Abstract
Selenocysteine (Sec or U) is encoded by UGA, a stop codon reassigned by a Sec-specific elongation factor and a distinctive RNA structure. To discover possible code variations in extant organisms we analyzed 6.4 trillion base pairs of metagenomic sequences and 24,903 microbial genomes for tRNASec species. As expected, UGA is the predominant Sec codon in use. We also found tRNASec species that recognize the stop codons UAG and UAA, and ten sense codons. Selenoprotein synthesis programmed by UAG in Geodermatophilus and Blastococcus, and by the Cys codon UGU in Aeromonas salmonicida was confirmed by metabolic labeling with 75Se or mass spectrometry. Other tRNASec species with different anticodons enabled Escherichia coli to synthesize active formate dehydrogenase H, a selenoenzyme. This illustrates the ease by which the genetic code may evolve new coding schemes, possibly aiding organisms to adapt to changing environments. Our results reveal that the genetic code is much more flexible than previously thought.
Keywords: genetic code, selenocysteine, sense codon recoding, metagenome, synthetic biology
Graphical Abstract
The micronutrient selenium is present in proteins in the form of the versatile 21st amino acid, selenocysteine, in which the thiol moiety of cysteine (Cys) is replaced by a selenol group [1]. Selenoproteins are present in organisms from all domains of life [2]; such proteins are essential in mammalian cells [3], yet plants and fungi lack this amino acid. Sec is present in the active site of many redox enzymes [4]. The codon for Sec is UGA which is normally a translational stop signal [5]. During translation of selenoprotein mRNAs, UGA is recoded by the interaction of a specialized elongation factor SelB (in bacteria) with a downstream Sec insertion sequence [5–6]. Recently, a synthetic biology study succeeded in reassigning Sec to a large number of sense and stop codons in Escherichia coli [7] demonstrating that alterations to the genetic code can be tolerated. This prompted the question, whether deviations of the standard UGA Sec assignment may naturally occur.
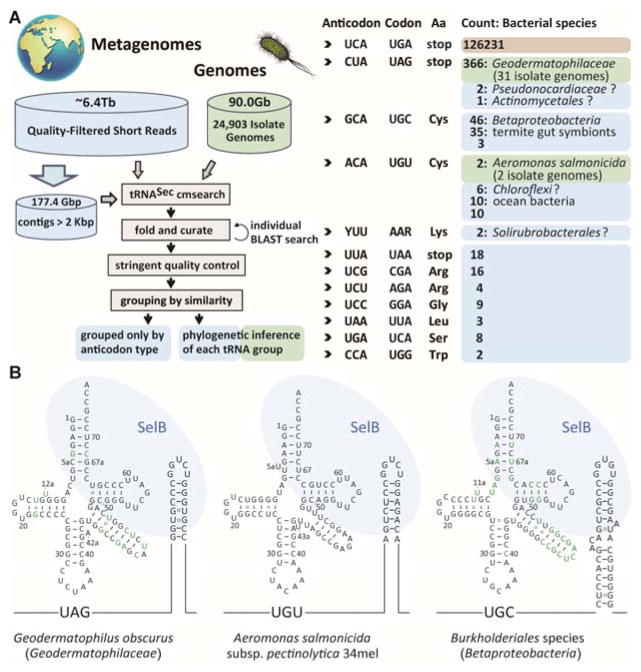
A computational study scanning several trillion base pairs of metagenomic data revealed a large number of stop codon reassignments in bacteria and bacteriophages [8]. This inspired us to perform a comprehensive search of the available metagenomic and microbial genomic sequence data for anticodon variants of the typical tRNASecUCA, the longest tRNA [9] with a tertiary structure quite different from that of canonical tRNAs [10]. A BLAST search of the tRNADB-CE database [11] revealed four tRNASecCUA sequences, suggesting UAG codon recognition. Searching all public microbial genomes in the National Center for Biotechnology Information (NCBI) and all assembled metagenome data in the Integrated Microbial Genomes (IMG) system [12] yielded a tRNASecGCA group, indicating UGC codon recognition. We then developed a general computational pipeline that scanned ~6.4 Tb of unassembled short reads, ~180 Gb of assembled contigs (> 2 kb), and 24,903 microbial genomes in IMG (Figure 1A). The results affirmed UGA as the predominant Sec codon. In addition, 12 different tRNASec anticodon variants capable of recognizing the stop codons UAG and UAA, and 10 sense codons were discovered (Figure 1A). Further sequence validations (see Supplementary Information) ascertained these tRNASec variants not to be sequencing artifacts.
Figure 1. Non-canonical selenocysteine assignments in nature.
(A) The tRNASec search pipeline and the manually curated output. The non-canonical tRNASec sequences are grouped by codon recognition; their numbers are given with their (putative) bacterial origins from phylogenetic inference. Green color indicates results from whole genomes, while blue represents results from only metagenomic data. For comparison the number of canonical tRNASecUCA sequences is shown (most of them were not curated). “Y” and “R” denote “C or U” and “A or G”, respectively. (B) Inferred cloverleaf structures of non-canonical tRNASec species. The nucleotide polymorphism among the same tRNASec group is indicated with green letters. The Sec codons and the SECIS elements of formate dehydrogenase mRNAs are shown.
We grouped these non-canonical tRNASec species by anticodon type, sequence, and structural similarity (Figure 1A, and Supplementary Information). The largest group (anticodon CUA) contains 366 nearly identical tRNASec sequences from the actinobacterial Geodermatophilaceae family [13] (Figure 1A, 1B left panel), while the other 3 tRNASecCUA species may be of rhizosphere bacterial origin (Figures S1, S2). The tRNASecCUA species amounted to 3% of the total tRNASec species found in a soil metagenome (3300001205).
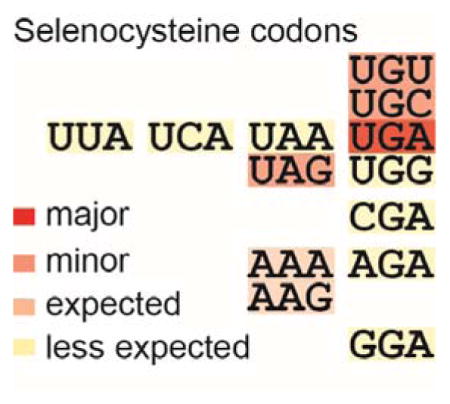
The next group contains tRNASec species (with anticodon GCA able to decode Cys) from Betaproteobacteria and termite gut symbionts (Figure 1A, Figure S3). Two Aeromonas salmonicida genomes contain a tRNASecACA, able to decode the UGU Cys codon (Figure 1B, middle panel). The other 16 members of this group may originate from Chloroflexi (Figure S4) and ocean bacteria (Figure S5). The 2 tRNASecUUU/CUU species (recognizing AAA/AAG Lys codons) may derive from the Solirubrobacterales (Figure S6). Additional tRNASec variants able to recognize the stop codon UAA and 6 sense codons (CGA, AGA, GGA, UUA, UCA, UGG) were also found (Figure S7).
We then wanted to confirm the coding properties of these non-canonical tRNASec species. For proof of selenoprotein synthesis three strategies were possible: (i) metabolic labeling of the organisms with 75Se, (ii) replacing parts of the E. coli selenoprotein synthesis machinery with genes and tRNASec from our genomic or metagenomic findings, and (iii) replacing E. coli tRNASec with the newly discovered tRNASec species. In the last two strategies E. coli formate dehydrogenase H (FDHH, encoded by the fdhF gene) would serve as reporter [7].
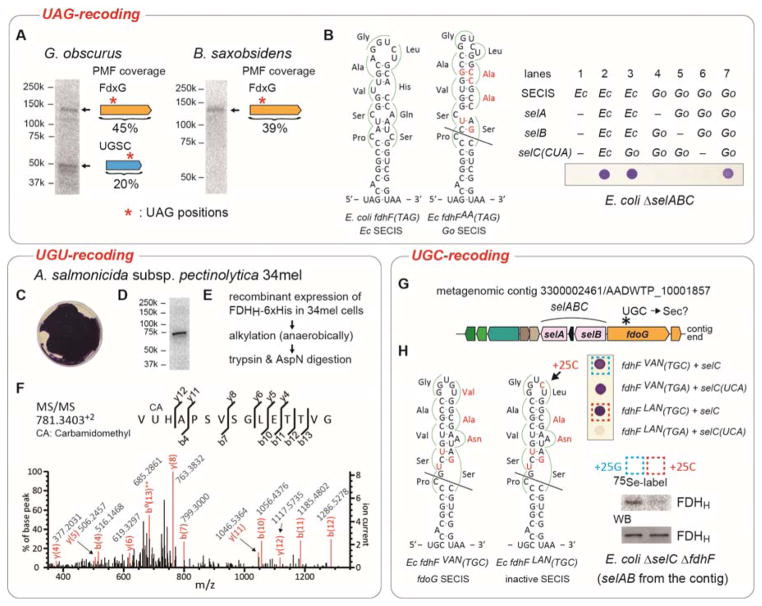
To confirm UAG-directed Sec incorporation we grew Geodermatophilus obscurus G-20 and Blastococcus saxobsidens cells in the presence of [75Se]selenite and detected radiolabeled selenoproteins of 140 and 50 kDa size. The genome sequences predict formate dehydrogenases (FDHs) (Figure S8, Tables S1 & S2) and UGSC-motif proteins [14], (Figures S1, S8, Table S3). This was the first indication for UAG read-through by Sec to form the expected FDH (FdxG) and UGSC-motif protein products (Figure 2A). Crude cell extracts were then resolved by SDS-PAGE; the proteins in the gel slices corresponding to 140 and 50 kDa were trypsinized for subsequent liquid chromatography (LC) coupled with tandem mass spectrometry (LC-MS/MS) analysis. The 140 kDa gel slices from G. obscurus and B. saxobsidens harbored full-length FdxG, while the 50 kDa gel slice from G. obscurus contained a UGSC-motif protein (Figure 2A & Table S4).
Figure 2. Recoding of UAG and cysteine codons to selenocysteine.
(A) Metabolic 75Se labeling of G. obscurus and B. saxobsidens cells. Crude extracts were resolved by SDS-PAGE, and their putative selenoproteins were visualized by PhosphorImager analysis. The results of peptide mass fingerprinting (PMF) analyses of the proteins in the excised gel bands are shown to the right of the bands. (B) FDHH expression in E. coli ΔselABC ΔfdhF cells with the G. obscurus selABC genes and a chimeric fdhF(140TAG) gene variant having a G. obscurus SECIS element with a few nucleotide modifications shown in red. The selC(CUA) genes express tRNASecCUA. The expressed selenoprotein FDHH reduced benzyl viologen, resulting in a purple color. (C) FDHH activity of A. salmonicida subsp. pectinolytica 34mel cells. (D) Metabolic 75Se labeling of the FDHH of 34mel. (E) The procedure of sample preparation for the LC-MS/MS analysis of the FDHH selenoprotein. (F) PMF confirms Sec incorporation at codon 140 in the recombinant FDHH. (G) tRNASecGCA gene (selC) locus in a metagenomic contig. (H) In vivo FDHH assays in E. coli ΔselABC ΔfdhF cells with the selABC genes of the metagenomic contig and a chimeric fdhF(140TGC) gene carrying the contig’s SECIS element with a few nucleotide modifications shown in red. The two transformed strains boxed were metabolically labeled with 75Se, and the radioactive FDHH proteins were analyzed by SDS-PAGE and autoradiography or western blotting (WB).
Like E. coli, the G. obscurus genome [13b] encodes a Sec incorporation machinery consisting of the selA, selB, selC and selD genes. In bacteria, SelD produces the Se donor selenophosphate, SelA converts Ser-tRNASec (the tRNA is the product of the selC gene) to Sec-tRNASec, and SelB carries Sec-tRNASec to the ribosome in a SECIS-dependent manner. To test their functionality, the E. coli ΔselABC ΔfdhF strain ME6 was complemented with the G. obscurus selABC genes. The product of the E. coli fdhF gene is the selenoenzyme FDHH (Table S1) whose activity requires Sec at position 140 [15], if replaced by Cys the activity drops 300-fold [16]. FDHH is readily detected by the reduction of benzyl viologen resulting in a purple color [17]. To serve as a reporter, the plasmid-encoded E. coli fdhF gene transformed into strain ME6 was modified to have a TAG codon in position 140, followed by a G. obscurus-type SECIS element leading to an FDHH variant with two amino acid changes (see Figure 2B). Expression of the G. obscurus selABC genes in this modified strain produced active FDHH (Figure 2B). These data confirm that in this E. coli FDHH variant the UAG140 codon is recoded to Sec.
UGU (Cys) recoding was confirmed in Aeromonas salmonicida subsp. pectinolytica 34mel, the type strain of the γ-proteobacterium A. salmonicida subspecies [18]. Unlike other Aeromonas species the pectinolytica subspecies and strain Y577 [19] pair tRNASecACA with a UGU Cys codon in fdhF. In addition to the in vivo FDH activity in pectinolytica cells (Figure 2C), their anaerobic metabolic labeling with 75Se produced radioactive Sec-containing FDHH (Figure 2D). We confirmed Sec incorporation encoded by UGU140 in FDHH by overexpressing the protein from a plasmid (Figure 2E) and LC-MS/MS analysis (Table S4). The recoded selenopeptide (LC retention time 24.82) had the correct mass (Figure S9) and the appropriate secondary fragmentation pattern (Figure 2F). The co-eluting Cys- and Sec-peptides were detected through their different masses with ion intensities of ~100:1, respectively (Figure S9A). The mass peaks of the Sec-peptides were absent in protein samples obtained from pectinolytica cells expressing a fdhF variant lacking the SECIS element (Table S4). Thus, the Cys140 codon of the fdhF gene is translated as Sec in a SECIS-dependent manner in A. pectinolytica 34mel.
UGC Cys→Sec recoding by tRNASecGCA may be a common trait in the Burkholderiales. In some long metagenomic Burkholderiales contig the selC gene is flanked by selB and selA genes, and the selABC operon is located next to a formate dehydrogenase (fdoG) gene whose active site UGC Cys codon is followed by a putative SECIS element (Figures 1B right panel & 2G). The E. coli ΔselABC strain was complemented with the Burkholderiales contig selAB and selC-opal variant, and an E. coli fdhF variant harboring the Burkholderiales contig SECIS element. This strain produced active FDHH (purple color) (Figure 2H, the 2nd row), but an inactive SECIS element (with a G25C mutation) did not form FDHH and the cells were colorless (Figure 2H, the 4th row). In combination with the Burkholderiales contig selC, we changed UGA140 to UGC for the chimeric fdhF variants that carried functional or inactive SECIS elements. As the FDHH Cys140 enzyme produced a purple color (Figure 2H, the 1st & 3rd rows), 75Se-labeling was used to demonstrate that the functional SECIS element led to a clear signal (Figure 2H). Thus, the Burkholderiales contig selA, selB, tRNASecGCA, and SECIS enabled UGC-recoding in E. coli.
The metagenomic tRNASec variants that recognize other stop and sense codons were also tested for Sec reassignment. We selected one representative metagenomic tRNASec species for each anticodon type and expressed them in an E. coli ΔselC ΔfdhF strain, together with the E. coli fdhF variants that carry the proper cognate codons at position 140 [7]. Surprisingly, all but tRNASecUCC of the tested tRNASec species recoded the respective codons for Sec, as they supported the expression of active FDHH in their host E. coli cells (Figure S10). It should be mentioned that GGA was also poorly recoded in our earlier Sec recoding strategy [7]. The different recoding efficiencies may result from distortions of the ideal SECIS element structure by the nature of the upstream codon [20]. In light of these results we believe that these tRNASec species may be used for recoding sense codons in the organisms they originate from.
What about eukaryotic organisms? Although we found 9 tRNASec variants of algal origin (2 are shown in Figure S7), they need further validation, because they are almost identical to canonical tRNASec species. A similar search of 92 mammalian genomes (215 Gbp) and of the Drosophila melanogaster genome (139 Mbp) showed no exception to the use of UGA as the Sec codon. Whether this is related to the necessity of selenoproteins in high-level redox signaling pathways [21] or due to the sophisticated backup systems [22] remains to be investigated. However, in the lower eukaryote Euplotes crassus UGA serves both as a Cys and also as a SECIS-dependent Sec codon [23].
Natural reassignment of sense codons has not been seen in bacteria. But it is known in mitochondrial genomes, where a particular codon lost its original assignment and now leads to insertion of another amino acid [24]. Our case here is different; Sec insertion is mediated by a SECIS element and thus gives rise to dual use of the codon for another amino acid (through pairing with tRNASec variants carrying the proper anticodon).
What might account for this facile recoding to Sec? It is pertinent to note that Sec incorporation is different from that of all other amino acids; it is facilitated by its own ‘orthogonal’ system [5] consisting of a different elongation factor (SelB), a required SECIS RNA element, a structurally unusual tRNA (tRNASec) [10], a dual meaning stop codon (UGA), and the use of release factor 2. Therefore, Sec recoding events may not have as general an effect on the protein translation machinery as one might expect from recoding canonical sense codons [25]. The ease of Cys to Sec recoding may be a consequence of the often desirable properties of selenoenzymes and selenoproteins with novel redox functions and increased enzyme activity [4, 26], while still allowing the expression of useful Cys-proteins and Cys-enzymes. Our finding of facile Sec recoding also opens our minds to the possible existence of other coding schemes. It also underscores the limitations of the current computational programs to predict selenoproteins from genome sequences, as these algorithms rest on UGA as the sole Sec codon.
Overall our approach provides new evidence of a limited but unequivocal plasticity of the genetic code whose secrets still lie hidden in the majority of unsequenced organisms.
Supplementary Material
Acknowledgments
We thank Hans Aerni and Jesse Rinehart for advice on LC-MS/MS and Jean Kanyo (Yale University) for the dedicated efforts on the MS analyses. We also thank Andreas Brune, Filipa Gody-Vitorino, Hans-Peter Klenk, Ryan Lynch, Katherine McMahon, Daniel Marcus, William Mohn, Len Pennacchio, and Ameet Pinto for permission to use unpublished sequence data produced through the DOE-JGI’s community sequencing program. We are grateful to Patrick O’Donoghue, Oscar Vargas-Rodriguez, Jiqiang Ling for enlightened discussions and Daniel Drell and Robert Stack for encouragement. This work was supported by grants from the National Institute for General Medical Sciences (GM22854 to D.S.) and from the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the Department of Energy (DE-FG02-98ER20311 to D.S.; for funding the genetic experiments). The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, was supported under Contract No. DE-AC02-05CH11231.
Footnotes
Supporting information for this article is given via a link at the end of the document.
Contributor Information
Dr. Takahito Mukai, Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520 (USA)
Dr. Markus Englert, Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520 (USA)
Dr. H. James Tripp, Department of Energy Joint Genome Institute (DOE JGI), Walnut Creek, CA 94598 (USA)
Dr. Corwin Miller, Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520 (USA)
Dr. Natalia N. Ivanova, Department of Energy Joint Genome Institute (DOE JGI), Walnut Creek, CA 94598 (USA)
Dr. Edward M. Rubin, Department of Energy Joint Genome Institute (DOE JGI), Walnut Creek, CA 94598 (USA)
Dr. Nikos C. Kyrpides, Department of Energy Joint Genome Institute (DOE JGI), Walnut Creek, CA 94598 (USA)
Prof. Dieter Söll, Email: dieter.soll@yale.edu, Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520 (USA). Department of Chemistry, Yale University, New Haven, CT 06520 (USA)
References
- 1.Cone JE, Del Rio RM, Davis JN, Stadtman TC. Proc Natl Acad Sci USA. 1976;73:2659–2663. doi: 10.1073/pnas.73.8.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambrogelly A, Palioura S, Söll D. Nat Chem Biol. 2007;3:29–35. doi: 10.1038/nchembio847. [DOI] [PubMed] [Google Scholar]
- 3.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 4.Metanis N, Hilvert D. Curr Opin Chem Biol. 2014;22:27–34. doi: 10.1016/j.cbpa.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 5.Böck A, Thanbichler M, Rother M, Resch A. In: Aminoacyl-tRNA Synthetases. Ibba M, Francklyn CS, Cusack S, editors. Landes Bioscience; Georgetown, TX: 2005. pp. 320–327. [Google Scholar]
- 6.Heider J, Baron C, Böck A. EMBO J. 1992;11:3759–3766. doi: 10.1002/j.1460-2075.1992.tb05461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bröcker MJ, Ho JM, Church GM, Söll D, O’Donoghue P. Angew Chem Int Ed Engl. 2014;53:319–323. doi: 10.1002/anie.201308584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivanova NN, Schwientek P, Tripp HJ, Rinke C, Pati A, Huntemann M, Visel A, Woyke T, Kyrpides NC, Rubin EM. Science. 2014;344:909–913. doi: 10.1126/science.1250691. [DOI] [PubMed] [Google Scholar]
- 9.Schön A, Böck A, Ott G, Sprinzl M, Söll D. Nucleic Acids Res. 1989;17:7159–7165. doi: 10.1093/nar/17.18.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Palioura S, Sherrer RL, Steitz TA, Söll D, Simonovic M. Science. 2009;325:321–325. doi: 10.1126/science.1173755. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Itoh Y, Chiba S, Sekine S, Yokoyama S. Nucleic Acids Res. 2009;37:6259–6268. doi: 10.1093/nar/gkp648. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Itoh Y, Sekine S, Suetsugu S, Yokoyama S. Nucleic Acids Res. 2013;41:6729–6738. doi: 10.1093/nar/gkt321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abe T, Inokuchi H, Yamada Y, Muto A, Iwasaki Y, Ikemura T. Front Genet. 2014;5:114. doi: 10.3389/fgene.2014.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Markowitz VM, Chen IM, Chu K, Szeto E, Palaniappan K, Pillay M, Ratner A, Huang J, Pagani I, Tringe S, Huntemann M, Billis K, Varghese N, Tennessen K, Mavromatis K, Pati A, Ivanova NN, Kyrpides NC. Nucleic Acids Res. 2014;42:D568–573. doi: 10.1093/nar/gkt919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a) Luedemann GM. J Bacteriol. 1968;96:1848–1858. doi: 10.1128/jb.96.5.1848-1858.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ivanova N, et al. Stand Genomic Sci. 2010;2:158–167. doi: 10.4056/sigs.711311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Gladyshev VN. PLoS Genet. 2008;4:e1000095. doi: 10.1371/journal.pgen.1000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zinoni F, Birkmann A, Leinfelder W, Böck A. Proc Natl Acad Sci USA. 1987;84:3156–3160. doi: 10.1073/pnas.84.10.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Axley MJ, Böck A, Stadtman TC. Proc Natl Acad Sci USA. 1991;88:8450–8454. doi: 10.1073/pnas.88.19.8450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandrand-Berthelot MA, Wee MYK, Haddock BA. FEMS Microbiology Lett. 1978;4:37–40. [Google Scholar]
- 18.Pavan ME, Pavan EE, Lopez NI, Levin L, Pettinari MJ. Genome Announc. 2013;1 doi: 10.1128/genomeA.00675-13. pii: e00675–00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vincent AT, Trudel MV, Freschi L, Nagar V, Gagne-Thivierge C, Levesque RC, Charette SJ. BMC Genomics. 2016;17:44. doi: 10.1186/s12864-016-2381-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshizawa S, Rasubala L, Ose T, Kohda D, Fourmy D, Maenaka K. Nat Struct Mol Biol. 2005;12:198–203. doi: 10.1038/nsmb890. [DOI] [PubMed] [Google Scholar]
- 21.Lu J, Holmgren A. Free Radic Biol Med. 2014;66:75–87. doi: 10.1016/j.freeradbiomed.2013.07.036. [DOI] [PubMed] [Google Scholar]
- 22.a) Lin HC, Ho SC, Chen YY, Khoo KH, Hsu PH, Yen HC. Science. 2015;349:91–95. doi: 10.1126/science.aab0515. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Turanov AA, Everley RA, Hybsier S, Renko K, Schomburg L, Gygi SP, Hatfield DL, Gladyshev VN. PLoS One. 2015;10:e0140353. doi: 10.1371/journal.pone.0140353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turanov AA, Lobanov AV, Fomenko DE, Morrison HG, Sogin ML, Klobutcher LA, Hatfield DL, Gladyshev VN. Science. 2009;323:259–261. doi: 10.1126/science.1164748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.a) Yokobori S. Viva Origino. 2014;42:47–49. [Google Scholar]; b) Ling J, O’Donoghue P, Söll D. Nat Rev Microbiol. 2015;13:707–721. doi: 10.1038/nrmicro3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.a) Ho JM, Reynolds NM, Rivera K, Connolly M, Guo LT, Ling J, Pappin DJ, Church GM, Söll D. ACS Synth Biol. 2015 doi: 10.1021/acssynbio.1025b00197. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Mukai T, Yamaguchi A, Ohtake K, Takahashi M, Hayashi A, Iraha F, Kira S, Yanagisawa T, Yokoyama S, Hoshi H, Kobayashi T, Sakamoto K. Nucleic Acids Res. 2015;43:8111–8122. doi: 10.1093/nar/gkv787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.a) Beld J, Woycechowsky KJ, Hilvert D. Oxidative Folding of Peptides and Proteins, The Royal Society of Chemistry. 2009:253–273. [Google Scholar]; b) Snider GW, Ruggles E, Khan N, Hondal RJ. Biochemistry. 2013;52:5472–5481. doi: 10.1021/bi400462j. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.