Abstract
Through an international multi-center collaboration, 13 individuals from nine unrelated families and affected by likely pathogenic biallelic variants in TBC1-domain-containing kinase (TBCK) were identified through whole-exome sequencing. All affected individuals were found to share a core phenotype of intellectual disability and hypotonia, and many had seizures and showed brain atrophy and white-matter changes on neuroimaging. Minor non-specific facial dysmorphism was also noted in some individuals, including multiple older children who developed coarse features similar to those of storage disorders. TBCK has been shown to regulate the mammalian target of rapamycin (mTOR) signaling pathway, which is also stimulated by exogenous leucine supplementation. TBCK was absent in cells from affected individuals, and decreased phosphorylation of phospho-ribosomal protein S6 was also observed, a finding suggestive of downregulation of mTOR signaling. Lastly, we demonstrated that activation of the mTOR pathway in response to L-leucine supplementation was retained, suggesting a possible avenue for directed therapies for this condition.
Main Text
Intellectual disability (ID) is a common diagnosis with few specific disease-directed therapeutic options.1 In some specific cases when a molecular etiology for ID has been discovered, especially in the setting of inborn errors of metabolism, there has been vast improvement in the outcomes of those individuals.2 The genetic heterogeneity of ID has traditionally complicated both diagnosis and gene discovery, leading to a large proportion of affected individuals who never receive a molecular diagnosis. The introduction of agnostic testing, such as genome-wide arrays and whole-exome sequencing (WES), has accelerated both discovery of and diagnosis for individuals with ID. However, such acceleration has also highlighted the need to identify independent pathogenic variants in the same gene before a causal link to ID can be established. Using WES, in nine unrelated families we were able to identify 13 individuals who are affected by ID and hypotonia and harbor biallelic pathogenic variants in TBC1-domain-containing kinase (TBCK), a gene recently implicated in the etiology of ID in a single family.3
All affected individuals were recruited under research protocols approved by the institutional review boards (IRBs) of their respective institutions after informed consent was obtained. We sequenced DNA from 13 affected individuals from nine families and their healthy parents to identify the etiology of their ID and hypotonia. We performed WES if the variant was unknown in the family or Sanger sequencing if the variant had been identified in another family member (Table S1). After identifying TBCK variants in each family, we assembled the cohort through personal collaborations and the assistance of GeneMatcher. Each subject sequenced provided written consent, and all work was performed either in a clinical laboratory or under an IRB-approved protocol. Variants previously reported in dbSNP, the 1000 Genomes Project, and the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project (ESP) Exome Variant Server with a minor allele frequency > 1% were excluded. Anticipating that synonymous variants are far less likely to be pathogenic, we focused our variant analysis primarily on nonsynonymous variants, splice-acceptor and donor-site mutations, and coding insertions/deletions (indels). When applicable, emphasis was placed on identifying homozygous or compound-heterozygous variants shared between the two affected siblings. In all cases, filtering WES data for compound-heterozygous, X-linked, or heterozygous de novo variants did not provide any additional candidate variants. Pathogenic homozygous or compound-heterozygous variants in TBCK were confirmed by Sanger sequencing. Parents were found to be heterozygous for each of the variants found in their children, consistent with an autosomal-recessive model. The one homozygous missense variant was computationally predicted to have an functional effect, given that it contains a highly conserved nucleotide (phyloP score = 8.35 [−20.0;10.0]), encodes an amino acid highly conserved up to C. elegans (considering 13 species), has a moderate physicochemical difference between Leu and Pro (Grantham distance = 98 [0–215]), is found in the Rab-GTPase-TBC domain, and is predicted to be deleterious by SIFT (score = 0.01, median = 3.35) and disease causing by MutationTaster (p value = 1).
These 13 affected individuals all share a core phenotype of ID and hypotonia in addition to variable expressivity of other features as summarized in Table 1 (see also Figure 1A). A few individuals demonstrated facial features reminiscent of a storage disease (e.g., macroglossia and coarse facies), but it was not a universal feature and might develop as the children age. Several individuals demonstrated other mild facial anomalies, but none that appeared diagnostic (Figures 1B–1D and 1G–1I). Seven individuals from five families had seizures, which were often refractory to pharmacologic management. Development ranged from profound to mild developmental delay, and only one individual demonstrated regression. Brain MRI showed a thin corpus callosum and brainstem, ex vacuo dilation of the lateral ventricles, periventricular leukomalacia, and white-matter changes ranging from non-specific to a picture of leukodystrophy (Figures 1E, 1F, 1J, and 1K). The following features were found in a minority of individuals, so their relevance to the phenotype remains unclear: hyperthyroidism, hypothyroidism, pernicious anemia, suspected immune deficiency, corneal clouding, scoliosis, central adrenal insufficiency, growth hormone deficiency, and profound bilateral hearing loss.
Table 1.
Characteristics of Individuals with Biallelic Variants in TBCK
|
Affected Individuals |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1-1a | 1-2a | 2-1 | 3-1 | 4-1 | 4-2 | 5-1 | 6-1 | 6-2 | 7-1 | 8-1 | 9-1 | 9-2 | |
| Variants (GenBank: NM_001163435.2) | Hom c.1897+1G>A | Hom c.1897+1G>A | Hom c.831_832insTA (p.Pro278Tyrfs∗18) | Hom c.1652T>C (p.Leu551Pro) | Het c.[2060−2A>G];[803_806delTGAA], p.[=];[Met268fsArg∗26] | Het c.[2060−2A>G];[803_806delTGAA], p.[=];[Met268fsArg∗26] | Hom c.376C>T (p.Arg126∗) | Hom c.1370delA (p.Asn457Thrfs∗15) | Hom c.1370delA (p.Asn457Thrfs∗15) | Hom c.455+4 C>G | Hom c.376C>T (p.Arg126∗) | Het c.[(658+1_ 659−1)_(2059+1_2060−1)del];[376C>T], p[=];[Arg126∗] | Het c.[(658+1_ 659−1)_(2059+1_2060−1)del];[376C>T], p[=];[Arg126∗] |
| Mutation type | splice site and frameshift | splice site and frameshift | insertion and frameshift | missense | splice site and frameshift; frameshift | splice site and frameshift; frameshift | nonsense | frameshift | frameshift | splice, skipping of exons 3 and 4 | nonsense | deletion of exons 7–22; nonsense | deletion of exons 7–22; nonsense |
| Consanguinity reported | + | + | − | + | − | − | + | + | + | + | − | − | − |
| Age at last exam | died at 5 years | 11 years | 5 years | 11 years | 4 years | 2 years | 10 years | 12 years | 3 years | 6 years | 12 years | 10 years | 2 years |
| Sex | male | female | male | male | female | female | female | male | female | male | female | female | male |
| Ethnicity | Saudi | Saudi | Syrian | Pakistani | mixed European | mixed European | Hispanic | Algerian | Algerian | Palestinian | Puerto Rican and Dominican | Puerto Rican and Mexican | Puerto Rican and Mexican |
| Prenatal findings | decreased fetal movement, oligohydramnios | none reported | premature contractions | none reported | ventriculomegaly | none reported | none reported | none reported | none reported | none reported | none reported | renal pyelectasis and hepatic calcifications | none reported |
| Development | |||||||||||||
| Motor | no sitting | rolling at 11 years | no sitting | mild delay | independent walking | sitting | severe delay | sitting | walking at 2.5 years | sitting | sitting | no sitting | no sitting |
| Speech | few words | babbling, no words | non-verbal | mild delay | few single words | many words | no words | no words | few words | few words | no words | non-verbal | non-verbal |
| Cognitive | severe delay | severe delay | severe delay | mild delay | severe delay | mild delay | severe delay | severe delay | moderate delay | severe delay | severe delay | severe delay | severe delay |
| Regression | − | − | − | − | − | − | + | − | − | − | − | − | − |
| Ventilator dependence | − | − | tracheostomy only | − | − | − | + | − | − | − | tracheostomy only | − | − |
| MRI findings | diffuse brain atrophy, abnormal white-matter signal intensity | diffuse brain atrophy | posterior thinning of the corpus callosum and brainstem, ex vacuo dilation of the ventricles, periventricular leukomalacia | mild prominence of the lateral ventricles | none reported | none reported | central and cortical volume loss with atrophy | discrete abnormalities of the periventricular and bilateral parietal white matter | discrete abnormalities of the periventricular and bilateral parietal white matter | discrete abnormalities of the periventricular and bilateral parietal white matter | abnormal white-matter intensities | bilateral frontal white-matter hyperintensities on T2-weighted FLAIR | overall paucity of white matter, mild hyperintensity and central volume loss on T2-weighted FLAIR |
| Hypotonia | severe | severe | severe | mild to moderate | moderate | severe | severe | moderate | moderate | severe | severe | progressively severe | progressively severe |
| Reflexes | delayed | reduced | absent in lower extremities | normal | reduced | reduced | globally decreased | present | present | absent in lower extremities | reduced | not reported | not reported |
| Seizures | + | + | + | − | − | − | + | − | − | − | + | − | − |
| Unusual facial features | sloped forehead, bulbous nose, tented upper lip, upward slant of palpebral fissures | tented upper lip, sloped forehead, bulbous nose | macrocephaly | epicanthal folds, broad nasal bridge, deep-set eyes | − | − | mildly coarse facies, synophrys, long eyelashes, macroglossia, microcephaly, hirsuitism | wide forehead, bulbous nose, open mouth, thick lips, deep palate, prognathic mandible | macrocephaly, wide forehead, short neck | − | macrocephaly, very coarse facial features, bushy eyebrows, macroglossia | metopic ridge, thick bushy eyebrows, mildly high-arched palate | mildly coarse facial features, hirsute, epicanthal folds, long philtrum, tented upper lip |
| Other findings | abnormal eye movements, asthma, eczema | hypothyroidism, pernicious anemia, recurrent candidiasis | strabismus, nystagmus | autism, bipolar disorder, high-arched palate, broad fingers and toes, mild scoliosis | mild macrocephaly | − | corneal clouding, central adrenal insufficiency and growth hormone deficiency, scoliosis | hearing loss, behavioral difficulties, delayed bone age | delayed bone age | pectus excavatum, pulmonic stenosis | possible mitochondrial complex III deficiency, severe scoliosis, neurogenic bladder, hyperthyroidism | small pectus excavatum, mild scoliosis, diastasis recti, clinodactyly, partial syndactyly of toes 2 and 3 | cryptorchidism, 11 rib pairs, resolved pelviectasis |
Abbreviations are as follows: Hom, homozygous; and Het, heterozygous.
These individuals were previously reported by Alazami et al.3
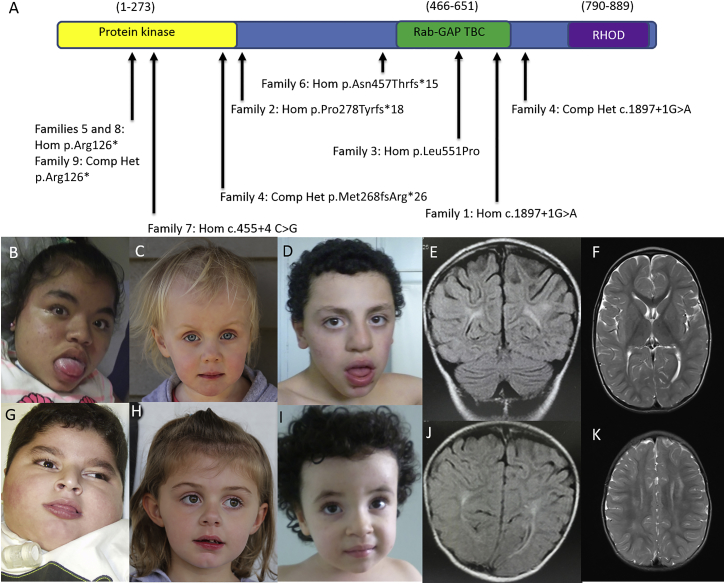
Figure 1.
Diagram of TBCK with Reported Variants and Facial Features and Brain Imaging of Affected Individuals
(A) A diagram of TBCK includes known domains and the variants present in the individuals described in this report. Abbreviations are as follows: Hom, homozygous; and Comp Het, compound heterozygous.
(B–D and G–I) Facial features of individuals 8-1 (B) and 2-1 (G), affected siblings 4-1 (C) and 4-2 (H), and affected siblings 6-1 (D) and 6-2 (I). Individuals 8-1 and 2-1 have coarse features with significant macroglossia, and individuals 4-1 and 4-2 have no significant facial dysmorphisms. Note the mild dysmorphic features, including a wide forehead, bulbous nose, open mouth with macroglossia, thick lips, deep palate, and prognathic mandible, in individual 6-1 and the wide forehead and short neck in individual 6-2.
(E, F, and I–K) Brain MRI of individuals 6-1 (E), 6-2 (J), and 7-1 (F and K) demonstrates discrete abnormalities of the periventricular and parietal white matter in both studies.
Although all 13 individuals demonstrated developmental delays, the least-affected individual (individual 3-1) had the only missense variant in our cohort and showed only mild delays and minimal MRI abnormalities. The other individuals all had variants suspected to lead to significant protein truncation from splice-site substitutions, small insertions, small deletions, or multi-exonic deletions and subsequently appeared to have a more severe phenotype. It is likely that additional individuals with pathogenic missense variants in the population have yet to be identified given that they would have a milder phenotype, and their variant would be less easily recognized as disease causing. Additionally, in one family (individuals 9-1 and 9-2), the combination of a nonsense variant and a multi-exon deletion required both exome sequencing and a genome-wide array to confirm the diagnosis.
Clinical diagnosis was also complicated by the significant variability in presentation, even within the sibling pairs we identified, and the lack of a clear environmental or therapeutic intervention as an explanation. Also, although multiple individuals in this series showed dysmorphic features, there does not appear to be a clearly identifiable facial gestalt associated with this syndrome. Coarse facial features did appear to develop in middle childhood in some individuals, but this was also non-specific. Similarly, brain MRI appeared non-specific and might have mimicked a leukodystrophy process in the more severely affected individuals. These features of the syndrome highlight the need for a clinically agnostic diagnostic tool, such as exome sequencing or gene panels, to confirm the diagnosis.4 Previous diagnoses considered for these individuals included static encephalopathy, lysosomal-storage disease, leukodystrophy, and mitochondrial disorder, but no single alternate differential diagnosis was clearly favored throughout the cohort. Also complicating diagnosis, this disease is not restricted to one geographic region, given that these affected individuals have diverse ethnic backgrounds. However, there does appear to be a common variant, c.376C>T (p.Arg126∗) (GenBank: NM_001163435.2), reported in the three Hispanic families, suggesting a founder effect in this population.
TBCK was named for the Tre-2, Bub2p, and Cdc16p (TBC) 1 domain and the protein kinase domain found within the encoded protein. Interestingly, TBCK has recently been implicated in the regulation of mammalian target of rapamycin (mTOR) signaling, given that knockdown of TBCK decreases the transcription of mTOR complex (mTORC) proteins and downregulates mTOR signaling.5 mTOR is a proline-directed serine threonine protein kinase; activation of the mTOR pathway leads to phosphorylation of ribosomal protein S6. Many developmental disorders, including tuberous sclerosis (MIM: 191100) and PTEN-related disorders (MIM: 158350), are associated with aberrant mTOR signaling, and mTOR signaling has been shown to play a role in structural brain malformations, epilepsy, autism, and ID.6, 7 One of the many activators of the mTOR pathway has been shown to be leucine, one of the essential amino acids, through mTORC1.8 The effects of leucine supplementation on mTOR activation have been studied in the context of adipogenesis, increased muscle mass, and diabetes control.9, 10
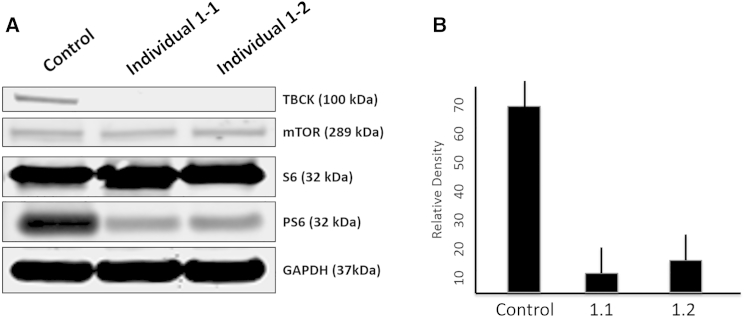
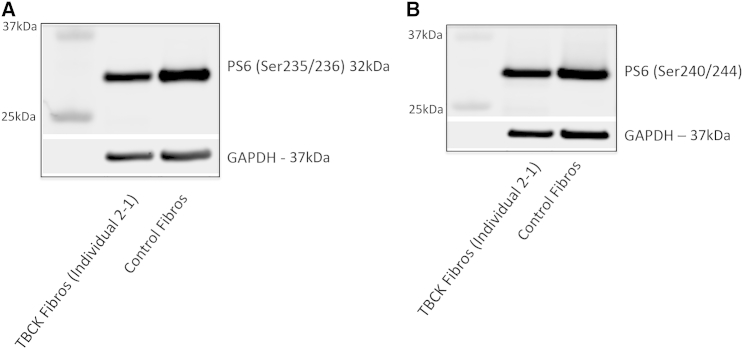
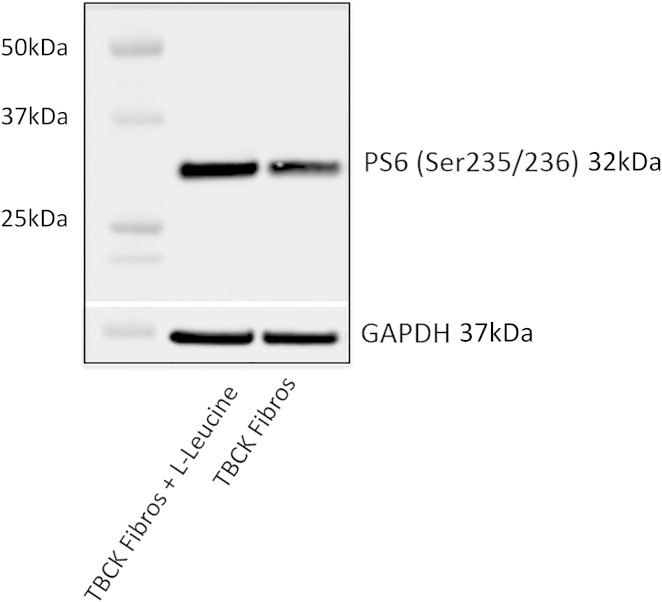
To evaluate the functional consequences of the TBCK variants, we used western blotting to assess the amount of TBCK in immortalized lymphoblastoid cell lines (LCLs) from one healthy control subject and two individuals (1-1 and 1-2) with homozygous TBCK variants predicted to lead to a frameshift. Blots were probed with a polyclonal anti-TBCK antibody raised against the C-terminal region within amino acids 457–694. Whereas TBCK was clearly detectable as a single band in control LCLs, no protein was found in cell extracts from the affected individuals’ LCLs (Figure 2A). Western blotting demonstrated mTOR-pathway signaling in LCLs and in cultured fibroblasts from one other individual (2-1) with a different predicted frameshift TBCK variant. Levels of non-phosphorylated and phosphorylated S6 (PS6) isoform Ser235/236 were assessed in triplicate. Densitometry of the blots revealed that PS6 levels were, on average, 78% (range = 76%–82%, SEM ± 6) lower in LCLs harboring TBCK mutations than in control LCLs (p < 0.05; Figure 2B) and that PS6 Ser235/236 levels were, on average, 36% (range = 31%–41%, SEM ± 5) lower in fibroblasts harboring mutant TBCK than in control fibroblasts (p < 0.05; Figure 3A); a decrease in PS6 (Ser240/244) was observed as well (Figure 3B). There was no change in the level of non-phosphorylated S6 or mTOR proteins in either LCLs or fibroblasts containing TBCK mutations. The addition of leucine (600 μg/ml), a known nutrient stimulator of mTOR activation, to the culture media of fibroblasts from individual 2-1 induced an upregulation of basal mTOR signaling, as evidenced by increased levels of PS6 (24% ± 4%; p < 0.05; Figure 4).
Figure 2.
Western Blot Demonstrates Absent TBCK and Decreased mTOR Signaling in Lympoblastic Cells of Affected Individuals
Western blots of immortalized lymphocytes (LCLs) from a control individual and individuals 1-1 and 1-2; both affected individuals have homozygous frameshift variants.
(A) Top: TBCK levels were lower in proband LCLs (from individual 1-1 and 1-2) than in control LCLs. Middle: levels of mTOR and non-phosphorylated S6 were unchanged, but levels of PS6 isoform Ser235/236 were lower in LCLs with mutant TBCK than in control cells. Bottom: GAPDH was used as a loading control.
(B) Relative densitometry graph shows the levels of PS6 isoform Ser235/236 in fibroblasts with mutant TBCK (from individuals 1-1 and 1-2) and control fibroblasts across independent western blots in triplicate. Lines depict the SE. Cells were provided by Fowzan S. Alkuraya and colleagues of the King Faisal Specialist Hospital and Research Center.
Figure 3.
Western Blots Demonstrate Decreased mTOR Signaling in Fibroblasts of Affected Individuals
Levels of PS6 isoforms Ser235/236 (A) and Ser240/244 (B) were lower in fibroblasts harboring mutant TBCK (from individual 2-1) than in control cells. The blot demonstrates that levels of PS6 isoform Ser235/236 were, on average, 36% (range 31%–41%, SEM ± 5) lower in fibroblasts with mutant TBCK than in control fibroblasts (p < 0.05). There was no change in the level of non-phosphorylated S6 or mTOR in either LCLs or fibroblasts containing TBCK mutations. GAPDH was used as a loading control.
Figure 4.
Western Blot Demonstrates that Leucine Normalizes mTOR Signaling in Fibroblasts of Affected Individuals
Enhanced mTOR signaling in TBCK mutant fibroblasts (individual 2-1) with the addition of exogenous L-leucine (600 μg/ml). The western blot demonstrates increased levels of PS6 (24 ± 4%; p < 0.05). GAPDH was used as a loading control.
The precise function of TBCK has not been fully elucidated, but previous studies have shown that TBCK is downregulated by rapamycin and that RNAi directed against TBCK increases STAT3 phosphorylation and decreases ERK1/2 phosphorylation.11, 12 In addition, depleting TBCK at the cellular level leads to decreased cell proliferation, smaller cells, and aberrant actin organization.5 Given that we have demonstrated the absence of TBCK in multiple affected individuals’ cells, it is likely that disruption of these processes causes their phenotypes. We were able to show that cells from these individuals demonstrated reduced phosphorylation of the Ser235/236 isoform of PS6, suggesting that reduced mTOR signaling is a pathogenic mechanism. This would add TBCK-related ID syndrome to the list of other neurodevelopmental disorders that have been associated with aberrant mTOR signaling: neurofibromatosis type 1 (MIM: 162200), tuberous sclerosis, PTEN-related disorders, and fragile X syndrome (MIM: 300624).6, 13
There are few targeted therapies for neurodevelopmental disorders, but by elucidating the molecular and cellular pathology in these individuals, we were able to hypothesize a targeted therapeutic for restoring mTOR signaling. When leucine was added to the cells of the affected individual, it was able to increase the mTOR signaling by using increased phosphorylation of PS6 Ser235/236 as a proxy. This shows that there is a TBCK-independent pathway for leucine to stimulate the mTOR pathway, which could be utilized for therapeutic effect in these individuals. Leucine supplementation is already clinically available for certain metabolic disorders and has a wide therapeutic range (as reviewed in Pedroso et al.14).
In conclusion, we have reported a series of 13 individuals who are from nine unrelated families, harbor biallelic mutations in TBCK, and display overlapping features of ID and hypotonia. We propose that this condition be named TBCK-related ID syndrome. A potential avenue for therapeutic intervention was demonstrated by TBCK-independent upregulation of mTOR signaling with the addition of leucine to the culture media in affected cells. Future work will include initiating a registry for individuals with TBCK-related ID syndrome to further delineate the natural history of the syndrome and test targeted therapies for the condition.
Conflicts of Interest
P.C. is a shareholder and a member of the scientific advisory board of Evogen. J.J., M.G., and G.D. are employees of GeneDx.
Acknowledgments
We thank the families in this study for their enthusiastic participation. The contributions of Mais Hashem, Niema Ibrahim, and Fowzan S. Alkuraya of the King Faisal Specialist Hospital and Research Center included providing cells from family 1 and were invaluable to the development of this project. We would also like to acknowledge the very helpful assistance of GeneMatcher and Megan Cho of GeneDx in the identification of additional affected individuals. It was supported in part by grants from the National Research Agency (ANR-10-IAHU-01) and the Fondation pour la Recherche Médicale (DEQ20120323702) to L.C. This work was supported in part by the Institute for Translational Medicine and Therapeutics of the University of Pennsylvania and by the NIH National Center for Advancing Translational Sciences under award number UL1TR000003. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Published: March 31, 2016
Footnotes
Supplemental Data include one table and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.03.016.
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes, http://browser.1000genomes.org/index.html
dbSNP, http://www.ncbi.nih.gov/SNP
GeneMatcher, http://genematcher.org
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://OMIM.org
Supplemental Data
References
- 1.Harris J.C. Oxford University Press; 2005. Intellectual Disability: Understanding Its Development, Causes, Classification, Evaluation, and Treatment; p. 448. [Google Scholar]
- 2.van Karnebeek C.D., Stockler S. Treatable inborn errors of metabolism causing intellectual disability: a systematic literature review. Mol. Genet. Metab. 2012;105:368–381. doi: 10.1016/j.ymgme.2011.11.191. [DOI] [PubMed] [Google Scholar]
- 3.Alazami A.M., Patel N., Shamseldin H.E., Anazi S., Al-Dosari M.S., Alzahrani F., Hijazi H., Alshammari M., Aldahmesh M.A., Salih M.A. Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families. Cell Rep. 2015;10:148–161. doi: 10.1016/j.celrep.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 4.Saudi Mendeliome G., Saudi Mendeliome Group Comprehensive gene panels provide advantages over clinical exome sequencing for Mendelian diseases. Genome Biol. 2015;16:134. doi: 10.1186/s13059-015-0693-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y., Yan X., Zhou T. TBCK influences cell proliferation, cell size and mTOR signaling pathway. PLoS ONE. 2013;8:e71349. doi: 10.1371/journal.pone.0071349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crino P.B. mTOR: A pathogenic signaling pathway in developmental brain malformations. Trends Mol. Med. 2011;17:734–742. doi: 10.1016/j.molmed.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 7.Takei N., Nawa H. mTOR signaling and its roles in normal and abnormal brain development. Front. Mol. Neurosci. 2014;7:28. doi: 10.3389/fnmol.2014.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynch C.J. Role of leucine in the regulation of mTOR by amino acids: revelations from structure-activity studies. J. Nutr. 2001;131:861S–865S. doi: 10.1093/jn/131.3.861S. [DOI] [PubMed] [Google Scholar]
- 9.Combaret L., Dardevet D., Rieu I., Pouch M.N., Béchet D., Taillandier D., Grizard J., Attaix D. A leucine-supplemented diet restores the defective postprandial inhibition of proteasome-dependent proteolysis in aged rat skeletal muscle. J. Physiol. 2005;569:489–499. doi: 10.1113/jphysiol.2005.098004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalogeropoulou D., Lafave L., Schweim K., Gannon M.C., Nuttall F.Q. Leucine, when ingested with glucose, synergistically stimulates insulin secretion and lowers blood glucose. Metabolism. 2008;57:1747–1752. doi: 10.1016/j.metabol.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Chen X.G., Liu F., Song X.F., Wang Z.H., Dong Z.Q., Hu Z.Q., Lan R.Z., Guan W., Zhou T.G., Xu X.M. Rapamycin regulates Akt and ERK phosphorylation through mTORC1 and mTORC2 signaling pathways. Mol. Carcinog. 2010;49:603–610. doi: 10.1002/mc.20628. [DOI] [PubMed] [Google Scholar]
- 12.Komurov K., Padron D., Cheng T., Roth M., Rosenblatt K.P., White M.A. Comprehensive mapping of the human kinome to epidermal growth factor receptor signaling. J. Biol. Chem. 2010;285:21134–21142. doi: 10.1074/jbc.M110.137828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inoki K., Corradetti M.N., Guan K.L. Dysregulation of the TSC-mTOR pathway in human disease. Nat. Genet. 2005;37:19–24. doi: 10.1038/ng1494. [DOI] [PubMed] [Google Scholar]
- 14.Pedroso J.A., Zampieri T.T., Donato J., Jr. Reviewing the Effects of L-Leucine Supplementation in the Regulation of Food Intake, Energy Balance, and Glucose Homeostasis. Nutrients. 2015;7:3914–3937. doi: 10.3390/nu7053914. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.