Abstract
The innate immune system provides the first line of defense against pathogens through the recognition of non-specific patterns in RNA to protect the cell in a generalized way. The human RNA-activated protein kinase, PKR, is a dsRNA binding protein and an essential sensor in the innate immune response, which recognizes viral and bacterial pathogens through their RNAs. Upon activation via RNA-dependent autophosphorylation, PKR phosphorylates the eukaryotic initiation factor eIF2α leading to termination of translation. PKR has a well-characterized role in recognizing viral RNA, where it binds long stretches of double-stranded RNA non-sequence specifically to promote activation; however, the mechanism by which bacterial RNA activates PKR and the mode by which self RNA avoids activating PKR are unknown. We characterized activation of PKR by three functional bacterial RNAs with pseudoknots and extensive tertiary structure: the cyclic-di GMP riboswitch, the glmS riboswitch-ribozyme, and the twister ribozyme, two of which are ligand-activated. These RNAs were found to activate PKR with comparable potency to long dsRNA. Enzymatic structure mapping in the absence and presence of PKR reveals a clear PKR footprint and provides a structural basis for how these bacterial RNAs activate PKR. In the case of the cyclic di-GMP riboswitch and the glmS riboswitch-ribozyme, PKR appears to dimerize on the peripheral double-stranded regions of the native RNA tertiary structure. Overall, these results provide new insights into how PKR acts as an innate immune signaling protein for the presence of bacteria and suggest a reason for the apparent absence of protein-free riboswitches and ribozymes in the human genome.
The first line of defense to invading organisms is the innate immune system, which involves pattern recognition receptors (PRR’s) that detect molecular patterns in order to discriminate between self and non-self.1 Many proteins are involved in the innate immune system including the RNA-activated Protein Kinase, PKR.2 This essential sensor is comprised of an N-terminal double-stranded RNA binding domain (dsRBD) that contains two tandem double-stranded RNA binding motifs (dsRBMs), as well as a C-terminal catalytic kinase domain.3 The dsRBM is found in a number of key proteins, including Dicer, Drosha, and ADAR, and binds dsRNA in a non-sequence-specific fashion primarily in the minor groove.3,4 The classical model of PKR activation involves its binding to long stretches of dsRNA, where at least 16 bp are needed for PKR to bind and 30 bp for it to dimerize and become active.5–7 Upon dimerization, PKR can autophosphorylate followed by phosphorylation of eIF2α leading to inhibition of translation initiation.8 Because PKR must dimerize to become active, too much RNA shuts off PKR activity by stranding PKR monomers onto RNA islands.7
PKR is known to be classically activated by long dsRNA from viruses, but it is also known to be activated by other structured RNAs.9 More recently, PKR has been shown to be activated by bacterial RNA, both from total RNA in vivo as well as a discrete bacterial RNA leader in vitro.10,11 In vivo concentrations of Mg2+ have a substantial effect on RNA folding and function.12 Bacteria have physiological Mg2+ concentrations of ~2–3 mM,12,13 while human cells have physiological Mg2+ concentrations of ~0.5–1 mM14 suggesting that bacterial RNAs could refold in eukaryotic cells depending on local Mg2+ concentrations. In addition, standard PKR activation assays in the literature have been performed typically over the range of 4–10 mM Mg2+.6 As such, the effects of prokaryotic and eukaryotic Mg2+ levels on PKR activation are important to test.
Key functional RNAs discovered in the last few decades include riboswitches and ribozymes. Riboswitches are aptamers that control gene function by binding a metabolite, which can be anything from a protein to a small molecule or ion.15 Ribozymes are catalytic RNAs that can make and break chemical bonds under certain cellular conditions, typically to regulate downstream gene function. Riboswitches and ribozymes are known primarily in prokaryotes.16–18 An outstanding question is the reason for the apparent absence of riboswitches and ribozymes in eukaryotes.
Three representative functional bacterial RNAs are studied here: a 110 nt cyclic di-GMP (cdiGMP) riboswitch from Vibrio cholerae, a 145 nt glmS riboswitch-ribozyme from Bacillus anthracis, and a 61 nt twister ribozyme from Clostridia bolteae. These RNAs span different classes and sizes of functional RNAs. In addition, we tested PKR activation by the full-length 5′UTR of the cdiGMP riboswitch, which is 350 nt in length and contains both the aptamer and expression domains. All of these RNAs have pseudoknots and both of the ribozymes are double-pseudoknotted, allowing the importance of RNA tertiary structure in activating PKR to be tested.19–21 These particular bacterial RNAs were also chosen in part on the basis of their diversity and their pathogenesis to humans: V. cholerae is a gram-negative bacterium that causes cholera, B. anthracis is a gram-positive bacterium that causes anthrax, and C. bolteae is a gram-positive bacterium that causes bowel-related issues associated with autism.
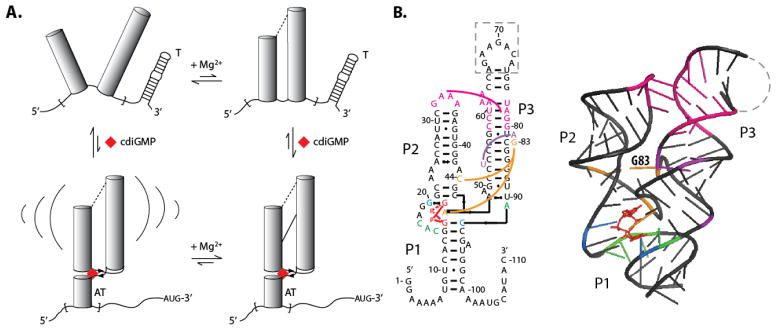
Cyclic-diGMP is a bacterial second messenger that activates extracellular polysaccharide production and biofilm formation in addition to regulating motility and virulence.22 Classically, cdiGMP was known to perform this function by binding protein enzymes, but more recently it has been shown to bind certain riboswitches.23,24 The cdiGMP riboswitch from V. cholerae is a class-I cdiGMP riboswitch that controls the tfoX gene by an antitermination mechanism (Figure 1).24 The cdiGMP signaling pathway is important in controlling expression of the gene in a host cell, as cdiGMP levels are often decreased upon infection allowing the bacteria to express virulence factors necessary to survive.25 Both cdiGMP and Mg2+ affect the fold of the riboswitch (Figure 1). In limiting Mg2+ conditions (0.5 mM Mg2+) in the absence of cdiGMP, the aptamer adopts an undocked state.26,27 In excess Mg2+ conditions, (4.0 mM Mg2+) the aptamer assumes a pre-docked, but not fully formed fold that upon addition of saturating cdiGMP forms a docked structure with an antiterminator that allows the downstream gene to turn on.26,27 The glmS riboswitch-ribozyme regulates levels of glucosamine-6 phosphate (GlcN6P),28 which is the ligand that it binds. The twister ribozyme was recently discovered and is one of the fastest self-cleaving ribozymes, found in a large range of organisms (see structures below). Crystal structures are available for both of these RNAs.19,21
Figure 1.
Full length 5′UTR (Vc2FL) and aptamer-only (Vc2) constructs of the cdiGMP riboswitch from Vibrio cholerae. a) Effect of Mg2+ and cdiGMP on docking of the aptamer region of the cdiGMP riboswitch leading to the formation of an antiterminator, ‘AT’, from a terminator, ‘T’. Structure mapping and prediction of the terminator and antiterminator are provided in Supplemental Figure 4. The upper left, upper right and lower right states are referred to in the literature as ‘undocked’, ‘pre-docked’, and ‘docked’, respectively;26 the latter two states contain the pseudoknot. Brackets in lower left figure indicate structural dynamics. b) Secondary and tertiary structure models of Vc2.20,26 Nucleotides in the dashed region of the secondary structure were replaced with the U1A binding site during crystallization.
In the present study, we conducted activation assays on the V. cholerae cyclic diGMP riboswitch aptamer domain (Vc2) and its full-length 5′UTR (Vc2FL) in the absence and presence of cyclic-diGMP. We also removed key tertiary interactions in the Vc2 and Vc2FL riboswitches by a G83C mutation to test whether tertiary RNA interactions play a role in activation of PKR. Lastly we tested activation of PKR by the post-cleavage product of the glmS riboswitch-ribozyme and twister ribozyme. To understand the potent activation of PKR by these riboswitches and ribozymes, we conducted structure mapping and PKR footprinting, which led to a structural model for how these functional bacterial RNAs interact with and activate PKR.
RESULTS AND DISCUSSION
PKR is activated by both the Vc2 and Vc2FL forms of the cdiGMP riboswitch
The first of the three functional bacterial RNAs that was tested for activation of PKR is the cdiGMP riboswitch from Vibrio cholerae. We tested both the aptamer domain alone (Vc2) and the full-length 5′-UTR (Vc2FL), which contains both Vc2 and the terminator and antiterminator hairpins, among other possible regulatory structure. These experiments were conducted under both standard activation conditions of 4 mM Mg2+, similar to prokaryotic cell conditions, and human cellular salt conditions of 0.5 mM Mg2+ in the presence and absence of saturating amounts of the metabolite, cdiGMP. These concentrations were chosen to populate the undocked, pre-docked, and docked states, as described in Figure 1.26 Results are summarized in Supplementary Table 1 and representative activation gels are provided in Figure 2.
Figure 2.
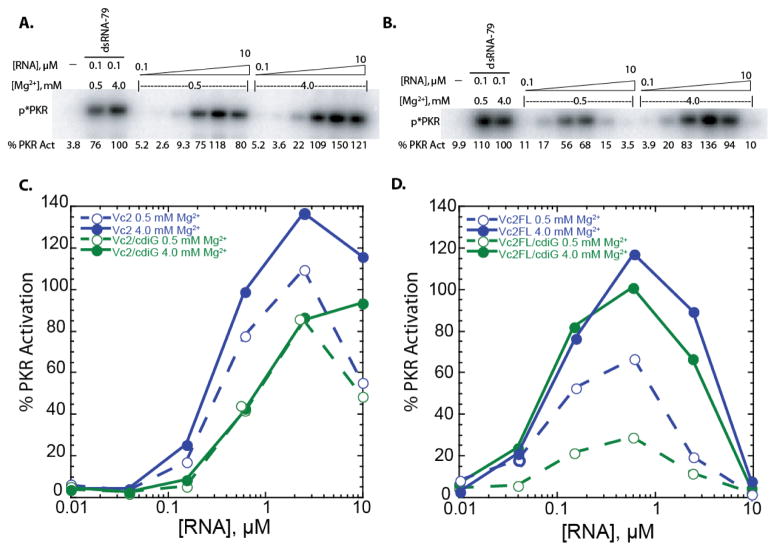
Activation of PKR by Vc2 & Vc2FL at 0.5 and 4 mM Mg2+ in the absence and presence of cdiGMP. a,b) SDS-PAGE gels for a) Vc2 at 0.5 and 4 mM Mg2+ and b) Vc2FL at 0.5 and 4 mM Mg2+. Bands indicate phosphorylated PKR, ‘p*PKR’. Percent PKR activation was normalized to that of 0.1 μM dsRNA-79 in 4 mM Mg2+ conditions. c,d) Percentage of PKR activation versus the concentration of RNA for c) Vc2 and d) Vc2FL at 0.5 mM and 4 mM Mg2+ and in the absence and presence of cdiGMP (denoted cdiG in legends). Plotted are the averages of two trials and all values can be viewed in the supplementary information. Note Vc2 +cdiGMP, 0.5 mM Mg2+ 2.5 and 0.625 μM points were offset slightly so they were not hidden behind the 4 mM Mg2+ data points.
Under standard activation conditions of 4 mM Mg2+, which populates the pre-docked state (Figure 1A), Vc2 potently activated PKR with a maximal activation level at ~140% that of 0.1 μM dsRNA-79 (Figure 2A). Activation was completely dependent on the presence of RNA. As the concentration of RNA was increased, activation increased and then decreased, with a bell-shaped dependence on RNA concentration, consistent with RNA-dependent dimerization of PKR (Figure 2C). Upon lowering the Mg2+ concentration to a physiological value of 0.5 mM, which populates primarily the undocked state, activation was still potent, at ~110% that of 0.1 μM dsRNA-79. Upon adding 20 μM cdiGMP, the activation decreased slightly for both 4 and 0.5 mM Mg2+ conditions, with both requiring more RNA to gain activation and having a somewhat lower maxima of ~90% dsRNA-79 but which was strong nonetheless. Note that cdiGMP alone does not activate PKR nor compete with activation by dsRNA-79 (Supplemental Figure 1). (For evidence of cdiGMP binding, Vc2 folding, and PKR binding to the natively folded riboswitch, see structure mapping data below.) These ligand containing results, especially those in 4 mM Mg2+, reveal that the fully docked state also activates PKR. Overall, these results indicate that the bacterial Vc2 riboswitch is a potent activator of PKR under prokaryotic and eukaryotic salt conditions and in the presence and absence of bound ligand.
Next, the ability of the full length 5′UTR (Vc2FL) to activate PKR was tested. As shown in Figure 2D, the results were quite similar to Vc2. Potent activation was observed in the absence of ligand and under standard activation conditions of 4 mM Mg2+, which populates the pre-docked state, with a bell-shaped dependence on RNA concentration. Lowering the Mg2+ concentration to 0.5 mM, which populates primarily the undocked state, decreased activation ~2-fold, somewhat more than for Vc2 but still with strong activation. Upon adding cdiGMP in the presence of 4 mM Mg2+ to force the fully docked state, activation was affected only slightly, with maximal activation at ~110% dsRNA-79. (For evidence of cdiGMP binding and accompanying changes in the secondary structure of the expression domain, see Supplemental Figure 4). At eukaryotic Mg2+ concentration, activation was affected more significantly by ligand, where activation fell to ~20% that in dsRNA-79. All four conditions for Vc2FL revealed bell-shaped activation curves that were ‘left-shifted’ compared to the aptamer alone: a lower molar concentration of RNA was needed to gain activation and a lower concentration of RNA led to loss of activation. The bell-shape to the RNA activation profile is again consistent with RNA-dependent dimerization of PKR, and the leftward shift of the profile supports the longer Vc2FL (350 nt) requiring fewer moles of transcript to both activate and inactivate PKR. We also note that both ligand-free and ligand-bound Vc2 and Vc2FL compete with dsRNA-79 for activation (Supplementary Figure 1), indicating that the riboswitches bind to the same, or overlapping, site(s) as dsRNA on PKR. Overall, in the majority of the cases tested, the Vc2 and Vc2FL 5′-UTR strongly activate PKR by RNA-dependent dimerization under diverse salt and ligand conditions and with several different RNA constructs.
Tertiary interactions in Vc2 and Vc2FL contribute to PKR activation
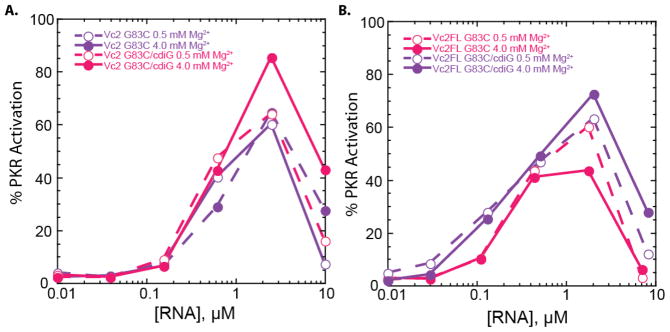
As described in the previous section, both the aptamer domain alone (Vc2) and full-length 5′-UTR (Vc2FL) of the cdiGMP riboswitch activate PKR potently under prokaryotic and eukaryotic Mg2+ conditions, in both the absence and presence of the cdiGMP ligand. The aptamer domain contains tertiary interactions that form to various extents in so-called undocked, pre-docked, and docked states under the Mg2+ and cdiGMP concentrations studied herein (Figure 1).26 In the previous section, we showed that under conditions in which the ribozyme is fully folded and ligand bound, PKR is strongly activated. We wished to test the extent to which PKR activation is affected by riboswitch tertiary interactions because they assemble three A-form helical segments. To achieve this, we prepared G83C mutants of Vc2 and Vc2FL and tested them for activation of PKR in both 0.5 and 4 mM Mg2+ and in the absence and presence of 20 μM cdiGMP. The G83 residue is located at the center of the aptameric domain (Figure 1B) and changing it to a C has been shown to prevent the pseudoknot-containing ligand-free pre-docked structure in the upper right-hand corner of Figure 1A from forming.20,26
As shown in Figure 3, the four combinations of Mg2+ concentration and cdiGMP for Vc2 and Vc2FL G83C constructs led to WT-like results of bell-shaped activation with maximal activation at ~60–90% that of 0.1 μM dsRNA-79. These results indicate that removing tertiary interactions does not enhance activation of PKR in either the aptamer alone or the full length 5′-UTR. On the contrary, activation is more potent with wild-type than G83C mutants for Vc2 and Vc2FL in 4 mM Mg2+ with or without ligand, conditions under which tertiary interactions are known to form in wild-type (Figure 2).
Figure 3.
Activation of PKR by Vc2 & Vc2FL G83C mutants at 0.5 and 4 mM Mg2+ in the absence and presence of cdiGMP. a,b) Percentage of PKR activation versus the concentration of RNA for a) Vc2 and b) Vc2FL at 0.5 mM and 4 mM Mg2+ and in the absence and presence of cdiGMP (denoted cdiG in legends). Plotted are the averages of two trials and all values are provided in Supplementary Table 1. Percent PKR activation was normalized to that of 0.1 μM dsRNA-79 in 4 mM Mg2+ conditions.
The glmS riboswitch-ribozyme activates PKR
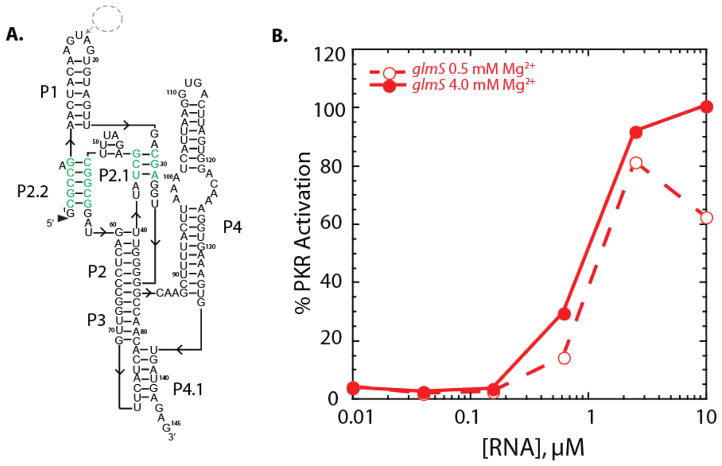
We next investigated regulation of PKR by two bacterial ribozymes, the larger glmS riboswitch-ribozyme and the smaller twister ribozyme. In both cases, one-piece versions were used since these represent the forms of the ribozymes as they occur in nature; in addition, multiple crystal structures are available for each ribozyme.19,21 Like the cdiGMP riboswitch, the glmS riboswitch-ribozyme binds a metabolite, here GlcN6P, to regulate expression, but the glmS riboswitch-ribozyme also self-cleaves to regulate gene expression.28 Since the one-piece version of the ribozyme self-cleaved to a significant extent during transcription (see Methods), we worked with the self-cleaved form of this RNA, which is missing just a small single-strand extension on the 5′-end. The secondary structure of the glmS riboswitch-ribozyme is shown in Figure 4A where the nucleotides involved in the double pseudoknot are highlighted, and long base-paired regions are apparent.
Figure 4.
Dependence of activation of PKR on Mg2+ concentration for the glmS ribozyme. a) Secondary structure of the 145 nt self-cleaved B. anthracis glmS ribozyme. The double pseudoknot interactions are shown in green. Nucleotides were inserted into the loop in P1 between nt 16–17 for a U1A binding site to aid in crystallization (dashed circle). b) Percentage of PKR activation versus concentration of glmS RNA at 0.5 and 4 mM Mg2+. Plotted are the averages of two trials and all values can be viewed in the supplementary information. Percent PKR activation was normalized to that of 0.1 μM dsRNA-79 in 4 mM Mg2+ conditions.
The self-cleaved glmS riboswitch-ribozyme activated PKR potently, at 100% the level of 0.1 μM dsRNA-79, and did so similarly at 0.5 and 4 mM Mg2+ (Figure 4). The dependence of activation on RNA concentration was bell-shaped, consistent with dimerization of PKR on the glmS RNA (Figure 4). The shapes of the PKR activation curves for the glmS riboswitch-ribozyme were similar to Vc2, with onset of activation at ~0.1 μM RNA, maximal activation ~95% of 0.1 μM dsRNA-79 at ~1 μM RNA, and loss of activation beginning at ~10 μM RNA (Figures 2A and 4B). Consistent with this similar behavior, the crystal structures of Vc2 and the glmS riboswitch-ribozyme reveal similar ‘Y-shaped’ overall architectures and extensive tertiary structure. (For folding of the glmS riboswitch-ribozyme and binding of PKR, see structure mapping data below.)
PKR is activated by the small self-cleaving twister ribozyme by extending its 3′-end
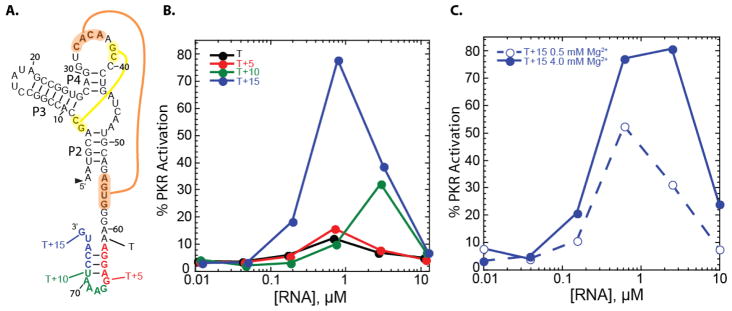
The third and final bacterial RNA investigated for activation of PKR is the recently discovered twister ribozyme. Unlike the prior two bacterial RNAs, twister is not a riboswitch; moreover, it is a smaller RNA of just 66 nt that does not form an extensive and homologous ‘Y-shaped’ tertiary structure. A one-piece version of the twister from Clostridium bolteae was studied, along with various 3′-end extensions. Like the glmS riboswitch-ribozyme, all of the twister ribozyme constructs cleaved during transcription. The secondary structure of the twister ribozyme is provided in Figure 5, and the double pseudoknot is highlighted.
Figure 5.
Activation of PKR by a twister ribozyme is dependent on flanking-tail length and concentration of Mg2+. a) Secondary structure of the C. bolteae twister ribozyme (black) with the double pseudoknot interactions shown in yellow and orange. b) Percentage of PKR activation versus concentration of twister RNA for varying length flanking tails under standard PKR activation conditions (4 mM Mg2+, 10 min). c) Percentage of PKR activation for the highest activating twister construct, T +15, at 0.5 and 4 mM Mg2+. Plotted are the averages of three trials, and all values can be viewed in the supplementary information. Percent PKR activation was normalized to that of 0.1 μM dsRNA-79 in 4 mM Mg2+ conditions.
We began with a self-cleaved version of twister with the minimal 3′-end needed to form the secondary structure of the active ribozyme and tested its ability to activate PKR in 4 mM Mg2+ (Figure 5A). This construct, which we denote as ‘T’, did not significantly activate PKR (Figure 5B). We then lengthened the 3′-end of the ribozyme by 5, 10, or 15 nucleotides. The T+5 RNA also did not significantly activate PKR. Results on T and T+5 indicate that not all RNAs activate PKR.
The T+10 RNA, on the other hand, showed significantly more activation, up to 30% the level of dsRNA-79, but at a high RNA concentration for maximal activity of ~3 μM. Strikingly, the T+15 RNA gave substantial activation, at ~80% that of 0.1 μM dsRNA-79 and a lower RNA concentration for maximal activation, of ~0.6 μM. This T+15 RNA has the ability to form a simple stem-loop at the 3′end.29 The T+15 RNA continues to activate PKR under eukaryotic Mg2+ conditions of 0.5 mM, albeit ~2-fold lower (Figure 5C). In sum, the majority of the riboswitches and ribozymes tested activate PKR, and do so using a variety of RNA secondary and tertiary structures, ranging from complex tertiary structures to simple stem-loops.
Structure mapping and PKR footprinting reveal binding modes of PKR
In an effort to gain insight into the structural basis for activation of PKR by functional RNAs, we performed structure mapping on the 5′-end labeled aptamer domain of the Vc2 riboswitch and the glmS riboswitch-ribozyme in the absence and presence of PKR. These two RNAs were chosen because crystal structures are available and they are large and complex enough to potentially lock PKR into specific sites to offer footprinting.
As shown in Figure 6, structure-mapping experiments on the Vc2 aptamer in the absence of cdiGMP and PKR were consistent with the Vc2 crystal structure. Single strand-specific RNases T1 and A largely cleaved in the expected single-stranded regions, whereas the double strand-specific RNase V1 cleaved expected double-stranded regions. Under native conditions, G-specific single-strand RNase T1 cleaved G67, G70, G105, and G110, which are either in L3 or at the 3′-single-strand overhang (Figure 1), consistent with the crystal structure whereas all of the other Gs are base paired. Pyrimidine-specific single-strand RNase A cleaved C17, C22, C46, and C72, along with several positions in the 3′-single-strand overhang. These cleavages are consistent with the crystal structure, except for C22 and C46, which are in the lower part of P2 in the crystal structure. However, this part of P2 is likely weak as it is only 2 base pairs in length and C22 and C46 are also cleaved by RNase V1 suggesting pairing in a fraction of the strands. The double-strand specific RNase V1 cleaved at C17, C22, C27, A28, C46, C59, C64–C65, C72, U90, C92, C93, and A100, which are in helical regions. Addition of cdiGMP gave rise to several changes in protection consistent with published effects; in particular, G32 and G45 became protected from RNase T1 cleavage, while A48 and C59 gained protection from RNase V1.27 These data support native folding of the Vc2 riboswitch aptamer domain and proper binding of its cdiGMP ligand.
Figure 6.
Structure mapping and PKR footprinting of Vc2. a) 5′-end labeled RNA at 4 mM Mg2+ was incubated alone or with cdiGMP or PKR, subjected to limited nuclease digestion, and fractionated by 12% denaturing PAGE. Leftmost lanes are controls in the absence of nuclease; underlined lanes were incubated at 37°C for 15 min to simulate the RNase activity. ‘OH−‘ is a limited alkaline hydrolysis ladder and ‘den T1’ is RNase T1 in denaturing conditions; these yield a ladder of all nucleotides and all Gs, respectively. The right side of the gel displays RNase digestion in native conditions for RNases T1, A and V1 for 15 min at the temperature noted. The following were probed: Vc2 RNA bound to cdiGMP (‘RNA+CDG’), RNA only, and RNA with PKR titrated from 0.44 to 7 μM. Gray dashed line depicts gel defect. Black dots denote PKR footprinting. b) Structure mapping and PKR footprinting data placed on the crystal structure of Vc2 (PDB ID: 3MXH). Spheres are 2′-hydroxyls that are either always protected (light pink) or PKR protected (black). cdiGMP is shown as a red stick model. Figure 1 provides the secondary structure. Dashed line represents the U1A binding site. c) 90° rotation.
Effects of PKR on cleavage by RNases T1, A, and V1 were tested. PKR conferred significant protection from RNase V1 cleavage, but only small protections from RNase T1 and no changes for RNase A, consistent with PKR’s known preference to bind dsRNA. For instance, PKR protected RNase V1 from cleavage at positions C27, A28, A48 C59, C64–C65, C72, U90, and U92, in pairings P1, P2, and P3. Protections from RNase T1 were found at G32, G45, and G67, while no protections from RNase A were observed. Moreover, there were no enhancements of RNA cleavage in the base pairing regions of the aptamer in the presence of PKR, consistent with absence of large-scale conformational changes or disruption of secondary structure, supporting the notion that PKR binds the natively folded Vc2 aptamer structure. Interestingly, a subset of the protections--those at G32, G45, A48, and C59--are the same as those with cdiGMP binding, suggesting that PKR also enforces RNA tertiary structure. PKR may thus clamp down on the native tertiary structure of the ribozyme. There are additional protections that are unique to PKR versus cdiGMP including those at A48, C64–C65, G67, C72, U90, U92, and A100. These results indicate that PKR footprints to the aptamer.
Given that there were no significant changes in tertiary structure, we modeled the RNase protections onto the crystal structure of the Vc2 riboswitch aptamer domain. Nucleotides that were protected from RNases in both the absence and presence of PKR, denoted with pink spheres on the 2′OH, are in the pseudoknotted region of helical docking, further supporting PKR binding the natively folded aptamer. Positions of RNase protection dependent on the presence of PKR are denoted with black spheres. These positions line the periphery of the riboswitch in the double-stranded regions (Figure 6B, C) and suggest several PKR footprints as depicted in Figure 8 and discussed below. Native RNA tertiary structure appears to be required for site binding of PKR, as the G83C mutant, which disrupts tertiary structure, has no clear PKR footprint (Supplemental Figures 2 and 3) even though it activates PKR (Figure 3). Apparently, loss of tertiary structure in G83C allows PKR to access multiple activating helical regions, the average of which reveals no distinct footprint.
Figure 8.
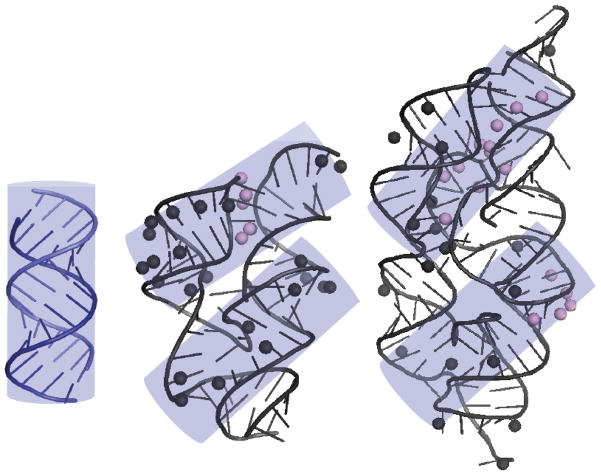
Footprints of PKR (lilac transparent cylinders) mapped onto 16 bp of A-form dsRNA, the Vc2 riboswitch, and the glmS riboswitch-ribozyme. Positions of PKR-mediated RNase protection for the Vc2 riboswitch and the glmS riboswitch-ribozyme are from the structure mapping experiments displayed in Figures 6B and 7C, respectively. The 16 bp of A-form dsRNA are modeled as a cylinder that is superposed onto the Vc2 and glmS structures covers the majority of PKR-mediated RNase protections. Two footprints are possible for the Vc2 riboswitch and glmS riboswitch-ribozyme which may drive PKR dimerization and activation. Note that glmS is large and rotating the bottom cylinder counterclockwise by ~45° may afford an additional helical binding site.
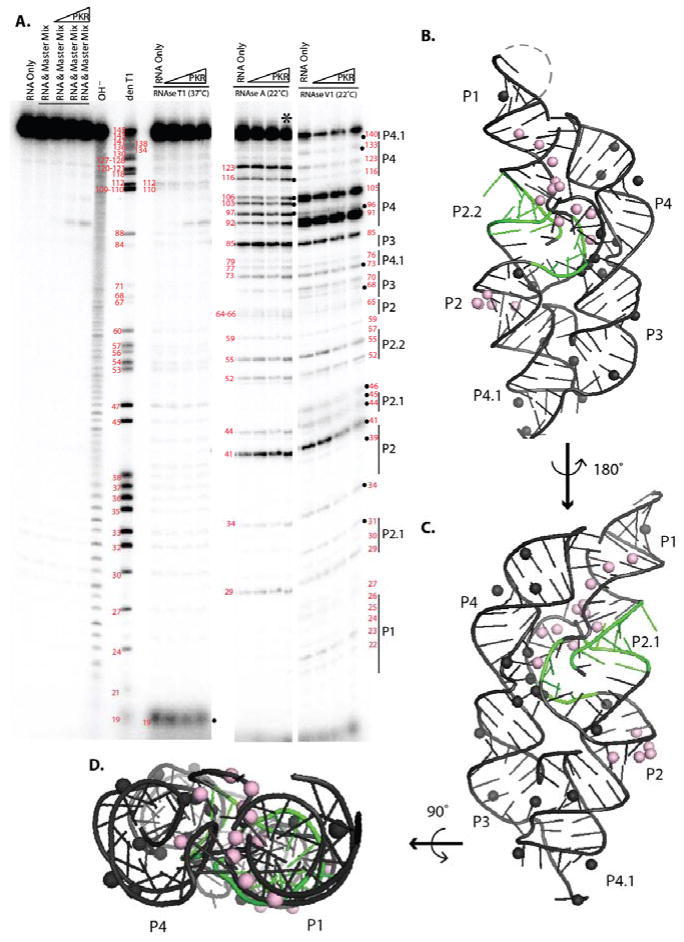
The glmS riboswitch-ribozyme was also subjected to structure mapping in the absence and presence of PKR (Figure 7). In the absence of PKR, the single and double strand-specific RNases cleaved in the single and double stranded region expected from the crystal structure. RNase T1 cleaved only two Gs significantly, G19 and G112, consistent with the crystal structure of this highly structured RNA (Refer to Figure 4 for secondary structure). RNase A strongly cleaved residues U41, C85, and C123, which are exposed in the crystal structure. In addition, RNase A cleaved the AU- and GU-rich P4 region to some extent, consistent with partial melting of P4. RNase V1 cleaved position C44, A46, G68, U70, C85, U91, and U105 in P2.1, P3, and P4, consistent with their base pairing in the crystal structure, and showing that the P4 is also folded at least some of the time. Overall, these structure mapping data support native folding of the glmS riboswitch-ribozyme.
Figure 7.
Structure mapping and PKR footprinting of the glmS ribozyme. a) 5′-end labeled RNA at 4 mM Mg2+ was incubated alone or with PKR, subjected to limited nuclease digestion, and fractionated by 12% denaturing PAGE. Leftmost lanes are controls in the absence of nuclease; underlined lanes were incubated at 37°C for 15 min to simulate the RNase activity. ‘OH− ‘ is a limited alkaline hydrolysis ladder and ‘den T1’ is RNase T1 in denaturing conditions; these yield a ladder of all nucleotides and all Gs respectively. The right side of the gel displays RNase digestion under native conditions for RNases T1, A and V1 for 15 min at the temperatures noted. The following were probed: glmS RNA only, and RNA with PKR titrated from 0.44 to 7 μM. The lane with an asterisk was not used in our data analysis because it seemed to be overdigested. Black dots denote PKR footprinting. b) Structure mapping and PKR footprinting data mapped onto the crystal structure of self-cleaved glmS (PDB ID: 3G9C) using the cleavage data. Spheres are 2′-hydroxyls that are either always protected (light pink) or PKR protected (black). Figure 4 provides the secondary structure. Dashed line represents the U1A binding site. c) A 180° rotation. d) 90° rotation. Absence of RNase V1 cleavage in P2 is consistent with the inability of this nuclease to cleave all dsRNA sites.51,52
Upon addition of PKR, significant protections from RNase V1 but not RNases T1 and A were observed, supporting PKR binding to double-stranded regions of this functional bacterial RNA. In particular, RNase V1 protections by PKR were found at C44 and A46 in P2.1, A68 and U70 in P3, and C85 in J3/4. There were also no clear enhancements of RNA cleavage in the presence of PKR, consistent with PKR binding to the fully folded glmS riboswitch-ribozyme. Modeling of the RNase protections onto the crystal structure of the glmS riboswitch-ribozyme was then performed as described above. Positions of protection in both the absence and presence of PKR are again found in helical docking area (pink spheres), while positions of protection dependent on PKR are found on the peripheral double-stranded regions of the riboswitch (Figure 7B-D, black spheres) suggesting several PKR footprints (Figure 8).
Conclusions
We tested three different functional bacterial RNAs for their ability to activate PKR: the Vc2 riboswitch, the glmS riboswitch-ribozyme, and the twister ribozyme. Most constructs derived from these RNAs activated PKR, provided they were long enough to form sufficient RNA structure. In the case of the Vc2 aptamer and glmS riboswitch-ribozyme, RNA folding and PKR protection were examined by structure mapping. Footprints of PKR are provided in Figure 8, along with a model of 16 bp dsRNA. Prior studies revealed that 16 bp is the minimal length of dsRNA required to bind one protomer of PKR, but that 30 bp is the minimal length needed for potent activation of PKR.5,6 Work from Cole and co-workers showed that 30 bp is the minimal length to dimerize PKR,7 consistent with a PKR dimerization model of autophosphorylation. Along these lines, the 23 bp hairpin inhibitor of PKR, HIV-1 TAR RNA, becomes an activator when it dimerizes, as it is then long enough to dimerize PKR.30
Two footprints of PKR are found on the Vc2 aptamer and glmS riboswitch-ribozyme that are A-form-like and similar in length to 16 bp dsRNA (Figure 8). This suggests that PKR dimerizes on functional bacterial RNAs, which leads to its activation, a notion further supported by the bell-shaped dependence of PKR activation on the concentration of these RNAs (Figures 2 and 4). The two PKR footprints on these functional RNAs are parallel to one another, or nearly so. Observation that PKR is activated by these two functionally distinct RNAs, which are structurally similar only in their ‘Y-shaped’ architecture, is consistent with PKR having a broad-spectrum response to accessible structured RNA. Moreover, activation of PKR by RNAs with unrelated tertiary structures—those from cdiGMP riboswitch, glmS ribozyme, and twister ribozyme in this study, and that of the trp 5′UTR from B. subtilis11—provides overwhelming support that a specific tertiary structure is not needed to activate PKR. Rather, it appears that duplexed regions, exposed upon tertiary folding, are the key elements for activating PKR. This is as expected for an innate immune sensor and consistent with the non-specific recognition of bacterial RNA by the dsRBD.10,11
Transcriptome-wide mapping of RNA structure has been recently developed in several labs.31–33 Comparison of in vitro and in vivo transcriptome-wide RNA structure mapping data sets in yeast and human reveals that much of the RNA in the human genome is either unwound by ATP-dependent helicases or bound with proteins.32,34 Other studies show that RNA secondary structures that are nearby on a transcript exchange faster in vivo than in vitro,35 and that RNA secondary structure is destabilized in the presence of many cosolutes, which may be related to the cosolute-rich cytoplasm of a cell.36,37 Despite extensive efforts, 17,18 no riboswitches have been identified in the human genome21,22 and the few known ribozymes are present either in RNPs, as in the case of the ribosome and spliceosome,16 poorly reactive, as in the CPEB3 HDV-like ribozyme with a one-base pair P1.1 pseudoknot,38 or discontinuous, as in the hammerhead ribozyme 39. Internal RNA modifications, including pseudouridine40,41 and m6A,41–43 have been identified transcriptome-wide in numerous mRNA and ncRNAs. In particular, m6A modification, which is the most common modification of mRNA and ncRNA, has been shown to destabilize model RNA duplexes,44 affect mRNA stability,45 destabilize mRNA and ncRNA secondary structure in vivo to facilitate protein binding,46 and abrogate PKR activation both in vitro and in vivo.11–15,47–49 External modifications, such as the 7mG cap, also prevent PKR activation by the default 5′-triphosphate present in certain viral and bacterial transcripts.50 The innate immune system may thus distinguish self RNA from non-self RNA by a combination of unwinding cellular RNAs, destabilizing them with modifications and cellular conditions, and cloaking structured self-RNAs with proteins and modifications. Non-self RNAs may then be detected as those naked RNAs with appreciable secondary and tertiary structure. This may account, at least in part, for the apparent absence of protein-free riboswitches and ribozymes in the human genome.
METHODS
Design of RNAs; PCR primers, hemiduplex templates, and RNA sequences; RNA preparation and purification; protein expression and purification; PKR activation assays; and structure mapping and PKR footprinting are available as Supporting Information online.
Supplementary Material
Acknowledgments
We thank S. Strobel, K. Smith and R. Meehan for cdiGMP riboswitch constructs and M. Been for the glmS plasmid as well as J. Bingaman for providing purified cleaved glmS RNA. We also thank J. Bingaman, K. Leamy, E. Frankel and D. Mitchell for helpful comments on the manuscript. This research was supported by National Institutes of Health Grant R01GM110237.
Footnotes
Notes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supplementary methods of sequences used, an Excel file of activation assay data, and 1 figure of activation data and 3 figures of structure mapping. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Creagh EM, O’Neill LA. TLRs, NLRs and RLRs: a trinity of pathogen sensors that co-operate in innate immunity. Trends Immunol. 2006;27:352–357. doi: 10.1016/j.it.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Hunter T, Hunt T, Jackson RJ, Robertson HD. The characteristics of inhibition of protein synthesis by double-stranded ribonucleic acid in reticulocyte lysates. J Biol Chem. 1975;250:409–417. [PubMed] [Google Scholar]
- 3.Tian B, Bevilacqua PC, Diegelman-Parente A, Mathews MB. The double-stranded-RNA-binding motif: interference and much more. Nat Rev Mol Cell Biol. 2004;5:1013–1023. doi: 10.1038/nrm1528. [DOI] [PubMed] [Google Scholar]
- 4.Bevilacqua PC, Cech TR. Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry. 1996;35:9983–9994. doi: 10.1021/bi9607259. [DOI] [PubMed] [Google Scholar]
- 5.Manche L, Green SR, Schmedt C, Mathews MB. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol Cell Biol. 1992;12:5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng X, Bevilacqua PC. Activation of the protein kinase PKR by short double-stranded RNAs with single-stranded tails. RNA. 2004;10:1934–1945. doi: 10.1261/rna.7150804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemaire PA, Anderson E, Lary J, Cole JL. Mechanism of PKR Activation by dsRNA. J Mol Biol. 2008;381:351–360. doi: 10.1016/j.jmb.2008.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu S, Kaufman RJ. A model for the double-stranded RNA (dsRNA)-dependent dimerization and activation of the dsRNA-activated protein kinase PKR. J Biol Chem. 1997;272:1291–1296. doi: 10.1074/jbc.272.2.1291. [DOI] [PubMed] [Google Scholar]
- 9.Nallagatla SR, Toroney R, Bevilacqua PC. Regulation of innate immunity through RNA structure and the protein kinase PKR. Curr Opin Struct Biol. 2011;21:119–127. doi: 10.1016/j.sbi.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bleiblo F, Michael P, Brabant D, Ramana CV, Tai T, Saleh M, Parrillo JE, Kumar A, Kumar A. Bacterial RNA induces myocyte cellular dysfunction through the activation of PKR. J Thor Dis. 2012;4:114–125. doi: 10.3978/j.issn.2072-1439.2012.01.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hull CM, Bevilacqua PC. Mechanistic Analysis of Activation of the Innate Immune Sensor PKR by Bacterial RNA. J Mol Biol. 2015;427:3501–3515. doi: 10.1016/j.jmb.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Truong DM, Sidote DJ, Russell R, Lambowitz AM. Enhanced group II intron retrohoming in magnesium-deficient Escherichia coli via selection of mutations in the ribozyme core. Proc Natl Acad Sci USA. 2013;110:E3800–3809. doi: 10.1073/pnas.1315742110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tyrrell J, McGinnis JL, Weeks KM, Pielak GJ. The cellular environment stabilizes adenine riboswitch RNA structure. Biochemistry. 2013;52:8777–8785. doi: 10.1021/bi401207q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD. Mol Biol Cell. 3. Garland; New York: 1994. [Google Scholar]
- 15.Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doudna JA, Cech TR. The chemical repertoire of natural ribozymes. Nature. 2002;418:222–228. doi: 10.1038/418222a. [DOI] [PubMed] [Google Scholar]
- 17.Breaker RR. Prospects for riboswitch discovery and analysis. Mol Cell. 2011;43:867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vu MM, Jameson NE, Masuda SJ, Lin D, Larralde-Ridaura R, Luptak A. Convergent evolution of adenosine aptamers spanning bacterial, human, and random sequences revealed by structure-based bioinformatics and genomic SELEX. Chem Biol. 2012;19:1247–1254. doi: 10.1016/j.chembiol.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cochrane JC, Lipchock SV, Smith KD, Strobel SA. Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme. Biochemistry. 2009;48:3239–3246. doi: 10.1021/bi802069p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith KD, Lipchock SV, Livingston AL, Shanahan CA, Strobel SA. Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch. Biochemistry. 2010;49:7351–7359. doi: 10.1021/bi100671e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eiler D, Wang J, Steitz TA. Structural basis for the fast self-cleavage reaction catalyzed by the twister ribozyme. P Natl Acad Sci USA. 2014;111:13028–13033. doi: 10.1073/pnas.1414571111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tischler AD, Camilli A. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol Microbiol. 2004;53:857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 24.Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamayo R, Pratt JT, Camilli A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu Rev Microbiol. 2007;61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood S, Ferre-D’Amare AR, Rueda D. Allosteric tertiary interactions preorganize the c-di-GMP riboswitch and accelerate ligand binding. ACS Chem Biol. 2012;7:920–927. doi: 10.1021/cb300014u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kulshina N, Baird NJ, Ferre-D’Amare AR. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat Struct Mol Biol. 2009;16:1212–1217. doi: 10.1038/nsmb.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 29.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heinicke LA, Wong CJ, Lary J, Nallagatla SR, Diegelman-Parente A, Zheng X, Cole JL, Bevilacqua PC. RNA dimerization promotes PKR dimerization and activation. J Mol Biol. 2009;390:319–338. doi: 10.1016/j.jmb.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 32.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wan Y, Qu K, Zhang QC, Flynn RA, Manor O, Ouyang Z, Zhang J, Spitale RC, Snyder MP, Segal E, Chang HY. Landscape and variation of RNA secondary structure across the human transcriptome. Nature. 2014;505:706–709. doi: 10.1038/nature12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung JW, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, Chang HY. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015;519:486–490. doi: 10.1038/nature14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahen EM, Watson PY, Cottrell JW, Fedor MJ. mRNA secondary structures fold sequentially but exchange rapidly in vivo. PLoS Biol. 2010;8:e1000307. doi: 10.1371/journal.pbio.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakano S, Karimata H, Ohmichi T, Kawakami J, Sugimoto N. The effect of molecular crowding with nucleotide length and cosolute structure on DNA duplex stability. J Am Chem Soc. 2004;126:14330–14331. doi: 10.1021/ja0463029. [DOI] [PubMed] [Google Scholar]
- 37.Lambert D, Draper DE. Effects of osmolytes on RNA secondary and tertiary structure stabilities and RNA-Mg2+ interactions. J Mol Biol. 2007;370:993–1005. doi: 10.1016/j.jmb.2007.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salehi-Ashtiani K, Luptak A, Litovchick A, Szostak JW. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science. 2006;313:1788–1792. doi: 10.1126/science.1129308. [DOI] [PubMed] [Google Scholar]
- 39.Martick M, Horan LH, Noller HF, Scott WG. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature. 2008;454:899–902. doi: 10.1038/nature07117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, Leon-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, Fink G, Regev A. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159:148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, Sorek R, Rechavi G. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 43.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roost C, Lynch SR, Batista PJ, Qu K, Chang HY, Kool ET. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J Am Chem Soc. 2015;137:2107–2115. doi: 10.1021/ja513080v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nallagatla SR, Toroney R, Bevilacqua PC. A brilliant disguise for self RNA: 5′-end and internal modifications of primary transcripts suppress elements of innate immunity. RNA Biol. 2008;5:140–144. doi: 10.4161/rna.5.3.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nallagatla SR, Bevilacqua PC. Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. RNA. 2008;14:1201–1213. doi: 10.1261/rna.1007408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuen KC, Xu B, Krantz ID, Gerton JL. NIPBL Controls RNA Biogenesis to Prevent Activation of the Stress Kinase PKR. Cell Rep. 2016;14:93–102. doi: 10.1016/j.celrep.2015.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nallagatla SR, Hwang J, Toroney R, Zheng X, Cameron CE, Bevilacqua PC. 5′-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science. 2007;318:1455–1458. doi: 10.1126/science.1147347. [DOI] [PubMed] [Google Scholar]
- 51.Lockard RE, Kumar A. Mapping tRNA structure in solution using double-strand-specific ribonuclease V1 from cobra venom. Nucleic Acids Res. 1981;9:5125–5140. doi: 10.1093/nar/9.19.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lowman HB, Draper DE. On the recognition of helical RNA by cobra venom V1 nuclease. J Biol Chem. 1986;261:5396–5403. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.