Abstract
Sepsis following surgical injury remains a growing and worrisome problem following both emergent and elective surgery. Although early resuscitation efforts and prompt antibiotic therapy have improved outcomes in the first 24–48 hours, late onset sepsis is now the most common cause of death in modern intensive care units. This time shift may be, in part, a result of prolonged exposure of the host to the stressors of critical illness which, over time, erode the health promoting intestinal microbiota and allow for virulent pathogens to predominate. Colonizing pathogens can then subvert the immune system and contribute to the deterioration of the host response. Here we posit that novel approaches integrating the molecular, ecological and evolutionary dynamics of the evolving gut microbiome/pathobiome during critical illness are needed to understand and prevent the late onset sepsis that develops following prolonged critical illness.
Keywords: Late onset sepsis, intestinal microbiota, pathobiota, pathoadaptive immune response, non- resolving inflammation
Sepsis incidence and severity is increasing among surgical patients
Sepsis following surgical injury is a growing problem with persistently high mortality rates and long-term complications [1,2]. A 2009 report from investigators at Massachusetts General Hospital analyzing 2,039,776 admissions from the Nationwide Inpatient Sample, the largest in-patient dataset available in the United States, indicated that severe sepsis has increased following elective surgery from 0.3% to 0.9% [3]. Although in- hospital mortality rates declined from 44 to 34% during the 10 year interval, postoperative and post- injury sepsis continue to impose significant disease burden, costs and high death rates despite advances in care and newer antibiotics [3,4]. Across virtually all surgical injuries (major surgical intervention, trauma, burns) the majority of sepsis- related deaths have now shifted to occur late in the course of hospitalization and have been shown to be associated with immune-suppression and multi- drug resistant pathogens [5–9]. We now face a new challenge in the care of surgical patients, “late onset sepsis” which occurs as a result of more invasive and aggressive surgical therapies being applied to an ever- aging population subjected to extreme medical and surgical interventions. Not unexpectedly, the microbes that are most commonly associated with late onset post-injury sepsis include Staphylococcus spp., Pseudomonas aeruginosa, Acinetobacter baumannii, Enterococcus faecalis, and Candida albicans to name a few. In many cases the mere presence of the pathogen itself is an independent predictor of mortality regardless of its location, density, or genotype [10]. In this clinical scenario, the pathogenesis of infection- related sepsis is framed as a simple matter of an exhausted immune system unable to contain a given microbial burden [11]. Yet microbial burden is often imprecisely characterized and defined primarily by the isolation of a given pathogen cultured from a site that is considered to be normally sterile (lung, urine, central line, blood) that contemporaneously correlates with the onset of sepsis and progressive organ failure [4]. This definition falls short in establishing the causality between the originally identified pathogen and the progression or resolution of the septic response [12]. When the organism from the original site of isolation is no longer able to be cultured, yet the patient’s organ failure continues to progress, current dogma explains this as “runaway inflammation” – a process no longer driven by the initiating stimulus (i.e. the pathogen) but now controlled by programmed inflammation [13]. Conversely when organ failure resolves and the inciting pathogen is cleared from its original site of culture, causality is invoked. Yet, over the long course of care of a critically ill patient undergoing dialysis, multiple operations, ventilator support, etc, clear evidence for either scenario is often weak at best.
More precisely defining the putative pathogen or pathogen community that initiates and drives late onset sepsis and the attendant organ failure that ensues continues to present a major challenge. Two isolates of the same microbial species can differ widely in behavior as they may have acquired new genes over their short life- history [14]. Bacteria can express highly variable phenotypes depending on how the local environment has shaped their genotype and thus, “microbial burden” can no longer be simply defined by microbial abundance or species alone [15–21]. As such the potential threat of microbes present on mucosal surfaces such as the gut, lung, and wound cannot be understood by their mere isolation and antibiotic sensitivities [22]. At a given colonization site microbe- microbe interactions and host- microbe interactions are being constantly shaped by one another, the net result of which leads to emergent properties in both bacteria and host cell responses which themselves exert downward causality (Figure 1). If microbial burden were to be defined by the net output of the interactions between the entire body’s biomass of microbes (skin, gut, and lung) and the host’s immune response then the sepsis response could be understood in a more system’s biology manner [23,24]. As microbes respond to host immunity and as host immunity in turn responds to dynamic microbial phenotype expression, new states of equilibrium emerge and the sepsis course can be better modeled. Koch’s postulates no longer suffice in human critical illness and have now been replaced by the molecular Koch’s postulates to incorporate the dynamic virulence expression in bacteria that occurs in response to a specific local context [15]. In this review we posit that microbial burden, defined by number of microbes, their community structure, and the context- dependent virulence response that occurs at all points at which they engage the immune and inflammatory system, has a profound and underappreciated influence on the course and outcome of surgical injury. This is especially true in the gut, one of the most diverse microbial environments of the human body [25,26]. Bacteria in this environment are likely to face social dilemmas more frequently which then impose strong selection pressure on their growth rates, motility, cooperation by quorum-sensing, and virulence expression [27–29]. The sheer enormity of the available surface area of the gut, its massive concentration of immune cells, and its constant exposure to an infinite number of spatially nested host- pathogen interactions defines the limitless potential for opportunistic pathogens to colonize, inflame, and cause harm to the host. In most cases this occurs without ever isolating the offending pathogen or pathogen community, without bacterial translocation or bacteremia, and without a clear understanding of the mechanisms by which human critical illness and its treatment shift the virulence trajectory of these pathogens to enhance their fitness and subvert the immune system [30–32]. Yet advances in metagenomics, metatranscriptomics, and proteomics have the potential to allow for a more informed interrogation of these events as they are happening [33]. The current challenge is to be able to harness and interpret the massive amount of information generated by such a meta- omics analysis. Finally, the fact that the most important information on host- pathogen interaction is contained within clinically unavailable spatially nested micro- sites (i.e. ileal crypts, third order bronchi, etc), presents yet another challenge. The use of serum based or whole expelled mucosal material (stool, sputum) is done simply as a matter of convenience and their predictive power has fallen short in a number of studies [4,12]. The future holds great promise that we will be able to understand how the microbial-host interface within nested colonization sites drives immune responsiveness.
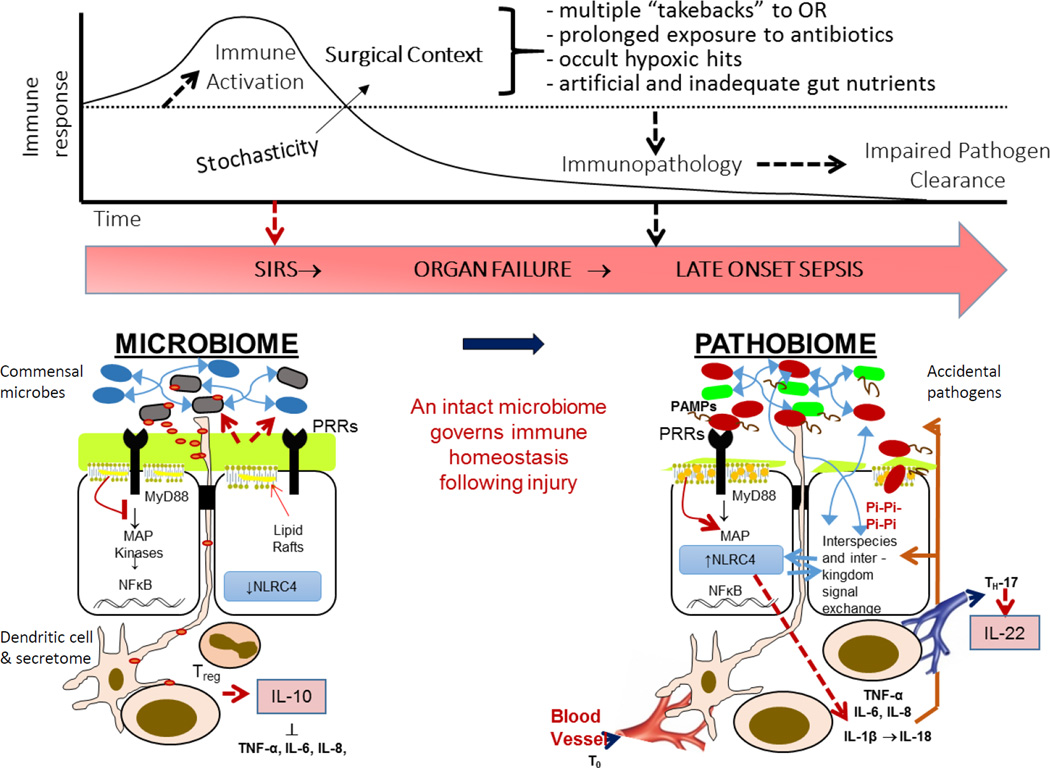
Figure 1. Proposed interactome between the intestinal microbiome versus pathobiome and the immune system.
The often unavoidable transition of the normal gut microbiome to a pathobiome during the course of critical illness has the potential to subvert the immune system when pathogens activate their virulence and express quorum sensing signaling molecules that lead to pathoadaptive inflammation. A major hypothesis to test is whether maintaining the intestinal microbiota through the course of critical illness will more appropriately direct the immune response toward recovery and accelerate organ recovery.
Pi= phosphate, PRR= pathogen recognition molecule, PAMP= pathogen associated molecular pattern.
The disappearing microbiome and its replacement by a pathobiome- role in late onset sepsis
It is now well established that the mammalian intestine harbors a complex microbial community (microbiota) that provides numerous health benefits. In fact recent studies have demonstrated that the normal microbiota are the single most important factor protecting the intestinal mucosal epithelium from pathogen invasion; even more critical than the overlying mucus or immune cells [30,34]. Yet paradoxically, within hours of a sudden insult in both humans and mice, key protective elements of the intestinal microbiota become disrupted both compositionally and functionally [35,36]. These changes are highly predictive of sepsis- associated mortality in critically ill patients [37]. We have recently shown by 16SrRNA analysis and culture, that the gut of critically ill septic patients undergoes a nearly complete ecologic collapse with emergence of ultra-low diversity pathogen communities of two to four multi-drug resistant healthcare associated pathogens [35]. These pathogens can express virulence in response to host factors such as inorganic phosphate depletion or the release of opioids which are known to transduce the virulence circuitry of highly problematic bacteria such as Pseudomonas aeruginosa [20]. Our laboratory has shown that limitations of phosphate availability in the intestine results in induction of P. aeruginosa and other pathogens to express a lethal phenotype by triggering phosphosensory and regulatory pathways that connect directly to virulence pathways that in turn become activated [38]. These pathways are widely conserved across pathogens of clinical relevance to sepsis in surgical patients [39]. Other such environmental cues that trigger pathogen virulence include endogenous and exogenous opioids which have been shown to be increased in human sepsis and play an important role in its pathogenesis [40,41]. We were able to demonstrate in our laboratory that the endogenous k- opioid peptide dynorphin, a host tissue activating cue released into the gut during injury, triggers virulence expression in P. aeruginosa leading to its transformation to a lethal phenotype [42]. Interestingly, there appears to be an important “interference” interplay between incoming host signals such as opioids and how bacterial phosphosensory systems dampen or amplify the transduction of these signals depending on the local concentration of phosphate. For example when local phosphate conditions are abundant, bacteria cancel out or dampen incoming host signals such as opioids. Conversely when local phosphate conditions are depleted, bacteria amplify the opioid signal [43]. These findings speak to a high fidelity fine-tuned virulence response of microbes to local environmental cues whereby when key elemental nutrients such as phosphate are abundant and bacterial growth is supported, bacteria do not respond to stress signals from their host, perhaps reasoning that nutrient availability to support growth trumps the need to express virulence. On the other hand, an environment which lacks phosphate is not likely to be nutrient rich and not supportive of growth and thus triggering virulence to allow for host invasion (or even host death if necessary) makes sense. While it may seem counterintuitive that an intestinal microbe would kill the very host upon which it survival depends, if the host is no longer hospitable to its commensal lifestyle (i.e parenteral nutrition, dialysis, pressor use, multiple antibiotics), killing the host and feeding off its carcass makes sense [44]. Alternatively a strategy to kill the host allows the microbe to jump to a new host as other hosts (animals, flies, worms, ants, etc) feed off the carcass [45].
Bacterial Telesensing- molecular details on the bidirectional chemical dialogue between pathogen and host
Michael Gilmore at Harvard coined the term “telesensing” to describe how bacteria intercept host signals as a mechanism to capture changes in host physiologic state and respond accordingly. Our group was the first to show that intestinal pathogens sense host stress through the release of soluble compounds such as norepinephrine, interferon gamma, dynorphin and end-products of ischemia such as adenosine. We elucidated the molecular pathways by which microbial pathogens dynamically express virulence in response to each of these host factors. We used the prototype opportunistic pathogen, Pseudomonas aeruginosa, as a model pathogen in which to carry out detailed molecular experiments to demonstrate telesensing in response to surgical injury. In the case of interferon gamma (INF-γ), we demonstrated that mice secrete INF-γ into the gut lumen during stress, and that P. aeruginosa, via an outer membrane porin, OprF, binds INF-γ and transduces its quorum sensing signaling system via C4 HSL, a key regulator quorum sensing signaling molecule in P. aeruginosa [46]. Next we demonstrated in mice that intestinal ischemia-reperfusion injury (i.e. intestinal I/R) caused the release of an endogenous κ-opioid peptide dynorphin into the intestinal tract [47]. When we introduced P. aeruginosa into the mouse intestine and cause intestinal I/R, we observed dynorphin to be present within the cytoplasm of intestinal bacteria. Dynorphin is a quaternary amine so it is able to cross membranes quite readily. Reductionist experiments with a synthetic κ-opioid and several reported and mutant strains of P. aeruginosa demonstrated that κ-opioids diffuse into the cytoplasm of P. aeruginosa, bind to the multiple virulence factor regulator MvfR and activate a series of virulence genes that cause P. aeruginosa to express a hypervirulent phenotype leading to disruption of the epithelial barrier, sepsis and mortality in mice. Finally we continued with the intestinal I/R model and performed reductionist experiments to show that in cultured intestinal epithelial cells (Caco-2) exposed to hypoxia and reoxygenation, they release adenosine as a cytoprotectant to bind to the AR2 receptor and preserve their barrier function [48,49]. When cells were co-incubated with P. aeruginosa, we observed that P. aeruginosa took up the adenosine, and via adenosine deaminase, converted it to inosine which directly activated the quorum sensing signaling pathway in P. aeruginosa leading to enhance virulence against the epithelial barrier. The intriguing aspect of this observation was that P. aeruginosa intercepted an epithelial protectant, adenosine that not only deprived epithelial cells of their endogenous mechanism to activate tight junctional proteins and preserve their barrier function, but at the same time served the P. aeruginosa to activate its own virulence mechanism. This evolutionarily contrived process shows the highly sophisticated nature of P. aeruginosa that can understand the host environment and subvert it to its own end for tissues invasion and if needed, for it to kill its host. While it may seem counterintuitive that an intestinal pathogen would want to kill its host, this actually occurs in nature all the time. In the case of critical illness, it could be argued that once a highly virulent pathogen such as P. aeruginosa perceives that its host cannot survive sufficiently to meet its needs, it has the capacity to kill its host, feed off its carcass and jump to a new host during co-predation such as when other animals feed of the dying host. Thus the capacity of opportunistic pathogens to be so finely tuned to perturberances in host physiology should make us take pause when we impose prolonged critical illness on our patients depriving the gut microbiota of food, oxygen, phosphate and other key factors that promote a healthy gut ecosystem. This is a particular problem when multiple antibiotics are used that cause loss of microbiome diversity and allow predator-type pathogens to predominate.
Loss of intestinal microbiota diversity during critical illness is a rapid development and its precise mechanism (besides being due to antibiotic use) remains unknown. Various studies have now documented the loss of the protective microflora during stress [17,37,50]. However, the mechanism for this phenomenon remains unknown. Lactobacilli and anaerobes as well as their metabolites such as the cytoprotective short chain fatty acids, decrease by more than 90% within as little as 6 hours of a major insult, which suggests that it is not simply due to lack of nutrients but rather a result of some type of global chemosignal that shifts the microbiota both compositionally and functionally [36]. As a consequence, acute loss of the colonization resistance of the microbiota results in a greater opportunity for unrestrained growth of coliforms (i.e. Escherichia coli, Klebsiella pneumoniae), gram- positive bacteria (i.e. Enterococcus, Staphylococcus aureus), and fungi to bind pathogen recognition receptors (PRR) and activate the inflammasome [51]. In a teleological sense, this may represent a normal physiologic response of the host to stimulate and inform the immune system to activate compensatory host clearance mechanism. Such a compensatory mechanism may have previously offered a survival advantage to our ancestral hosts when medical care was absent and exposure to hospital associated pathogens did not exist. However today, hospital confinement following injury could be viewed as a “microbial minefield” in which the threat of colonization by highly pathogenic and resistant microbes is omnipresent. As such, the gut immune system may respond in a pathoadaptive manner that can result in non- resolving inflammation and multiple organ failure. To illustrate this, a recent study has shown that immune activation in response to Salmonella enterica infection results in the production of antimicrobial peptides that greatly enhance the competitive advantage of Salmonella enterica by allowing it to obtain nutrients at the expense of the commensal microbiota [52]. The process by which microbes shift their phenotype and develop competitive strategies to overcome the normal microbiota may result in a pathoadaptive immune response. How yet-to-be-identified environmental and host chemosignals drive these emergent phenotypes in the colonizing pathobiome over the course of critical illness remains a major question to address.
There is now compelling evidence that this type of response need not necessarily be initiated and driven by a healthcare associated pathogens (HAPs) per se as even symbiotic (commensal) microbes can be transformed to express a hyper- virulent pro-inflammatory response and thus behave as pathogens (i.e pathobionts) [29]. Multiple lines of evidence now suggest that a microbiont (commensal microbe considered part of the normal microflora) is transformed to a pathobiont in response to host chemosignals released during surgical injury. For example our laboratory demonstrated that commensal E. coli can be transformed to express a pathologic phenotype capable of increased adherence to the intestinal epithelium when they express type 1 fimbriae in response to the host environment of surgical injury [53]. A more recent report in mice demonstrated that E. coli can be transformed in vivo to express a lethal phenotype when it is exposed to both broad spectrum antibiotics and epithelial injury [54]. In this study, mice fed several oral antibiotics were administered dextran sodium sulfate to cause colitis. Within a few days, E. coli symbionts became transformed into multi-drug resistant pathobionts that caused severe lethal gut- derived sepsis via activation of the Naip5-Nlrc4 inflammasome [55]. Along similar lines, our laboratory has shown that limitation of inorganic phosphate availability in the gut shifts intestinal C. albicans to express a lethal phenotype by inducing hyphae formation [21,56]. Thus a more complete understanding of the mechanisms of phase transition from commensal immune-stimulatory organisms to barrier disrupting lethal pathobionts will require more in- depth molecular elucidation.
Yet, in contrast to the mouse gut, the human gut during late onset sepsis harbors a pathobiome consisting of multi-drug resistant healthcare acquired pathogens, beyond commensal E. coli, whose evolutionary trajectories and virulence mechanisms have been shaped not only by their unusual life- histories (passage through multiple hosts and environments), but also more recently by antibiotics, hypoxia, and other environmental cues unique to the acute and chronic human condition [35,57,58]. Here we assert that if we are to more completely elucidate and control the mechanism by which microbial burden overpowers immune clearance mechanism during late sepsis, mouse models must incorporate the unique virulence strategies of the real human pathogens that are present in the gut during the care of the critically ill.
Intriguing human studies from Japan have provided insight into the disappearing microbiome and its replacement by a pathobiome in patients following a variety of sudden insults including trauma [36]. Within as little as 6 hours of a sudden insult (i.e. myocardial infarction, stroke, acute surgical illness, etc.), there is a profound loss of the normal protective microbiota and their important cytoprotective exoproducts such as short chain fatty acids. These studies were performed by culturing the stool of patients at the time they entered the hospital and shortly thereafter. The presence of pathogenic species and a shift to a more basic fecal pH (presumed to be due to loss of short chain fatty acid secretion) correlated to the disappearance of lactobacilli, firmicutes, and other anaerobes. In addition to showing that loss of the protective microbiota and predominance of gamma proteobacteria developed acutely and over the course of confinement, the investigators importantly demonstrated that this microbial pattern and fecal pH predicted the development of the systemic inflammatory response syndrome and mortality [36]. Although much remains to be understood by these findings, they provide a clear message that our health promoting microbial partners disappear through the course of injury and may have a profound effect on the outcome of sepsis. The normal microbiota not only provide colonization resistance against invading pathogens, but they also inform the immune system in a way that may attenuate the organ damaging inflammation that occurs during critical illness and its treatment. The extent to which loss of the microbiota participates in the inflammatory process during injury remains unknown and will require extensive study. Yet the promiscuous use of antibiotics through the course of injury with the subsequent repopulation of the gut by healthcare associated multi-drug resistant pathogens presents a compelling case that the emerging pathobiome may drive the septic response in a manner which is counterproductive to homeostasis and recovery [35,59]. Careful attention to developing technology to understand these mechanisms with high resolution molecular detail will be necessary to advance our understanding of the protective role of the microbiome in the catabolic response to injury and the real and present danger that the emerging pathobiome poses on the immune response and long term outcome of the patient.
Commensal microbes stimulate immunity and suppress inflammation. Pathogens subvert immunity and activate inflammation
There is a growing body of evidence that the intestinal epithelium and its underlying immune cells discriminate between commensal organisms (symbionts) and pathogenic microbes (pathobionts) [60,61]. As mentioned, normal commensal microbiota and their PAMPs may be highly immunostimulatory during acute stress whereas “accidental” pathogens, i.e. those that have not co-evolved with their hosts, are more likely to subvert the immune system and cause inflammation. Several studies have shown this by co-culturing intestinal epithelial cells (IECs) with dendritic cells (DCs) while inoculating commensal versus pathogenic bacteria onto the apical surface of the IECs [60,61]. Although such studies do not account for how local microenvironmental cues present during physiologic stress (pH, redox, compensatory host tissue factors) might differentially shape the various microbes to express emergent properties that will change the epithelial- immune output, they do inform us that indeed there is discrimination between microbial friend or foe [50, 62–64]. Lack of accounting for the full integration of the host microbial interactome has allowed for the relative dismissal of the possibility that a gut microbiome transitioning to a gut pathobiome could be a driving force for the immunosuppression seen along the sepsis continuum. This notion raises the possibility that the emerging pathobiome itself may be the cause of the immunosuppression seen during late sepsis (Figure 1). One mechanism among the many that may be a control point by which gut microbes affect systemic immune regulation is through the control of TH17 cells [65]. Circulating TH17 cells pass through the intestinal wall and are affected by luminal microbial composition and function. A pro-inflammatory TH17 immune response could be beneficial in clearing infection or immunopathogenic if expressed excessively. TH17 cells are capable of orchestrating an immune- suppressive or immune- enhancing response and the resulting immune phenotype may be significantly affected by the emerging pathobiome [66–68].
Yet current paradigms characterize the progression from injury to severe sepsis as a failing and exhausted immune system that may be developing independent of the gut pathobiome [13]. Eventually, the impaired immune system becomes incapable of containing the threatening microbial burden. Here we posit that a potentially overlooked control point is the effect of the emerging gut pathobiome to directly subvert the immune clearance mechanisms via dysregulation of the immune system. This process need not occur via translocation or bacteremia; pathogenic microbes are fully capable of affecting the immune system at the very borders of the epithelial surface without the need for invasion.
In favor of this notion are recent studies that have demonstrated that the two most important variables that determine the infectivity of a microbe are its ability to activate its quorum sensing system and its ability to subvert the host immune system [69]. Quorum sensing is a dynamic system of microbial virulence regulation originally described as a mechanism by which bacteria can sense their population density and respond in a manner which allows them to coordinate complex assemblage behavior [70–72]. Thus sensing a quorum of bacteria allows them to understand their population density and determine that amount that is necessary to overcome the host. Bacteria accomplish this by secreting and taking up highly diffusible small molecules (quorum sensing (QS) molecules) which are highly variable and species specific [73]. A given bacterial species may have hundreds of its own QS molecules to do this, many of which have been shown to have variable effects on neighboring community microbes as well as host cells [74,75]. Yet our work and the work of others have demonstrated that bacteria sense more than just a quorum [16–21,37]. For example we have shown that the QS system can also sense and be activated by host compensatory molecules such as immune elements, end- products of hypoxia, and both endogenous and exogenous opioids [18,20]. Accidental pathogens, i.e. those that have not co-evolved with their hosts, such as carbapenem- resistant Pseudomonas aeruginosa, Acinetobacter baumannii, Klebsiella pneumoniae carbapenemase (KPC) appear to be especially triggered by these host compensatory signals resulting in the expression of highly virulent phenotypes [76–78]. Many of the dynamically expressed virulence determinants in pathogens cause immune subversion. In fact there are many examples where the quorum sensing signals released into the local environment are those that suppress the immune response. In this manner microbes can intercommunicate and induce a highly coordinated, regulated, and measured response [79]. Microbes need to make decisions as they are attaching, invading, expressing toxins, and subverting and hijacking host proteins [80]. They accomplish this through the QS system [81]. This affords microbes, depending on the tissues and context, to calibrate their strategy, even sometime to retreat, depending on the particulars of the chemical dialogue [82]. It is important to appreciate the dynamism of microbial virulence regulation to recognize that virulence expression is not a constitutive property of a pathogen and as such presents a major challenge to study when modeling events in the environmentally neutral and ideal conditions that are present in vitro [83]. One technique that has been used has been to prepare bacterial lysates in an effort to “freeze” dynamic virulence expression [84]. This approach is limited by the recognition that all stored and available virulence factors are potentially present in the lysates and are not actually secreted, externally expressed (as appendages), or injected (i.e. type III secretion machinery- molecular syringe) in vivo. Nonetheless, some interesting observations have been made.
Mechanistic and therapeutic opportunities to preserve the microbiome and constrain the pathobiome: toward ecosystem engineering as a strategy for sepsis prevention
The development and testing of novel hypotheses integrating the molecular, ecological, and evolutionary dynamics within complex environments such as the gut during surgical injury hold real promise for accelerating our understanding of the importance of virulence expression in the sepsis response and its potential as a target for evolutionarily robust anti-virulence drugs [85,86]. While the role of the intestinal microbiota in health and disease has received significant attention during the last decade, strategies to preserve their composition and function have largely failed among critically ill patients. Although the use of single strain probiotic microorganisms such as Lactobacilli spp. seems to be a rational approach to replace the function of the disappearing microbiota during surgical injury, its clinical failure in human trails in critically ill patients should be no surprise given that one species of bacteria is not likely to supplant the entire spectrum of function of the lost microbiota [87–89]. Furthermore how these supplemented microbes survive and function within the critically ill gut has never been fully addressed. There is essentially no information regarding why catabolic stress causes the disappearance of the microbiota in the first place. Also, simply providing prebiotic fuels to preserve or “rebloom” the normal microbiota is likely to be inadequate as these chemically defined compounds’ bioavailability at specific microniches in the gut remain unknown [90]. First we need to understand the ecological changes in the gut to understand how we might preserve this highly diverse ecosystem through the course of injury. Second, as antibiotics use is practically unavoidable when caring for the critically ill and injured, we need to understand how antibiotics affect the normal composition and function of the protective microbiota. Finally, we must recognize that continuously developing agents which eliminate all offending pathogens is not a sustainable strategy that will remain ahead of the evolutionary curve of our most feared hospital associated pathogens. We need to constrain their virulence tactics rather than eliminate them entirely to affect the tipping point at which they are forced to transition to harm or kill their host [91]. As an example of such a virulence- directed agent, our laboratory developed a phosphorylated high-molecular-weight polyethylene glycol (Pi-PEG) molecule, which has the capacity to leverage the strength of normal microbiota to directly suppress virulence of the pathobiota by embedding critical resources (phosphate) into the local biologic exchange market and directly affect the multidimensional molecular dialogue within the complex gut [43,92]. While it is important that research continues on host- based immune- mediated mechanisms of sepsis, equally important is research to define the role of compositional and functional changes in the gut microbiome associated with sepsis. Novel approaches to prevent sepsis following injury might include efforts to restrain inflammation using immune directed therapy within the first 24–48 hours while later employing agents that preserve the gut microbiome as well as constrain the emerging pathobiome in an effort to prevent late onset sepsis and its effect on overall immune function.
In summary here we posit that sepsis pathogenesis following surgical injury is as much as function of microbial virulence regulation as it is a function of immune regulation. Techniques are now available to define the ever- changing gut microbiome/pathobiome through the course of injury and determine how it influences the immune system [93]. A more holistic, cognitive, and computational view of the host pathogen interaction will be necessary to test novel hypotheses and inspire therapeutic approaches that can contain rather than eliminate gut microbes and the way they sense and respond to an unprecedented array of host chemosignals to adapt and survive prolonged critical illness and the extreme medical intervention that is often necessary to heal a severe injury [94–97]. Such an approach will require an iterative workflow between computational modeling such as agent based modeling and evolving knowledge in experimental biology to be able to fully harness and manage the overwhelming amount of information that is needed to more completely understand the natural history of, and therapeutic opportunities in post- injury sepsis [98].
Acknowledgments
NIH grant 2R01GM062344-13A1 (JCA)
References
- 1.Walkey AJ, Lagu T, Lindenauer PK. Trends in sepsis and infection sources in the United States. A population-based study. Ann Am Thorac Soc. 2015;12(2):216–220. doi: 10.1513/AnnalsATS.201411-498BC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donnelly JP, Hohmann SF, Wang HE. Unplanned Readmissions After Hospitalization for Severe Sepsis at Academic Medical Center-Affiliated Hospitals. Crit Care Med. 2015 doi: 10.1097/CCM.0000000000001147. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bateman BT, Schmidt U, Berman MF, Bittner EA. Temporal trends in the epidemiology of severe postoperative sepsis after elective surgery: a large, nationwide sample. Anesthesiology. 2010;112(4):917–925. doi: 10.1097/ALN.0b013e3181cea3d0. [DOI] [PubMed] [Google Scholar]
- 4.Tulloch LG, Chan JD, Carlbom DJ, Kelly MJ, Dellit TH, Lynch JB. Epidemiology and Microbiology of Sepsis Syndromes in a University-Affiliated Urban Teaching Hospital and Level-1 Trauma and Burn Center. J Intensive Care Med. 2015 doi: 10.1177/0885066615592851. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 5.Otto GP, Sossdorf M, Claus RA, Rodel J, Menge K, Reinhart K, Bauer M, Riedemann NC. The late phase of sepsis is characterized by an increased microbiological burden and death rate. Crit Care. 2011;15(4):R183. doi: 10.1186/cc10332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hotchkiss RS, Karl IE. The pathophysiology and treatment of sepsis. N Engl J Med. 2003;348(2):138–150. doi: 10.1056/NEJMra021333. [DOI] [PubMed] [Google Scholar]
- 7.Venet F, Davin F, Guignant C, Larue A, Cazalis MA, Darbon R, Allombert C, Mougin B, Malcus C, Poitevin-Later F, Lepape A, Monneret G. Early assessment of leukocyte alterations at diagnosis of septic shock. Shock. 2010;34(4):358–363. doi: 10.1097/SHK.0b013e3181dc0977. [DOI] [PubMed] [Google Scholar]
- 8.Samuelsson A, Isaksson B, Hanberger H, Olhager E. Late-onset neonatal sepsis, risk factors and interventions: an analysis of recurrent outbreaks of Serratia marcescens, 2006–2011. J Hosp Infect. 2014;86(1):57–63. doi: 10.1016/j.jhin.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 9.Carl MA, Ndao IM, Springman AC, Manning SD, Johnson JR, Johnston BD, Burnham CA, Weinstock ES, Weinstock GM, Wylie TN, Mitreva M, Abubucker S, Zhou Y, Stevens HJ, Hall-Moore C, Julian S, Shaikh N, Warner BB, Tarr PI. Sepsis from the gut: the enteric habitat of bacteria that cause late-onset neonatal bloodstream infections. Clin Infect Dis. 2014;58(9):1211–1218. doi: 10.1093/cid/ciu084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Struelens MJ. Detection of microbial DNAemia: does it matter for sepsis management? Intensive care medicine. 2010;36(2):193–195. doi: 10.1007/s00134-009-1710-2. [DOI] [PubMed] [Google Scholar]
- 11.Hotchkiss RS, Coopersmith CM, McDunn JE, Ferguson TA. The sepsis seesaw: tilting toward immunosuppression. Nature medicine. 2009;15(5):496–497. doi: 10.1038/nm0509-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Culshaw N, Glover G, Whiteley C, Rowland K, Wyncoll D, Jones A, Shankar-Hari M. Healthcare-associated bloodstream infections in critically ill patients: descriptive cross-sectional database study evaluating concordance with clinical site isolates. Ann Intensive Care. 2014;4:34. doi: 10.1186/s13613-014-0034-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sertaridou E, Papaioannou V, Kolios G, Pneumatikos I. Gut failure in critical care: old school versus new school. Ann Gastroenterol. 2015;28(3):309–322. [PMC free article] [PubMed] [Google Scholar]
- 14.Stecher B, Maier L, Hardt WD. Blooming' in the gut: how dysbiosis might contribute to pathogen evolution. Nat Rev Microbiol. 2013;11(4):277–284. doi: 10.1038/nrmicro2989. [DOI] [PubMed] [Google Scholar]
- 15.Seal JB, Morowitz M, Zaborina O, An G, Alverdy JC. The molecular Koch's postulates and surgical infection: a view forward. Surgery. 2010;147(6):757–765. doi: 10.1016/j.surg.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 16.Mekalanos JJ. Environmental signals controlling expression of virulence determinants in bacteria. J Bacteriol. 1992;174(1):1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rowley G, Spector M, Kormanec J, Roberts M. Pushing the envelope: extracytoplasmic stress responses in bacterial pathogens. Nat Rev Microbiol. 2006;4(5):383–394. doi: 10.1038/nrmicro1394. [DOI] [PubMed] [Google Scholar]
- 18.Fink D, Romanowski K, Valuckaite V, Babrowski T, Kim M, Matthews JB, Liu D, Zaborina O, Alverdy JC. Pseudomonas aeruginosa potentiates the lethal effect of intestinal ischemia-reperfusion injury: the role of in vivo virulence activation. J Trauma. 2011;71(6):1575–1582. doi: 10.1097/TA.0b013e31821cb7e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Babrowski T, Romanowski K, Fink D, Kim M, Gopalakrishnan V, Zaborina O, Alverdy JC. The intestinal environment of surgical injury transforms Pseudomonas aeruginosa into a discrete hypervirulent morphotype capable of causing lethal peritonitis. Surgery. 2013;153(1):36–43. doi: 10.1016/j.surg.2012.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Babrowski T, Holbrook C, Moss J, Gottlieb L, Valuckaite V, Zaborin A, Poroyko V, Liu DC, Zaborina O, Alverdy JC. Pseudomonas aeruginosa virulence expression is directly activated by morphine and is capable of causing lethal gut-derived sepsis in mice during chronic morphine administration. Annals of surgery. 2012;255(2):386–393. doi: 10.1097/SLA.0b013e3182331870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Romanowski K, Zaborin A, Valuckaite V, Rolfes RJ, Babrowski T, Bethel C, Olivas A, Zaborina O, Alverdy JC. Candida albicans isolates from the gut of critically ill patients respond to phosphate limitation by expressing filaments and a lethal phenotype. PLoS One. 2012;7(1):e30119. doi: 10.1371/journal.pone.0030119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lisboa T, Waterer G, Rello J. We should be measuring genomic bacterial load and virulence factors. Critical care medicine. 2010;38(10 Suppl):S656–S662. doi: 10.1097/CCM.0b013e3181f2453a. [DOI] [PubMed] [Google Scholar]
- 23.Vodovotz Y, Csete M, Bartels J, Chang S, An G. Translational systems biology of inflammation. PLoS Comput Biol. 2008;4(4):e1000014. doi: 10.1371/journal.pcbi.1000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Namas R, Zamora R, Namas R, An G, Doyle J, Dick TE, Jacono FJ, Androulakis IP, Nieman GF, Chang S, Billiar TR, Kellum JA, Angus DC, Vodovotz Y. Sepsis: Something old, something new, and a systems view. J Crit Care. 2012;27(3):314.e1-311. doi: 10.1016/j.jcrc.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salyers AA, Gupta A, Wang Y. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends in microbiology. 2004;12(9):412–416. doi: 10.1016/j.tim.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 26.Ackerman J. The ultimate social network. Sci Am. 2012;306(6):36–43. doi: 10.1038/scientificamerican0612-36. [DOI] [PubMed] [Google Scholar]
- 27.Nogueira T, Rankin DJ, Touchon M, Taddei F, Brown SP, Rocha EP. Horizontal gene transfer of the secretome drives the evolution of bacterial cooperation and virulence. Current biology : CB. 2009;19(20):1683–1691. doi: 10.1016/j.cub.2009.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hol FJ, Galajda P, Nagy K, Woolthuis RG, Dekker C, Keymer JE. Spatial structure facilitates cooperation in a social dilemma: empirical evidence from a bacterial community. PLoS One. 2013;8(10):e77042. doi: 10.1371/journal.pone.0077042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stecher B, Denzler R, Maier L, Bernet F, Sanders MJ, Pickard DJ, Barthel M, Westendorf AM, Krogfelt KA, Walker AW, Ackermann M, Dobrindt U, Thomson NR, Hardt WD. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc Natl Acad Sci U S A. 2012;109(4):1269–1274. doi: 10.1073/pnas.1113246109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alverdy JC, Chang EB. The re-emerging role of the intestinal microflora in critical illness and inflammation: why the gut hypothesis of sepsis syndrome will not go away. Journal of leukocyte biology. 2008;83(3):461–466. doi: 10.1189/jlb.0607372. [DOI] [PubMed] [Google Scholar]
- 31.Clark JA, Coopersmith CM. Intestinal crosstalk: a new paradigm for understanding the gut as the "motor" of critical illness. Shock. 2007;28(4):384–393. doi: 10.1097/shk.0b013e31805569df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mittal R, Coopersmith CM. Redefining the gut as the motor of critical illness. Trends Mol Med. 2014;20(4):214–223. doi: 10.1016/j.molmed.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wren BW. Microbial genome analysis: insights into virulence, host adaptation and evolution. Nature reviews Genetics. 2000;1(1):30–39. doi: 10.1038/35049551. [DOI] [PubMed] [Google Scholar]
- 34.Lawley TD, Bouley DM, Hoy YE, Gerke C, Relman DA, Monack DM. Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infection and immunity. 2008;76(1):403–416. doi: 10.1128/IAI.01189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zaborin A, Smith D, Garfield K, Shakhsheer B, Kade M, Tirrell M, Tiedje J, Gilbert J, Zaborina O, Alverdy JC. Membership and behavior of ultra-low-diversity pathogen communities present in the gut of humans during prolonged critical illness. MBio. 2014;5(5):1–14. doi: 10.1128/mBio.01361-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayakawa M, Asahara T, Henzan N, Murakami H, Yamamoto H, Mukai N, Minami Y, Sugano M, Kubota N, Uegaki S, et al. Dramatic changes of the gut flora immediately after severe and sudden insults. Digestive diseases and sciences. 2011;56(8):2361–2365. doi: 10.1007/s10620-011-1649-3. [DOI] [PubMed] [Google Scholar]
- 37.Shimizu K, Ogura H, Hamasaki T, Goto M, Tasaki O, Asahara T, Nomoto K, Morotomi M, Matsushima A, Kuwagata Y, et al. Altered gut flora are associated with septic complications and death in critically ill patients with systemic inflammatory response syndrome. Digestive diseases and sciences. 2011;56(4):1171–1177. doi: 10.1007/s10620-010-1418-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zaborin A, Romanowski K, Gerdes S, Holbrook C, Lepine F, Long J, Poroyko V, Diggle SP, Wilke A, Righetti K, Morozova I, Babrowski T, Liu DC, Zaborina O, Alverdy JC. Red death in Caenorhabditis elegans caused by Pseudomonas aeruginosa PAO1. Proc Natl Acad Sci U S A. 2009;106(15):6327–6332. doi: 10.1073/pnas.0813199106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamarche MG, Wanner BL, Crépin S, Harel J. The phosphate regulon and bacterial virulence: a regulatory network connecting phosphate homeostasis and pathogenesis. FEMS Microbiol Rev. 2008;32(3):461–473. doi: 10.1111/j.1574-6976.2008.00101.x. [DOI] [PubMed] [Google Scholar]
- 40.Glattard E, Welters ID, Lavaux T, Muller AH, Laux A, Zhang D, Schmidt AR, Delalande F, Laventie BJ, Dirrig-Grosch S, Colin DA, Van Dorsselaer A, Aunis D, Metz-Boutigue MH, Schneider F, Goumon Y. Endogenous morphine levels are increased in sepsis: a partial implication of neutrophils. PLoS One. 2010;5(1):e8791. doi: 10.1371/journal.pone.0008791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams JP, Thompson JP, Young SP, Gold SJ, McDonald J, Rowbotham DJ, Lambert DG. Nociceptin and urotensin-II concentrations in critically ill patients with sepsis. Br J Anaesth. 2008;100(6):810–814. doi: 10.1093/bja/aen093. [DOI] [PubMed] [Google Scholar]
- 42.Zaborina O, Lepine F, Xiao G, Valuckaite V, Chen Y, Li T, Ciancio M, Zaborin A, Petrof EO, Turner JR, Rahme LG, Chang E, Alverdy JC. Dynorphin activates quorum sensing quinolone signaling in Pseudomonas aeruginosa. PLoS Pathog. 2007;3(3):e35. doi: 10.1371/journal.ppat.0030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zaborin A, Gerdes S, Holbrook C, Liu DC, Zaborina OY, Alverdy JC. Pseudomonas aeruginosa overrides the virulence inducing effect of opioids when it senses an abundance of phosphate. PLoS One. 2012;7(4):e34883. doi: 10.1371/journal.pone.0034883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Köhler T, Buckling A, van Delden C. Cooperation and virulence of clinical Pseudomonas aeruginosa populations. Proc Natl Acad Sci U S A. 2009;106(15):6339–6344. doi: 10.1073/pnas.0811741106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang M, Schaefer AL, Dandekar AA, Greenberg EP. Quorum sensing and policing of Pseudomonas aeruginosa social cheaters. Proc Natl Acad Sci U S A. 2015;112(7):2187–2191. doi: 10.1073/pnas.1500704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu L1, Estrada O, Zaborina O, Bains M, Shen L, Kohler JE, Patel N, Musch MW, Chang EB, Fu YX, Jacobs MA, Nishimura MI, Hancock RE, Turner JR, Alverdy JC. Recognition of host immune activation by Pseudomonas aeruginosa. Science. 2005;29(5735):774–777. doi: 10.1126/science.1112422. 309. [DOI] [PubMed] [Google Scholar]
- 47.Zaborina O, Lepine F, Xiao G, Valuckaite V, Chen Y, Li T, Ciancio M, Zaborin A, Petroff E, Turner IR, Rahme LG, Chang E, Alverdy JC. Dynorphin Activates Quorum Sensing Quinolone Signaling in Pseudomonas aeruginosa. PLoS Pathog. 2007;3(3):e35. doi: 10.1371/journal.ppat.0030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kohler JE, Zaborina O, Wu L, Wang Y, Bethel C, Chen Y, Shapiro J, Turner JR, Alverdy JC. Components of intestinal epithelial hypoxia activate the virulence circuitry of Pseudomonas. Am J Physiol Gastrointest Liver Physiol. 2005;288(5):G1048–G1054. doi: 10.1152/ajpgi.00241.2004. [DOI] [PubMed] [Google Scholar]
- 49.Patel NJ, Zaborina O, Wu L, Wang Y, Wolfgeher DJ, Valuckaite V, Ciancio MJ, Kohler JE, Shevchenko O, Colgan SP, Chang EB, Turner JR, Alverdy JC. Recognition of intestinal epithelial HIF-1alpha activation by Pseudomonas aeruginosa. Am J Physiol Gastrointest Liver Physiol. 2007;292(1):G134–G142. doi: 10.1152/ajpgi.00276.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morowitz MJ, Babrowski T, Carlisle EM, Olivas A, Romanowski KS, Seal JB, Liu DC, Alverdy JC. The human microbiome and surgical disease. Ann Surg. 2011;253(6):1094–1101. doi: 10.1097/SLA.0b013e31821175d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stecher B, Chaffron S, Käppeli R, Hapfelmeier S, Freedrich S, Weber TC, Kirundi J, Suar M, McCoy KD, von Mering C, Macpherson AJ, Hardt WD. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog. 2010;6(1):e1000711. doi: 10.1371/journal.ppat.1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Behnsen J, Jellbauer S, Wong CP, Edwards RA, George MD, Ouyang W, Raffatellu M. The cytokine IL-22 promotes pathogen colonization by supressening related commensal bacteria. Immunity. 2014;40:262–273. doi: 10.1016/j.immuni.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hendrickson BA, Guo J, Laughlin R, Chen Y, Alverdy JC. Increased type 1 fimbrial expression among commensal Escherichia coli isolates in the murine cecum following catabolic stress. Infection and immunity. 1999;67(2):745–753. doi: 10.1128/iai.67.2.745-753.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ayres JS, Trinidad NJ, Vance RE. Lethal inflammasome activation by a multidrug-resistant pathobiont upon antibiotic disruption of the microbiota. Nature medicine. 2012;18(5):799–806. doi: 10.1038/nm.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 2011;477(7366):592–595. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zaborina O, Zaborin A, Romanowski K, Babrowski T, Alverdy J. Host stress and virulence expression in intestinal pathogens: development of therapeutic strategies using mice and C. elegans. Curr Pharm Des. 2011;17(13):1254–1260. doi: 10.2174/138161211795703771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brown SP, Cornforth DM, Mideo N. Evolution of virulence in opportunistic pathogens: generalism, plasticity, and control. Trends in microbiology. 2012;20(7):336–342. doi: 10.1016/j.tim.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, Schnabl B, DeMatteo RP, Pamer EG. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455(7214):804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zoumpopoulou G, Tsakalidou E, Dewulf J, Pot B, Grangette C. Differential crosstalk between epithelial cells, dendritic cells and bacteria in a co-culture model. International journal of food microbiology. 2009;131(1):40–51. doi: 10.1016/j.ijfoodmicro.2008.12.037. [DOI] [PubMed] [Google Scholar]
- 61.Bahrami B, Macfarlane S, Macfarlane GT. Induction of cytokine formation by human intestinal bacteria in gut epithelial cell lines. Journal of applied microbiology. 2011;110(1):353–363. doi: 10.1111/j.1365-2672.2010.04889.x. [DOI] [PubMed] [Google Scholar]
- 62.Franklin BS, Latz E. For gut's sake: NLRC4 Inflammasomes distinguish friend from foe. Nat Immunol. 2012;18(5):429–431. doi: 10.1038/ni.2289. 13. [DOI] [PubMed] [Google Scholar]
- 63.Franchi L, Kamada N, Nakamura Y, Burberry A, Kuffa P, Suzuki S, Shaw MH, Kim YG, Núñez G. NLRC4-driven production of IL-1β discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat Immunol. 2012;13(5):449–456. doi: 10.1038/ni.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Srinivasan N. Telling apart friend from foe: discriminating between commensals and pathogens at mucosal sites. Innate Immun. 2010;16(6):391–404. doi: 10.1177/1753425909357577. [DOI] [PubMed] [Google Scholar]
- 65.Esplugues E, Huber S, Gagliani N, Hauser AE, Town T, Wan YY, O'Connor W, Jr, Rongvaux A, Van Rooijen N, Haberman AM, et al. Control of TH17 cells occurs in the small intestine. Nature. 2011;475(7357):514–518. doi: 10.1038/nature10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, Fouser LA. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203(10):2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sonnenberg GF, Nair MG, Kirn TJ, Zaph C, Fouser LA, Artis D. Pathological versus protective functions of IL-22 in airway inflammation are regulated by IL-17A. J Exp Med. 2010;207(6):1293–1305. doi: 10.1084/jem.20092054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kolls JK, McCray PB, Jr, Chan YR. Cytokine-mediated regulation of antimicrobial proteins. Nat Rev Immunol. 2008;8(11):829–835. doi: 10.1038/nri2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gama JA, Abby SS, Vieira-Silva S, Dionisio F, Rocha EP. Immune subversion and quorum-sensing shape the variation in infectious dose among bacterial pathogens. PLoS pathogens. 2012;8(2):e1002503. doi: 10.1371/journal.ppat.1002503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kendall MM, Sperandio V. Quorum sensing by enteric pathogens. Curr Opin Gastroenterol. 2007;23(1):10–15. doi: 10.1097/MOG.0b013e3280118289. [DOI] [PubMed] [Google Scholar]
- 71.Antunes LC, Ferreira RB. Intercellular communication in bacteria. Crit Rev Microbiol. 2009;35(2):69–80. doi: 10.1080/10408410902733946. [DOI] [PubMed] [Google Scholar]
- 72.Antunes LC, Ferreira RB, Buckner MM, Finlay BB. Quorum sensing in bacterial virulence. Microbiology. 2010;156:2271–2282. doi: 10.1099/mic.0.038794-0. [DOI] [PubMed] [Google Scholar]
- 73.Roux A, Payne SM, Gilmore MS. Microbial telesensing: probing the environment for friends, foes, and food. Cell Host Microbe. 2009;6(2):115–124. doi: 10.1016/j.chom.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smalley NE, An D, Parsek MR, Chandler JR, Dandekar AA. Quorum sensing protects Pseudomonas aeruginosa against cheating by other species in a laboratory co-culture model. J Bacteriol. 2015 doi: 10.1128/JB.00482-15. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dandekar AA, Chugani S, Greenberg EP. Bacterial quorum sensing and metabolic incentives to cooperate. Science. 2012;338(6104):264–266. doi: 10.1126/science.1227289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. The Lancet infectious diseases. 2009;9(4):228–236. doi: 10.1016/S1473-3099(09)70054-4. [DOI] [PubMed] [Google Scholar]
- 77.Khuntayaporn P, Montakantikul P, Mootsikapun P, Thamlikitkul V, Chomnawang MT. Prevalence and genotypic relatedness of carbapenem resistance among multidrug-resistant P. aeruginosa in tertiary hospitals across Thailand. Annals of clinical microbiology and antimicrobials. 2012;11(1):25. doi: 10.1186/1476-0711-11-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clinical microbiology reviews. 2008;21(3):538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kravchenko VV, Kaufmann GF, Mathison JC, Scott DA, Katz AZ, Grauer DC, Lehmann M, Meijler MM, Janda KD, Ulevitch RJ. Modulation of gene expression via disruption of NF-kappaB signaling by a bacterial small molecule. Science. 2008;321(5886):259–263. doi: 10.1126/science.1156499. [DOI] [PubMed] [Google Scholar]
- 80.Wu F, Menn DJ, Wang X. Quorum-sensing crosstalk-driven synthetic circuits: from unimodality to trimodality. Chem Biol. 2014;21(12):1629–1638. doi: 10.1016/j.chembiol.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boyle KE, Monaco H, van Ditmarsch D, Deforet M, Xavier JB. Integration of Metabolic and Quorum Sensing Signals Governing the Decision to Cooperate in a Bacterial Social Trait. PLoS Comput Biol. 2015;11(5):e1004279. doi: 10.1371/journal.pcbi.1004279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Antunes LC, McDonald JA, Schroeter K, Carlucci C, Ferreira RB, Wang M, Yurist-Doutsch S, Hira G, Jacobson K, Davies J, Allen-Vercoe E, Finlay BB. Antivirulence activity of the human gut metabolome. MBio. 2014;5(4):e01183–e01114. doi: 10.1128/mBio.01183-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gallie J, Libby E, Bertels F, Remigi P, Jendresen CB, Ferguson GC, Desprat N, Buffing MF, Sauer U, Beaumont HJ, Martinussen J, Kilstrup M, Rainey PB. Bistability in a metabolic network underpins the de novo evolution of colony switching in Pseudomonas fluorescens. PLoS Biol. 2015;13(3):e1002109. doi: 10.1371/journal.pbio.1002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Morandi B, Agazzi A, D'Agostino A, Antonini F, Costa G, Sabatini F, Ferlazzo G, Melioli G. A mixture of bacterial mechanical lysates is more efficient than single strain lysate and of bacterial-derived soluble products for the induction of an activating phenotype in human dendritic cells. Immunol Lett. 2011;138(1):86–91. doi: 10.1016/j.imlet.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 85.Alverdy J, Holbrook C, Rocha F, Seiden L, Wu RL, Musch M, Chang E, Ohman D, Suh S. Gut-derived sepsis occurs when the right pathogen with the right virulence genes meets the right host: evidence for in vivo virulence expression in Pseudomonas aeruginosa. Ann Surg. 2000;232(4):480–489. doi: 10.1097/00000658-200010000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sansonetti PJ. War and peace at mucosal surfaces. Nat Rev Immunol. 2004;4(12):953–964. doi: 10.1038/nri1499. [DOI] [PubMed] [Google Scholar]
- 87.Petrof EO, Dhaliwal R, Manzanares W, Johnstone J, Cook D, Heyland DK. Probiotics in the critically ill: A systematic review of the randomized trial evidence. Critical care medicine. 2012 doi: 10.1097/CCM.0b013e318260cc33. [DOI] [PubMed] [Google Scholar]
- 88.Morrow LE, Gogineni V, Malesker MA. Probiotics in the intensive care unit. Nutr Clin Pract. 2012;27(2):235–241. doi: 10.1177/0884533612440290. [DOI] [PubMed] [Google Scholar]
- 89.Jacobi CA, Schulz C, Malfertheiner P. Treating critically ill patients with probiotics: Beneficial or dangerous? Gut Pathog. 2011;3(1):2. doi: 10.1186/1757-4749-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Morrow LE, Gogineni V, Malesker MA. Probiotic, prebiotic, and synbiotic use in critically ill patients. Curr Opin Crit Care. 2012;18(2):186–191. doi: 10.1097/MCC.0b013e3283514b17. [DOI] [PubMed] [Google Scholar]
- 91.Clatworthy AE, Pierson E, Hung DT. Targeting virulence: a new paradigm for antimicrobial therapy. Nat Chem Biol. 2007;3(9):541–548. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 92.Zaborin A, Defazio JR, Kade M, Kaiser BL, Belogortseva N, Camp DG, 2nd, Smith RD, Adkins JN, Kim SM, Alverdy A, Goldfeld D, Firestone MA, Collier JH, Jabri B, Tirrell M, Zaborina O, Alverdy JC. Phosphate-containing polyethylene glycol polymers prevent lethal sepsis by multidrug-resistant pathogens. Antimicrob Agents Chemotherapy. 2014;58(2):966–977. doi: 10.1128/AAC.02183-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, Gordon JI. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci U S A. 2011;108(15):6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mellbye B, Schuster M. The sociomicrobiology of antivirulence drug resistance: a proof of concept. mBio. 2011;2(5) doi: 10.1128/mBio.00131-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Baquero F, Coque TM, de la Cruz F. Ecology and evolution as targets: the need for novel eco-evo drugs and strategies to fight antibiotic resistance. Antimicrobial agents and chemotherapy. 2011;55(8):3649–3660. doi: 10.1128/AAC.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Little AE, Robinson CJ, Peterson SB, Raffa KF, Handelsman J. Rules of engagement: interspecies interactions that regulate microbial communities. Annu Rev Microbiol. 2008;62:375–401. doi: 10.1146/annurev.micro.030608.101423. [DOI] [PubMed] [Google Scholar]
- 97.Vayssier-Taussat M, Albina E, Citti C, Cosson JF, Jacques MA, Lebrun MH, Le Loir Y, Ogliastro M, Petit MA, Roumagnac P, Candresse T. Shifting the paradigm from pathogens to pathobiome: new concepts in the light of meta-omics. Front Cell Infect Microbiol. 2014;4:29. doi: 10.3389/fcimb.2014.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Seal JB, Alverdy JC, Zaborina O, An G. Agent-based dynamic knowledge representation of Pseudomonas aeruginosa virulence activation in the stressed gut: Towards characterizing host-pathogen interactions in gut-derived sepsis. Theor Biol Med Model. 2011;8:33. doi: 10.1186/1742-4682-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]