Abstract
The human molecular chaperones heat shock protein 70 (Hsp70) and heat shock cognate protein 70 (Hsc70) bind to the hepatitis C viral nonstructural protein 5A (NS5A) and regulate its activity. Specifically, Hsp70 is involved in NS5A-augmented internal ribosomal entry site (IRES)-mediated translation of the viral genome, whilst Hsc70 appears to be primarily important for intracellular infectious virion assembly. To better understand the importance of these two chaperones in the viral life cycle, infected human cells were treated with allosteric Hsp70/Hsc70 inhibitors (AHIs). Treatment with AHIs significantly reduced the production of intracellular virus at concentrations that were non-toxic to human hepatoma Huh7.5 cells. The supernatant of treated cultures was then used to infect naïve cells, revealing that AHIs also lowered levels of secreted virus. In contrast to their effects on virion assembly, AHIs did not impact the stability of NS5A or viral protein translation in IRES assays. These results suggest that Hsc70 plays a particularly important and sensitive role in virion assembly. Indeed, it was found that combination of AHIs with a peptide-based viral translation inhibitor exhibited additive antiviral activity. Together these results suggest that the host Hsc70 is a new antiviral target and that its inhibitors utilise a new mechanism of action.
Keywords: Hsp70, Hsc70, Allosteric heat shock protein inhibitors, Hepatitis C virus, Viral assembly, Viral translation
1. Introduction
Hepatitis C virus (HCV) infects 3% of the world’s population and is mainly responsible for liver transplantation in patients with cirrhosis in developed countries [1]. Furthermore, HCV is the most common chronic blood-borne pathogen in the USA, affecting 1.8% of the population, and is the major aetiological factor responsible for the recent doubling in the incidence of hepatocellular carcinoma [2].
Very recently, interferon-free therapies utilising combinations of direct-acting antivirals (DAAs) with or without ribavirin (RBV) are being used successfully to achieve sustained virological response (SVR) in the majority of cases. Besides the very high costs associated with these treatments, other issues include variability in activity across different genotypes, such as genotype 3 that can result in failure to achieve SVR [3]. If RBV is required, significant side effects can occur, such as haemolytic anaemia [4]. Treatment with DAAs can also result in resistant viruses as targeting viral proteins puts direct selective pressure for resistant mutants. Furthermore, a small percentage of patients are infected with intergenotypic recombinant strains of HCV that may not respond optimally to the current standard treatments [5]. Therefore, there is still a need for new compounds to use in drug cocktails to improve efficacy, to avert resistance, to reduce costs, to reduce potential adverse events (if RBV is used) and to fight novel recombinant viral strains.
HCV is an RNA virus classified in the genus Hepacivirus in the Flaviviridae family. It possesses an ca. 9.6-kb positive-sense RNA genome that is translated as a single polypeptide of ca. 3000 amino acids in length. It is subsequently proteolytically cleaved into 10 viral proteins including the structural proteins core, E1, E2 and the integral membrane ion channel p7 as well as the nonstructural (NS) proteins NS2, NS3, NS4A, NS4B, NS5A and NS5B [6]. The 5′ non-coding region of the viral genome possesses an internal ribosomal entry site (IRES). The HCV viral life cycle in a cell can be divided into six phases: (i) binding and internalisation; (ii) cytoplasmic release and uncoating; (iii) viral polyprotein translation and processing; (iv) RNA genome replication; (v) packaging and assembly; and (vi) virus morphogenesis and secretion. NS5A, a 56–59 kDa multifunctional phosphoprotein, is a component of the viral replicase complex and has been implicated in regulation of HCV genome replication, IRES-mediated viral protein translation, virion assembly and infectious virion secretion [7].
Cellular heat shock proteins (HSPs) have been shown by our group and others to play important roles in the replication of RNA viruses. HSPs play many roles in normal protein homeostasis, including protein folding and degradation [8]. In addition, heat shock cognate protein 70 (Hsc70) and Hsp70 are also known to play important roles in the assembly and disassembly of multiprotein complexes. For example, Hsc70 plays an essential role in the movement of clathrin triskelions during endocytosis [9]. Indeed, members of the HSP70 family were first identified in genetic screens for proteins required for lambda phage replication through their effects on assembly of viral transcriptional complexes [10]. We have previously shown that Hsc70, Hsp70 and Hsp40 (a cofactor of Hsp70) assemble with NS5A [11]. These complexes were essential for NS5A-augmented IRES-mediated translation, virus production and assembly of infectious virions [12–14]. Consistent with these results, (i) Hsp70 and/or Hsc70 knockdown, (ii) the HSP synthesis inhibitor quercetin (a bioflavonoid) and (iii) a small hairpin peptide (HCV4) from NS5A domain I that is capable of blocking the interaction between NS5A and Hsp70 inhibited virus production. These results suggested that inhibiting Hsc70 or Hsp70 might provide a new way to create antivirals.
Members of the HSP70 family of chaperones are highly conserved. They consist of a nucleotide-binding domain (NBD) that hydrolyses ATP, which is attached to a substrate-binding domain (SBD) that binds to protein ‘clients’ [8]. ATP hydrolysis in the NBD regulates the structure of the SBD such that repeated ATPase cycles allow the chaperone to reversibly interact with clients. Recently, small molecules have been discovered that interrupt this cycle. For example, YM-01, JG-40 and JG-98, derivatives of the parental compound MKT-077, bind to an allosteric site in the NBD to limit ATPase activity and stabilise SBD/client interactions. This allosteric site is highly conserved between Hsc70 and Hsp70; thus, the molecules inhibit both chaperones equally [15–17]. Still, these compounds have proven to be powerful chemical probes, revealing the roles of Hsc70 and Hsp70 in regulating the stability of proteins involved in cancer, Alzheimer’s disease and Listeria infection [16–21]. Likewise, we reasoned that these inhibitors might illuminate the roles of Hsp70 and Hsc70 in the HCV life cycle, while also exploring whether these chaperones could be new targets for the development of antivirals. Here we report that allosteric Hsp70/Hsc70 inhibitors (AHIs) indeed block HCV infectious virion assembly in infected cells.
2. Materials and methods
2.1. Plasmid constructs
Hsc70, Hsp70 and their NBDs and SBDs were cloned in the pET-28b plasmid (EMD Millipore, Billerica, MA). The HCV IRES reporter plasmid has been previously described [11]. An intragenotype 2 chimeric monocistronic reporter virus (pNRLFC), based on pJ6/JFH-C parental virus, has been described previously [22]. For the current study, chemically synthesised plasmids pFNX-HCV and pFNX-RLuc (having similar sequences to pNRLFC) were used for construction of recombinant virus [23].
2.2. Cell culture
The cell lines Huh7.5 and 293T were maintained in a humidified atmosphere containing 5% CO2 at 37°C in Dulbecco’s Modified Eagle Medium (Corning Life Sciences, Tewksbury, MA) supplemented with 10% fetal bovine serum (Omega Scientific, Tarzana, CA) and 2 mM L-glutamine (Life Technologies, Grand Island, NY). Huh7.5 cells were a kind gift from Charles Rice (The Rockefeller University, New York, NY). 293T cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA).
2.3. Cell viability
Cell viability was determined using the MTT Cell Proliferation Assay (ATCC) according to the manufacturer’s instructions.
2.4. Inhibitors
YM-01, JG-40, JG-98 and JG-258 were synthesised and characterised as reported previously [20] and were dissolved in dimethyl sulphoxide (DMSO). Purity was >95% by nuclear magnetic resonance (NMR) and high-performance liquid chromatography (HPLC). HCV4 was synthesised as previously described [12].
2.5. Infectious virus production
pFNX-HCV and pFNX-RLuc were in vitro transcribed and the purified RNA was electroporated into Huh7.5 cells to generate infectious viral supernatant as previously described [22].
2.6. Viral assays
The reporter and wild-type viruses were utilised as described previously [12]. All infection assays were performed with a multiplicity of infection (MOI) of 1.0.
2.6.1. Intracellular virus production
Huh7.5 cells were infected for 3 h, at which point the medium was replaced, the cells were treated with AHIs and the infection was allowed to proceed. Cells were harvested 48 h later and luciferase activity was measured using a Renilla Luciferase Assay System (Promega, Madison, WI). For wild-type virus, Western analyses for viral core protein were performed.
2.6.2. Viral entry
Huh7.5 cells were treated with AHIs and were immediately infected with the Renilla reporter virus for 3 h, at which point the medium was replaced and the infection was allowed to proceed for 18 h, followed by harvesting the cells and measuring luciferase activity.
2.6.3. Intracellular viral protein production
Huh7.5 cells were infected for 3 h, at which point the medium was replaced and the infection was allowed to proceed for 30 h. The cells were then treated with AHIs, were harvested at the indicated time points and Renilla luciferase activity was measured.
2.6.4. Infectious virion secretion
2.6.4.1. Long-term assay
The supernatants from the intracellular virus production cultures were concentrated ca. 30-fold to remove ca. 97% of the AHIs, were brought back to the original volume with fresh medium and were used to infect naïve cells for 3 h, at which point the medium was replaced and the infection was allowed to proceed. Amicon® Ultra 0.5 mL Centrifugal Filters (EMD Millipore) were used to remove the AHIs. Cells were harvested 72 h after the infection and Renilla luciferase activity was measured. For wild-type virus, Western analyses for viral proteins were performed.
2.6.4.2. Short-term assay
Huh7.5 cells were infected for 3 h, at which point the medium was replaced and the infection was allowed to proceed for 30 h. Supernatants were removed from the original cultures, cells were washed with phosphate-buffered saline (PBS), fresh medium was added and the cells were treated with AHIs. At the indicated time points, the supernatants were collected, were concentrated to remove the AHIs as above and were used to infect naïve cells for 3 h, at which point the medium was replaced. Cells were harvested 72 h after infection and luciferase activity was measured.
2.6.5. Intracellular infectious virion assembly
Huh7.5 cells were infected for 3 h, at which point the medium was replaced, the cells were treated with brefeldin A and the infection was allowed to proceed. Supernatants were removed 30 h after infection, the cells were washed with PBS and fresh medium was added. The cultures were subjected to three cycles of freeze–thaw to release assembled viral particles. These suspensions were cleared of cellular debris and were used to infect naïve cells for 3 h, at which point the medium was replaced and the infection was allowed to proceed. Cells were harvested 72 h after infection and Renilla luciferase activity was assayed. For wild-type virus, Western analyses for viral proteins were performed.
2.7. IRES reporter assay
293T cells were treated with 1μM AHIs and 2 h later were transfected with the HCV IRES reporter plasmid and either pMSCV-NS5A-FLAG or pMSCV-GFP. All transfections were done using X-tremeGENE 9 Transfection Reagent (Roche, Indianapolis, IN). Seventy-two hours post-transfection, Renilla and firefly luciferase activities were determined using a Dual Luciferase Assay System (Promega).
2.8. Production and purification of recombinant proteins
Hsc70, Hsp70 and their NBDs and SBDs were expressed and purified as described previously [13]. Briefly, protein expression was induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) in BL21(DE3) Escherichia coli cells with shaking at 250 rpm for 5 h at 37°C. Cells were centrifuged for 5 min at 6000 rpm and were stored at −20°C. Cells were re-suspended in 50 mM Tris–HCl (pH 7.6), 300 mM NaCl and 20 mM imidazole with Protease Inhibitor Cocktail (Sigma-Aldrich, St Louis, MO) and were lysed by sonication. Cells were spun down in a SS-34 rotor at 16 500 rpm for 30 min, were filtered through a 0.2-μm filter and were applied to a Hi-trap Nickel column by syringe at room temperature. Proteins were eluted in one step with 50 mM Tris–HCl (pH 7.6), 300 mM NaCl and 400 mM imidazole.
2.9. Surface plasmon resonance (SPR)
Binding studies were performed on a Biacore® 3000 instrument (Biacore AB, Uppsala, Sweden). Proteins were immobilised on CM5 sensor chips by amine coupling. The solution phase analytes were dissolved in HBS-EP buffer, which contained 0.15 M NaCl, 10 mM HEPES (pH 7.4), 3 μM ethylene diamine tetra-acetic acid (EDTA) and 0.005% polysorbate 20. The solutions traversed the sensors at a flow rate of 50 μL/min. Costar® low-retention polypropylene tubes (Corning Life Sciences) were used throughout.
2.10. Statistical analysis
Error bars reflect the standard deviation. P-values were determined by Student’s t-test. All viral assays were done with six biological replicates for each individual assay.
2.11. Calculation of the half-maximal inhibition concentration (IC50)
IC50 values were calculated based on the formula derived from a third-degree polynomial curve fit in Microsoft Excel (Microsoft Corp., Redmond, WA).
3. Results
3.1. Allosteric Hsp70/Hsc70 inhibitors are well tolerated in Huh7.5 cells
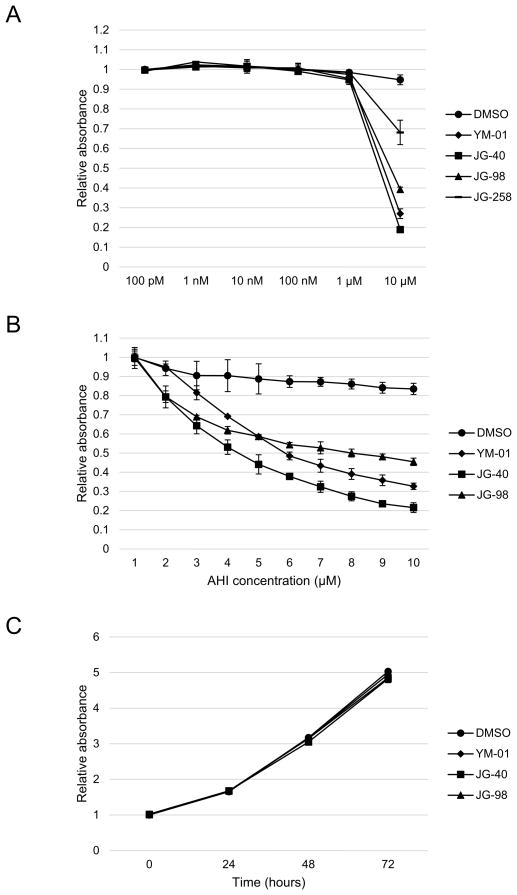
Despite the central roles played by Hsc70 and Hsp70 in protein homeostasis, AHIs have proven to be surprisingly non-toxic in many cellular and animal models [19]. Toxicity is typically observed at concentrations above 5 μM, but anti-chaperone activity occurs at much lower levels (between 200 nM and 1000 nM). To establish these parameters in our system, we first tested YM-01, JG-40 and JG-98 for their cytotoxicity in Huh7.5 cells. JG-258, an isostere of JG-98 that does not bind HSP70 family proteins and is inactive, was also included as a negative control. After 72 h of treatment, MTT assays were performed. In the first experiments, a broad range of compound concentrations was used (Fig. 1A). This result was then used to guide the calculation of a half maximal effective concentration (EC50) value within the range of 1–10 μM. From these experiments, the EC50 values were calculated to be ca. 5 μM (Fig. 1B), consistent with the results in other cultured cells. No toxicity was observed at 1 μM, even when cell viability was examined at intervals up to 72 h (Fig. 1C).
Fig. 1.
Cytotoxicity profile of allosteric Hsp70/Hsc70 inhibitors (AHIs). (A) Cellular toxicity of the AHIs in the concentration range of 100 pM to 10 μM. Huh7.5 cells were treated with the indicated concentrations of the AHIs and MTT assays were then performed every 24 h up to 72 h. The 72-h data points are shown. (B) Cellular toxicity of the AHIs in the concentration range of 1μM to 10 μM. Huh7.5 cells were treated with the indicated concentrations of the AHIs and MTT assays were then performed as above. The 72-hour data points are shown. (C) Cellular toxicity of the AHIs at 1 μM concentration over a time course of 72 h. Huh7.5 cells were treated with 1 μM of the AHIs and MTT assays were then performed as above. DMSO, dimethyl sulphoxide.
To confirm the interaction of AHIs with Hsc70 and Hsp70 in our hands, SPR was utilised. Consistent with previous reports [15,24,25], it was found that YM-01 bound to immobilised human Hsp70 and Hsc70 (data not shown). As expected, binding was observed to either full-length protein or truncated NBD, but not to truncated SBD (data not shown).
3.2. Allosteric Hsp70/Hsc70 inhibitors significantly attenuate virus production in a dose-dependent manner
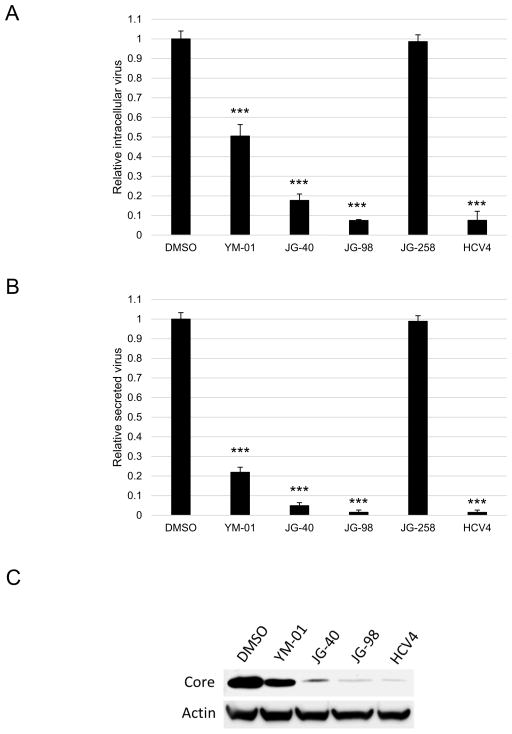
The HCV cell culture system was subsequently utilised to explore the roles of Hsp70 and Hsc70 in viral replication. Initially, compounds were tested at the highest safe dose of 1 μM in 48-h intracellular infectious virus production assays using the Renilla reporter virus. As shown in Fig. 2A, all three AHIs displayed significant antiviral activity. JG-98 was the most potent, consistent with its relatively tight affinity for chaperones [26]. Subsequently, the levels of secreted virus in these cultures were determined through long-term infectious virion secretion assays. It was found that infectious virus production was significantly reduced, particularly when JG-98 was used (>20-fold reduction in virus) (Fig. 2B). The previously reported HCV4 peptide (IC50 = 452 pM), which inhibits viral translation, was used as a positive control [12,27]. To confirm the antiviral activity of AHIs, the wild-type virus was utilised in intracellular infectious virus production assays followed by Western analyses to measure the levels of viral proteins. As shown in Fig. 2C, the levels of core were significantly (>90%) diminished by AHI treatment, especially in response to JG-98. This result is consistent with the reduced luciferase activity of the reporter virus. In these initial studies, MAL3-101, a different allosteric Hsp70 inhibitor, and VER-155008, an orthosteric Hsp70 inhibitor, were also tested; whilst VER-155008 had slight antiviral activity, the MKT-077 family of compounds were the only chaperone modulators that were effective below their toxic doses (data not shown).
Fig. 2.
The allosteric Hsp70/Hsc70 inhibitors (AHIs) YM-01, JG-40 and JG-98 significantly attenuate virus production. (A) Intracellular virus production assay indicates that YM-01, JG-40 and JG-98 significantly block virus production. (B) Viral secretion is significantly attenuated by AHIs as demonstrated by a long-term infectious virion secretion assay. (C) AHIs significantly block virus production. The intracellular virus production assay was performed as in (A) using wild-type virus and Western analyses were performed with antibodies against the viral core and cellular actin proteins. *** P < 0.0005. DMSO, dimethyl sulphoxide.
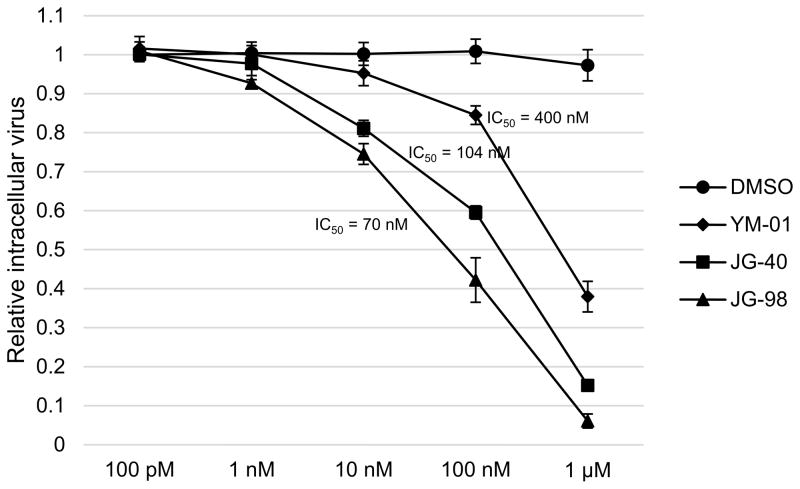
Next we determined the dose dependence of the antiviral activity using cells infected with the reporter virus. After 48 h, the antiviral activity of YM-01, JG-40 and JG-98 was found to be dose-dependent (Fig. 3). The IC50 value of JG-98 was ca. 70 nM, which is >70-fold lower than its cytotoxic level.
Fig. 3.
Allosteric Hsp70/Hsc70 inhibitors (AHIs) block virus production in a dose-dependent manner. Dose response of the AHIs in the concentration range of 100 pM to 1 μM as determined by intracellular virus production assays. IC50, half-maximal inhibition concentration.
3.3. Allosteric Hsp70/Hsc70 inhibitors do not affect viral entry
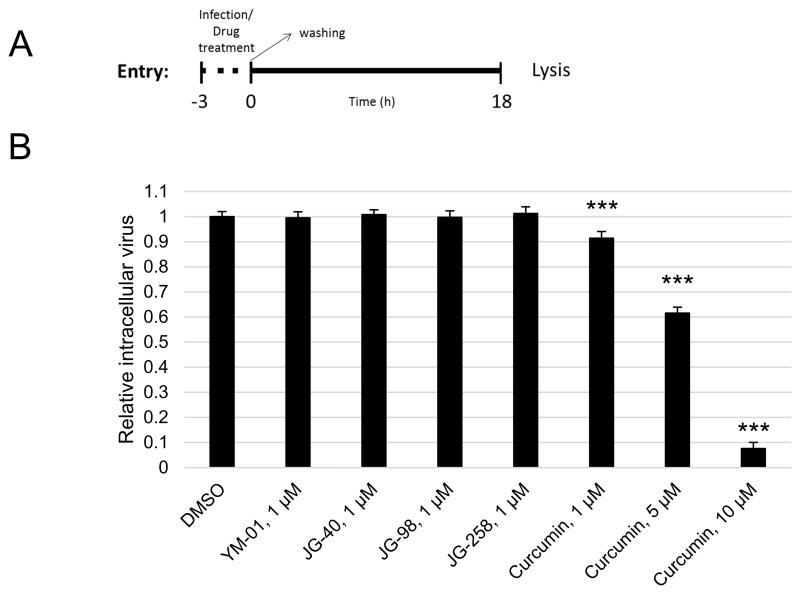
Having established the effective doses and safety of the AHIs, we proceeded to explore which specific phase(s) of the viral life cycle might be affected. Initially, viral entry was tested utilising curcumin, a previously reported viral entry inhibitor, as a positive control (Fig. 4A). Whilst curcumin significantly decreased intracellular viral levels in a dose-dependent manner, AHIs had no effect on viral entry at their maximum safe dose of 1 μM (Fig. 4B).
Fig. 4.
Allosteric Hsp70/Hsc70 inhibitors (AHIs) do not affect viral entry. (A) Schematic of the entry assay. (B) Huh7.5 cells were treated with the indicated compounds and were immediately infected for 3 h, at which point, the medium was replaced and the infection was allowed to proceed for 18 h. The cells were then lysed and luciferase activity was measured. *** P < 0.0005. DMSO, dimethyl sulphoxide.
3.4. Allosteric Hsp70/Hsc70 inhibitors do not affect viral protein translation
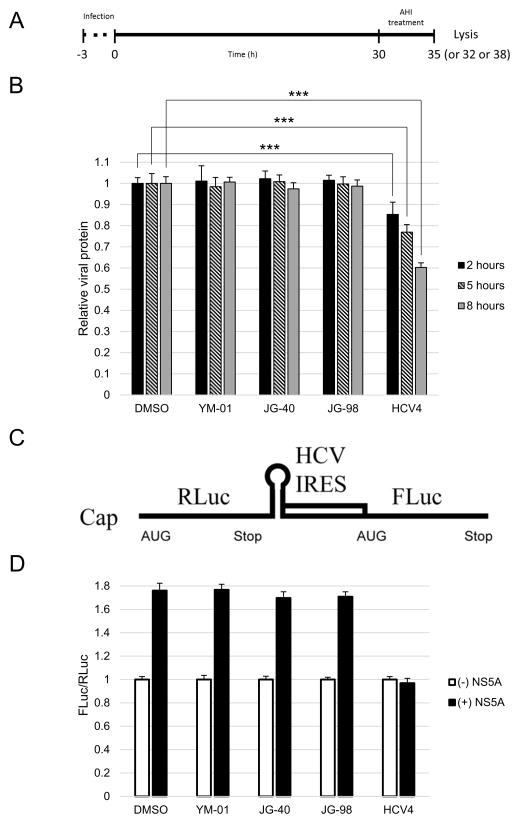
As viral entry was not affected by the AHIs, we proceeded to determine whether viral translation could by a potential target by performing intracellular viral protein production assays (Fig. 5A). Surprisingly, the AHIs did not have any effect on viral protein levels at any of the time points assayed (Fig. 5B), whereas the positive control (HCV4) inhibited viral translation.
Fig. 5.
Allosteric Hsp70/Hsc70 inhibitors (AHIs) do not affect viral protein translation and internal ribosomal entry site (IRES) activity. (A) Schematic of the intracellular viral protein production assay. (B) Intracellular viral protein production assay demonstrates that the AHIs do not affect viral protein translation. (C) Schematic of the bicistronic construct utilised for determining the levels of NS5A-augmented IRES-mediated translation. (D) NS5A-augmented IRES-mediated translation is not affected by the AHIs as determined by IRES reporter assays. *** P < 0.0005.
3.5. Allosteric Hsp70/Hsc70 inhibitors do not affect NS5A-augmented IRES-mediated translation
To further confirm that AHIs do not affect viral translation, a previously reported bicistronic reporter system was utilised. In this assay, viral IRES-mediated translation is measured. Specifically, the firefly luciferase (FLuc) open reading frame (ORF) is driven by a 5′ cap, whilst the Renilla luciferase (RLuc) ORF is driven by the HCV IRES (Fig. 5C). The ratio of FLuc/RLuc then provides a measure of HCV IRES-mediated translation. To make this reporter sensitive to NS5A, it is cotransfected with either an NS5A- or GFP-expressing construct. As shown in Fig. 5D, NS5A-augmented IRES-mediated translation is significantly blocked by the HCV4 control peptide, in agreement with a previous report [12], whereas none of the AHIs had any effect.
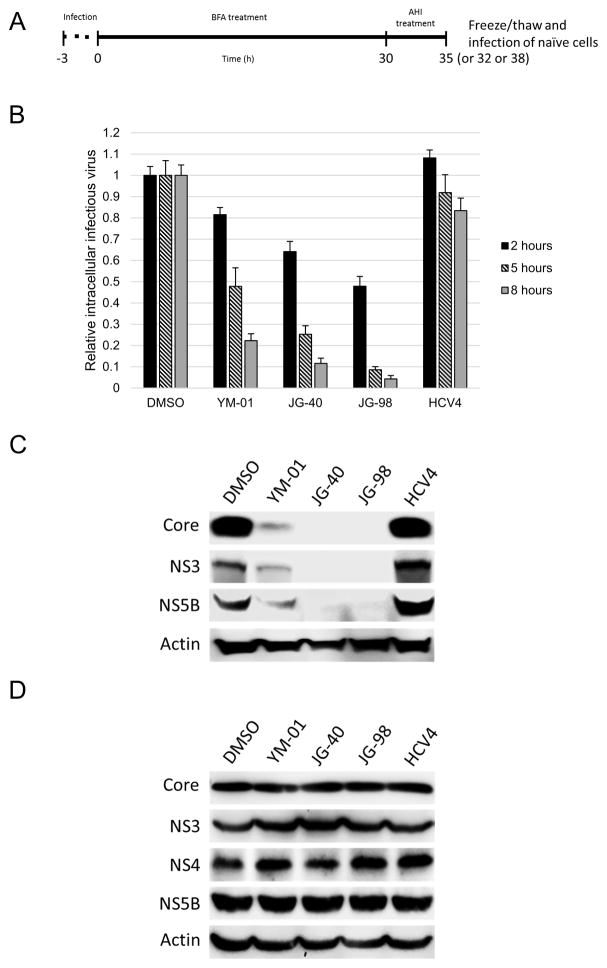
3.6. Allosteric Hsp70/Hsc70 inhibitors block intracellular assembly of infectious virions
Next we determined the effect of AHIs on the assembly of infectious virions inside the cell through intracellular infectious virion assembly assays (Fig. 6A). As shown in Fig. 6B, YM-01, JG-40 and JG-98 significantly blocked intracellular infectious virion assembly. JG-98 (1 μM) reduced virion levels by >20 fold at the 8-h time point. As expected, HCV4 was inactive in this platform (Fig. 6B).
Fig. 6.
Allosteric Hsp70/Hsc70 inhibitors (AHIs) block intracellular infectious virion assembly. (A) Schematic of the assembly assay. BFA, brefeldin A. (B) Intracellular infectious virion assembly assays performed with the Renilla reporter virus indicate that YM-01, JG-40 and JG-98 significantly block infectious virion assembly. (C) AHIs block intracellular assembly of infectious virus. Intracellular infectious virion assembly assays was performed as in (A) and (B) using wild-type virus and Western analyses were performed on the cells infected with intracellular virus with antibodies against the viral core, NS3 and NS5B as well as cellular actin proteins. (D) AHIs do not affect viral protein levels. Intracellular infectious virion assembly assay was performed as in (A) and (B) using wild-type virus and Western analyses were performed on the cells treated with AHIs for 8 h. DMSO, dimethyl sulphoxide.
To confirm this activity, 8-h intracellular infectious virion assembly assays were performed with the wild-type virus. As shown in Fig. 6C, levels of the core protein were nearly undetectable after treatment with JG-40 or JG-98. This was not due to a direct effect of AHIs on the stability of viral proteins in the original host cells as these levels were not impacted (Fig. 6D).
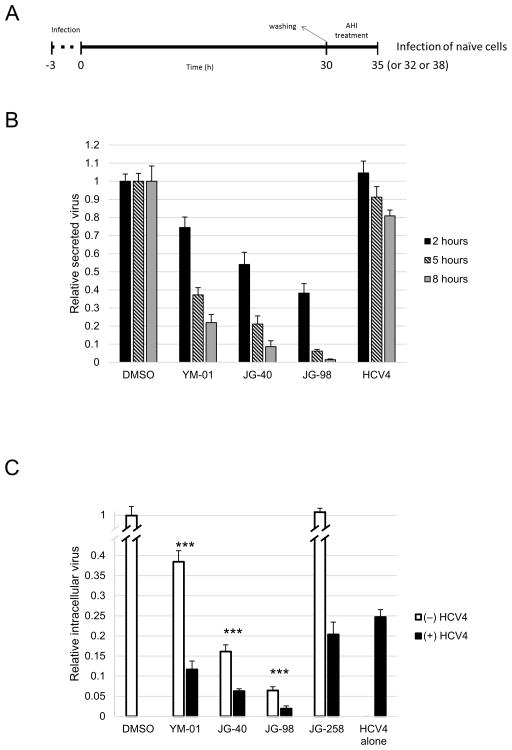
Short-term infectious virion secretion assays were also performed (Fig. 7A), which demonstrated similar results to the assembly assays (Fig. 7B).
Fig. 7.
Allosteric Hsp70/Hsc70 inhibitors (AHIs) significantly affect infectious virion secretion. (A) Schematic of the secretion assay. (B) Short-term infectious virion secretion assays demonstrate the significant effect of YM-01, JG-40 and JG-98 on the levels of infectious virion secretion. (C) The combination of the AHIs and the HCV4 peptide displays significantly improved antiviral activity as demonstrated by the intracellular virus production assays. *** P < 0.0005. DMSO, dimethyl sulphoxide.
3.7. Combination of allosteric Hsp70/Hsc70 inhibitors with the HCV4 peptide displays higher antiviral activity
We hypothesised that since the AHIs and the HCV4 peptide block different stages of the viral life cycle, combined administration of both inhibitors should result in more potent antiviral activity. To test this idea, the intracellular virus production assays were performed in combination with the HCV4 peptide. As shown in Fig. 7C, the combination of AHIs and HCV4 peptide (low dose) dramatically improved antiviral activity in intracellular infectious virus production assays, with the JG-98/HCV4 combination almost abolishing virus proliferation. This result is consistent with the idea that the inhibitors act at distinct stages of the viral life cycle.
4. Discussion
In this study, we explored the roles of Hsp70 and Hsc70 in the HCV life cycle. We had previously reported that Hsc70 and Hsp70 play distinct roles in this process [13], with Hsp70 regulating NS5A-augmented IRES-mediated translation of the viral genome [12,13] and Hsc70 being involved in intracellular infectious virion assembly [13]. The inhibitors used in this study bind to both Hsc70 and Hsp70, so we used them to explore which chaperone role might be most critical in viral processing. We found that the AHIs preferentially affected infectious virion assembly, but not viral translation. Whilst viral secretion also appears to be affected (Fig. 7B), we believe this to be secondary to the effects on virion assembly.
There may be several possibilities for the effect on assembly. First, the AHIs are known to block ATP cycling by the chaperones. Some activities of Hsc70 and Hsp70 require ATP cycling, such as assembly of multiprotein complexes, whereas others do not, such as blocking protein aggregation. Thus, one possibility is that active ATP cycling is preferentially required for the activity of Hsc70 in virion assembly. Another possibility is that AHIs disrupt virion assembly by impacting clathrin-mediated processes. Active nucleotide cycling is known to be required for Hsc70 to mediate the conformational changes associated with clathrin cycling [28]. Thus, AHIs could block virion assembly and maturation, in part through effects on vesicle trafficking. This model is supported by observations that disruption of the interactions between HCV core protein and clathrin leads to impaired assembly, as marked by decreased co-localisation of E2 and core around lipid droplets [29]. Thus, it may be possible that treatment of infected cells with AHIs leads to disruption of clathrin-mediated assembly/secretion at one or more points during the process. Finally, it is also known that YM-01 can activate Hsp70- and Hsc70-mediated degradation of denatured proteins [16,17]. This pathway appears to be less important in the HCV system because we found that 8 h of AHI treatment did not reduce the levels of several viral proteins, including core. It may be possible that longer treatment times are required, but the initial results suggest that client degradation is not a major pathway invoked by AHIs.
Why did the AHIs not reduce viral protein translation? Our previous findings demonstrated that the interaction between NS5A and Hsp70 is mediated by the NBD of Hsp70 rather than the SBD [12]. Thus, NS5A is a non-canonical client of the Hsp70 system. Therefore, inhibition of Hsp70 by the AHIs may not affect viral protein translation because they do not impact non-canonical client interactions. Alternatively, NS5A might directly compete with AHIs for binding to Hsp70, thereby eliminating their inhibitory effects on Hsp70 in this context. Regardless, these results are important for understanding the roles of Hsp70 and Hsc70 in the HCV viral life cycle. They point to a specific vulnerability around ATP cycling by Hsc70.
5. Conclusions
Most approved antivirals bind to viral proteins and, accordingly, they favour the generation of resistant strains. Thus, there is great interest in targeting host factors. Along these lines, it is attractive to consider Hsc70 and Hsp70 as a potential new class of therapeutic targets. One major concern with such a strategy is safety because these chaperones play central roles in normal protein homeostasis. However, we found that JG-98 was toxic at 100-fold higher concentrations than required to block HCV virion assembly. This impressive selectivity might be a product of the allosteric mechanism of the AHIs, which likely do not block all chaperone functions. Thus, the use of these AHIs as antivirals seems promising. It is especially promising to consider AHIs in combination with DAAs or molecules with other mechanisms of action. Finally, other viruses, in addition to HCV, may co-opt Hsc70 and Hsp70 to enable their life cycles, so the insights provided here may also allow for the development of therapies for other viruses.
Highlights.
Allosteric Hsp70/Hsc70 inhibitors (AHIs) block hepatitis C virus proliferation, with no associated cytotoxicity.
Viral protein production and IRES activity are not affected by AHIs.
AHIs significantly block intracellular infectious virion assembly.
Combination of AHIs with a peptide inhibitor of viral translation displays an additive effect.
Acknowledgments
The authors thank David Dawson for helpful discussions and Charles Rice (The Rockefeller University, New York, NY) for providing Huh7.5 cells.
Funding: This study was funded by the National Institutes of Health [grant NIH R01DK090794 to SWF; grant NIH NS059690 to JEG; and grant NIH R21AI084090 to AD], the California Center for Antiviral Drug Discovery, University of California Office of the President [grant MRPI # 143226 to AD] and the Cedars-Sinai Programmatic Award (to VA).
Footnotes
Competing interests: None declared.
Ethical approval: Not required.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–67. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB. Hepatocellular carcinoma: an epidemiologic view. J Clin Gastroenterol. 2002;35(5 Suppl 2):S72–8. doi: 10.1097/00004836-200211002-00002. [DOI] [PubMed] [Google Scholar]
- 3.Donaldson EF, Harrington PR, O’Rear JJ, Naeger LK. Clinical evidence and bioinformatics characterization of potential hepatitis C virus resistance pathways for sofosbuvir. Hepatology. 2015;61:56–65. doi: 10.1002/hep.27375. [DOI] [PubMed] [Google Scholar]
- 4.Shiffman ML. What future for ribavirin? Liver Int. 2009;29(Suppl 1):68–73. doi: 10.1111/j.1478-3231.2008.01936.x. [DOI] [PubMed] [Google Scholar]
- 5.Hedskog C, Doehle B, Chodavarapu K, Gontcharova V, Crespo Garcia J, De Knegt R, et al. Characterization of hepatitis C virus intergenotypic recombinant strains and associated virological response to sofosbuvir/ribavirin. Hepatology. 2015;61:471–80. doi: 10.1002/hep.27361. [DOI] [PubMed] [Google Scholar]
- 6.Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436:933–8. doi: 10.1038/nature04077. [DOI] [PubMed] [Google Scholar]
- 7.Ross-Thriepland D, Harris M. Hepatitis C virus NS5A: enigmatic but still promiscuous 10 years on! J Gen Virol. 2015;96:727–38. doi: 10.1099/jgv.0.000009. [DOI] [PubMed] [Google Scholar]
- 8.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–84. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xing Y, Bocking T, Wolf M, Grigorieff N, Kirchhausen T, Harrison SC. Structure of clathrin coat with bound Hsc70 and auxilin: mechanism of Hsc70-facilitated disassembly. EMBO J. 2010;29:655–65. doi: 10.1038/emboj.2009.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yochem J, Uchida H, Sunshine M, Saito H, Georgopoulos CP, Feiss M. Genetic analysis of two genes, dnaJ and dnaK, necessary for Escherichia coli and bacteriophage lambda DNA replication. Mol Gen Genet. 1978;164:9–14. doi: 10.1007/BF00267593. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez O, Fontanes V, Raychaudhuri S, Loo R, Loo J, Arumugaswami V, et al. The heat shock protein inhibitor quercetin attenuates hepatitis C virus production. Hepatology. 2009;50:1756–64. doi: 10.1002/hep.23232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khachatoorian R, Arumugaswami V, Ruchala P, Raychaudhuri S, Maloney EM, Miao E, et al. A cell-permeable hairpin peptide inhibits hepatitis C viral nonstructural protein 5A-mediated translation and virus production. Hepatology. 2012;55:1662–72. doi: 10.1002/hep.25533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khachatoorian R, Ganapathy E, Ahmadieh Y, Wheatley N, Sundberg C, Jung CL, et al. The NS5A-binding heat shock proteins HSC70 and HSP70 play distinct roles in the hepatitis C viral life cycle. Virology. 2014;454–455:118–27. doi: 10.1016/j.virol.2014.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khachatoorian R, Arumugaswami V, Raychaudhuri S, Yeh GK, Maloney EM, Wang J, et al. Divergent antiviral effects of bioflavonoids on the hepatitis C virus life cycle. Virology. 2012;433:346–55. doi: 10.1016/j.virol.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rousaki A, Miyata Y, Jinwal UK, Dickey CA, Gestwicki JE, Zuiderweg ER. Allosteric drugs: the interaction of antitumor compound MKT-077 with human Hsp70 chaperones. J Mol Biol. 2011;411:614–32. doi: 10.1016/j.jmb.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang AM, Miyata Y, Klinedinst S, Peng HM, Chua JP, Komiyama T, et al. Activation of Hsp70 reduces neurotoxicity by promoting polyglutamine protein degradation. Nat Chem Biol. 2013;9:112–8. doi: 10.1038/nchembio.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abisambra J, Jinwal UK, Miyata Y, Rogers J, Blair L, Li X, et al. Allosteric heat shock protein 70 inhibitors rapidly rescue synaptic plasticity deficits by reducing aberrant tau. Biol Psychiatry. 2013;74:367–74. doi: 10.1016/j.biopsych.2013.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis MJ, Gregorka B, Gestwicki JE, Swanson JA. Inducible renitence limits Listeria monocytogenes escape from vacuoles in macrophages. J Immunol. 2012;189:4488–95. doi: 10.4049/jimmunol.1103158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colvin TA, Gabai VL, Gong J, Calderwood SK, Li H, Gummuluru S, et al. Hsp70–Bag3 interactions regulate cancer-related signaling networks. Cancer Res. 2014;74:4731–40. doi: 10.1158/0008-5472.CAN-14-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du NN, Peng ZG, Bi CW, Tang S, Li YH, Li JR, et al. N-substituted benzyl matrinic acid derivatives inhibit hepatitis C virus (HCV) replication through down-regulating host heat-stress cognate 70 (Hsc70) expression. PLoS One. 2013;8:e58675. doi: 10.1371/journal.pone.0058675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, Colvin T, Rauch JN, Acosta-Alvear D, Kampmann M, Dunyak B, et al. Validation of the Hsp70–Bag3 protein–protein interaction as a potential therapeutic target in cancer. Mol Cancer Ther. 2015;14:642–8. doi: 10.1158/1535-7163.MCT-14-0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arumugaswami V, Remenyi R, Kanagavel V, Sue EY, Ngoc Ho T, Liu C, et al. High-resolution functional profiling of hepatitis C virus genome. PLoS Pathog. 2008;4:e1000182. doi: 10.1371/journal.ppat.1000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chu D, Ren S, Hu S, Wang WG, Subramanian A, Contreras D, et al. Systematic analysis of enhancer and critical cis-acting RNA elements in the protein-encoding region of the hepatitis C virus genome. J Virol. 2013;87:5678–96. doi: 10.1128/JVI.00840-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deocaris CC, Widodo N, Shrestha BG, Kaur K, Ohtaka M, Yamasaki K, et al. Mortalin sensitizes human cancer cells to MKT-077-induced senescence. Cancer Lett. 2007;252:259–69. doi: 10.1016/j.canlet.2006.12.038. [DOI] [PubMed] [Google Scholar]
- 25.Miyata Y, Li X, Lee HF, Jinwal UK, Srinivasan SR, Seguin SP, et al. Synthesis and initial evaluation of YM-08, a blood–brain barrier permeable derivative of the heat shock protein 70 (Hsp70) inhibitor MKT-077, which reduces tau levels. ACS Chem Neurosci. 2013;4:930–9. doi: 10.1021/cn300210g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, Srinivasan SR, Connarn J, Ahmad A, Young ZT, Kabza AM, et al. Analogs of the allosteric heat shock protein 70 (Hsp70) inhibitor, MKT-077, as anti-cancer agents. ACS Med Chem Lett. 2013;4 doi: 10.1021/ml400204n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khachatoorian R, Ruchala P, Waring A, Jung CL, Ganapathy E, Wheatley N, et al. Structural characterization of the HSP70 interaction domain of the hepatitis C viral protein NS5A. Virology. 2015;475:46–55. doi: 10.1016/j.virol.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newmyer SL, Schmid SL. Dominant-interfering Hsc70 mutants disrupt multiple stages of the clathrin-coated vesicle cycle in vivo. J Cell Biol. 2001;152:607–20. doi: 10.1083/jcb.152.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neveu G, Barouch-Bentov R, Ziv-Av A, Gerber D, Jacob Y, Einav S. Identification and targeting of an interaction between a tyrosine motif within hepatitis C virus core protein and AP2M1 essential for viral assembly. PLoS Pathog. 2012;8:e1002845. doi: 10.1371/journal.ppat.1002845. [DOI] [PMC free article] [PubMed] [Google Scholar]