Abstract
Participating in the repair of nuclear DNA is one mechanism by which p53 suppresses tumorigenesis, but there is growing evidence that p53 also helps maintain the mitochondrial genome through its translocation into mitochondria and interactions with mtDNA repair proteins. Because of the susceptibility of mtDNA to oxidative damage and replication errors, it is vital to protect mtDNA genomic stability to preserve health and fitness. Here, we focus on reviewing the evidence for the involvement of p53 in maintaining the integrity of mtDNA through its activities in both the nucleus and the mitochondria.
Keywords: p53, mitochondria, translocation, mtDNA, DNA repair, reactive oxygen species
1. Introduction
The p53 protein, encoded by the TP53 gene in humans, is commonly referred to as the “guardian of the genome” due to its activities directed at maintaining genomic stability through the repair of damaged DNA and the integration of cell birth/death signaling pathways with DNA damage checkpoints [1–3]. The best known function of p53 is as a transcription factor that binds to specific p53 responsive elements in the promoter of target genes, but its interaction with the genome is complex and includes enhancer-like functions. The modulation of target gene expression by p53 depends on various factors including its post-translational modification and interaction with nuclear co-factors, indicating that its activities are highly context dependent. Under physiological or various cellular stress conditions, p53 can regulate diverse cellular processes including DNA repair, cell cycle, apoptosis, redox homeostasis and metabolism [4–7].
The array of biological processes regulated by p53 also includes mitochondrial function [8]. The mitochondrion is a conduit of many catabolic and anabolic pathways essential for cell function, maintenance and proliferation. Besides contributing to metabolism, the mitochondrion is engaged in other important roles such as signal transduction through reactive oxygen species (ROS) generation [9]. Thus, despite the original Warburg hypothesis that cancer cells have dysfunctional mitochondria, it is now generally accepted that cancer cells are as equally reliant on mitochondrial metabolism as they are on aerobic glycolysis [10,11].
The initial report of an association between p53 and respiration was followed by the identification of Synthesis of Cytochrome C Oxidase 2 (SCO2) as a p53 transcriptional target gene [12,13]. Cells deficient in SCO2 cannot respire due to defective cytochrome c oxidase assembly, and notably, they display high ROS levels and oxidative DNA damage [14]. This observation demonstrated that the controlled transfer of high energy reducing equivalents (NADH) generated by glycolysis to molecular oxygen during respiration can serve to decrease total cellular ROS levels. Thus, the promotion of respiration by p53 is consistent with previous work demonstrating that p53 can have potent antioxidant activities to protect the genome [15,16]. This antioxidant effect further supports the symbiotic theory of the mitochondrion that posited protection from oxygen toxicity (ROS) as a major evolutionary driving force for the incorporation of primordial mitochondrion into eukaryotes [5,17]. Thus, because mtDNA is necessary for mitochondrial function, there is reason to predict that p53 also participates in maintaining its integrity in addition to the other mechanisms by which it regulates the mitochondria [8,18]. In this review, we summarize what is known about the role of p53 as guardian of the mitochondrial genome but the details of mtDNA repair mechanisms can be found in other focused reviews [19–21].
2. The mitochondrial genome and its quality control
2.1. mtDNA
Reflecting its symbiotic origin, most of the proto-mitochondrion bacterial genome was transferred to the eukaryotic chromosomes so that it now encodes only a small subset of proteins and RNAs necessary for mitochondrial biogenesis [22]. The vast majority of the mitochondrial proteome (~1500 proteins) are nuclear-encoded and require translocation into the mitochondrion through specific mechanisms that will be briefly reviewed as pertinent to the import of p53 [23]. The circular 16,569 bp human mtDNA contains the genes for 13 proteins involved in oxidative phosphorylation (OxPhos): 7 complex I (NADH dehydrogenase) subunits MTND1, 2, 3, 4L, 4, 5, 6; 1 complex III subunit MTCYB; 3 complex IV subunits MTCO1, 2, 3; and 2 complex V subunits MTATP6, 8. In addition to these structural genes of the respiratory complexes, it contains 2 rRNAs and 22 tRNAs needed for intra-mitochondrial protein synthesis. These genes have neither introns nor noncoding sequences, and they are polycistronically transcribed from a promoter located in the control region of the two strands of the mtDNA (G-rich heavy (H) and C-rich light (L) strands). The resulting transcripts are then cleaved and polyadenylated.
Unlike the diploid genome in the nucleus which undergoes Mendelian segregation during mitosis, the mitochondrial genome is maternally inherited and therefore can be considered to be clonal in nature [22]. The cell has multiple mitochondria, each containing multiple mtDNA copies, which undergo replicative segregation into two daughter cells. Another phenomenon unique to the mitochondrial genome is the concept of heteroplasmy which arises due to the multiplicity of mtDNA copy number in each cell that can in turn give rise to a mixture of variant mtDNAs [24]. The composition of variant mtDNAs can change over time and can differ by cell/tissue type within an individual or between individuals from the same mother, adding another dimension to mtDNA homeostasis.
The details of how two spatially separated genomes in the nucleus and the mitochondrion orchestrate their gene expression, replication and maintenance remain poorly understood, but it is likely that the mechanisms have been finely tuned by evolution. The high mutation rate of mtDNA (~10-fold higher than nuclear DNA) and the resulting variant haplotypes may have exerted a profound influence on human radiation across the world [25]. mtDNA variants found in different populations (haplogroups) that affect the coupling efficiency of OxPhos and ROS generation have been postulated to have had determinative effects on human migration and survival under the bioenergetic, climatic, and immunologic demands of new environments. The translation of mitochondrial genetics will require further insights such as understanding the consequences of mismatched mitochondrial-nuclear DNA, a crucial component in preventing mtDNA diseases by pronucleus transfer into an enucleated normal mtDNA-containing zygote of a “third parent” [26]. Nonetheless, it is an exciting era for biomedical research as genes such as p53 and ATM, largely associated with the nuclear genome, expand their function into the mitochondria [27].
2.2. mtDNA repair mechanisms
A growing body of work indicates that p53 plays an important role in mtDNA homeostasis with at least three different research groups reporting decreased mtDNA content in various cells and mouse tissues deficient in p53 [28–30]. p53 is involved in almost all nuclear DNA repair pathways including nucleotide-excision repair (NER), base-excision repair (BER), mismatch repair (MMR), non-homologous end-joining (NHEJ) and homologous recombination (HR) through its transcriptional or non-transcriptional activities [3,31]. With the exception of NER, components of these nuclear DNA repair pathways are also shared in mtDNA maintenance, and one of the best studied and major repair mechanism in the mitochondria is BER [19,20]. Oxidative DNA damage is largely repaired by the BER pathway, so it is fitting that this pathway predominates in an organelle thought to have evolved to protect against oxygen toxicity [5,19].
Generally, the quality control mechanisms of the mitochondria and its genome can be organized into organellar (fusion-fission, autophagy/mitophagy) and mtDNA (DNA replication/transcription, DNA repair) maintenance programs [32]. The role of p53 in regulating the homeostasis of mtDNA can also be divided into pathways that involve: 1) p53 transcriptional target genes; and 2) the non-nuclear effects of p53 protein in the cytosol and mitochondria. Some of these pathways are overlapping, but they serve to structure this review about the role of p53 in maintaining mtDNA integrity.
3. p53 transcriptional target genes involved in mtDNA maintenance
An early clue of its involvement in mtDNA maintenance came from the report that p53 regulates the transcription of Ribonucleotide Reductase M2 B gene (RRM2B, also known as p53R2), which when mutated was found to cause a severe mtDNA depletion syndrome in the skeletal muscle of humans [33,34]. Accordingly, p53R2−/− mice have decreased mtDNA content in kidney, muscle and liver, albeit with different levels of depletion for unclear reasons. Furthermore, reduction in p53R2 expression is associated with mtDNA depletion and decreased mitochondrial mass in p53 deficient primary human and mouse cell lines [28]. Thus, although p53R2 was initially observed to translocate to the nucleus upon DNA damage and contribute to maintaining deoxyribonucleotide supplies for nuclear DNA repair, depletion of mtDNA in the absence of p53R2 indicate that it also plays an important role in mtDNA maintenance [33,34].
Transcription Factor A, Mitochondrial (TFAM), a gene necessary for both transcription and replication of the mitochondrial genome, has been reported to be another transcriptional target of p53 in mouse models [30,35]. TFAM mRNA and protein levels are reduced in the skeletal muscle of p53−/− mice which may contribute to their lower aerobic exercise capacity [30]. In contrast to the p53 null state, higher aerobic exercise capacity and increased TFAM expression were observed in the p53 R172H mutation knockin mouse model of Li-Fraumeni syndrome (LFS), an autosomal dominant premature cancer condition due to various germline mutations of the TP53 gene [36,37]. Furthermore, in this specific mouse model, exercise endurance, TFAM mRNA, and TFAM protein were increased in a mutant allele dose-dependent manner [37]. Cells derived from patients with two other LFS mutations also showed increased TFAM expression compared to their family members who did not carry the mutations [37]. However, the generalizability of this finding to other LFS patients or LFS mouse models remains to be determined. These observations demonstrate that the mitochondrial and cell cycle regulatory activities of p53 can be dissociated in the setting of LFS mutations, but not all p53 mutations are likely to be equivalent in their mitochondrial activity. Analysis of the TFAM gene sequence in mice has revealed putative p53 responsive elements, one of which was shown to transactivate a luciferase reporter and to interact with p53 protein by chromatin immunoprecipitation assay [30]. It should be noted here that p53 has also been shown to interact with mitochondrial matrix-localized TFAM protein as will be discussed later in this review [38]. Therefore, the regulation of this critical mtDNA factor by p53 appears to span both the nuclear and mitochondrial subcellular compartments.
4. Non-nuclear roles of p53 in mtDNA maintenance
4.1. Role in mitophagy
Rather than repairing damaged mtDNA, the cell can eliminate the entire organelle by mitophagy, a specific autophagic process that engulfs and creates a double-membrane autophagosome around the mitochondrion which then fuses with the lysosome for proteolysis [39]. Autophagy represents a cellular process which is known to involve both the transcriptional and non-nuclear activities of p53 and thus is a fitting process to start describing its activities outside the nucleus. First, in the nucleus, p53 has been reported to promote autophagy by transactivating genes encoding: DRAM1, which is induced by DNA damage and localized to lysosomes with autophagosome accumulation; and sestrin 1 and 2, AMPK β1 and β2, and TSC2, which act on the AMPK pathway to inhibit the autophagy suppressor mTOR, thus activating autophagy [40,41]. A recent study has more comprehensively identified an array of autophagy-related genes as targets of p53 transactivation [42]. In contrast to p53 promotion of autophagy through its transcriptional activity, wild-type and mutant p53 in the cytoplasm have both been shown to inhibit autophagy in a series of studies using different model systems [40]. Generally, the non-transcriptional effects of p53 on autophagy are inhibitory [41]. They occur under basal conditions where p53 is not activated and are independent of its wild-type DNA binding domain. As an elegant demonstration of the potential role of mitophagy in maintaining mtDNA integrity, the expression of mitophagy mediator E3 ubiquitin ligase parkin which can interact with p53 in the cytoplasm, selectively eliminated a deleterious mtDNA mutation through autophagy, resulting in the enrichment of functional wild-type mtDNA mitochondria (Table 2) [43–45]. The positive and negative effects of p53 on mitophagy/autophagy and how this might impact mtDNA maintenance under different cellular conditions remains to be further clarified.
Table 2.
Mitochondrial role of p53 in mtDNA maintenance
| p53 binding partner | Process/function | References |
|---|---|---|
| CHCHD4 | mtDNA repair; translocates p53 into the mitochondrial intermembrane space via the disulfide relay system | {Zhuang, 2013 #1976} |
| OGG1/APE1 | BER; p53 senses DNA damage and stimulates OGG1/APE1 to remove 8-oxoG through glycosylase and endonuclease activities | {Achanta, 2004 #2514} |
| Parkin | Mitophagy; p53 inhibits mitophagy through interaction with parkin in cytosol and also inhibits its gene PARK2 expression | {Zhang, 2011 #2209;Hoshino, 2013 #2073;Hoshino, 2014 #2551} |
| POLG | mtDNA maintenance; p53 enhances mtDNA replication and BER | {de Souza-Pinto, 2004 #1543;Achanta, 2005 #719;Chen, 2006 #698;Bakhanashvili, 2008 #725} |
| RECQL4 | BER; p53-RECQL4 complex increases POLG activity | {Gupta, 2014 #2088} |
| SSBP1 | Exonuclease activity; p53 interacts with SSBP1 through TAD and enhances its exonuclease activity | {Wong, 2009 #2071} |
| TFAM | BER; p53 increases mtDNA damage recognition and repair by TFAM | {Yoshida, 2003 #697;Wong, 2009 #1353} |
APE1, apurinic endonuclease 1; BER, base excision repair; CHCHD4, coiled-coil-helix-coiled-coil-helix domain containing 4; mtDNA, mitochondrial DNA; OGG1, 8-oxoguanine DNA glycosylase; POLG, DNA polymerase γ; RECQL4, RecQ helicase-like protein 4; SSBP1, single stranded DNA-binding protein 1; TAD, transactivation domain; TFAM, mitochondrial transcription factor A
4.2. Translocation of p53 to the mitochondria
Much of the work on the mitochondrial pool of p53 has been done in cells undergoing some form of cell death which are listed for reference (Table 1) [41]. p53 was first reported to translocate to the mitochondria and to cause mitochondrial membrane depolarization and caspase activation during apoptosis independent of its transcriptional activity [46,47]. Subsequent studies have shown that p53-transactivated PUMA expression displaces cytoplasmic p53 bound to anti-apoptotic protein BCLXL and allows it to activate BAX during apoptosis [41,48,49]. For the purposes of this review, we will mainly focus on what is known about the import of p53 protein into mitochondria under non-cell death conditions where it would be expected to play a role in mtDNA homeostasis.
Table 1.
Mitochondrial role of p53 in cell death
| p53 binding partner | Process/function | References |
|---|---|---|
| BCL2 family proteins (BCL2, BCLXL, BAX, BAK, BAD, BID) | Apoptosis; p53 promotes mitochondrial outer membrane permeabilization (MOMP) through interactions with BCL2 family proteins | {Mihara, 2003 #2068;Leu, 2004 #2193;Jiang, 2006 #2194;Moll, 2005 #2195} |
| Cyclophilin D | Necrosis; p53 promotes MOMP by binding with cyclophilin D in mitochondrial matrix | {Vaseva, 2012 #1549} |
| HAUSP | Apoptosis; deubiquitylation of p53 on the mitochondrial outer membrane results in p53-BCLXL, p53-BCL2 and p53-BAK complexes | {Marchenko, 2007 #748} |
| MDM2 | Apoptosis; mono-ubiquitylation of p53 by MDM2 increases protein stability and mitochondrial translocation | {Marchenko, 2007 #748} |
| MnSOD | Apoptosis; p53 inhibits MnSOD enzymatic activity and increases ROS level | {Zhao, 2005 #1544} |
| mtHsp70 (mortalin-2) | Apoptosis; cytosolic p53 retention (or mitochondrial trafficking) by mtHsp70 increases MOMP | {Iosefson, 2010 #2569;Marchenko, 2000 #1546} |
| Tid-1 (mtHsp40) | Apoptosis; p53 forms mtHsp70/Tid-1/p53 ternary complex on mitochondrial outer membrane | {Ahn, 2010 #1574;Trinh, 2010 #2341} |
BCL2, B-cell lymphoma 2; HAUSP, herpes virus-associated ubiquitin-specific protease; mtHsp40, mitochondrial heat-shock protein 40kDa; mtHsp70, mitochondrial heat-shock protein 70kDa; MDM2, mouse double minute 2 homolog; MnSOD, manganese superoxide dismutase; ROS, reactive oxygen species; Tid-1, tumorous imaginal discs 1
Because mtDNA is located on the matrix side of the inner mitochondrial membrane, p53 may regulate mtDNA repair only after translocating to the proximity of the mitochondrial matrix. Significant advances have been made in understanding the molecular basis of protein import into the mitochondria. In yeast, five different pathways have been identified that form a cooperative network to direct the import of proteins utilizing: 1) presequence targeting to the inner membrane and matrix; 2) carrier translocase of the inner membrane 22 (TIM22) to the inner membrane; 3) redox carrier/respiration mediated import into the intermembrane space (IMS); 4) β-barrel insertion into the outer membrane; and 5) α-helical insertion into the outer membrane [50,51]. The two classical import systems utilize mitochondrial targeting sequences (MTS) or carriers (cytosolic chaperones) to translocate cytosolic proteins to the inner membrane or matrix while the mechanism that involves redox/respiration dependent import into the intermembrane space has recently been shown to translocate p53 [52].
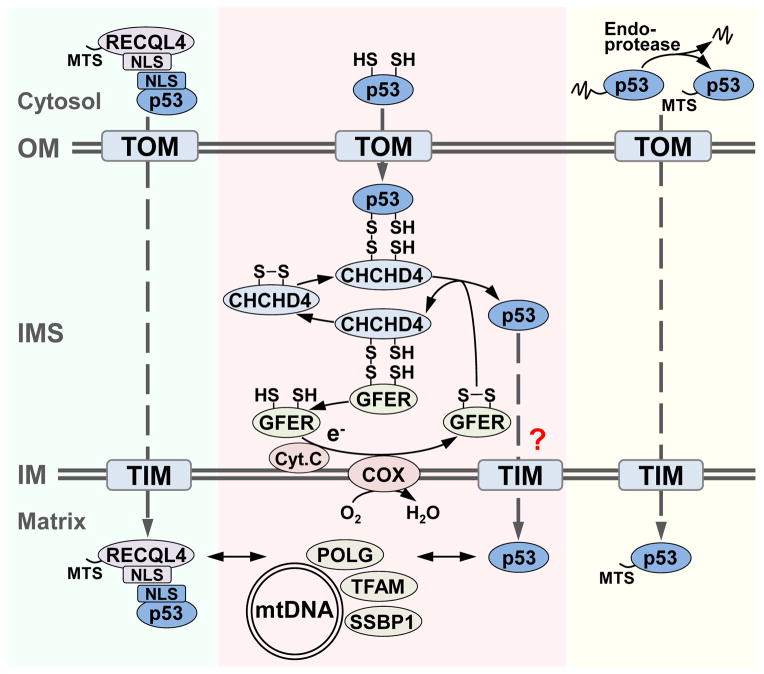
To date, at least three different mechanisms have been described by which p53 translocates into mitochondria under a non-cell death context. p53 has been shown to interact with RecQ helicase-like protein 4 (RECQL4), a nuclear DNA helicase that has an N-terminal MTS and binds to the translocase of the outer membrane 20 (TOM20) (Figure 1, left) [53]. The interaction between RECQL4 and p53 has been mapped to amino acid positions 270-400 of RECQL4 and 293-362 of p53, which contain the nuclear localization signals (NLS) for the respective proteins. The formation of this complex has been proposed to mask the NLS1 motif of p53 thereby allowing its translocation into the mitochondria [53]. Another mechanism of p53 mitochondrial localization is through the unveiling of a cryptic MTS through the action of a cytosolic protease that recognizes conserved serine endoprotease consensus sites present in mouse, rat and human p53 (Figure 1, right) [54]. The proteolytic cleavage of p53 results in a ~40 kDa fragment that localizes to the mitochondria. It should be noted that the cleavage of the N-terminal 156 amino acids of human p53 results in the loss of transactivation domains 1 and 2 (TAD1 and TAD2) which have been shown to mediate protein-protein interactions in the mitochondria as will be discussed later.
Figure 1. Model of p53 import into mitochondrion under non-cell death conditions.
Schematized are mechanisms of p53 import into mitochondria via RECQL4 binding and targeting (left), CHCHD4-mediated respiration-driven disulfide-relay system (middle), and N-terminal cleavage and exposure of cryptic MTS (right). Double arrowheads indicate mitochondrial proteins that have been shown to interact with p53, and dashed arrows indicate the tentative pathways of p53 translocation. The question mark indicates that p53 is speculated to enter into matrix via the CHCHD4-mediated transfer to TIM complex. COX, cytochrome c oxidase; IM, mitochondrial inner membrane; IMS, intermembrane space; MTS, mitochondrial targeting sequence; NLS, nuclear localization signal; OM, mitochondrial outer membrane; TIM, translocase of inner mitochondrial membrane complex; TOM, translocase of outer mitochondrial membrane complex.
A third mechanism for the import of p53 utilizes a well-characterized redox carrier that depends on both active respiration and maintenance of the mitochondrial membrane potential (Figure 1, middle) [52,55]. The typical substrate for this system possesses cysteine-rich motifs capable of forming two intramolecular disulfide bonds which have previously been reported to be present in p53 (Cys-135/141 and Cys-275/277) [56]. In yeast, the import receptor Mia40, an IMS-localized oxidoreductase, covalently binds to its substrate through intermolecular disulfide bonds upon its translocation across the TOM complex [55,57]. Mia40 is maintained in an oxidized state by coupling with the sulfhydryl oxidase Erv1, and they constitute a “disulfide relay” that essentially transfers electrons from the imported substrate to molecular oxygen via cytochrome c and the cytochrome c oxidase (COX) complex [55]. Upon release from Mia40, the substrate with newly created intramolecular disulfide bonds is effectively trapped in the IMS where it resides or can be targeted for further translocation into the inner membrane or matrix in coordination with other import machineries.
The mammalian homolog of Mia40, coiled-coil-helix-coiled-coil-helix domain containing 4 (CHCHD4) protein, has been found to covalently bind p53 at the Cys-135 residue (disulfide Cys-135/141 pair) in a respiration-dependent manner, providing an additional level of specificity to this interaction [52]. Although other substrates of Mia40 such as TIM22 and mitochondrial ribosome Mrp10 are known to translocate to the inner membrane and matrix, respectively, it remains to be clarified how p53 through its interaction with CHCHD4 might gain access to the matrix space where mtDNA resides [51]. It is also possible that the three different pathways may be cross-utilized in the import of p53 in the cytosolic and mitochondrial compartments, but the endo-protease cleaved p53 that lacks Cys-135/141 would not be able to interact with the CHCHD4 pathway (Figure 1) [54]. Importantly, utilization of this respiration-driven mechanism by p53 supports the homeostatic nature by which it regulates redox and metabolic functions [58]. Under this scenario, higher mitochondria respiratory activity, possibly resulting in increased localized ROS production and oxidative mtDNA damage, would increase the translocation of p53 to the mitochondria where there may be increased demand for its repair activities [9,52]. Furthermore, the translocation of p53 into the mitochondria results in the subcellular partitioning of its activity and thus the modulation of its nuclear activity [52].
4.3. A role for p53 in mtDNA repair
In addition to participating in maintaining mtDNA through mitophagic and transcriptional activities, the translocation of p53 into mitochondria permits its binding and interaction with mtDNA and proteins involved in its maintenance which are listed in Table 2. Wild-type p53 protein itself was first reported to have intrinsic 3′–5′ exonuclease activity localized to its core domain (amino acids 80-290) that excises mismatched nucleotides from the replicating DNA strand [59,60]. This activity is enhanced in the presence of human mitochondrial single-stranded DNA-binding protein 1 (SSBP1), a factor involved in mtDNA replication that can interact with amino acids 1-61 of p53 (transactivation domains TAD1 and TAD2) [61].
Base excision repair, involved in the repair of oxidative DNA damage, is the best characterized and main repair mechanism known in the mitochondria [19,62]. In fact, BER is the only repair pathway to which p53 has mechanistically been linked in the mitochondria. An elegant series of studies by different investigators identified the stimulatory role of p53 in BER both in the nucleus and the mitochondria [63–67]. Early work using whole cell extracts showed that p53 can be pulled down with two BER proteins, 8-oxoguanine glycosylase (OGG1) and apurinic endonuclease (APE), and can promote the removal of oxidized DNA bases (Table 2) [63,64]. However, a concurrent study using mitochondrial extracts reported that p53 regulates the mitochondrial BER pathway through the repair synthesis incorporation step, which involves POLG, but not the DNA glycosylase and AP endonuclease steps [65]. While another group confirmed parts of both reports, p53 has been shown to interact with POLG and to promote its activity and accuracy [63,65,66,68,69]. A review of the literature only revealed the BER components Flap Structure-Specific Endonuclease 1 (FEN1) and 8-Oxoguanine DNA Glycosylase (OGG1) as p53 transcriptional target genes, suggesting that non-nuclear mechanisms of p53 play a predominant role in mtDNA repair [70,71].
As mentioned earlier, the p53 protein interacts with TFAM which is also involved in the structural organization of mtDNA into nucleoids (Table 2) [72]. In addition, TFAM and p53 bind more selectively to damaged mtDNA. Both the N- and C-terminal domains (amino acids 1-93 and 363-376, respectively) of p53 have been shown to interact with TFAM through its high mobility group-box (HMG-box) motifs [38,73]. The N-terminus of p53 interfaces with TFAM via its TAD1 and TAD2 domains [73]. Functionally, TFAM binding to mtDNA inhibits the incision activities of the BER enzymes OGG1 and APE which can be alleviated in the presence of p53 [67]. Recent work has shown that RECQL4 and p53, which co-translocate into the mitochondria (Figure 1), potentiate the exonuclease and polymerase activities of POLG to maintain mtDNA integrity (Table 2) [74]. These findings support earlier work showing the importance of RECQL4 in maintaining mtDNA integrity [75]. Accordingly, fibroblasts obtained from patients with hereditary cancer syndromes due to mutations in either RECQL4 (Rothmund-Thomson syndrome) or TP53 (Li-Fraumeni syndrome) share mtDNA mutations and sequence variants, suggesting converging activities of these two genes in the mitochondria [74].
Using a sensitive “long” PCR-based assay to quantify DNA damage, CHCHD4-binding and translocation of p53 into mitochondria has been shown to enhance the BER mediated repair of oxidative mtDNA damage including base modification and strand breaks (Table 2) [19,52,76]. Although cells deficient in respiration due to mtDNA depletion (Rho0) were reported to have a complete mitochondrial BER pathway, the mitochondrial BER activity of Rho0 cells could not be complemented to wild-type levels, suggesting an unknown/missing factor [77]. Given these observations, it is tempting to speculate that part of the missing factor may be attributable to the absence of p53 in the mitochondria of Rho0 cells caused by the inability to use the disulfide relay protein system for its import in the absence of respiration. The localization of BER components including mtDNA to the particulate fraction of the inner membrane also suggests that targeting p53 to the inner membrane via CHCHD4 may be sufficient for p53 activities [78,79]. Finally, the mutant p53 R175H retains its ability to repair oxidative mtDNA damage however this mutant is known to be missing exonuclease activity, effectively dissociating these two activities of p53 in mtDNA homeostasis [52,59,69].
5. Conclusions and perspectives
Further elucidation of how p53 participates in maintaining the mitochondrial genome is likely to reveal new insights into the fundamental aspects of aerobic life and impact our understanding of disease pathogenesis and tumorigenesis. There is much that we do not know, but there are at least two basic areas with respect to p53 and mtDNA which could benefit from additional focus: 1) the characterization of p53 protein inside the mitochondria; and 2) the potential role of p53 in heteroplasmy. First, the interpretation of p53 mitochondrial localization data obtained using purification, immuno-precipitation, and visualization methods can be limited by their respective technical issues. Also, the absence of p53 in mitochondrial proteomic databases is equivocal because even established mitochondrial protein may not be detected. Because the level of p53 in unstressed mitochondria is likely very low, it is not surprising that p53 cannot be readily detected in vivo using most conventional techniques. Emerging techniques that can sensitively detect molecular interactions with minimal perturbations such as Bimolecular Fluorescence Complementation (BiFC) may be one consideration [80]. Another area that could be of interest is exploring whether p53 plays a role in heteroplasmy as it has already been linked to some of the determinants of heteroplasmy such as mtDNA repair and mitophagy. In addition, p53 can play a role in mitochondrial dynamics by regulating genes such as DRP1 and OPA1, and can interact with TFAM, which affects the segregation and distribution of mtDNA into daughter mitochondria [81–85]. Another clue implicating p53 in heteroplasmy is the observation that p53 null female mice are essentially infertile unlike their male siblings that have preserved reproductive capacity [86]. As the sole donor of mtDNA to progeny, oogenesis in female mice requires a stringent purifying selection process that is being elucidated in fruit flies and mammals [87,88]. Exploring how p53 interacts with the mitochondria and maintains its genomic integrity may provide new insights into the biology of aerobic life.
Acknowledgments
We wish to thank William M. Kamp, Ju-Gyeong Kang and Ping-yuan Wang for helpful discussions and critical review of the manuscript. J.H.P. is a recipient of the Korean Biomedical Scientist Fellowship Program (KBSFP) administered by the Korea Research Institute of Bioscience and Biotechnology (KRIBB). This work was supported by the intramural program of National Heart, Lung and Blood Institute (NHLBI), NIH.
References
- 1.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–6. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–10. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 3.Sengupta S, Harris CC. p53: traffic cop at the crossroads of DNA repair and recombination. Nat Rev Mol Cell Biol. 2005;6:44–55. doi: 10.1038/nrm1546. [DOI] [PubMed] [Google Scholar]
- 4.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–31. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 5.Zhuang J, Ma W, Lago CU, Hwang PM. Metabolic regulation of oxygen and redox homeostasis by p53: Lessons from evolutionary biology? Free Radic Biol Med. 2012 doi: 10.1016/j.freeradbiomed.2012.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Zhang C, Hu W, Feng Z. Tumor suppressor p53 and its mutants in cancer metabolism. Cancer Lett. 2015;356:197–203. doi: 10.1016/j.canlet.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kruiswijk F, Labuschagne CF, Vousden KH. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol. 2015;16:393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- 8.Lago CU, Sung HJ, Ma W, Wang PY, Hwang PM. p53, aerobic metabolism, and cancer. Antioxid Redox Signal. 2011;15:1739–48. doi: 10.1089/ars.2010.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell. 2012;48:158–67. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warburg O. On the origin of cancer cells. Science. 1956;123:309–14. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 11.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–37. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 12.Zhou S, Kachhap S, Singh KK. Mitochondrial impairment in p53-deficient human cancer cells. Mutagenesis. 2003;18:287–92. doi: 10.1093/mutage/18.3.287. [DOI] [PubMed] [Google Scholar]
- 13.Matoba S, et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–3. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 14.Sung HJ, et al. Mitochondrial respiration protects against oxygen-associated DNA damage. Nat Commun. 2010;1:1–8. doi: 10.1038/ncomms1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Budanov AV, Sablina AA, Feinstein E, Koonin EV, Chumakov PM. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304:596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- 16.Sablina AA, Budanov AV, Ilyinskaya GV, Agapova LS, Kravchenko JE, Chumakov PM. The antioxidant function of the p53 tumor suppressor. Nat Med. 2005;11:1306–13. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Margulis L. Symbiosis in Cell Evolution. W.H. Freeman and Company; New York: 1993. [Google Scholar]
- 18.Galluzzi L, Morselli E, Kepp O, Vitale I, Pinti M, Kroemer G. Mitochondrial liaisons of p53. Antioxid Redox Signal. 2011;15:1691–714. doi: 10.1089/ars.2010.3504. [DOI] [PubMed] [Google Scholar]
- 19.Kazak L, Reyes A, Holt IJ. Minimizing the damage: repair pathways keep mitochondrial DNA intact. Nat Rev Mol Cell Biol. 2012;13:659–71. doi: 10.1038/nrm3439. [DOI] [PubMed] [Google Scholar]
- 20.Sykora P, Wilson DM, 3rd, Bohr VA. Repair of persistent strand breaks in the mitochondrial genome. Mech Ageing Dev. 2012;133:169–75. doi: 10.1016/j.mad.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Copeland WC, Longley MJ. Mitochondrial genome maintenance in health and disease. DNA Repair (Amst) 2014;19:190–8. doi: 10.1016/j.dnarep.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calvo SE, Mootha VK. The mitochondrial proteome and human disease. Annu Rev Genomics Hum Genet. 2010;11:25–44. doi: 10.1146/annurev-genom-082509-141720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stewart JB, Chinnery PF. The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat Rev Genet. 2015;16:530–42. doi: 10.1038/nrg3966. [DOI] [PubMed] [Google Scholar]
- 25.Wallace DC. Mitochondrial DNA Variation in Human Radiation and Disease. Cell. 2015;163:33–8. doi: 10.1016/j.cell.2015.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunham-Snary KJ, Ballinger SW. GENETICS. Mitochondrial-nuclear DNA mismatch matters. Science. 2015;349:1449–50. doi: 10.1126/science.aac5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valentin-Vega YA, et al. Mitochondrial dysfunction in ataxia-telangiectasia. Blood. 2012;119:1490–500. doi: 10.1182/blood-2011-08-373639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lebedeva MA, Eaton JS, Shadel GS. Loss of p53 causes mitochondrial DNA depletion and altered mitochondrial reactive oxygen species homeostasis. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbabio.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kulawiec M, Ayyasamy V, Singh KK. p53 regulates mtDNA copy number and mitocheckpoint pathway. J Carcinog. 2009;8:8. doi: 10.4103/1477-3163.50893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park JY, et al. p53 improves aerobic exercise capacity and augments skeletal muscle mitochondrial DNA content. Circ Res. 2009;105:705–12. 11–712. doi: 10.1161/CIRCRESAHA.109.205310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helton ES, Chen X. p53 modulation of the DNA damage response. J Cell Biochem. 2007;100:883–96. doi: 10.1002/jcb.21091. [DOI] [PubMed] [Google Scholar]
- 32.Scheibye-Knudsen M, Fang EF, Croteau DL, Wilson DM, 3rd, Bohr VA. Protecting the mitochondrial powerhouse. Trends Cell Biol. 2015;25:158–70. doi: 10.1016/j.tcb.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, Takei Y, Nakamura Y. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–9. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 34.Bourdon A, et al. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat Genet. 2007;39:776–80. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- 35.Shadel GS. Expression and maintenance of mitochondrial DNA: new insights into human disease pathology. Am J Pathol. 2008;172:1445–56. doi: 10.2353/ajpath.2008.071163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lang GA, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–72. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 37.Wang PY, et al. Increased oxidative metabolism in the Li-Fraumeni syndrome. N Engl J Med. 2013;368:1027–32. doi: 10.1056/NEJMoa1214091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshida Y, Izumi H, Torigoe T, Ishiguchi H, Itoh H, Kang D, Kohno K. P53 physically interacts with mitochondrial transcription factor A and differentially regulates binding to damaged DNA. Cancer Res. 2003;63:3729–34. [PubMed] [Google Scholar]
- 39.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maiuri MC, Galluzzi L, Morselli E, Kepp O, Malik SA, Kroemer G. Autophagy regulation by p53. Curr Opin Cell Biol. 2010;22:181–5. doi: 10.1016/j.ceb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 41.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–30. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kenzelmann Broz D, Spano Mello S, Bieging KT, Jiang D, Dusek RL, Brady CA, Sidow A, Attardi LD. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev. 2013;27:1016–31. doi: 10.1101/gad.212282.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suen DF, Narendra DP, Tanaka A, Manfredi G, Youle RJ. Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proc Natl Acad Sci U S A. 2010;107:11835–40. doi: 10.1073/pnas.0914569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoshino A, et al. Cytosolic p53 inhibits Parkin-mediated mitophagy and promotes mitochondrial dysfunction in the mouse heart. Nat Commun. 2013;4:2308. doi: 10.1038/ncomms3308. [DOI] [PubMed] [Google Scholar]
- 45.Hoshino A, et al. Inhibition of p53 preserves Parkin-mediated mitophagy and pancreatic beta-cell function in diabetes. Proc Natl Acad Sci U S A. 2014;111:3116–21. doi: 10.1073/pnas.1318951111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marchenko ND, Zaika A, Moll UM. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J Biol Chem. 2000;275:16202–12. doi: 10.1074/jbc.275.21.16202. [DOI] [PubMed] [Google Scholar]
- 47.Vaseva AV, Marchenko ND, Ji K, Tsirka SE, Holzmann S, Moll UM. p53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell. 2012;149:1536–48. doi: 10.1016/j.cell.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell. 2001;7:673–82. doi: 10.1016/s1097-2765(01)00213-1. [DOI] [PubMed] [Google Scholar]
- 49.Chipuk JE, Bouchier-Hayes L, Kuwana T, Newmeyer DD, Green DR. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science. 2005;309:1732–5. doi: 10.1126/science.1114297. [DOI] [PubMed] [Google Scholar]
- 50.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–44. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wenz LS, Opalinski L, Wiedemann N, Becker T. Cooperation of protein machineries in mitochondrial protein sorting. Biochim Biophys Acta. 2015;1853:1119–29. doi: 10.1016/j.bbamcr.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 52.Zhuang J, Wang PY, Huang X, Chen X, Kang JG, Hwang PM. Mitochondrial disulfide relay mediates translocation of p53 and partitions its subcellular activity. Proc Natl Acad Sci U S A. 2013;110:17356–61. doi: 10.1073/pnas.1310908110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De S, et al. RECQL4 is essential for the transport of p53 to mitochondria in normal human cells in the absence of exogenous stress. J Cell Sci. 2012;125:2509–22. doi: 10.1242/jcs.101501. [DOI] [PubMed] [Google Scholar]
- 54.Boopathi E, Srinivasan S, Fang JK, Avadhani NG. Bimodal protein targeting through activation of cryptic mitochondrial targeting signals by an inducible cytosolic endoprotease. Mol Cell. 2008;32:32–42. doi: 10.1016/j.molcel.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Riemer J, Fischer M, Herrmann JM. Oxidation-driven protein import into mitochondria: Insights and blind spots. Biochim Biophys Acta. 2011;1808:981–9. doi: 10.1016/j.bbamem.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 56.Hainaut P, Mann K. Zinc binding and redox control of p53 structure and function. Antioxid Redox Signal. 2001;3:611–23. doi: 10.1089/15230860152542961. [DOI] [PubMed] [Google Scholar]
- 57.Mesecke N, Terziyska N, Kozany C, Baumann F, Neupert W, Hell K, Herrmann JM. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell. 2005;121:1059–69. doi: 10.1016/j.cell.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 58.Olovnikov IA, Kravchenko JE, Chumakov PM. Homeostatic functions of the p53 tumor suppressor: regulation of energy metabolism and antioxidant defense. Semin Cancer Biol. 2009;19:32–41. doi: 10.1016/j.semcancer.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mummenbrauer T, Janus F, Muller B, Wiesmuller L, Deppert W, Grosse F. p53 Protein exhibits 3′-to-5′ exonuclease activity. Cell. 1996;85:1089–99. doi: 10.1016/s0092-8674(00)81309-4. [DOI] [PubMed] [Google Scholar]
- 60.Huang P. Excision of mismatched nucleotides from DNA: a potential mechanism for enhancing DNA replication fidelity by the wild-type p53 protein. Oncogene. 1998;17:261–70. doi: 10.1038/sj.onc.1201946. [DOI] [PubMed] [Google Scholar]
- 61.Wong TS, Rajagopalan S, Townsley FM, Freund SM, Petrovich M, Loakes D, Fersht AR. Physical and functional interactions between human mitochondrial single-stranded DNA-binding protein and tumour suppressor p53. Nucleic Acids Res. 2009;37:568–81. doi: 10.1093/nar/gkn974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Croteau DL, Stierum RH, Bohr VA. Mitochondrial DNA repair pathways. Mutat Res. 1999;434:137–48. doi: 10.1016/s0921-8777(99)00025-7. [DOI] [PubMed] [Google Scholar]
- 63.Zhou J, Ahn J, Wilson SH, Prives C. A role for p53 in base excision repair. EMBO J. 2001;20:914–23. doi: 10.1093/emboj/20.4.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Achanta G, Huang P. Role of p53 in sensing oxidative DNA damage in response to reactive oxygen species-generating agents. Cancer Res. 2004;64:6233–9. doi: 10.1158/0008-5472.CAN-04-0494. [DOI] [PubMed] [Google Scholar]
- 65.de Souza-Pinto NC, Harris CC, Bohr VA. p53 functions in the incorporation step in DNA base excision repair in mouse liver mitochondria. Oncogene. 2004;23:6559–68. doi: 10.1038/sj.onc.1207874. [DOI] [PubMed] [Google Scholar]
- 66.Chen D, Yu Z, Zhu Z, Lopez CD. The p53 pathway promotes efficient mitochondrial DNA base excision repair in colorectal cancer cells. Cancer Res. 2006;66:3485–94. doi: 10.1158/0008-5472.CAN-05-4103. [DOI] [PubMed] [Google Scholar]
- 67.Canugovi C, Maynard S, Bayne AC, Sykora P, Tian J, de Souza-Pinto NC, Croteau DL, Bohr VA. The mitochondrial transcription factor A functions in mitochondrial base excision repair. DNA Repair (Amst) 2010;9:1080–9. doi: 10.1016/j.dnarep.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Achanta G, Sasaki R, Feng L, Carew JS, Lu W, Pelicano H, Keating MJ, Huang P. Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma. Embo J. 2005;24:3482–92. doi: 10.1038/sj.emboj.7600819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bakhanashvili M, Grinberg S, Bonda E, Simon AJ, Moshitch-Moshkovitz S, Rahav G. p53 in mitochondria enhances the accuracy of DNA synthesis. Cell Death Differ. 2008;15:1865–74. doi: 10.1038/cdd.2008.122. [DOI] [PubMed] [Google Scholar]
- 70.Christmann M, Tomicic MT, Origer J, Kaina B. Fen1 is induced p53 dependently and involved in the recovery from UV-light-induced replication inhibition. Oncogene. 2005;24:8304–13. doi: 10.1038/sj.onc.1208994. [DOI] [PubMed] [Google Scholar]
- 71.Chatterjee A, Mambo E, Osada M, Upadhyay S, Sidransky D. The effect of p53-RNAi and p53 knockout on human 8-oxoguanine DNA glycosylase (hOgg1) activity. FASEB J. 2006;20:112–4. doi: 10.1096/fj.04-3423fje. [DOI] [PubMed] [Google Scholar]
- 72.Kukat C, et al. Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proc Natl Acad Sci U S A. 2015;112:11288–93. doi: 10.1073/pnas.1512131112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wong TS, Rajagopalan S, Freund SM, Rutherford TJ, Andreeva A, Townsley FM, Petrovich M, Fersht AR. Biophysical characterizations of human mitochondrial transcription factor A and its binding to tumor suppressor p53. Nucleic Acids Res. 2009;37:6765–83. doi: 10.1093/nar/gkp750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gupta S, De S, Srivastava V, Hussain M, Kumari J, Muniyappa K, Sengupta S. RECQL4 and p53 potentiate the activity of polymerase gamma and maintain the integrity of the human mitochondrial genome. Carcinogenesis. 2014;35:34–45. doi: 10.1093/carcin/bgt315. [DOI] [PubMed] [Google Scholar]
- 75.Croteau DL, et al. RECQL4 localizes to mitochondria and preserves mitochondrial DNA integrity. Aging Cell. 2012;11:456–66. doi: 10.1111/j.1474-9726.2012.00803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Santos JH, Meyer JN, Mandavilli BS, Van Houten B. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol Biol. 2006;314:183–99. doi: 10.1385/1-59259-973-7:183. [DOI] [PubMed] [Google Scholar]
- 77.Stuart JA, Hashiguchi K, Wilson DM, 3rd, Copeland WC, Souza-Pinto NC, Bohr VA. DNA base excision repair activities and pathway function in mitochondrial and cellular lysates from cells lacking mitochondrial DNA. Nucleic Acids Res. 2004;32:2181–92. doi: 10.1093/nar/gkh533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stuart JA, Mayard S, Hashiguchi K, Souza-Pinto NC, Bohr VA. Localization of mitochondrial DNA base excision repair to an inner membrane-associated particulate fraction. Nucleic Acids Res. 2005;33:3722–32. doi: 10.1093/nar/gki683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boesch P, Ibrahim N, Dietrich A, Lightowlers RN. Membrane association of mitochondrial DNA facilitates base excision repair in mammalian mitochondria. Nucleic Acids Res. 2010;38:1478–88. doi: 10.1093/nar/gkp1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miller KE, Kim Y, Huh WK, Park HO. Bimolecular Fluorescence Complementation (BiFC) Analysis: Advances and Recent Applications for Genome-Wide Interaction Studies. J Mol Biol. 2015;427:2039–55. doi: 10.1016/j.jmb.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li J, Donath S, Li Y, Qin D, Prabhakar BS, Li P. miR-30 regulates mitochondrial fission through targeting p53 and the dynamin-related protein-1 pathway. PLoS Genet. 2010;6:e1000795. doi: 10.1371/journal.pgen.1000795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kasashima K, Sumitani M, Endo H. Human mitochondrial transcription factor A is required for the segregation of mitochondrial DNA in cultured cells. Exp Cell Res. 2011;317:210–20. doi: 10.1016/j.yexcr.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 83.Wallace DC, Chalkia D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb Perspect Biol. 2013;5:a021220. doi: 10.1101/cshperspect.a021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kong B, Wang Q, Fung E, Xue K, Tsang BK. p53 is required for cisplatin-induced processing of the mitochondrial fusion protein L-Opa1 that is mediated by the mitochondrial metallopeptidase Oma1 in gynecologic cancers. J Biol Chem. 2014;289:27134–45. doi: 10.1074/jbc.M114.594812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mishra P, Chan DC. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat Rev Mol Cell Biol. 2014;15:634–46. doi: 10.1038/nrm3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hu W, Feng Z, Teresky AK, Levine AJ. p53 regulates maternal reproduction through LIF. Nature. 2007;450:721–4. doi: 10.1038/nature05993. [DOI] [PubMed] [Google Scholar]
- 87.Fan W, et al. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science. 2008;319:958–62. doi: 10.1126/science.1147786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hill JH, Chen Z, Xu H. Selective propagation of functional mitochondrial DNA during oogenesis restricts the transmission of a deleterious mitochondrial variant. Nat Genet. 2014;46:389–92. doi: 10.1038/ng.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]