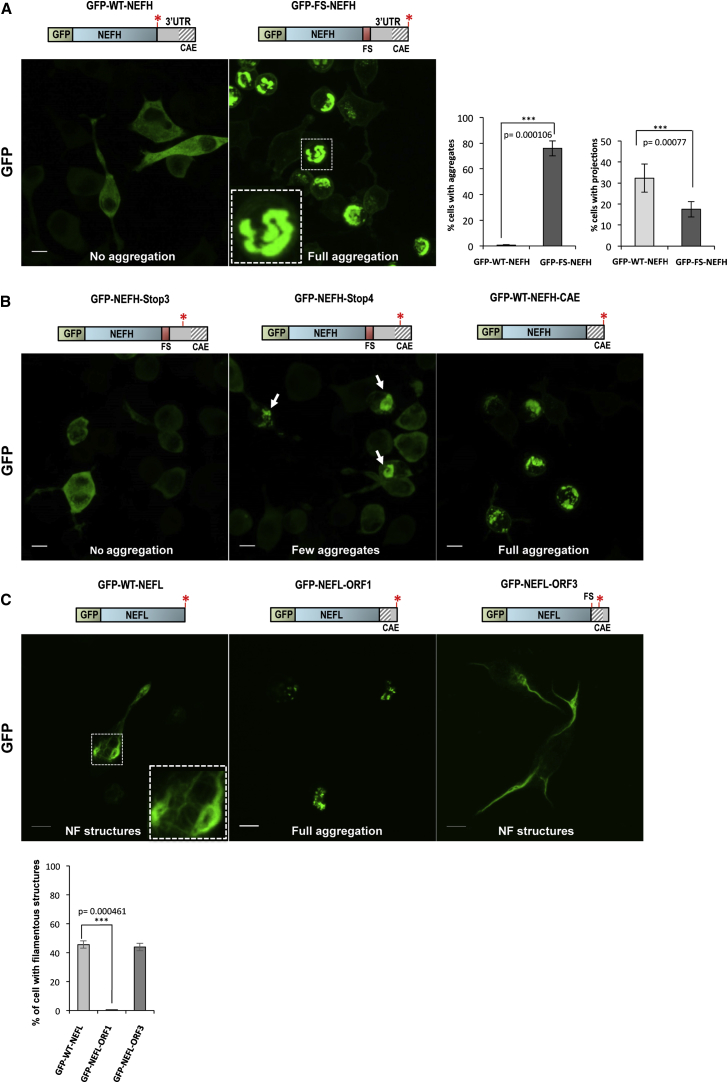
After our in silico analysis, we investigated the potential of the 3′ UTR-encoded CAEs identified in affected individuals to cause aggregation in Neuro-2a cells. GFP-WT-NEFH transfection led to evenly distributed expression in the cytoplasm of Neuro-2a cells (Figure 3A). Expression of GFP-FS-NEFH revealed prominent abnormal perinuclear aggregation 24 hr after transfection (Figure 3A). Quantification showed that over 75% of cells transfected with GFP-FS-NEFH contained aggregates, whereas less than 1% of GFP-WT-NEFH-transfected cells did (Figure 3A). Cells transfected with wild-type NEFH retained their typical Neuro-2a cell morphology with “axon-like” projections extending from the cell body. By contrast, GFP-FS-NEFH-expressing cells were round, and their axon-like projections were significantly reduced (Figure 3A).
Figure 3.
Protein Aggregation in Cultured Neuro-2a Cells Expressing NF Genes with CAE-Encoding 3′ UTRs
(A) Perinuclear aggregates in GFP-FS-NEFH-transfected cells after 24 hr. Graphs show the number of transfected cells containing NEFH aggregates and the percentage of cells with neuronal projections from six independent experiments. Error bars represent the SD, and the scale bar represents 10 μm.
(B) A small proportion of cells transfected with the truncated construct, GFP-NEFH-Stop4, showed protein aggregation, and cells transfected with GFP-WT-NEFH-CAE showed high levels of aggregation. Scale bars represent 10 μm.
(C) Prominent protein aggregation in cells transfected with GFP-NEFL-ORF. The graph shows the number of cells expressing NF-like structures. Error bars represent the SD, and scale bars represent 10 μm.
In order to experimentally identify which of the additional 40 amino acids are responsible for the aggregation, we created a series of truncated constructs harboring stop codons (Stop1–Stop4) at different positions throughout the NEFH frameshift mutation sequence (Figure S3). Cells transfected with constructs Stop1, Stop2, and Stop3 did not contain aggregates (Figure S3 and Figure 3B), and their subcellular distribution pattern was identical to that of cells transfected with GFP-WT-NEFH. We observed aggregates in a small proportion of cells (20%) transfected with the longest construct, GFP-Stop4-NEFH (Figure S3 and Figure 3B); however, aggregation was still more severe in the full-length extension described above (GFP-FS-NEFH). To challenge the predicted CAE, we cloned the most distal 22 amino acids predicted to cause aggregation (SSRIRVTQFSLFLSLCKKKLLR) directly in frame with the C-terminal end of wild-type NEFH (GFP-NEFH-CAE construct). As expected, cells transfected with the GFP-NEFH-CAE construct caused prominent aggregation at the same level observed in cells transfected with GFP-FS-NEFH (Figure 3B). These results prove that the most distal 22 amyloidogenic amino acids are sufficient and necessary for the formation of aggregates.
We also tested the ability of the 3′ UTR-encoded CAE in NEFL to cause aggregation in cells. Neuro-2a cells were transfected with the following constructs: GFP-tagged NEFL (GFP-WT-NEFL); NEFL without a stop codon and fused with NEFL 3′ UTR ORF1, encoding the predicted CAE (GFP-NEFL-ORF1); and NEFL fused with the 3′ UTR ORF3, which was not predicted to encode CAEs (GFP-NEFL-ORF3). Although a few cells transfected with GFP-WT-NEFL presented aggregates because of the self-assembling nature of NEFL, about 45% of transfected cells adopted NF-like structures, an indication of NF assembly (Figure 3C). These filamentous structures can also be explained by the ability of NEFL to self-assemble; in contrast, NEFH is an obligate heteropolymer and requires interaction with either NEFL or NEFM in order to assemble into NF structures.19 However, all cells transfected with GFP-NEFL-ORF1 formed prominent aggregation and adopted a rounded shape without forming filamentous structures (Figure 3C). In contrast, cells transfected with GFP-NEFL-ORF3 resulted in NF structures comparable with those of cells transfected with GFP-WT-NEFL. These results suggest that specific translation of ORF1 in the 3′ UTR is required for the formation of aggregates.
Evaluation of Cells Expressing CAEs Encoded by the NEFH 3′ UTR
To further characterize the NEFH aggregation structures, we stained cells with thioflavin T, a dye commonly used to stain amyloid fibrils with β sheet structures in individual tissues.32 Confocal fluorescent imaging showed strong thioflavin T staining of NEFH aggregates, suggesting a fibrillary amyloid-like structure (Figure 4F). In order to further understand the composition of the aggregates, we performed transmission electron microscopy (EM) in transfected Neuro-2a cells. In cells transiently expressing GFP-FS-NEFH, we observed a disordered array of filaments of approximately 10 nm in diameter, consistent with NF size (Figure 4E). Similar disordered NF inclusion structures were previously reported in the anterior horn cells in an ALS-affected individual harboring a SOD1 (MIM: 147450) mutation.33 These disordered arrays of filaments were observed in 12/100 images of cells expressing GFP-FS-NEFH in no fewer than five different cells and were never observed in 0/40 GFP-WT-NEFH images or 0/20 images of untransfected cells. Although we could not distinguish untransfected from transfected cells in the EM images, transfection efficiency was ∼70%, and we analyzed close to 20 rounded cells transfected with GFP-FS-NEFH, which is a feature of cells containing severe aggregates. On the basis of the frequency of this feature in cells expressing GFP-FS-NEFH and its absence in cells expressing GFP-WT-NEFH or untransfected cells, we trust that these structures are the aggregates.
Figure 4.
Evaluation of Cultured Neuro-2a Cells Expressing the NEFH 3′ UTR-Encoded CAE
(A) Western blot shows comparable levels of GFP in cells transfected with GFP-WT-NEFH and GFP-FS-NEFH.
(B–D) Sample time-lapse phase-contrast and GFP live-cell images obtained from IncuCyte imager system. (B) Calculation of the average area (μM2) of green (GFP) objects per cell at different time points. (C) Quantification of GFP-positive cell confluence. (D) Quantification of the average eccentricity of green objects measures object roundness from 0 to 1 (a perfect circle has a value of 0). Error bars represent the SD.
(E) Fibers of approximately 10 nm in diameter in a Neuro-2a cell transfected with GFP-FS-NEFH. Filaments are visible in the cytoplasm in longitudinal sections (blue box) and cross-sections (red box). Approximately 10% of cells transfected with GFP-FS-NEFH showed filaments. Scale bars represent 200 nm.
(F) Aggregates from GFP-FS-NEFH cells co-localized with thioflavin T staining. Scale bars represent 10 μm.
Presence of Mutant NEFH Disrupts the NF Network in Cultured Neuro-2a Cells
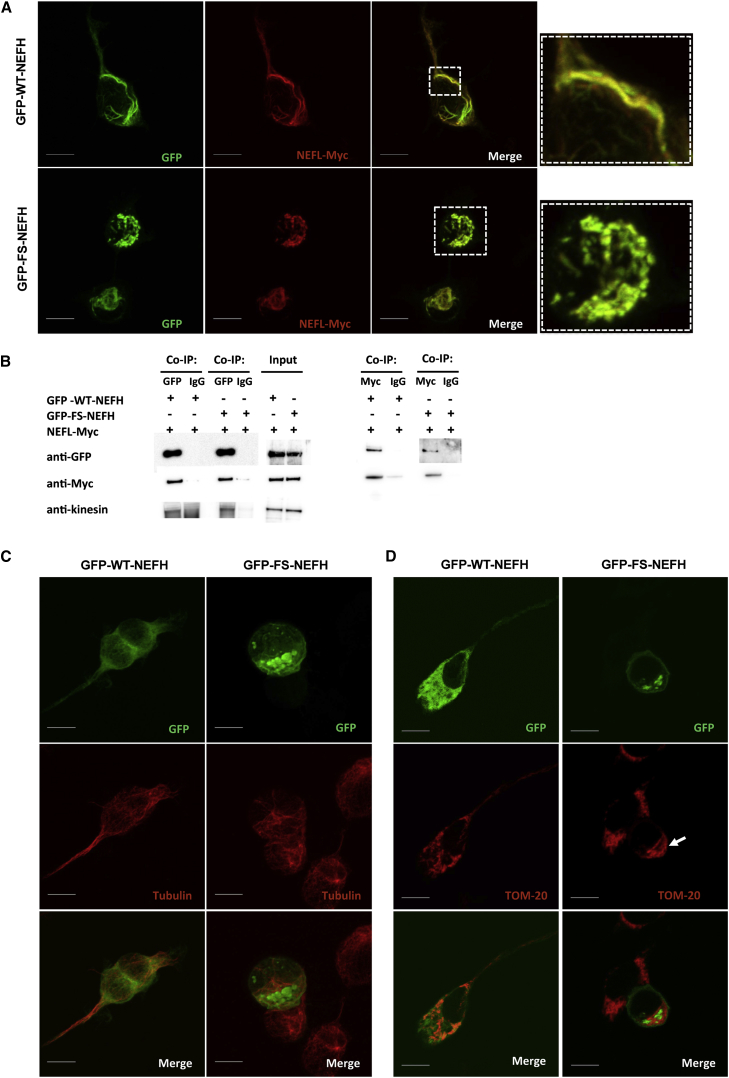
In order to see the effect of the mutant NEFH on the NF network, we co-transfected GFP-NEFH constructs with a plasmid encoding NEFL fused to a Myc tag at the C terminus (NEFL-Myc). As expected, the GFP-WT-NEFH construct co-localized with NEFL-Myc and assembled into organized NF-like structures (Figure 5A). The GFP-FS-NEFL construct also co-localized with NEFL-Myc but did so within the massive aggregates, suggesting arrest and co-aggregation of NEFL and consequently disruption of the NF network (Figure 5A). We performed co-immunoprecipitation experiments to confirm interaction between the mutant NEFH and NEFL. We immunoprecipitated cell lysates with an anti-GFP antibody to pull down NEFH. Western blot confirmed that NEFL-Myc was co-immunoprecipitated in cells transfected with either GFP-WT-NEFH or GFP-FS-NEFH (Figure 5B). The reverse co-immunoprecipitation with NEFL-Myc pull-down also confirmed the interaction. We further detected kinesin in the co-immunoprecipitate in cells transfected with either wild-type or mutant NEFH (Figure 5B). These results suggest that the mutant NEFH traps NEFL, kinesins, and possibly other interacting proteins in the aggregates and consequently blocks these proteins from performing their proper functions. Hence, this stop-loss NEFH mutation found in individuals with CMT2 is most likely a toxic gain-of-function mutation. To determine whether the aggregates exclusively affect the NF network or additional cytoskeleton components, we stained cells with a tubulin antibody. The microtubule network was normally distributed and assembled in cells transfected with either GFP-WT-NEFH or GFP-FS-NEFH, indicating that it was not affected by the aggregates (Figure 5C).
Figure 5.
Confocal Microscopy Shows NEFH Subcellular Co-localization
(A) Cells co-transfected with GFP-WT-NEFH and NEFL-Myc showed co-localization of NEFH and NEFL in NF network structures. Cells co-transfected with GFP-FS-NEFH and NEFL-Myc showed co-localization in the aggregates. Scale bars represent 10 μm.
(B) Co-immunuoprecipitation assay: cell lysates were immunoprecipitated with an antibody against either GFP or Myc and IgG as a negative control. Both GFP-WT-NEFH and GFP-FS-NEFH were co-immunoprecipitated with NEFL-Myc. Kinesin was also present in the co-immunoprecipitate.
(C) Cells transfected with either GFP-WT-NEFH or GFP-FS-NEFH showed normal distribution of the microtubule network in cells stained with anti-tubulin. Scale bars represent 10 μm.
(D) Subcellular localization of mitochondria: cells transfected with GFP-WT-NEFH showed an even cytoplasmic distribution of mitochondria, whereas GFP-FS-NEFH-transfected cells showed mitochondria accumulation next to aggregates. Scale bars represent 10 μm.
The NF network has been shown to be important for the spatial subcellular distribution of mitochondria;34 therefore, cells were stained with an antibody against the mitochondrial outer-membrane protein TOM20. Mitochondria were evenly distributed in cells expressing GFP-WT-NEFH; however, in cells expressing GFP-FS-NEFH, mitochondria accumulated adjacently to the NEFH aggregates (Figure 5D). Similarly, it has been shown that NEFL mutations linked to CMT cause altered mitochondrial distribution and co-localization with aggregates.35 Our results support the importance of NF integrity in proper mitochondrial distribution.
The NEFH Frameshift Variant Disrupts Motor Neuron Development in Zebrafish
In order to assess the effects of the NEFH frameshift variant in vivo, we injected RNA into 1-cell-stage zebrafish embryos. Equal amounts of RNA encoding either GFP-WT-NEFH or GFP-FS-NEFH were injected into transgenic Tg(Olig2:DsRed)36 embryos at a dosage at which there was no apparent effect on body morphology (Figure 6A) but a measurable difference in motor neuron outgrowth (Figure 6B). We assessed the common path of the caudal anterior primary motor neurons at 48 hpf. We found that embryos injected with GFP-FS-NEFH RNA had significantly shorter axon lengths than did both GFP-WT-NEFH-injected and uninjected larvae, whereas there was no significant difference between the motor neurons lengths of the uninjected embryos and the embryos injected with GFP-WT-NEFH RNA (Figure 6C). This supports the pathogenicity of NEFH mutations and indicates that the addition of the 3′ UTR-encoded CAE can function through a toxic gain-of-function mechanism.
Figure 6.
Phenotypic Analysis in Zebrafish Embryos Injected with Either Wild-Type or Mutant RNAs
(A) Zebrafish embryos injected with RNA encoding either GFP-WT-NEFH or GFP-FS-NEFH did not show major morphological defects at 48 hpf.
(B) Motor neurons labeled in the transgenic line Tg(Olig2:Dsred). Embryos injected with GFP-WT-NEFH showed normal motor neuron development, whereas zebrafish injected with GFP-FS-NEFH showed examples of stunted axons. The scale bar represents 100 μm.
(C) Quantification of the average axon length shows shorter lengths in GFP-FS-NEFH-injected fishes than in both GFP-WT-NEFH-injected and uninjected larvae. Axon length was not significantly different between GFP-WT-NEFH-injected fishes and uninjected control fish. The average axon length per fish was calculated from the first four myotomes. Data were compiled from three independent experiments, and significance was determined by a one-way ANOVA with a Bonferroni post-test. p values are ∗0.024 and ∗∗0.014.
(D) Confocal images depict relative GFP fluorescence of the tagged NEFH proteins. Scale bars represent 100 μm.
(E) Semiquantitative assessment of relative fluorescence intensity as mean gray values 0–255.
(F) Western blot shows the presence of both wild-type and mutant NEFH proteins at 24 hpf. GFP-FS-NEFH-injected larvae were also incubated in 50 μM chloroquine (Chq) in an attempt to increase the amount of the mutant protein.
(G) The presence of both GFP-WT-NEFH and GFP-FS-NEFH mRNAs was confirmed by RT-PCR.
To correlate the observed phenotype with the presence of the NEFH proteins, we assessed GFP fluorescence at 8 hpf (data not shown) and 24 hpf (Figures 6D and 6E) and quantitatively by western blot at 24 hpf (Figure 6F). Both methods showed markedly decreased amounts of the mutant NEFH, most likely as a result of the toxicity of the protein. We further confirmed that the decreased amount of the mutant NEFH occurred at the protein level by verifying the presence of microinjected RNA (Figure 6G). We speculate that the protein quality-control mechanisms are efficient in larval zebrafish and that toxic misfolded proteins are rapidly degraded. Because motor neuron disease is a progressive degeneration, it is possible that aggregates can form during aging as protein turnover slows down. The effect of the NEFH frameshift variants even among the decreased protein amounts further confirms the pathogenicity of the translation readthrough of the 3′ UTR of NEFH.
Genome-wide Analysis of the Human 3′ UTR-Encoded CAE
In order to investigate additional potential candidate genes whose 3′ UTRs code for CAEs causing aggregation, we performed a bioinformatics aggregation-prediction analysis of all human 3′ UTR sequences. Human 3′ UTR sequences were acquired from the UTRef section of UTRdb, a curated collection of eukaryotic 5′ and 3′ UTRs. The UTRef section contains 34,619 3′ UTR sequences from genes retrieved from RefSeq transcripts.37 We translated these sequences into the three forward reading frames to simulate stop-loss mutations caused by either missense (frame 1) or frameshift (frames 2 and 3) mutations. After we filtered out amino acid sequences with over 90% similarity and genes of uncertain function (LOC symbols), approximately 12,400 genes per reading frame were annotated with the aggregation-prediction programs TANGO and PASTA. Next, sequences were filtered for highly stringent threshold aggregation scores: above 200 for TANGO and below −4 for PASTA. These score cutoffs were based on the aggregation-prediction scores obtained for the NEFH 3′ UTR. Sequences that lack an alternative stop codon were filtered out because they would most likely be degraded by the nonstop-decay mechanism. It has been reasoned that the stability of stop-loss mRNAs and/or proteins decreases as the distance between the mutated stop codon and the next alternative stop codon increases.38 Therefore, only sequences containing an alternative stop codon within a stretch coding for 50 amino acids were considered. After applying these filter criteria, we obtained 4,861 genes (approximately 1,600 genes per reading frame) containing a 3′ UTR sequence with a high potential for aggregation if translated (Figure S4).
Although our results suggest that a large number of genes have the potential to cause aggregation as a result of a stop-loss mutation, several physiological factors that vary in different intracellular micro environments (such as temperature, pH, pressure, and protein concentration) influence aggregation.39 Moreover, the frequency of stop loss caused by a missense mutation within the stop codon (frame 1) is very low: 0.027% (609 of 2,207,918 variants) as observed in the NHLBI ESP Exome Variant Server (ESP6500). Finally, in order to cause significant protein-aggregation disease, the protein must be present in cells that can be negatively affected by aggregations (such as postmitotic neurons), and the presence of the protein must overwhelm the cell’s ability to clear aggregations.
Next, we filtered for disease-associated genes that had previously been reported to cause aggregation and that encode a predicted CAE in any 3′ UTR reading frame. We obtained a list with the top 21 high-risk aggregation genes (Table S2). Although variants in these genes have already been shown to cause aggregation, stop-loss mutations resulting in translation of a CAE from a 3′ UTR have not been reported yet. Because the genes listed encode proteins prone to aggregation, they might be more susceptible to aggregation by our proposed mechanism. We decided to experimentally validate three known ALS-associated genes from that list: FUS, SOD1, and TARDBP (MIM: 605078). Neuro-2a cells transfected with FUS fused in frame with its predicted 3′ UTR-ORF1-encoded CAE (GFP-FUS-CAE) (Figures 7A and 7B) showed prominent aggregation in the cytoplasm and neuronal projections of transfected cells (Figure 7C). In comparison, wild-type FUS (GFP-WT-FUS) localized to the nucleus. Cells transfected with SOD1 and TARDBP fused with their respective 3′ UTR-encoded CAEs did not result in protein aggregation (Figure S5). These results show that aggregation induced by translation of the 3′ UTR-encoded CAE is not a NF-exclusive phenomenon; however, it is important to validate the bioinformatics aggregation prediction of the 3′ UTR-encoded CAE given that other intrinsic protein factors might interfere with protein structure.
Figure 7.
Protein Aggregation in Cultured Neuro-2a Cells Expressing FUS Fused in Frame with Its Predicted 3′ UTR-ORF1-Encoded CAE
(A) TANGO score of ORF1 in the FUS 3′ UTR.
(B) Consensus sequence of positive residues (asterisks) for all aggregation predictors tested for ORF1 in the FUS 3′ UTR.
(C) GFP-WT-FUS localized to the nucleus, whereas GFP-FUS-CAE aggregated in the cytoplasm and neuronal-like projections. A magnified view of the data (M) is shown in the white box. Scale bars represent 10 μm.
Discussion
We describe a protein-aggregation-inducing mechanism triggered by translation of a CAE from the 3′ UTR of NF genes. We report frameshift variants in NEFH in two multigenerational families affected by autosomal-dominant CMT2. Despite distinct stop-loss mutations in each family, both lead to an alternative ORF and translation of an additional 40 amino acids encoded by the 3′ UTR. Computer algorithms used for predicting aggregation identified a 3′ UTR-encoded CAE comprising the last 20 amino acids of the mutant extension. Interestingly, analysis of the 3′ UTR of NEFL, a previously described CMT-associated gene encoding an aggregation-prone protein in ALS, revealed a CAE encoded by ORF1 of the 3′ UTR. Neuro-2a cells transfected with both NEFH and NEFL expressing their predicted amyloidogenic regions resulted in rounded cells with prominent perinuclear aggregates and a lack of axon-like projections. A series of expression constructs for NEFH and NEFL demonstrated that aggregation is specifically caused by translation of the reading frame predicted to contain an amyloidogenic sequence, and it is independent of the length of the extension of the peptides. For NEFL, the presence of a CAE encoded by ORF1 of the NEFL 3′ UTR could induce aggregation if a stop-loss missense mutation or translation termination readthrough occurs. NEFL is terminated by the UGA codon, which has been demonstrated to be the least efficient termination codon during aminoglycoside-induced readthrough.40 Therefore, prolonged use of aminoglycoside antibiotics, such as gentamicin, could potentially induce aggregation of NEFL. Interestingly, neurotoxic effects, including peripheral neuropathy, have been associated with antibiotic use.41 Further studies of NEFL aggregation induced by drugs activating translational readthrough will be required.
Amyloid fibrils and neurofibrillary tangles are associated with severe neurodegenerative disease due to accumulation of toxic protein aggregates, including NF aggregation in motor neuron diseases.9, 18 Although variants in NEFH have been associated with increased susceptibility to ALS, NEFH does not represent a high-penetrance ALS-associated gene.13, 42 Further, the nature of the mutations in our CMT individuals is distinct from that of the coding deletions or insertions of NEFH KLS repeats previously described in ALS individuals.13, 42 Our functional studies demonstrated that the presence of NEFH containing additional amino acids encoded by the 3′ UTR is detrimental to transfected Neuro-2a cells and results in morphologically abnormal rounded cells with fewer projections, reduced viability, and prominent NEFH aggregates. In addition, cells expressing the CMT2 NEFH mutant (GFP-FS-NEFH) revealed co-aggregation with NEFL and disruption of the NF network, sequentially altering cell morphology and mitochondrial distribution. Moreover, zebrafish embryos injected with mRNA encoding the mutant NEFH showed significantly decreased lengths of motor neuron axons than did those injected with the wild-type. Our results are in line with previous studies demonstrating that mutations in NEFL and variants in associated proteins, such as HSPB1 and KIF1A, cause NF aggregation in motor neuron diseases, including axonal CMT, distal motor neuropathy, and hereditary spastic paraplegia (MIM: 604187).18, 43 Combined, these results make it evident that motor neurons are particularly susceptible to NF dysfunction. Therefore, these overlapping but distinct diseases caused by NF abnormalities might share similar mechanisms of pathogenesis. Our genetic, in vitro, and in vivo studies confirm previous hypotheses that aggregation of NF proteins, and especially NEFH, is a powerful cytotoxic event that has detrimental effects on motor neurons. Because the stop-loss mutant NEFH studied retains all major domains intact, the mutant protein most likely retains the ability to interact with other proteins. Yet, we have shown that these interacting proteins are trapped in the aggregates and are thus removed from their normal function. Aggregation of NFs in the cell body might consequently impede transport along axons, given that NFs are major components of the axonal cytoskeleton. The fact that NF proteins have long half-lives and slow turnover rates probably contributes to aggregate accumulation over time in vivo. Therefore, studies aiming to help minimize or clear the aggregates could be beneficial for treating affected individuals.
Finally, our bioinformatics aggregation-prediction analysis of the entire human 3′ UTR collection showed that a large number of genes could potentially be affected if they are translated into a specific reading frame encoding a CAE. We experimentally validated aggregation induced by translation of a CAE from the FUS 3′ UTR, confirming that this is not a NF-exclusive phenomenon. It is commonly implied that stop-loss mutations are associated with a clinical phenotype through a loss-of-function mechanism; however, our studies have revealed the importance of investigating protein aggregation propensity due to CAEs encoded by the 3′ UTR as a toxicity-inducing mechanism.
Acknowledgments
We deeply appreciate the commitment of the families who participated in this study. This work was supported by the NIH (R01NS075764, U54NS065712, and U54NS092091 to S.Z.), the Charcot-Marie-Tooth Association, the Austrian Science Fund (FWF P23223-B19 and P27634FW), and the Muscular Dystrophy Association. We also thank the Inherited Neuropathy Consortium for advice and general support. M.M.R. is grateful to the Medical Research Council (MRC Centre grant G0601943) and the National Institutes of Neurological Diseases and Stroke Office of Rare Diseases (U54NS065712) for their support. Part of this work was undertaken at University College London Hospitals, which received a proportion of funding from the Department of Health’s National Institute for Health Research Biomedical Research Centres.
Published: March 31, 2016
Footnotes
Supplemental Data include five figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.02.022.
Web Resources
The URLs for data presented herein are as follows:
OMIM, http://www.omim.org/
Picard, http://picard.sourceforge.net
UTRDb, http://utrdb.ba.itb.cnr.it/
Supplemental Data
References
- 1.Ross C.A., Poirier M.A. Protein aggregation and neurodegenerative disease. Nat. Med. 2004;10(Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 2.Aguzzi A., O’Connor T. Protein aggregation diseases: pathogenicity and therapeutic perspectives. Nat. Rev. Drug Discov. 2010;9:237–248. doi: 10.1038/nrd3050. [DOI] [PubMed] [Google Scholar]
- 3.Li S.H., Li X.J. Aggregation of N-terminal huntingtin is dependent on the length of its glutamine repeats. Hum. Mol. Genet. 1998;7:777–782. doi: 10.1093/hmg/7.5.777. [DOI] [PubMed] [Google Scholar]
- 4.Gibb W.R., Lees A.J. The relevance of the Lewy body to the pathogenesis of idiopathic Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry. 1988;51:745–752. doi: 10.1136/jnnp.51.6.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LaFerla F.M., Oddo S. Alzheimer’s disease: Abeta, tau and synaptic dysfunction. Trends Mol. Med. 2005;11:170–176. doi: 10.1016/j.molmed.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Blokhuis A.M., Groen E.J., Koppers M., van den Berg L.H., Pasterkamp R.J. Protein aggregation in amyotrophic lateral sclerosis. Acta Neuropathol. 2013;125:777–794. doi: 10.1007/s00401-013-1125-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin H., Schlaepfer W.W. Role of neurofilament aggregation in motor neuron disease. Ann. Neurol. 2006;60:399–406. doi: 10.1002/ana.20965. [DOI] [PubMed] [Google Scholar]
- 8.Johnson-Kerner B.L., Ahmad F.S., Diaz A.G., Greene J.P., Gray S.J., Samulski R.J., Chung W.K., Van Coster R., Maertens P., Noggle S.A. Intermediate filament protein accumulation in motor neurons derived from giant axonal neuropathy iPSCs rescued by restoration of gigaxonin. Hum. Mol. Genet. 2015;24:1420–1431. doi: 10.1093/hmg/ddu556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Llorens J. Toxic neurofilamentous axonopathies -- accumulation of neurofilaments and axonal degeneration. J. Intern. Med. 2013;273:478–489. doi: 10.1111/joim.12030. [DOI] [PubMed] [Google Scholar]
- 10.Brownlees J., Ackerley S., Grierson A.J., Jacobsen N.J., Shea K., Anderton B.H., Leigh P.N., Shaw C.E., Miller C.C. Charcot-Marie-Tooth disease neurofilament mutations disrupt neurofilament assembly and axonal transport. Hum. Mol. Genet. 2002;11:2837–2844. doi: 10.1093/hmg/11.23.2837. [DOI] [PubMed] [Google Scholar]
- 11.Hoffman P.N., Cleveland D.W., Griffin J.W., Landes P.W., Cowan N.J., Price D.L. Neurofilament gene expression: a major determinant of axonal caliber. Proc. Natl. Acad. Sci. USA. 1987;84:3472–3476. doi: 10.1073/pnas.84.10.3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carter J., Gragerov A., Konvicka K., Elder G., Weinstein H., Lazzarini R.A. Neurofilament (NF) assembly; divergent characteristics of human and rodent NF-L subunits. J. Biol. Chem. 1998;273:5101–5108. doi: 10.1074/jbc.273.9.5101. [DOI] [PubMed] [Google Scholar]
- 13.Figlewicz D.A., Krizus A., Martinoli M.G., Meininger V., Dib M., Rouleau G.A., Julien J.P. Variants of the heavy neurofilament subunit are associated with the development of amyotrophic lateral sclerosis. Hum. Mol. Genet. 1994;3:1757–1761. doi: 10.1093/hmg/3.10.1757. [DOI] [PubMed] [Google Scholar]
- 14.Al-Chalabi A., Andersen P.M., Nilsson P., Chioza B., Andersson J.L., Russ C., Shaw C.E., Powell J.F., Leigh P.N. Deletions of the heavy neurofilament subunit tail in amyotrophic lateral sclerosis. Hum. Mol. Genet. 1999;8:157–164. doi: 10.1093/hmg/8.2.157. [DOI] [PubMed] [Google Scholar]
- 15.Berciano J., García A., Peeters K., Gallardo E., De Vriendt E., Pelayo-Negro A.L., Infante J., Jordanova A. NEFL E396K mutation is associated with a novel dominant intermediate Charcot-Marie-Tooth disease phenotype. J. Neurol. 2015;262:1289–1300. doi: 10.1007/s00415-015-7709-4. [DOI] [PubMed] [Google Scholar]
- 16.Jordanova A., De Jonghe P., Boerkoel C.F., Takashima H., De Vriendt E., Ceuterick C., Martin J.J., Butler I.J., Mancias P., Papasozomenos S.Ch. Mutations in the neurofilament light chain gene (NEFL) cause early onset severe Charcot-Marie-Tooth disease. Brain. 2003;126:590–597. doi: 10.1093/brain/awg059. [DOI] [PubMed] [Google Scholar]
- 17.Yum S.W., Zhang J., Mo K., Li J., Scherer S.S. A novel recessive Nefl mutation causes a severe, early-onset axonal neuropathy. Ann. Neurol. 2009;66:759–770. doi: 10.1002/ana.21728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhai J., Lin H., Julien J.P., Schlaepfer W.W. Disruption of neurofilament network with aggregation of light neurofilament protein: a common pathway leading to motor neuron degeneration due to Charcot-Marie-Tooth disease-linked mutations in NFL and HSPB1. Hum. Mol. Genet. 2007;16:3103–3116. doi: 10.1093/hmg/ddm272. [DOI] [PubMed] [Google Scholar]
- 19.Evgrafov O.V., Mersiyanova I., Irobi J., Van Den Bosch L., Dierick I., Leung C.L., Schagina O., Verpoorten N., Van Impe K., Fedotov V. Mutant small heat-shock protein 27 causes axonal Charcot-Marie-Tooth disease and distal hereditary motor neuropathy. Nat. Genet. 2004;36:602–606. doi: 10.1038/ng1354. [DOI] [PubMed] [Google Scholar]
- 20.Ackerley S., James P.A., Kalli A., French S., Davies K.E., Talbot K. A mutation in the small heat-shock protein HSPB1 leading to distal hereditary motor neuronopathy disrupts neurofilament assembly and the axonal transport of specific cellular cargoes. Hum. Mol. Genet. 2006;15:347–354. doi: 10.1093/hmg/ddi452. [DOI] [PubMed] [Google Scholar]
- 21.Link V., Shevchenko A., Heisenberg C.P. Proteomics of early zebrafish embryos. BMC Dev. Biol. 2006;6:1. doi: 10.1186/1471-213X-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez M., Falk M.J., Gai X., Postrel R., Schüle R., Zuchner S. Innovative genomic collaboration using the GENESIS (GEM.app) platform. Hum. Mutat. 2015;36:950–956. doi: 10.1002/humu.22836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalez M.A., Lebrigio R.F., Van Booven D., Ulloa R.H., Powell E., Speziani F., Tekin M., Schüle R., Züchner S. GEnomes Management Application (GEM.app): a new software tool for large-scale collaborative genome analysis. Hum. Mutat. 2013;34:842–846. doi: 10.1002/humu.22305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Q., Xie F., Siedlak S.L., Nunomura A., Honda K., Moreira P.I., Zhua X., Smith M.A., Perry G. Neurofilament proteins in neurodegenerative diseases. Cell. Mol. Life Sci. 2004;61:3057–3075. doi: 10.1007/s00018-004-4268-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kovach M.J., Campbell K.C., Herman K., Waggoner B., Gelber D., Hughes L.F., Kimonis V.E. Anticipation in a unique family with Charcot-Marie-Tooth syndrome and deafness: delineation of the clinical features and review of the literature. Am. J. Med. Genet. 2002;108:295–303. doi: 10.1002/ajmg.10223. [DOI] [PubMed] [Google Scholar]
- 26.Steiner I., Gotkine M., Steiner-Birmanns B., Biran I., Silverstein S., Abeliovich D., Argov Z., Wirguin I. Increased severity over generations of Charcot-Marie-Tooth disease type 1A. J. Neurol. 2008;255:813–819. doi: 10.1007/s00415-008-0693-1. [DOI] [PubMed] [Google Scholar]
- 27.Fernandez-Escamilla A.M., Rousseau F., Schymkowitz J., Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 28.de Groot N.S., Castillo V., Graña-Montes R., Ventura S. AGGRESCAN: method, application, and perspectives for drug design. Methods Mol. Biol. 2012;819:199–220. doi: 10.1007/978-1-61779-465-0_14. [DOI] [PubMed] [Google Scholar]
- 29.Garbuzynskiy S.O., Lobanov M.Y., Galzitskaya O.V. FoldAmyloid: a method of prediction of amyloidogenic regions from protein sequence. Bioinformatics. 2010;26:326–332. doi: 10.1093/bioinformatics/btp691. [DOI] [PubMed] [Google Scholar]
- 30.Walsh I., Seno F., Tosatto S.C., Trovato A. PASTA 2.0: an improved server for protein aggregation prediction. Nucleic Acids Res. 2014;42:W301–W307. doi: 10.1093/nar/gku399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bidou L., Allamand V., Rousset J.P., Namy O. Sense from nonsense: therapies for premature stop codon diseases. Trends Mol. Med. 2012;18:679–688. doi: 10.1016/j.molmed.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 32.Groenning M. Binding mode of Thioflavin T and other molecular probes in the context of amyloid fibrils-current status. J. Chem. Biol. 2010;3:1–18. doi: 10.1007/s12154-009-0027-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kokubo Y., Kuzuhara S., Narita Y., Kikugawa K., Nakano R., Inuzuka T., Tsuji S., Watanabe M., Miyazaki T., Murayama S., Ihara Y. Accumulation of neurofilaments and SOD1-immunoreactive products in a patient with familial amyotrophic lateral sclerosis with I113T SOD1 mutation. Arch. Neurol. 1999;56:1506–1508. doi: 10.1001/archneur.56.12.1506. [DOI] [PubMed] [Google Scholar]
- 34.Wagner O.I., Lifshitz J., Janmey P.A., Linden M., McIntosh T.K., Leterrier J.F. Mechanisms of mitochondria-neurofilament interactions. J. Neurosci. 2003;23:9046–9058. doi: 10.1523/JNEUROSCI.23-27-09046.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pérez-Ollé R., López-Toledano M.A., Goryunov D., Cabrera-Poch N., Stefanis L., Brown K., Liem R.K. Mutations in the neurofilament light gene linked to Charcot-Marie-Tooth disease cause defects in transport. J. Neurochem. 2005;93:861–874. doi: 10.1111/j.1471-4159.2005.03095.x. [DOI] [PubMed] [Google Scholar]
- 36.Kucenas S., Snell H., Appel B. nkx2.2a promotes specification and differentiation of a myelinating subset of oligodendrocyte lineage cells in zebrafish. Neuron Glia Biol. 2008;4:71–81. doi: 10.1017/S1740925X09990123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grillo G., Turi A., Licciulli F., Mignone F., Liuni S., Banfi S., Gennarino V.A., Horner D.S., Pavesi G., Picardi E., Pesole G. UTRdb and UTRsite (RELEASE 2010): a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2010;38:D75–D80. doi: 10.1093/nar/gkp902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamby S.E., Thomas N.S., Cooper D.N., Chuzhanova N. A meta-analysis of single base-pair substitutions in translational termination codons (‘nonstop’ mutations) that cause human inherited disease. Hum. Genomics. 2011;5:241–264. doi: 10.1186/1479-7364-5-4-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang L., Liu X., Cheng B., Huang K. How our bodies fight amyloidosis: effects of physiological factors on pathogenic aggregation of amyloidogenic proteins. Arch. Biochem. Biophys. 2015;568:46–55. doi: 10.1016/j.abb.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 40.Howard M.T., Shirts B.H., Petros L.M., Flanigan K.M., Gesteland R.F., Atkins J.F. Sequence specificity of aminoglycoside-induced stop condon readthrough: potential implications for treatment of Duchenne muscular dystrophy. Ann. Neurol. 2000;48:164–169. [PubMed] [Google Scholar]
- 41.Grill M.F., Maganti R.K. Neurotoxic effects associated with antibiotic use: management considerations. Br. J. Clin. Pharmacol. 2011;72:381–393. doi: 10.1111/j.1365-2125.2011.03991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skvortsova V., Shadrina M., Slominsky P., Levitsky G., Kondratieva E., Zherebtsova A., Levitskaya N., Alekhin A., Serdyuk A., Limborska S. Analysis of heavy neurofilament subunit gene polymorphism in Russian patients with sporadic motor neuron disease (MND) Eur. J. Hum. Genet. 2004;12:241–244. doi: 10.1038/sj.ejhg.5201144. [DOI] [PubMed] [Google Scholar]
- 43.Wang L., Brown A. A hereditary spastic paraplegia mutation in kinesin-1A/KIF5A disrupts neurofilament transport. Mol. Neurodegener. 2010;5:52. doi: 10.1186/1750-1326-5-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.