Summary
Obesity-induced inflammation mediated by immune cells in adipose tissue appears to participate in the pathogenesis of insulin resistance. We show that natural killer (NK) cells in adipose tissue play an important role. High fat diet (HFD) increases NK cell numbers and the production of pro-inflammatory cytokines, notably TNFα, in epididymal, but not subcutaneous, fat depots. When NK cells were depleted either with neutralizing antibodies or genetic ablation in E4bp4+/− mice, obesity-induced insulin resistance improved in parallel with decreases in both adipose tissue macrophage (ATM) numbers and ATMs and adipose tissue inflammation. Conversely, expansion of NK cells following IL-15 administration or reconstitution of NK cells into E4bp4−/−mice increased both ATM numbers and adipose tissue inflammation and exacerbated HFD-induced insulin resistance. These results indicate that adipose NK cells control ATMs as an upstream regulator potentially by producing pro-inflammatory mediators including TNFα and thereby contribute to the development of obesity-induced insulin resistance.
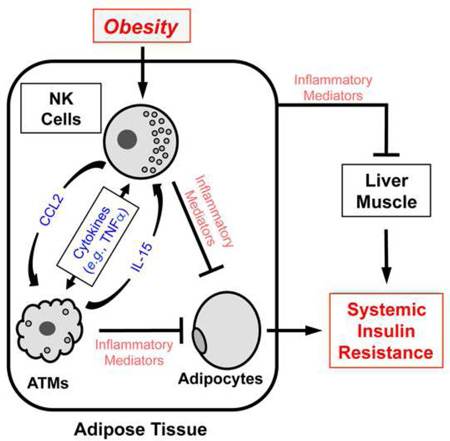
Graphical Abstract
Introduction
The rising prevalence of obesity (Flegal et al., 2012) is elevating the prevalence of associated conditions, including cardiovascular disease and certain cancers and immunological disorders. Obesity is also a major driver of insulin resistance and type 2 diabetes. Recent studies suggest that obesity-induced inflammation underlies the development of insulin resistance and type 2 diabetes [reviewed in McNelis and Olefsky (2014)]. This notion is supported by the observation that obesity increases inflammation, especially in adipose tissue, by elevating pro-inflammatory gene expression levels (Xu et al., 2003).
Obesity-induced inflammation in adipose tissue is mainly mediated by resident immune cells. ATMs comprise 40–60% of the immune cells in adipose tissue and are thought to be the primary effector cells in obesity-induced insulin resistance (Weisberg et al., 2003; Xu et al., 2003). Obesity dramatically increases the number of ATMs, in particular CD11c+ ATMs. It has also been suggested that obesity induces ATM polarization, where anti-inflammatory M2 phenotypes are superseded by pro-inflammatory M1 phenotypes that are often represented by CD11c+ ATMs (Lumeng et al., 2008). These observations are supported by the fact that many pharmacological inflammation inhibitors that improve glycemic control [including thiazolidinediones (TZDs)] also decrease total and CD11c+ ATM numbers and the expression levels of pro-inflammatory genes (e.g., TNFα) in ATMs and adipose tissue (Fujisaka et al., 2009; Cipolletta et al., 2012; Kim et al., 2013). Moreover, studies with genetically altered mouse models in which pro-inflammatory pathways in myeloid cells are suppressed show that obesity-induced inflammation in adipose tissue and obesity-induced insulin resistance are reduced in parallel (McNelis and Olefsky, 2014).
How does obesity affect ATMs? Mounting evidence suggests that neighboring adipose tissue immune cells may regulate ATM numbers and inflammation. For example, in obesity, CD8 T cells can promote a more inflammatory phenotype in ATMs (Nishimura et al., 2009), whereas in lean conditions, regulatory T cells (Tregs) may check ATM inflammation (Feuerer et al., 2009). We have found that NK cells play a critical role in the development of obesity-induced insulin resistance and that this role partly involves the regulation of ATM numbers and activation. NK cells are a specialized subset of lymphocytes with two major functions (Di Santo, 2006). The first is to kill tumor or infected cells via the cytolytic activities of released perforin and granzymes. The second NK cell function is to regulate neighboring immune cells by secreting a vast array of active substances, including both pro- or anti-inflammatory cytokines (e.g., TNFα, IFNγ, and IL-10), that are known to play key roles in the migration, maturation, activation, and polarization of other immune cells. For example, NK cell-derived IFNγ and TNFα induce macrophage polarization towards more pro-inflammatory M1 phenotypes (Biswas and Mantovani, 2010; Kroner et al., 2014). TNFα has also been shown to promote insulin resistance in obese rodents (Hotamisligil et al., 1993; Kirchgessner et al., 1997), although these studies did not clarify which cells produced the TNFα. We have found that the loss of epididymal NK cells improves obesity-induced insulin resistance, while NK cell expansion exacerbates the condition. These metabolic outcomes were associated with alterations in ATM and adipose tissue inflammation. Thus, epididymal NK cells have a critical role in controlling local ATMs and adipose tissue inflammation, thereby regulating systemic insulin resistance in obesity.
Results
Obesity increases NK cell numbers and activation in epididymal fat
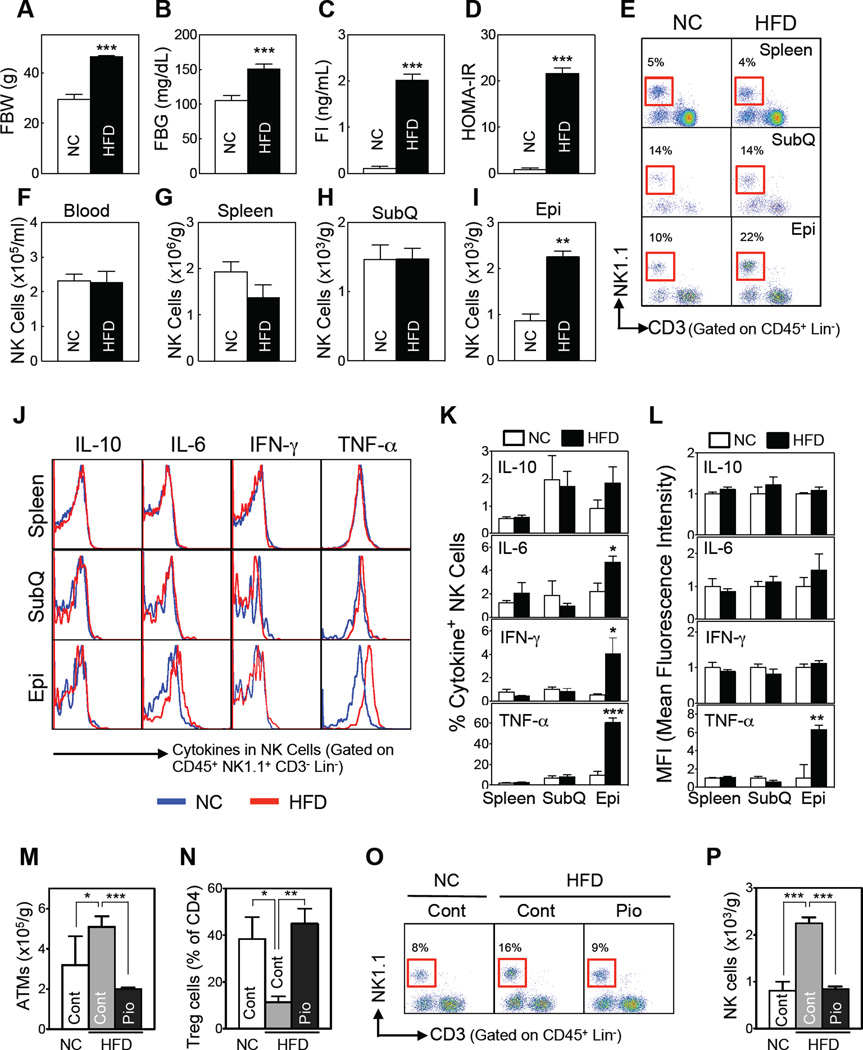
When C57Bl/6 mice were fed high fat diet (HFD) for 12 weeks, their body weight (Figure 1A) and subcutaneous and epididymal fat pad and spleen weights rose significantly (Figure S1A). They also developed insulin resistance, as shown by increased fasting glucose and insulin levels and consequent homeostatic model assessment-insulin resistance (HOMA-IR) indices (Figure 1B – D). Flow cytometric analyses showed that compared to normal chow (NC)-fed mice, HFD significantly increased the numbers of macrophages, CD4 and CD8 T cells, and B cells in epididymal fat, a murine surrogate for abdominal fat (Figure S1E). By contrast, immune cell numbers were unchanged in the blood, spleen, and subcutaneous fat of the HFD-fed mice (Figure S1B–S1E). Of note, NK cell numbers in the epididymal fat were 2.5-fold higher after 12 weeks HFD than in NC-fed controls (Figure 1E and I). NK cell number changes were not observed in subcutaneous fat or other tissues (Figure 1E – H).
Figure 1. HFD-induced obesity increases adipose tissue NK cell numbers and activation.
(A–L) C57BL/6 male mice were fed a HFD or NC for 12 weeks. (A) Fasting body weight. (B) Fasting blood glucose levels. (C) Fasting serum insulin levels. (D) HOMA-IR (n=5–7/group). (E–L) HFD-induced changes in NK cell phenotypes. (E) Representative flow cytometric plots of the NK cells (red box). The lineage markers (Lin) were TER-119, CD19, and GR-1 for blood; F4/80 was added for the tissue immune cell analyses. (F–I) NK cell numbers (n=3–4/group) in the blood (F), spleen (G), subcutaneous fat (SubQ) (H), and epididymal fat (Epi) (I). (J–L) Flow cytometric analysis of the ex vivo cytokine production by tissue NK cells (n=6/group). (J) Representative cytokine expression. (K) Frequencies of cytokine+ NK cells. (L) Mean fluorescence intensity. (M–P) Pioglitazone (Pio)-induced changes in epididymal fat immune cell numbers. Seven-week-old C57BL/6 mice (n = 8) were simultaneously treated with a HFD (or NC) and Pio (100 mg/kg diet) for 8 weeks. After overnight fasting, epididymal ATMs, Tregs, and NK cells were analyzed by flow cytometry. (M) ATM numbers. (N) Treg numbers. (O) Representative flow cytometry plots of NK cells (red box). (P) NK cell numbers. The data are presented as mean ± S.E.M. *p<0.05, *p<0.01, and ***p<0.001. See also Figure S1.
Similar tissue-specific patterns were evident when NK cell activity was assessed by measuring ex vivo cytokine production (Figure 1J). HFD significantly elevated the frequencies of IL-6-, IFNγ-, and TNFα-producing NK cells in epididymal fat but not in subcutaneous fat or spleen (Figure 1K). In particular, ~60% of the epididymal NK cells from HFD-fed mice were TNFα+, i.e., they produced TNFα, compared to ~10% of NK cells from NC-fed mice. Moreover, individual epididymal NK cells from HFD-fed mice expressed ~6-fold more TNFα than NK cells from NC-fed mice, as indicated by mean fluorescence intensity (Figure 1L). Therefore, three distinct measures showed that epididymal NK cells were both amplified and activated by 12 weeks of HFD: a 2.5-fold greater overall number, ~6-fold greater frequencies of TNFα+ NK cells, and 6-fold greater TNFα protein production by individual NK cells. Although there were also greater frequencies of IL-6+ and IFNγ+ epididymal NK cells in the HFD-fed mice than in the NC-fed mice, HFD did not increase the production of either IL-6 or IFNγ by the individual cells (Figure 1L). HFD did not affect the frequency of IL-10-producing NK cells or the amount of IL-10 produced per cell, regardless of the tissue examined (Figure 1J – L).
Since NK cells are responsive to cytokines and chemokines produced by other immune cells, we assessed the expression of known NK cell chemoattractants in sorted ATMs. The ATMs from 12-week HFD- vs. NC-fed mice expressed greater amounts of Ccl3, Ccl4, and Cxcl10, but not Ccl5, Ccl7, Cxcl9, Cxcl1 or Cxcl11 (Figure S1F). Elevated chemokine expression was not apparent when whole fat samples were analyzed (Figure S1G), suggesting that adipocytes and other cells do not contribute significantly to chemoattractant production.
NK cells may also be activated by cytokines produced by other cells. We found that both sorted ATMs and epididymal fat from HFD- vs. NC-fed mice expressed more IL-15, which plays key roles in NK cell proliferation/activation (Ma et al., 2006). Other cytokines that are known to drive NK cell proliferation/activation were not upregulated (Figure S1H) and IL-15 gene expression in other tissues was unchanged (Figure S1I). This suggests that IL-15 is responsible for epididymal fat-specific NK cell activation in obesity.
TZDs are known to improve obesity-induced insulin resistance in parallel with suppressing adipose tissue inflammation (Fujisaka et al., 2009; Cipolletta et al., 2012; Kim et al., 2013). Pioglitazone treatment of HFD-fed animals decreased NK cell numbers in epididymal fat (Figures 1O and P), which parallels the known changes in ATMs (Figure 1M) and Tregs (Figure 1N).
NK cell depletion improves HFD-induced inflammation and insulin resistance
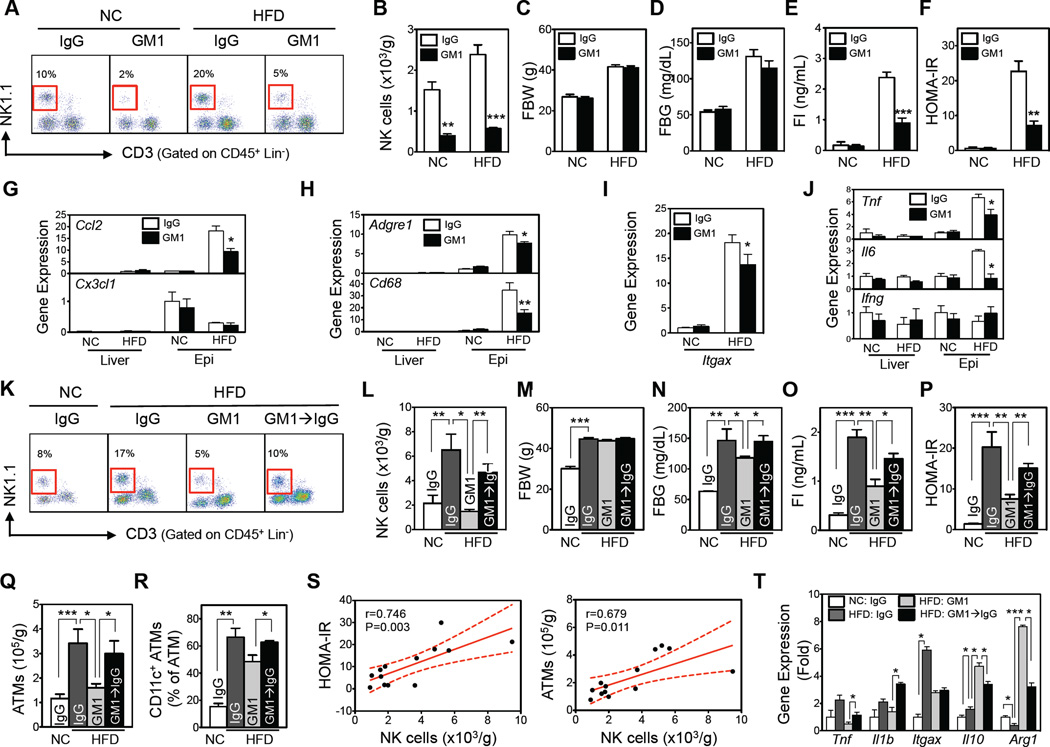
Based on these findings, we asked whether gain or loss of NK cell function affects adipose tissue inflammation and metabolic parameters in obese mice. Anti-asialo GM1 (GM1) antibody has often been used for NK cell depletion, although a subpopulation of CD8 T cells may also be affected (Trambley et al., 1999). After obesity was established in 8-week HFD mice, treatment with GM1 antibodies for 4 weeks depleted >80% of the NK cells in all tissues tested (Figures 2A, 2B,S2A, and S2B). The GM1 antibody also decreased CD8 T-cell numbers in the epididymal fat of HFD-fed mice and the spleen of NC-fed mice (Figure S2B and S2C). The injections did not affect CD4 or invariant natural killer T (iNKT)-cell numbers in any tissue under either diet (Figure S2A–S2C).
Figure 2. Depletion of NK cells by anti-asialo GM1 antibody improves HFD-induced insulin resistance and recovery of NK cell numbers worsens it.
(A–J) NK cell depletion by GM1 or isotype-control (IgG) antibodies (n=4–5/group). (A) Representative flow cytometric plots of NK cells (red box). The lineage markers (Lin) were TER-119, Gr-1, CD19, and F4/80. (B) Epididymal fat NK cell numbers. (C) Fasting body weight. (D) Fasting blood glucose levels. (E) Fasting serum insulin levels. (F) HOMA-IR. (G–J) Gene expression of macrophage chemokines (G), macrophage markers (H), CD11c gene expression (in epididymal fat) (I), and inflammatory mediator genes (J), as determined by qRT-PCR (n=4–5/group). (K–T) C57BL/6 mice fed NC or a HFD for 10 weeks were injected with GM1 or control (IgG) antibodies for 3 weeks, after which the GM1-injected HFD mice were divided into two groups: one group continued to be injected with GM1 while the other group was injected with the control antibody (GM1→IgG) to allow the depleted NK cell compartment to recover (see Figure S2E). After 2 weeks of NK cell recovery, the immunological and metabolic phenotypes were measured (n=4–5/group). (K) Representative flow cytometric plots of epididymal fat NK cells (red box). The lineage markers (Lin) were TER119, Gr-1, CD19, and F4/80. (L) Epididymal fat NK cell numbers. (M) Fasting body weight. (N) Fasting blood glucose levels. (O) Fasting serum insulin levels. (P) HOMA-IR. (Q, R) ATM inflammation in epididymal fat (n=4/group). (Q) Total epididymal ATM numbers. (R) Epididymal CD11c+ ATM frequencies. (S) Correlation between epididymal NK cell numbers and HOMA-IR and ATM numbers. (T) Gene expression of FACS-sorted epididymal ATMs, as measured by qRT-PCR. The data are presented as mean ± S.E.M. *p<0.05, **p<0.01, and ***p<0.001. See also Figure S2.
NK cell depletion had no effect on metabolic parameters in NC-fed mice (Figure 2C – F). By contrast, in HFD-fed mice, NK cell depletion significantly improved insulin sensitivity, as indicated by the reduced fasting insulin and HOMA-IR (Figure 2D – F), without affecting either total body or fat pad weights (Figures 2C and S2D). NK cell depletion also suppressed HFD-induced adipose tissue inflammation, including 1) the macrophage chemoattractant Ccl2, 2) macrophage-specific markers Adgre1 (F4/80) and Cd68, 3) an M1 marker in ATMs, Itgax (CD11c), and 4) pro-inflammatory cytokines Tnf and Il6 (Figures 2G – J). Like CCL2, CX3CL1 has major effects on macrophages. However, Cx3cl1 gene expression was unaffected by either HFD or NK cell depletion. Moreover, none of these manipulations affected these genes in liver (Figures 2G, 2H and 2J). Although NK cell production of IFNγ is a suggested mediator of obesity-induced insulin resistance (Wensveen et al., 2015), neither HFD nor NK cell depletion altered Ifng gene expression in either liver or epididymal fat (Figure 2J).
The effect of restoration of function was studied by ceasing GM1 antibody treatment (Figure S2E). After 2 weeks of recovery with control antibody, NK cell numbers recovered in all tissues (Figures 2K, 2L, and S2F–S2H). By contrast, CD8 T-cell numbers in epididymal fat or spleen did not recover (Figures S2H and S2I). The switch from GM1 to control antibody did not alter body or tissue weights (Figures 2M and S2J) or numbers of other tissue lymphocytes, including iNKT cells (Figure S2G–S2I). The recovery of NK cells associated with an exacerbation of insulin resistance (Figure 2N – P). Notably, NK cell depletion and recovery also affected epididymal ATM numbers. The HFD-induced increases in both total and CD11c+ ATMs were decreased during NK cell depletion and restored during NK cell recovery (Figures 2Q, 2R, and S2K). Epididymal NK cell and ATM numbers during depletion and restoration correlated with each other (Figure 2S). Since the only variable in this experiment was an increase or decrease in NK cell numbers, these data indicate that changes in ATM numbers are likely to result from the changes in NK cell numbers. NK cell numbers also correlated significantly with fasting glucose and insulin concentrations and HOMA-IR, but not with fasting body and fat weights (Figures 2S and S2L).
NK cell depletion and recovery also predictably affected pro- (TNFα and IL-1β) and anti-(IL-10) inflammatory cytokines and macrophage M1/M2 markers (CD11c and Arg1). All of these markers were elevated in FACS-sorted epididymal ATMs from HFD- vs. NC-fed mice, except Arg1. NK cell depletion reduced both pro-inflammatory markers and further increased IL-10 and Arg1 expression (Figure 2T). NK cell recovery reversed all of these changes, without affecting CD11c expression.
The fact that the CD8 T-cell numbers did not recover when the GM1 antibody was stopped suggests that this selective GM1 positive subset of CD8 T cells are not responsible for the inflammatory and metabolic changes that associated with GM1 antibody treatment (Figures S2H and S2I). However, since the entire pool of CD8 T cells was previously shown to play a role in obesity-induced inflammation and insulin resistance (Nishimura et al., 2009), we assessed the effects of a second NK cell-depleting antibody.
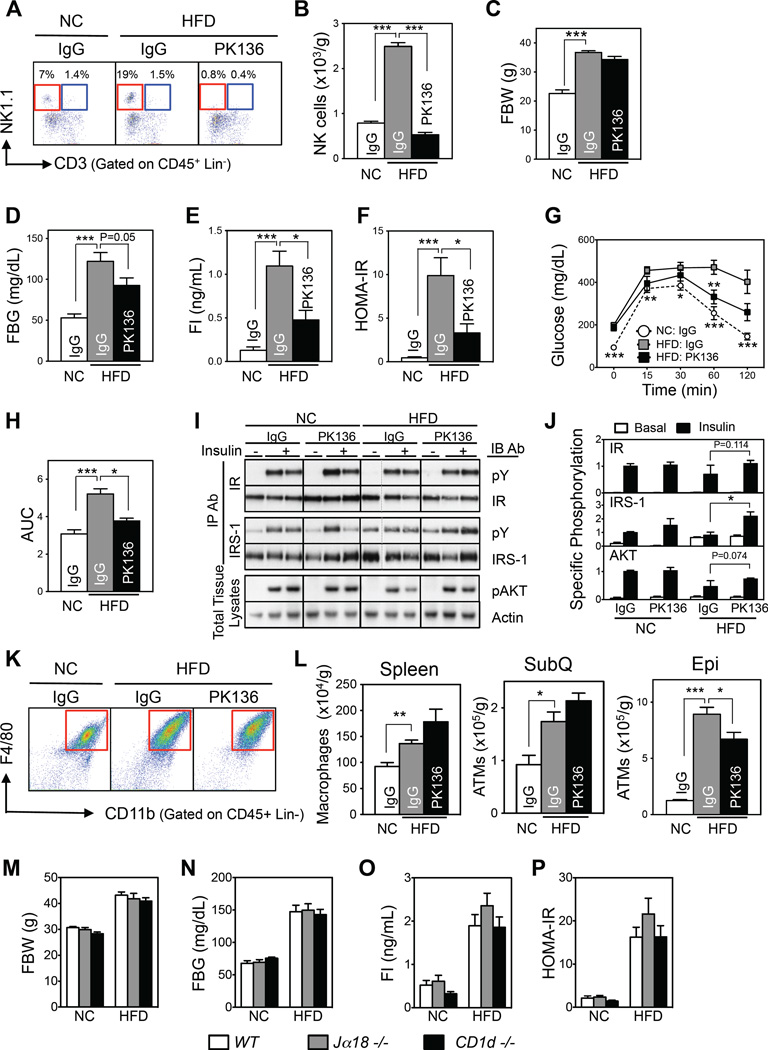
Anti-NK1.1 antibody (PK-136) depletes both NK and iNKT cells without affecting CD8 T-cell numbers (Kitaichi et al., 2002). PK-136 administration decreased NK cell numbers in HFD-fed mice by ~80% in all tissues tested (Figures 3A, 3B, S3A, and S3B). It also decreased iNKT cell numbers in epididymal fat and spleen without affecting CD8 T cells or other lymphocytes (Figure S3C–S3E). The depletion did not change total body and tissue weights (Figures 3C and S3F) but improved insulin sensitivity, as indicated by the decreased fasting glucose and insulin levels and HOMA-IR (Figure 3D – F). NK cell depletion also improved glucose tolerance (Figures 3G and H) and insulin signaling in the liver and muscle (Figures 3I, 3J and S3G). Consistent with the depletion of NK cells, PK-136 treatment also reduced ATM numbers in the epididymal fat without altering macrophage numbers in the spleen or subcutaneous fat (Figures 3K and 3L).
Figure 3. Depletion of NK cells by anti-NK1.1 antibody also improves HFD-induced insulin resistance.
(A–L) NK cell depletion by PK-136 antibody (n=4–5/group). (A) Representative flow cytometric plots of NK (red box) and iNKT (blue box) cells. The lineage markers (Lin) were TER-119, Gr-1, CD19, and F4/80. (B) Epididymal NK cell numbers. (C) Fasting body weight. (D) Fasting blood glucose levels. (E) Fasting serum insulin levels. (F) HOMA-IR. (G) Glucose tolerance test (GTT). (H) Area under the curve (AUC) of GTT. (I) Representative Western blot of the insulin signaling pathway in liver. (J) Quantitation of (I). (K) Representative flow cytometry plots of epididymal ATMs. (L) Macrophage numbers in the spleen, subcutaneous fat (SubQ), and epididymal fat (Epi). (M–P) iNKT cell-deficient homozygous knockout mice (Jα18−/− and CD1d−/−) (n=8–11/group). (M) Fasting body weight. (N) Fasting blood glucose levels. (O) Fasting serum insulin levels. (P) HOMA-IR. The data are presented as mean ± S.E.M. *p<0.05, **p<0.01, and ***p<0.001. See also Figure S3.
Because PK-136 depletes iNKT cells along with NK cells, we further investigated the potential roles of iNKT cells in obesity-induced insulin resistance using Jα18−/− and CD1d−/− mice, which are iNKT cell- but not NK cell-deficient (Exley et al., 2003; Leadbetter et al., 2008). These gene deletions did not affect any of the metabolic parameters in either NC- or HFD-fed mice, including body weight, fasting glucose or insulin, HOMA-IR, or glucose or insulin tolerance test results (Figures 3M – P and S3H–S3J). We therefore concluded that iNKT cells do not participate in the regulation of either body weight or metabolism. This supports the notion that NK cells are specifically responsible for the PK-136-induced improvements in obesity-induced ATM inflammation and insulin resistance.
The studies so far indicate that loss of NK cells and their restoration regulate both adipose tissue inflammation and systemic insulin resistance. While each individual study could suggest that other cells might be involved, e.g., the GM1+ subset of CD8 T cells or the PK-136-targeted iNKT cells, the additional results essentially eliminated these possibilities.
NK cell expansion with IL-15 exacerbates HFD-induced insulin resistance
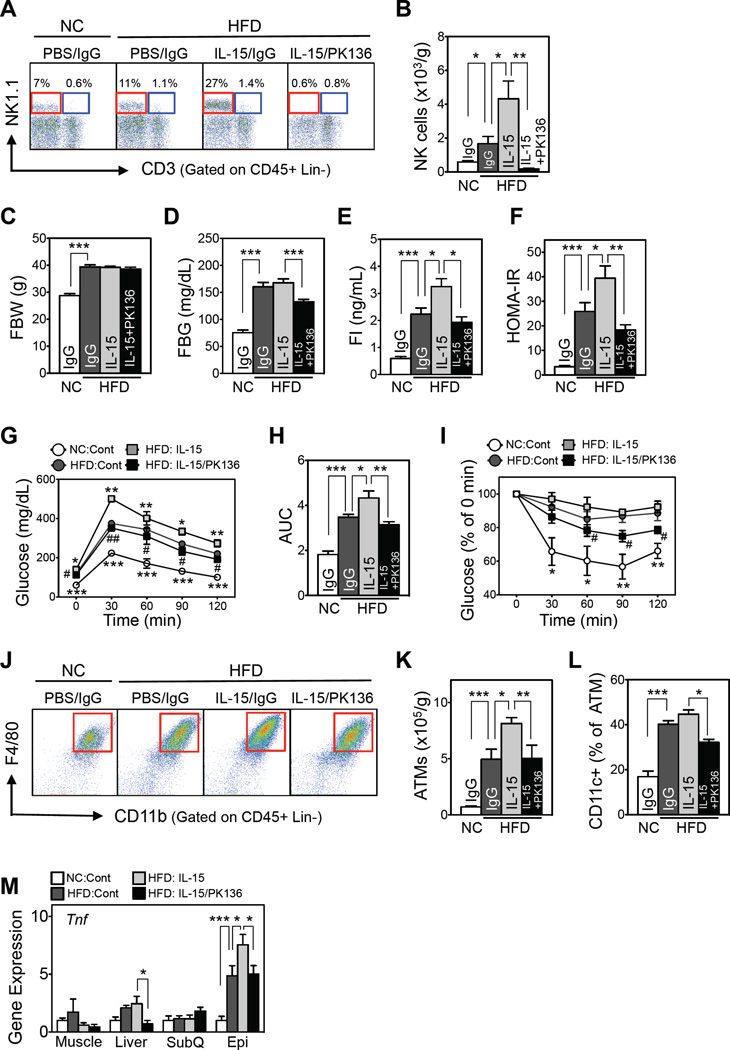
Since NK cell depletion improves obesity-induced inflammation and insulin resistance, we asked whether the opposite manipulation, i.e., NK cell expansion, concordantly exacerbates these conditions. IL-15 is an established method for expanding NK cells (Rubinstein et al., 2006). HFD increased NK cell numbers in epididymal fat ~2-fold, and this was further doubled during IL-15 administration (Figures 4A and B). NK cell numbers in the spleen rose marginally (Figure S4C) and not at all in the circulation (Figure S4B). As has been reported previously, IL-15 also increased CD8 T-cell numbers in the spleen (Rubinstein et al., 2006) but not in the circulation or epididymal fat (Figure S4B–S4D). IL-15 did not affect numbers of iNKT cells or other lymphocytes in other tissues.
Figure 4. NK cell expansion by IL-15 injections exacerbates HFD-induced insulin resistance.
(A) Representative flow cytometric plots of epididymal NK (red box) and iNKT (blue box) cells. The lineage markers (Lin) were TER-119, Gr-1, CD19, and F4/80. (B) Epididymal adipose NK cell numbers. (C) Fasting body weight. (D) Fasting blood glucose levels. (E) Fasting serum insulin levels. (F) HOMA-IR. (G) GTT. (H) AUC of GTT. (I) Insulin tolerance test (ITT). For Figure 4B–F and H: *p<0.05, **p<0.01, and ***p<0.001. For Figure 4G and I: *p<0.05, **p<0.01, and ***p<0.001 vs. HFD:Cont; #p<005 and ##p<0.01 vs. HFD:IL-15. (J) Representative flow cytometric plots of epididymal ATMs (red box). The lineage markers were TER-119, CD3, CD19, and NK1.1. (K) Total epididymal ATM numbers (n=5/group). (L) Epididymal CD11c+ ATM frequencies. (M) Tnf expression levels, as determined by qRT-PCR. For Figure 4K–M: *p<0.05, **p<0.01, and ***p<0.001. n=5–7/group. The data are presented as mean ± S.E.M. See also Figure S4.
Because IL-15 affects other immune cells in addition to NK cells, we administered IL-15 together with NK cell-depleting PK-136. Co-administration of PK-136 and IL-15 in HFD-fed mice decreased both NK and iNKT cells in all tissues without affecting CD8 T-cell or other lymphocyte numbers (Figure S4B–S4D). Total body or adipose tissue weights were unaffected by IL-15 treatment with and without PK-136 administration (Figures 4C and S4E). However, IL-15-driven NK cell expansion worsened insulin resistance and reduced glucose and insulin tolerance. This effect was partly reversed by reducing NK cell numbers by PK-136 co-administration (Figure 4D – I). IL-15-driven NK cell expansion also increased total and CD11c+ ATM numbers and epididymal Tnf expression, whereas concomitant NK cell depletion with PK-136 reversed these changes (Figures 4J – M). While IL-15 treatment did not increase Tnf expression in liver, the liver expression of this gene was reduced by PK-136; no other tissues were affected. These results together indicate that NK cell expansion exacerbates obesity-induced inflammation and insulin resistance.
Genetic depletion of NK cells improves adipose tissue inflammation and insulin resistance
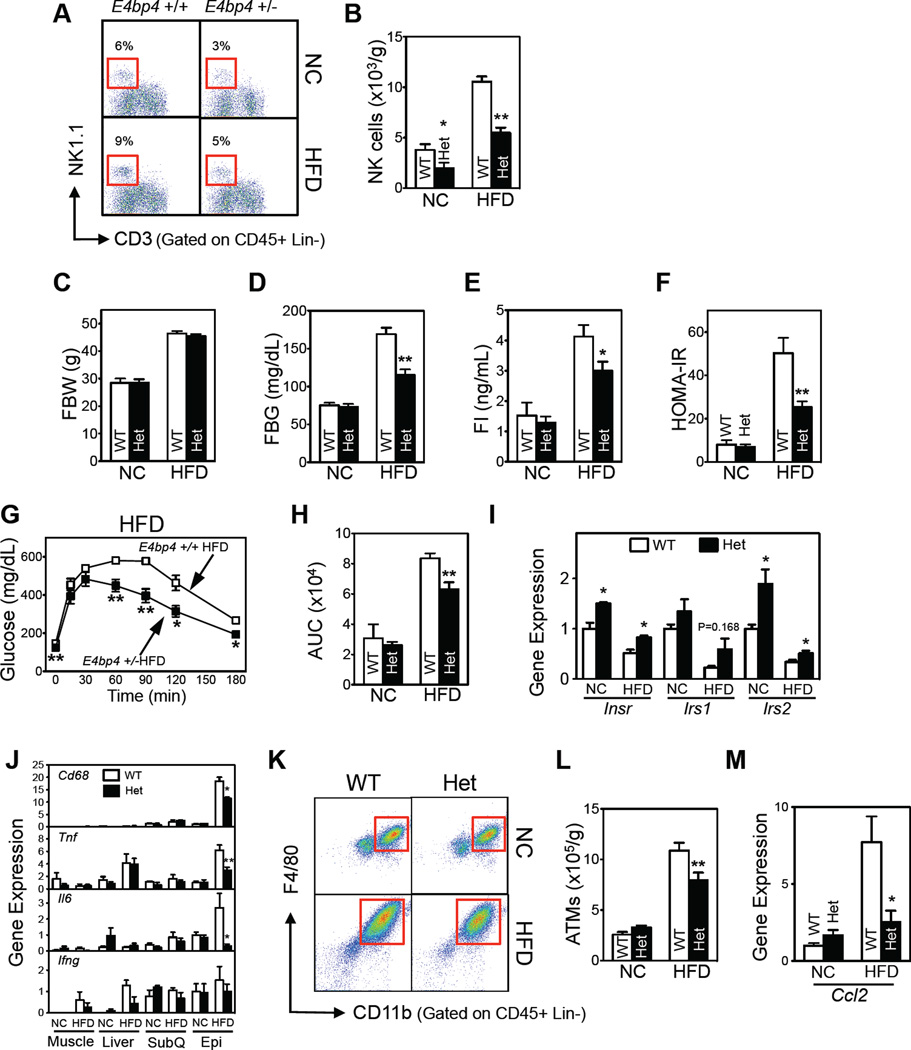
The effect of loss of NK cell function was further studied using NK cell-deficient E4bp4−/− mice (Gascoyne et al., 2009). Homozygous E4bp4−/− mice had 15% of the epididymal NK cells seen in wild-type (WT) controls (Figure S5A and S5B). However, compared to heterozygous and WT mice, both NC- and HFD-fed E4bp4−/− mice had significantly lower body and fat pad weights (Figure S5C and S5D). This accounts for improved metabolic parameters compared to the WT controls (Figure S5E and S5F). By contrast, the body and fat pad weights of heterozygous E4bp4+/− mice were indistinguishable from those of WT mice (Figures 5C, S5D and S5E). E4bp4+/− mice had ~50% of the NK cells seen in WT mice (Figures 5A and 5B). However, the numbers of iNKT and CD8 and CD4 T cells in spleen or epididymal fat were unaffected by the heterozygous deletion (Figures S5G and S5H). Glucose tolerance and insulin sensitivity were both improved in HFD-fed E4bp4+/− mice compared to the WT controls (Figures 5D–H, S5E and S5F). Thus, the metabolic changes in E4bp4+/− mice can be ascribed to lower NK cell numbers (Figures 5A and B). Consistent with the insulin sensitization data, the epididymal fat expression of insulin signaling genes (Insr, Irs1, and Irs2), which was decreased by HFD, was partly restored in E4bp4+/− mice (Figure 5I). Unlike the HFD mice, NC-fed E4bp4+/− mice had indistinguishable metabolic phenotypes compared to the WT controls (Figures 5D–H, S5E, and S5F).
Figure 5. Genetic ablation of NK cells in E4bp4 knockout mice improves HFD-induced insulin resistance.
E4bp4+/− heterozygous knockout mice. (A) Representative flow cytometric plots of the epididymal NK cells. The lineage markers (Lin) were TER-119, Gr-1, CD19, and F4/80. (B) Epididymal NK cell numbers. (C) Fasting body weight. (D) Fasting blood glucose levels. (E) Fasting serum insulin levels. (F) HOMA-IR. (G) GTT. (H) AUC of GTT. (I–J) Gene expression levels of insulin signaling (I) and inflammatory mediators (J), as determined by qRT-PCR. (K) Representative flow cytometric plots of ATMs. The lineage markers were TER-119, CD3, CD19, and NK1.1. (L) Total epididymal ATM numbers. (M) Ccl2 gene expression levels in epididymal fat, as determined by qRT-PCR. The data are presented as mean ± S.E.M. *p<0.05, **p<0.01, and ***p<0.001. n=5–6/group. See also Figure S5.
HFD in WT mice increases epididymal expression of Il6, Tnf, and Cd68 (Figure 5J). The expression of these genes was reduced in the E4bp4+/− mice. By contrast, the expression of these genes in subcutaneous fat, muscle, and liver was not affected (Figure 5J). Ifng expression was unaffected by HFD or E4bp4 heterozygosity in any of the tested tissues (Figure 5J). ATM numbers paralleled these changes: HFD-induced increases in ATMs were suppressed in epididymal fat of E4bp4+/− mice (Figures 5K and L). Poor recruitment of ATMs into epididymal fat may have mediated this suppression since the HFD-induced increase in adipose Ccl2 expression in WT mice was reversed in E4bp4+/− mice (Figure 5M).
Reconstitution of NK cells in E4bp4−/− mice
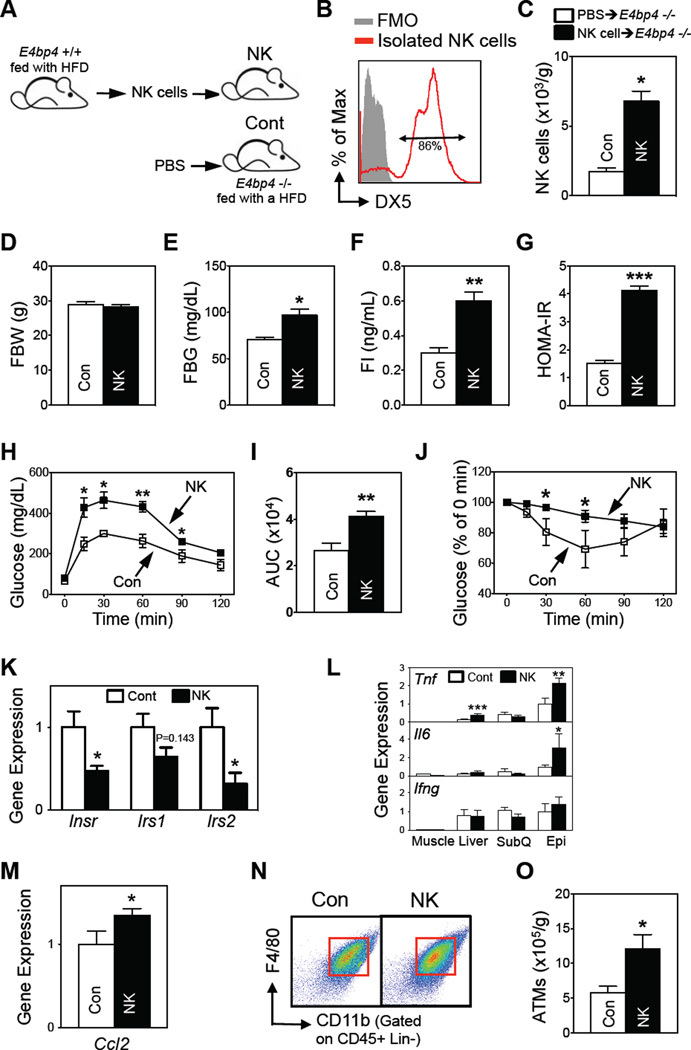
E4BP4 also plays roles in the development of other immune cells (Male et al., 2012) and in the regulation of metabolic genes, including Fgf21 (Tong et al., 2010). While E4bp4+/− and WT mice did not differ in terms of liver expression of Fgf21 (Figure S5I), we reconstituted E4bp4−/−mice with NK cells (>85% pure) to confirm that NK cell numbers, not other factors, were responsible for shaping the metabolism of E4bp4 knockout mice (Figures 6A and 6B). Reconstitution increased NK cell numbers in spleen and subcutaneous and epididymal fat but did not affect other lymphocytes, including CD8 T cells and iNKT cells, body weight, or adiposity (Figures 6C, 6D and S6A–S6E). Importantly, NK cell reconstitution did not affect liver Fgf21 expression (Figure S6F). However, it did increase fasting glucose and insulin and HOMA-IR (Figures 6E – G), worsened GTT and ITT responses (Figure 6H – J), and decreased epididymal expression of insulin signaling genes (Figure 6K). NK cell reconstitution also increased the epididymal expression of Il6 and Tnf and liver expression of Tnf without affecting inflammatory gene expression in other tissues (Figure 6L). Reconstitution did not change Ifng expression in any tissue (Figure 6L). However, epididymal fat Ccl2 expression and ATM numbers were increased in the NK cell-reconstituted mice (Figures 6M – O). Thus, gain of NK cell function appears to aggravate adipose tissue inflammation and insulin resistance.
Figure 6. Reconstitution of NK cells in E4bp4 knockout mice increases HFD-induced insulin resistance.
(A) Experimental scheme for reconstitution experiments (n=4–5/group). (B) Purity of the transferred NK cells. (C) Epididymal NK cell numbers. (D) Fasting body weight. (E) Fasting blood glucose levels. (F) Fasting serum insulin levels. (G) HOMA-IR. (H) GTT. (I) AUC of GTT. (J) ITT. (K–M) Epididymal fat gene expression of insulin signaling molecules (K), inflammatory mediators (L), and Ccl2 (M), as determined by qRT-PCR. (N) Representative flow cytometric plots of epididymal ATMs. The lineage markers were TER-119, CD3, CD19, and NK1.1. (O) Total epididymal ATM numbers (n=3–4/group). The data are presented as mean ± S.E.M. *p<0.05, **p<0.01, and ***p<0.001. See also Figure S6.
Discussion
Multiple lines of evidence in this study support the notion that epididymal NK cells play important roles in the metabolic derangements associated with obesity. First, during the induction of obesity, NK cell numbers and activation increased in the epididymal fat, and NK cell numbers correlated significantly with fasting glucose and insulin concentrations, HOMA-IR, and ATM numbers (Figures 1, 2S and S2L). Second, selective suppression of NK cell numbers reduced ATM numbers, decreased epididymal fat pad expression of pro-inflammatory genes, and improved systemic insulin resistance and glucose tolerance (Figures 2, 3, and 5). This was true regardless of whether NK cell suppression was achieved using mechanistically distinct GM1 and PK-136 neutralizing antibodies or heterozygous E4bp4+/− mice. Third, manipulations that expanded NK cells secondarily increased ATM numbers, elevated epididymal fat expression of pro-inflammatory genes, and exacerbated insulin resistance and glucose intolerance (Figures 4 and 6). This too was true regardless of whether NK cell expansion was through IL-15 administration or reconstitution of NK cells into NK cell-deficient mice. Our findings suggest that obesity increases NK cell numbers and activation in epididymal fat, which promotes local inflammation. It is often the case in metabolic studies that alterations in one tissue affect other tissues. Indeed, we found that although the inflammatory effects of NK cell depletion were observed primarily in epididymal fat, the depletion also improved insulin resistance in liver and muscle, despite having minimal direct effects on these tissues. The consistent correlations between epididymal NK cell numbers and their effects on ATMs, epididymal adipose tissue inflammation, and metabolic endpoints suggest that epididymal NK cells are novel regulators of both immunological and metabolic homeostasis in this adipose tissue depot.
In general, NK cells either kill other immune cells or regulate them through inflammatory mediator production. Obesity does not affect NK cell cytotoxicity, as indicated by CD107a expression (Wensveen et al., 2015). Moreover, in adipose tissue, the perforin-granzyme cell death pathway in CD8 T cells appears to play a more important role than the same pathway in NK cells (Revelo et al., 2015). Thus, epididymal NK cells are more likely to alter ATM functions and metabolic homeostasis by secreting inflammatory modulators. We found that epididymal NK cells produce two key inflammatory mediators, the macrophage chemokine CCL2 (Figures 2G, 5M, and 6M) and the pro-inflammatory cytokine TNFα (Figure 1J – L). Since NK cell depletion decreased HFD-induced Ccl2 expression in epididymal fat, and NK cell reconstitution into E4bp4 knockout mice increased it (Figures 2G, 5M, and 6M), it appears that NK cells may regulate ATM numbers by shaping CCL2-mediated ATM recruitment. Our findings also suggest that NK cells regulate the inflammatory status of ATMs, because NK cell depletion suppressed the expression of pro-inflammatory genes (e.g., Tnf and Itgax) and promoted the expression of anti-inflammatory or M2 genes (e.g., Il10 and Arg1) in sorted ATMs (Figure 2T). TNFα, IFNγ, and IL-6 are signature cytokines of NK cells (Vivier et al., 2008). Our results show that TNFα produced by epididymal NK cells is an important regulator of ATM inflammation as HFD increased both the frequencies of TNFα+ epididymal NK cells (6-fold) and the amount of TNFα produced by individual NK cells (>6-fold) (Figure 1). The corresponding effects on IFN-γ and IL-6 were marginal (Figure 1J–L).
Our results also showed that in addition to NK cell regulation of epididymal ATMs, the ATMs also regulate epididymal NK cells (Figure S1). With the induction of obesity, ATMs increased their production of NK cell chemoattractants such as CCL3, CCL4, and CXCL10 (Figure S1F), which are likely to promote the recruitment of NK cells in obesity. This notion is supported by the observation that when NK cells were transferred from obese WT mice into HFD-fed E4bp4−/− mice, they homed to adipose tissue (Figure 6C). Our findings also suggest that ATMs promote the local proliferation of NK cells by producing IL-15 (Figure S1H) (Ma et al., 2006). This possibility is supported by the study of Wensveen et al. (2015) showing that adipose BrdU+ and Ki67.2+ NK cell numbers increase with obesity. Therefore, the obesity-associated increases in NK cell numbers appear to be related to ATM-mediated effects on both NK cell migration and local proliferation.
We speculate that CCL3, CCL4, CXCL10, and IL-15 secreted by obesity-activated ATMs increase NK cell numbers and activation, while conversely, activated NK cells produce CCL2 and TNFα, which recruit and activate ATMs, respectively. The ATMs also produce pro-inflammatory cytokines, including IL-1β and TNFα (Kim et al., 2013), which can activate adipocytes, NK cells, and other immune cells. It therefore appears that NK cells and ATMs together create a self-amplifying pro-inflammatory adipose tissue milieu that promotes insulin resistance. In relation to this, it is notable that NK cell numbers markedly influence metabolic parameters in obese mice, yet NK cell gain or loss has little effect in lean mice (Figures 2 and 5). This is probably because the NK cells must be primed by obesity-activated ATMs and possibly adipocytes before they can exert their effects on ATMs.
Both NK and iNKT cell numbers were reduced by PK-136 antibody-mediated depletion in parallel with improvements in metabolic parameters. To distinguish whether the metabolic improvements were due to depletion of NK cells vs. iNKT cells, we assessed the role of iNKT cells using mice that lack iNKT cells. The results for both Jα18−/− and Cd1d−/− mice showed that the absence of iNKT cells had no influence on either body weight or metabolic phenotype (Figures 3M-3P and S3). Previous studies with Jα18−/− and Cd1d−/− mice yielded widely disparate conclusions about the potential metabolic roles of iNKT cells [reviewed in (Mathis, 2013; Lynch, 2014)]. However, the body weights of mice in these different studies also varied widely. We do not know why the same iNKT cell knockout mouse lines have different body weights and adiposity in different facilities. It is possible that environmental settings activate iNKT cells, which in turn reduce body weight and adiposity, thereby influencing metabolism (Lee and Lee, 2014). In any case, our studies with the iNKT knockout mouse lines indicated that the metabolic effects of PK-136 depletion were due to changes in NK cell numbers rather than iNKT cell numbers.
Recent studies show that NK and innate lymphoid cells (ILCs) are closely related (Eberl et al., 2015). For example, E4BP4 regulates NK cell and ILC development (Geiger et al., 2014; Seillet et al., 2014). Since NK cells and ILC Type 1 (ILC1) share some cell surface markers, including NKp46, it is formally possible that the NK cell populations we studied contained ILC1, and that ILC1 may have thus contributed to the regulation of the immunological and metabolic phenotypes in our studies. Weighing against this possibility, the frequencies of ILC1 in most tissues are extremely rare (Daussy et al., 2014; Robinette et al., 2015). Furthermore, Wensveen et al. (2015) showed that adipose NK cells gated according to CD3- and NK1.1+ do not express IL-7Rα (CD127) and c-Kit (CD117), the two markers that distinguish ILC1 and NK cells, which led them to conclude that the adipose NK cells do not contain ILC1. Our studies support this conclusion. When we reconstituted E4bp4−/− mice with NK cells isolated according to a similar gating strategies (Wensveen et al., 2015), obesity-induced inflammation and insulin resistance worsened (Figure 6). While relations between ILC1 and obesity should certainly be investigated, our results and those of Wensveen et al., support roles for roles for adipose NK cells, and not ILC1, in the development of obesity-induced inflammation and insulin resistance.
Our findings and conclusions are consistent with but extend and elaborate previous findings. Obese humans and animal models have both been shown to have greater adipose NK cell numbers than lean controls (Nave et al., 2008; Duffaut et al., 2009; O'Rourke et al., 2009; Xu et al., 2013). Other studies have also shown that NK cell reduction, either by diphtheria toxin depletion (O'Rourke et al., 2014) or PK-136 antibody neutralization (Wensveen et al., 2015), improved metabolic parameters. Wensveen et al. also used additional approaches to conclude as we have that NK cells regulate ATMs and thereby influence glucose tolerance and insulin resistance. However, our studies also differed in terms of kinetics and potential cytokine mediators. Their carefully conducted experiments led them to conclude that in obesity adipocytes upregulate ligands for the NK cell-activating receptor NCR1, which triggers epididymal NK cells to proliferate and produce IFN-γ. They therefore concluded that INFγ stimulates ATMs and eventually promotes insulin resistance. Our results may appear to be at odds with these observations, as we found that obesity only slightly increased the frequencies of IFNγ-producing epididymal NK cells, without increasing IFNγ production in individual NK cells (Figure 1J–L). Furthermore, neither HFD nor NK cell number changes affected Ifng gene expression in epididymal fat or other tissues (Figures 2J, 5J, and 6L). By contrast, we found that obesity strongly promoted both TNFα production in epididymal fat and the numbers of TNFα-producing NK cells (60% vs 4% for IFNγ). Moreover, increases and decreases in NK cell numbers elevated and reduced epididymal TNFα gene expression in tandem. These results led us to conclude that NK cell-derived TNFα, and not IFNγ, is a primary driver of ATM activation.
We reconcile this apparent discrepancy as follows. Wensveen et al generally assessed metabolic responses after 12 weeks of HFD, while their immunological analyses were after shorter, 2–6 week, periods of HFD feeding. By contrast, we assessed both immunological and metabolic outcomes at the same time, typically after 12 weeks of HFD. Thus, the immunological observations of Wensveen et al. reflect early events during the induction of insulin resistance, whereas ours reveal the immunological status at a later stage of obesity. Together our studies show that NK cells play important roles both early and later in obesity, with an initial IFNγ-centric mechanism that then transitions into a TNFα-centric mechanism. Notably, the metabolic phenotypes of WT and E4bp4+/− mice only differed after 9 weeks of HFD (Figure S5), suggesting that NK cell-mediated metabolic regulation may start at a later stage of obesity, when NK cells produce TNFα. We speculate that the early NK cell-mediated events (in which IFNγ plays a critical role) may be important for establishing a noxious environment that leads to ATM stimulation and downstream immune events. Later in obesity, NK cells start to produce TNFα, which may then promote insulin resistance.
The kinetics of PK-136 antibody-mediated NK cell depletion represented a second noteworthy difference between our experimental designs. Wensveen et al. initiated their PK-136 antibody depletions concurrently with the initiation of HFD. Since we intended to mimic treatment as opposed to prevention strategies, we initiated PK-136 antibody depletion (and other manipulations to induce the loss or gain of NK cells) after 8 weeks of HFD, at which point obesity was firmly established. Nevertheless, the two experimental protocols showed similar metabolic improvements, suggesting that either prevention or intervention might prove to be beneficial.
It is likely that as our understanding of the potential roles of inflammation in the pathogenesis of obesity-induced insulin resistance evolves, new concepts and even new cell types will be considered. Epididymal NK cells and their regulation of neighboring ATMs in epididymal fat are important new additions to the field. Moreover, since we showed that therapeutic manipulations of NK cells altered metabolic outcomes in obese animals, adipose NK cells may represent a new therapeutic target for treating insulin resistance and type 2 diabetes.
Experimental Procedures
Animals
All animal experiments were conducted in accordance with the NIH guidelines under protocols approved by the Institutional Animal Care and Use Committee of the Joslin Diabetes Center. C57BL/6 mice were purchased from the Jackson Lab. Jα18 and CD1d knockout mice were kindly provided by Dr. Michael Brenner (Harvard Medical School). The Jα18, CD1d, and E4bp4 knockout mice are fully backcrossed onto the C57BL/6 background (>10 generations). Only male animals were used. The wild-type (WT) controls in all experiments were littermate controls. All animals were maintained under a standard light cycle (12 h light/dark) and allowed free access to water and food. To induce obesity and insulin resistance, mice were fed a high fat diet (HFD, 60% fat, D12492, Research Diets, Inc.) for the indicated time. Mice fed normal chow (NC, 10% fat, D12450B, Research Diets, Inc.) served as controls.
NK cells
To deplete NK cells, C57BL/6 mice fed NC or HFD for 8 weeks were injected intraperitoneally with anti-asialo GM1 (550 µg/mouse, Wako) (Godeny and Gauntt, 1987) or anti-mouse NK-1.1 antibody (PK-136; 150 µg/mouse, BioLegend) (Andoniou et al, 2005) every 3–4 days for another 4 weeks while continuing their diet. Host species-matched isotype-control antibodies were injected as controls. Metabolic and immunological analyses were performed 2 days after the last injections. To expand NK cells in vivo, mice fed NC or HFD for 6 weeks were intraperitoneally injected with IL-15 (0.5 µg/mouse, R&D system) every 3 days for 4 weeks. Metabolic and immunological analyses were performed 3 days after the last injections. To reconstitute E4bp4−/− homozygous knockout mice with NK cells, splenic NK cells were isolated from C57BL/6 WT mice that had been fed HFD for 8 weeks by negative sorting using a mouse NK cell isolation kit (Miltenyi Biotec). The preparations consisted predominantly (>85%) of NK cells. The E4bp4−/− homozygous knockout mice were then injected retroorbitally on three occasions with 1.3×107 NK cells at 10-day intervals. Metabolic and immunological analyses were performed 7 days after the last NK cell injection.
Metabolic phenotype measurements
Fasting blood glucose and insulin levels were measured after overnight fasting. Fasting blood glucose levels were determined by using a Contour glucose meter (Bayer). Fasting insulin levels were assayed by using an ELISA kit (Crystal Chem). The insulin resistance index HOMA-IR was calculated on the basis of the fasting glucose and insulin levels (Matthews et al., 1985). The GTT was performed by intraperitoneally injecting glucose (1.5 g/kg body weight) after overnight fasting. Insulin tolerance tests (ITT) were performed by intraperitoneally injecting insulin (0.75 U/kg body weight) after 6 h of fasting. Blood samples were drawn from the tail vein before and 15, 30, 60, and 120 min after the glucose or insulin injection, and the glucose levels were measured by using a Contour glucose meter (Bayer).
Measurements of in vivo insulin signaling
For in vivo stimulation of insulin signaling, mice were fasted for 16 h and then anesthetized with an i.p. injection of pentobarbital (100 mg/kg). Insulin was injected (5 U/Kg) through the inferior vena cava. After 5 min, Liver and skeletal muscles from the hind limbs were harvested. Liver and muscles were homogenized and lysed at 4 °C, and lysates were subjected to western blotting to assess insulin-stimulated phosphorylation cascades as described previously (Yuan et al., 2001).
Isolation of adipose stromal vascular cells (SVCs) and splenocytes
SVCs were isolated from adipose tissues by using a well-established collagenase-based method (Herrero et al., 2010; Kim et al., 2013). Briefly, epididymal and subcutaneous fat pads were harvested from mice, weighed, and chopped into small pieces in PBS containing 2% (wt/vol) BSA. Collagenase type 2 (10 mg/ml, Worthington) and deoxyribonuclease I (2 mg/ml, Sigma-Aldrich) were added to the tissue suspensions, which were then incubated for 20 min at 37°C with shaking. The cell suspensions were then filtered through a 250 µm nylon mesh and centrifuged at 200 × g for 3 min to separate the floating adipocyte and pelleted SVC fractions. To collect any remaining adipocyte-associated SVCs, the floating adipocytes were washed twice with PBS containing 2% BSA and 5 mM EDTA. The two SVC pellets were then combined, resuspended in PBS containing 2% (vol/vol) FBS, and subjected to flow cytometric analyses. To obtain splenocytes, spleens were excised, weighed, and minced in 1 ml of PBS containing 2% (vol/vol) FBS. The RBCs were lysed by adding 4.5 ml of Ack lysis buffer (Lonza) into the splenocyte suspension. The splenocytes were then collected by centrifugation at 300 × g for 5 min at 4°C. After washing, the splenocytes were filtered through a 70-µm mesh and subjected to flow cytometric analyses.
Flow cytometric analyses
Blood was collected from the tail vein in the presence of 5 mM EDTA, incubated with BD Fc Block, stained with fluorophore-conjugated antibodies specific for cell-surface makers (Tables S1 and S2), and lysed with FACS Lysing Solution (BD Biosciences). The SVC and splenocyte preparations were incubated with BD Fc Block and then stained with antibodies. Cell viability was determined by staining with propidium iodide. The stained cells were then analyzed by LSRII flow cytometer (BD Biosciences) or sorted by using the FACSAria instrument (BD Biosciences). The data were further analyzed by using the FlowJo software (Flow Jo). For the NK cell analyses, NK cells were defined as CD45+, TER119−, F4/80−, CD19−, CD3−, and NK1.1+ in lymphocyte/monocyte gating. iNKT cells were defined as CD45+, TER119−, F4/80−, CD19−, CD3+, and NK1.1+ in lymphocyte/monocyte gating. Macrophages were defined as CD45+, TER119−, CD19−, CD3−, NK1.1−, F4/80+, and CD11b+. To identify the obesity-specific ATM subpopulation, antibodies against CD11c were used as well as the antibodies employed for macrophage identification. The antibodies used to identify CD4 and CD8 T cells, and B cells are listed in Table S1. Total cell numbers were calculated by using a BD Cell viability kit (BD Biosciences) according to the manufacturer’s instructions.
Measurements of ex vivo cytokine production
Isolated splenocytes or SVCs from fat pads were stimulated with PMA (50 ng/ml, Sigma-Aldrich) and ionomycin (1 nM, Sigma-Aldrich) for 4 hours. Golgistop (BD) (4 µl) was added during the last 3 hours. The cells were then stained with antibodies against cell-surface markers, fixed, and permeabilized by using a Cytofix/CytoPerm kit (BD), followed by intracellular staining of IFN-γ, TNF-α, IL-10, or IL-6 (Feuerer et al., 2009). The cells were then analyzed by using LSRII instruments and FlowJo software (Flow Jo).
Quantitative real-time RT-PCR analysis
Total RNA from tissues and ATMs was prepared by using Trizol and RNeasy Lipid Tissue Mini Kit (Qiagen). The RNA from ATMs was amplified by using a MessageAmpII aRNA kit (Ambion), after which cDNA was generated by using an Advantage RT-PCR kit (Clontech). Gene expression levels were determined by real-time RT-PCR. The probes used in the experiments were for CD68 (Cd68, Mm00839636_g1), F4/80 (Adgre1, Mm00802530_m1), CD11c (Itgax, Mm00498698_m1), TNFα (Tnf, Mm00443258_m1), IL-1β (Il1b, Mm00434227_g1), IL-6 (Il6, Mm00446190_m1), IFNγ (Ifng, Mm01168134_m1)m, IL-10 (Il10, Mm00439614_m1), Arg1 (Arg1, Mm00475988_m1), IL-15 (Il15, Mm00434310_m1), IL-12a (Il12a, Mm00434169_m1), IL-12b (Il12b, Mm01288989_m1), IL-13 (Il13, Mm00434204_m1), IL18 (Il18, Mm00434226_m1), CCL2 (Ccl2, Mm00441242_m1), CX3CL1 (Cx3cl1, Mm00436454_m1), CCL3 (Ccl3, Mm00441259_g1), CCL4 (Ccl4, Mm00443111_m1), CCL5 (Ccl5, Mm01302427_m1), CCL7 (Ccl7, Mm00443113_m1), CXCL9 (Cxcl9, Mm00434946_m1), CXCL10 (Cxcl10, Mm00445235_m1), CXCL11 (Cxcl11, Mm00444662_m1), IR (Insr, Mm01211875_m1), IRS-1 (Irs1, Mm01278327_m1), IRS-2 (Irs2, Mm03038438_m1), and FGF21 (Fgf21, Mm00840165_g1). GAPDH (Gapdh, Mm99999915_g1) served as a housekeeping control gene (Applied Biosystems).
Statistical analysis
Statistical significance was examined by using two-tailed unpaired Student’s t-tests or one-way ANOVA, followed by Fisher’s least significant difference test. The variance of each group was analyzed by F test or Bartlett’s test. The data are presented as mean ± S.E.M. P-values <0.05 were considered to indicate statistical significance.
Supplementary Material
Acknowledgments
We thank Drs. Diane Mathis (Harvard Medical School) and Lydia Lynch (Brigham and Women's Hospital/Harvard Medical School) for helpful discussions. We also thank Drs. E. Leadbetter, M.B. Brenner, M.A. Exley, and S. Balk (Harvard Medical School) for providing the Jα18 and Cd1d knockout mice. We thank J. LaVecchio and G. Buruzula at Joslin Diabetes Research Center Flow Cytometry Core. This work was supported by grants from the Korea Food Research Institute E0150302-01 and E0080202-08 (M.-S.K.); the Helen and Morton Adler Chair (S.E.S.); National Institutes of Health (NIH) grants R21 DK80380 (J.L.), R01 DK95327 (S.E.S.), and P30 DK36836 (Joslin Diabetes Research Center); and the American Diabetes Association grants RA 1-10-BS-97 and 1-15-BS-111 (J.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
B.-C.L., M.-S.K., S.E.S., and J.L. designed the studies. B.-C.L., M.-S.K., M.P., Y.Y., N.K., T.S., and J.Y., W.M. performed the experiments and analyzed the data. D.E. performed the iNKT cell knockout mouse experiments. H.P. performed the qRT-PCR analysis. S.S., and J.W. performed the insulin signaling experiment. H.B. provided the E4bp4 knockout mice. B.-C.L., M.-S.K., S.E.S., and J.L. wrote the manuscript.
References
- Andoniou CE, van Dommelen SL, Voigt V, Andrews DM, Brizard G, Asselin-Paturel C, Delale T, Stacey KJ, Trinchieri G, Degli-Esposti MA. Interaction between conventional dendritic cells and natural killer cells is integral to the activation of effective antiviral immunity. Nat Immunol. 2005;6:1011–1019. doi: 10.1038/ni1244. [DOI] [PubMed] [Google Scholar]
- Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11:889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- Cipolletta D, Feuerer M, Li A, Kamei N, Lee J, Shoelson SE, Benoist C, Mathis D. PPAR-γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486:549–553. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daussy C, Faure F, Mayol K, Viel S, Gasteiger G, Charrier E, Bienvenu J, Henry T, Debien E, Hasan UA, et al. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J Exp Med. 2014;211:563–577. doi: 10.1084/jem.20131560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Santo JP. Natural killer cell developmental pathways: a question of balance. Annu Rev Immunol. 2006;24:257–286. doi: 10.1146/annurev.immunol.24.021605.090700. [DOI] [PubMed] [Google Scholar]
- Duffaut C, Galitzky J, Lafontan M, Bouloumie A. Unexpected trafficking of immune cells within the adipose tissue during the onset of obesity. Biochem Biophys Res Commun. 2009;384:482–485. doi: 10.1016/j.bbrc.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Eberl G, Di Santo JP, Vivier E. The brave new world of innate lymphoid cells. Nat Immunol. 2015;16:1–5. doi: 10.1038/ni.3059. [DOI] [PubMed] [Google Scholar]
- Exley MA, Bigley NJ, Cheng O, Shaulov A, Tahir SM, Carter QL, Garcia J, Wang C, Patten K, Stills HF, et al. Innate immune response to encephalomyocarditis virus infection mediated by CD1d. Immunology. 2003;110:519–526. doi: 10.1111/j.1365-2567.2003.01779.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerer M, Herrero L, Cipolletta D, Naaz A, Wong J, Nayer A, Lee J, Goldfine AB, Benoist C, Shoelson S, et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat Med. 2009;15:930–939. doi: 10.1038/nm.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegal KM, Carroll MD, Kit BK, Ogden CL. Prevalence of obesity and trends in the distribution of body mass index among US adults, 1999–2010. JAMA. 2012;307:491–497. doi: 10.1001/jama.2012.39. [DOI] [PubMed] [Google Scholar]
- Fujisaka S, Usui I, Bukhari A, Ikutani M, Oya T, Kanatani Y, Tsuneyama K, Nagai Y, Takatsu K, Urakaze M, et al. Regulatory mechanisms for adipose tissue M1 and M2 macrophages in diet-induced obese mice. Diabetes. 2009;58:2574–2582. doi: 10.2337/db08-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoyne DM, Long E, Veiga-Fernandes H, de Boer J, Williams O, Seddon B, Coles M, Kioussis D, Brady HJ. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat Immunol. 2009;10:1118–1124. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- Geiger TL, Abt MC, Gasteiger G, Firth MA, O'Connor MH, Geary CD, O'Sullivan TE, van den Brink MR, Pamer EG, Hanash AM, et al. Nfil3 is crucial for development of innate lymphoid cells and host protectiohogens. J Exp Med. 2014;211:1723–1731. doi: 10.1084/jem.20140212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godeny EK, Gauntt CJ. Murine natural killer cells limit coxsackievirus B3 replication. J Immunol. 1987;139:913–918. [PubMed] [Google Scholar]
- Herrero L, Shapiro H, Nayer A, Lee J, Shoelson SE. Inflammation and adipose tissue macrophages in lipodystrophic mice. Proc Natl Acad Sci U S A. 2010;107:240–245. doi: 10.1073/pnas.0905310107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-α: direct role in obesity-linked insulin resistance. Science. 1993;259:87–91. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
- Kim MS, Yamamoto Y, Kim K, Kamei N, Shimada T, Liu L, Moore K, Woo JR, Shoelson SE, Lee J. Regulation of diet-induced adipose tissue and systemic inflammation by salicylates and pioglitazone. PLoS One. 2013;8:e82847. doi: 10.1371/journal.pone.0082847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchgessner TG, Uysal KT, Wiesbrock SM, Marino MW, Hotamisligil GS. Tumor necrosis factor-α contributes to obesity-related hyperleptinemia by regulating leptin release from adipocytes. J Clin Invest. 1997;100:2777–2782. doi: 10.1172/JCI119824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaichi N, Kotake S, Morohashi T, Onoe K, Ohno S, Taylor AW. Diminution of experimental autoimmune uveoretinitis (EAU) in mice depleted of NK cells. J Leukoc Biol. 2002;72:1117–1121. [PubMed] [Google Scholar]
- Kroner A, Greenhalgh AD, Zarruk JG, Passos Dos Santos R, Gaestel M, David S. TNF and increased intracellular iron alter macrophage polarization to a detrimental M1 phenotype in the injured spinal cord. Neuron. 2014;83:1098–1116. doi: 10.1016/j.neuron.2014.07.027. [DOI] [PubMed] [Google Scholar]
- Leadbetter EA, Brigl M, Illarionov P, Cohen N, Luteran MC, Pillai S, Besra GS, Brenner MB. NK T cells provide lipid antigen-specific cognate help for B cells. Proc Natl Acad Sci U S A. 2008;105:8339–8344. doi: 10.1073/pnas.0801375105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BC, Lee J. Cellular and molecular players in adipose tissue inflammation in the development of obesity-induced insulin resistance. Biochim Biophys Acta. 2014;1842:446–462. doi: 10.1016/j.bbadis.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumeng CN, DelProposto JB, Westcott DJ, Saltiel AR. Phenotypic switching of adipose tissue macrophages with obesity is generated by spatiotemporal differences in macrophage subtypes. Diabetes. 2008;57:3239–3246. doi: 10.2337/db08-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch L. Adipose invariant natural killer T cells. Immunology. 2014;142:337–346. doi: 10.1111/imm.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma A, Koka R, Burkett P. Diverse functions of IL-2, IL-15, and IL-7 in lymphoid homeostasis. Annu Rev Immunol. 2006;24:657–679. doi: 10.1146/annurev.immunol.24.021605.090727. [DOI] [PubMed] [Google Scholar]
- Male V, Nisoli I, Gascoyne DM, Brady HJ. E4BP4: an unexpected player in the immune response. Trends Immunol. 2012;33:98–102. doi: 10.1016/j.it.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Mathis D. Immunological goings-on in visceral adipose tissue. Cell Metab. 2013;17:851–859. doi: 10.1016/j.cmet.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- McNelis JC, Olefsky JM. Macrophages, immunity, and metabolic disease. Immunity. 2014;41:36–48. doi: 10.1016/j.immuni.2014.05.010. [DOI] [PubMed] [Google Scholar]
- Nave H, Mueller G, Siegmund B, Jacobs R, Stroh T, Schueler U, Hopfe M, Behrendt P, Buchenauer T, Pabst R, et al. Resistance of Janus kinase-2 dependent leptin signaling in natural killer (NK) cells: a novel mechanism of NK cell dysfunction in diet-induced obesity. Endocrinology. 2008;149:3370–3378. doi: 10.1210/en.2007-1516. [DOI] [PubMed] [Google Scholar]
- Nishimura S, Manabe I, Nagasaki M, Eto K, Yamashita H, Ohsugi M, Otsu M, Hara K, Ueki K, Sugiura S, et al. CD8+ effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity. Nat Med. 2009;15:914–920. doi: 10.1038/nm.1964. [DOI] [PubMed] [Google Scholar]
- O'Rourke RW, Metcalf MD, White AE, Madala A, Winters BR, Maizlin II, Jobe BA, Roberts CT, Slifka MK, Marks DL., Jr Depot-specific differences in inflammatory mediators and a role for NK cells and IFN-γ in inflammation in human adipose tissue. Int J Obes (Lond) 2009;33:978–990. doi: 10.1038/ijo.2009.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revelo XS, Tsai S, Lei H, Luck H, Ghazarian M, Tsui H, Shi SY, Schroer S, Luk CT, Lin GH, et al. Perforin is a novel immune regulator of obesity-related insulin resistance. Diabetes. 2015;64:90–103. doi: 10.2337/db13-1524. [DOI] [PubMed] [Google Scholar]
- Robinette ML, Fuchs A, Cortez VS, Lee JS, Wang Y, Durum SK, Gilfillan S, Colonna M, Immunological Genome C. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol. 2015;16:306–317. doi: 10.1038/ni.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein MP, Kovar M, Purton JF, Cho JH, Boyman O, Surh CD, Sprent J. Converting IL-15 to a superagonist by binding to soluble IL-15Rα. Proc Natl Acad Sci U S A. 2006;103:9166–9171. doi: 10.1073/pnas.0600240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seillet C, Rankin LC, Groom JR, Mielke LA, Tellier J, Chopin M, Huntington ND, Belz GT, Carotta S. Nfil3 is required for the development of all innate lymphoid cell subsets. J Exp Med. 2014;211:1733–1740. doi: 10.1084/jem.20140145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X, Muchnik M, Chen Z, Patel M, Wu N, Joshi S, Rui L, Lazar MA, Yin L. Transcriptional repressor E4-binding protein 4 (E4BP4) regulates metabolic hormone fibroblast growth factor 21 (FGF21) during circadian cycles and feeding. J Biol Chem. 2010;285:36401–36409. doi: 10.1074/jbc.M110.172866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trambley J, Bingaman AW, Lin A, Elwood ET, Waitze SY, Ha J, Durham MM, Corbascio M, Cowan SR, Pearson TC, et al. Asialo GM1(+) CD8(+) T cells play a critical role in costimulation blockade-resistant allograft rejection. J Clin Invest. 1999;104:1715–1722. doi: 10.1172/JCI8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9:503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensveen FM, Jelencic V, Valentic S, Sestan M, Wensveen TT, Theurich S, Glasner A, Mendrila D, Stimac D, Wunderlich FT, et al. NK cells link obesity-induced adipose stress to inflammation and insulin resistance. Nat Immunol. 2015;16:376–385. doi: 10.1038/ni.3120. [DOI] [PubMed] [Google Scholar]
- Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, Sole J, Nichols A, Ross JS, Tartaglia LA, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112:1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Grijalva A, Skowronski A, van Eijk M, Serlie MJ, Ferrante AW., Jr Obesity activates a program of lysosomal-dependent lipid metabolism in adipose tissue macrophages independently of classic activation. Cell Metab. 2013;18:816–830. doi: 10.1016/j.cmet.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Konstantopoulos N, Lee J, Hansen L, Li ZW, Karin M, Shoelson SE. Reversal of obesity- and diet-induced insulin resistance with salicylates or targeted disruption of Ikkβ. Science. 2001;293:1673–1677. doi: 10.1126/science.1061620. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.