Abstract
Oxysterol binding protein-related protein 2 (ORP2) is a lipid binding protein that has been implicated in various cellular processes, including lipid sensing, cholesterol efflux, and endocytosis. We recently identified ORP2 as a member of a protein complex that regulates glucocorticoid biosynthesis. Herein, we examine the effect of silencing ORP2 on adrenocortical function and show that the ORP2 knockdown cells exhibit reduced amounts of multiple steroid metabolites, including progesterone, 11-deoxycortisol, and cortisol, but have increased concentrations of androgens, and estrogens. Moreover, silencing ORP2 suppresses the expression of most proteins required for cortisol production and reduces the expression of steroidogenic factor 1 (SF1). ORP2 silencing also increases cellular cholesterol, concomitant with decreased amounts of 22-hydroxycholesterol and 7-ketocholesterol, two molecules that have been shown to bind to ORP2. Further, we show that ORP2 binds to liver X receptor (LXR) and is required for nuclear LXR expression. LXR and ORP2 are recruited to the CYP11B1 promoter in response to cAMP signaling. Additionally, ORP2 is required for the expression of other LXR target genes, including ABCA1 and the LDL receptor (LDLR). In summary, we establish a novel role for ORP2 in regulating steroidogenic capacity and cholesterol homeostasis in the adrenal cortex.
Keywords: ORP2, steroid hormones, lipid transfer proteins, cholesterol, LXR, cortisol
1. Introduction
In mammalian cells, cholesterol homeostasis is contingent upon the distribution of cholesterol and its oxygenated derivatives (oxysterols) along an intracellular gradient and their interaction with regulatory components such as lipid binding proteins (Ridgway N, 2010, Ridgway ND et al., 1998). The oxysterol binding protein OSBP/ORPs family of transport proteins has been implicated in the energy-independent transport of oxysterols and cholesterol between organelles. Members of the mammalian 12-member family all contain cholesterol/oxysterol binding motifs that are adjacent to domains with regulatory and membrane targeting functions (Ngo MH et al., 2010, Olkkonen VM and Li S, 2013). The family can further be grouped into six subfamilies based on sequence identity, with each individual member having distinct functions including the regulation of lipid metabolism, signal transduction, vesicular transport and cytoskeletal regulation (Olkkonen and Levine, 2004, Schultz et al., 2009). ORPs have variable N terminal regions with most members containing a FFAT motif (two phenylalanines in an acidic tract), pleckstrin homology (PH) domains and ankyrin repeats, as well as a C terminus composed of an oxysterol recognition domain (Fairn and McMaster, 2008, Jansen et al., 2011).
Members of the OSBP/ORP family participate in a wide variety of molecular processes (Olkkonen VM and Li S, 2013). For example, overexpression of OSBP in Chinese hamster ovary (CHO) cells increases expression of sterol regulated genes as well as de novo cholesterol synthesis, and suppress acyl-CoA:cholesterol acyltransferase (ACAT) activity when bound to 25-hydroxycholesterol via modulation of sphingolipid and cholesterol synthesis and trafficking (Storey, 1998). Studies by Ridgway et al. revealed that OSBP functions as a cytosolic protein that, when bound to 25-hydroxycholesterol, localizes to the Golgi apparatus, with possible regulatory functions in addition to that of oxysterol transport (Ridgway, 1992). Additionally, the ORP subfamily III, which includes ORP3, ORP6 and ORP7, have been found to be dually localized on the ER and the plasma membrane, and it has been suggested that they may have the function of facilitating lipid transport between the two compartments (M. Lehto, Tienari, J., Lehtonen, S., Lehtonen, E., Olkkonen, V., 2004, Pichler, 2001).
ORP2 encodes a 55 kDa protein that is expressed ubiquitously in mammalian tissues (Laitinen et al., 2002). It is homologous to the sterol binding domain of OSBP, yet lacks the N terminal extension that contains a PH domain required for Golgi localization and the functional effects of OSBP (Laitinen et al., 2002). Despite the absence of the PH domain, ORP2 has been found to be distributed between the cytosol and Golgi apparatus, and is still capable of Golgi association, which has been attributed to a resemblance of the Golgi targeting mechanism of yeast Osh4p/Kes1p described in Lehto, et al. (M. Lehto and Olkkonen, 2003). Studies on CHO cells have shown that ORP2 localizes to the surface of lipid droplets (LD), and binds several oxysterols with different affinities, with the highest affinity ligand to date being 22(R)-hydroxycholesterol (Hynynen et al., 2005). Overexpression of ORP2 in CHO cells has been associated with a decrease in cholesteryl esters as well as a dispersal of the droplets throughout the cytoplasm (Hynynen et al., 2005, Hynynen et al., 2009). Also, stably transfected CHO cell expressing ORP2 have been shown to display a decrease in free cholesterol within cells, as well as an enhanced cholesterol efflux to all acceptors and a reduction in ACAT activity (Fairn and McMaster, 2008, Kakela et al., 2005). ORP2 is also capable of binding 25-hydroxycholesterol via a binding pocket similar to that of its yeast homolog Osh4p, as well as interacting with phospholipid vesicles containing PIPs (Suchanek, 2007).
In the adrenal gland, steroid hormones are produced from cholesterol through a metabolic pathway that is dependent on the inter-organelle transfer of its oxygenated derivatives between the mitochondrion and the endoplasmic reticulum (ER). We have recently identified a protein complex containing multiple proteins including RhoA, and the RhoA effector diaphanous 1 (DIAPH1), that regulates microtubule-dependent mitochondrial movement and cortisol production (Li D et al., 2013). Interestingly, components of this complex include two members of the ORP family, ORP2 and ORP10. Based on the established role of ORP2 in cholesterol homeostasis and its identification as a component of a macromolecular protein complex implicated in steroid hormone metabolism, the aim of this study was to determine the effect of silencing ORP2 on adrenocortical steroidogenic gene expression and hormone production.
2. Materials and Methods
2.1. Reagents
Dibutyryl cAMP, 22(R)-hydroxycholesterol, 22(S)-hydroxycholesterol, 25-hydroxycholesterol, progesterone, and pregnenolone were purchased from Sigma-Aldrich (St. Louis, MO).
2.2. Cell culture
H295R adrenocortical cells were obtained from Dr. William E. Rainey (University of Michigan, Ann Arbor, MI) and cultured in Dulbecco's Modified Eagle's/F12 medium (Hyclone, Logan, UT) supplemented with 10% Nu-Serum I (BD Biosciences, Palo Alto, CA), 1 % ITS Plus (BD Biosciences), antibiotics, and antimycotics. Stable ORP2 knockdown cell lines (ORP2kd) were generated by transfecting H295R cells with shRNA plasmids (pGFP-V-RS HuSH vector; Origene, Rockville, MD) and clones selected by culturing in 10 µg/ml puromycin, and cultured in the DME/F12 medium and supplements described for wild type cells. Clones from cells transfected with two different shRNAs (clone 1: 5’-AAC ATA ATG AAG CCT ACA CCT GGA CCA AC (ORP2 shRNA#1) and clone 2 5’-AGT GCG TTC CAC TCG GAA GGT CTC AAC CA (ORP2 shRNA#2) were tested and all yielded similar results. Stable cell lines transfected with a scrambled shRNA or the empty vector were generated as negative controls. ORP2 overexpressing cells (ORP2UP) were generated by transfecting H295R cells with pCMV6-AC-GFP-ORP2 (Origene) and stable clones selected by incubation in 100 ng/ml G418. The pCMV6-AC-GFP empty vector was used as a negative control. CV1 cells were obtained from the American Type Culture Collection (Manassas, VA) and cultured in MEM (Cellgro, Manassas, VA) containing 10% fetal bovine serum, antibiotics, and antimycotics.
2.3. Cortisol and DHEA assays
H295R wild type, scrambled shRNA, ORP2KD (ORP2 shRNA#1 and ORP2 shRNA#2), or ORP2UP cells were subcultured into 6-well plates and treated with 0.4 mM Bt2cAMP (Sigma Aldrich, St. Louis, MO) for 48 h. Cortisol and dehydroepiandrosterone (DHEA) released into the media were determined in triplicate against standards prepared in DMEM/F12 medium using a 96-well plate enzyme linked immune DHEA and cortisol assays (Assay Designs, Inc., Ann Arbor, MI). Steroid hormone amount was normalized to the total cellular protein content, as determined using a bicinchoninic acid (BCA) assay (Pierce, Rockford, IL).
2.4. Sterol and steroid Metabolite Analysis
Wild type and ORP2KD cells (ORP2 shRNA#1 and ORP2 shRNA#2) were treated with 0.4 mM Bt2cAMP for 48 h and the media and cells collected for quantification of sterol and steroid metabolites by ultra performance liquid chromatography tandem mass spectrometry. Analysis was performed by the West Coast Metabolomics Center at the University of California Davis Metabolomics Facility (Supported by NIH 1U24DK097154). Briefly, Ultra Performance Liquid Chromatography (UPLC) tandem Mass Spectroscopy (MS-MS) was carried out with a Waters Acquity UPLC system connected with the Xevo TQ triple quadrupole mass spectrometer (Gaikwad NW, 2013). The analyses were performed using Electro Spray Ionization (ESI) in positive ion mode, capillary voltage of 3.0 kV, extractor cone voltage of 3 V and detector voltage of 500 V. Desolvation gas flow was maintained at 600 L/h. Source temperature and desolvation temperatures were set at 150 and 350 °C, respectively. The collision energy was varied to optimize daughter ions. Analytical separations on the UPLC system were conducted using an Acquity UPLC BEH C18 1.7 µm column (1 × 50 mm) at a flow rate of 0.2 ml/min. The gradient started with 100% A (0.1% formic acid in H2O) and 0% B (0.1% formic acid in CH3CN) and changed to 20% A over 5 min, then 0 % A over 2 min. Finally, over 1 min it was changed back to original 100% A and column was equilibrated at 100% A for 2 min, resulting in a total separation time of 10 min. The elutions from the UPLC column were introduced to the mass spectrometer and resulting data were analyzed and processed using MassLynx 4.1 software. Reference standards were used to optimize the UPLC/MS-MS conditions prior to analysis.
2.5. RNA isolation, quantitative RT-PCR, and microarray
H295R wild type, scrambled shRNA, ORP2KD(shRNA#1), and ORP2UP cells were subcultured into 12-well plates and treated with 0.4 mM Bt2cAMP for 18 h. Total RNA was isolated using RNeasy Mini Kit (Qiagen, Germantown, MD), and amplified in triplicate using a One-Step SYBR Green RT-PCR Kit (Quanta Biosciences, Gaithersburg, MD) and the primer sets listed in Table 1. Gene expression was normalized to β-actin mRNA content and calculated using the ΔΔ cycle threshold (ΔΔCT) method. mRNA expression of ORP2KD shRNA#2 is shown in Supplemental Figure 2. Microarray analysis was carried out by Phalanx Biotech Group, Inc. (Palo Alto, CA) using the Human Whole Genome OneArray™ DNA Microarray (HOA_004) on RNA isolated from wild type, scrambled shRNA, and ORP2 shRNA#1 cell lines. Three technical replicate hybridizations were performed per sample. Hybridization intensities were analyzed by normalizing to the wild type cell line and principal component, gene ontology, and pathway enrichment analyses carried out to identify subsets of genes which share a common molecular function or biological process that were differentially expressed.
Table 1.
Primer sets and antibodies used for real time RT-PCR and western blotting, respectively.
| RT-PCR Primers | ||
|---|---|---|
| Gene | Forward (5′–3′) | Reverse (5′–3′) |
| β-Actin | ACGGCTCCGGCATGTGCAAG | TGACGATGCCGTGCTGCATG |
| ORP2 | AGAGGTGACCACCTGAGAAAGG | GTTGATCCTCCAGAGCAGCTTG |
| CYP17A1 | CTCTTGCTGCTTCACCTA | TCAAGGAGATGACATTGGTT |
| CYP11A1 | CGTGGAGTCGGTTTATGTC | CTCTGGTAATACTGGTGATAGG |
| CYP11B1/2 | ACGGCGACAACTGTATCC | AGAGCGTCATCAGCAAGG |
| HSD3B2 | CCAGTAGCATAGAGGTAGCC | TCAGATTCCACCCGTTAGC |
| CYP21A2 | TGTGGAACTGGTGGAAGC | GGTGGAGCCTGTAGATGG |
| StAR | GCTCTCTACTCGGTTCTC | GCTGACTCTCCTTCTTCC |
| HSL | CACTACAAACGCAACGAGAC | CCAGAGACGATAGCACTTCC |
| SCARBI | CCATCCTCACTTCCTCAAC | CCACAGGCTCAATCTTCC |
| LDLR | ACGGTGGAGATAGTGACAATG | AGACGAGGAGCACGATGG |
| ABCA1 | CAGGCTACTACCTGACCTTGGT | CTGCTCTGAGAAACACTGTCCTC |
| NR5A1 | GGAGTTTGTCTGCCTCAAGTTCA | CGTCTTTCACCAGGATGTGGTT |
| Western blotting antibodies | ||
| Target protein Catalog no. and vendor | Dilution | |
| ORP2 | 17217-1-AP, Proteintech Inc. | 1:2500 |
| GAPDH | sc-25778, Santa Cruz Biotechnology, Inc. | 1:5000 |
| CYP17A1 | sc-66849, Santa Cruz Biotechnology, Inc. | 1:2000 |
| CYP11A1 | sc-292456, Santa Cruz Biotechnology, Inc. | 1:2500 |
| CYP11B | sc-28205, Santa Cruz Biotechnology, Inc. | 1:1000 |
| HSD3B2 | ab80500, Abcam | 1:1000 |
| CYP21A2 | ab80208, Abcam | 1:1000 |
| StAR | sc-25806, Santa Cruz Biotechnology, Inc. | 1:2000 |
| HSL | sc-25843, Santa Cruz Biotechnology, Inc. | 1:1000 |
| SR-BI | sc-67098, Santa Cruz Biotechnology, Inc. | 1:1000 |
| LDLR | ab30532, Abcam | 1:1000 |
| SF1 | 07-618, Millipore | 1:5000 |
| lamin A/C | sc-376248, Santa Cruz Biotechnology, Inc. | 1:5000 |
| F1-ATPase | sc-33618, Santa Cruz Biotechnology, Inc. | 1:2500 |
2.6. Western blotting
H295R wild type, scrambled and ORP2KD(shRNA #1 and shRNA#2) and ORP2UP were subcultured onto 6-well plates treated with Bt2cAMP (0.4 mM) for 48 h, and whole cell lysates harvested in radioimmunoprecipitation assay (RIPA) buffer [50nM Tris, Cl (pH 7.4), 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 nM NaCl, 0.4 mM EDTA, 150 nM aprotinin, 1 mM leupeptin, 1 mM E-64, 500 mM 4-(2-aminoethyl) benzenesulfonylfluoride]. Cells were then lysed by sonication (one 2 s burst) and incubated on ice for 30 min. Lysates were centrifuged at 12,000 rpm for 15 min at 4°C, and supernatant was collected. Aliquots of each sample containing 40 µg protein were separated on 8% SDS-PAGE gels, and transferred to polyvinylidine difluoride (PVDF) membranes (Millipore, Billerica, MA). Blots were probed with antibodies listed in Table 1, as well as alkaline phosphate-conjugated secondary antibody (1:5000; ECF Western Blotting Kit; GE Healthcare). Blots were imaged on a VersaDoc 4000 (Bio-Rad Laboratories, Hercules, CA) and densitometric analysis was carried out using Quantity One software (Bio-Rad Laboratories).
In some experiments H295R negative shRNA or ORP2KD cells were sub-cultured into 100-mm dishes and cytoplasmic and nuclear fractions were purified using the NE-PER Nuclear and Cytoplasmic Extraction Kit (Pierce, Rockford, IL) following the manufacturer’s instructions. Aliquots (30 µg of protein) of cytoplasmic and nuclear fractions were separated by SDS-PAGE and transferred to PVDF membranes and incubated with anti-ORP2, anti-GAPDH, or anti-lamin A/C. Blots were washed and incubated with secondary antibody and imaged as described above.
2.7. Immunofluorescence Staining
Immunostaining and microscopy were performed as described previously (Li et al., 2007). Briefly, H295R cells were plated on glass cover slips, fixed, permeabilized, blocked with 1% BSA for 1 h, and then incubated with anti-ORP2 (14751-1-AP, Proteintech), anti-ORP2 (SAB2500724, Sigma), anti-ORP10 (GTX108097, GeneTex, Inc, Irvine, CA), or anti StAR (sc-25806, Santa Cruz Biotechnology) antibodies (diluted 1:500 in 1% BSA) overnight, followed by incubation with Alexa Fluor-conjugated IgG (Life Technologies, Grand Island, NY) for 1 h. Cover slips were stained with 4,6’-diamino-2-phenylindole dihydrochloride (DAPI; Life Technologies) or MitoTracker Red (Life Technologies) and then mounted onto slides using Fluoromount G (Southern Biotech, Birmingham, AL). Images were captured using a fluorescence microscope (Nikon Eclipse Ti) and processed using NIS-Elements BR imaging software (Nikon).
2.8. Chromatin immunoprecipitation (ChIP)
ChIP assay was performed as previously described (Dammer et al., 2007). Briefly, H295R cells were sub-cultured into 150-mm dishes and treated with 0.4 mM Bt2cAMP for 1 h. After cross-linking with 1% formaldehyde, cells were and chromatin isolated using a Magna ChIP A/G Chromatin Immunoprecipitation Kit (Millipore). The purified chromatin was immunoprecipitated overnight at 4°C on a tube rotator using 5 µg of primary antibody [anti-acetyl (Lys9) histone H3 (07-352, Millipore), anti-SF1 (07-618, Millipore), anti-LXRB (HPA005468, Sigma), anti-ORP2 (14751-1-AP, Proteintech)], or anti-rabbit IgG (Millipore) and 30 µL protein A/G Plus agarose (Santa Cruz Biotechnology). Real Time PCR was carried out using the iTaq Universal SYBR Green Supermix (Bio-Rad Laboratories Inc, Hercules, CA) and the following primer set: CYP11B1 (forward, 5’-CAG GAA TGA AAC AGG TTG GAG G -3’ and reverse, 5’-GAG ACG TGA TTA GTT GAT GGC TC-3’). PCR reactions were carried out per the manufacturer’s instructions and output ΔCt values were normalized to input values.
2.9. Co-immunoprecipitation assay (coIP)
CV1 cells were plated onto 10 cm dishes and transfected with 10 µg pCMV6-myc/FLAG vector, pCMV6-myc/FLAG-LXRB, pCMV6-myc/FLAG-SF1, and pCMV6-GFP-ORP2 (all plasmids purchased from Origene) for 48 h. Cells were washed twice with PBS and harvested into RIPA buffer. Lysates were then sonicated 6 times for 5 sec followed by centrifugation at 12,000 rpm for 15 min at 4°C. Five % of the lysates were retained for analysis of the input protein expression by western blotting and the remaining lysates pre-cleared with 20 µL BSA (20 mg/mL) and immunoprecipitated overnight at 4°C on a tube rotator using protein A/G Plus agarose beads (Santa Cruz Biotechnology, Inc) and anti-FLAG (Sigma) antibody. The immobilized protein complexes were washed twice in RIPA buffer and twice in PBS, and then separated by SDS-PAGE. Western blots were probed with anti-GFP (Origene) antibody and expression was detected using an ECF Western Blotting Kit (GE Biosciences) and visualized using a VersaDoc 4000 imager (Bio-Rad).
2.10. Statistical Analysis
One-way ANOVA, Tukey-Kramer multiple comparison, and unpaired student t-tests were performed using Prism 6 (GraphPad Software Inc., San Diego, CA). Significant differences from a compared value were defined as p < 0.05 and denoted by asterisks (*) or carats (^).
3. Results
3.1. Silencing ORP2 diminishes cortisol secretion
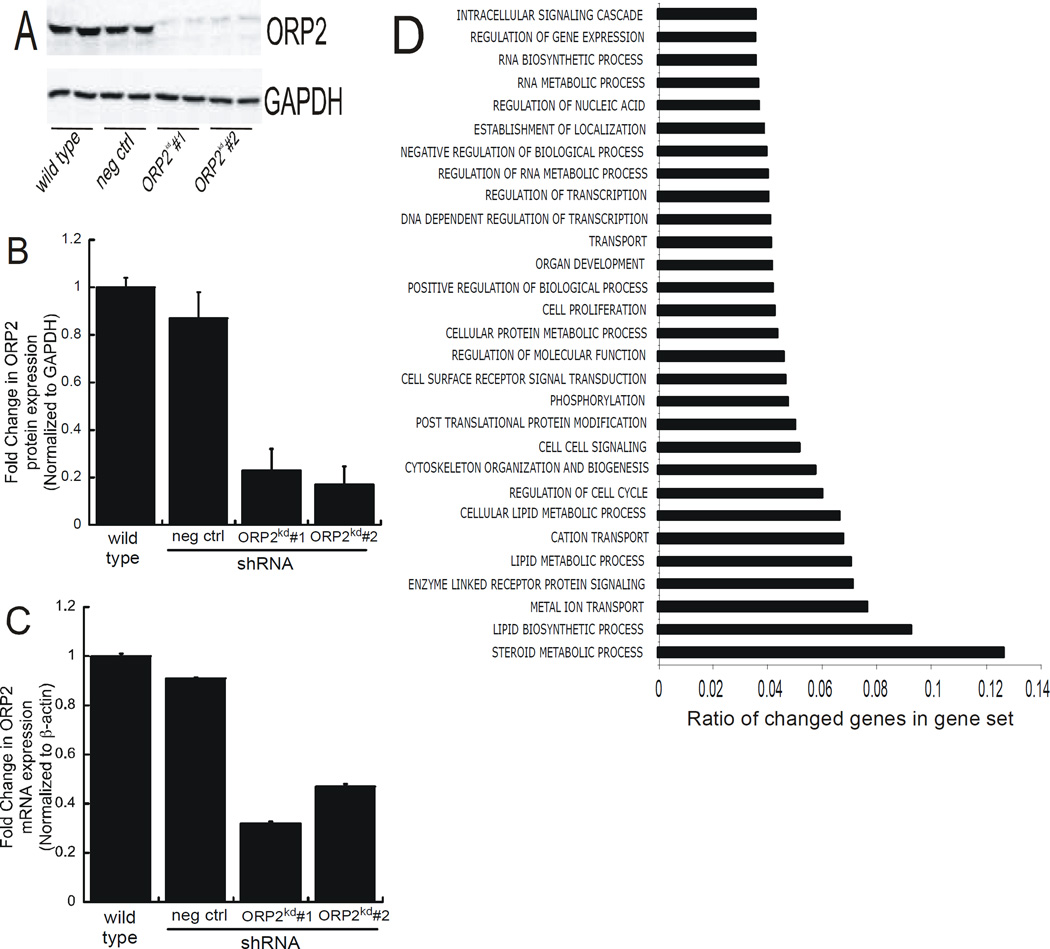
Previous studies reported from our lab identified the RhoA effector, DIAPH1, as component of a protein complex that mediated cortisol biosynthesis (Li D et al., 2013). ACTH/cAMP stimulated the interaction between RhoA and DIAPH1 and also increased the rate of mitochondrial movement (Li D and Sewer MB, 2010). Given that these studies identified ORP2 as a component of this macromolecular protein complex, we sought to define the functional significance of ORP2 in regulating the steroidogenic pathway by generating a cell line where ORP2 was stably suppressed via the expression of shRNAs against the lipid binding protein. As shown in Figure 1 stable expression of two different ORP2 shRNAs resulted in a greater than 75% reduction in ORP2 protein expression (1A and 1B). Consistent with a decrease in ORP2 protein expression, the levels of ORP2 transcript were reduced by 67% and 49% in clone #1 and clone #2, respectively (Figure 1C). Microarray studies were performed on wild type, scrambled shRNA, and ORP2 shRNA#1 RNA to assess the effect of silencing the expression of ORP2 on global gene expression. These studies revealed that 2561 genes were differentially expressed (log2 ratio ≥ 1.0 or ≤−1.0 and p < 0.05). Gene ontology (GO) analysis showed significant changes in several biological processes, including lipid biosynthetic process, enzyme-linked receptor protein signaling, and transport. Notably, the most altered cluster of genes were involved in steroid metabolic processes (Figure 1D). Representative genes in the lipid biosynthetic process cluster that changed significantly include lipoprotein lipase, 7-dehydrocholesterol reductase, and SULT2A1 (sulfotransferase family, cytosolic 2A dehydrogenase), whereas the low density lipoprotein receptor (LDLR) and the scavenger receptor class B, member 1 (SCARB1) were genes in the transport cluster that exhibited significantly different expression in the ORP2kd cells.
Figure 1. Generation of an ORP2 knockdown cell line.
(A) Whole cell lysates were harvested from wild type, scrambled negative shRNA control, ORP2 shRNA#1 and ORP shRNA#2 cells and analyzed by SDS-PAGE and western blotting using antibodies against ORP2 (top) and GAPDH (bottom). Shown is a representative blot from experiments performed on at least 6 different occasions, each time in at least duplicate. (B) Densitometric analysis of protein isolated from wild type, scrambled shRNA, ORP2 shRNA#1, and ORP2 shRNA#2, cells that were subjected to western blotting. The expression of ORP2 protein is normalized to GAPDH and the data graphed represent the mean ± SEM of five separate experiments, each performed in at least duplicate. (C) RNA from untreated wild type, scrambled shRNA, ORP2 shRNA#1, or ORP2 shRNA#2 was isolated for analysis of ORP2 expression by real time RT-PCR. Data are graphed as fold change in mRNA expression over wild type and normalized to the mRNA expression of β-actin and represent the mean ± SEM of three separate experiments, each performed in triplicate. (D) RNA isolated from wild type, scrambled and ORP2 shRNA#1 was subjected to microarray analysis. The graph data represent multilevel gene ontology (GO) analysis of the biological processes that were significantly changed in ORP2kd cells when compared to the wild type. The vertical axis represents the GO terminology and the x-axis represents the proportion of genes within each category that were significantly changed.
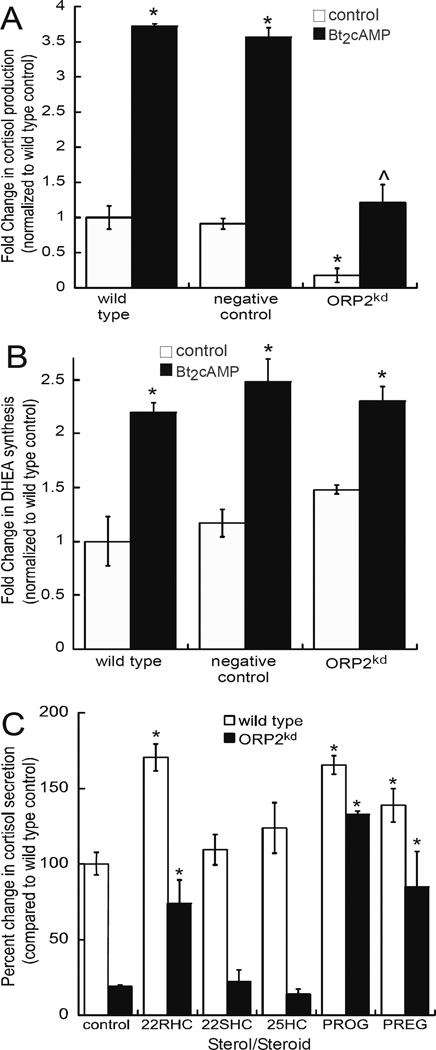
Next we analyzed the effects of the ORP2 knockdown on cortisol and DHEA secretion. Interestingly, ORP2kd cells exhibited a 90% decrease in basal cortisol secretion and a 68% decrease in cortisol secretion in dibutyryl cAMP (Bt2cAMP)-stimulated cells, when compared to the levels observed in the wild type or scrambled shRNA expressing cells (Fig. 2A). Despite the reduction in basal cortisol secretion, there was still a 5.5-fold increase in cortisol secretion in the ORP2kd cells in response to Bt2cAMP, which compares to a 3.7-fold increase in cortisol production in wild type cells treated with Bt2cAMP. In contrast, basal levels of DHEA secretion was increased by 1.5-fold in the ORP2 knockdown cell line (Fig. 2B). Given the decreased amount of cortisol in the ORP2kd cell line, we next sought to determine if the ORP2kd cells still maintain the capacity to produce the glucocorticoid. We treated wild type and ORP2kd cells with different substrates and assessed the amount of cortisol secreted into the media by ELISA. As shown in Figure 2C, supplementation with 22R-hydroxycholesterol, progesterone and pregnenolone all increased the production of cortisol to levels comparable to the wild type cells. However, neither 22S-hydroxycholesterol nor 25-hydroxycholesterol was able to increase cortisol secretion from the ORP2kd cell line.
Figure 2. Silencing ORP2 diminishes cortisol secretion.
Cortisol (A) and DHEA (B) concentrations was measured in medium collected from wild type, scrambled shRNA, or ORP2kd H295R cells that were treated for 48 h with 0.4 mM Bt2cAMP by ELISA and normalized to total cellular protein content. ORP2kd hormone levels reflect the average of data obtained from both ORP2 shRNA#1 and ORP2 shRNA#2 clones. Data are graphed as fold change normalized to the metabolite secreted from untreated wild type cells and represent the mean ± SEM of four separate experiments, each performed in triplicate. Asterisks (*) and carats (^) indicate a statistically significant difference (p < 0.05) compared to the wild type control and wild type Bt2cAMP-treated cells, respectively. (C) ELISA assays were used to determine the amount of cortisol secreted into media that was collected from wild type or ORP2kd H295R cells that were treated for 24 h with 10 µM of 22(R)-hydroxycholesterol (22RHC), 22(S)-hydroxycholesterol (22SHC), 25-hydroxycholesterol (25HC), progesterone (PROG) and pregnenolone (PREG). Cortisol amounts were normalized to the concentration of cellular protein and data graphed represent the mean ± STD of three separate experiments, each performed in triplicate. Asterisks denote a statistically significant difference (p < 0.05) when compared to the wild type control cells.
3.2. Steroid metabolite amounts in ORP2kd cells
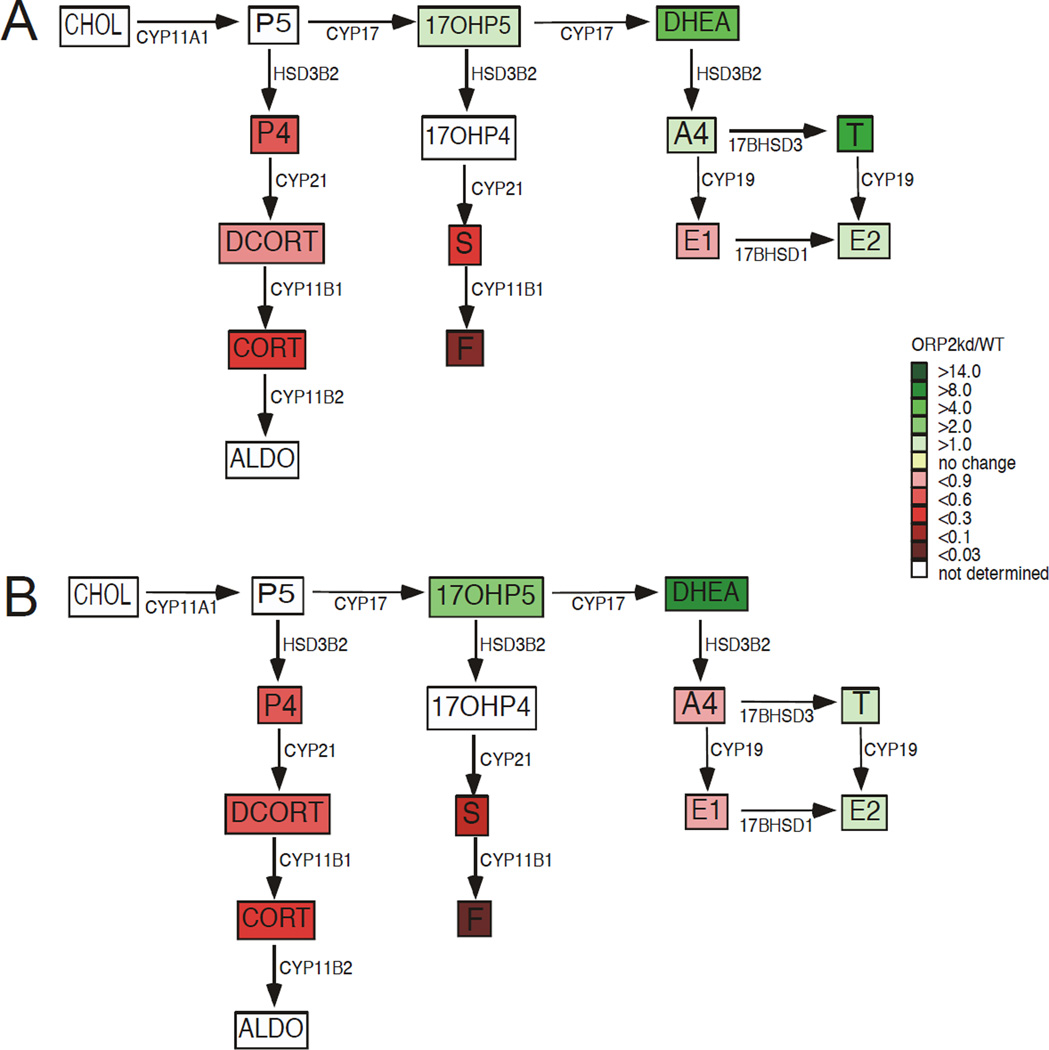
Based on our findings identifying a role for ACTH/cAMP-stimulated mitochondrial movement (Li D and Sewer MB, 2010) and a DIAPH1-containing complex (Li D et al., 2013) in regulating adrenocortical steroidogenesis, we hypothesized that ORP2 may act to facilitate the movement of substrates between organelles. Moreover, since we observed increased DHEA secretion and reduced cortisol production, we speculated that ORP2 might play a role in moving the 11-deoxycortisol produced in the ER to mitochondria for conversion to cortisol. Though our ELISA data in Figure 2 support this hypothesis, we assessed the concentrations of steroid metabolites by mass spectrometry to determine if the ORP2kd cells were accumulating 11-deoxycortisol. Consistent with the ELISA data (Figures 2A and 2B) cortisol levels were substantially decreased in both untreated (Figure 3A) and Bt2cAMP-stimulated cells (Figure 3B). In contrast to our prediction, basal levels of progesterone and 11-deoxycortisol were decreased in both the cells (Supplementary Figure 1) and in the cell culture media (Figure 3). Of note, our findings in the wild type cells are consistent with the work of Xing et al. (Xing Y et al., 2011), who have demonstrated that activation of the cAMP signaling pathway results in an increase in glucocorticoid production, concomitant with a decrease in androgen biosynthesis. While silencing ORP2 impaired glucocorticoid production, the knockdown cells harbored an increased capacity to produce androgens and estrogens, with increased DHEA, testosterone, estrone, and estradiol (Figure 3B). The changes in steroid metabolites secreted into the media by ORP2kd cells were mirrored by similar changes in the cellular amounts of the metabolites when compared to wild type cells (Supplemental Figure 1). Significantly, while the levels of corticosteroids were decreased, the ORP2kd cells were still responsive to activation of the cAMP signaling pathway, albeit with a reduction in the magnitude of the response.
Figure 3. Suppressing ORP2 alters cellular steroid metabolite concentrations.
H295R WT and ORP2kd cells were cultured into 100 mm dishes treated for 48 h with 0.4 mM Bt2cAMP media collected and the amounts of steroids quantified by ultra performance liquid chromatography tandem mass spectrometry as described in Materials and Methods. Data are displayed as a heat map of the steroid hormone biosynthetic pathway, with steroid metabolites in untreated cells shown in A and Bt2cAMP metabolites depicted in B. Fold change in metabolite secretion ORP2kd is normalized to the wild type and data represent the mean ± SD of two separate experiments (n=6 per experiment). Steroid metabolite amounts are normalized to the cellular protein concentration. Enzymes catalyzing each reaction are denoted adjacent to the arrows. Abbreviations are as follows: cholesterol, CHOL; P5, pregnenolone; 17-OHP5, 17-hydroxypregnenolone; DHEA, dehydroepiandrosterone; P4, progesterone; 17-OHP4, 17-hydroxyprogesterone; A4, androstenedione; T, testosterone; DCORT, deoxycorticosterone; DCRT, 11-deoxycortisol; E1, estrone; E2, estradiol; CORT, corticosterone; S, 11-deoxycortisol; F, cortisol; ALDO, aldosterone.
3.3. Down-regulation of steroidogenic genes contributes to decreased glucocorticoid output
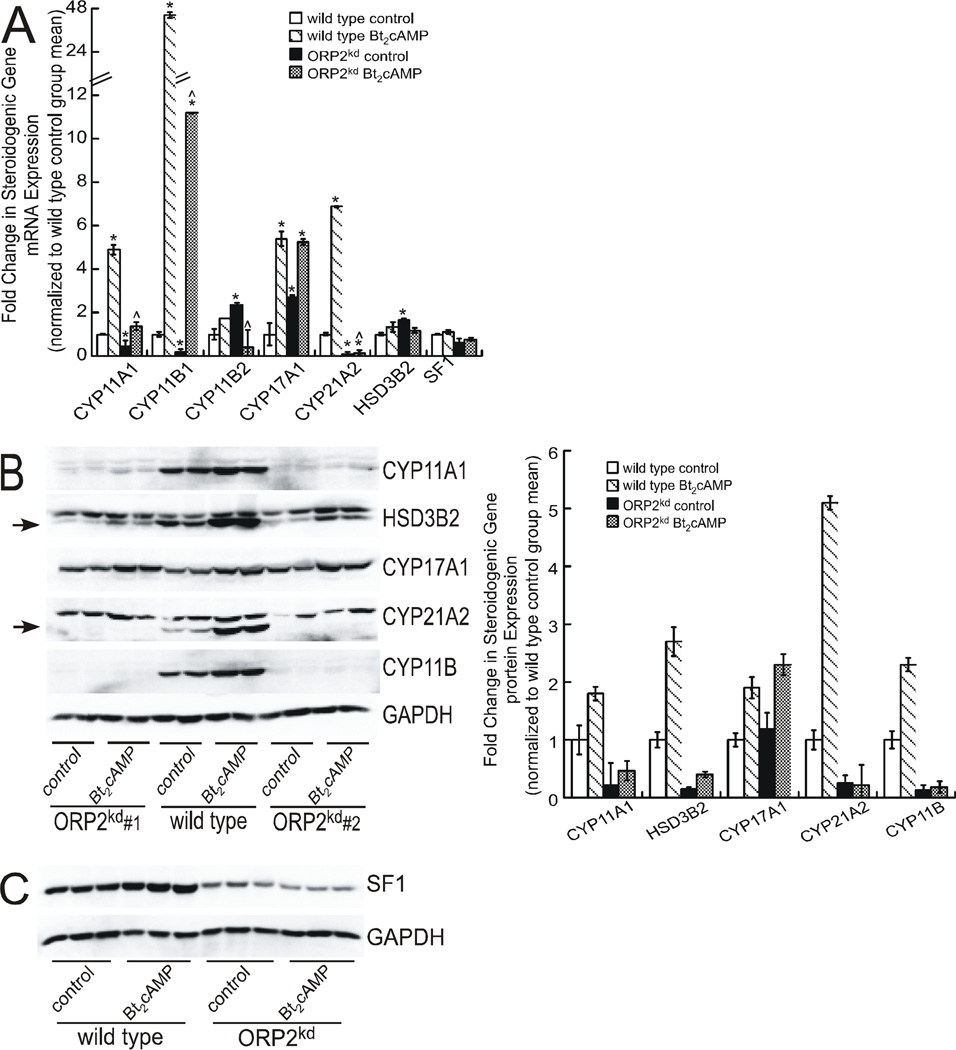
The changes in hormone secretion and steroid metabolite concentrations suggest that ORP2 may negatively regulate basal DHEA secretion and play a key role in both constitutive and cAMP-stimulated cortisol production; therefore, we sought to determine the effect of silencing OPR2 on the expression of genes required for DHEA and cortisol biosynthesis. Real-time RT-PCR revealed that the ORP2 knockdown (ORP2kd) cells marked changes in the mRNA expression of the genes that are required for cortisol biosynthesis (Figure 4A). As we have previously shown (Li D et al., 2007, Lucki NC et al., 2012), activation of the cAMP signal transduction cascade induces the expression of all steroidogenic enzymes required for the conversion of cholesterol to cortisol in wild type H295R cells, with CYP11A1 and CYP11B1 exhibiting a 4.9- and 45.7-fold increase in transcript levels, respectively (Figure 4A). However, both the constitutive and Bt2cAMP-stimulated expression of these two cytochrome P450 isoforms was reduced in the ORP2kd cell line. Moreover, the mRNA expression of CYP21A2 was almost completely diminished in the ORP2kd cell line. In contrast to the negative effect of ORP2 knockdown on most steroidogenic enzymes, silencing ORP2 increased the constitutive expression of CYP17A1 mRNA by 2.7-fold (Figure 4A), which is consistent with the increase in adrenal androgen production in the ORP2kd cell line (Figures 2B and 3). Similarly, the mRNA expression of 3β-HSD type II (HSD3B2) was increased by 1.7-fold in unstimulated ORP2kd cells. Similar findings were observed in the ORP2KD shRNA#2 cell line (Supplementary Figure 2).
Figure 4. ORP2 suppression alters the expression of multiple steroidogenic genes.
(A) Total RNA from H295R wild type and ORP2kd cells (shRNA #1) that were treated with 0.4 mM Bt2cAMP for 24 h was isolated using RNeasy Mini Kit, standardized to 10 ng/µl, and amplified in triplicate using a One-Step SYBR Green RT-PCR Kit for CYP11A1, CYP11B1, CYP11B2, CYP17A1, CYP21A2, HSD3β2 and SF1. Gene expression was normalized to β-actin mRNA content and calculated using the ΔΔ cycle threshold (ΔΔCT) method. Data are graphed as fold change over untreated control and represent the mean ± SEM of five separate experiments, each performed in triplicate. Asterisks and carats denote statistically significant differences when compared to wild type control and wild type Bt2cAMP-treated, respectively. (B) and (C) Wild type and ORP2kd H295R cells were treated with 0.4 mM Bt2cAMP for 48 h, and whole cell lysates harvested and separated by SDS-PAGE followed by western blotting using antibodies against CYP11A1, HSD3B2 (specific band denoted by arrow), CYP17A1, CYP21A2 (specific band denoted by arrow), CYP11B and GAPDH (B) or SF1 and GAPDH (C). Shown in B and C are representative blots for assays that were performed at least four separate times, in at least duplicate. Data graphed in B represent the mean ± SEM of densitometric analysis of protein expression of shRNA#1 normalized to GAPDH. Western blot in panel C is representative data from wild type and ORPkd shRNA#1 H295R cells.
In agreement with the decrease in the mRNA expression of steroidogenic genes, western blotting, analysis of CYP11A1, CYP21A2, and CYP11B1/2 showed that the proteins were all suppressed in both basal and Bt2cAMP-stimulated ORP2kd cells when compared to wild type H295R cells (Figure 4B). CYP17A1 was the only steroidogenic enzyme that exhibited no change in protein expression in the ORP2kd cell lines (shRNA#1 and shRNA#2). Although we observed an increase in the basal expression of HSD3B2 transcript in ORP2kd cells, this was not found at the protein level, where the expression levels of the enzyme were reduced in both basal and Bt2cAMP-stimulated ORP2kd cells. Since the nuclear receptor steroidogenic factor 1 (SF1) is essential for maintaining the transcription of most genes required for steroid hormone biosynthesis (Schimmer BP and White PC, 2010), we examined the expression of SF1 in the ORP2kd cell line. Notably, the protein expression of SF1 was decreased by approximately 38% (Figure 4C).
3.4. ORP2 knockdown impairs efficient cholesterol mobilization
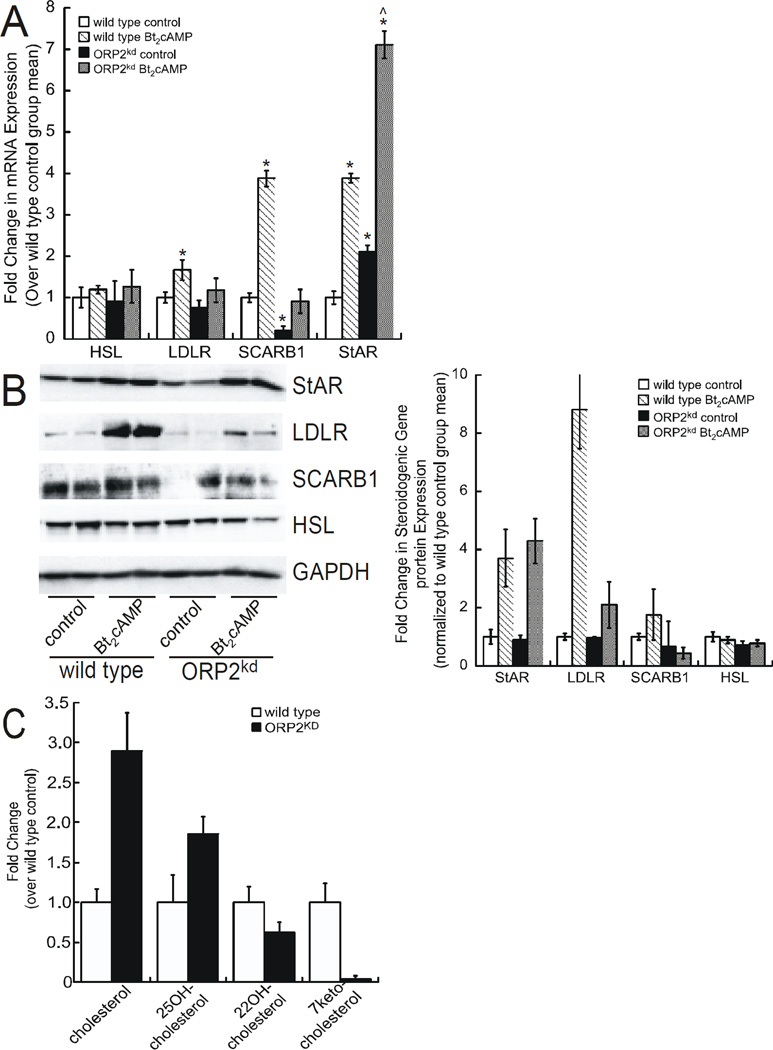
Given that the rate-limiting step in steroidogenesis is the delivery of cholesterol to the inner mitochondrial membrane, we next assessed the effect of ORP2 silencing on the expression of proteins that are involved in cholesterol uptake, transport, and desterification. Of the proteins involved in cholesterol transport, ORP2 silencing had the greatest stimulatory effect on the steroidogenic acute regulatory protein (StAR), a cholesterol binding protein that is required for the movement of cholesterol from the outer mitochondrial membrane to the inner mitochondrial membrane (Stocco, 1996). StAR mRNA expression was increased by 2.1-fold in untreated knockdown cells and by 7.1-fold in Bt2cAMP-stimulated ORP2kd cells (Figure 5A). In contrast, silencing ORP2 prevented the Bt2cAMP-stimulated increase in LDLR mRNA expression and reduced the basal expression of the SCARB1 to 21% of wild type controls. Additionally, the cAMP-stimulated protein expression of LDLR and SCARB1 was reduced in the ORP2kd cells (Figure 5B). No significant effect was seen on the mRNA expression of hormone sensitive lipase (HSL). Consistent with these findings, suppression of ORP2 decreased the cAMP-dependent protein expression of LDLR, but had no significant effect on StAR or HSL (Figure 5B). The lack of an effect on StAR protein is in contrast to the increase in the transcript levels of the gene in the ORP2kd cells.
Figure 5. ORP2 silencing down-regulates genes required for cholesterol mobilization.
(A) Total RNA from control and Bt2cAMP-treated H295R wild type and ORP2kd cells was isolated using RNeasy Mini Kit, standardized to 10 ng/µl, and amplified in triplicate using a One-Step SYBR Green RT-PCR Kit for HSL, LDLR, SCARB1 and StAR. Gene expression was normalized to β-actin mRNA content and calculated using the ΔΔ cycle threshold (ΔΔCT) method. Data are graphed as fold change over untreated control and represent the mean ± SEM of five separate experiments, each performed in triplicate. Asterisks and carats denote statistically significant differences when compared to wild type control and wild type Bt2cAMP-treated, respectively. (B) Wild type and ORP2kd H295R cells were treated with Bt2cAMP (0.4 mM) for 48 h, and whole cell lysates harvested and separated by SDS-PAGE followed by western blotting using antibodies against StAR, LDLR, SRB1, HSL, and GAPDH. Shown are representative blots, where the protein expression of each gene was analyzed in at least duplicate. Data graphed in B represent the mean ± SEM of densitometric analysis of protein expression from four independent experiments. (C) Cholesterol, 25-hydroxycholesterol, 22-hydroxycholesterol, and 7-ketocholesterol levels in wild type and ORP2kd cells were quantified by mass spectrometry as described in the Materials and Methods. Data are expressed as pmol per 5 × 106 million cells.
Despite lack of altered StAR protein expression (Figure 5B), the effect of silencing ORP2 on the production of cortisol suggested that ORP2 might play a role in mediating efficient substrate processing through the steroidogenic pathway. Consistent with this hypothesis, mass spectrometric quantification of cholesterol demonstrated that silencing ORP2 led to a 2.9-fold and 1.9-fold increase in cellular free cholesterol and 25-hydroxycholesterol, respectively (Figure 5C). Concomitant with the increases in cholesterol and 25-hydroxycholesterol, the amounts of 22-hydroxycholesterol and 7-ketocholesterol were reduced, by 36% and 98%, respectively (Figure 5C).
3.5. ORP2 is expressed in the nucleus and cytoplasm of H295R cells
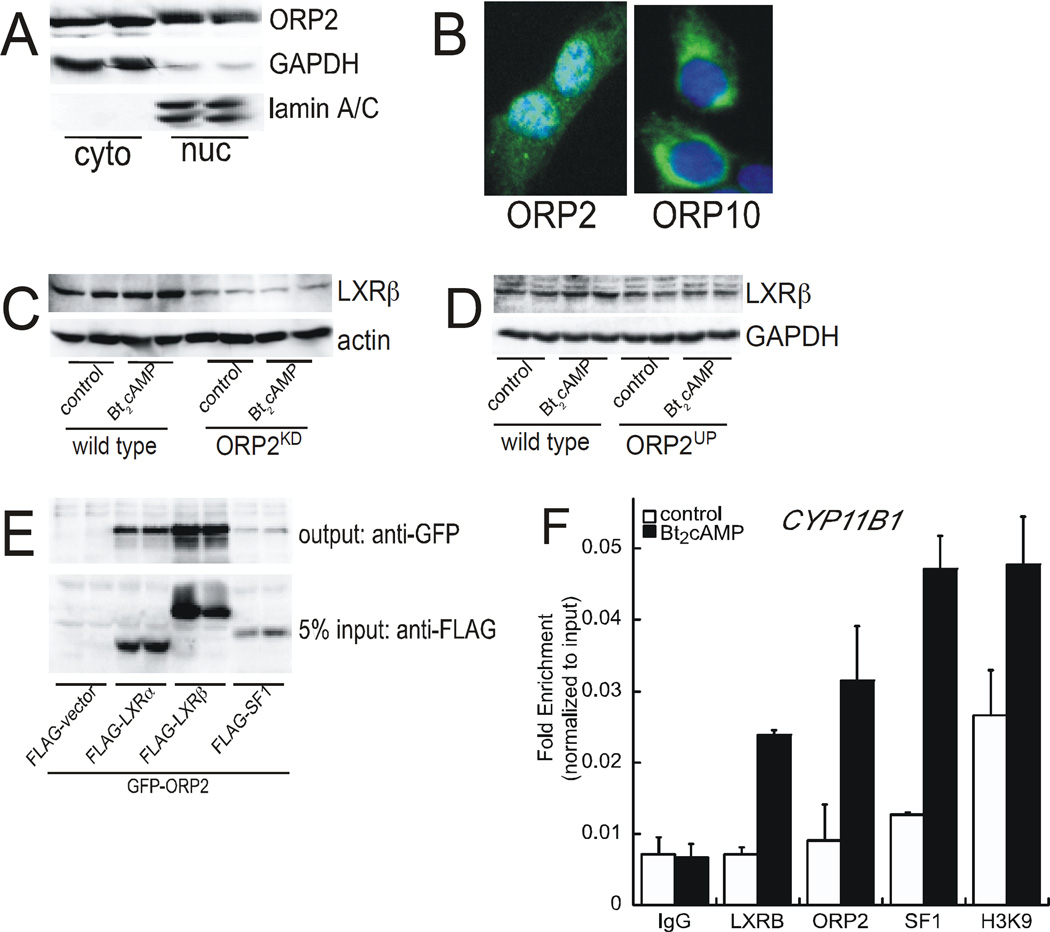
The effect of silencing ORP2 on the protein expression of SF1 (Figure 4C) and the expression of a subset of SF1 target genes (Figure 3B) suggest that the lipid binding protein may act to regulate gene transcription. Further, recent findings demonstrating that ORP1S translocates into the nucleus to facilitate ligand delivery of sterols to the nuclear receptor liver X receptor (LXR) (Lee S et al., 2012), suggest that ORP2 may act in the nuclear compartment. Thus, we examined the expression of ORP2 in cytoplasmic and nuclear fractions isolated from wild type untreated H295R cells and observed that ORP2 is expressed in the nucleus (Figure 6A). Consistent with these western blot data, immunofluorescence staining shows nuclear expression of ORP2 (Figure 6B). In contrast, ORP10 another member of the ORP family that we identified along with ORP2 as a DIAPH1 binding partner (Li D et al., 2013), is exhibits cytoplasmic expression (Figure 6B).
Figure 6. ORP2 binds to LXR and is required for nuclear import.
(A) Cytoplasmic and nuclear fractions were isolated from untreated wild type H295R cells and subjected to SDS-PAGE and western blotting for ORP2 (top), GAPDH (middle), and lamin A/C (bottom). (B) H295R cells plated onto coverslips were incubated with anti-ORP2 antibody or anti-ORP10 and DAPI and the coverslips imaged by immunofluorescence microscopy. (C) Nuclear extracts isolated from wild type or ORP2kd cells that were treated for 2 h with 0.4 mM Bt2cAMP were subjected to SDS-PAGE and western blotting using antibodies against LXRβ (top panel) and β-actin (bottom panel). (D) Wild type and ORP2kd cells were treated with 0.4 mM Bt2cAMP for 48 h and whole cell lysates isolated for SDS-PAGE and western blotting. Top panel LXRβ and bottom panel GAPDH. (E) Co-immunoprecipitation assays were performed as described in the Materials and Methods section by transfecting CV1 cells with expression plasmids for LXRα, LXRβ, SF1, and ORP2. Output blots were incubated with anti-GFP to detect GFPtagged ORP2 and input blots with anti-FLAG for the nuclear receptors. (F) H295R cells were treated with 0.4 mM Bt2cAMP for 1 h, crosslinked with formaldehyde and the isolated chromatin immunoprecipitated with antibodies against LXRβ, ORP2, SF1, or histone H3 acetylated lysine 9. IgG was used as a negative control. Data are graphed as fold enrichment and normalized to the delta Ct values of the input DNA.
The role of other nuclear receptors, notably LXR (Cummins CL and Mangelsdorf DJ, 2006, Cummins CL et al., 2007, Jefcoate CR, 2006, Nilsson M et al., 2007) in adrenocortical steroidogenesis, coupled with the recent findings demonstrating the ORP1S facilitates ligand delivery to LXR (Lee S et al., 2012), prompted us to examine the effect of silencing ORP2 on the nuclear localization of LXR. As shown in Figure 6C, Bt2cAMP stimulated an increase in the nuclear expression of LXRβ in wild type cells, where both basal and Bt2cAMP-stimulated LXRβ nuclear expression was reduced in the ORP2kd cell line. Despite the reduction in nuclear LXRβ expression, the expression of the receptor was not significantly affected in whole cell lysates, suggesting that ORP2 may play a role in nuclear import of LXRβ. Consistent with a role for the lipid binding protein in mediating LXR function, coimmunoprecipitation assays revealed that both LXRα and LXRβ interact (Figure 6E). This is in contrast to the lack of interaction with SF1. Next we performed chromatin immunoprecipitation assays were performed to determine if ORP2 is recruited to the CYP11B1 promoter, one of the steroidogenic genes that exhibited a substantial loss of cAMP-stimulated expression in ORP2kd cells (Figure 4). Consistent with the well-established role of SF1 in regulated CYP11B1 transcription (Morohashi K et al., 1992, Morohashi K et al., 1993, Wang X-L et al., 2000), Bt2cAMP increased the recruitment of the receptor to the promoter, concomitant with an increase in the acetylation of histone H3 lysine 9 (Figure 6F). Interestingly, both LXRβ and ORP2 were also enriched at the CYP11B1 promoter in response to activation of the cAMP signaling pathway.
3.6. ORP2 is required for StAR localization and ABCA1 expression
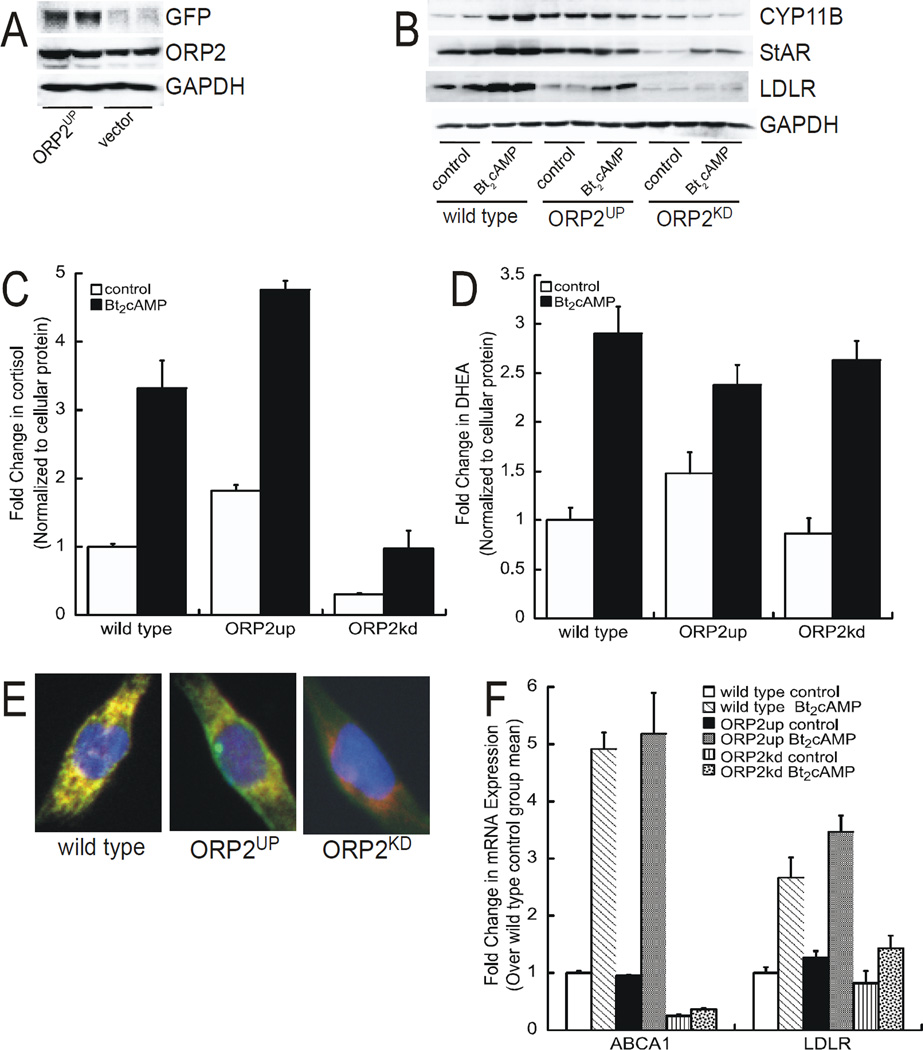
To further probe the role of ORP2 in regulating adrenocortical steroidogenesis we generated an H295R cell line where the protein was overexpressed (Figure 7A). As shown in Figure 7B, overexpression of did not result in a further increase in cAMP-stimulated CYP11B1/2 or StAR protein expression, but did increase the basal expression of CYP11B1/2. However, secretion of cortisol was higher in response to Bt2cAMP in the ORP2 overexpressing (ORP2up) cells compared to wild type (Figure 7C), with no significant effect on DHEA production (Figure 7D). Next we compared the localization of StAR (functional in mitochondria) in the ORP2up and ORP2kd cells and found that StAR colocalized with mitochondria in wild type and ORP2up cells, but did not in the ORP2kd cells (Figure 7E), suggesting that the ORP2 is required for efficient StAR-dependent mobilization of cholesterol. Finally, given the effect of ORP2 silencing on the nuclear expression of LXRβ (Figure 6C) we postulated that other LXR targets may be affected by reduced expression of the sterol binding protein. While the mRNA expression of ABCA1 and LDLR were increased in both wild type and ORP2 overexpressing cells, cAMP-stimulated expression of these plasma membrane receptors was abrogated (Figure 7F).
Figure 7. Overexpression of ORP2 increases cortisol production.
(A) Whole cell lysates isolated from H295R cells that were transfected with a GFP-tagged ORP2 expression plasmid were subjected to SDS-PAGE and western blotting for GFP (top), ORP2 (middle), or GAPDH (bottom). (B) Protein isolated from untreated or Bt2cAMP-treated (48 h, 0.4 mM) wild type, ORP2up, or ORP2kd cells was separated by SDS-PAGE and transferred to PVDF membranes for western blotting. Blots were incubated with antibodies against CYP11B1/2, StAR, LDLR, or GADPH. Shown are representative blots of experiments that were performed on four separate occasions, each time in duplicate. (C) and (D) Media was collected from wild type, ORP2up, or ORP2kd cells that were treated with 0.4 mM Bt2cAMP for 48 h and the levels of cortisol (C) and DHEA (D) determined by ELISA. Amounts of steroid metabolites were normalized to the amount of cellular protein. Graphs represent experiments that were performed on four separate occasions, each time in triplicate. (E) Wild type, ORP2up, or ORP2kd cells were plated onto coverslips and then incubated with anti-StAR and MitoTracker Red. Coverslips were imaged as described in the Materials and Methods section. (F) RNA isolated from wild type, ORP2up, or ORP2kd cells was subjected to quantitative RT-PCR using primers against ABCA1, LDLR, and β-actin. ABCA1 and LDLR mRNA expression was normalized to β-actin mRNA content and calculated using the ΔΔ cycle threshold (ΔΔCT) method. Data are graphed as fold change over untreated control and represent the mean ± SEM of three separate experiments, each performed in triplicate.
4. Discussion
Cortisol biosynthesis necessitates the inter-organelle transport of metabolites between mitochondria and the ER. We have previously shown that activation of the ACTH/cAMP signaling pathway rapidly increases the rate of mitochondrial movement, and that this movement and cortisol production is dependent on microtubules (Li D and Sewer MB, 2010). Efficient mitochondrial movement and cortisol secretion was also dependent on RhoA and the RhoA effector DIAPH1. We subsequently showed that DIAPH1 interacts with several proteins, including kinesin, vimentin, AKAP13 (A-kinase anchoring protein 13; AKAP-Lbc), and ORP2 (Li D et al., 2013). Given that in vitro studies done on other members of the ORP family have found that OSBP and ORP9L mediate phosphatidylinositol 4-phosphate-dependent cholesterol transport between liposomes and suggest that their primary in vivo function is sterol transfer between two intracellular organelles, the Golgi apparatus and the ER (Ngo and Ridgway, 2009), we hypothesized that ORP2 could also participate in the inter-organelle transfer of steroid metabolites. This hypothesis is supported by ELISA data showing that cortisol, but not DHEA production, is suppressed in ORP2kd cells (Figure 2).
Members of the ORP protein family are emerging as regulators of lipid metabolism by serving as transporters between organelles, signaling mediators, and sensors of cellular lipid concentrations (Fairn GD and McMaster CR, 2008, Lehto M et al., 2001, Olkkonen VM and Levine TP, 2004, Ridgway N, 2010). By virtue of the ability of several members of this protein family to bind to cholesterol and oxysterols, ORPs are implicated in cholesterol metabolism (Du X et al., 2011, Wang C et al., 2002). However, ORPs have also been shown to bind to phospholipids, notably phosphatidic acid and phosphatidylinositide phosphates (Stefan CJ et al., 2011, Xu Y et al., 2001). Several ORPs, including ORP2, bind to 25-hydroxycholesterol in in vitro assays (Suchanek M et al., 2007). ORP2 also binds to 22(R)-hydroxycholesterol, cholesterol, and 7-ketocholesterol, with 22(R)-hydroxycholesterol having the highest affinity for the lipid binding protein (Hynynen R et al., 2009). Significantly, silencing ORP2 leads to a reduction in the cellular levels of oxysterols that have been shown to bind to the protein, including 7-ketocholesterol and 22-hydroxycholesterol (Figure 5C), suggesting a feedback mechanism whereby the concentrations of ligands are regulated in response to the expression levels of the cognate lipid binding protein.
The structures of several ORP homologs in the Osh family have been solved, with some structures revealing a role for the proteins in binding phosphatidylinositol phosphates (Kajwara K et al., 2014, Mesmin B et al., 2013, Moser von Filseck J et al., 2015, Moser von Filseck J et al., 2014, Moser von Filseck J et al., 2015, Tong J et al., 2013, Weber-Boyvat M et al., 2015). Notably, Osh6 and Osh7 have been shown to bind to phosphatidylserine (Maeda K et al., 2013). Structural studies of a yeast ORP homolog (known as Osh4 and Kes1) support a mechanism wherein sterols bind in a hydrophobic tunnel and phospholipids interact on the surface of the protein, probably to facilitate membrane docking and sterol exchange (Im YJ et al., 2005). Data indicate that phosphatidylinositol-4-phosphate and sterol bind to the same cavity, leading to a mechanism whereby Osh4 exchanges sterol for phosphatidylinositol-4-phosphate to enable transport between membranes (de Saint-Jean M et al., 2011). Significantly, while studies of the interaction between ORPs and lipids have focused on sterols and phospholipids, the x-ray structure of Osh4 reveals that the binding of sterols to the hydrophobic pocket does not require direct protein-lipid interactions, but instead depends on interactions between water molecules and the hydroxyl groups at carbons 3, 7, and 25 (Im YJ et al., 2005). Moreover, recent structural studies have reported that Osh4 binds to 16, 22-diketocholesterol (Koag MC et al., 2013). Given the absence of direct contact sites between Osh4 and sterols, particularly at the 3-hydoxyl group, we initially postulated that ORP2 binds to 11-deoxycortisol and facilitates the delivery of this substrate from ER to mitochondria. However, rather than an accumulation of 11-deoxycortisol in ORP2kd cells, mass spectrometric analysis of steroid metabolites revealed decreases in several metabolites upstream of 11-deoxycortisol, notably progesterone (Figures 3A and 3B). These findings are consistent with a role for ORP2 in modulating the HSD3B2-catalyzed conversion of pregnenolone and 17-hydroxypregnenolone to progesterone and 17-hydroxyprogesterone, respectively. Although there is support for a role for lipid binding proteins in facilitating cellular processes by selectively targeting substrates to receptors and enzymes (Lathe R and Kotelevtsev Y, 2014), a role for ORP2 in modulating HSD3B2 function is an area of ongoing investigation.
We also unexpectedly find that the silencing of ORP2 primarily decreases the constitutive expression of steroidogenic genes (Figure 4A). Most steroidogenic genes, except CYP21A2, are responsive to stimulation with Bt2cAMP, albeit at a lower magnitude of induction. However, with the exception of CYP17A1, the cAMP-dependent protein expression of the steroidogenic enzymes was significantly reduced (Figure 4B). The decreased expression of most steroidogenic enzymes in the ORP2kd cell line could be due to a down-regulation in the protein expression of the nuclear receptor SF1 (Figure 4C). However, the lack of an effect of reduced SF1 expression on CYP17A1 and StAR, both of which are SF1 targets, suggests that ORP2 may act in a gene-specific manner, or that the reduction in SF1 protein expression may not elicit a direct effect on steroidogenic gene transcription. It is equally plausible that ORP2 may regulate another transcription factor that selectively induces the expression of CYP11A1, CYP11B1, and CYP21A2. Since LXR has been implicated in adrenocortical steroidogenesis (Cummins CL et al., 2007), we investigated the role of ORP2 in regulating the function of this nuclear receptor in the H295R cell line, and found that while the levels of LXRβ were unchanged in whole cell lysates (Figure 6D), the nuclear expression of LXRβ was reduced in cells where the expression of ORP2 was silenced (Figure 6C). We also show that LXRα and LXRβ interact with ORP2 (Figure 6E). These findings are in agreement with the findings of Lee and colleagues demonstrating that LXR binds to ORP1S (Lee S et al., 2012). Consistent with reduced LXRβ nuclear expression, the transcript levels of known LXR targets are reduced in the ORP2 knockdown cell line (Figure 7F).
Given that SF1 is predominantly expressed in the nucleus, the mechanism by which ORP2-dependent lipid sensing regulates SF1 protein expression is unclear. However, we show herein that ORP2 is expressed in both the nucleus and cytoplasm of H295R adrenocortical cells (Figure 6A). Thus, nuclear ORP2 and/or the lipid that it binds to may regulate SF1 stability, and by extension the expression of select SF1 target genes. Interestingly, overexpression of a variant of ORP1, ORP1L, increases the transactivation of the nuclear receptor LXR (Johansson M et al., 2003). Moreover, as mentioned above, recent studies by Lee and colleagues, demonstrate that sterols such as 22(R)-hydroxycholesterol, promote the nuclear import of ORP1S, which binds to and facilitates activation of the apoE gene (Lee S et al., 2012). Of note, initial studies on ligand binding to SF1 identified oxysterols as molecules that increase the activity of the receptor (Lala DS et al., 1997). While the role of oxysterols as bonafide ligands for the receptor has since been disputed (Mellon SH and Bair SR, 1998), our findings suggest that ORP2 may play a role in oxysterol-dependent regulation of SF1 function, perhaps by regulating the stability of the receptor. Studies are underway to identify the specific lipid to which ORP2 binds in adrenocortical cells, and to define the role of that lipid in controlling the protein expression of SF1.
The trafficking of esterified cholesterol and subsequent storage into lipid droplets is mediated by the integral membrane proteins acyl-CoA:cholesterol acyltransferase (ACAT). Moreover, cleavage of these esters by hormone sensitive lipase (HSL) in order to form free cholesterol capable of transportation to the mitochondrial membrane (Chang, 2000, Ikonen, 2008, Thiele, 2008). Previous studies have identified ORP2 as localizing on lipid droplets and capable of ligand binding 22(R)-hydroxycholesterol, where the overexpression of ORP2 in A431 cells led to reduced ACAT activity and cholesterol esterification as well as an increase on cholesterol efflux, and the silencing of ORP2 decreased the rate of triglyceride hydrolysis under lipid depletion conditions (Hynynen R et al., 2009). Our studies provide further support for the role of ORP2 in regulating cholesterol homeostasis, where silencing ORP2 leads to the accumulation of cellular cholesterol (Figure 5C). Notably, vimentin another protein which like ORP2 associates with DIAPH1 (Li D et al., 2013), plays an integral role in lipid droplet formation and the movement of cholesterol to mitochondria in adrenocortical cells (Shen W-J et al., 2012). The fact that ORP2kd cells produce adrenal androgens indicate that while cholesterol trafficking is not completely disrupted, cholesterol accumulation in the knockdown cell line suggests that cholesterol mobilization is not efficiently coupled. This hypothesis is further supported by our finding that the mitochondrial localization of StAR is reduced in the ORP2 knockdown cell line (Figure 7E). Taken together, our data lend support for roles for ORP2 in regulating LXR nuclear function and in the transport of cholesterol during steroid hormone biosynthesis. We conclude that ORP2 is critical for assuring optimal glucocorticoid output and posit that aberrant expression and/or function of this lipid binding protein is may contribute to disease states associated with adrenocortical dysfunction.
Supplementary Material
HIGHLIGHTS.
Silencing ORP2 results in cellular cholesterol accumulation.
Depletion of ORP2 reduces cortisol biosynthesis, without decreasing adrenal androgen production.
ORP2 binds to LXR in the nucleus of adrenocortical cells.
ORP2 acts as a novel lipid sensor in the adrenal cortex.
Acknowledgments
Funding support: NIH/NIDDK DK094151.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Chang C, Sakashita N, Ornvold K, Lee O, Chang E, Dong R, Lin S, Lee CG, Strom S, Kashyap R, Fung J, Farese R, Patoiseau JF, Delhon A, Chang T. Immunological quantitation and localization of ACAT-1 and ACAT-2 in human liver and small intestine. Journal of Biological Chemistry. 2000;275:28083–28092. doi: 10.1074/jbc.M003927200. [DOI] [PubMed] [Google Scholar]
- Cummins CL, Mangelsdorf DJ. Liver X receptors and cholesterol homeostasis: spotlight on the adrenal gland. Biochem Soc Trans. 2006;34:1110–1113. doi: 10.1042/BST0341110. [DOI] [PubMed] [Google Scholar]
- Cummins CL, Volle DH, Zhang Y, McDonald JG, Sion B, Lefrancois-Martinez AM, Caira F, Veyssiere G, Mangelsdorf DJ, Lobaccaro JM. Liver X receptors regulate adrenal cholesterol balance. J Clin Invest. 2007;116:1902–1912. doi: 10.1172/JCI28400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammer EB, Leon A, Sewer MB. Coregulator exchange and sphingosine-sensitive cooperativity of steroidogenic factor-1, general control nonderepressed 5, p54, and p160 coactivators regulate cyclic adenosine 3',5'-monophosphate-dependent cytochrome P450c17 transcription rate. Mol Endocrinol. 2007;21:415–438. doi: 10.1210/me.2006-0361. [DOI] [PubMed] [Google Scholar]
- de Saint-Jean M, Delfosse V, Douguet D, Chicanne G, Payrastre B, Bourguet W, Antonny B, Drin G. Osh4p exchanges sterols for phosphatidyl 4-phosphate between lipid bilayers. J Cell Biol. 2011;195:965–978. doi: 10.1083/jcb.201104062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Kumar J, Ferguson C, Schulz TA, Ong YS, Hong W, Prinz WA, Parton RG, Brown AJ, Yang H. A role for oxysterol-binding protein-related protein 5 in endosomal cholesterol trafficking. J Cell Biol. 2011;192:121–135. doi: 10.1083/jcb.201004142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairn GD, McMaster CR. Emerging roles of the oxysterol-binding protein family in metabolism, transport, and signaling. Cell Mol Sci. 2008;65:228–236. doi: 10.1007/s00018-007-7325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairn GD, McMaster CR. Emerging roles of the oxysterol-binding protein family in metabolism, transport, and signaling. Cell Mol Life Sci. 2008;65:228–236. doi: 10.1007/s00018-007-7325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaikwad NW. Ultra performance liquid chromatography-tandem mass spectrometry method for profiling of steroid metabolome in human tissue. Anal Chem. 2013;85:4951–4960. doi: 10.1021/ac400016e. [DOI] [PubMed] [Google Scholar]
- Hynynen R, Suchanek M, Spandl J, Back N, Thiele C, Olkkonen VM. OSBP-related protein 2 is a sterol receptor on lipid droplets that regulates the metabolism of neutral lipids. J Lipid Res. 2009;50:1305–1315. doi: 10.1194/jlr.M800661-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakela R, Tanhuanpaa K, Laitinen S, Somerharju P, Olkkonen VM. Overexpression of OSBP-related protein 2 (ORP2) induces changes in cellular cholesterol metabolism and enhances endocytosis. Biochem J. 2005;390:273–283. doi: 10.1042/BJ20042082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynynen R, Suchanek M, Spandl J, Back N, Thiele C, Olkkonen VM. OSBP-related protein 2 is a sterol receptor on lipid droplets that regulates the metabolism of neutral lipids. Journal of Lipid Research. 2009;50:1305–1315. doi: 10.1194/jlr.M800661-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonen E. Cellular cholesterol trafficking and compartmentalization. Nature Reviews / Molecular Cell Biology. 2008;9:125–138. doi: 10.1038/nrm2336. [DOI] [PubMed] [Google Scholar]
- Im YJ, Raychaudhuri S, Prinz WA, Hurley JH. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature. 2005;437:154–158. doi: 10.1038/nature03923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen M, Ohsaki Y, Rega L, Bittman R, Olkkonen VM, Ikonen E. Role of ORPs in sterol Transport from plasma membrane to ER and lipid droplets in mammalian cells. Traffic. 2011;12:218–231. doi: 10.1111/j.1600-0854.2010.01142.x. [DOI] [PubMed] [Google Scholar]
- Jefcoate CR. Liver X receptor opens a new gateway to StAR and to steroid hormones. J Clin Invest. 2006;116:1832–1835. doi: 10.1172/JCI29160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M, Bocher V, Lehto M, Chinetti G, Kuismanen E, Ehnholm C, Staels B, Olkkonen VM. The Two Variants of Oxysterol Binding Protein-related Protein-1 Display Different Tissue Expression Patterns, Have Different Intracellular Localization, and Are Functionally Distinct. Mol Biol Cell. 2003;14:903–915. doi: 10.1091/mbc.E02-08-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajwara K, Ikeda A, Aguilera-Romero A, Castillon GA, Kagiwada S, Hanada K, Riezman H, Muñiz M, Funato K. Osh proteins regulate COPII-mediated vesicular transport of ceramide from the endoplasmic reticulum in budding yeast. J Cell Sci. 2014;127:376–387. doi: 10.1242/jcs.132001. [DOI] [PubMed] [Google Scholar]
- Kakela R, Tanhuanpaa K, Laitinen S, Somerharju P, Olkkonen VM. Overexpression of OSBP-related protein 2 (ORP2) in CHO cells induces alterations of phospholipid species composition. Biochem Cell Biol. 2005;83:677–683. doi: 10.1139/o05-056. [DOI] [PubMed] [Google Scholar]
- Koag MC, Cheun Y, Kou Y, Ouzan-Shubeita H, Min K, Monzingo AF, Lee S. Synthesis and structure of 16, 22-diketocholesterol bound to oxysterol-binding protein Osh4. Steroids. 2013;78:938–944. doi: 10.1016/j.steroids.2013.05.016. [DOI] [PubMed] [Google Scholar]
- Laitinen S, Lehto M, Lehtonen S, Hyvarinen K, Heino S, Lehtonen E, Ehnholm C, Ikonen E, Olkkonen VM. ORP2, a homolog of oxysterol binding protein, regulates cellular cholesterol metabolism. J Lipid Res. 2002;43:245–255. [PubMed] [Google Scholar]
- Lala DS, Syka PM, Lazarchik SB, Mangelsdorf DJ, Parker KL, Heyman RA. Activation of the orphan nuclear receptor steroidogenic factor 1 by oxysterols. Proc Natl Acad Sci U S A. 1997;94:4895–4900. doi: 10.1073/pnas.94.10.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathe R, Kotelevtsev Y. Steroid signaling: Ligand-binding promiscuity, molecular symmetry, and the need for gating. Steroids. 2014;82:14–22. doi: 10.1016/j.steroids.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Lee S, Wang P-Y, Jeong Y, Mangelsdorf DJ, Anderson RGW, Michaely P. Sterol-dependent nuclear import of ORP1S promotes LXR regulated transactivation of apoE. Exp Cell Res. 2012;318:2128–2142. doi: 10.1016/j.yexcr.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehto M, Laitinen S, Chinetti G, HJohansson M, Ehnholm C, Staels B, Ikonen, Olkkonen VM. The OSBP-related protein family in humans. J Lipid Res. 2001;42:1203–1213. [PubMed] [Google Scholar]
- Lehto M, Olkkonen VM. The OSBP-related proteins: a novel protein family involved in vesicle transport, cellular lipid metabolism, and cell signalling. Biochim Biophys Acta. 2003;1631:1–11. doi: 10.1016/s1388-1981(02)00364-5. [DOI] [PubMed] [Google Scholar]
- Lehto M, Tienari J, Lehtonen S, Lehtonen E, Olkkonen V. Subfamily III of mammalian oxysterol-binding protein (OSBP) homologues: the expression and intracellular localization of ORP3, ORP6 and ORP7. Cell Tissue Research. 2004;315:39–57. doi: 10.1007/s00441-003-0817-y. [DOI] [PubMed] [Google Scholar]
- Li D, Dammer EB, Lucki NC, Sewer MB. cAMP-stimulated phosphorylation of diaphanous 1 regulates protein stability and interaction with binding partners in adrenocortical cells. Mol Biol Cell. 2013;24:848–857. doi: 10.1091/mbc.E12-08-0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Sewer MB. RhoA and DIAPH1 Mediate Adrenocorticotropin-Stimulated Cortisol Biosynthesis by Regulating Mitochondrial Trafficking. Endocrinology. 2010;151:4313–4323. doi: 10.1210/en.2010-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Urs AN, Allegood J, Leon A, Merrill AH, Jr, Sewer MB. cAMP-Stimulated Interaction Between Steroidogenic Factor-1 and Diacylglycerol Kinase-Facilitates Induction of CYP17. Mol Cell Biol. 2007;27:6669–6685. doi: 10.1128/MCB.00355-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Urs AN, Allegood J, Leon A, Merrill AH, Jr, Sewer MB. Cyclic AMP-stimulated interaction between steroidogenic factor 1 and diacylglycerol kinase theta facilitates induction of CYP17. Mol Cell Biol. 2007;27:6669–6685. doi: 10.1128/MCB.00355-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucki NC, Bandyopadhyay S, Wang E, Merrill AH, Sewer MB. Acid ceramidase (ASAH1) is a global regulator of steroidogenic capacity and adrenocortical gene expression. Mol Endocrinol. 2012;26:228–243. doi: 10.1210/me.2011-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K, Anand K, Chiapparino A, Kumar A, Poletto M, Kaksonen M, Gavin AC. Interactome map uncovers phosphatidyl serine transport by oxysterol-binding proteins. Nature. 2013;501:257–261. doi: 10.1038/nature12430. [DOI] [PubMed] [Google Scholar]
- Mellon SH, Bair SR. 25-Hydroxycholesterol is not a ligand for the orphan nuclear receptor steroidogenic factor-1 (SF-1. Endocrinology. 1998;139:3026–3029. doi: 10.1210/endo.139.6.6129. [DOI] [PubMed] [Google Scholar]
- Mesmin B, Antonny B, Drin G. Insights into the mechanisms of sterol transport between organelles. Cell Mol Life Sci. 2013;70:3405–3421. doi: 10.1007/s00018-012-1247-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K, Honda S, Inomata Y, Handa H, Omura T. A common trans-acting factor, Ad4-binding protein, to the promoters of steroidogenic P-450s. J. Biol Chem. 1992;267:17913–17919. [PubMed] [Google Scholar]
- Morohashi K, Zanger UM, Honda S, Hara M, Waterman MR, Omura T. Activation of CYP11A and CYP11B gene promoters by the steroidogenic cell-specific transcription factor, Ad4BP. Mol. Endocrinol. 1993;7:1196–1204. doi: 10.1210/mend.7.9.8247022. [DOI] [PubMed] [Google Scholar]
- Moser von Filseck J, Copič A, Delfosse V, Vanni S, Jackson CL, Bourguet W, Drin G. INTRACELLULAR TRANSPORT. Phosphatidylserine transport by ORP/Osh proteins is driven by phosphatidylinositol 4-phosphate. Science. 2015;349:432–436. doi: 10.1126/science.aab1346. [DOI] [PubMed] [Google Scholar]
- Moser von Filseck J, Mesmin B, Bigay J, Antonny B, Drin G. Building lipid 'PIPelines' throughout the cell by ORP/Osh proteins. Biochem Soc Trans. 2014;42:1465–1470. doi: 10.1042/BST20140143. [DOI] [PubMed] [Google Scholar]
- Moser von Filseck J, VAnni S, Mesmin B, Antonny B, Drin G. A phosphatidylinositol-4-phosphate powered exchange mechanism to create a lipid gradient between membranes. Nat Commun. 2015;6:6671. doi: 10.1038/ncomms7671. [DOI] [PubMed] [Google Scholar]
- Ngo M, Ridgway ND. Oxysterol Binding Protein-related Protein 9 (ORP9) Is a Cholesterol Transfer Protein That Regulates Golgi Structure and Function. Molecular Biology of the Cell. 2009;20:1388–1399. doi: 10.1091/mbc.E08-09-0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo MH, Colbourne TR, Ridgway N. Functional implications of sterol transport by the oxysterol-binding protein gene family. Biochem J. 2010;429:13–24. doi: 10.1042/BJ20100263. [DOI] [PubMed] [Google Scholar]
- Nilsson M, Stulnig TM, Lin CY, Yeo AL, Nowotny P, Liu ET, Steffensen KR. Liver X receptors regulate adrenal steroidogenesis and hypothalamic-pituitary-adrenal feedback. Mol Endocrinol. 2007;21:126–137. doi: 10.1210/me.2006-0187. [DOI] [PubMed] [Google Scholar]
- Olkkonen VM, Levine TP. Oxysterol binding proteins: in more than one place at one time? Biochem Cell Biol. 2004;82:87–98. doi: 10.1139/o03-088. [DOI] [PubMed] [Google Scholar]
- Olkkonen VM, Li S. Oxysterol-binding proteins: Sterol and phosphoinositide sensors coordinating transport, signaling, and metabolism. Prog Lipid Res. 2013;52:529–538. doi: 10.1016/j.plipres.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Olkkonen VM, Levine T. Oxysterol binding proteins: in more than one place at one time? Biochem. Cell Biol. 2004;82:87–98. doi: 10.1139/o03-088. [DOI] [PubMed] [Google Scholar]
- Pichler H, Gaigg B, Hrastnik C, Achleitner G, Kohlwein SD, Zellnig G, Perktold A, Daum G. A subfraction of the yeast endoplasmic reticulum associates with the plasma membrane and has a high capacity to synthsize lipids. European Journal of Biochemistry. 2001;268:2351–2361. doi: 10.1046/j.1432-1327.2001.02116.x. [DOI] [PubMed] [Google Scholar]
- Ridgway N. Oxysterol-binding proteins. Subcell Biochem. 2010;51:159–182. doi: 10.1007/978-90-481-8622-8_6. [DOI] [PubMed] [Google Scholar]
- Ridgway ND, Lagace TA, Cook HW, Byers DM. Differential effects of sphingomyelin hydrolysis and cholesterol transport on oxysterol-binding protein phosphorylation and Golgi localization. J Biol Chem. 1998;273:31621–31628. doi: 10.1074/jbc.273.47.31621. [DOI] [PubMed] [Google Scholar]
- Ridgway ND, Dawson P, Ho YK, Brown M, Goldstein J. Translocation of Oxysterol Binding Protein to Golgi Apparatus Triggered by Ligand Binding. Journal of Cell Biology. 1992;116:307–319. doi: 10.1083/jcb.116.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmer BP, White PC. Minireview: steroidogenic factor 1: its roles in differentiation, development, and disease. Mol Endocrinol. 2010;24:1322–1337. doi: 10.1210/me.2009-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz T, Choi M, Raychaudhuri S, Mears J, Gihrlando R, Hinshaw J, Prinz W. Lipid-regulated sterol transfer between closely apposed membranes by oxysterol-binding protein homologues. Journal of Cell Biology. 2009;197:889–903. doi: 10.1083/jcb.200905007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W-J, Zaidi SK, Patel S, Cortez Y, Ueno M, Azhar R, Azhar S, Kraemer FB. Ablation of vimentin results in defective steroidogenesis. Endocrinology. 2012;153:3249–3257. doi: 10.1210/en.2012-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan CJ, Manford AG, Baird D, Yamada-Hanff, Mao Y, Emr SD. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 2011;144:389–401. doi: 10.1016/j.cell.2010.12.034. [DOI] [PubMed] [Google Scholar]
- Stocco D, Clark B. Regulation of the Acute Production of Steroids in Steroidogenic Cells. Endocrine Reviews. 1996;17:221–244. doi: 10.1210/edrv-17-3-221. [DOI] [PubMed] [Google Scholar]
- Storey M, Byers D, Cook H, Ridgway ND. Cholesterol regulates oxysterol binding protein (OSBP) phosphorylation and Golgi localization in Chinese hamster ovary cells: correlation with stimulation of sphingolmyelin synthesis by 25-hydroxycholesterol. Biochemical Journal. 1998;336:247–256. doi: 10.1042/bj3360247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchanek M, Hynynen R, Wohlfahrt G, Lehto M, Johansson M, Saarinen H, Radzikowska A, Thiele C, Olkkonen VM. The mammalian oxysterol-binding protein-related proteins (ORPs) bind 25-hydroxycholesterol in an evolutionarily conserved pocket. Biochem J. 2007;405:473–480. doi: 10.1042/BJ20070176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchanek M, Hynynen R, Wohlfahrt G, Lehto M, Johansson M, Saarinen H, Radzikowsz A, Thiele C, Olkkonen V. The mammalian oxysterol-binding protein-related proteins (ORPs) bind 25-hydroxycholesterol in an evolutionarily conserved pocket. Biochemical Journal. 2007;405:473–480. doi: 10.1042/BJ20070176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele C, Spandl J. Cell biology of lipid droplets. Current opinion in cell biology. 2008;20:378–385. doi: 10.1016/j.ceb.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Tong J, Yang H, Yang H, Eom SH, Im YJ. Structure of Osh3 reveals a conserved mode of phosphoinositide binding in oxysterol-binding proteins. Structure. 2013;21:1203–1213. doi: 10.1016/j.str.2013.05.007. [DOI] [PubMed] [Google Scholar]
- Wang C, JeBailey L, Ridgway N. Oxytsterol-binding-protein (OSBP)-related protein 4 binds 25-hydroxycholesterol and interacts with vimentin intermediate filaments. Biochem J. 2002;361:461–472. doi: 10.1042/0264-6021:3610461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X-L, Basett M, Zhang Y, Yin S, Clyne C, White PC, Rainey WE. Transcriptional regulation of human 11beta-hydroxylase (hCYP11B1) Endocrinology. 2000;141:3587–3594. doi: 10.1210/endo.141.10.7689. [DOI] [PubMed] [Google Scholar]
- Weber-Boyvat M, Kentala H, Peränen J, Olkkonen VM. Ligand-dependent localization and function of ORP-VAP complexes at membrane contact sites. Cell Mol Life Sci. 2015;72:1967–1987. doi: 10.1007/s00018-014-1786-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Edwards MA, Ahlem C, Kennedy M, Cohen A, Gomez-Sanchez CE, Rainey WE. The effects of ACTH on steroid metabolomic profiles in human adrenal cells. J Endocrinol. 2011;209:327–335. doi: 10.1530/JOE-10-0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Liu Y, Ridgway ND, McMaster CR. Novel members of the human oxysterol-binding protein family bind phospholipids and regulate vesicular transport. J Biol Chem. 2001;276:18407–18414. doi: 10.1074/jbc.M101204200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.