Abstract
Copy number variability at 16p13.11 has been associated with intellectual disability, autism, schizophrenia, epilepsy and attention-deficit hyperactivity disorder. Adolescent/adult- onset psychosis has been reported in a subset of these cases. Here, we report on two children with CNVs in 16p13.11 that developed psychosis before the age of 7. The genotype and neuropsychiatric abnormalities of these patients highlight several overlapping genes that have possible mechanistic relevance to pathways previously implicated in Autism Spectrum Disorders, including the mTOR signaling and the ubiquitin-proteasome cascades. A careful screening of the 16p13.11 region is warranted in patients with childhood onset psychosis.
INTRODUCTION
New candidate genes and chromosomal regions associated with psychotic disorders including schizophrenia are being discovered at a rapid rate [Abazyan et al., 2014; Abdolmaleky et al., 2014; Andreassen et al., 2014; Balan et al., 2014; Chen et al., 2014; Collins et al., 2014; Guella et al., 2014; Wirgenes et al., 2014; Won et al., 2014; Xu et al., 2014; Yao et al., 2014; Yuan et al., 2014; Zhao et al., 2014]. This family of illnesses is considered to be highly heritable with complex genetic etiology.
Copy number variants (CNVs) are chromosomal rearrangements involving large segments of DNA, ranging from 1000 to several million base pairs in length. CNVs are results of deletions or duplications, and are potentially accompanied by inversion or translocation events. While many CNVs are believed to be benign polymorphisms, others are associated with highly variable and pleiotropic phenotypes that often include both neurodevelopmental and somatic disruptions. For example, 22q11.2 deletions, the first CNVs found to be associated with schizophrenia, variably cause multiple congenital anomalies, intellectual disability, palatal and skeletal anomalies, and cardiac defects [Antshel et al., 2005].
Several additional large and rare CNVs have subsequently been implicated in the etiology of schizophrenia [Szatkiewicz et al., 2014], however, mechanistic insight into pathways leading from genotype to phenotype are not well understood. Many CNVs associated with schizophrenia have also been shown to increase risk for other disorders, such as autism spectrum disorders, intellectual deficit, developmental delay and epilepsy [McCarthy et al., 2014]. Thus, when a CNV is implicated in one mental health disorder, conviction of its causality is strengthened when it has already been implicated in another [Doherty and Owen 2014]. Several recurrent reciprocal 16p rearrangements have been implicated in psychiatric disorders, including 16p11.2 [McCarthy et al., 2009] and 16p13.11 [Ramalingam et al., 2011]. Patients bearing microdeletion and microduplications in 16p11.2 exhibit a range of neurocognitive phenotypes, macro- and microcephaly respectively and a high incidence of autism spectrum disorders, while microdeletions have also been associated with schizophrenia [McCarthy et al., 2009]. Patients bearing deletions within the 16p13.11 region have been reported to exhibit a spectrum of epilepsy disorders [Heinzen et al., 2010] in addition to microcephaly, developmental delay, and a range of psychiatric disorders [Nagamani et al., 2011] while duplications in this region have been reported in patients with intellectual disability, autism, seizure disorders, dysmorphic features and congenital anomalies [Ramalingam et al., 2011]. Both deletion and duplication of the 16p13.11 region have been implicated in adolescent and adult onset schizophrenia [Tropeano et al., 2013].
Here we describe two patients with childhood-onset psychosis who harbor CNVs at 16p13.11. The finding of childhood onset forms of a more typically young-adult onset condition further strengthens the evidence that perturbed copy number at this locus may contribute to the development of psychosis [Nagamani et al., 2011].
Clinical Reports
Patient #1
Patient 1 is a 9-year-old boy with childhood onset psychosis. His mother was treated with sertraline at the end of her pregnancy. He was born full term via vaginal delivery with a birth weight of 3.69 kilograms. No neonatal or perinatal complications were reported. At 5 years of age, the patient had an increased blood lead level of 9 mcg/dL, which subsequently normalized when the family moved residences. Variable intermittent exotropia was noted by his psychiatrist but has not been confirmed by an ophthalmologist. An electroencephalogram (EEG) done to assess staring spells was normal.
Patient 1 sat at 6 months, crawled at 8 months and took his first steps at 10 months. He has a history of motor dyscoordination and received occupational and physical therapy in school. He was trained for bowel and bladder continence at 4 years of age. He said his first word other than "mama" and "dada" at 3.5 years of age, spoke in phrases at 4.5 years of age and sentences at 5.5 to 6 years of age. At age 9 years he was found to be functioning in the below-average range in nonverbal and spatial reasoning (SS = 81 and 83, 10th and 13th centiles, respectively) and in the low range of verbal reasoning (SS = 71, 3rd centile).
Since earliest childhood, Patient 1 has had frequent tantrums with screaming episodes and physical aggression. Around 6 years of age, the patient’s psychotic symptoms presented with intermittent hallucinations and delusions progressing over the next several years to daily auditory, visual and tactile hallucinations with considerable distress. These included seeing aliens in trees, and multiple voices coming out of walls or the school intercom telling him to hurt himself and others. He would see the visual hallucinations with his eyes open or closed, and found it difficult to hear people speaking to him when he was experiencing auditory hallucinations. He was diagnosed with childhood onset schizophrenia. The hallucinations remitted within several weeks after starting risperidone.
Family history is significant for probable schizophrenia in Patient 1’s biological father. He was not available and not tested for CNV. The patient’s mother has history of depression and anxiety.
Social history was significant for physical abuse by the biological father to the biological mother during her pregnancy and to Patient 1 during the first year of his life. The biological father left the family when Patient 1 was one year of age.
Patient #2
Patient 2 is a 5-year-old girl diagnosed with Chiari type I malformation, seizures, autism spectrum disorder, and childhood onset psychosis. She was 4.17 kilograms at birth following a 38-week gestation pregnancy and was delivered by cesarean section due to concerns of maternal hypertension. Patient 2 began having staring spells at 1.5 years of age and had a witnessed generalized tonic clonic seizure at approximately 3 years of age. Her EEG was consistent with epilepsy of generalized onset, with sleep activated bioccipital slowing and a photoparoxysmal response with generalized spike and wave complexes. Oxcarbazepine failed to control her seizures and levetiracetam increased her behavioral problems. Her seizures are currently controlled with lamotrigine.
An MRI of the brain was significant only for a Chiari 1 malformation for which she has undergone two surgeries for decompression; the second procedure was conducted due to concerns for an increasing syrinx. Her mother has described intermittent exotropia of both eyes. An ophthalmologist did not observe an episode of exotropia during examination and noted that variable intermittent exotropia was likely. She has a history of cyclic neutropenia and episodes of hypoglycemia. She has no history of any hearing or vision problem.
Patient 2 began walking around 9 months. She started picking up objects with pincer grasp around 14 months. Early Intervention Services began at 18 months of age and the early intervention staff voiced concerns about behavioral dysregulation, speech and social skills, as well as hyperactivity. Her first words were at 2.5 years, including “please” and “thank you” and she had been using the signs for these beforehand. She did not have phrases until she was almost 36 months old. Patient 2 was diagnosed by a Psychologist with expertise in Autism Spectrum Disorders, using the DSM-IV-TR. She met criteria on the developmental history and the Autism Diagnostic Observation Schedule (Module 2), and using the comparison score, which is a measure of severity across modules [Gotham et al., 2009], she scored a 10 out of possible 10. Consistent with this rating, Patient 2 requires very substantial supports for social communication and fixated interests/repetitive behaviors. She received applied behavioral analysis treatment with good results. She started full day programming at 47 months, including half of her time in an integrated classroom and half of her time in a classroom for children with ASD, in addition to Occupational Therapy for 60 minutes a week. A Mental Health Worker, Family Partner, Intensive Care Coordinator, and TILL would also visit the house. Patient 2 now has good joint attention, which is the ability to share a common focus (on people, objects, a concept, an event, etc.) with someone else, and improved social interest and reciprocity. Her speech is still delayed, but testing at age 3 found her visual perception and fine motor performance to be in the average range.
Patient 2 began to report disturbing visual hallucinations at 4 years of age including monsters, a big black wolf, a man with blood on his face, and spiders crawling in her ears. She exhibited paranoia and suffered delusions that prevented her from sleeping in her bedroom. She would wipe at her ears and say, "Get them out!" On other occasions, she would scream and say that crackers where eating her hand. She additionally refused to sleep in her room anymore, because she said, "It is waiting for me." She also started talking about people staring at her. The intensity of the symptoms was reduced on medication, and the frequency fell from daily to 2 or 3 times per week on risperidone, and then to only once every few months on haloperidol.
There is a family history of idiopathic seizures on both sides of her family: her father had a history of photo-induced epilepsy, while her maternal grandmother was reported to have "petit mal-like seizures”.
Social history is unremarkable.
METHODS
Each patient and available first generation relatives underwent phlebotomy for chromosomal microarray analysis of peripheral blood lymphocytes (Claritas Genomics, Cambridge MA). Whole exome sequencing was provided by Yale University Centers for Mendelian Genomics on a Illumina HiSeq 2000 instrument with blood samples pooled 6 per lane. Libraries (TruSeq DNA v2 Sample Preparation kit; Illumina, San Diego, CA) and whole exome capture (EZ Exome 2.0, Roche) were performed according to standard protocols. FASTQs were aligned by Codified Genomics (proprietary algorithm, Houston, TX).
Medical records of the patients and their families were collected by The Manton Center for Orphan Disease Research, Gene Discovery Core under informed consent governed by the Institutional Review Board of Boston Children's Hospital. A standard assessment was performed, which documented physical features and recorded medical, developmental, psychiatric, and family illness history, supplemented by medical records (Table I).
Table 1.
Characteristics of 16p13.11 variations
| Parameter | Case #1 | Case #2 |
|---|---|---|
| Psychosis | yes | yes |
| Age of psychosis onset | 72 months | 48 months |
| Microcephaly | No | No |
| Macrocephaly | No | No |
| Epilepsy | No | Yes |
| Global DD | No | Borderline or No |
| Learning disability | Below average | Yes (borderline) |
| Language delay | Low average | Yes |
| Hypotonia | No | No |
| Failure to thrive | No | No |
| Autism Spectrum Disorder | No | Yes |
| Cardiac defect | No | No |
| Dysmorphism | clubfoot | No |
| Abnormal MRI or EEG | Normal EEG, MRI not done | Abnormal EEG with sleep activated midline and left parietal spikes. MRI with Chiari I malformation and holocord syrinx |
| Parent with mental health problems | Yes, father | No, but seizures and neurologic disorder |
| Strabismus | Variable Intermittent Exotropia | Variable Intermittent Exotropia |
| Other abnormality | club foot | Chiari I malformation |
| Height (on average) | 5th percentile | Between 75th and 90th percentile |
| Weight (on average) | 25th percentile | Above 97th percentile (due to medicine) |
RESULTS
Patient #1
Patient 1 has a copy loss at 16p13.11 (minimum coordinates 15132264–15147411, maximum coordinates 15048733–15179946). The size of the deletion is 15kb (min) to 131kb (max). This interval contains the genes PDXDC1, NTAN1, and RRN3. The patient’s mother does not have the copy loss. His father is thought to have schizophrenia, however, was not available for testing. Thus, the mode of inheritance is either paternal or de novo. Exome sequencing did not reveal any notable candidates in the patient or mother.
Patient #2
Patient 2 has a paternally inherited copy gain at 16p13.11 (min coordinates 14897761–16276117, max coordinates 14780303–16458270). The gain is between 1.4 (min) and 1.7Mb (max) in size. This interval contains the genes NOMO1, NPIP, NTAN1, PDXDC1, RRN3, FLJ00285, MPV17L, C16orf45, KIAA0430, NDE1, miR484, MYH11, FOPNL, ABCC1, and ABCC6. The patient also has a de novo copy gain at Xq28 (min coordinates 152955334–152961664, max coordinates 152951719–152986547), between 6kb and 35kb in size, containing SLC6A8 and possibly BCAP31. Three full siblings do not exhibit either CNV. Mother’s CMA was also normal. Father also had a small CNV at Xp22 (min coordinates 781,817–1,113,726, max coordinates 777,877–1,118,362, (between 332 and 340kb)), that has been seen frequently in patients of diverse phenotypes, likely a mismapping from the Y chromosome (Claritas Genomics, personal communication). Exome sequencing did not reveal any candidate mutations of interest for the patient’s condition.
DISCUSSION
Expansion of Previous Findings Related to the 16p13.11 Region
The 16p13.11 genomic region has been reported to be associated with developmental delay, microcephaly, epilepsy, short stature, facial dysmorphism, behavioral problems, and schizophrenia [Lipton and Sahin 2014; Mayer et al., 2004]. To our knowledge, this is the first description of childhood onset psychosis associated with a deletion or duplication of 16p13.11. Previous studies reported an onset of psychosis at 15 years with the deletion [Rees et al., 2014] and 12 years with the duplication at the broader locus 16p13.1 [Rees et al., 2014]. Our patients exhibited significantly younger onset, as we note onset at 6 years of age for the child with the deletion and 4 years for the child with the duplication.
A recent analysis of 16p13.11 CNVs in schizophrenia found 24 cases of 16p13.11 duplication or deletion out of 6882 in the case group (with schizophrenia) and 12 out of 6316 in the control group (without). This result reached borderline statistical significance, P = 0.056. The authors combined their results with data from an earlier study, and determined the statistical support for this locus in schizophrenia to P = 5.7×10−5 [Rees et al., 2014]. However, the strongest evidence for this CNV being associated with schizophrenia comes from a large sample study [Ingason et al., 2011] in which 4345 schizophrenia patients and 35,079 controls were analyzed for duplications and deletions at 16p13.1 (which contains the 16p13.11 region) by microarray. The authors found a threefold excess of duplications and deletions in schizophrenia cases compared with controls, with duplications present in 0.30% of cases versus 0.09% of controls (P = 0.007) and deletions in 0.12% of cases and 0.04% of controls (P > 0.05). The same study proposed NTAN1 and NDE1 as candidate genes for schizophrenia [Hosak 2013; Ingason et al., 2011]. NTAN1 copy number is altered in both patients detailed here, and NDE1 is duplicated in the second patient.
Implication of NTAN1, PDXDC1, and RRN3 in the Observed Phenotypes
The two patients in this report share in common altered copy number of NTAN1, PDXDC1 and RRN3. Preclinical and clinical data are reviewed below that implicate dysregulation of cellular pathways requiring these proteins in psychosis.
NTAN1 (N-terminal asparagine amidase) is a ubiquitously expressed enzyme that converts N-terminal asparagine residues to aspartate through enzymatic deamidation [Cantor et al., 2011]. The N-terminal aspartate residues are subsequently conjugated to arginine, which is recognized by specific E3 ubiquitin ligases and targeted to the proteasome for degradation. Genetically modified mice lacking the Ntan1 gene exhibit biochemical deficits in the asparagine branch of the N-end rule pathway responsible for maintaining a shortened half-life for select proteins with N-terminal asparagine residues [Kwon et al., 2000]. Ntan1 knockout mice(−/−) are fertile and outwardly unremarkable, showing no evidence of any abnormal motor function. However, neurobehavioral and cognitive testing has identified deficits that bear resemblance to preclinical models of cognitive domains affected in schizophrenia [Kwon et al., 2000]. Ntan1−/− mice have reduced exploratory behavior, deficits in conditioned avoidance learning, and deficits in spatial working memory tasks as the cognitive load is increased. These animals also display marked differences in social exploration compared to wild type mice [Kwon et al., 2000], although they have not been characterized in conventional social interaction or social approach tasks typically associated with autism animal models [Silverman et al., 2010]. Overactivity or underactivity of the ubiquitin-proteasome system (UPS) would be expected to impair a wide variety of cellular processes that contribute to synaptic function and plasticity including regulation of GABA receptors [Crider et al., 2014], and cellular mechanisms that alter synaptic strength such as dendritic spine dynamics [Jarome and Helmstetter 2013]. For example, it has been demonstrated that the activity-dependent remodeling of dendritic spines requires the local activity of the proteasome at activated synapses in order to support new dendritic spine formation [Hamilton et al., 2012]. Multiple components of the proteasome pathway have been additionally implicated in autism [Crider et al., 2014] and schizophrenia [Rubio et al., 2013]. Another gene in this pathway implicated in intellectual disability is UBE3A. This gene encodes the E3 ubiquitin ligase and contains causative mutations in Angelman’s syndrome patients who also exhibit motor dysfunction and symptoms of autism spectrum disorders.
PDXDC1 (Pyridoxal-Dependent Decarboxylase Domain-Containing Protein 1) has not been associated with schizophrenia or other mental health disorders, but rather has been associated with hearing loss [Haraksingh et al., 2014]. There is also evidence for the association of SNPs in PDXDC1 with changes in inflammatory markers [Kraja et al., 2014]. The role of this ubiquitously expressed gene has not been explored in the brain.
RRN3, RNA Polymerase Transcription Factor, (or TIA-IA), encodes an essential initiation factor for Pol I mediated transcription. The activity of this initiation factor is regulated by phosphorylation by the mammalian target of rapamycin (mTOR). RRN3 represents the primary target of the mTOR cascade involved in regulating transcription by limiting or enhancing activity of RNA Pol I [Buttgereit et al., 1985; Schnapp et al., 1990]. Thus, RRN3 initiation factor is a key mTOR sensitive hub that enables coordinate regulation of RNA and protein synthesis in response to the changing cellular environments [Lipton and Sahin 2014; Mayer et al., 2004]. Although the RRN3 gene has not previously been associated with psychiatric conditions, the dysregulation of the mTOR pathway has been implicated in a spectrum of neurological and psychiatric conditions [Lipton and Sahin 2014]. Notably, mutations in the Tuberous Sclerosis Complex (TSC), which provide regulated suppression of mTOR activity, will lead to the TSC brain disorders that are characterized by unrestrained mTOR activity at the cellular level and clinical presentations of aberrant cell growth, seizures, and autism spectrum disorders. The mTOR pathway is critical to neuronal synaptic plasticity by virtue of its role in coupling synapse specific activity to the local regulation of protein synthesis in synaptic compartments needed for synaptic remodeling that strengthen synapses and supports dendritic spine growth.
Thus, 2 of the 3 genes common across the 16p13.11 duplication/deletion region represent critical components of pathways that impinge on local synaptic plasticity signaling pathways at the synapse and have been previously associated with neurodevelopmental disorders. It therefore seems possible that the disruption of both RRN3 and NTAN1 could provide synergistic dysregulation of synaptic plasticity pathways previously been linked to neurodevelopmental disorders in patients with 16p13.11 CNVs.
The use of cellular reprogramming techniques to create inducible pluripotent stem cells from patients will provide future opportunities to functionally evaluate the potential consequences of duplications and deletions. Future studies will explore the phenotypic profile of neurons differentiated from iPSC lines and the functional integrity of these cellular pathways in preparations derived from patients bearing these mutations.
Additional Genes of Interest Duplicated in Patient #2
The duplicated region of 16p13.11 in Patient #2 includes the gene NDE1. NDE1 is thought to regulate a range of key neurodevelopmental processes including neuronal migration and differentiation. The cellular functions of NDE1 are regulated by its recruited binding partners, including LIS1. LIS1 is a dosage-sensitive gene crucial for neuronal migration and cerebral development that is known to underlie Miller–Dieker lissencephaly syndrome (OMIM 247200). A key function of NDE1 is to recruit LIS1 to dynein and promote movement of dynein along microtubules, while it while it inhibits dynein motility in the absence of LIS1 [McKenney et al., 2010; Shim et al., 2008].
Additional binding partners for NDE1 include the ‘Disrupted in Schizophrenia 1’ (DISC1) protein [Bradshaw et al., 2008]. DISC1 is a well-established risk factor for schizophrenia that acts as a scaffold for many other proteins and include additional schizophrenia risk genes such as NDE1, NDEL1, and PDE4B. These proteins can assemble as a complex at the post synaptic density to regulate function of NDE1 by virtue of PKA mediated phosphorylation events on the NDE1 protein that are triggered by local PDE4B regulation and will result in a loss of LIS1 from the regulated DISC1 complex. [Bradshaw et al., 2008].
DISC1 was first identified as a susceptibility gene for schizophrenia in a large Scottish family that demonstrated co-segregation of a balanced (1;11) (q42.1;q14.3) translocation with schizophrenia and related psychiatric disorders [Blackwood et al., 2001; St Clair et al., 1990]. Genetic association with DISC1 has been demonstrated in independent study samples for schizophrenia, neurocognitive and neuroimaging endophenotypes, suggesting that DISC1 is most likely to be related to impaired cognitive ability observed in individuals with schizophrenia and their families. Variants of NDE1 have been associated with schizophrenia in patients possessing a DISC1 haplotype that confers susceptibility to schizophrenia [Hennah et al., 2007].
Nde1-null mice display defects in neuronal proliferation, neuronal migration, and show microcephaly with thinning cortical layer and reduced numbers of neurons [Feng and Walsh 2004]. In the adult brain, Nde-1 is widely expressed in cortical neurons and subsets of astroglia. It is also found within the stem cells in the adult subventricular zone. It has been shown that the overexpression of Nde-1 in a hippocampal neural stem line will promote neuronal differentiation while inhibiting astroglial differentiation, suggesting a potential role for regulation of neuronal fate determination [Pei et al., 2014]. Thus, NDE1 and its interaction partners are known regulators of key cellular functions during development, and the altered expression levels of this gene during development could be hypothesized to contribute to a predisposition for psychiatric disorders.
Potential Role of Xq28 Duplication in Patient #2 with 16p13.11 Duplication
Interestingly, patient #2 with a 16p13.11 duplication also has a de novo duplication at Xq28. There is not enough evidence to conclude whether the Xq28 variant is clinically significant, and it is therefore classified as uncertain. This copy number variant (CNV) has not been reported in currently available literature/database resources. SLC6A8 (OMIM 300036), which is associated with cerebral creatine deficiency syndrome-1, is located within the duplicated interval. Carrier females may have mild neuropsychologic symptoms [Calhoun and Raymond 2014]. However, the effect of a duplication on this gene is unknown.
BCAP31 may be located in the duplicated interval, as it is contained in the “max” coordinates. However, it is not a good candidate for the patient’s condition. While hemizygous mutations in the BCAP31 gene are associated with deafness, dystonia, and central hypomyelination, phenotypes have not been reported in females [Cacciagli et al., 2013].
Most (~95%) CNVs are less than 500kb, and most CNVs are not known to cause disease, so this may be a benign variant. This, combined with the fact that 16p13.11 is known to affect mental health, leads us to conclude that the Xq28 duplication is not the root cause of the patient’s condition. However, it is possible that it exacerbates or otherwise contributes to the disorder in the patient. A recent study found that a second CNV greatly increases the probability of a patient having a syndromic phenotype [Grayton et al., 2012]. This study analyzed the genomes of 2312 children known to carry a copy-number variant associated with intellectual disability and congenital abnormalities. 10.1% of affected children carried a second large copy-number variant in addition to the primary genetic lesion. The authors also found that syndromic disorders could be distinguished from those with extreme phenotypic heterogeneity on the basis of the total number of copy-number variants [Grayton et al., 2012].
CONCLUSIONS
This report presented the cases of two children with CNVs in 16p13.11 that developed psychosis before the age of 7. The 16p13.11 deletion has been linked to typical adolescent/adult onset psychosis but not previously to childhood onset psychosis. It has also been linked to both intellectual disability, and autism spectrum disorders (ADHD) [Heinzen et al., 2010; Ullmann et al., 2007]. While not previously linked to psychosis before age 12 years, the broader 16p13.1 duplication containing 16p13.11 has been reported in association with schizophrenia, intellectual disability/developmental delay, autism spectrum disorders and attention-deficit hyperactivity (ADHD) [Heinzen et al., 2010; Ullmann et al., 2007]. NTAN1, RRN3 and NDE1 are mechanistically implicated in neurodevelopmental processes but as NTAN1 and RRN3 are affected in both patients, this strengthens the case for the role of these genes in childhood neurodevelopmental disturbances that could contribute to early onset psychosis.
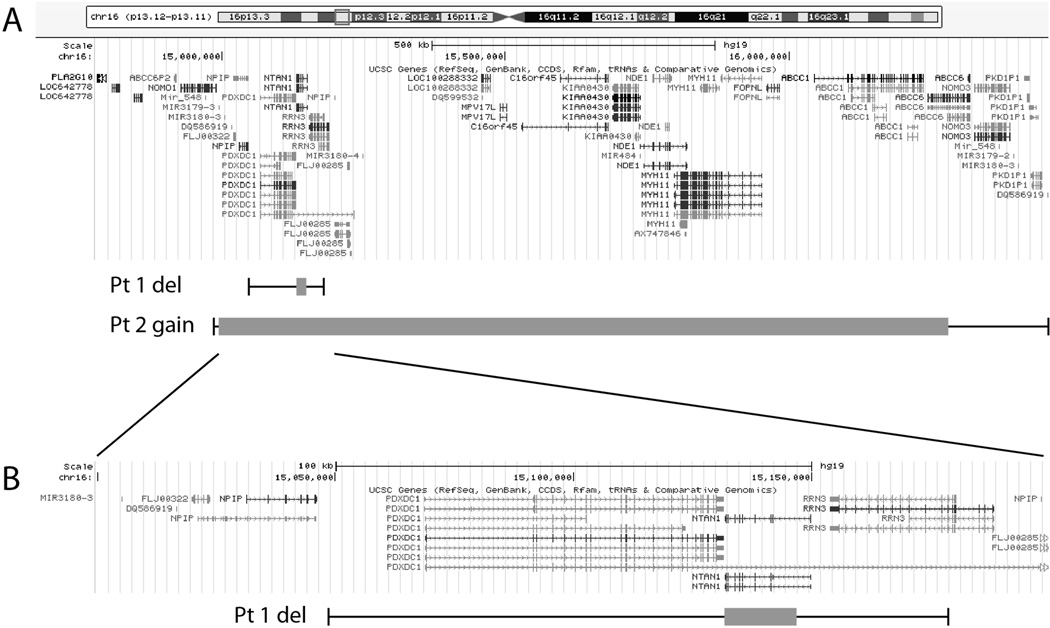
Figure 1.
Map of chromosome 16p13.11 illustrating locations of genes and extent of deleted and duplicated sequences in the region. (A) Ideogram of human chromosome 16 (above) and locations of transcripts for genes at 16p13.11 (middle) adapted from the UCSC Genome Browser (http://genome.ucsc.edu/) based on genome assembly GRCh37/hg19. Below are indicated the extent of deleted and duplicated sequences in patients 1 and 2 relative to positions of genes along the chromosome. Grey boxes encompass minimum regions of copy number variation and whiskers illustrate maximum potential areas of disruption. (B) Expanded view of the region deleted in patient 1 illustrating areas of minimum and maximum deleted sequences related to genes in the region.
Acknowledgments
The authors acknowledge assistance and support from The Manton Center for Orphan Disease Research. This work was funded by generous support from The Tommy Fuss Fund, GETTYLAB, and the Research Connection at Boston Children’s Hospital, with additional support from the Boston Children’s Hospital Intellectual and Developmental Disabilities Research Center funded by National Institutes of Health grant P30 HD18655. Whole exome sequencing was performed at the Yale Center for Genome Analysis supported by NIH U54 HG006504. Special thanks to Paula Maness, M.E. Cortizas, and Patrice Milos of Claritas Genomics.
CONFLICT OF INTEREST
Robin Kleiman is currently a consultant for Ironwood Pharmaceuticals and has consulted for En Vivo Pharmaceuticals in the past. She was formerly an employee of Pfizer for 12 years, and Selventa for 1 year.
In the past 3 years, Dr. Gonzalez-Heydrich has received grant support from the Tommy Fuss Fund, the Al Rashed Family, and Glaxo-SmithKline. He holds equity in Neuro'motion, a company that develops emotional regulation training technology. In previous years, he has served as a consultant to Abbott Laboratories, Pfizer Inc, Johnson & Johnson (Janssen, McNeil Consumer Health), Novartis, Parke-Davis, Glaxo-SmithKline, AstraZeneca, and Seaside Therapeutics; has been a speaker for Abbott Laboratories, Pfizer Inc, Novartis, Bristol-Meyers Squibb; and has received grant support from Abbott Laboratories, Pfizer Inc, Johnson & Johnson (Janssen, McNeil Consumer Health), Akzo-Nobel/Organon and the NIMH.
REFERENCES
- Abazyan B, Dziedzic J, Hua K, Abazyan S, Yang C, Mori S, Pletnikov MV, Guilarte TR. Chronic exposure of mutant DISC1 mice to lead produces sex-dependent abnormalities consistent with schizophrenia and related mental disorders: a gene-environment interaction study. Schizophr Bull. 2014;40:575–584. doi: 10.1093/schbul/sbt071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdolmaleky HM, Nohesara S, Ghadirivasfi M, Lambert AW, Ahmadkhaniha H, Ozturk S, Wong CK, Shafa R, Mostafavi A, Thiagalingam S. DNA hypermethylation of serotonin transporter gene promoter in drug naive patients with schizophrenia. Schizophr Res. 2014;152:373–380. doi: 10.1016/j.schres.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassen OA, Harbo HF, Wang Y, Thompson WK, Schork AJ, Mattingsdal M, Zuber V, Bettella F, Ripke S, Kelsoe JR, Kendler KS, O'Donovan MC, Sklar P, The Psychiatric Genomics Consortium Bipolar D, Schizophrenia Work G, The International Multiple Sclerosis Genetics C. McEvoy LK, Desikan RS, Lie BA, Djurovic S, Dale AM. Genetic pleiotropy between multiple sclerosis and schizophrenia but not bipolar disorder: differential involvement of immune-related gene loci. Mol Psychiatry. 2015;20:2017–2214. doi: 10.1038/mp.2013.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antshel KM, Kates WR, Roizen N, Fremont W, Shprintzen RJ. 22q11.2 deletion syndrome: genetics, neuroanatomy and cognitive/behavioral features keywords. Child Neuropsychol. 2005;11:5–19. doi: 10.1080/09297040590911185. [DOI] [PubMed] [Google Scholar]
- Balan S, Iwayama Y, Yamada K, Toyota T, Ohnishi T, Toyoshima M, Shimamoto C, Ide M, Iwata Y, Suzuki K, Kikuchi M, Hashimoto T, Kanahara N, Yoshikawa T, Maekawa M. Sequencing and expression analyses of the synaptic lipid raft adapter gene PAG1 in schizophrenia. J Neural Transm. 2014;122:477–485. doi: 10.1007/s00702-014-1269-0. [DOI] [PubMed] [Google Scholar]
- Blackwood DH, Fordyce A, Walker MT, St Clair DM, Porteous DJ, Muir WJ. Schizophrenia and affective disorders--cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet. 2001;69:428–433. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw NJ, Ogawa F, Antolin-Fontes B, Chubb JE, Carlyle BC, Christie S, Claessens A, Porteous DJ, Millar JK. DISC1, PDE4B, and NDE1 at the centrosome and synapse. Biochem Biophys Res Commun. 2008;377:1091–1096. doi: 10.1016/j.bbrc.2008.10.120. [DOI] [PubMed] [Google Scholar]
- Buttgereit D, Pflugfelder G, Grummt I. Growth-dependent regulation of rRNA synthesis is mediated by a transcription initiation factor (TIF-IA) Nucleic Acids Res. 1985;13:8165–8180. doi: 10.1093/nar/13.22.8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacciagli P, Sutera-Sardo J, Borges-Correia A, Roux JC, Dorboz I, Desvignes JP, Badens C, Delepine M, Lathrop M, Cau P, Levy N, Girard N, Sarda P, Boespflug-Tanguy O, Villard L. Mutations in BCAP31 cause a severe X-linked phenotype with deafness, dystonia, and central hypomyelination and disorganize the Golgi apparatus. Am J Hum Genet. 2013;93:579–586. doi: 10.1016/j.ajhg.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun AR, Raymond GV. Distal Xq28 microdeletions: clarification of the spectrum of contiguous gene deletions involving ABCD1, BCAP31, and SLC6A8 with a new case and review of the literature. Am J Med Genet A. 2014;164A:2613–2617. doi: 10.1002/ajmg.a.36661. [DOI] [PubMed] [Google Scholar]
- Cantor JR, Stone EM, Georgiou G. Expression and biochemical characterization of the human enzyme N-terminal asparagine amidohydrolase. Biochemistry. 2011;50:3025–3033. doi: 10.1021/bi101832w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SF, Chao YL, Shen YC, Chen CH, Weng CF. Resequencing and association study of the NFKB activating protein-like gene (NKAPL) in schizophrenia. Schizophr Res. 2014;157:169–174. doi: 10.1016/j.schres.2014.05.038. [DOI] [PubMed] [Google Scholar]
- Collins AL, Kim Y, Bloom RJ, Kelada SN, Sethupathy P, Sullivan PF. Transcriptional targets of the schizophrenia risk gene MIR137. Transl Psychiatry. 2014;4:e404. doi: 10.1038/tp.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crider A, Pandya CD, Peter D, Ahmed AO, Pillai A. Ubiquitin-proteasome dependent degradation of GABAAalpha1 in autism spectrum disorder. Mol Autism. 2014;5:45. doi: 10.1186/2040-2392-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty JL, Owen MJ. Genomic insights into the overlap between psychiatric disorders: implications for research and clinical practice. Genome Med. 2014;6:29. doi: 10.1186/gm546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Walsh CA. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron. 2004;44:279–293. doi: 10.1016/j.neuron.2004.09.023. [DOI] [PubMed] [Google Scholar]
- Grayton HM, Fernandes C, Rujescu D, Collier DA. Copy number variations in neurodevelopmental disorders. Prog Neurobiol. 2012;99:81–91. doi: 10.1016/j.pneurobio.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Guella I, Sequeira A, Rollins B, Morgan L, Myers RM, Watson SJ, Akil H, Bunney WE, Delisi LE, Byerley W, Vawter MP. Evidence of allelic imbalance in the schizophrenia susceptibility gene ZNF804A in human dorsolateral prefrontal cortex. Schizophr Res. 2014;152:111–116. doi: 10.1016/j.schres.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AM, Oh WC, Vega-Ramirez H, Stein IS, Hell JW, Patrick GN, Zito K. Activity-dependent growth of new dendritic spines is regulated by the proteasome. Neuron. 2012;74:1023–1030. doi: 10.1016/j.neuron.2012.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraksingh RR, Jahanbani F, Rodriguez-Paris J, Gelernter J, Nadeau KC, Oghalai JS, Schrijver I, Snyder MP. Exome sequencing and genome-wide copy number variant mapping reveal novel associations with sensorineural hereditary hearing loss. BMC Genomics. 2014;15:1155. doi: 10.1186/1471-2164-15-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzen EL, Radtke RA, Urban TJ, Cavalleri GL, Depondt C, Need AC, Walley NM, Nicoletti P, Ge D, Catarino CB, Duncan JS, Kasperaviciute D, Tate SK, Caboclo LO, Sander JW, Clayton L, Linney KN, Shianna KV, Gumbs CE, Smith J, Cronin KD, Maia JM, Doherty CP, Pandolfo M, Leppert D, Middleton LT, Gibson RA, Johnson MR, Matthews PM, Hosford D, Kalviainen R, Eriksson K, Kantanen AM, Dorn T, Hansen J, Kramer G, Steinhoff BJ, Wieser HG, Zumsteg D, Ortega M, Wood NW, Huxley-Jones J, Mikati M, Gallentine WB, Husain AM, Buckley PG, Stallings RL, Podgoreanu MV, Delanty N, Sisodiya SM, Goldstein DB. Rare deletions at 16p13.11 predispose to a diverse spectrum of sporadic epilepsy syndromes. Am J Hum Genet. 2010;86:707–718. doi: 10.1016/j.ajhg.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennah W, Tomppo L, Hiekkalinna T, Palo OM, Kilpinen H, Ekelund J, Tuulio-Henriksson A, Silander K, Partonen T, Paunio T, Terwilliger JD, Lonnqvist J, Peltonen L. Families with the risk allele of DISC1 reveal a link between schizophrenia and another component of the same molecular pathway, NDE1. Hum Mol Genet. 2007;16:453–462. doi: 10.1093/hmg/ddl462. [DOI] [PubMed] [Google Scholar]
- Hosak L. New findings in the genetics of schizophrenia. World J Psychiatry. 2013;3:57–61. doi: 10.5498/wjp.v3.i3.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingason A, Rujescu D, Cichon S, Sigurdsson E, Sigmundsson T, Pietilainen OP, Buizer-Voskamp JE, Strengman E, Francks C, Muglia P, Gylfason A, Gustafsson O, Olason PI, Steinberg S, Hansen T, Jakobsen KD, Rasmussen HB, Giegling I, Moller HJ, Hartmann A, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Bramon E, Kiemeney LA, Franke B, Murray R, Vassos E, Toulopoulou T, Muhleisen TW, Tosato S, Ruggeri M, Djurovic S, Andreassen OA, Zhang Z, Werge T, Ophoff RA, Investigators G. Rietschel M, Nothen MM, Petursson H, Stefansson H, Peltonen L, Collier D, Stefansson K, St Clair DM. Copy number variations of chromosome 16p13.1 region associated with schizophrenia. Mol Psychiatry. 2011;16:17–25. doi: 10.1038/mp.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarome TJ, Helmstetter FJ. The ubiquitin-proteasome system as a critical regulator of synaptic plasticity and long-term memory formation. Neurobiol Learn Mem. 2013;105:107–116. doi: 10.1016/j.nlm.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraja AT, Chasman DI, North KE, Reiner AP, Yanek LR, Kilpelainen TO, Smith JA, Dehghan A, Dupuis J, Johnson AD, Feitosa MF, Tekola-Ayele F, Chu AY, Nolte IM, Dastani Z, Morris A, Pendergrass SA, Sun YV, Ritchie MD, Vaez A, Lin H, Ligthart S, Marullo L, Rohde R, Shao Y, Ziegler MA, Im HK, Cross Consortia Pleiotropy G, Cohorts for Heart a, Aging Research in Genetic E, Genetic Investigation of Anthropometric Traits C, Global Lipids Genetics C, Meta-Analyses of G, Insulin-related traits C, Global BC, Consortium AD, Women's Genome Health S, Howard University Family S. Schnabel RB, Jorgensen T, Jorgensen ME, Hansen T, Pedersen O, Stolk RP, Snieder H, Hofman A, Uitterlinden AG, Franco OH, Ikram MA, Richards JB, Rotimi C, Wilson JG, Lange L, Ganesh SK, Nalls M, Rasmussen-Torvik LJ, Pankow JS, Coresh J, Tang W, Linda Kao WH, Boerwinkle E, Morrison AC, Ridker PM, Becker DM, Rotter JI, Kardia SL, Loos RJ, Larson MG, Hsu YH, Province MA, Tracy R, Voight BF, Vaidya D, O'Donnell CJ, Benjamin EJ, Alizadeh BZ, Prokopenko I, Meigs JB, Borecki IB. Pleiotropic genes for metabolic syndrome and inflammation. Mol Genet Metab. 2014;112:317–338. doi: 10.1016/j.ymgme.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YT, Balogh SA, Davydov IV, Kashina AS, Yoon JK, Xie Y, Gaur A, Hyde L, Denenberg VH, Varshavsky A. Altered activity, social behavior, and spatial memory in mice lacking the NTAN1p amidase and the asparagine branch of the N-end rule pathway. Mol Cell Biol. 2000;20:4135–4148. doi: 10.1128/mcb.20.11.4135-4148.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton JO, Sahin M. The neurology of mTOR. Neuron. 2014;84:275–291. doi: 10.1016/j.neuron.2014.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Zhao J, Yuan X, Grummt I. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 2004;18:423–434. doi: 10.1101/gad.285504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy SE, Gillis J, Kramer M, Lihm J, Yoon S, Berstein Y, Mistry M, Pavlidis P, Solomon R, Ghiban E, Antoniou E, Kelleher E, O'Brien C, Donohoe G, Gill M, Morris DW, McCombie WR, Corvin A. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol Psychiatry. 2014;19:652–658. doi: 10.1038/mp.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy SE, Makarov V, Kirov G, Addington AM, McClellan J, Yoon S, Perkins DO, Dickel DE, Kusenda M, Krastoshevsky O, Krause V, Kumar RA, Grozeva D, Malhotra D, Walsh T, Zackai EH, Kaplan P, Ganesh J, Krantz ID, Spinner NB, Roccanova P, Bhandari A, Pavon K, Lakshmi B, Leotta A, Kendall J, Lee YH, Vacic V, Gary S, Iakoucheva LM, Crow TJ, Christian SL, Lieberman JA, Stroup TS, Lehtimaki T, Puura K, Haldeman-Englert C, Pearl J, Goodell M, Willour VL, Derosse P, Steele J, Kassem L, Wolff J, Chitkara N, McMahon FJ, Malhotra AK, Potash JB, Schulze TG, Nothen MM, Cichon S, Rietschel M, Leibenluft E, Kustanovich V, Lajonchere CM, Sutcliffe JS, Skuse D, Gill M, Gallagher L, Mendell NR, Wellcome Trust Case Control C. Craddock N, Owen MJ, O'Donovan MC, Shaikh TH, Susser E, Delisi LE, Sullivan PF, Deutsch CK, Rapoport J, Levy DL, King MC, Sebat J. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney RJ, Vershinin M, Kunwar A, Vallee RB, Gross SP. LIS1 and NudE induce a persistent dynein force-producing state. Cell. 2010;141:304–314. doi: 10.1016/j.cell.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamani SC, Erez A, Bader P, Lalani SR, Scott DA, Scaglia F, Plon SE, Tsai CH, Reimschisel T, Roeder E, Malphrus AD, Eng PA, Hixson PM, Kang SH, Stankiewicz P, Patel A, Cheung SW. Phenotypic manifestations of copy number variation in chromosome 16p13.11. Eur J Hum Genet. 2011;19:280–286. doi: 10.1038/ejhg.2010.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z, Lang B, Fragoso YD, Shearer KD, Zhao L, McCaffery PJ, Shen S, Ding YQ, McCaig CD, Collinson JM. The expression and roles of Nde1 and Ndel1 in the adult mammalian central nervous system. Neuroscience. 2014;271:119–136. doi: 10.1016/j.neuroscience.2014.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalingam A, Zhou XG, Fiedler SD, Brawner SJ, Joyce JM, Liu HY, Yu S. 16p13.11 duplication is a risk factor for a wide spectrum of neuropsychiatric disorders. J Hum Genet. 2011;56:541–544. doi: 10.1038/jhg.2011.42. [DOI] [PubMed] [Google Scholar]
- Rees E, Walters JT, Georgieva L, Isles AR, Chambert KD, Richards AL, Mahoney-Davies G, Legge SE, Moran JL, McCarroll SA, O'Donovan MC, Owen MJ, Kirov G. Analysis of copy number variations at 15 schizophrenia-associated loci. Br J Psychiatry. 2014;204:108–114. doi: 10.1192/bjp.bp.113.131052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio MD, Wood K, Haroutunian V, Meador-Woodruff JH. Dysfunction of the ubiquitin proteasome and ubiquitin-like systems in schizophrenia. Neuropsychopharmacology. 2013;38:1910–1920. doi: 10.1038/npp.2013.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp A, Pfleiderer C, Rosenbauer H, Grummt I. A growth-dependent transcription initiation factor (TIF-IA) interacting with RNA polymerase I regulates mouse ribosomal RNA synthesis. EMBO J. 1990;9:2857–2863. doi: 10.1002/j.1460-2075.1990.tb07475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim SY, Samuels BA, Wang J, Neumayer G, Belzil C, Ayala R, Shi Y, Shi Y, Tsai LH, Nguyen MD. Ndel1 controls the dynein-mediated transport of vimentin during neurite outgrowth. J Biol Chem. 2008;283:12232–12240. doi: 10.1074/jbc.M710200200. [DOI] [PubMed] [Google Scholar]
- Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nat Rev Neurosci. 2010;11:490–502. doi: 10.1038/nrn2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Clair D, Blackwood D, Muir W, Carothers A, Walker M, Spowart G, Gosden C, Evans HJ. Association within a family of a balanced autosomal translocation with major mental illness. Lancet. 1990;336:13–16. doi: 10.1016/0140-6736(90)91520-k. [DOI] [PubMed] [Google Scholar]
- Szatkiewicz JP, O'Dushlaine C, Chen G, Chambert K, Moran JL, Neale BM, Fromer M, Ruderfer D, Akterin S, Bergen SE, Kahler A, Magnusson PK, Kim Y, Crowley JJ, Rees E, Kirov G, O'Donovan MC, Owen MJ, Walters J, Scolnick E, Sklar P, Purcell S, Hultman CM, McCarroll SA, Sullivan PF. Copy number variation in schizophrenia in Sweden. Mol Psychiatry. 2014;19:762–773. doi: 10.1038/mp.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropeano M, Ahn JW, Dobson RJ, Breen G, Rucker J, Dixit A, Pal DK, McGuffin P, Farmer A, White PS, Andrieux J, Vassos E, Ogilvie CM, Curran S, Collier DA. Male-biased autosomal effect of 16p13.11 copy number variation in neurodevelopmental disorders. PLoS One. 2013;8:e61365. doi: 10.1371/journal.pone.0061365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullmann R, Turner G, Kirchhoff M, Chen W, Tonge B, Rosenberg C, Field M, Vianna-Morgante AM, Christie L, Krepischi-Santos AC, Banna L, Brereton AV, Hill A, Bisgaard AM, Muller I, Hultschig C, Erdogan F, Wieczorek G, Ropers HH. Array CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Hum Mutat. 2007;28:674–682. doi: 10.1002/humu.20546. [DOI] [PubMed] [Google Scholar]
- Wirgenes KV, Tesli M, Inderhaug E, Athanasiu L, Agartz I, Melle I, Hughes T, Andreassen OA, Djurovic S. ANK3 gene expression in bipolar disorder and schizophrenia. Br J Psychiatry. 2014;205:244–245. doi: 10.1192/bjp.bp.114.145433. [DOI] [PubMed] [Google Scholar]
- Won S, Kwon MS, Mattheisen M, Park S, Park C, Kihara D, Cichon S, Ophoff R, Nothen MM, Rietschel M, Baur M, Uitterlinden AG, Hofmann A, Investigators G. Lange C. Efficient strategy for detecting gene×gene joint action and its application in schizophrenia. Genet Epidemiol. 2014;38:60–71. doi: 10.1002/gepi.21779. [DOI] [PubMed] [Google Scholar]
- Xu XM, Ding M, Pang H, Wang BJ. TPH2 gene polymorphisms in the regulatory region are associated with paranoid schizophrenia in Northern Han Chinese. Genet Mol Res. 2014;13:1497–1507. doi: 10.4238/2014.March.12.1. [DOI] [PubMed] [Google Scholar]
- Yao J, Ding M, Xing J, Xuan J, Pang H, Pan Y, Wang B. Genetic association between the dopamine D1-receptor gene and paranoid schizophrenia in a northern Han Chinese population. Neuropsychiatr Dis Treat. 2014;10:645–652. doi: 10.2147/NDT.S61227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Jin C, Sha W, Zhou Z, Zhang F, Wang M, Wang J, Li J, Feng X, Yu S, Wang J. A competitive PCR assay confirms the association of a copy number variation in the VIPR2 gene with schizophrenia in Han Chinese. Schizophr Res. 2014;156:66–70. doi: 10.1016/j.schres.2014.04.004. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Ding M, Pang H, Xu XM, Wang BJ. Relationship between genetic polymorphisms in the DRD5 gene and paranoid schizophrenia in northern Han Chinese. Genet Mol Res. 2014;13:1609–1618. doi: 10.4238/2014.March.12.13. [DOI] [PubMed] [Google Scholar]