Abstract
Linkage and association of Tourette Syndrome (TS) and Attention-Deficit/Hyperactivity Disorder (ADHD) have previously been reported in the 11q24 chromosomal region. To identify the risk gene within the region we studied the potassium inwardly-rectifying channel J5 (KCNJ5) gene in a sample of 170 nuclear families with TS. We genotyped eight markers across the gene and observed biased transmission of haplotypes from parents to probands in this sample. We then tested these markers in an independent sample of 242 nuclear families with ADHD and found the same haplotype was significantly over transmitted to ADHD probands. Screening of the coding region of KCNJ5 in 48 probands with TS did not identify any variation that could explain the association of the haplotype. We also genotyped two microsatellite markers, one in the promoter and the other in the 3′ region and found no evidence for association for either marker for TS, however, we found significant evidence for association with the 3′ repeat and ADHD. A small gene (c11orf45) of unknown function lies within the first intron of KCNJ5 that is transcribed in the opposite orientation and this gene may regulate the expression of KCNJ5. We studied the correlation of the expression of KCNJ5 and the antisense transcript in brain tissues from control individuals and found that the antisense transcript and the short KCNJ5 isoform are co-expressed in three brain regions. The results of this study indicate that KCNJ5 is associated with TS and ADHD in our samples, however, the functional variant(s) remain to be identified.
Keywords: ADHD, antisense transcription, association, chromosome 11, genetics, KCNJ5, Tourette Syndrome
Tourette Syndrome (TS) is a neuropsychiatric disorder characterized by motor and vocal tics with onset in childhood (American Psychiatric Association 2013). Family studies have demonstrated that genetic factors play an important role in the manifestation of TS and segregation analysis have led to the consensus that TS is a genetically complex multigenic disorder involving a high degree of locus and allelic heterogeneity (Barr 2005; Pauls 2003; State 2010). Linkage and association studies of TS have suggested a risk locus in the 11q24 chromosomal region (Barr 2005). We note that this region was originally identified as 11q23, however, the current genome annotation (hg19) indicates the location of these markers as 11q24. Linkage to the chromosome 11q24 region was reported in a single large multigenerational pedigree (127 members) from the French Canadian population (Merette et al. 2000). That study focused on the 24 markers that were previously identified by Simonic and colleagues as being significantly associated in the South African Afrikaner population (Simonic et al. 1998, 2001). The most significant result in the French Canadian family was in the 11q24 region, found using multipoint analysis (LOD score of 3.24, or 3.18 after correction for multiple testing) across the markers D11S1377 (Mfd316) and D11S933 (11q24.1–24.2). Interestingly, one of the markers in the linked region (D11S933) is located 7 cM from the marker D11S912 (11q24.3) that resulted in a LOD score greater than 1 in the first Tourette Syndrome Association (TSA) linkage genome scan (The Tourette Syndrome Association International Consortium for Genetics 1999). This region has therefore been implicated by studies using independent TS samples using association (case–control and family based controls) and linkage. Also of interest is suggestive evidence for linkage from two, independent genome scans for ADHD in this region overlapping in 11q24 (Arcos-Burgos et al. 2004; Ogdie et al. 2003).
A strong candidate gene on chromosome 11q24 is the gene for the potassium inwardly-rectifying channel J5 (KCNJ5) selected based on its function. Potassium channels are membrane-spanning proteins that selectively conduct K+ ions across the cell membrane. These channels play an important role in cellular signaling processes and are critical to neurotransmission. The inward rectifier K+ channels (Kirs) belong to a superfamily of channels with four subunits each containing two transmembrane segments with a pore loop in between (Ho et al. 1993; Kubo et al. 1993; Luscher & Slesinger 2010). These channels conduct K+ currents more in the inward direction than outward, and they are important in setting the resting membrane potential.
In this study we examined the association of the KCNJ5 gene with TS based on the location and biology of the gene. We tested for association in a sample of 170 nuclear families (228 affected children) with TS and identified trends for association with a haplotype of markers selected to tag the major haplotypes. On the basis of this association finding, and because of the previous linkage findings for ADHD to this region (Arcos-Burgos et al. 2004; Ogdie et al. 2003), we also examined a subset of the markers for association to ADHD in an independent sample of 242 nuclear families with 277 children diagnosed with ADHD. In the ADHD sample, we observed association for ADHD with the same haplotype showing trends for biased transmission for TS in the TS sample. On the basis of this association findings, we then sought to identify the functional DNA changes contributing to risk. We screened the coding regions of the gene for non-synonymous coding region changes. We then genotyped two microsatellites markers, either of which could influence gene expression. One of these is located in the promoter and the other located in the 3′ region. Considering the data showing regulatory roles of natural antisense transcripts (NATs) in the gene expression of the corresponding sense transcript, we also screened the antisense transcript (c11orf45) that is located within the first intron of the KCNJ5 gene. We then studied the correlation between expression of both transcripts in three different regions of the human brain to determine if the two transcripts were co-expressed. We found they were co-expressed in the three brain regions examined.
Materials and methods
Subjects
Tourette Syndrome sample
The sample consisted of 170 nuclear families from Ontario, Canada with one or more affected siblings for a total of 228 affected children. All families were recruited from The Tourette Syndrome Clinic at The Toronto Western Hospital. This study was approved by the research ethics of the University Health Network. Written informed parental consent and verbal assent for younger children or written patient consent was obtained for all participants.
The diagnostic assessment of subjects for this study has been previously described (The Tourette Syndrome Association International Consortium for Genetics 1999). Briefly, information about symptoms associated with TS and obsessive–compulsive disorder was collected using a self- and family-report based on the tic inventory and ordinal severity scales of the Yale Global Tic Severity Scale (Leckman et al. 1989) and the symptom checklist and ordinal scales of the Yale-Brown Obsessive–Compulsive scale (Goodman et al. 1989). The information was checked by an experienced neuropsychiatrist and complemented by the direct examination of subjects using the same scales.
The majority of the sample (92%) described their ethnicity by self report as European Caucasian, 3% as non-European and 5% as mixed European and Non-European.
ADHD sample
The sample consisted of 242 nuclear families from Toronto, Canada recruited following referral to the Child Development or Neuropsychiatry Clinics at the Hospital for Sick Children in Toronto. It included 242 probands and 35 affected siblings.
The diagnostic assessment has been described in previous studies of this sample (Couto et al. 2009). Briefly, probands and their siblings between 7 and 16 years old were included if they met DSM-IV criteria for one of the three ADHD subtypes (predominantly inattentive, predominantly hyperactive/impulsive or combined) based on semi-structured parent (Ickowicz et al. 2006) and teacher (Tannock et al. 2002) interviews. Children with TS or chronic tics were excluded from this sample.
The majority of the sample (90%) describe their ethnicity as European ancestry, with the remaining 10% describing their ethnicity as either African, Chinese, Indian, native Canadian or of mixed descent.
This protocol was approved by the Hospital for Sick Children’s Research Ethics Board and informed written consent or verbal assent was obtained for all participants.
Genotyping
DNA was extracted from blood using a high salt method (Miller et al. 1988). The single nucleotide polymorphism (SNP) assays were manufactured by Applied Biosystems Inc., Life Technologies (Foster City, CA, USA) as either Assays-On-Demand (predesigned) or as Assays-by-Design (made to order). The 10 μl polymerase chain reaction (PCR) reactions contained 30 ng of genomic DNA, 10 μmol of TaqMan® Universal PCR Master Mix (Applied Biosystems Inc., Life Technologies) and 0.25 μl of the allelic discrimination mix which is a premade mix containing the specific primers (18 μm) and probes (4 μm; Applied Biosystems Inc., Life Technologies). The thermal cycling conditions were 95° C for 10 min followed by 40 cycles of 94°C for 15 seconds and an annealing temperature of 59°C for 1 min. Included on each 96-well plate were two negative controls. The end-point data, for each plate, were collected using the ABI PRISM 7900HT Sequence Detection System (SDS; Applied Biosystems Inc. Life Technologies) with the allelic discrimination analysis mode of the SDS software package version 2.0 (Applied Biosystems Inc., Life Technologies).
A total of six SNPs were genotyped across the KCNJ5 gene in the TS sample. The initial panel of SNPs was selected to tag the major haplotypes using Tagger Pairwise Method (de Bakker et al. 2005) as implemented on the International HapMap Project Browser (www.hapmap.org). With this selection of markers we captured 70% of all markers in the region at r2 ≥ 0.80 and minor allele frequency (MAF) ≥0.10 in the Centre d’Etude du Polymorphisme Humain (CEU) population. Two additional SNPs in the intergenic region shared with P53S1P1 were also genotyped in the TS sample.
Five of the tag SNPs were genotyped in the ADHD sample after finding trends for biased transmission of haplotypes in the TS sample. After finding association in the ADHD sample, we screened the gene for DNA variants (below) and genotyped two additional SNPs and two repeat polymorphisms. The non-synonymous SNP, rs7102584, was genotyped after we detected the least frequent allele present in three of the 48 probands that were resequenced for the coding region of the gene. An additional marker (rs11221507) in the 5′ untranslated region (UTR) of the antisense transcript was genotyped in the TS sample after we found that the frequency of the C allele in the screened probands was twice that of the reported frequency for this marker in the Caucasian population (0.133).
Two repeat polymorphisms, a (CA)n(GA)n (D11S4150) repeat in the promoter region and a (GTTTT)n repeat motif in the 3′ UTR of the gene, were tested after association was identified. These variants were genotyped using the ABI PRISM® 3100-Avant Genetic Analyzer (Applied Biosystems Inc., Life Technologies).
Resequencing
The promoter region, the three exons of the KCNJ5 gene and the entire antisense transcript (c11orf45) were resequenced in 48 TS probands. Primers were designed using Primer Express Software, version 3.0 (PE Applied Biosystems, Life Technologies, Carlsbad CA) for a total of 22 fragments, 15 of them covering the antisense transcript (7091 bp). For the PCR amplification we used 60 ng of genomic DNA, 1.5 mM MgSO4, 1× PCR Enhancer solution (Invitrogen Corporation, Life Technologies, Carlsbad, CA, USA) and the PCR buffer provided. The PCR reactions were carried out with an initial 5 min denaturing step at 95°C followed by 35 cycles of 95°C for 1 min, annealing temperatures between 59 and 70°C for 1 min according to the pair of primers for each fragment, 72°C for 1 min, and a final extension phase of 72°C for 10 min. Amplification products were sequenced using the Big Dye Terminator v3.0 Cycle Sequencing System (Applied Biosystems Inc., Life Technologies). Sequences were analyzed using the ContigExpress program provided in the Vector NTI Advance 9.0 software package (Invitrogen Corporation, Life Technologies).
Expression analysis
Postmortem brain tissues from 40 control individuals (without documented psychiatric disorder) were used for the expression analysis of KCNJ5 and the antisense transcript (c11orf45). Brain tissues were obtained from the NICHD Brain Bank for Developmental Disorders. Ethics approval was obtained from the University Health Network and The Hospital for Sick Children for use of the tissues. RNA was extracted from three different regions of the brain, dorsolateral pre-frontal cortex (DLPFC), caudate nucleus and hippocampus. Complementary DNA (cDNA) was synthesized using iScript Reverse Transcription Supermix (Bio-Rad Laboratories Inc., Hercules, CA, USA) for Quantitative reverse transcription PCR (RT-qPCR) from 1 μg of total RNA using random primers. We determined the transcript copy number for KCNJ5 and c11orf45 by absolute quantification with real time quantitative PCR using the SYBR Green PCR Master Mix and primers designed with Primer Express Software. One endogenous control, RPLPO, was used for normalization. All reactions were performed in two replicates on the ABI PRISM 7900HT Sequence Detection System. To compare the expression of the two different KCNJ5 isoforms (Fig. 1) relative to each other and to the expression of the antisense transcript, we also designed primers for exon 1 of KCNJ5. In total, four different fragments in three different regions of the KCNJ5 locus (KCNJ5 3′ region, KCNJ5 exon 1, antisense transcript 3′ region) and the RPLPO 3′ region were amplified for each sample.
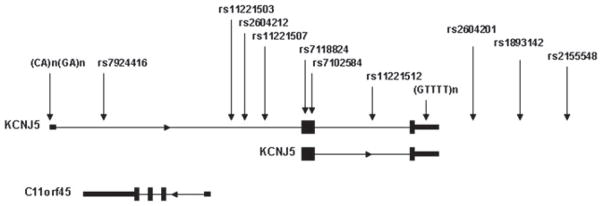
Figure 1.
Ideogram of KCNJ5 and c11orf45 showing the relative positions of the markers genotyped.
Statistical analysis
Genotyping errors were checked by first identifying Mendelian errors using the PedStats program (http://www.sph.umich.edu/csg/abecasis/PedStats/). Further data checking was performed using the error option of Merlin (http://www.sph.umich.edu/csg/abecasis/Merlin/) (Abecasis et al. 2002) to identify potential double recombinants as a sensitive check for genotyping errors. All Mendelian errors and double recombinants were either resolved or removed from the analyses. The TDTphase program from the UNPHASED, version −2.404 package (Dudbridge 2003) was used to test for the biased transmission of alleles for single markers and the TRANSMIT program, version 2.5.4 (Clayton 1999) for the transmission of haplotypes. The robust estimate of the variance option was used which is robust to the inclusion of affected siblings and to prior linkage. Haplotypes with frequencies less than 0.10 were pooled for the analyses. The degree of linkage disequilibrium (LD) between marker alleles in this study was evaluated using Haploview v3.2 (http://www.broad.mit.edu/mpg/haploview) (Barrett et al. 2005).
Correction for multiple testing was performed using Single Nucleotide Polymorphism Spectral Decomposition (SNPSpD) (http://gump.qimr.edu.au/general/daleN/SNPSpD/). This calculates the number of independent SNPs using the LD information (Nyholt 2004). The results showed that the effective number of independent marker loci for the single SNPs analysis in the ADHD sample is 5.4 and the experiment-wide significance threshold required to keep Type I error rate at 5% is 0.009.
For the correlation of gene expression between the log-transformed expression levels for the sense and the antisense transcripts we used SAS version 9.3.
Results
To test for association of the KCNJ5 gene in TS, we genotyped six SNPs covering this gene in a sample of 170 nuclear families with 228 affected children. Transmission disequilibrium test (TDT) analysis showed no significant association for any of the single SNPs tested in this sample (Table 1). Haplotype analysis for three markers in a region of high LD in this gene identified biased transmission from parents to probands for one haplotype [rs7118824 (G), rs11221512 (G), rs2604201 (G); P-value = 0.022, global P value for haplotypes > 0.10 = 0.028] (Table 2). This result would not meet the correction for the number of tests performed. However, based on this trend and the previous evidence for linkage of ADHD to this region, we then tested the relationship of this gene to ADHD. We genotyped five of the six markers covering the KCNJ5 gene in an independent sample of nuclear families with ADHD probands and affected siblings (277 affected children) and tested for association to ADHD. The results from the single marker TDT analysis in this sample showed association of the marker rs2604201 (Table 3) in the 3′ region of the gene (P = 0.019) that would not meet the calculated experiment-wide significance threshold required to keep Type I error rate at 5% (0.009). However, haplotype analysis revealed biased transmission of the same haplotype observed with TS for ADHD (P = 0.0006, global P value for haplotypes > 0.10 = 0.0016, Table 4).
Table 1.
Transmission disequilibrium test analyses of TS in the TS sample
| Gene | Location* | Marker | Allele | Allele frequency | Transmissions† | Non-transmissions | χ2 | P value |
|---|---|---|---|---|---|---|---|---|
| KCNJ5 | Intron 1 | rs7924416‡ | C | 0.755 | 69 | 76 | 0.338 | 0.561 |
| T | 0.245 | 76 | 69 | |||||
| Intron 1 | rs11221503‡ | C | 0.802 | 60 | 52 | 0.571 | 0.449 | |
| T | 0.198 | 52 | 60 | |||||
| Intron 1 | rs2604212‡ | C | 0.570 | 94 | 90 | 0.087 | 0.768 | |
| G | 0.430 | 90 | 94 | |||||
| Intron 1 | rs11221507 | T | 0.888 | 43 | 34 | 1.052 | 0.305 | |
| C | 0.112 | 34 | 43 | |||||
| Exon 2 (syn) | rs7118824‡ | G | 0.834 | 60 | 46 | 1.854 | 0.173 | |
| T | 0.166 | 46 | 60 | |||||
| Exon 2 (nonsyn) | rs7102584 | G | 0.012 | 3 | 5 | 0.505 | 0.477 | |
| C | 0.988 | 5 | 3 | |||||
| Intron 2 | rs11221512‡ | A | 0.115 | 36 | 36 | 0.000 | 1.000 | |
| G | 0.885 | 36 | 36 | |||||
| 3′ | rs2604201‡ | A | 0.279 | 69 | 81 | 0.961 | 0.327 | |
| G | 0.721 | 81 | 69 | |||||
| Intergenic | rs1893142 | G | 0.307 | 72 | 85 | 1.078 | 0.299 | |
| A | 0.693 | 85 | 72 | |||||
| Intergenic | rs2155548 | A | 0.770 | 82 | 73 | 0.523 | 0.469 | |
| C | 0.230 | 73 | 82 |
Based on NCBI Build 36.1, the March 2006 UCSC human reference sequence.
Number of times the alleles were transmitted from heterozygous parents to affected offspring in this sample.
Single nucleotide polymorphism selected using Tagger Pairwise Method (de Bakker et al. 2005).
Table 2.
Haplotype analysis of TS in the TS sample
| Gene | rs7118824 | rs11221512 | rs2604201 | Freq | Obs | Exp | Var (O-E) | χ2 (1 df) | P value |
|---|---|---|---|---|---|---|---|---|---|
| KCNJ5 | G | A | A | 0.102 | 41 | 42.7 | 16.7 | 0.165 | 0.685 |
| G | G | A | 0.005 | 0.001 | 3.081 | 3.082 | 3.078 | 0.079 | |
| T | G | A | 0.163 | 67 | 74.1 | 26.5 | 1.893 | 0.169 | |
| G | A | G | 0.004 | 1 | 1.531 | 0.763 | 0.370 | 0.543 | |
| G | G | G | 0.722 | 328 | 313.6 | 39.4 | 5.263 | 0.022 | |
| T | G | G | 0.005 | 1 | 3.043 | 2.533 | 1.648 | 0.199 |
χ2 on 3 df (common haplotypes, freq > 0.1) = 9.0705, P = 0.028.
Table 3.
Transmission disequilibrium test analyses of ADHD in the ADHD sample
| Gene | Location* | Marker | Allele | Allele frequency | Transmissions† | Non-transmissions | χ2 | P value |
|---|---|---|---|---|---|---|---|---|
| KCNJ5 | Intron 1 | rs11221503 | C | 0.813 | 60 | 69 | 0.628 | 0.428 |
| T | 0.187 | 69 | 60 | |||||
| Intron 1 | rs2604212 | C | 0.546 | 96 | 100 | 0.082 | 0.775 | |
| G | 0.454 | 100 | 96 | |||||
| Exon 2 (syn) | rs7118824 | G | 0.812 | 72 | 51 | 3.603 | 0.058 | |
| T | 0.188 | 51 | 72 | |||||
| Exon 2 (nonsyn) | rs7102584 | G | 0.018 | 5 | 9 | 1.159 | 0.282 | |
| C | 0.982 | 9 | 5 | |||||
| Intron 2 | rs11221512 | A | 0.095 | 24 | 38 | 3.189 | 0.074 | |
| G | 0.905 | 38 | 24 | |||||
| 3′ | rs2604201 | A | 0.275 | 67 | 97 | 5.519 | 0.019 | |
| G | 0.725 | 97 | 67 |
Based on NCBI Build 36.1, the March 2006 UCSC human reference sequence.
Number of times the alleles were transmitted from heterozygous parents to affected offspring in this sample.
P-values shown are not corrected for multiple testing. Experiment-wide significance threshold required to keep Type I error rate at 5%: 0.009.
Table 4.
Haplotype results for ADHD in the ADHD sample
| Gene | rs7118824 | rs11221512 | rs2604201 | Freq | Obs | Exp | Var (O-E) | χ2 (1 df) | P value |
|---|---|---|---|---|---|---|---|---|---|
| KCNJ5 | G | A | A | 0.077 | 32.5 | 40.32 | 16.5 | 3.703 | 0.054 |
| G | G | A | 0.011 | 6.004 | 6.784 | 3.297 | 0.185 | 0.667 | |
| T | G | A | 0.170 | 77.49 | 90.54 | 41.09 | 4.14 | 0.042 | |
| G | A | G | 0.005 | 1 | 2.14 | 1.075 | 1.208 | 0.272 | |
| G | G | G | 0.727 | 409 | 383.4 | 55.13 | 11.817 | 0.0006 | |
| T | G | G | 0.009 | 2.002 | 4.746 | 2.878 | 2.616 | 0.106 |
χ2 on 2 df (common haplotypes, freq > 0.1) = 12.906, P = 0.0016; χ2 on 3 df (common haplotypes, freq > 0.05) = 12.999, P = 0.0046.
On the basis of these positive association findings, we screened the coding regions of the gene for DNA changes in 48 probands from families with TS. The sequenced regions included the putative promoter region, the three coding exons and the 3′UTR. Screening of these regions identified a single non-synonymous DNA change (Gln282Glu, rs7102584) in exon 2. This change was present in 3 of the 48 screened probands (allele frequency = 0.03). The frequency reported in public databases for this marker is 0.017 for the CEU sample. The Gln to Glu change was predicted as benign by PolyPhen, however, the change may impact other aspects of gene function. Because of the higher frequency in the screened probands, we genotyped this marker in both samples, however, the frequency of the minor allele is uncommon in the parental chromosomes and there were few informative transmissions in either sample (Tables 1 and 3).
Two microsatellites have been identified in the KCNJ5 gene, both of which could influence gene function. The first one is located in the 5′ region covering part of the promoter region and first untranslated exon (D11S4150). Microsatellites have been implicated in the regulation of transcription (Gebhardt et al. 1999; Hamada et al. 1984; Naylor & Clark 1990; Rothenburg et al. 2001; Vinces et al. 2009). The second microsatellite is located in the 3′ UTR and could influence transcript stability. No significant association of the alleles of either microsatellite marker was observed in the TS sample (Tables 5 and 6). No association of the alleles for the marker D11S4150 was detected in the ADHD sample, however, the (GTTTT)n polymorphism was significantly associated (P = 0.004), with the two most common alleles in the (GTTTT)n repeat polymorphism significantly biased in transmission (Tables 7 and 8).
Table 5.
Transmission disequilibrium test analysis of the (GTTTT)n repeat polymorphism in the KCNJ5 gene TS sample
| Allele | MAF | Transmitted | Non-transmitted | χ2 | P value |
|---|---|---|---|---|---|
| 1 | 0.692 | 68 | 62 | 0.277 | 0.599 |
| 2 | 0.079 | 16 | 25 | 1.976 | 0.160 |
| 3 | 0.219 | 49 | 47 | 0.042 | 0.838 |
| 4 | 0.004 | 2 | 1 | ||
| 5 | 0.005 |
χ2 on 3 df = 2.924, P = 0.4035.
Table 6.
Transmission disequilibrium test analysis of the D11S4150 repeat polymorphism in the KCNJ5 gene TS sample
| Allele (bp) | MAF | Transmitted | Non-transmitted | χ2 | P value |
|---|---|---|---|---|---|
| 232 | 0.035 | 8 | 5 | 0.692 | 0.405 |
| 234 | 0.028 | 25 | 27 | 0.077 | 0.782 |
| 236 | 0.137 | 25 | 22 | 0.191 | 0.662 |
| 240 | 0.035 | 31 | 22 | 1.528 | 0.216 |
| 242 | 0.165 | 9 | 18 | 3.000 | 0.083 |
| 244 | 0.080 | 9 | 8 | 0.059 | 0.808 |
| 246 | 0.047 | 11 | 19 | 2.133 | 0.144 |
| 248 | 0.080 | 22 | 23 | 0.022 | 0.882 |
| 250 | 0.097 | 16 | 14 | 0.133 | 0.715 |
| 254 | 0.026 | 7 | 5 | 0.333 | 0.564 |
χ2 on 17 df = 18.118, P = 0.382.
Table 7.
Transmission disequilibrium test analysis of the (GTTTT)n repeat polymorphism in the KCNJ5 gene ADHD sample
| Allele | MAF | Transmitted | Non-transmitted | χ2 | P value |
|---|---|---|---|---|---|
| 1 | 0.696 | 86 | 51 | 8.942 | 0.003 |
| 2 | 0.084 | 16 | 28 | 3.273 | 0.071 |
| 3 | 0.213 | 45 | 70 | 5.435 | 0.019 |
| 4 | 0.004 | 2 | 0 |
χ2 on 3 df = 13.2871, P = 0.0040.
Table 8.
Transmission disequilibrium test analysis of the D11S4150 repeat polymorphism in the KCNJ5 gene ADHD sample
| Allele (bp) | MAF | Transmitted | Non-transmitted | χ2 | P value |
|---|---|---|---|---|---|
| 234 | 0.035 | 19 | 12 | 1.581 | 0.209 |
| 236 | 0.167 | 13 | 10 | 0.391 | 0.532 |
| 238 | 0.092 | 5 | 8 | 0.692 | 0.405 |
| 240 | 0.052 | 15 | 18 | 0.273 | 0.602 |
| 242 | 0.170 | 10 | 9 | 0.053 | 0.819 |
| 244 | 0.064 | 5 | 8 | 0.692 | 0.405 |
| 246 | 0.050 | 7 | 7 | 0.000 | 1.000 |
| 248 | 0.061 | 11 | 15 | 0.615 | 0.433 |
| 250 | 0.094 | 12 | 9 | 0.429 | 0.513 |
| 252 | 0.068 | 3 | 7 | 1.600 | 0.206 |
χ2 on 18 df = 22.924, P = 0.1941.
We screened the entire sequence of the antisense transcript (c11or45) present in the first intron in 48 probands with TS. Of the 33 variants identified, we genotyped one, rs11221507, because the frequency in the screened probands was higher than that reported in public databases for the Caucasian population. However, no significant association of this marker with TS was detected in our sample (Table 1).
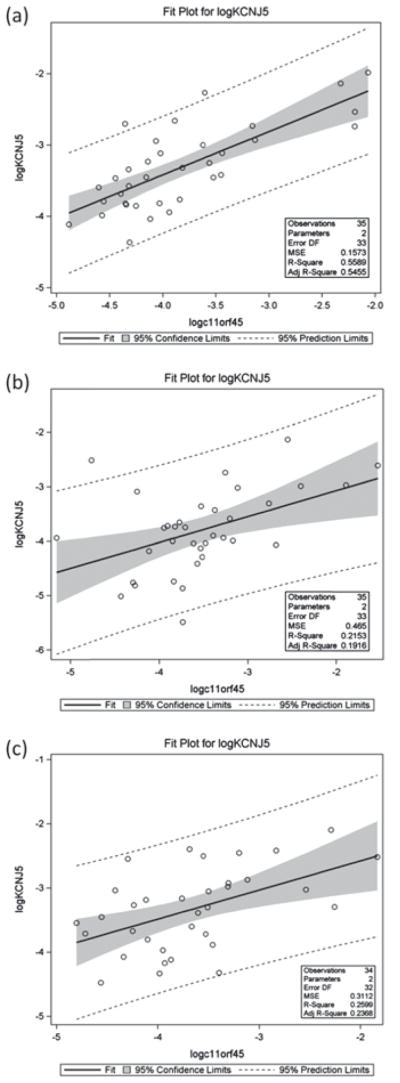
We then investigated the possibility of co-regulation of KCNJ5 and the antisense transcript c11orf45 by examining expression in three different regions of the brain (Fig. 2). The KCNJ5 gene has two different isoforms, a short and a long isoform with the exon 2 and the 3′ UTR common to both of them (Fig. 1). Two different fragments were amplified to quantify the transcripts from these isoforms, one in exon 1 for the long isoform and one in the 3′ region. The quantity of amplified cDNA or copy number detected for the first exon (long isoform) was very low in the three different tissues for all the samples. The Ct standard deviation for the replicates was higher than 0.5 in many cases. Because the expression was low with poor reproducibility, we did not include these data in the analysis for gene expression. We then assumed that the amplification that we detected for the fragment in the 3′ region of KCNJ5 (common to both isoforms, Fig. 1) corresponds to the expression of the short isoform. Analysis of KCNJ5 and c11orf45 expression revealed that KCNJ5 and c11orf45 were positively and statistically significant correlated in DLPFC (r = 0.7476, P = .0001), caudate nucleus (r = 0.4641, P = 0.0045) and hippocampus (r = 0.5098, P = 0.0017). The fitted regression lines for each of the correlations, along with 95% confidence intervals are shown in Fig. 2a–c. Using the Brain Explorer application to mine the Allen Brain Atlas (http://human.brain-map.org/static/brainexplorer) (Lein et al. 2007) we found that, as expected, KCNJ5 and c11orf45 are co-expressed in the three brain regions we studied, but the co-expression profile is not uniform throughout the human brain with the genes independently expressed in some regions.
Figure 2. Relationship of KCNJ5 and c11orf45 expression in three brain regions.
(a) KCNJ5 and c11orf45 expression in DLPFC. (b) KCNJ5 and c11orf45 expression in caudate nucleus. (c) KCNJ5 and c11orf45 expression in hippocampus.
In an effort to identify regulatory SNPs that could explain the association, we mined the Braincloud database (Colantuoni et al. 2011) testing for correlation of expression of the KCNJ5 mRNA in human prefrontal cortex with cis SNPs (100 Kb upstream or downstream from the gene) as well as checked for correlation with SNPs genome wide. None of the SNPs covered in the Braincloud data set met the genome-wide statistically significant cut off set by that study (P < 10−12). The probe used for measuring the expression of KCNJ5 is in the 3′ region. From our results, we could speculate that it is measuring the expression of the short transcript, since our data indicate that exon 1 has very low expression in prefrontal cortex. The arrays used for that study did not include probes that cover exon 1 or the antisense transcript thus we cannot determine if there are any SNPs correlated with expression of these transcripts.
Discussion
In this study we tested KCNJ5 as the risk gene for TS and ADHD in the 11q24 linkage region. The notable finding from our study was association of the haplotype in ADHD families (P-value = 0.0006) that showed a trend in the TS families (P-value = 0.022). The genetic relationship of TS and ADHD is unclear (Mathews & Grados 2011; O’Rourke et al. 2011; Stewart et al. 2006), however, ADHD is very common in individuals with TS (~50%) (Robertson 2000) and in families with TS (Stewart et al. 2006). Further, previous studies have identified genes overlapping between the disorders (State 2011). Our finding of biased transmission of the same haplotype in both samples to both phenotypes suggests that this gene could also be a common genetic risk factor to both disorders. However, we cannot rule out that the relationship is due to the presence of comorbid ADHD in the TS children.
Screening the coding region of this gene we found a non-synonymous variant (rs7102584) present in three of 48 probands. The allele frequency of this variant was very low in the parental chromosomes and there were very few informative transmissions, thus could not explain the association. In addition, the amino acid change was predicted to be benign. No additional non-synonymous variants in the coding region were detected in the screening. Likewise, we found no change in the promoter region, other than a previously reported complex microsatellite repeat consisting of (CA)n(GA)n in the putative promoter/first untranslated exon. Because of its localization in the gene, this DNA variant could influence splicing or alternative transcription start site usage, or influence transcription. Microsatellites are found in the 5′ region of genes more often than expected by chance and are thought to be involved in rapid gene evolution and responsiveness to changing environmental conditions (King et al. 1997). One predicted mechanism of gene regulation is the capacity of long stretches of alternating purines and pyrimidines to create Z DNA which influences RNA polymerase activity and chromatin remodeling (Liu et al. 2006). However, we did not observe evidence of association in either the TS or ADHD samples.
Within the 3′ UTR there are 6–10 repeats of a (GTTTT)n motif that could influence RNA stability. We genotyped this microsatellite marker in both samples to determine if this repeat was associated and could perhaps explain the association. This polymorphism was associated in the ADHD sample, however, not in the TS sample, thus does not explain the association in both samples.
An interesting characteristic of the KCNJ5 gene is the presence of c11orf45, a transcript within the first intron that is transcribed in the opposite orientation. This antisense transcript has no ortholog in other species and no known function. The evidence for the existence of this gene is at the transcript level, which suggests that it could be an anti-sense intronic noncoding RNA. Also known as NATs, this is a genome-wide phenomenon present in between 5% and 30% of the protein coding loci in diverse eukaryotes and in 15% of human protein-encoding genes (Lapidot & Pilpel 2006). Previous studies found that sense-antisense pair transcripts tend to be co-expressed and/or inversely expressed more frequently than expected by chance, suggesting shared mechanisms regulating gene expression (Chen et al. 2005; Katayama et al. 2005). Indeed, our studies of the expression profiles of the short isoform of KCNJ5 and c11orf45 in three different brain regions (DLPFC, caudate nucleus and hippocampus) indicated that they are co-expressed in these three tissues. However, in silico mining of the Allen Brain Atlas showed independent expression in other brain regions. Complex patterns of expression that show brain regional differences between genes and their intronic transcripts have been documented for a number of genes (Mercer et al. 2008). For example, the non-coding RNA (ncRNA) transcript located within an intron of Odz3 showed differential spatial expression than Odz3 with the ncRNA strongly expressed throughout the hippocampus, whereas the Odz3 gene is expressed only in the CA1 subfield (Mercer et al. 2008).
A confound when interpreting co-expression in tissues is that expression is measured over all of the cells in the sample and cannot distinguish if transcripts are co-expressed within the same cell in the tissues. Gene expression profiles can differ dramatically among different cells within the same tissue and region and even between adjacent cells (Lein et al. 2007).
We are left with several possible mechanisms that could explain the observed haplotype associations that cannot be explained by the single SNPs or the microsatellite markers genotyped, nor variations in the coding sequence. Variations in the c11orf45 antisense transcript could affect the transcription levels of KCNJ5. However, predictions of function from the sequence changes in the non-coding transcripts are difficult based on sequence alone and functional studies are required. We screened the proximal promoter for DNA variants, however, remote regulatory regions (outside the promoter) could dysregulate the transcription of KCNJ5 or c11orf45. The promoter of the antisense transcript c11orf45 is immediately upstream of the promoter for the short iso-form of KCNJ5. Expression of c11orf45 may influence the switch to the expression of the short isoform by favoring transcription of the short isoform and repressing the long iso-form of KCNJ5. In the three brain regions we examined, we observed expression of c11orf45 and low expression of the first exon of the long isoform, supporting this possibility.
In summary, we identified biased transmission of the same haplotypes for TS and ADHD in independent samples, supporting this gene as a possible susceptibility locus for both disorders. However, we were unable to identify functional risk alleles that could account for the association in both samples. This highlights the difficulties in identifying functional variants in the non-coding genome.
Acknowledgments
Support for this study was provided by grants from the Ontario Mental Health Foundation and the Canadian Institutes of Health Research (MOP-84323). The collection of families for this study was supported by grants from The Tourette Syndrome Association of America, N.I.H. grant MS40024-01, the Ontario Mental Health Foundation, and The Tourette Syndrome Foundation of Canada.
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin – rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association. American Psychiatric Association: Diagnostic and Statistical Manual of Mental Disorders: DSM-5 2013 [Google Scholar]
- Arcos-Burgos M, Castellanos FX, Pineda D, Lopera F, Palacio JD, Palacio LG, Rapoport JL, Berg K, Bailey-Wilson JE, Muenke M. Attention-deficit/hyperactivity disorder in a population isolate: linkage to loci at 4q13.2, 5q33.3, 11q22, and 17p11. Am J Hum Genet. 2004;75:998–1014. doi: 10.1086/426154. Epub 2004 Oct 1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- Barr CL. Progress in gene localization. In: Kurlan R, editor. Handbook of Tourette’s Syndrome and Related Tic and Behavioral Disorders. Marcel Dekker Inc; New York, NY: 2005. pp. 379–398. [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Chen J, Sun M, Hurst LD, Carmichael GG, Rowley JD. Genome-wide analysis of coordinate expression and evolution of human cis-encoded sense-antisense transcripts. Trends Genet. 2005;21:326–329. doi: 10.1016/j.tig.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Clayton D. A generalization of the transmission/disequilibrium test for uncertain-haplotype transmission. Am J Hum Genet. 1999;65:1170–1177. doi: 10.1086/302577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, Colantuoni EA, Elkahloun AG, Herman MM, Weinberger DR, Kleinman JE. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto JM, Gomez L, Wigg K, Ickowicz A, Pathare T, Malone M, Kennedy JL, Schachar R, Barr CL. Association of attention-deficit/hyperactivity disorder with a candidate region for reading disabilities on chromosome 6p. Biol Psychiatry. 2009;66:368–375. doi: 10.1016/j.biopsych.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F. Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol. 2003;25:115–121. doi: 10.1002/gepi.10252. [DOI] [PubMed] [Google Scholar]
- Gebhardt F, Zanker KS, Brandt B. Modulation of epidermal growth factor receptor gene transcription by a polymorphic dinucleotide repeat in intron 1. J Biol Chem. 1999;274:13176–13180. doi: 10.1074/jbc.274.19.13176. [DOI] [PubMed] [Google Scholar]
- Goodman WK, Price LH, Rasmussen SA, Mazure C, Fleischmann RL, Hill CL, Heninger GR, Charney DS. The Yale-brown obsessive compulsive scale. I. Development, use, and reliability. Arch Gen Psychiatry. 1989;46:1006–1011. doi: 10.1001/archpsyc.1989.01810110048007. [DOI] [PubMed] [Google Scholar]
- Hamada H, Seidman M, Howard BH, Gorman CM. Enhanced gene expression by the poly(dT-dG).poly(dC-dA) sequence. Mol Cell Biol. 1984;4:2622–2630. doi: 10.1128/mcb.4.12.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho K, Nichols CG, Lederer WJ, Lytton J, Vassilev PM, Kanazirska MV, Hebert SC. Cloning and expression of an inwardly rectifying ATP-regulated potassium channel. Nature. 1993;362:31–38. doi: 10.1038/362031a0. [DOI] [PubMed] [Google Scholar]
- Ickowicz A, Schachar R, Sugarman R, Chen S, Millette C, Cook L. The Parent Interview for Child Symptoms (PICS): a situation-specific clinical-research interview for attention deficit hyperactivity and related disorders. Can J Psychiatry. 2006;50:325–328. doi: 10.1177/070674370605100508. [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- King DG, Soller M, Kashi Y. Evolutionary tuning knobs. Endeavour. 1997;21:36–40. [Google Scholar]
- Kubo Y, Baldwin TJ, Jan YN, Jan LY. Primary structure and functional expression of a mouse inward rectifier potassium channel. Nature. 1993;362:127–133. doi: 10.1038/362127a0. [DOI] [PubMed] [Google Scholar]
- Lapidot M, Pilpel Y. Genome-wide natural antisense transcription: coupling its regulation to its different regulatory mechanisms. EMBO Rep. 2006;7:1216–1222. doi: 10.1038/sj.embor.7400857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leckman JF, Riddle MA, Hardin MT, Ort SI, Swartz KL, Stevenson J, Cohen DJ. The Yale Global Tic Severity Scale: initial testing of a clinician-rated scale of tic severity. J Am Acad Child Adolesc Psychiatry. 1989;28:566–573. doi: 10.1097/00004583-198907000-00015. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Liu H, Mulholland N, Fu H, Zhao K. Cooperative activity of BRG1 and Z-DNA formation in chromatin remodeling. Mol Cell Biol. 2006;26:2550–2559. doi: 10.1128/MCB.26.7.2550-2559.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscher C, Slesinger PA. Emerging roles for G protein-gated inwardly rectifying potassium (GIRK) channels in health and disease. Nat Rev Neurosci. 2010;11:301–315. doi: 10.1038/nrn2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews CA, Grados MA. Familiality of Tourette syndrome, obsessive-compulsive disorder, and attention-deficit/ hyperactivity disorder: heritability analysis in a large sib-pair sample. J Am Acad Child Adolesc Psychiatry. 2011;50:46–54. doi: 10.1016/j.jaac.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merette C, Brassard A, Potvin A, Bouvier H, Rousseau F, Emond C, Bissonnette L, Roy MA, Maziade M, Ott J, Caron C. Significant linkage for Tourette syndrome in a large French Canadian family. Am J Hum Genet. 2000;67:1008–1013. doi: 10.1086/303093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor LH, Clark EM. d(TG)n.d(CA)n sequences upstream of the rat prolactin gene form Z-DNA and inhibit gene transcription. Nucleic Acids Res. 1990;18:1595–1601. doi: 10.1093/nar/18.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogdie MN, Macphie IL, Minassian SL, Yang M, Fisher SE, Francks C, Cantor RM, McCracken JT, McGough JJ, Nelson SF, Monaco AP, Smalley SL. A genomewide scan for attention-deficit/hyperactivity disorder in an extended sample: suggestive linkage on 17p11. Am J Hum Genet. 2003;72:1268–1279. doi: 10.1086/375139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke JA, Scharf JM, Platko J, Stewart SE, Illmann C, Geller DA, King RA, Leckman JF, Pauls DL. The familial association of Tourette’s disorder and ADHD: the impact of OCD symptoms. Am J Med Genet Part B Neuropsychiatr Genet. 2011;156B:553–560. doi: 10.1002/ajmg.b.31195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauls DL. An update on the genetics of Gilles de la Tourette syndrome. J Psychosom Res. 2003;55:7–12. doi: 10.1016/s0022-3999(02)00586-x. [DOI] [PubMed] [Google Scholar]
- Robertson MM. Tourette syndrome, associated conditions and the complexities of treatment. Brain. 2000;123(Pt 3):425–462. doi: 10.1093/brain/123.3.425. [DOI] [PubMed] [Google Scholar]
- Rothenburg S, Koch-Nolte F, Rich A, Haag F. A polymorphic dinucleotide repeat in the rat nucleolin gene forms Z-DNA and inhibits promoter activity. Proc Natl Acad Sci USA. 2001;98:8985–8990. doi: 10.1073/pnas.121176998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonic I, Gericke GS, Ott J, Weber JL. Identification of genetic markers associated with Gilles de la Tourette syndrome in an Afrikaner population. Am J Hum Genet. 1998;63:839–846. doi: 10.1086/302002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonic I, Nyholt DR, Gericke GS, Gordon D, Matsumoto N, Ledbetter DH, Ott J, Weber JL. Further evidence for linkage of Gilles de la Tourette syndrome (GTS) susceptibility loci on chromosomes 2p11, 8q22 and 11q23-24 in South African Afrikaners. Am J Med Genet. 2001;105:163–167. doi: 10.1002/ajmg.1192. [DOI] [PubMed] [Google Scholar]
- State MW. The genetics of child psychiatric disorders: focus on autism and Tourette syndrome. Neuron. 2010;68:254–269. doi: 10.1016/j.neuron.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- State MW. The genetics of Tourette disorder. Curr Opin Genet Dev. 2011 doi: 10.1016/j.gde.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SE, Illmann C, Geller DA, Leckman JF, King R, Pauls DL. A controlled family study of attention-deficit/hyperactivity disorder and Tourette’s disorder. J Am Acad Child Adolesc Psychiatry. 2006;45:1354–1362. doi: 10.1097/01.chi.0000251211.36868.fe. [DOI] [PubMed] [Google Scholar]
- Tannock R, Hum M, Masellis M, Humphries T, Schachar R. Teacher Telephone Interview for Children’s Academic Performance, Attention, Behavior and Learning: DSM-IV Version (TTI-IV) The Hospital for Sick Children; Toronto, Canada: 2002. Unpublished Document. [Google Scholar]
- The Tourette Syndrome Association International Consortium for Genetics. A complete genome screen in sib pairs affected by Gilles de la Tourette Syndrome. Am J Hum Genet. 1999;65:1428–1436. doi: 10.1086/302613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinces MD, Legendre M, Caldara M, Hagihara M, Verstrepen KJ. Unstable tandem repeats in promoters confer transcriptional evolvability. Science. 2009;324:1213–1216. doi: 10.1126/science.1170097. [DOI] [PMC free article] [PubMed] [Google Scholar]