Abstract
Background
Prenatal alcohol exposure (PAE) is associated with alterations in numerous physiological systems, including the stress and immune systems . We have previously shown that PAE increases the course and severity of arthritis in an adjuvant-induced arthritis (AA) model. While the molecular mechanisms underlying these effects are not fully known, changes in neural gene expression are emerging as important factors in the etiology of PAE effects. As the prefrontal cortex (PFC) and hippocampus (HPC) play key roles in neuroimmune function, PAE-induced alterations to their transcriptome may underlie abnormal steady-state functions and responses to immune challenge. The current study examined brains from adult PAE and control females from our recent AA study to determine whether PAE causes long-term alterations in gene expression and whether these mediate the altered severity and course of arthritis in PAE females
Methods
Adult females from PAE, pair-fed [PF], and ad libitum-fed control [C]) groups were injected with either saline or complete Freund’s adjuvant. Animals were terminated at the peak of inflammation or during resolution (days 16 and 39 post-injection, respectively); cohorts of saline-injected PAE, PF and C females were terminated in parallel. Gene expression was analyzed in the PFC and HPC using whole genome mRNA expression microarrays.
Results
Significant changes in gene expression in both the PFC and HPC were found in PAE compared to controls in response to ethanol exposure alone (saline-injected females), including genes involved in neurodevelopment, apoptosis, and energy metabolism. Moreover, in response to inflammation (adjuvant-injected females), PAE animals showed unique expression patterns, while failing to exhibit the activation of genes and regulators involved in the immune response observed in control and pair-fed animals.
Conclusions
These results support the hypothesis that PAE affects neuroimmune function at the level of gene expression, demonstrating long-term effects of PAE on the CNS response under steady-state conditions and following an inflammatory insult.
Keywords: prenatal alcohol exposure (PAE), ethanol, inflammation, arthritis, gene expression, rat
Introduction
The prevalence of fetal alcohol spectrum disorders (FASD) in North America is estimated at 2–5% of live births, making prenatal alcohol exposure (PAE) a leading cause of neurodevelopmental disorders (May et al. 2009). In addition to lasting neurocognitive deficits, impairments in self-regulation, and deficits in adaptive functioning, children with FASD also display changes in a number of physiological systems, including the immune system, with adverse impacts on both innate and adaptive immunity (S. Johnson et al. 1981; Streissguth et al. 1985; Gauthier et al. 2005).
Animal models have corroborated clinical findings, with PAE animals displaying behavioural and cognitive deficits, including delays in learning and memory, and altered responsivity to stressors (reviewed in Hellemans et al., 2010). Moreover, PAE animals also exhibit altered development of the thymus, decreased lymphocyte proliferative responses to mitogens, increased susceptibility to infections, and greater vulnerability to immune and inflammatory challenges compared to controls (reviewed in Bodnar & Weinberg, 2013). PAE animals also show larger increases in plasma levels of pro-inflammatory cytokines, as well as reduced proliferative responses of B cells to lipopolysaccharide (LPS), and splenic T cells and T lymphoblasts to Concanavalin A and/or interleukin-2 (Zhang et al., 2005; Weinberg & Jerrells, 1991) Likewise, in an adjuvant-induced arthritis (AA) paradigm, we recently demonstrated that PAE animals show increased severity of joint inflammation and a prolonged course of disease (39 days post-injection, higher incidence of arthritis in PAE compared pair-fed [PF] and control [C] animals) (Zhang et al., 2012). These findings suggest that although PAE causes deficits in adaptive immunity, PAE offspring show increased responses to some immune/inflammatory challenges.
The immune, neuroendocrine and central nervous systems have extensive bidirectional communication, sharing numerous ligands and receptors. Brain regions, such as the prefrontal cortex (PFC) and hippocampus (HPC) not only play a role in the regulation of neuroendocrine function, but also respond to immune/inflammatory molecules, including cytokines and neuropeptides (Crofford et al. 1992). For example, adjuvant injection induces c-Fos expression in the hippocampus for up to 4 months, suggesting a role for this region in AA (Carter et al. 2011). Thus, long-term changes in gene expression may modulate AA manifestation and progression. Indeed, mounting evidence suggests a role for altered gene expression in the etiology of FASD (Kobor & Weinberg, 2011). Widespread changes to gene expression levels in fetal and neonatal brains following PAE, as well as long-lasting alterations to the neural transcriptome following alcohol exposure during the neonatal (third-trimester equivalent) period or across all three trimesters have been reported (Green et al. 2007; Hard et al. 2005; Zhou et al., 2011; Kleiber et al., 2012, 2013).
Using steady-state animals (saline-injected) as a baseline, the current study examined brains from adult PAE and control females from our recent AA study to determine whether long-term alterations in gene expression mediate the altered severity and course of arthritis observed in PAE females (Zhang et al., 2012). Since the PFC and HPC play key roles in both neuroendocrine and neuroimmune processes and show altered function following PAE, PAE-induced alterations in the transcriptome of these regions could result in marked downstream effects, including dysregulation of the immune response and neuroendocrine-neuroimmune interactions (Norman et al. 2009). Whole genome microarrays were utilized to assess gene expression in the PFC and HPC of adult PAE, PF and C females terminated at the peak or during resolution of inflammation (days 16 and 39 post-adjuvant injection, respectively); cohorts of saline-injected PAE, PF and C females were terminated in parallel. Under steady-state condition, we identified changes in gene expression and altered activation states of upstream regulators specific to PAE. Furthermore, at the peak of inflammation, we found not only changes in genes related to PAE, but also, a failure of PAE animals to mount appropriate responses to the immune challenge, showing no change in the activation or inhibition of inflammation-related genes and upstream regulators identified in controls.
Materials and Methods
Breeding and prenatal ethanol exposure
All animal protocols were approved by the University of British Columbia Animal Care Committee and are consistent with the NIH Guide for the Care and Use of Laboratory Animals (National Research Council 2011). Details of the breeding and feeding procedures have been published (Zhang et al, 2012). Briefly, male and female Sprague-Dawley rats (Animal Care Center, University of British Columbia) were paired; presence of a vaginal plug indicated gestation day (GD) 1. Pregnant dams were singly housed and assigned to experimental groups: Prenatal ethanol exposure (PAE; ad libitum access to liquid ethanol diet, 36% ethanol-derived calories); Pair-fed (PF; liquid-control diet, maltose-dextrin isocalorically substituted for ethanol, in the amount consumed by a PAE partner, g/kg body weight/GD); or Ad libitum-fed control (C; laboratory chow, ad libitum). All animals had ad libitum access to water. Experimental diets (Weinberg/Kiever Ethanol Diet #710324, Weinberg/Kiever Control Diet #710109, Dyets Inc., Bethlehem, PA) were fed from GD 1–21, then replaced with laboratory chow. Litters were weighed and culled at birth to 5 males and 5 females, when possible. Following weaning (postnatal day 22), offspring were group-housed by litter and sex. Female offspring were used in the present study due to their increased susceptibility to arthritis (Whitacre, 2001).
Induction of arthritis and termination of animals
Details of the adjuvant-induced arthritis (AA) paradigm have been published (Zhang et al. 2012). Female offspring (50–65 days of age) from C, PF, and PAE groups received an intradermal injection of 0.1 ml of a 12 mg/ml suspension of complete Freund’s adjuvant (CFA) or 0.1 ml physiological saline at the base of the tail. Animals were single-housed post-injection, and monitored for clinical signs of arthritis under light anesthesia with isofluorane. Paws were scored individually for redness and swelling on days 7, 10, and every other day thereafter until day 39 following injection (Zhang et al., 2012).
Animals were terminated by decapitation, following brief exposure to CO2, in two cohorts: day 16 post-injection or day 39 post-injection (peak or resolution phase of AA, respectively). Each cohort contained 9 adjuvant-injected animals and 5 saline-injected animals for each group (C, PF, and PAE). Brains were rapidly removed, immediately frozen on dry ice, and stored at −70 °C.
Tissue dissection and RNA extraction
Brains were thawed to 4 °C, and the PFC and HPC were dissected, placed in RNAlater, and stored at −20 °C. Total RNA and DNA were simultaneously extracted from the tissues (Qiagen AllPrep DNA/RNA Mini kit). RNA integrity was determined using the Agilent BioAnalyzer mRNA Nano assay.
Microarray assay of whole genome gene expression and quality control
The Ambion Illumina TotalPrep RNA Amplification kit was used to generate cRNA (750 ng) from total RNA (250 ng) for each sample. Expression data were obtained using the Illumina RatRef-12 Expression BeadChip microarray with the Illumina iScan, which provides probe-level data for all expressed genes (~ 1 probe per gene). Datasets were filtered to remove control probes and probes with a detection p-value >0.05 in comparison to negative control probes. After filtering, 20215 and 20069 probes remained in the PFC and HPC, respectively (out of a total 23350 probes). The filtered, log2-transformed gene expression profiles were quantile-normalized within each tissue.
Differential gene expression analysis
Gene expression analysis utilized the sva and limma packages in the statistical program R (Leek & Storey 2007; Smyth 2005). Using sva, surrogate variables representative of heterogeneity from sources other than experimental treatments (e.g. batch effects) were generated. These were included in linear modeling of gene expression with limma, which uses moderated F- and t-statistics to identify significant differences. Gene expression changes were modeled in two ways using separate sample means: effects of prenatal treatment alone on steady state levels of gene expression (saline-injected animals, n=5 per C, PF, PAE group), and interaction of prenatal treatment with an inflammatory challenge (adjuvant- versus saline-injected animals; n=5 for saline, n=9 for adjuvant per C, PF, PAE group). Each probe received a moderated F-statistic, and their p-values were corrected for multiple testing using Benjamini-Hochberg correction. The false-discovery rate (FDR) was controlled at <25% due to the moderate alcohol-exposure paradigm and its relatively subtle effects. Significant changes in PAE compared to controls had a moderated t-statistic p-value <0.05. Sequences for significant probes were queried against the RefSeq database for Rattus norvegicus to identify target transcripts.
Verification of microarray results
Differentially expressed genes were verified using reverse-transcription quantitative real time PCR (RT-qPCR) on the Corbett Rotorgene 6000 for both PFC and HPC, with the same RNA used for microarray analysis (n=4 in both tissues for each C, PF, and PAE). Primers were designed using well-established guidelines to obtain gene-level data and multiple reference genes were used to normalize expression data (Nolan et al. 2006). Three reference genes across a spectrum of expression levels and no evidence for differences across groups (F-statistic p-value >0.05) were selected for each tissue (Table S4). The normalization factor for each sample was calculated using the geometric mean of cycle threshold (Ct) values (Vandesompele et al. 2002). Expression levels relative to the factor were determined, and analysis of variance (ANOVA) was conducted to test for significant differences between groups (Schmittgen & Livak 2008).
Gene Ontology and Pathway analysis
Gene ontology (GO) analysis was conducted to identify “Biological Processes” enriched for the effects of prenatal treatment and adjuvant exposure using the gene-score resampling (GSR) method in ermineJ (Lee et al. 2005). The set of candidate FASD genes from the curated Neurocarta database was included in the analysis as a custom GO term (Portales-Casamar et al. 2013; Table S1). Benjamini-Hochberg correction was used with an FDR of 1% within single brain regions and 10% when comparing overlapping effects. Where many GO categories were identified, these were mapped to their parent GO Slim terms using CateGOrizer to determine common categories of altered function (Zhi-Liang et al. 2008). Following GO analysis, the Ingenuity© Upstream Regulator Analysis tool (URA, Ingenuity Systems Inc., Redwood City, CA) was used to predict master transcriptional regulators that explain the observed expression changes within the dataset. Genes with a fold-change ≥ 1.2 and p < 0.05 between treatments were analyzed for effects of PAE and adjuvant injection. For steady-state effects of PAE, prenatal groups were compared, while adjuvant effects were assessed by comparing adjuvant- to saline-injected animals in each prenatal group. Significantly activated and inhibited genes were identified through a Z-score > 2 or < -2 respectively, as well as an overlap p-value ≤ 0.1, calculated by Fisher’s Exact test.
Results
Developmental Data
As expected, body weights of PAE dams were lower than those of controls (p<0.001) by the end of pregnancy (GD21) [Group x Day interaction, F(6,99)=17.2, p<0.0001], with PF dams intermediate to PAE and C; dams no longer differed in weight by lactation day 8. At birth, PAE (5.7± 0.17 g) females weighed less than their C (6.5± 0.18 g) counterparts (main effect of group, F(2,66)=7.02, p<0.01), which persisted until weaning (PAE, 51.2±1.4 g; PF, 55.3±1.6 g; C, 55.2±1.5 g) (group x day, F(6,99)=1.96, p=0.079). Blood ethanol levels for dams in this paradigm typically average ~100–150 mg/dl (Sliwowska al. 2008; Lan et al. 2009).
Prenatal ethanol exposure altered steady-state levels of gene expression in PFC and HPC
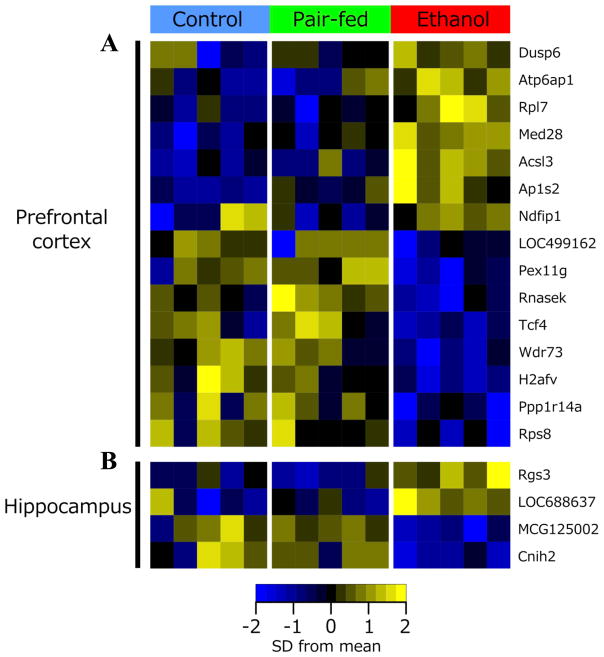
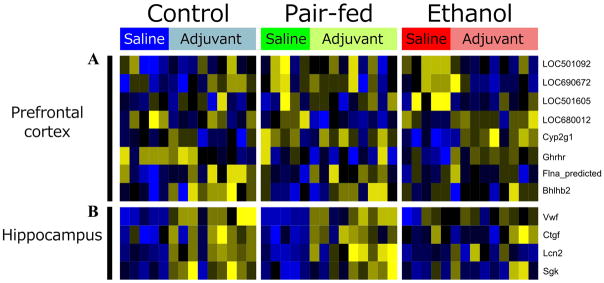
PAE effects on steady-state levels of gene expression were examined in saline-injected females on Days 16 and 39 post-injection (~ PND 75 and 95, respectively). On Day 16, p-value distributions were skewed towards zero for contrasts of PAE vs C and PAE vs PF, suggesting gene expression differences in PAE compared to C and PF females (Figure S1). Following Benjamini-Hochberg correction, significant effects of prenatal treatment were found for 80 and 30 genes in the PFC and HPC, respectively, at 25% FDR (Figure 1). While many genes (43% in PFC, 37% in HPC) showed significant effects of ethanol exposure against both control groups, only a subset (15 in PFC, 4 in HPC; p <0.05) showed changes specific to PAE, in that levels were similar between C and PF animals (Table 1, Figure 2). These had a number of annotated functions in common, including neurodevelopment, differentiation, neuronal signaling, and regulation of cell death and transcription.
Figure 1. Prenatal treatment alters gene expression patterns under steady-state conditions.
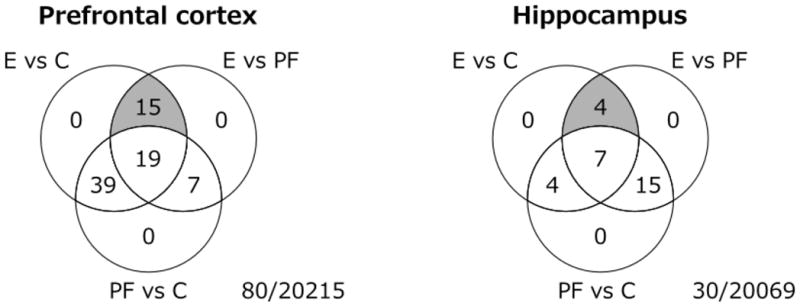
Venn diagram of the number of the number of probes significantly altered in each contrast at Day 16 post-saline injection, with moderated F-statistic q <0.25 and moderated t-statistic p <0.05 (80 in the PFC, 30 in the HPC). The number of probes with unique effects in PAE versus both PF and C animals are highlighted in grey, and listed in Table 1. The center of each Venn diagram shows the number of probes differentially expressed among all three prenatal treatment groups. The intersection on the left of each diagram shows the number of probes with a common effect of prenatal ethanol exposure and pair-feeding. The intersection on the right of each diagram shows the number of probes with a unique effect of pair-feeding.
Table 1.
Genes with a significantly expression under steady-state conditions in PAE compared to both C and PF animals (p <0.05) in the PFC (a) and HPC (b) at D16 post-saline injection.
| Table 1a: Genes differentially expressed in PFC of Ethanol-exposed vs. both PF and C animals under steady-state conditions in the microarray.
| ||||||||
|---|---|---|---|---|---|---|---|---|
| Gene Symbol | Gene Name | Average Expression | F | p-value | q-value | Fold change
|
||
| Ethanol/Control | Ethanol/Pairfed | Pairfed/Control | ||||||
| H2afv | Rattus norvegicus similar to H2A histone family, member V isoform 1 (LOC685909) | 10.6 | 18.7 | 4.8E-05 | 0.11 | 0.65 | 0.76 | 0.86 |
| Tcf4 | transcription factor 4 | 11.2 | 11.4 | 7.0E-04 | 0.23 | 0.67 | 0.66 | 1.01 |
| Rnasek | ribonuclease, RNase K | 13.2 | 11.1 | 8.0E-04 | 0.23 | 0.68 | 0.57 | 1.19 |
| Ppp1r14a | protein phosphatase 1, regulatory (inhibitor) subunit 14A | 10.0 | 12.6 | 4.1E-04 | 0.23 | 0.68 | 0.64 | 1.05 |
| Rps8 | ribosomal protein S8 | 13.0 | 11.1 | 7.9E-04 | 0.23 | 0.69 | 0.74 | 0.93 |
| ILMN_1372701 | na | 9.4 | 11.3 | 7.3E-04 | 0.23 | 0.71 | 0.79 | 0.90 |
| ILMN_1374168 | na | 9.1 | 10.7 | 9.4E-04 | 0.25 | 0.77 | 0.73 | 1.05 |
| Pex11g | peroxisomal biogenesis factor 11 gamma | 7.0 | 11.5 | 6.7E-04 | 0.23 | 0.82 | 0.71 | 1.16 |
| Ndfip1 | Nedd4 family interacting protein 1 | 11.4 | 12.1 | 5.1E-04 | 0.23 | 1.32 | 1.37 | 0.97 |
| Acsl3 | acyl-CoA synthetase long-chain family member 3 | 10.2 | 12.2 | 4.9E-04 | 0.23 | 1.36 | 1.36 | 1.00 |
| Dusp6 | dual specificity phosphatase 6 | 9.9 | 12.5 | 4.4E-04 | 0.23 | 1.41 | 1.21 | 1.17 |
| Rpl7 | ribosomal protein L7 | 11.6 | 13.7 | 2.7E-04 | 0.22 | 1.44 | 1.36 | 1.05 |
| Med28 | mediator complex subunit 28 | 9.2 | 11.1 | 7.9E-04 | 0.23 | 1.48 | 1.29 | 1.15 |
| Atp6ap1 | ATPase, H+ transporting, lysosomal accessory protein 1 | 11.0 | 10.6 | 9.8E-04 | 0.25 | 1.50 | 1.35 | 1.11 |
| Ap1s2 | adaptor-related protein complex 1, sigma 2 subunit | 9.7 | 12.4 | 4.6E-04 | 0.23 | 1.60 | 1.35 | 1.19 |
| Table 1b: Genes differentially expressed in HPC of Ethanol-exposed vs. both PF and C animals under steady-state conditions in the microarray. | ||||||||
|---|---|---|---|---|---|---|---|---|
| Gene Symbol | Gene Name | Average Expression | F | p-value | q-value | Fold change | ||
| Ethanol/Control | Ethanol/Pairfed | Pairfed/Control | ||||||
| Cnih2 | cornichon homolog 2 (Drosophila) | 11.1 | 16.0 | 8.1E-05 | 0.14 | 0.61 | 0.60 | 1.01 |
| Caap1 | caspase activity and apoptosis inhibitor 1 | 9.2 | 15.2 | 1.1E-04 | 0.14 | 0.68 | 0.71 | 0.95 |
| LOC688637 | similar to WD repeat domain 36 | 8.8 | 15.4 | 1.0E-04 | 0.14 | 1.46 | 1.36 | 1.08 |
| Rgs3 | regulator of G-protein signaling 3 | 9.1 | 14.6 | 1.4E-04 | 0.15 | 1.71 | 1.83 | 0.93 |
Bold = p <0.05. na = probe had no specific alignment to current RefSeq RNA database.
Figure 2. Prenatal alcohol exposure alters steady-state gene expression at Day 16 post-saline injection.
In the prefrontal cortex (a), 15 genes were differentially expressed in response to ethanol. In the hippocampus (b), 4 genes were differentially expressed in response to ethanol. F-statistic q-value <0.25 for all genes identified.
By contrast, on day 39 post-injection, no relationship between gene expression and PAE was apparent in either brain region, according to p-value distributions (Figure S1). Moreover, only 2 probes met a 25% FDR, but were not specific to PAE effects (data not shown). Thus, subsequent analyses focused on brains from Day 16 post-injection.
Verification of results related to prenatal ethanol exposure with RT-qPCR
Of 19 probes showing differential expression due to PAE (Table 1), 17 aligned to a sequence in the Rattus norvegicus RefSeq database (ILMN_1372701 and ILMN_1374168 were the exceptions). Specific RT-qPCR primers were successfully designed for 15 of the 17 genes (Table S3; Rps8 and Rpl7 were not analyzed).
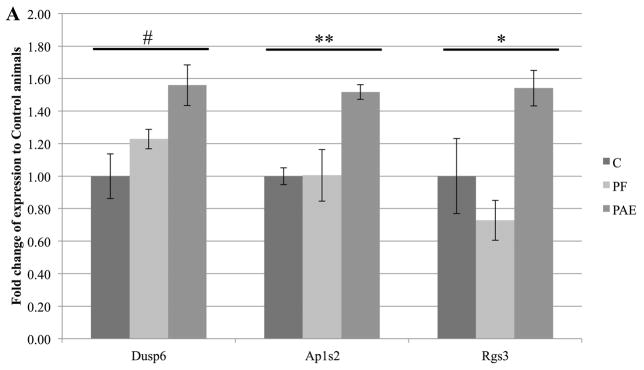
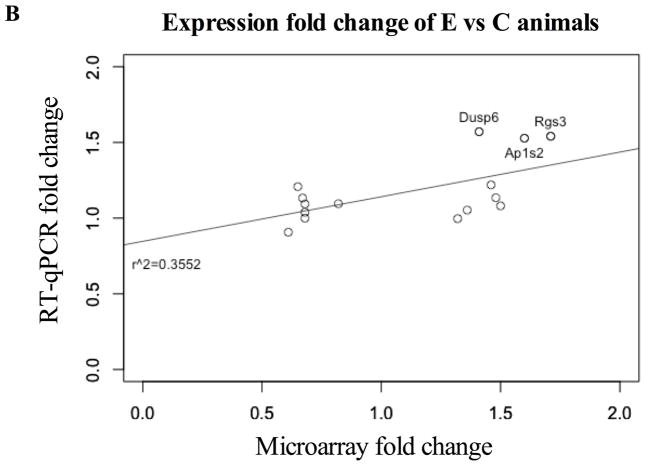
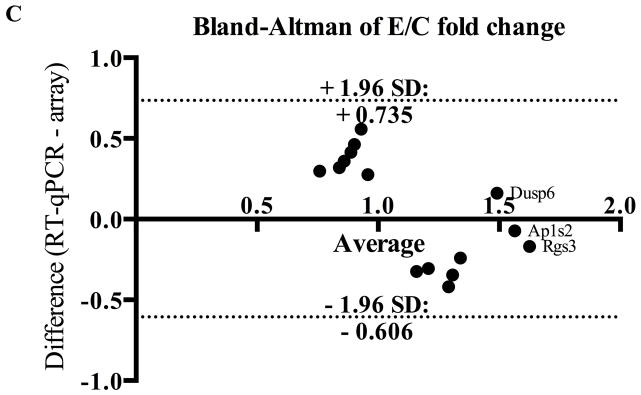
Despite differences with microarray technology, RT-qPCR verified two genes in the PFC (Ap1s2, Dusp6) and one in the HPC (Rgs3), all of which showed increased expression (p <0.1; Figure 3a). Moreover, for 7 significantly up-regulated genes in the microarray, changes trended in the same direction by RT-qPCR (Figure 3b). No down-regulated genes from microarray analysis showed significantly differences in PAE animals by RT-qPCR, but one gene (Cnih2) also trended downward. Importantly, positive correlation between microarray and RT-qPCR data was obtained for PAE effects (r2=0.35, p<0.02 Figure 3b), and significant genes were corroborated by the small differences between methods shown in the Bland-Altman plot (Figure 3c). No correlation was found for PF animals (Figure S4). Collectively, the general agreement between qPCR and microarray data suggested that PAE caused persistent alterations to gene expression in the PFC and HPC.
Figure 3. RT-qPCR verification of genes altered by prenatal alcohol exposure.
(a) Three genes were significantly upregulated in PAE animals (Dusp6 and Ap1s2 in PFC; Rgs3 in HPC). Graphs were plotted as fold change to control animals (where C animals expression = 1) ± SEM. ** = p<0.01, * = p<0.05, # = p<0.1. (b) Fold-changes in expression were positively correlated between microarray and RT-qPCR results for E vs C animals (r2=0.3552, p<0.02). Annotated data points represent genes identified as significant in both methods. (c) Bland-Altman plot of genes identified by microarray analysis. Dotted lines represent the 95% limits of agreement (Bias = 0.06467) and annotated data points represent genes identified as significant in both methods.
Gene Ontology and Upstream Regulator Analysis of PAE effects under steady-state conditions
GO analysis was performed to ascertain the broad functional impact of PAE-induced changes in gene expression. Following multiple test correction, 6 processes were altered in the PFC of PAE compared to PF and C animals at a 1% FDR (Figure S2a): positive regulation of cell projection organization, chemical/ion homeostasis, response to virus, and regulation of intracellular transport. In the HPC, gene-score resampling (GSR) identified 79 processes specific to PAE, which were involved in metabolism (24%), cell communication (18%), development (18%), transport (15%), and signal transduction (10%) (Figure S2a). At a 10% FDR, several PAE-specific biological processes overlapped between brain regions: positive regulation of neuron differentiation, dorsal/ventral pattern formation, circadian rhythm, regulation of lymphocyte differentiation, and regulation of lipase activity (Fig S2b). Moreover, GSR also identified the NeuroCarta candidate gene list for FASD in the PFC of PAE females (Portales-Casamar et al. 2013).
As noted, gene sets were then analyzed using Ingenuity’s Upstream Regulator Analysis (URA) to predict master regulators driving the observed expression changes within the dataset. In the PFC, a significant activation of Gast and an activation of Lep that approached statistical significance were identified in PAE compared to PF and C animals (Table 2a), whereas in the HPC, significant differential activation of Laminin and Ifng was observed (Table 2b).
Table 2.
Genes identified using Ingenuity Pathway Analysis Upstream Regulator in the PFC (a) and HPC (b) of steady-state animals.
| Table 2a – Upstream Regulator Analysis in the PFC of animals under steady-state conditions
| ||||||||
|---|---|---|---|---|---|---|---|---|
| Gene Symbol | Gene Name | Predicted status | Z-score | Overlap p-value | ||||
| PAEvC | PAEvPF | PFvC | PAEvC | PAEvPF | PFvC | |||
| Gast | Gastrin | Activated | 2.1 | 2.2 | NA | 0.05 | 0.009 | 1.00 |
| Lep | Leptin | Activated | 2.5 | 2.6 | NA | 0.12 | 0.04 | 1.00 |
| Table 2b – Upstream Regulator Analysis in the HPC of animals under steady-state conditions
| ||||||||
|---|---|---|---|---|---|---|---|---|
| Gene Symbol | Gene Name | Predicted status | Z-score | Overlap p-value | ||||
| PAEvC | PAEvPF | PFvC | PAEvC | PAEvPF | PFvC | |||
| Ifng | Interferon-gamma | Activated | 3.8 | 2.5 | NA | 0.05 | 0.04 | 1.00 |
| Laminin | Laminin | Activated | 2.0 | 2.0 | NA | 0.01 | 0.04 | 1.00 |
Genes with a Z-score ≥2 or ≤-2 and an overlap p-value ≤0.1 are considered significant (bold). Those with no overlap had a p-value of 1 and no Z-score (NA).
Prenatal treatments resulted in common, graded, and differential effects under steady state conditions
A number of prenatal group effects not specific to PAE were observed in the microarray analysis (Figure 1). Of the probes affected by prenatal treatment, many showed the same levels of expression in PAE and PF compared to C animals (Table S5), while a handful were altered in opposite directions by ethanol exposure and pair-feeding (Table S6). Conversely, several genes exhibited graded effects of prenatal treatment, with effects of ethanol greater than those of pair-feeding (PAE>PF>C), or vice versa (PF>PAE>C) (Table S6). Pair-feeding also had some unique effects, particularly in the HPC (Table S7), on genes involved in small molecule metabolism, transport, signal transduction, and stress responses. At a 10% FDR, GSR identified two PF-related processes overlapping between the PFC and HPC: negative regulation of neuron projection development and positive regulation of epithelial cell migration (Figure S2c). Moreover, the curated list of candidate FASD genes from NeuroCarta was also identified in the HPC of the PF group (Portales-Casamar et al. 2013).
PAE altered neural gene expression in response to an inflammatory challenge
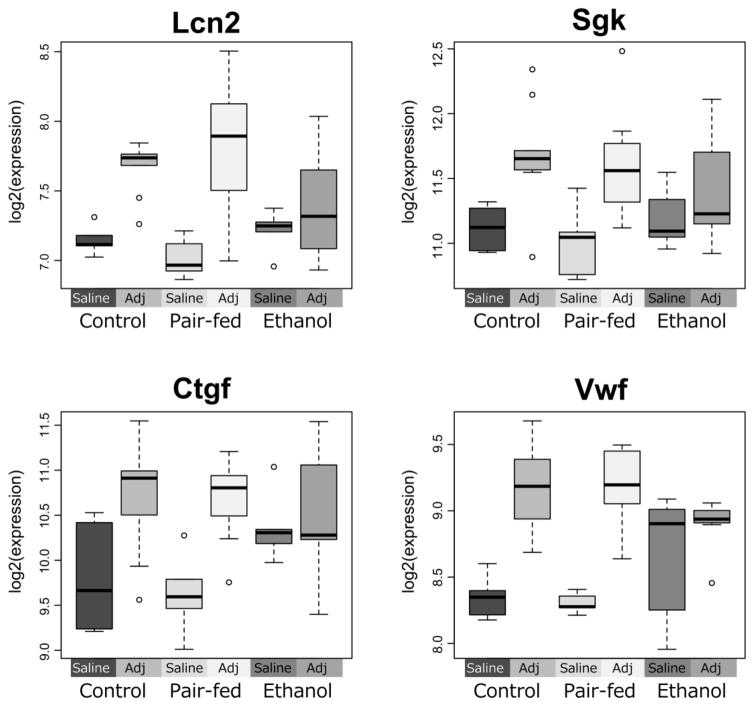
Consistent with the findings on steady state gene expression, the greatest effects of immune challenge were observed on Day 16 post-injection (peak of inflammation). The dominant neural response to adjuvant across prenatal treatments was an up-regulation of mRNA levels. However, some genes (8 in PFC, and 4 in HPC) were differentially expressed in PAE compared to PF and C animals (Table 3 and Figure 4). For all hippocampal genes identified, C and PF animals showed a significant up-regulation of expression, while PAE animals showed no change in expression levels between the saline and adjuvant conditions (Figure 5). These genes (Ctgf, Lcn2, Sgk, Vwf) were multifunctional, with roles in growth, proliferation, adhesion, structural organization, and cellular response to immunological or stressful stimuli.
Table 3.
Genes with a significantly different response to Adjuvant in E compared to both C and PF animals (p <0.05) in PFC (a) and HPC (b) at the peak of inflammation (D16).
| Table 3a: Genes differentially expressed in PFC of Ethanol-exposed animals in response to adjuvant.
| ||||||||
|---|---|---|---|---|---|---|---|---|
| Gene Symbol | Gene Name | Average Expression | F | p-value | q-value | Fold change (Adjuvant/Saline)
|
||
| Control | Pair-fed | Ethanol | ||||||
| ILMN_1351665 | na | 7.0 | 7.9 | 3.4E-04 | 0.17 | 0.80 | 0.80 | 1.12 |
| Ghrhr | growth hormone releasing hormone receptor | 7.0 | 8.2 | 2.5E-04 | 0.14 | 0.87 | 0.78 | 1.23 |
| ILMN_1354124 | na | 6.9 | 7.1 | 7.0E-04 | 0.24 | 0.94 | 0.99 | 1.34 |
| ILMN_1364624 | na | 8.4 | 7.2 | 6.3E-04 | 0.24 | 1.22 | 1.06 | 0.51 |
| ILMN_1372588 | na | 11.1 | 8.7 | 1.7E-04 | 0.13 | 1.38 | 1.10 | 0.67 |
| ILMN_1351971 | na | 11.9 | 9.8 | 6.5E-05 | 0.08 | 1.40 | 1.23 | 0.71 |
| Flna | filamin A, alpha | 8.6 | 7.1 | 7.1E-04 | 0.24 | 1.33 | 1.27 | 0.99 |
| Bhlhe40 | basic helix-loop-helix family, member e40 | 9.5 | 8.1 | 2.8E-04 | 0.15 | 1.42 | 1.45 | 1.02 |
| Table 3b: Genes differentially expressed in HPC of Ethanol-exposed animals in response to adjuvant.
| ||||||||
|---|---|---|---|---|---|---|---|---|
| Gene Symbol | Gene Name | Average Expression | F | p-value | q-value | Fold change (Adjuvant/Saline)
|
||
| Control | Pair-fed | Ethanol | ||||||
| Sgk1 | serum/glucocorticoid regulated kinase 1 | 11.4 | 9.1 | 1.1E-04 | 0.18 | 1.63 | 1.67 | 1.01 |
| Vwf | von Willebrand factor | 8.9 | 15.6 | 7.3E-07 | 0.00 | 1.76 | 1.70 | 1.06 |
| Lcn2 | lipocalin 2 | 7.4 | 18.6 | 1.1E-07 | 0.00 | 1.55 | 1.92 | 1.03 |
| Ctgf | connective tissue growth factor | 10.4 | 11.4 | 1.6E-05 | 0.05 | 1.77 | 2.14 | 0.85 |
Bold = p <0.05. na = probe had no specific alignment to current RefSeq RNA database.
Figure 4. Adjuvant exposure alters gene expression at Day 16 post-injection.
8 genes showed significant changes in expression among treatment groups in prefrontal cortex (a). 4 genes demonstrated significant changes among treatment groups in the hippocampus (b). F-statistic q-value <0.25 for all genes identified.
Figure 5. Ethanol-exposed animals show altered response to adjuvant.
In a subset of genes, Ethanol-exposed animals showed no response to Adjuvant, although pair-fed and control animals responded with an upregulation of the gene (Lcn2, Sgk). In others, gene expression levels in ethanol animals were already elevated compared to pair-feds and controls, but did not change in response to the extent of their control counterparts (Ctgf, Vwf).
Gene Ontology and Upstream Regulator Analysis of PAE effects in response to adjuvant
GSR identified numerous biological processes altered in response to adjuvant at a 1% FDR. In both the PFC and HPC, PAE animals had the fewest uniquely altered categories (8% in PFC, and 11% in HPC), while C animals had the most (25% in PFC and 30% in HPC) (Figure S3a). Four PAE-specific processes overlapped between brain regions (Figure S3b): regulation and positive regulation of epithelial cell proliferation, cellular protein complex assembly, and regulation of hormone level. In categories identified only in PF and C (normal response to adjuvant exposure), 6 overlapped between the PFC and HPC: response to organic nitrogen, actin filament-based process, actin cytoskeleton organization, regulation of cell morphogenesis, developmental growth, and mRNA metabolic process (Figure S3c).
Moreover, URA of gene sets for both the PFC and HPC predicted several master regulators of PAE-specific response to adjuvant, as well as some present only in PF and C animals. In the PFC, 2 PAE-specific genes (Fn1, Dicer1) and 4 PF/C-specific genes (Agt, Foxo3, P38 Mapk, Osm) were significantly activated, while a single PAE-specific gene, Calmodulin, was significantly inhibited (Table 4a). In the HPC, 2 PAE-specific genes (Adcyap1, Prl) showed significant inhibition and one, Nr1i3, showed marginally significant activation (Table 4b). As well, PF/C-specific effects were found for Adamts12 (inhibited) and Foxo4 (activated). Of note, Foxo3 approached significance in the HPC of PF and C animals, representing the only overlap between brain regions.
Table 4.
Genes identified using Ingenuity Pathway Analysis Upstream Regulator in the PFC (a) and HPC (b) of adjuvant versus control animals.
| Table 4a – Upstream Regulator Analysis of the PFC in adjuvant VS saline animals
| ||||||||
|---|---|---|---|---|---|---|---|---|
| Gene Symbol | Gene Name | Predicted status | Z-score | Overlap p-value | ||||
| C | PF | PAE | C | PF | PAE | |||
| PAE-specific | ||||||||
| Calmodulin | Calmodulin | Inhibited | NA | 0.4 | −2.0 | 1.00 | 0.02 | 0.02 |
| Dicer1 | Dicer 1, ribonuclease type III | Activated | NA | NA | 2.0 | 1.00 | 1.00 | 0.09 |
| Fn1 | Fibronectin 1 | Activated | 1.3 | 1.1 | 2.1 | 0.03 | 0.0002 | 0.03 |
| NON-PAE | ||||||||
| Agt | Angiotensinogen | Activated | 2.5 | 2.2 | NA | 0.02 | 0.002 | 1.00 |
| Foxo3 | Forkhead box O3 | Activated | 2.3 | 3.1 | 0.2 | 0.02 | 0.002 | 1.00 |
| Osm | Oncostatin M | Activated | 2.9 | 2.7 | NA | 0.1 | 0.07 | 1.00 |
| P38 Mapk | p38 mitogen-activated protein kinase | Activated | 2.0 | 3.2 | NA | 0.04 | 0.005 | 1.00 |
| Table 4b – Upstream Regulator Analysis in the HPC of adjuvant VS saline animals
| ||||||||
|---|---|---|---|---|---|---|---|---|
| Gene Symbol | Gene Name | Predicted status | Z-score | Overlap p-value | ||||
| C | PF | PAE | C | PF | PAE | |||
| PAE-specific | ||||||||
| Adcyap1 | Adenylate cyclase activating polypeptide 1 | Inhibited | NA | NA | −2.2 | 1.00 | 1.00 | 0.09 |
| Nr1i3 | Nuclear receptor subfamily 1, group I, member 3 | Activated | NA | NA | 2.2 | 1.00 | 1.00 | 0.12 |
| Prl | Prolactin | Inhibited | 2.3 | NA | −2.0 | 0.24 | 1.00 | 0.05 |
| NON-PAE | ||||||||
| Adamts12 | ADAM metallopeptidase with thrombospondin type 1 motif, 12 | Inhibited | −2.4 | −2.0 | NA | 0.0003 | 0.001 | 1.00 |
| Foxo4 | Forkhead box O4 | Activated | 2.0 | 2.0 | NA | 0.07 | 0.04 | 1.00 |
| Foxo3 | Forkhead box O3 | Activated | 2.6 | 2.6 | NA | 0.13 | 0.13 | 1.00 |
Genes with a Z-score ≥2 or ≤-2 and an overlap p-value ≤0.1 are considered significant (bold). Those with no overlap had a p-value of 1 and no Z-score (NA).
Discussion
Prenatal ethanol exposure altered patterns of neural gene expression under both steady-state and immune challenge conditions. In saline-injected females, we identified PAE-induced changes in the expression of Rgs3, Dusp6, and Ap1s2, as well as activation of upstream regulators involved in metabolism and immune function. At the peak of inflammation, adjuvant injection caused PAE-specific changes in gene expression, and uncovered a failure to mount appropriate responses to inflammatory challenge in PAE animal, as evidenced by the absence of changes in inflammation-related genes and upstream regulators identified in controls.
Prenatal ethanol exposure altered neural gene expression under steady-state conditions
Microarray analysis identified unique effects of PAE on 15 and 4 genes in the PFC and HPC, respectively. These had roles in neurodevelopment, cell death, differentiation, transcriptional regulation, and neuronal signaling. Using RT-qPCR, we successfully verified the significant up-regulation of Dusp6 and Ap1s2 in the PFC, as well as Rgs3 in the HPC. Furthermore, the majority of genes not verified by RT-qPCR trended in the same direction as the microarray. The discrepancy in technical verification may arise from the different methods of measurement between the technologies and the underpowered analysis resulting from a relatively low number of samples. Additional large-scale experiments will be required to fully validate these results at the biological level. It is tempting to speculate that these genes play important roles in the cognitive and behavioural deficits observed in FASD. Ap1s2 is involved in neurodevelopment and associated with intellectual disability and autism spectrum disorder, while Dusp6 promotes apoptosis and is linked to bipolar disorder (Bork et al., 2008; Kim et al., 2012). Activation of Laminin could also be involved in the altered neuronal migration patterns observed in PAE brains (Ozer et al., 2000). Moreover, inappropriate feeding behaviour in children with FASD, as well as altered glucose metabolism and insulin tolerance in PAE animals have been reported (Werts et al., 2014; Harper et al. 2014). As Rgs3 negatively regulates glucose output via cAMP production in hepatic cells, it may also play a role in altered energy metabolism within the brain when combined with the activation of gastrin and leptin in the PFC (Raab et al., 2005). Furthermore, the activation of interferon- γ in the HPC supports a role for this cytokine in the altered immune system activity and response to challenge in PAE offspring.
Previous studies on fetal and neonatal brains have uncovered ethanol-induced alterations in the expression of genes related to energy metabolism, adhesion, cytoskeletal remodeling, cell cycle, proliferation, differentiation, apoptosis, as well as neuronal growth and survival (Green et al. 2007; Hard et al. 2005; Zhou et al., 2011). Long-term PAE studies in brains of adult male mice identified networks related to cellular development, free radical scavenging, and small molecule metabolism, as well as genes involved in cognitive function, anxiety, ADHD, and mood disorders (Kleiber et al., 2012). Interestingly, none of the genes found here directly overlapped with those previously identified. These disparities are likely due to species- and sex-specific effects, differences between exposure paradigms, and different gene expression patterns in whole brains versus specific regions. As such, these discrepancies highlight the importance of examining both sexes and targeted brain regions to gain deeper insight into PAE effects. It is also possible that immediate changes in gene expression in response to PAE may not persist or that environmental influences cause alterations over the course of development. Moreover, the relatively moderate levels of ethanol exposure (BALs ~120–150 mg/dl) in this paradigm are consistent with those reported for children with FASD who show functional and cognitive deficits (Mattson et al., 2011). Perhaps most importantly, the genes identified here have not previously been examined in gene expression studies, suggesting that we have uncovered novel candidates for the effects of PAE in females. Whether our specific changes are mediated through epigenetic mechanisms remains to be investigated (Kobor & Weinberg, 2011).
A limitation of this study is that estrous stages were not determined at the time of termination. We have previously shown that PAE induces changes in basal levels of hippocampal glucocorticoid and serotonin Type 1A (5-HT1A) receptor mRNA as a function of estrous stage, which likely have widespread effects on global expression patterns in the brain (Sliwowska et al., 2008). While most females in the present study were likely in diestrus, estrous cycle variation might partially explain intra-group differences in gene expression (Lan et al., 2009).
Prenatal ethanol exposure altered the gene expression response to adjuvant
PAE-specific responses to adjuvant were found for 8 and 4 genes in the PFC and HPC, respectively. These had roles in growth, proliferation, adhesion, structural organization, and cellular response to immunological or stressful stimuli. Across all prenatal treatments, adjuvant caused a global increase in gene expression compared to saline-injected animals. Importantly, PAE animals failed to exhibit the up-regulation in expression observed in controls for genes related to immune and cellular responses to stressful stimuli (Ctgf, Lcn2, Sgk, Vwf). Up-regulation of immune-related genes normally occurs in the CNS in response to peripheral inflammatory stimuli or neuroinflammation, which occurs in AA (reviewed in Ousman & Kubes 2012; Liu et al. 2012). PAE animals may fail to detect these immune changes, and/or launch the appropriate neuroendocrine/neuroimmune response, which could contribute to the prolonged inflammation observed in our previous AA study (Zhang et al., 2012). Consistent with this finding, most master regulators identified in the Upstream Regulator Analysis were involved in the immune response. For example, P38 Mapk plays a role in signal transduction within the normal inflammatory cascade and is only activated in PF and C animals (Cuadrado & Nebreda 2010). Moreover, Adamts12 modulates neutrophil apoptosis during inflammation, while Osm attenuates the inflammatory response (Moncado-Pazos et al., 2012; Dumas et al., 2012). Thus, inhibition of Adamts12 and activation of Osm in control animals may blunt their responses to adjuvant. Furthermore, Adcyap1 modulates anti-inflammatory responses and is neuroprotective in neurons following inflammation (Waschek, 2013). Its inhibition in PAE animals suggests a lower level of protection against inflammation than the one that would occur in controls. In turn, as Prl promotes pro-inflammatory responses, its PAE-specific activation suggests an altered response to adjuvant (Brand et al., 2004). Failure of PAE animals to activate Foxo-related pathways may also play a role in their unique response to adjuvant, as knockdown of Foxo3 or Foxo4 increases inflammatory responses (Hwang et al., 2011; Zhou et al., 2009). The possibility that Foxo3 is already up-regulated in PAE animals, and thus may not change further after adjuvant injection remains to be investigated (Kleiber et al., 2013). The activation of fibronectin in PAE animals is interesting, as it is involved in the development of inflammatory arthritis (Barilla & Carsons, 2000). Greater production or sensitivity to this protein could underlie the altered course and severity of AA in PAE animals. Finally, activation of Dicer1 in PAE animals suggests alterations to microRNA processing under stress conditions, previously demonstrated following PAE (Guo et al., 2011).
Effects of pair-feeding on neural gene expression: Pair-feeding is a treatment in itself
A number of genes were similarly altered, or showed graded and differential effects in PAE and PF compared to C animals (Figure 2). These may respond to common effects of ethanol exposure and pair-feeding, such as reduced caloric availability or altered stress system regulation. While both PAE and PF animals receive the same number of calories, PAE dams eat ad libitum whereas PF dams receive a reduced ration, likely resulting in hunger and stress (Harris & Seckl 2011). Moreover, PF dams tend to consume their daily ration within a few hours and are deprived until the next feeding, which may have unique metabolic effects associated with “disordered” eating. Our results suggest that the HPC may be susceptible to fetal programming in response to energy-, and stress-related environmental factors. Interestingly, the curated list of candidate FASD genes from NeuroCarta was identified in the HPC of PF animals, suggesting that these genes are potentially related to common mechanisms underlying prenatal alcohol exposure, nutrition, and stress (Portales-Casamar et al. 2013). Studies such as ours are critical to separate the effects of prenatal stress and prenatal alcohol exposure at the level of gene expression.
Summary and Conclusions
Our results support the hypothesis that PAE has long-term effects on gene expression patterns in the brain, as well as on the response to a systemic inflammatory insult. As both the PFC and HPC play important roles in cognitive, neuroendocrine, and immune function, the identified changes in steady-state and activated expression likely contribute to immune-related alterations, as well as cognitive and behavioural deficits arising from PAE. Moreover, an inability to mount appropriate response to immune/inflammatory challenges may contribute to the increased vulnerability of individuals with FASD to infections and immune problems. These findings extend our previous data demonstrating that PAE animals exhibit increased susceptibility to and impaired recovery from an inflammatory challenge (Zhang et al. 2012), and suggest that the adverse impact of prenatal ethanol exposure on the neural transcriptome may underlie long-term health and developmental outcomes observed in individuals with FASD.
Supplementary Material
Acknowledgments
This work is supported by National Institute on Mental Health grant R24 MH081797 and NeuroDevNet NCE funding to MSK and JW, as well as by R37 AA007789 from the National Institute on Alcohol Abuse Alcoholism to JW. KAS was supported by the NeuroDevNet NCE/CFRI Graduate Studentship. MSK is a Senior Fellow of the Canadian Institute for Advanced Research and a Scholar of the Mowafaghian Foundation. We would like to thank Xingqi Zhang, Linda Ellis, Wayne Yu, Tamara Bodnar, and Parker Holman for their assistance in this study.
Contributor Information
Katarzyna A. Stepien, Department of Medical Genetics, University of British Columbia.
Alexandre A. Lussier, Department of Medical Genetics, University of British Columbia.
Sarah M. Neumann, Centre for Molecular Medicine and Therapeutics, Child and Family Research Institute, University of British Columbia.
Paul Pavlidis, Centre for High Throughput Biology, Department of Psychiatry, University of British Columbia.
Michael S. Kobor, Department of Medical Genetics, Centre for Molecular Medicine and Therapeutics, Child and Family Research Institute, Human Early Learning Partnership, University of British Columbia.
Joanne Weinberg, Department of Cellular and Physiological Sciences, University of British Columbia.
References
- Barilla ML, Carsons SE. Fibronectin fragments and their role in inflammatory arthritis. Seminars in Arthritis and Rheumatism. 2000;29(4):252–65. doi: 10.1016/s0049-0172(00)80012-8. [DOI] [PubMed] [Google Scholar]
- Bodnar T, Weinberg J. Prenatal Alcohol Exposure: Impact on Neuroendocrine – Neuroimmune Networks. In: Cui C, et al., editors. Neural-Immune Interactions in Brain Function. Springer Science+Business Media; New York: 2013. pp. 307–357. [Google Scholar]
- Borck G, Mollà-Herman A, Boddaert N, Encha-Razavi F, Philippe A, Robel L, Desguerre I, Brunelle F, Benmerah A, Munnich A, Colleaux L. Clinical, cellular, and neuropathological consequences of AP1S2 mutations: further delineation of a recognizable X-linked mental retardation syndrome. Human Mutation. 2008;29(7):966–74. doi: 10.1002/humu.20531. [DOI] [PubMed] [Google Scholar]
- Brand JM, Frohn C, Cziupka K, Brockmann C, Kirchner H, Luhm J. Prolactin triggers pro-inflammatory immune responses in peripheral immune cells. European Cytokine Network. 2014;15(2):99–104. [PubMed] [Google Scholar]
- Carter JL, Lubahn C, Lorton D, Osredkar T, Der TC, Schaller J, Evelsizer S, Flowers S, Ruff N, Reese B, Bellinger DL. Adjuvant-induced arthritis induces c-Fos chronically in neurons in the hippocampus. Journal of Neuroimmunology. 2011;230(1–2):85–94. doi: 10.1016/j.jneuroim.2010.09.005. [DOI] [PubMed] [Google Scholar]
- Crofford LJ, Sano H, Karalis K, Webster EL, Goldmuntz EA, Chrousos GP, Wilder RL. Local secretion of corticotropin-releasing hormone in the joints of Lewis rats with inflammatory arthritis. Journal of Clinical Investigation. 1992;90(6):2555–64. doi: 10.1172/JCI116150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado A, Nebreda AR. Mechanisms and functions of p38 MAPK signalling. Biochemical Journal. 2010;429(3):403–17. doi: 10.1042/BJ20100323. [DOI] [PubMed] [Google Scholar]
- Dumas A, Lagarde S, Laflamme C, Pouliot M. Oncostatin M decreases interleukin-1 β secretion by human synovial fibroblasts and attenuates an acute inflammatory reaction in vivo. Journal of Cellular and Molecular Medicine. 2012;16(6):1274–85. doi: 10.1111/j.1582-4934.2011.01412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier TW, Drews-Botsch C, Falek A, Coles C, Brown LA. Maternal Alcohol Abuse and Neonatal Infection. Alcoholism: Clinical & Experimental Research. 2005;29(6):1035–1043. doi: 10.1097/01.alc.0000167956.28160.5e. [DOI] [PubMed] [Google Scholar]
- Gauthier TW, Manar MH, Brown LA. Is maternal alcohol use a risk factor for early-onset sepsis in premature newborns? Alcohol (Fayetteville, NY) 2004;33(2):139–45. doi: 10.1016/j.alcohol.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Green ML, Singh AV, Zhang Y, Nemeth KA, Sulik KK, Knudsen TB. Reprogramming of genetic networks during initiation of the Fetal Alcohol Syndrome. Developmental Dynamics. 2007;236(2):613–631. doi: 10.1002/dvdy.21048. [DOI] [PubMed] [Google Scholar]
- Guo Y, Chen Y, Carreon S, Qiang M. Chronic intermittent ethanol exposure and its removal induce a different miRNA expression pattern in primary cortical neuronal cultures. Alcoholism: Clininal and Experimental Research. 2012;36(6):1058–66. doi: 10.1111/j.1530-0277.2011.01689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hard ML, Abdolell M, Robinson BH, Koren G. Gene-expression analysis after alcohol exposure in the developing mouse. Journal of Laboratory and Clinical Medicine. 2005;145(1):47–54. doi: 10.1016/j.lab.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Harper KM, Tunc-Ozcan E, Graf EN, Redei EE. Intergenerational effects of prenatal ethanol on glucose tolerance and insulin response. Physiological Genomics. 2014;46(5):159–68. doi: 10.1152/physiolgenomics.00181.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A, Seckl J. Glucocorticoids, prenatal stress and the programming of disease. Hormones and behavior. 2011;59(3):279–89. doi: 10.1016/j.yhbeh.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Hellemans KG, Sliwowska JH, Verma P, Weinberg J. Prenatal alcohol exposure: fetal programming and later life vulnerability to stress, depression and anxiety disorders. Neuroscience and Biobehavioral Reviews. 2010;34(6):791–807. doi: 10.1016/j.neubiorev.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J, Rajendrasozhan S, Yao H, Chung S, Sundar IK, Huyck HL, Pryhuber GS, Kinnula VL, Rahman I. FOXO3 deficiency leads to increased susceptibility to cigarette smoke-induced inflammation, airspace enlargement, and chronic obstructive pulmonary disease. Journal of Immunology. 2011;187(2):987–998. doi: 10.4049/jimmunol.1001861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S, Knight R, Marmer DJ, Steele RW. Immune deficiency in fetal alcohol syndrome. Pediatric research. 1981;15(6):908–11. doi: 10.1203/00006450-198106000-00005. [DOI] [PubMed] [Google Scholar]
- Kim SH, Shin SY, Lee KY, Joo EJ, Song JY, Ahn YM, Lee YH, Kim YS. The genetic association of DUSP6 with bipolar disorder and its effect on ERK activity. Progress in Neuropsychopharmacology & Biological Psychiatry. 2012;37(1):41–9. doi: 10.1016/j.pnpbp.2011.11.014. [DOI] [PubMed] [Google Scholar]
- Kleiber ML, Laufer BI, Wright E, Diehl EJ, Singh SM. Long-term alterations to the brain transcriptome in a maternal voluntary consumption model of fetal alcohol spectrum disorders. Brain research. 2012;1458:18–33. doi: 10.1016/j.brainres.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Kleiber ML, Mantha K, Stringer RL, Singh SM. Neurodevelopmental alcohol exposure elicits long-term changes to gene expression that alter distinct molecular pathways dependent on timing of exposure. Journal of Neurodevelopmental Disorders. 2013;5(1):6. doi: 10.1186/1866-1955-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobor MS, Weinberg J. Focus on: epigenetics and fetal alcohol spectrum disorders. Alcohol Research & Health. 2011;34(1):29–37. [PMC free article] [PubMed] [Google Scholar]
- Lan N, Yamashita F, Halpert AG, Sliwowska JH, Viau V, Weinberg J. Effects of prenatal ethanol exposure on hypothalamic-pituitary-adrenal function across the estrous cycle. Alcoholism: Clinical and Experimental Research. 2009;33(6):1075–88. doi: 10.1111/j.1530-0277.2009.00929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Braynen W, Keshav K, Pavlidis P. ErmineJ: tool for functional analysis of gene expression data sets. BMC bioinformatics. 2005;6(1):269. doi: 10.1186/1471-2105-6-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS genetics. 2007;3(9):1724–35. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wu Z, Hayashi Y, Nakanishi H. Age-dependent neuroinflammatory responses and deficits in long-term potentiation in the hippocampus during systemic inflammation. Neuroscience. 2012;216:133–42. doi: 10.1016/j.neuroscience.2012.04.050. [DOI] [PubMed] [Google Scholar]
- Mattson SN, Crocker N, Nguyen TT. Fetal alcohol spectrum disorders: neuropsychological and behavioral features. Neuropsychology Review. 2011;21(2):81–101. doi: 10.1007/s11065-011-9167-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May PA, Gossage JP, Kalberg WO, Robinson LK, Buckley D, Manning M, Hoyme HE. Prevalence and epidemiologic characteristics of FASD from various research methods with an emphasis on recent in-school studies. Developmental disabilities research reviews. 2009;15(3):176–92. doi: 10.1002/ddrr.68. [DOI] [PubMed] [Google Scholar]
- Moncada-Pazos A, Obaya AJ, Llamazares M, Heljasvaara R, Suárez MF, Colado E, Noël A, Cal S, López-Otín C. ADAMTS-12 metalloprotease is necessary for normal inflammatory response. Journal of Biological Chemistry. 2012;287(47):39554–63. doi: 10.1074/jbc.M112.408625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council. Guide for the care and use of laboratory animals. 8. Washington, DC: The National Academies Press; 2011. [Google Scholar]
- Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nature protocols. 2006;1(3):1559–82. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- Norman AL, Crocker N, Mattson SN, Riley EP. Neuroimaging and fetal alcohol spectrum disorders. Developmental disabilities research reviews. 2009;15(3):209–17. doi: 10.1002/ddrr.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ousman SS, Kubes P. Immune surveillance in the central nervous system. Nature Neuroscience. 2012;15(8):1096–1101. doi: 10.1038/nn.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozer E, Sarioglu S, Güre A. Effects of prenatal ethanol exposure on neuronal migration, neuronogenesis and brain myelination in the mice brain. Clinical Neuropathology. 2000;19(1):21–5. [PubMed] [Google Scholar]
- Portales-Casamar E, Ch'ng C, Lui F, St-Georges N, Zoubarev A, Lai AY, Lee M, Kwok C, Kwok W, Tseng L, Pavlidis P. Neurocarta: aggregating and sharing disease-gene relations for the neurosciences. BMC genomics. 2013;14(1):129. doi: 10.1186/1471-2164-14-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab RM, Bullen J, Kelleher J, Mantzoros C, Stephanopoulos G. Regulation of mouse hepatic genes in response to diet induced obesity, insulin resistance and fasting induced weight reduction. Nutrition & Metabolism. 2005;2:15. doi: 10.1186/1743-7075-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nature Protocols. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Sliwowska JH, Lan N, Yamashita F, Halpert AG, Viau V, Weinberg J. Effects of prenatal ethanol exposure on regulation of basal hypothalamic-pituitary-adrenal activity and hippocampal 5-HT1A receptor mRNA levels in female rats across the estrous cycle. Psychoneuroendocrinology. 2008;33(8):1111–23. doi: 10.1016/j.psyneuen.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. limma: Linear Models for Microarray Data. In: Gentleman R, et al., editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- Streissguth AP, Clarren SK, Jones KL. Natural history of the fetal alcohol syndrome: a 10-year follow-up of eleven patients. Lancet. 1985;2(8446):85–91. doi: 10.1016/s0140-6736(85)90189-8. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome biology. 2002;3(7):RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waschek JA. VIP and PACAP: neuropeptide modulators of CNS inflammation, injury, and repair. British Journal of Pharmacology. 2013;169(3):512–23. doi: 10.1111/bph.12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg J, Jerrells TR. Suppression of immune responsiveness: sex differences in prenatal ethanol effects. Alcoholism: Clinical and Experimental Research. 1991;15(3):525–31. doi: 10.1111/j.1530-0277.1991.tb00554.x. [DOI] [PubMed] [Google Scholar]
- Werts RL, Van Calcar SC, Wargowski DS, Smith SM. Inappropriate feeding behaviors and dietary intakes in children with fetal alcohol spectrum disorder or probable prenatal alcohol exposure. Alcoholism: Clinical and Experimental Research. 2014;38(3):871–8. doi: 10.1111/acer.12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitacre CC. Sex differences in autoimmune disease. Nature immunology. 2001;2(9):777–80. doi: 10.1038/ni0901-777. [DOI] [PubMed] [Google Scholar]
- Zhang X, Lan N, Bach P, Nordstokke D, Yu W, Ellis L, Meadows GG, Weinberg J. Prenatal alcohol exposure alters the course and severity of adjuvant-induced arthritis in female rats. Brain, behavior, and immunity. 2012;26(3):439–450. doi: 10.1016/j.bbi.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Sliwowska JH, Weinberg J. Prenatal alcohol exposure and fetal programming: effects on neuroendocrine and immune function. Experimental biology and medicine (Maywood, NJ) 2005;230(6):376–88. doi: 10.1177/15353702-0323006-05. [DOI] [PubMed] [Google Scholar]
- Zhi-Liang H, Bao J, Reecy J. CateGOrizer: a web-based program to batch analyze gene ontology classification categories. Online J Bioinformatics 2008 [Google Scholar]
- Zhou FC, Zhao Q, Liu Y, Goodlett CR, Liang T, McClintick JN, Edenberg HJ, Li L. Alteration of gene expression by alcohol exposure at early neurulation. BMC genomics. 2011;12(1):124. doi: 10.1186/1471-2164-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Cao Q, Peng Y, Zhang QJ, Castrillon DH, DePinho RA, Liu ZP. FoxO4 inhibits NF-kappaB and protects mice against colonic injury and inflammation. Gastroenterology. 2009;137(4):1403–14. doi: 10.1053/j.gastro.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.