Summary
The transcription factor HOXA10 is an important regulator of myelopoiesis. Engineered over-expression of Hoxa10 in mice results in a myeloproliferative disorder that progresses to acute myeloid leukaemia (AML) over time, and in humans over-expression is associated with poor outcomes in AML. Here, we report that loss of Hoxa10 expression in mice results in reduced platelet count and platelet production, but does not affect clotting efficiency. About 40% fewer platelets were found in Hoxa10 null animals in comparison to wild type littermates. We found a nearly 50% reduction in the percentage of reticulated platelets in Hoxa10 null mice, suggesting deficient platelet production. Furthermore, Hoxa10 null animals recovered less efficiently from induced thrombocytopenia, supporting our hypothesis of defective platelet production. This also correlated with reduced colony formation potential of stem and progenitor cells seeded in megakaryocyte-enhancing conditions in vitro. Together, our results indicate that HOXA10 is important for megakaryopoiesis and platelet biogenesis.
Keywords: Platelets, HOXA10, Hoxa10, megakaryopoiesis, in vivo, thrombopoiesis
Introduction
HOXA10 is a transcription factor known to govern haematopoietic cell development through direct gene regulation and is associated with a number of functions during myelopoiesis, including expansion of progenitor cells and acquisition of phagocyte effector functions (Eklund, et al 2002, Eklund, et al 2000, Lindsey, et al 2005, Lu, et al 2003). HOXA10 is highly expressed in human CD34+ bone marrow cells, markedly down-regulated in maturing CD34− bone marrow cells, and expressed in myeloid, erythroid and megakaryocyte (Mk) precursors and cell lines (Sauvageau, et al 1994). HOXA10 overexpression has been studied extensively due to its effects on progenitor cell expansion and its role in leukemogenesis. Reported transplantation models suggest that overexpression of Hoxa10 in murine haematopoietic cells perturbs myeloid and lymphoid differentiation leading to development of acute myeloid leukaemia (AML) (Magnusson, et al 2007, Wang, et al 2009). Graded expression of Hoxa10 is critical for normal development of erythropoiesis and megakaryopoiesis (Thorsteinsdottir, et al 1997). High levels of HOXA10 have been reported to block Mk and erythroid differentiation–resulting in accumulation of megakaryocytic/erythroid progenitors (MEPs), as well as an increase the number of bone marrow megakaryocyte-blast colony-forming cells (Magnusson, et al 2007, Thorsteinsdottir, et al 1997). Importantly, Magnusson et al (2007) found that low/intermediate levels of Hoxa10 overexpression did not induce the same effects on Mk/erythroid differentiation, but instead increased in vitro expansion and reconstitution potential of lineage negative, Sca1+, kit+ (LSK) cells.
In previous work to elucidate mechanisms by which Hoxa10 overexpression influences myeloid leukaemogenesis, the Eklund laboratory identified several genes that are regulated by Hoxa10 in myeloid cells. These genes include Fgf2 (fibroblast growth factor 2), the phagocyte effector genes Cybb and Ncf2, Dusp4 (mitogen-activated protein kinase phosphatase 2), Tgfb2 (transforming growth factor β2) and Itgb3 (β3-integrin) (Bei, et al 2007, Lindsey, et al 2005, Shah, et al 2012, Shah, et al 2011, Wang, et al 2007). Hoxa10 has also been reported to regulate Itgb3 on endometrial cells to enhance implantation of ovaries in females (Daftary, et al 2002). ITGB3 is also part of a key fibrinogen receptor expressed on circulating platelets. Proper fibrinogen signalling and aggregation of platelets is important for normal haemostasis, suggesting that transcriptional regulation of the Itgb3 by Hoxa10 may influence proper platelet functions (Palumbo, et al 2004).
From the literature, it is evident that HOXA10 plays an important role in haematopoietic stem and progenitor cell (HSPC) expansion, megakaryopoiesis and erythropoiesis. However, the role of HOXA10 in megakaryocyte differentiation and thrombopoiesis has not been well characterized. We aimed to address this deficiency by investigating megakaryopoiesis in Hoxa10 null, Hoxa10+/− and wild type mice. Our complete blood count (CBC) results indicate that Hoxa10 null animals have significantly lower steady-state platelet counts in comparison to Hoxa10+/− and wild type animals. Platelet counts were not affected by age, but were lower in females than in males. We also found that Hoxa10 null bone marrow cells undergo less efficient Mk colony (CFU-Mk) formation, exhibit decreased numbers of newly formed platelets, and recover less efficiently from induced thrombocytopenia.
Methods
Mice
This study was carried out under protocols approved by the Northwestern University Animal Care and Use Committee. Hoxa10+/− mice of the C57Bl/6 background (Satokata, et al 1995) were bred in a barrier facility. Hoxa10 null animals were a limiting factor due to defective reproduction; Hoxa10 null animals cannot reproduce, and breeding heterozygous mice yields null animals in a less than Mendelian ratio.
Blood analysis
For CBC data, blood (~100 μl) was collected from anesthetized mice into ethylenediaminetetraacetic acid (EDTA)-coated Microvette collection tubes (Sarstedt; Nümbrecht, Germany) and analysed on a HemaVet 950 (Drew Scientific, Dallas, TX). Mice were categorized into three age groups for analysis (4–16, 17–24 or 25–36 weeks old). Although we found no statistically significant difference for blood counts between age groups or over time, the trends in blood counts suggested the CBC parameters might not be completely age-independent. Therefore, when comparing blood counts between genotypes, mice of all age groups were pooled together to provide the most comprehensive comparison. Each age group contained 6 mice (3 male and 3 female), thus allowing for a large sample size (n=18) for each genotype. To achieve this complete sample set for the limiting Hoxa10 null animals, blood counts from five null mice (2 male and 3 female) were sampled at two separate ages for inclusion in the analysis.
Tail-bleeding assays were performed as described (Apostolidis, et al 2012). Briefly, mice were anesthetized, the tails were pre-warmed at 37°C for 5 min to allow more efficient blood flow, 3-mm of the distal end was cut off using a scalpel, and the blood was allowed to drip (without submerging the tail) into fresh saline pre-warmed to 37°C. The duration of the bleed was reported as the total bleeding time. If the total bleeding time exceeded 10 min, the assay was stopped and pressure was applied to stop the bleeding to avoid losing too much blood. The total volume of blood lost in the saline was estimated by measuring the absorbance at 560 nm with volume adjustment for the amount of blood collected during bleeding, as described by Jirouskova et al (2007).
For studies of induced thrombocytopenia, 36- to 52-week-old mice were anesthetized by ketamine/xylazine and blood was sampled from the tail vein daily for 7 days. Thrombocytopenia was induced by intraperitoneal injection of anti-platelet serum (Absorbed Rabbit Anti-Mouse Thrombocyte, Accurate Chem and Sci Corp.; Westbury, NY) on day 0.
Colony assays
Mice were sacrificed by CO2 inhalation and cervical dislocation as primary and secondary methods of euthanasia, respectively. Bone marrow cells were harvested by flushing femurs with Hank’s balanced salt solution containing 1% penicillin-streptomycin. Red cells were lysed with erythrocyte lysis buffer (0.15 M NH4Cl, 1.0 mM KHCO3, 0.1 mM Na2EDTA, pH 7.2–7.4). Selected samples were also lineage-depleted using lineage-depletion kit (Catalogue number 130–090–858, Miltenyi Biotec; Auburn, CA). CFU-Mk were assayed using MegaCult collagen-based medium (Stem Cell Technologies; Vancouver, BC, Canada) supplemented with 100 ng/mL thrombopoietin (THPO) (Peprotech, Rocky Hill, NJ), and completed according to the manufacturer’s instructions. Colonies were identified as small (1–3 clusters, about 200 cells), medium (4–8 clusters, about 200–1000 cells) and large Mk colonies (more than 8 clusters with thousands of cells in the clusters).
Histology
Sternum samples were fixed in formalin, paraffin-embedded, cut for haematoxylin and eosin (H&E) staining by the Northwestern University Mouse Histology and Phenotyping Laboratory, and imaged using a Nikon (Tokyo, Japan) Eclipse TE2000-U inverted microscope with a 20x/NA 0.45 objective or a 4x/NA 0.13 objective. ImageJ (National Institutes of Health, Bethesda, MD; http://imagej.nih.gov/ij/index.html) was used to quantify the bone area of sternum section images (4 areas per mouse) taken at 4X magnification for 3 mice per genotype.
Flow Cytometry
Data were acquired on a LSRII flow cytometer and analysed using FACSDiva software (BD Biosciences, San Jose, CA).
Reticulated platelets
Briefly, 1 μl EDTA-anticoagulated whole blood was incubated with 10 μg/ml thiazole orange (Sigma-Aldrich; St. Louis, MO) and allophycocyanin (APC)-conjugated anti-CD41 antibody (eBioscience, San Diego, CA) in 60 μl calcium-and-magnesium-free phosphate-buffered saline (PBS) for 15 min in the dark at room temperature, fixed with 1 ml 1% paraformaldehyde in PBS for 30 min at room temperature, and then acquired. The percentage of reticulated platelets was estimated among CD41+ events within the forward- and side-scatter-gated platelet population.
Surface staining and ploidy
Bone marrow mononuclear cells (MNCs) were first labelled with fluorescein isothiocyanate (FITC)-conjugated rat anti-mouse CD41 (BD). For ploidy analysis, murine Mks labelled with anti-CD41-FITC were fixed for 15 min at room temperature in 0.5% paraformaldehyde in PBS, permeabilized for 1 h at 4°C in 70% methanol, and stained with propidium iodide (PI). Polyploid murine Mks were those events with high forward scatter, positive CD41 expression and ≥16N DNA content.
Platelet activation by flow cytometry
Blood from anesthetized mice was obtained by cardiac puncture and mixed with acid citrate dextrose (ACD; 120 mM sodium citrate, 110 mM glucose, 80 mM citric acid) at a 9:1 ratio. Platelet-rich plasma (PRP) was obtained by centrifugation at 500 x g for 4 min. PRP was gently transferred to a new tube and centrifuged again at 300 x g for 8 min to remove remaining red blood cells. The resulting PRP was transferred to a new tube and platelets were pelleted by centrifugation at 1300 x g for 5 min. Platelets were washed once with Tyrode’s buffer (10 mM HEPES, 137 mM NaCl, 2.8 mM KCl, 1 mM MgCl2, 12 mM NaHCO3, 0.4 mM Na2HPO4, pH 7.4), and diluted with Tyrode’s buffer containing 1 mM CaCl2. Washed platelets were activated with 5 U/ml thrombin (Sigma-Aldrich) and incubated with anti-CD41-FITC (BD Biosciences) and anti-CD62P-PE (eBioscience) antibodies for 15 min at room temperature, fixed with 1% paraformaldehyde, and acquired.
Statistics
Differences between treatments were evaluated using a 2-tailed Student’s t test of equal variance to compare differences between two genotypes or samples. In addition, analysis of variance (ANOVA) was used to evaluate the statistical significance of values between all three genotypes, samples or groups. P values ≤0.05 were considered significant. The boxplot graphically depicts groups of numerical data through their quartiles. The quartiles of a ranked set of data are the three points that divide the data set into four equal groups. The first quartile (Q1) is defined as the middle number between the smallest number and the median of the data set. The second quartile (Q2) is the median of the data. The third quartile (Q3) is the middle value between the median and the highest value of the data set. Box plots have lines extending vertically from the boxes (whiskers) indicating variability outside the upper and lower quartiles with 95% confidence intervals. Outliers are plotted as individual points.
Results
Complete blood cell counts
To characterize CBC counts we obtained a large, unbiased pool of peripheral blood samples from males and females of various ages. Mice 10–24 weeks of age are generally used for haematological studies. Comparing younger and older mice is particularly appealing with respect to graded haematopoietic development. We found no statistical difference in the CBC counts between the designated age groups in the wild type, Hoxa10+/− or Hoxa10 null cohorts and also no difference for a subset of mice age-tracked over time (Fig S1). For comparing CBCs between genotypes, mice of all age groups were pooled together for analysis. We found no significant difference in the total number of white blood cells (WBC), neutrophils, lymphocytes or monocytes between wild type, Hoxa10+/− and Hoxa10 null mice (Fig S2). There was a reduction in red blood cell counts in Hoxa10-deficient mice (both Hoxa10+/− and Hoxa10 null animals; Fig S3A). There was also a reduction in haemoglobin and haematocrit in Hoxa10+/− and Hoxa10 null animals (Fig S3B–C), but there was no effect on mean corpuscular volume (MCV) (Fig S3D).
Importantly, there was a notable difference in the platelet counts for the different genotypes (Fig 1A). The platelet counts for wild type mice ranged between 600–900 × 109/l. Hoxa10+/− animals had a reduced number of platelets with a range from 400–700 × 109/l in comparison to the wild type animals ((a)p=0.0007). Finally, Hoxa10 null animals had 40% lower platelet counts than wild type animals ((b)p=0.0005) and 10% lower platelet counts than Hoxa10 haploinsufficient littermates ((c)p=0.03).
Fig 1.
Platelet counts in wild type, Hoxa10+/− and Hoxa10 null mice. Box plots represent median and quartiles of platelet counts, whiskers represent the 95% confidence interval and black dots represent outliers. (A) Platelet counts between genotype cohorts (n=18). Wild type animals exhibited higher platelet counts than Hoxa10+/− mice (T-test (a)p=0.0007) and Hoxa10 null animals (T-test (b)p=0.0005). Platelet reduction between Hoxa10+/− and Hoxa10 null was also significant (T-test (c)p=0.03). (B) Platelet counts represented by sex (n=9 males and n=9 females). Females tended to exhibit lower platelet counts in all genotypes. Statistical difference was observed between male and female of Hoxa10+/− genotype (T-test (e)p=0.011), males of wild type vs. Hoxa10 null (T-test (d)p=0.017) and females of wild type vs. Hoxa10 null (T-test (f) p<0.001). (C) Total platelet volume of animals in the 3 experimental groups (n=18). There was a significant reduction in total platelet volume when comparing Hoxa10 null samples to wild type ((h)p<0.001) but not Hoxa10+/− ((i)p=0.06), and in Hoxa10+/− samples in comparison to wild type ((g)p=0.002). (D) Percentage of thiazole-positive reticulated platelets. Hoxa10 null animals exhibited a reticulated platelet percentage nearly 50% lower than for wild type ((k)T-test p=0.003) and Hoxa10 +/− ((l)T-test p=0.002) animals, while the difference between wild type and Hoxa10+/− was not significant ((j)T-test p=0.45). Values represent the mean ± SEM for n=9.
The C57Bl/6 strain exhibited lower platelet counts in females than males for all Hoxa10 genotypes (Fig 1B). Differences in platelet numbers between males and females are not uncommon in several mouse strains (Balb/C and CH3) (McDonald, et al 1992). The lower platelet counts were significant between Hoxa10+/− males and females ((e)p=0.01), but not between males and females of the other two genotypes. The most striking observation was the lower platelet counts for Hoxa10 null vs. wild type mice for both males ((d)p=0.01), and females ((f)p=0.00005).
Characteristics of Hoxa10 null platelets
It is possible that reduced platelet counts were due to altered platelet morphology. To eliminate the possibility that Hoxa10 null mice produced fewer but larger platelets, we examined the platelet size. Hoxa10 null mice had a wider distribution of platelet size and a tendency toward larger platelet size in comparison to wild type and haploinsufficient genotypes, but the difference in size was not statistically significant (Fig S4). Furthermore, there was a significant reduction in total platelet volume when comparing Hoxa10 null samples to wild type ((h)p<0.001), but not Hoxa10+/− ((i)p=0.06), and also in Hoxa10+/− samples in comparison to wild type ((g)p=0.002) (Fig 1C). To determine whether the reduced platelet counts stemmed from lower platelet production in vivo, we performed thiazole orange assays to detect reticulated (newly synthesized) platelets in the peripheral blood. Hoxa10 null mice had a lower fraction of reticulated platelets compared to wild type and Hoxa10+/− animals ((k)p=0.003 and (l)p=0.002, respectively), but there was no significant difference between wild type and Hoxa10+/− animals ((j)p=0.45) [Fig 1D]. Given that the Hoxa10 null mice have a reduced number of platelets and also a reduced percentage of reticulated platelets, the absolute number of reticulated platelets in these mice would be reduced by an even greater amount.
Platelet recovery after thrombocytopenia
Platelet depletion studies were performed to verify platelet production deficiency in Hoxa10 null animals. The platelet count was highest for wild type animals and lowest for Hoxa10 null animals (*p=0.005) at the start of the depletion study. By day 1 after intraperitoneal injection of anti-platelet serum, the platelet counts reached their nadir, which was similar for all genotypes. Given the difference in starting platelet counts, this represents a 90% decrease from baseline counts for wild type mice and an 80% decrease for Hoxa10 null mice. The platelet counts increased steadily over five days for all genotypes [Fig 2A]. Platelet recovery was most efficient for wild type animals and least efficient for Hoxa10 null littermates at the end of the time course (#p=0.05) [Fig 2A]. Platelet recovery for each animal in the cohorts is shown in Fig S5.
Fig 2.
Recovery from anti-platelet-serum-induced thrombocytopenia in mice. Time course carried out by sampling mouse blood daily for 7 days. Six mice at least 30 weeks of age were randomly selected for wild type, Hoxa10+/− and Hoxa10 null cohorts. (A) Platelet levels obtained immediately before and daily after thrombocytopenia induction. Wild type mice are represented by black diamonds, Hoxa10+/− animals by grey squares and Hoxa10 null animals by white triangles. Prior to treatment, wild type mice exhibited significantly higher levels of platelets in comparison to Hoxa10+/− and Hoxa10 null animals (*ANOVA p ≤ 0.002, *paired T-test p ≤ 0.005 wild type vs. Hoxa10+/− or Hoxa10 null). Recovery of platelet pool at the end of the time course was most effective in wild type animals (#ANOVA p ≤ 0.02, #paired T-test p ≤ 0.05 wild type vs. Hoxa10+/− or Hoxa10 null). (B) The average slope of platelet recovery was calculated by taking the average platelet counts for days 2–5 and fitting a regression curve. The regression line for each mouse cohort has an R2 ≥0.98. (C) Platelet reticulation spikes at day 1 after induced thrombocytopenia. The percentage of reticulated platelets was significantly different prior to treatment on day 0 (* ANOVA p ≤ 0.001, paired T-test p ≤0.02) for all cohorts. The most significant difference in new platelet production occurred on day 1, with Hoxa10+/− and Hoxa10 null mice exhibiting a significantly lower percentage of reticulated platelets in comparison to wild type littermates (# ANOVA p ≤ 0.016, paired T-test p ≤0.03). Values represent the mean ± SEM (n=6).
The slope of platelet count between day 2 and day 5 was highest for wild type animals and lowest in Hoxa10 null animals [Fig 2B]. Platelet recovery in wild type animals was 43% faster than in Hoxa10+/− animals and 80% faster than in Hoxa10 null animals. Platelet recovery in Hoxa10+/− animals was 25% faster than in Hoxa10 null animals. Reticulated platelet data support this observation. Fig 2C illustrates that Hoxa10 null animals started with the lowest percentage of reticulated platelets on day 0. Following platelet depletion with anti-platelet serum, new platelet formation was significantly higher in wild type animals on day 1 in comparison to Hoxa10+/− and Hoxa10 null animals [Fig 2C]. Together, our results suggest that platelet production is also decreased by HOXA10 deficiency after induction of thrombocytopenia.
Platelet function is normal in Hoxa10 null animals
In light of the slightly increased platelet size and lower reticulation frequency of Hoxa10 null platelets, we examined the clotting capacity of Hoxa10 null platelets. Using a tail vein bleed assay we found that, although Hoxa10 null animals had lower platelet count and more dispersed platelet size, the clotting efficiency in Hoxa10 mice was comparable to that in other littermates, as indicated by the length of time it took for clots to form (Fig 3A). In addition, the volume of blood lost was not significantly different between wild type, Hoxa10+/− and Hoxa10 null animals (Fig 3B). Furthermore, preliminary experiments in which platelets were collected and activated ex vivo, indicated no differences in the extent of platelet activation between the genotypes, as evidenced by CD62P (P-selectin, SELP) expression (Fig S6).
Fig 3.
Tail bleed assay and platelet function. Ten mice of each genotype were subjected to tail injury. (A) Bleeding time length was determined by tracking the amount of time it took for clots to form. The assay was discontinued and bleeding was stopped by pressure if bleeding time was longer than 10 minutes. (B) Blood volume loss was determined by weighing the amount of blood collected during the assay and by factoring in the volume of blood gained in the collection tube and measuring the absorbance at an optical density (OD) of 560 nm.
Reduced Mk progenitor proliferation potential accounts for lower platelet counts
Reticulated platelets are formed from proplatelet extensions of polyploid megakaryocytes. Polyploidization is a cell cycle process specific to Mks involving multiple rounds of DNA replication without cell division. Polyploidization is an essential step in terminal Mk differentiation that precedes proplatelet formation and increases the number of platelets released from each Mk, and to this end we examined the fraction of CD41+ cells within bone marrow mononuclear cells and the steady-state polyploidization of megakaryocytes freshly isolated from bone marrow via flow cytometry. The Hoxa10 null mice had an ~20% lower fraction of CD41+ cells than wild type mice (p=0.01; Fig. 4A), which is consistent with the lower platelet counts. Megakaryocytes from Hoxa10 null mice had a similar ploidy distribution as Mks from their wild type counterparts and also a similar percentage of Mks with high ploidy content (>16N) [Fig 4B].
Fig 4.
Reduced platelet counts in Hoxa10 null mice may be attributed to lower Mk progenitor proliferation potential. (A) CD41 expression among mononuclear cells freshly harvested from bone marrow for in vitro cultures. Data represents mean ± SEM for n=3. (B) Representative ploidy distributions of bone marrow megakaryocytes for each genotype. The bar graph represents the fraction of high-ploidy bone marrow Mks harvested from mice in the 3 experimental groups. Data represents mean ± SEM for n=6. (C) Total number of colonies in CFU-Mk assay. Data represents mean ± SEM for n=3. Small (3 Mks), medium (4–8 Mks) and large (≥9 Mks) colonies were enumerated and combined. (D) Percentage of medium and large colonies with respect to total colony counts. Larger colonies represent more primitive Mk progenitors and better expansion potential. Statistically significant reduction in the percentage of medium plus large colonies formed was observed for Hoxa10 null lineage-depleted cells (*T-test p=0.02). Data represent mean ± SEM for n=2.
To determine whether there are defects in the development of Mks, we performed colony formation assays. We obtained bone marrow cells from wild type, Hoxa10+/− and Hoxa10 null mice, depleted lineage-positive cells, seeded 5,000 lineage-negative progenitors into MegaCult medium, and cultured them for two weeks in the presence of 100 ng/ml THPO. Similar numbers of total colonies were obtained for wild type, Hoxa10+/− and Hoxa10 null cells [Fig 4C]. However, strikingly, the percentage of medium plus large colonies was reduced by half for Hoxa10 null cells (*p=0.02) and also tended to be lower for Hoxa10+/− cells (p=0.07) [Fig 4D].
Hoxa10 null mice have altered bone marrow structure
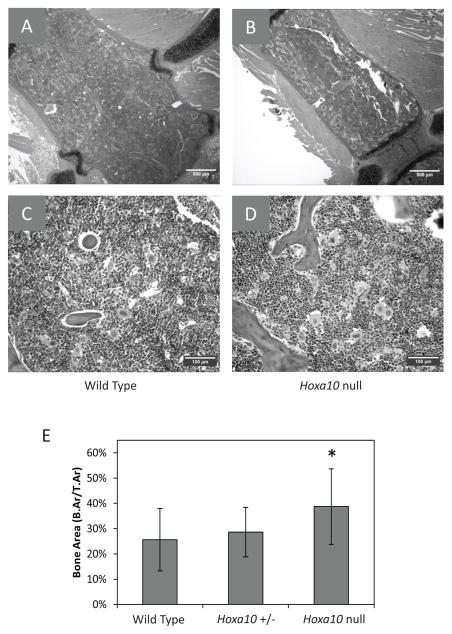
Histological screening indicated an altered structure of the Hoxa10 null sternum sections at steady state. Hoxa10 null sternums exhibited a tendency for increased cortical and trabecular bone as compared to those of wild type mice [Fig 5A–D]. There is substantial variability in the amount of bone area throughout each section and between animals; therefore to support our visual observation, we utilized an image analysis program, ImageJ, to quantify the bone area per tissue area (B.Ar/T.Ar) in the two-dimensional images of the sections. This analysis showed a significant increase in bone area for the sternums of the Hoxa10 null mice compared with that of wild type mice (p=0.03) [Fig 5E]. The null mice also had increased bone area compared to Hoxa10+/− sections, though this increase had lower statistical confidence (p=0.06), and bone areas in the wild type and Hoxa10+/− sections were not statistically different (p=0.52). The intervening haematopoietic marrow in these sternum sections appeared grossly normal [Fig 5C–D] and contained similar numbers of large Mks (data not shown).
Fig 5.
Hoxa10 null mice exhibit increased cortical and trabecular bone area in sternum sections. (A-D) Haematoxylin and eosin stained sternum sections. (A and C) Representative images of wild type mouse sterna at 4x and 20x, respectively. (B and D) Representative images of Hoxa10 null mouse sterna at 4x and 20x, respectively. Images were taken at 4x magnification for 3 mice per genotype with 4 viewing areas per mouse. ImageJ was used to perform area measurements of the bone area (B.Ar) and the tissue area (T.Ar, which includes the bone area) of these two-dimensional images. Scale bar = 500 μm in (A, B) and 100 μm in (C, D). (E) Hoxa10 null sternum sections have increased B.Ar/T.Ar compared to wild type (T-test p=0.03). Values represent mean ± SD.
Discussion
Complete blood cell counts revealed a significant and substantial difference in steady-state platelet counts between wild type, Hoxa10+/− and Hoxa10 null mice. We also showed that the lower platelet counts in Hoxa10 null mice may be attributed to decreased platelet production, as evidenced by a lower fraction of CD41+ cells in the marrow, lower percentages of reticulated platelets, slower platelet recovery after induced thrombocytopenia in vivo, and lower proliferative potential of Mk progenitors in colony formation assays. Our observation that Hoxa10 loss decreased Mk progenitor expansion is consistent with previous studies documenting increased Mk progenitor expansion with Hoxa10 overexpression (Magnusson, et al 2007, Thorsteinsdottir, et al 1997).
The ITGB3 subunit is part of a key fibrinogen receptor ITGA2B/ITGB3 (integrin αIIbβ3) expressed on Mks and platelets, and is essential for the platelet-clotting cascade to occur in vivo. The importance of ITGB3 in thrombopoiesis is highlighted by the haematological malignancy Glanzmann thrombasthenia, which is characterized by a mutation in either ITGA2B or ITGB3 and results in significantly impaired platelet function. Bei, et al (2007) previously reported that HOXA10 activates ITGB3, the gene encoding ITGB3 in myeloid cells and others have reported that ITGB3 gene expression is directly regulated by HOXA10 in endometrial cells (Daftary, et al 2002). In light of these reports we hypothesized that HOXA10 may influence platelet function. However, we found that Hoxa10 null mice, despite their dramatically reduced platelet numbers, had similar clotting efficiency as wild type mice after tail injury. The ITGB3 subunit (CD61) is present in two distinct integrin heterodimers, ITGAV/ITGB3 (CD51/CD61) and ITGA2B/ITGB3 (CD41/CD61). ITGAV/ITGB3 is found on several cell types, while ITGA2B/ITGB3 is found exclusively on Mks and platelets. Given that our model system involves systemic Hoxa10 loss, there may be phenotypic abnormalities in cell types beyond our immediate interest of Mks and platelets. Intriguingly, Itgb3 knockout mice display altered bone marrow pathology that is reminiscent of our observations in the Hoxa10 null mice; histological sections of femurs from Itgb3 null mice display a striking visual increase in cortical and trabecular bone mass (McHugh, et al 2000). It is also possible that Hoxa10 null mice could have aberrant development in vasculature and other organs that constrain normal physiology of the bone marrow compartments (Stokes and Granger 2012). Developmental defects are suggested by hampered reproduction, smaller Hoxa10+/− litter sizes in comparison to wild type litters, reduced survival of neonates and smaller size of adult mice (Satokata, et al 1995, Satokata and Uchiyama 1995, Vitiello, et al 2008). As the bone marrow microenvironment may be globally altered by Hoxa10 loss, it may be worthwhile to examine Hoxa10 loss in conditional knockout mice wherein the gene knockout can be restricted to either Mk/platelets or osteoclasts.
Given that moderately increased expression of Hoxa10 has been clearly demonstrated to enhance LSK cell proliferation and reconstitution potential (Magnusson, et al 2007), it would be reasonable to extrapolate the possibility that loss of Hoxa10 impairs HSPC proliferation. However, Hoxa10 null mice have been reported to have rather normal haematopopiesis and Hoxa10 deficient HSPCs have been shown to be competitive in repopulating assays (Lawrence, et al 2005). In agreement with this, we also observed normal blood cell counts across many lineages with the exception of platelet numbers and, to a lesser extent, RBCs. Since HOX family proteins share similar binding elements, it is possible that other HOX proteins are able to compensate for the role of HOXA10 in HSPC proliferation in the knockout mice. The reduced platelet counts and reduced Mk progenitor expansion ex vivo imply that HOXA10 may exert a unique function highly specific for Mk progenitors. There is evidence that Gata1 is a downstream target of HOXA10 (Magnusson, et al 2007), and the significance of GATA1 as a key regulator of megakaryopoiesis and erythropoiesis has been well documented (Colin, et al 1990, Orkin, et al 1998, Shivdasani, et al 1997).
The NAD+-dependent deacetylase enzyme, sirtuin 2 (SIRT2), has been documented to interact with HOXA10 (Bae, et al 2004). We previously showed that inhibitors of sirtuin enzymes enhance the polyploidization and proplatelet formation of megakaryocytes in vitro (Giammona, et al 2006, Giammona, et al 2009). Whether or not SIRT2 and HOXA10 have an important relationship in megakaryopoiesis remains to be investigated. Further studies evaluating the interactions of HOXA10 with ITGB3, GATA1, or SIRT2 may elucidate mechanisms regulating megakaryopoiesis. HOXA10 regulation of platelet-specific genes such as Itga2b/Itgb3 may also shed light on platelet-specific function.
In summary, we provide comprehensive evidence that loss of Hoxa10 decreases platelet production and steady-state platelet counts, but does not appear to affect platelet function. Our data also supports previous work outlining the importance of HOXA10 as a regulator of haematopoiesis.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. John Crispino for equipment use; Dr. Alaina Schlinker for expertise in flow cytometry setup of reticulated platelet and activated platelet assays; Dr. Pani Apostolidis and Dr. E. Terry Papoutsakis for advice and guidance on the tail bleed assay; Dylan Fogel for assisting with some of the animal experiments; and Ryan LeGraw for ImageJ analysis of sternum sections. We would also like to acknowledge the Northwestern University Mouse Histology Core and Phenotyping laboratory for histology staining of mouse specimens and the RLCCC Flow Cytometry Core Facility. This work was supported by grants from the National Institutes of Health (HL 093083) and the National Science Foundation (CBET 0853603). TAD was supported in part by NIH/NCI training grant T32CA09560.
Footnotes
Author Contributions
WMM was the principal investigator and takes primary responsibility for the paper. WMM directed the research. Co-investigator EAE provided guidance on the study design and data interpretation and provided the knockout mice. IMK prepared the animal protocols. IMK and TAD designed and performed the experiments and statistical analysis. IMK, TAD and WMM wrote the paper with input from EAE. The authors report no potential conflicts of interest.
Disclosure of potential conflict of interest
The authors declare no competing nancial interests.
References
- Apostolidis PA, Woulfe DS, Chavez M, Miller WM, Papoutsakis ET. Role of tumor suppressor p53 in megakaryopoiesis and platelet function. Experimental Hematology. 2012;40:131–142. e134. doi: 10.1016/j.exphem.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae NS, Swanson MJ, Vassilev A, Howard BH. Human histone deacetylase SIRT2 interacts with the homeobox transcription factor HOXA10. Journal of Biochemistry. 2004;135:695–700. doi: 10.1093/jb/mvh084. [DOI] [PubMed] [Google Scholar]
- Bei L, Lu Y, Bellis SL, Zhou W, Horvath E, Eklund EA. Identification of a HoxA10 activation domain necessary for transcription of the gene encoding beta3 integrin during myeloid differentiation. Journal of Biological Chemistry. 2007;282:16846–16859. doi: 10.1074/jbc.M609744200. [DOI] [PubMed] [Google Scholar]
- Colin Y, Joulin V, Le Van Kim C, Romeo PH, Cartron JP. Characterization of a new erythroid/megakaryocyte-specific nuclear factor that binds the promoter of the housekeeping human glycophorin C gene. Journal of Biological Chemistry. 1990;265:16729–16732. [PubMed] [Google Scholar]
- Daftary GS, Troy PJ, Bagot CN, Young SL, Taylor HS. Direct regulation of beta3-integrin subunit gene expression by HOXA10 in endometrial cells. Molecular Endocrinology. 2002;16:571–579. doi: 10.1210/mend.16.3.0792. [DOI] [PubMed] [Google Scholar]
- Eklund EA, Jalava A, Kakar R. Tyrosine phosphorylation of HoxA10 decreases DNA binding and transcriptional repression during interferon gamma-induced differentiation of myeloid leukemia cell lines. Journal of Biological Chemistry. 2000;275:20117–20126. doi: 10.1074/jbc.M907915199. [DOI] [PubMed] [Google Scholar]
- Eklund EA, Goldenberg I, Lu Y, Andrejic J, Kakar R. SHP1 protein-tyrosine phosphatase regulates HoxA10 DNA binding and transcriptional repression activity in undifferentiated myeloid cells. Journal of Biological Chemistry. 2002;277:36878–36888. doi: 10.1074/jbc.M203917200. [DOI] [PubMed] [Google Scholar]
- Giammona LM, Fuhrken PG, Papoutsakis ET, Miller WM. Nicotinamide (vitamin B3) increases the polyploidisation and proplatelet formation of cultured primary human megakaryocytes. British Journal of Haematology. 2006;135:554–566. doi: 10.1111/j.1365-2141.2006.06341.x. [DOI] [PubMed] [Google Scholar]
- Giammona LM, Panuganti S, Kemper JM, Apostolidis PA, Lindsey S, Papoutsakis ET, Miller WM. Mechanistic studies on the effects of nicotinamide on megakaryocytic polyploidization and the roles of NAD+ levels and SIRT inhibition. Experimental Hematology. 2009;37:1340–1352. e1343. doi: 10.1016/j.exphem.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirouskova M, Shet AS, Johnson GJ. A guide to murine platelet structure, function, assays, and genetic alterations. Journal of Thrombosis and Haemostasis. 2007;5:661–669. doi: 10.1111/j.1538-7836.2007.02407.x. [DOI] [PubMed] [Google Scholar]
- Lawrence HJ, Christensen J, Fong S, Hu YL, Weissman I, Sauvageau G, Humphries RK, Largman C. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood. 2005;106:3988–3994. doi: 10.1182/blood-2005-05-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey S, Zhu C, Lu YF, Eklund EA. HoxA10 represses transcription of the gene encoding p67phox in phagocytic cells. Journal of Immunology. 2005;175:5269–5279. doi: 10.4049/jimmunol.175.8.5269. [DOI] [PubMed] [Google Scholar]
- Lu Y, Goldenberg I, Bei L, Andrejic J, Eklund EA. HoxA10 represses gene transcription in undifferentiated myeloid cells by interaction with histone deacetylase 2. Journal of Biological Chemistry. 2003;278:47792–47802. doi: 10.1074/jbc.M305885200. [DOI] [PubMed] [Google Scholar]
- Magnusson M, Brun AC, Miyake N, Larsson J, Ehinger M, Bjornsson JM, Wutz A, Sigvardsson M, Karlsson S. HOXA10 is a critical regulator for hematopoietic stem cells and erythroid/megakaryocyte development. Blood. 2007;109:3687–3696. doi: 10.1182/blood-2006-10-054676. [DOI] [PubMed] [Google Scholar]
- McDonald TP, Swearingen CJ, Cottrell MB, Clift RE, Bryant SE, Jackson CW. Sex- and strain-related differences in megakaryocytopoiesis and platelet production in C3H and BALB/c mice. Journal of Laboratory and Clinical Medicine. 1992;120:168–173. [PubMed] [Google Scholar]
- McHugh KP, Hodivala-Dilke K, Zheng MH, Namba N, Lam J, Novack D, Feng X, Ross FP, Hynes RO, Teitelbaum SL. Mice lacking beta3 integrins are osteosclerotic because of dysfunctional osteoclasts. Journal of Clinical Investigation. 2000;105:433–440. doi: 10.1172/JCI8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Shivdasani RA, Fujiwara Y, McDevitt MA. Transcription factor GATA-1 in megakaryocyte development. Stem Cells. 1998;16(Suppl 2):79–83. doi: 10.1002/stem.5530160710. [DOI] [PubMed] [Google Scholar]
- Palumbo JS, Zogg M, Talmage KE, Degen JL, Weiler H, Isermann BH. Role of fibrinogen- and platelet-mediated hemostasis in mouse embryogenesis and reproduction. Journal of Thrombosis Haemostasis. 2004;2:1368–1379. doi: 10.1111/j.1538-7836.2004.00788.x. [DOI] [PubMed] [Google Scholar]
- Satokata I, Benson G, Maas R. Sexually dimorphic sterility phenotypes in Hoxa10-deficient mice. Nature. 1995;374:460–463. doi: 10.1038/374460a0. [DOI] [PubMed] [Google Scholar]
- Satokata I, Uchiyama M. Mice carrying null mutations of the homeotic genes Hoxa10 and Msx1. Tanpakushitsu Kakusan Koso. 1995;40:2134–2143. [PubMed] [Google Scholar]
- Sauvageau G, Lansdorp PM, Eaves CJ, Hogge DE, Dragowska WH, Reid DS, Largman C, Lawrence HJ, Humphries RK. Differential expression of homeobox genes in functionally distinct CD34+ subpopulations of human bone marrow cells. Proceedings of the National Academy of Sciences USA. 1994;91:12223–12227. doi: 10.1073/pnas.91.25.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah CA, Wang H, Bei L, Platanias LC, Eklund EA. HoxA10 regulates transcription of the gene encoding transforming growth factor beta2 (TGFbeta2) in myeloid cells. Journal of Biological Chemistry. 2011;286:3161–3176. doi: 10.1074/jbc.M110.183251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah CA, Bei L, Wang H, Platanias LC, Eklund EA. HoxA10 protein regulates transcription of gene encoding fibroblast growth factor 2 (FGF2) in myeloid cells. Journal of Biological Chemistry. 2012;287:18230–18248. doi: 10.1074/jbc.M111.328401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivdasani RA, Fujiwara Y, McDevitt MA, Orkin SH. A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet development. EMBO Journal. 1997;16:3965–3973. doi: 10.1093/emboj/16.13.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes KY, Granger DN. Platelets: a critical link between inflammation and microvascular dysfunction. Journal of Physiology. 2012;590:1023–1034. doi: 10.1113/jphysiol.2011.225417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsteinsdottir U, Sauvageau G, Hough MR, Dragowska W, Lansdorp PM, Lawrence HJ, Largman C, Humphries RK. Overexpression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Molecular Cell Biology. 1997;17:495–505. doi: 10.1128/mcb.17.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitiello D, Pinard R, Taylor HS. Gene expression profiling reveals putative HOXA10 downstream targets in the periimplantation mouse uterus. Reproductive Sciences. 2008;15:529–535. doi: 10.1177/1933719108316911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lu Y, Huang W, Papoutsakis ET, Fuhrken P, Eklund EA. HoxA10 activates transcription of the gene encoding mitogen-activated protein kinase phosphatase 2 (Mkp2) in myeloid cells. Journal of Biological Chemistry. 2007;282:16164–16176. doi: 10.1074/jbc.M610556200. [DOI] [PubMed] [Google Scholar]
- Wang H, Lindsey S, Konieczna I, Bei L, Horvath E, Huang W, Saberwal G, Eklund EA. Constitutively active SHP2 cooperates with HoxA10 overexpression to induce acute myeloid leukemia. Journal of Biological Chemistry. 2009;284:2549–2567. doi: 10.1074/jbc.M804704200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.