Abstract
Deficits in the basal ganglia pathways modulating cortical motor activity underlie both Parkinson disease (PD) and Huntington disease (HD). Phosphodiesterase 10A (PDE10A) is enriched in the striatum, and animal data suggest that it is a key regulator of this circuitry. Here, we report on germline PDE10A mutations in eight individuals from two families affected by a hyperkinetic movement disorder due to homozygous mutations c.320A>G (p.Tyr107Cys) and c.346G>C (p.Ala116Pro). Both mutations lead to a reduction in PDE10A levels in recombinant cellular systems, and critically, positron-emission-tomography (PET) studies with a specific PDE10A ligand confirmed that the p.Tyr107Cys variant also reduced striatal PDE10A levels in one of the affected individuals. A knock-in mouse model carrying the homologous p.Tyr97Cys variant had decreased striatal PDE10A and also displayed motor abnormalities. Striatal preparations from this animal had an impaired capacity to degrade cyclic adenosine monophosphate (cAMP) and a blunted pharmacological response to PDE10A inhibitors. These observations highlight the critical role of PDE10A in motor control across species.
Introduction
Cyclic adenosine monophosphate (cAMP) and cyclic guanosine monophosphate (cGMP) are essential second messengers regulating multiple signaling pathways in virtually all cell types.1 Intracellular levels are finely regulated by cyclic nucleotide phosphodiesterases (PDEs), which degrade cAMP and cGMP by hydrolysis of both cAMP and cGMP to the corresponding nucleoside 5′ monophosphate.2 PDE10A (MIM: 610652) encodes phosphodiesterase 10A, a dual cAMP-cGMP phosphodiesterase enriched in the medium spiny neurons (MSNs) of the corpus striatum.3 The underlying pathology of the classic movement disorders Parkinson disease (PD [MIM: 168600]) and Huntington disease (HD [MIM: 143100]) focuses on a loss of dopamine-producing cells in the substantia nigra in PD and striatal MSNs in HD.4, 5 This results in dysregulated cortical striatal-thalamic connections, contributing to both motor and cognitive symptoms.6, 7 PDE10A modulates G-protein-coupled signaling, including that due to activation of dopamine receptors D1 and D2, by degrading cyclic nucleotides and in animal models regulates the basal ganglia circuitry.8, 9 These observations have stimulated extensive research into PDE10A as a therapeutic target,10 despite little physiological data in humans. By a combination of homozygosity mapping and whole-exome sequencing (WES), we identified biallelic missense variants in two families affected by a hyperkinetic movement disorder. Cellular modeling of the variants revealed lower PDE10A levels in cells expressing the mutant than in cells expressing the wild-type (WT). Crucially, this recapitulated our observations in one of the individuals, who showed a marked loss of striatal PDE01A by positron-emission-tomography (PET) scanning. In a knock-in (KI) mouse model, we recapitulated the loss of striatal PDE10A, and the mice also displayed a motor phenotype. This suggests that there is strong back-translation of the human gene-phenotype findings in individuals with biallelic PDE10A mutations.
Subjects and Methods
Subject Evaluation
The families provided signed informed consent to participate in studies approved by the Leeds East research ethics committee (07/H1306/113; family 1) and by the ethics committee of the Oulu University Hospital (EETTMK 51/2008; family 2).
Two individuals underwent video recordings for further documentation of their movement disorders (Movies S1 and S2).
Two families affected by a distinct movement disorder were independently ascertained and investigated (Figure 1A). Individual IV:2 (family 1) underwent a PET scan with [11C]IMA107, a selective PDE10A PET radiotracer, allowing a robust quantification of PDE10A levels.
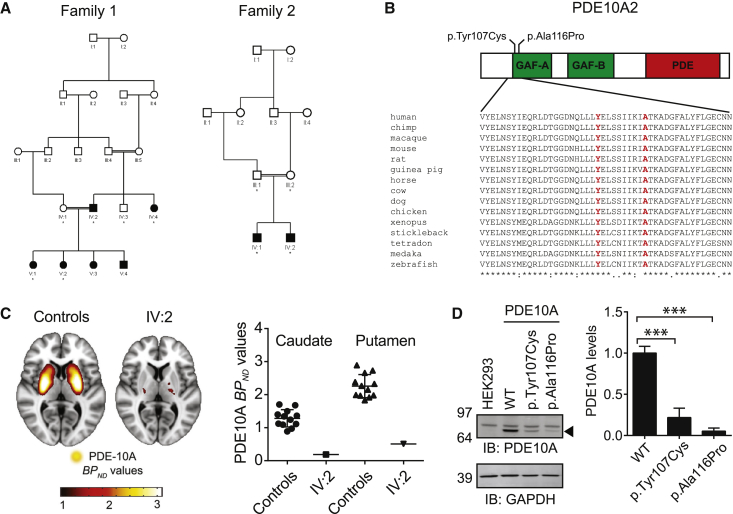
Figure 1.
Clinical Data on the Two Affected Families and Data on PDE10A Levels in HEK293 Cells
(A) Pedigrees of the two families. Affected individuals are shaded, and asterisks indicate that DNA was obtained for genetic analysis.
(B) Schematic diagram of PDE10A. The variants in the GAF-A domain are highlighted, and high conservation of the amino acids is shown across species.
(C) The level of PDE10A was reduced in an individual with a PDE10A mutation. 11C-IMA107 BPND parametric images derived from 12 healthy control individuals (left) and family 1 individual IV:2 (right) in stereotaxic space are overlaid onto the T1-weighted Montreal Neurological Institute template (representing a brain of average shape) and show significant loss of striatal and pallidal PDE10A signal in IV:2. The scatterplot shows 11C- IMA107 BPND values in the caudate and putamen for healthy control individuals (n = 12) and family 1 individual IV:2. Solid lines represent mean 11C-IMA107 BPND values + 1 SD for healthy control individuals and IV:2.
(D) PDE10A levels compared among WT, p.Tyr107Cys, and p.Ala116Pro proteins in HEK293 cells (right) and representative immunoblots from PDE10A transfection experiments (left). Levels of WT and mutant PDE10A were measured in HEK293 cells by immunoblotting using a specific PDE10A polyclonal antibody (Rbg426v2). The PDE10A band is highlighted with an arrowhead. The amount of mutant proteins was lower than that of the WT. Quantification of PDE10A in transfected HEK293 cells was normalized to GAPDH levels. Experiments were replicated four times with four technical replicates per experiment. Data are presented as a group mean ± SEM. ∗∗∗p < 0.001.
Neuroimaging Evaluation
Image Acquisition
PET and MRI scans were performed at Imanova. After providing written informed consent, participants were instructed to refrain from caffeine, tobacco, and alcohol for at least 12 hr before scanning. Healthy control individuals were recruited as part of an ongoing study.11 Subjects were positioned supine, and head position was maintained as previously described.11 All participants were scanned on a Siemens Biograph HI-REZ 6 PET-CT scanner after the injection of an intravenous bolus of ∼250 MBq [11C]IMA107. Dynamic emission data were acquired continuously for 90 min after the injection of [11C]IMA107. The dynamic images were reconstructed into 26 frames (8 × 15 s, 3 × 60 s, 5 × 120 s, 5 × 300 s, and 5 × 600 s) with in-house software and a filtered back-projection algorithm (direct inverse Fourier transform) with a 128 × 128 matrix and 2.6× zoom, producing images with isotropic voxel size of 2 × 2 × 2 mm3 and a transaxial Gaussian filter of 5 mm.
MRI scans were acquired with a 32-channel head coil on a Siemens MAGNETOM Verio 3T MRI scanner and included (1) a T1-weighted magnetization-prepared rapid gradient-echo (MPRAGE) sequence (time repetition [TR] = 2,300 ms, time echo [TE] = 2.98 ms, flip angle = 9°, time to inversion [TI] = 900 ms, and matrix = 240 × 256) for co-registration with the PET images and (2) fast GM T1 inversion recovery (FGATIR; TR = 3,000 ms, TE = 2.96 ms, flip angle = 8°, TI = 409 ms, and matrix = 240 × 256) and fluid and WM suppression (FLAWS; TR = 5,000 ms, TE = 2.94 ms, flip angle = 5°, TI = 409/1,100 ms, and matrix = 240 × 256) sequences for improving delineation of subcortical brain regions. All sequences used a 1 mm3 voxel size, anteroposterior phase-encoding direction, and symmetric echo.
Processing of [11C]IMA107 PET Data
Data analysis was performed with the MIAKAT software package (v.3.3.8, Imanova). Image processing and kinetic modeling of [11C]IMA107 PET data were blind to subject groups and performed with a standard template of regions of interest (see Niccolini et al.12 for a full description). The primary outcome measure was the non-displaceable binding potential (BPND) of [11C]IMA107 in the striatum, which is proportional to the ratio of the Bmax for PDE10A to the dissociation constant of [11C]IMA107 for PDE10A.
Genetic Analysis
In view of the consanguinity in both families, genetic analysis was performed under a recessive model. In family 1, we performed a genome-wide homozygosity scan by using the Affymetrix Human SNP Array 6.0 on DNA from four affected and two unaffected individuals. Genotype data were analyzed with AutoSNPa and IBDfinder software.13, 14
Two affected individuals from family 2 were investigated by chromosomal microarray on DNA extracted from fibroblasts with a 50-mer oligochip (HumanCytoSNP-12 v.2.1 BeadChip, Illumina). Analysis was performed with both GenomeStudio (hg19) and KaryoStudio (hg18) programs (Illumina).
WES
Analyses for Family 1
Alignment and Variant Calling. Target capture was performed with the Agilent SureSelect Human All Exon v.4 exome enrichment kit according to the manufacturer’s standard protocol. Sequencing of 150-bp paired-end reads was performed with an Illumina MiSeq. Reads were aligned to GRCh37 with Novoalign (Novocraft Technologies) and processed with the Genome Analysis Toolkit (GATK) and Picard (see Web Resources) for realignment of short indels and removal of duplicate reads. Depth of coverage of the consensus coding sequence (CCDS) was assessed with GATK, which showed that >94% of CCDS bases were covered by at least five good-quality reads (minimum Phred-like base quality of 17 and minimum mapping quality of 20). Single-nucleotide variants (SNVs) and indels were called with the GATK UnifiedGenotyper feature.15, 16
Filtering. Custom Perl scripts were used for removing variants present in dbSNP132 or with a minor allele frequency ≥ 0.1% and for annotating functional consequences. We called variants in the autozygous region only with a minimum Phred-like genotype quality of 30 and selected for indels within coding regions, non-synonymous SNVs, and splice-site variants. Variants present in the 1000 Genomes dataset (November 2011), the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project (ESP) Exome Variant Server, and another 2,500 ethnically matched in-house exomes were also removed.
Analyses for Family 2
Target capture was performed with the Agilent SureSelect Human All Exon v.4.1 according to the manufacturer’s standard protocol. Sequencing was performed on the Illumina GAIIx instrument, generating 100 bp paired-end reads. In brief, the Burrows-Wheeler Aligner (v.0.5.9) was used to align raw reads against the reference human genome (NCBI Genome build GRCh37).17 Local realignment around indels and coverage assessment were performed with GATK,16 and removal of duplicate reads was performed with Picard Tools (v.1.53). A mean coverage of 127× was obtained for all CCDS exons. The positions of variants on the genome were detected by SAMtools (v.0.1.17).18 Only variants covered with at least five reads and a quality score higher than 20 were considered. Subsequently, ANNOVAR was used for assigning annotations to the list of detected variants.19 To determine potential candidate variants, we focused on likely protein-damaging variants (frameshift, indel, nonsense, missense, and canonical splice-site variants) and discarded any variants with an allele frequency > 1% in the ExAC Browser or our in-house exome database (∼1,000 exomes). We also removed all variants seen as homozygous in the ExAC Browser.
HEK293 Cell Transfection Experiments
HEK293 cells were transfected with human PDE10A (WT, c.320A>G, or c.346G>C) in the pcDNA3.1 vector with the use of Lipofectamine 2000 (Life Technologies 11668-019) according to the manufacturer’s instructions. Samples for protein analysis were harvested after 24 hr. For every million cells plated on a 10 cm dish, 2 ug of plasmid and 30 μl of Lipofectamine 2000 were used. Lysates were analyzed by immunoblot as described below.
Construction of a PDE10A p.Tyr97Cys Targeting Vector and Targeted Murine Embryonic Stem Cells
We constructed a murine Pde10a (MGI: 1345143) targeting vector to create the p.Tyr97Cys variant, which is homologous to the human p.Tyr107Cys variant. Red/ET recombineering technology20 was used to target a loxP-flanked neomycin cassette into intron 4 of the Pde10a locus along with the TAT:TGT codon mutation in exon 4 of a murine bacterial artificial clone (BAC) (RP23-67C3, Invitrogen). Again via Red/ET recombineering, we cloned approximately 11 kb of the Pde10a locus, including the newly introduced neomycin cassette along with the p.Tyr97Cys genetic modification, from the BAC into a pUC57 plasmid to generate the final sequence-confirmed targeting vector. 5′ and 3′ homology arms were 5.4 and 5.5 kb, respectively (Figure S4).
The linearized PDE10A p.Tyr97Cys targeting vector was electroporated into murine Bruce4 embryonic stem (ES) cells (from Colin Stewart, National Cancer Institute), which were grown according to standard procedures previously described.21 After G418 selection, correctly targeted homologous recombinant ES cell clones were identified by Southern blot analysis. A probe outside of the 5′ homology arm was used in conjunction with MscI-digested ES cell genomic DNA. With this strategy, the endogenous WT allele yielded a band of ∼11 kb, whereas the targeted allele revealed a predicted RFLP of ∼8 kb as a result of the introduction of a novel MscI site from the PDE10A p.Tyr97Cys targeting vector. To ensure that correct homologous recombination occurred at the 3′ end, we confirmed targeting by Southern with BamHI-digested ES cell genomic DNA in combination with a probe external to the 3′ homology arm.
Generating KI Mice Carrying PDE10A p.Tyr97Cys
Targeted ES cells carrying the PDE10A p.Tyr97Cys variant with a normal 40XY karyotype (Coriell Institute for Medical Research) were microinjected into BALB/c blastocyst-stage embryos (Charles River Laboratories) according to standard procedures previously described.22 Resulting chimeric males were mated to EIIa-Cre transgenic females for the generation of offspring heterozygous for PDE10A p.Tyr97Cys in the germline and free of the neomycin selection cassette. Offspring from these and all subsequent matings were genotyped by qPCR analysis at Transnetyx; offspring with homozygous KI of PDE10A p.Tyr97Cys along with WT controls were then generated from heterozygous mating pairs. The Pfizer Institutional Animal Care and Use Committee reviewed and approved the animal use in these studies. The animal care and use program is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International.
Behavioral Phenotyping of KI Mice Carrying p.Tyr97Cys
Upon arrival at the animal facility, separate cohorts of young (4–5 weeks) and adult (7–10 weeks) WT/WT (WT), WT/KI (HET), and KI/KI (KI) male mice (n = 7–13 per genotype) were individually housed in standard ventilated caging on a 12/12 hr light/dark cycle (lights on at 6:00 a.m.) and were provided with food and water ad libitum. Mice were acclimatized for a minimum of 5 days prior to testing. Mice were subjected to a battery of behavioral tests during the light cycle with a minimum 2 day inter-test interval (ITI). Mice from the young cohort were maintained in the facility at the conclusion of initial phenotyping until they reached 24–25 weeks of age, at which time they were re-evaluated as aged mice. The phenotyping battery consisted of the SHIRPA protocol23 and then the following tests in order.
Rotarod
Balance and coordination were evaluated in test mice with an automated accelerating rotarod (Rotamex; Columbus Instruments). The rotarod consisted of four lanes separated by a visual barrier with a rotating spindle (3.0 cm diameter) elevated 44.5 cm from the floor. Infrared beams placed at the level of the spindle detected the presence or absence of the animal on the spindle. Mice were placed on the rotarod at 4 RPM, which was accelerated at a rate of 1 RPM per 8 s throughout a 5 min trial period. Mice were subjected to five consecutive trials separated by a 30 ± 5 min ITI. Trials ended at the conclusion of the 5 min trial period or when the mouse fell from the rod. Latency to fall (s) was recorded and analyzed by a two-way repeated-measures ANOVA (genotype × trial) with GraphPad Prism v.5.0 and Bonferroni post hoc tests as appropriate.
Open-Field Activity
VersaMax chambers for monitoring animal activity (AccuScan Instruments) were used for assessing alterations in general exploratory behavior. Chambers were housed in a testing room with environmental conditions similar to those of the housing room (∼400 lux and 60–70 dB background noise). Mice were habituated to the testing room for a minimum 60 min habituation period prior to testing. After habituation, mice were placed into individual open-field chambers, and behavior was recorded in the open field (40 × 40 × 40 cm) for a 60 min period. Infrared beams captured total distance traveled (cm) and rearing behavior (vertical activity measured by beam breaks). Data were analyzed by two-way repeated-measures ANOVA (genotype × time) with GraphPad Prism v.5.0 and Bonferroni post hoc tests as appropriate.
Dynamic Weight Bearing
The dynamic weight-bearing (DWB) test is a type of incapacitance test used to measure postural equilibrium in freely moving rodents (BioSeb). The apparatus consists of a Plexiglas enclosure (22 × 22 × 30 cm) with a floor instrument cage and video-camera interface that can independently measure the weight borne by each limb of the freely moving subject. Mice were placed individually in the apparatus for a 3–5 min period (no acclimation period). A trained observer blind to genotype mapped left and right hindpaws and forepaws according to the video. Raw paw-pressure values interfaced with video data were analyzed by DWB software. Data for individual measures were analyzed by one-way ANOVA with Dunnett’s post hoc test as appropriate.
Immunoblotting
Mouse striatal tissue or HEK293 cells were lysed in RIPA buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1.0% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS supplemented with protease and phosphatase inhibitor cocktails) and homogenized by brief sonication. Equal amounts of protein were separated in a reducing 4%–12% Tris-Glycine gel (Invitrogen) and blotted onto a nitrocellulose membrane (Hybond-ECL, GE). Protein levels were determined by fluorescent or ECl-based immunoblotting followed by densitometric analysis (Li-Cor-Odyssey, ImageJ). The following antibodies were used: mouse-anti-PDE10A-24F3.F11,24 rabbit-anti-PDE10A-426v2,25 mouse-anti-GAPDH-MAB374 (Millipore), goat-anti-Rabbit-IRDye680 (Li-Cor), goat-anti-Mouse-IRDye800 (Li-Cor), and donkey-anti-Mouse-HRP (Jackson). Data were analyzed by one-way ANOVA with GraphPad Prism v.5.0.
Results
Clinical Characterization of Families
Family 1 is a consanguineous UK family of Pakistani origin (Figure 1A, left). All affected individuals presented in childhood (mean age of 3 months) with axial hypotonia and a generalized hyperkinetic movement disorder. This was characterized by dyskinesia of the limbs and trunk. At times, the hyperkinetic movements had a jerky quality, and intermittent chorea and ballismus were also present. Facial involvement was evident with orolingual dyskinesia, drooling, and dysarthria. The severity of the hyperkinesis varies considerably within the family, and the oldest individual is the least affected. Cognitive performance is preserved, and all affected individuals graduated from high school.
Family 2 is from northern Finland (Figure 1A, right). Two affected boys were born to healthy first-cousin parents. Both boys show mild cognitive delay and severe axial hypotonia. From early infancy, they presented with a generalized hyperkinetic movement disorder characterized by dyskinesia affecting all four limbs and the trunk and face (Movies S1 and S2). Expressive language was not evident until 7 years of age, and both individuals have persistent dysarthria.
IV:2 is more severely affected; he has had focal epilepsy since 3.5 years of age and feeds via a gastrostomy tube. Further clinical details can be found in Table S1.
In both families, array comparative genomic hybridization, muscle biopsy, and, crucially, brain MRIs in individuals IV:2, V:2, and V:3 from family 1 and both of the affected individuals from family 2 were normal. Further details are provided in the Supplemental Note.
Genetic Analyses Reveal Missense Mutations in PDE10A
In family 1, genome-wide SNP analysis revealed a single 2.6 Mb region of concordant homozygosity on 6q26. WES revealed one potentially pathogenic variant within the homozygous region: c.320A>G (p.Tyr107Cys) in exon 4 of PDE10A (GenBank: NM_001130690.2) (Figure 1B). In family 2, there were 16 common autozygous regions, the three largest of which clustered on 6q26. WES revealed a single potentially deleterious mutation within the homozygous segments: c.346G>C (p.Ala116Pro), also in exon 4 of PDE10A (Figure 1B). Neither variant was present in dbSNP, 1000 Genomes, the NHLBI ESP, or our in-house exome databases. Both individual variants are fully conserved back to Danio rerio (Figure 1B), lie within a region of very high conservation, and are predicted to be deleterious.26 Sanger sequencing confirmed the presence of the variants and that they segregate with the phenotype in the families.
PDE10A Levels Are Decreased in an Individual with the p.Tyr107Cys Variant
Given that there are now well-characterized PET ligands for PDE10A, we wanted to understand the impact of the mutations on PDE10A levels in the human brain. Because of ethical considerations, we were able to study only individual IV:2 from family 1 in this manner. PDE10A PET signal was robustly lower in this individual than in healthy control individuals (Figure 1C), and quantitative analysis revealed a 70% reduction in PDE10A in all basal ganglia regions (Figure 1C and Figure S1). An accompanying structural MRI showed that this loss of PDE10A signal was not due to loss of striatal volume (see Figure S2).
Effect of PDE10A Mutations on PDE10A Levels in HEK293 Cells
To evaluate the functional consequence of the PDE10A mutations, we expressed WT and mutant PDE10A (c.320A>G and c.346G>C) in HEK293 cells. PDE10A levels were investigated by immunoblotting using validated antibodies (see Figure S3). A significant decrease in PDE10A levels was associated with both mutations (Figure 1D), consistent with the human PET study above. The reduction of PDE10A p.Ala116Pro was greater than that of PDE10A p.Tyr107Cys, perhaps reflecting the more severe phenotype seen in the Finnish family.
A Mouse Model Carrying PDE10A p.Tyr97Cys, Homologous to Human PDE10A p.Tyr107Cys, Displays Deficits in Motor Control
We next investigated KI mice carrying the PDE10A variant p.Tyr97Cys, which is homologous to the human p.Tyr107Cys variant (Figure S4). SHIRPA phenotyping of the HET and KI mice did not reveal any behavioral phenotypes.23 We therefore investigated locomotor activity in young (4–5 weeks of age), adult (7–10 weeks of age), and old animals (24–25 weeks of age). Data for adult mice are shown in Figures 2A–2C, and data for young and old mice are presented in Figure S5.
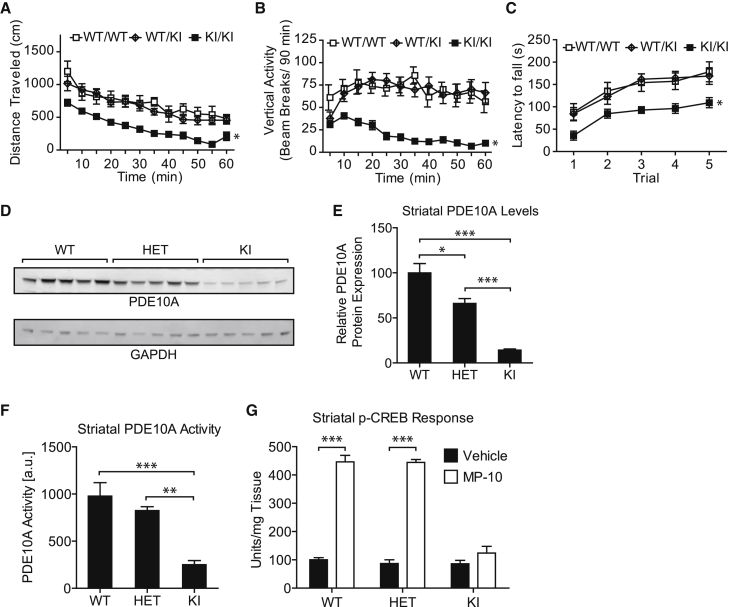
Figure 2.
Adult KI Mice with PDE10A p.Tyr97Cys Demonstrate Motor Abnormalities and Reduced Striatal PDE10A Levels and Function
(A and B) Total distance traveled (A) and rearing behavior (B) as measured by vertical activity were recorded and analyzed by a two-way repeated-measures ANOVA (genotype × trial) with GraphPad Prism v.5.0 and Bonferroni post hoc tests as appropriate. All data are presented as the group mean ± SEM. n = 13, 12, and 11 for WT (WT/WT), HET (WT/KI), and KI (KI/KI) mice, respectively. All adult mice were 7–10 weeks old. ∗p < 0.05.
(C) Latency to fall (s) was recorded and analyzed by a two-way repeated-measures ANOVA (genotype × trial) with GraphPad Prism v.5.0 and Bonferroni post hoc tests as appropriate. KI mice carrying the homologous p.Tyr97Cys variant showed a hypokinetic phenotype, deficits in rearing activity, and deficits in performance on the rotorod. ∗p < 0.05.
(D) The PDE10A p.Tyr97Cys variant led to a reduction in PDE10A. Representative immunoblots of striatal protein lysates from adult mice from each genotype (n = 5 for WT, HET, and KI) were analyzed by immunoblotting for levels of PDE10A with a specific polyclonal PDE10A antibody (Rbg426v2). GAPDH was used as a loading control.
(E) Quantification of PDE10A levels in striatal lysates. Data are presented as the group mean ± SEM. PDE10A levels were normalized to GAPDH levels. Normalized data were analyzed by one-way ANOVA with GraphPad Prism v.5.0. PDE10A levels were measured in the striatum from adult mice of all three genotypes (n = 5 for WT, HET, and KI). Levels of PDE10A p.Tyr97Cys were lower than WT protein levels. ∗p < 0.05, ∗∗∗p < 0.001.
(F) The PDE10A p.Tyr97Cys variant led to a reduction in enzymatic activity. PDE10A enzyme activity was measured in striatal lysates from adult mice of all three genotypes (n = 5 for WT, HET, and KI). Enzyme activity was measured by means of a scintillation proximity assay modified from an Amersham Biosciences protocol (TRKQ7090). The assay measures levels of 5′ AMP produced when cAMP is exposed to phosphodiesterase. Data are presented as the group mean ± SEM and were analyzed by one-way ANOVA with GraphPad Prism v.5.0. KI mice showed a reduction in PDE10A activity. ∗∗p < 0.01, ∗∗∗p < 0.001.
(G) The PDE10A p.Tyr97Cys variant led to a reduction in downstream signaling. Downstream PDE10A signaling was measured indirectly by quantifying pCREB levels in striatum from adult mice of all three genotypes (n = 5 for WT, HET, and KI), which had been dosed with either the selective PDE10A inhibitor MP-10 or vehicle. pCREB levels were higher in both WT and HET animals dosed with the PDE10A inhibitor than in vehicle-treated animals. Data are presented as the group mean ± SEM and were analyzed by a paired t test with GraphPad Prism v.5.0. ∗∗∗p < 0.001.
KI mice showed significantly less total distance traveled (Figure 2A and Figure S5, left) and rearing behavior (Figure 2B and Figure S5, middle) than did age-matched HET and WT controls. Relative to age-matched HET and WT controls, KI mice also demonstrated a reduced ability to maintain their balance on an accelerating rotarod (Figure 2C and Figure S5, right). In a DWB assay, relative to both HET and WT littermate controls, KI mice demonstrated statistically significant increases in forepaw bearing weight, whereas hindpaw weights were reduced (Figure S6). Together, these data show that KI mice display motor abnormalities.
p.Tyr97Cys KI Mice Have Reduced Levels of PDE10A in the Striatum
Consistent with the PET data on human PDE10A and the protein studies in HEK293 cells, KI mice showed lower striatal protein levels than did age-matched WT (p < 0.001) and HET animals (p < 0.001; Figures 2D and 2E). Striatal tissue from KI mice showed lower levels of PDE10A activity than did that of age-matched WT (p < 0.001) and HET animals (p < 0.01; Figure 2F). Inhibition of PDE10A is known to lead to an increase in levels of phosphorylated CREB (pCREB). After administration of the potent, selective PDE10A inhibitor MP-10, WT and HET mice showed 400% more (p < 0.001) striatal pCREB than did vehicle-treated mice. No such increase in pCREB was observed in the KI mice, confirming that the biallelic PDE10A variant reduces downstream pCREB signaling (Figure 2G).
Discussion
We have described eight individuals from two families affected by an early-onset hyperkinetic movement disorder associated with mutations in PDE10A. Expression of the mutant variants of human PDE10A in HEK293 cells revealed lower levels of both mutant proteins than of the WT. Critically, these observations were reflected in one of the affected individuals, who had a significant loss of striatal PDE10A signal when investigated with a specific PDE10A PET ligand in the absence of abnormalities on MRI. PDE10A levels in striatal tissue lysates from a KI model were also decreased, leading to a reduced ability to degrade cyclic nucleotides. In KI mice, downstream signaling by PDE10A was decreased, demonstrated by an absent pCREB response after dosage with a PDE10A inhibitor.
PDE10A is specifically found at high levels in the striatal MSNs,3 which receive input from the cortex and thalamus. MSNs can be divided into two cell types: “direct pathway” MSNs (dMSNs) express dopamine receptor D1 and project to the globus pallidus internal segment and substantia nigra pars reticulate, whereas “indirect pathway” MSNs (iMSNs) express dopamine receptor D2 and project indirectly to the same nuclei via the globus pallidus external segment and subthalamic nuclei. The classic model predicts that dMSN activation promotes movement, whereas iMSN activation leads to inhibition of movement.6 PDE10A is expressed in both cell types, but pharmacological data indicate that it might regulate them differentially.27 Pharmacological inhibition of PDE10A is consistent with the preferential activation of the indirect pathway,27 which should result in hypokinetic symptoms. Both Pde10a-knock-out (KO) mice8 and the Pde10a-KI mice described here are indeed hypokinetic. However, the affected individuals experience significant hyperkinetic motor symptoms. This is reminiscent of the phenotypes observed in mouse models of HD. Individuals affected by HD experience an initial hyperkinetic phase followed by a hypokinetic phase.28 However, only some HD models have a hyperkinetic phase, and it is generally short;29 the lack of recapitulation of motor symptoms between the KI animal and the affected individuals is thus not unprecedented.
Pharmacological inhibitors of PDE10A have been shown to reverse a number of phenotypes in HD mouse models.9 However PDE10A levels are reduced in animal models of HD.30 Furthermore, recent PET studies have shown that individuals with pathogenic HTT (MIM: 613004) expansions exhibit reduced levels of striatal PDE10A.31, 32 This correlates with disease severity, although significant decreases in PDE10A have also been seen in asymptomatic individuals carrying pathogenic expansions.31, 32 The phenotype described here results from loss of PDE10A activity in the striatum, differentiating the effects of selective PDE10A deficiency from those accompanying striatal cell loss. Our observations confirm that PDE10A plays a key role in regulating striato-cortical movement control. Genetic depletion of PDE10A appears to alter cyclic nucleotide signaling in the MSNs in humans and thus result in an activation of motor activity and a hyperkinetic movement disorder.
These data accord with the recent observation of activating mutations in adenylyl cyclase 5, ADCY5 (MIM: 600293).33, 34 The encoded enzyme converts ATP to pyrophosphate and cAMP and is also found at high levels in the striatum. A hyperkinetic movement disorder was reported in affected individuals, whereas cellular modeling of the mutations confirmed an increase in cAMP levels. The variants we report result in a reduced capacity to degrade cAMP in a cellular model. This is recapitulated in the animal model we report. These observations suggest that modulation of cyclic nucleotide levels in the striatum plays a key role in movement control.
Our observations, particularly the lack of pCREB response to PDE10A inhibition in our KI mouse model, suggest that the dose of PDE10A inhibitors might need to be modified in conditions characterized by reduced PDE10A levels. The Amaryllis study, a phase II clinical study of a PDE10A inhibitor, is presently underway in HD. The outcome of these trials will further inform the clinical relevance and impact of our observations.
In conclusion, we report a primary loss of PDE10A due to biallelic PDE10A mutations in humans; this results in striatal dysfunction, leading to a hyperkinetic movement disorder. The effects are independent of any other disease process, including evidence of cell loss. These observations confirm a key role for PDE10A in the physiology of movement control and a central role for modulation of striatal cyclic nucleotide levels in this process. They suggest cellular dysfunction rather than cell death as a cause of movement disorders.
Conflicts of Interest
S.J.S.R., E.C., M.A., J.H., and C.S. were full-time paid employees of Pfizer Inc. at the inception of this work. M.P., V.R., and M.A.V.-F. are full-time paid employees at Pfizer Inc. L.C.J. and C.J.S. are employees and shareholders at Pfizer Inc. N.J.B. is a full-time employee and shareholder at AstraZeneca and was previously a full-time employee at Pfizer Inc. He does not hold Pfizer shares. J.-P.S. is an employee and shareholder of AstraZeneca.
Acknowledgments
The authors are grateful to Ted Simon and Michael Roos for assistance with the generation of the transgenic knock-in mice and Edwin Berryman, Bonnie Deschenes, Tabitha Jones, Mona Sisodia, Ms. Pirjo Keränen, Ms. Anja Mattila, and Ms. Riitta Vuento for their expert assistance. This work was supported by the following: Wellbeing of Women (grant RG992), the University of Leeds Biomedical Health Research Centre (grant PSF42 to E.S.), the Academy of Finland Research Council for Health (decision numbers 138566 to J.U. and 266498 and 273790 to R.H.), the Sigrid Juselius Foundation (J.U. and R.H.), the Finland Foundation for Pediatric Research (J.U.), the Alma and K.A. Snellman Foundation (J.U.), the Emil Aaltonen Foundation (R.H.), the European Union’s Seventh Framework Programme (Marie Curie International Outgoing Fellowship under grant 273669 [BioMit] to R.H.), and the Department of Pediatrics and Adolescence at Oulu University Hospital (Special State Grants for Health Research).
Published: April 7, 2016
Footnotes
Supplemental Data include a Supplemental Note, six figures, one table, and two movies and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.03.015.
Contributor Information
Johanna Uusimaa, Email: johanna.uusimaa@oulu.fi.
Eamonn Sheridan, Email: e.sheridan@leeds.ac.uk.
Nicholas J. Brandon, Email: nick.brandon@azneuro.com.
Web Resources
The URLS for the data presented herein are as follows:
1000 Genomes, http://www.1000genomes.org/
ClinicalTrials.gov, the Amaryllis Study, https://clinicaltrials.gov/show/NCT02197130
AutoSNPa, http://dna.leeds.ac.uk/autosnpa/
Burrows-Wheeler Aligner, http://bio-bwa.sourceforge.net/
Exome Aggregation Consortium (ExAC) Browser, http://exac.broadinstitute.org/
Genome Analysis Toolkit (GATK), https://www.broadinstitute.org/gatk/
IBDfinder, http://dna.leeds.ac.uk/ibdfinder/
NCBI Genome build GRCh37, ftp://ftp.ncbi.nlm.nih.gov/genomes/Homo_sapiens/ARCHIVE/ANNOTATION_RELEASE.105/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
Novoalign, http://www.novocraft.com/products/novoalign/
OMIM, http://www.omim.org
SAMtools, http://www.htslib.org/
Transnetyx, http://www.transnetyx.com/
Supplemental Data
There are two videos of this individual. The first video shows him at a younger age, sitting in a high back chair attempting to kick a ball. He has obvious poor truncal tone and head control indicative of axial hypotonia. He has a generalized jerky hyperkinetic movement disorder with involuntary dyskinetic movements affecting all four limbs, trunk and face. Intermittently he displays dystonic posturing of his hands and also foot-clawing. During this video, he is sitting at a table, and underneath, bilateral dystonic foot clawing posturing of his feet is visible. In the second video he seems a few years older and is now in a wheelchair. He has an ongoing movement disorder, with generalized hyperkinesis and dyskinesia particularly prominent in the hands and feet. He continues to have low axial tone, and some athetoid truncal movements are evident when sitting. He intermittently has an open mouth, possible suggestive of jaw dystonia. On attempted supported walking, he shows high stepping gait, with manifestation of lower limb dystonic postures.
There are two videos of this individual. In the first (younger) he also has evidence of a prominent generalized hyperkinetic movement disorder affecting mainly all four limbs but also the face, especially the perioral region with orolingual dyskinesia. There is a jerky nature to his movement disorder. There is evidence of intermittent bilateral upper limb posturing with arms extended and pronated and wrists flexed and episodic foot clawing. He has reduced axial tone. In the second video, he appears older, and is sitting in a wheelchair. There is ongoing generalized dyskinesia mainly affected the limbs but also the trunk and face especially periorally. There seems to have been some improvement in his truncal tone. Intermittent choreathetoid movements of the trunk are evident. Intermittently he seems to have almost ballistic upper limb movements with some stereotypical choreiform upper limb involuntary movements. He has a high stepping gait and appears to toe-walk and has a tendency to walk with extended legs. On walking, some upper limb posturing was evident with arm extension, pronation and wrist flexion. Hand/finger dyskinesia seems to be worse on intention/action, e.g., when playing with the shape sorter.
References
- 1.Beavo J.A., Brunton L.L. Cyclic nucleotide research -- still expanding after half a century. Nat. Rev. Mol. Cell Biol. 2002;3:710–718. doi: 10.1038/nrm911. [DOI] [PubMed] [Google Scholar]
- 2.Conti M., Beavo J. Biochemistry and physiology of cyclic nucleotide phosphodiesterases: essential components in cyclic nucleotide signaling. Annu. Rev. Biochem. 2007;76:481–511. doi: 10.1146/annurev.biochem.76.060305.150444. [DOI] [PubMed] [Google Scholar]
- 3.Coskran T.M., Morton D., Menniti F.S., Adamowicz W.O., Kleiman R.J., Ryan A.M., Strick C.A., Schmidt C.J., Stephenson D.T. Immunohistochemical localization of phosphodiesterase 10A in multiple mammalian species. J. Histochem. Cytochem. 2006;54:1205–1213. doi: 10.1369/jhc.6A6930.2006. [DOI] [PubMed] [Google Scholar]
- 4.Jellinger K.A. The pathology of Parkinson’s disease. Adv. Neurol. 2001;86:55–72. [PubMed] [Google Scholar]
- 5.Walker F.O. Huntington’s disease. Lancet. 2007;369:218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- 6.DeLong M.R., Wichmann T. Circuits and circuit disorders of the basal ganglia. Arch. Neurol. 2007;64:20–24. doi: 10.1001/archneur.64.1.20. [DOI] [PubMed] [Google Scholar]
- 7.Gratwicke J., Jahanshahi M., Foltynie T. Parkinson’s disease dementia: a neural networks perspective. Brain. 2015;138:1454–1476. doi: 10.1093/brain/awv104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt C.J., Chapin D.S., Cianfrogna J., Corman M.L., Hajos M., Harms J.F., Hoffman W.E., Lebel L.A., McCarthy S.A., Nelson F.R. Preclinical characterization of selective phosphodiesterase 10A inhibitors: a new therapeutic approach to the treatment of schizophrenia. J. Pharmacol. Exp. Ther. 2008;325:681–690. doi: 10.1124/jpet.107.132910. [DOI] [PubMed] [Google Scholar]
- 9.Giampà C., Laurenti D., Anzilotti S., Bernardi G., Menniti F.S., Fusco F.R. Inhibition of the striatal specific phosphodiesterase PDE10A ameliorates striatal and cortical pathology in R6/2 mouse model of Huntington’s disease. PLoS ONE. 2010;5:e13417. doi: 10.1371/journal.pone.0013417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson L.S., Brandon N.J. Emerging biology of PDE10A. Curr. Pharm. Des. 2015;21:378–388. doi: 10.2174/1381612820666140826114744. [DOI] [PubMed] [Google Scholar]
- 11.Marques T.R., Natesan S., Niccolini F., Politis M., Gunn R.N., Searle G.E., Howes O., Rabiner E.A., Kapur S. Phosphodiesterase 10A in Schizophrenia: A PET Study Using [(11)C]IMA107. Am. J. Psychiatry. 2016 doi: 10.1176/appi.ajp.2015.15040518. p201515040518. [DOI] [PubMed] [Google Scholar]
- 12.Niccolini F., Haider S., Reis Marques T., Muhlert N., Tziortzi A.C., Searle G.E., Natesan S., Piccini P., Kapur S., Rabiner E.A. Altered PDE10A expression detectable early before symptomatic onset in Huntington’s disease. Brain. 2015;138:3016–3029. doi: 10.1093/brain/awv214. [DOI] [PubMed] [Google Scholar]
- 13.Carr I.M., Flintoff K.J., Taylor G.R., Markham A.F., Bonthron D.T. Interactive visual analysis of SNP data for rapid autozygosity mapping in consanguineous families. Hum. Mutat. 2006;27:1041–1046. doi: 10.1002/humu.20383. [DOI] [PubMed] [Google Scholar]
- 14.Carr I.M., Sheridan E., Hayward B.E., Markham A.F., Bonthron D.T. IBDfinder and SNPsetter: tools for pedigree-independent identification of autozygous regions in individuals with recessive inherited disease. Hum. Mutat. 2009;30:960–967. doi: 10.1002/humu.20974. [DOI] [PubMed] [Google Scholar]
- 15.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muyrers J.P., Zhang Y., Benes V., Testa G., Rientjes J.M., Stewart A.F. ET recombination: DNA engineering using homologous recombination in E. coli. Methods Mol. Biol. 2004;256:107–121. doi: 10.1385/1-59259-753-X:107. [DOI] [PubMed] [Google Scholar]
- 21.Roach M.L., Stock J.L., Byrum R., Koller B.H., McNeish J.D. A new embryonic stem cell line from DBA/1lacJ mice allows genetic modification in a murine model of human inflammation. Exp. Cell Res. 1995;221:520–525. doi: 10.1006/excr.1995.1403. [DOI] [PubMed] [Google Scholar]
- 22.Longenecker G., Kulkarni A.B. Generation of gene knockout mice by ES cell microinjection. Curr. Protoc. Cell Biol. 2009;Chapter 19 doi: 10.1002/0471143030.cb1914s44. 14, 1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rogers D.C., Peters J., Martin J.E., Ball S., Nicholson S.J., Witherden A.S., Hafezparast M., Latcham J., Robinson T.L., Quilter C.A., Fisher E.M. SHIRPA, a protocol for behavioral assessment: validation for longitudinal study of neurological dysfunction in mice. Neurosci. Lett. 2001;306:89–92. doi: 10.1016/s0304-3940(01)01885-7. [DOI] [PubMed] [Google Scholar]
- 24.Seeger T.F., Bartlett B., Coskran T.M., Culp J.S., James L.C., Krull D.L., Lanfear J., Ryan A.M., Schmidt C.J., Strick C.A. Immunohistochemical localization of PDE10A in the rat brain. Brain Res. 2003;985:113–126. doi: 10.1016/s0006-8993(03)02754-9. [DOI] [PubMed] [Google Scholar]
- 25.Charych E.I., Jiang L.X., Lo F., Sullivan K., Brandon N.J. Interplay of palmitoylation and phosphorylation in the trafficking and localization of phosphodiesterase 10A: implications for the treatment of schizophrenia. J. Neurosci. 2010;30:9027–9037. doi: 10.1523/JNEUROSCI.1635-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Threlfell S., Sammut S., Menniti F.S., Schmidt C.J., West A.R. Inhibition of Phosphodiesterase 10A Increases the Responsiveness of Striatal Projection Neurons to Cortical Stimulation. J. Pharmacol. Exp. Ther. 2009;328:785–795. doi: 10.1124/jpet.108.146332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berardelli A., Noth J., Thompson P.D., Bollen E.L., Currà A., Deuschl G., van Dijk J.G., Töpper R., Schwarz M., Roos R.A. Pathophysiology of chorea and bradykinesia in Huntington’s disease. Mov. Disord. 1999;14:398–403. doi: 10.1002/1531-8257(199905)14:3<398::aid-mds1003>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 29.Pouladi M.A., Morton A.J., Hayden M.R. Choosing an animal model for the study of Huntington’s disease. Nat. Rev. Neurosci. 2013;14:708–721. doi: 10.1038/nrn3570. [DOI] [PubMed] [Google Scholar]
- 30.Hebb A.L.O., Robertson H.A., Denovan-Wright E.M. Striatal phosphodiesterase mRNA and protein levels are reduced in Huntington’s disease transgenic mice prior to the onset of motor symptoms. Neuroscience. 2004;123:967–981. doi: 10.1016/j.neuroscience.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 31.Niccolini F., Haider S., Reis Marques T., Muhlert N., Tziortzi A.C., Searle G.E., Natesan S., Piccini P., Kapur S., Rabiner E.A. Altered PDE10A expression detectable early before symptomatic onset in Huntington’s disease. Brain. 2015;138:3016–3029. doi: 10.1093/brain/awv214. [DOI] [PubMed] [Google Scholar]
- 32.Russell D.S., Barret O., Jennings D.L., Friedman J.H., Tamagnan G.D., Thomae D., Alagille D., Morley T.J., Papin C., Papapetropoulos S. The phosphodiesterase 10 positron emission tomography tracer, [18F]MNI-659, as a novel biomarker for early Huntington disease. JAMA Neurol. 2014;71:1520–1528. doi: 10.1001/jamaneurol.2014.1954. [DOI] [PubMed] [Google Scholar]
- 33.Chen D.H., Méneret A., Friedman J.R., Korvatska O., Gad A., Bonkowski E.S., Stessman H.A., Doummar D., Mignot C., Anheim M. ADCY5-related dyskinesia: Broader spectrum and genotype-phenotype correlations. Neurology. 2015;85:2026–2035. doi: 10.1212/WNL.0000000000002058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y.Z., Friedman J.R., Chen D.H., Chan G.C., Bloss C.S., Hisama F.M., Topol S.E., Carson A.R., Pham P.H., Bonkowski E.S. Gain-of-function ADCY5 mutations in familial dyskinesia with facial myokymia. Ann. Neurol. 2014;75:542–549. doi: 10.1002/ana.24119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
There are two videos of this individual. The first video shows him at a younger age, sitting in a high back chair attempting to kick a ball. He has obvious poor truncal tone and head control indicative of axial hypotonia. He has a generalized jerky hyperkinetic movement disorder with involuntary dyskinetic movements affecting all four limbs, trunk and face. Intermittently he displays dystonic posturing of his hands and also foot-clawing. During this video, he is sitting at a table, and underneath, bilateral dystonic foot clawing posturing of his feet is visible. In the second video he seems a few years older and is now in a wheelchair. He has an ongoing movement disorder, with generalized hyperkinesis and dyskinesia particularly prominent in the hands and feet. He continues to have low axial tone, and some athetoid truncal movements are evident when sitting. He intermittently has an open mouth, possible suggestive of jaw dystonia. On attempted supported walking, he shows high stepping gait, with manifestation of lower limb dystonic postures.
There are two videos of this individual. In the first (younger) he also has evidence of a prominent generalized hyperkinetic movement disorder affecting mainly all four limbs but also the face, especially the perioral region with orolingual dyskinesia. There is a jerky nature to his movement disorder. There is evidence of intermittent bilateral upper limb posturing with arms extended and pronated and wrists flexed and episodic foot clawing. He has reduced axial tone. In the second video, he appears older, and is sitting in a wheelchair. There is ongoing generalized dyskinesia mainly affected the limbs but also the trunk and face especially periorally. There seems to have been some improvement in his truncal tone. Intermittent choreathetoid movements of the trunk are evident. Intermittently he seems to have almost ballistic upper limb movements with some stereotypical choreiform upper limb involuntary movements. He has a high stepping gait and appears to toe-walk and has a tendency to walk with extended legs. On walking, some upper limb posturing was evident with arm extension, pronation and wrist flexion. Hand/finger dyskinesia seems to be worse on intention/action, e.g., when playing with the shape sorter.