Abstract
Microfluidic devices can deliver soluble factors to cell and tissue culture microenvironments with precise spatiotemporal control. However, enclosed microfluidic environments often have drawbacks such as the need for continuous culture medium perfusion which limits the duration of experiments, incongruity between microculture and macroculture, difficulty in introducing cells and tissues, and high shear stress on cells. Here, we present an open-chamber microfluidic device that delivers hydrodynamically focused streams of soluble reagents to cells over long time periods (i.e., several hours). We demonstrate the advantage of the open chamber by using conventional cell culture techniques to induce the differentiation of myoblasts into myotubes, a process that occurs in 7–10 days and is difficult to achieve in closed chamber microfluidic devices. By controlling the flow rates and altering the device geometry, we produced sharp focal streams with widths ranging from 36 μm to 187 μm. The focal streams were reproducible (∼12% variation between units) and stable (∼20% increase in stream width over 10 h of operation). Furthermore, we integrated trenches for micropatterning myoblasts and microtraps for confining single primary myofibers into the device. We demonstrate with finite element method (FEM) simulations that shear stresses within the cell trench are well below values known to be deleterious to cells, while local concentrations are maintained at ∼22% of the input concentration. Finally, we demonstrated focused delivery of cytoplasmic and nuclear dyes to micropatterned myoblasts and myofibers. The open-chamber microfluidic flow-focusing concept combined with micropatterning may be generalized to other microfluidic applications that require stringent long-term cell culture conditions.
I. INTRODUCTION
In the past two decades, microfluidic devices have shown great potential in cell culture applications, with advantages in recapitulating physiological conditions, parallelizing experiments for large-scale data acquisition, minimizing reagent consumption for reducing costs, and reducing human error due to their high reproducibility.1–3 Microfluidics offers a high degree of control of the cell microenvironment because transport phenomena such as diffusion and laminar flow can be mathematically modeled, allowing for simulations of local concentrations and flow velocities that can be used to optimize device design. Micropatterning and microfabrication allow single cells to be confined within well-defined geometries, allowing for precise interrogation.4,5 Together, microfluidic and micropatterning techniques provide a means to precisely control both the soluble and insoluble microenvironments of cells in in vitro platforms. Therefore, microfluidic devices provide key advantages over traditional cell culture platforms in their ability to recapitulate the physiological conditions at the microscale.2,3,6,7 Microfluidic manipulation has been utilized for the focal stimulation of microdomains of single cells, subcellular compartments, or tissues in a wide range of applications including: in vitro models for tissue development that necessitate spatiotemporal presentation of soluble and physical signals,8,9 single cell analysis for multiplexed drug testing,10,11 localized chemical stimulation of tissue slices for neural electrophysiology and cancer drug screening,12–14 and localized neurochemical stimulation of muscle cells.15–17
Classical microfluidic approaches for localized stimulation in cellular assays have utilized pressure-driven laminar flow in enclosed channels. Laminar flow occurs in the large Peclet number regime, where advective transport dominates over diffusive transport.18,19 Takayama et al. developed a microfluidic technique, “partial treatment of cells using laminar flows” or PARTCELL, to deliver reagents at subcellular resolution to study phenomena such as mitochondrial movement and cytoskeletal changes.20,21 Conventionally, laminar flow-based microfluidic devices have operated in a configuration where cells are cultured within enclosed micro-chambers, leading to a number of drawbacks.10,22 First, the high surface area-to-volume ratio of microchannels renders cells susceptible to nutrient shortage and buildup of metabolic waste.23 Therefore, this susceptibility necessitates continuous media replenishment using perfusion to maintain nutrient levels, pH, and gas concentrations.23,24 Microfluidic cell culture systems that require constant perfusion on long timescales on the order of days are often cumbersome and unreliable. For example, C2C12 myotubes require 7–10 days for single myoblasts to fuse into mature myotubes in low-serum medium conditions. Second, due to the inherent differences between microculture and macroculture environments, biological data obtained from closed channel microfluidic devices are difficult to compare with conventional cell culture experiments.25,26 Conditions for sensitive cell types for long-term experiments need to be adjusted specifically for microfluidic devices, creating another barrier for use by life scientists outside of the microfluidics community.25 Third, closed channel microfluidic devices suffer from disadvantages such as high hydraulic resistance, difficulty to introduce cells and tissues, and clogging by particulates and air bubbles.27 Fourth, flow-induced shear stresses may confound the biological observation of interest and have deleterious effects on cells, e.g., affecting ion channel activation,28 motility,29 or in the case of sensitive cell types such as neurons, even causing cell death.30
To address the numerous drawbacks of closed microfluidic systems, several groups have developed “open” devices that operate within an open architecture without sealed channels and chambers.8,10,31,32 A central goal is for devices to deliver localized chemical stimulation to large cell culture surface areas and tissues in open architectures. Many designs seek to create a user-friendly experience for the researcher and a more benign microenvironment for the cells.22,32–34 In open-chamber designs, in which cells are bathed in the bulk media bath like in a petri dish, results can be directly compared with conventional cell cultures. Open-chamber microfluidic devices circumvent the media-replenishing problem of closed devices by operating within an open reservoir, allowing for easy media exchange. Furthermore, open-chamber devices allow straightforward cell seeding by simple top-loading with a pipette. The authors' group has developed a class of open-chamber gradient-generating devices known as “microjets.” The device operates at low flow rates, which applies minimal shear stresses and is benign to mammalian neurons.22,35 Delamarche et al. developed a class of devices using hydrodynamic focusing known as microfluidic probes (MFPs). These devices combine the concepts of laminar-flow microfluidics and scanning probes to perform noncontact localized chemistries on surfaces.36,37 MFPs have sufficient resolution to individually address small groups of cells.38
Hydrodynamic focusing, the confinement of a central stream by faster flowing side streams, is a promising method to achieve focused microfluidic flows, having been used in applications such as protein gradient generation in hydrogels,39 miniaturized biochemical assays,40 and cell sorting.41 The flow rates of the central stream and the flanking streams in microfluidic devices can be adjusted to define the width of the focusing stream and the diffusion patterns at the stream interface.42 Accordingly, many biological events can be closely mimicked based on the simple generation of hydrodynamic focusing streams in microfluidic devices. Previously, Wang et al.43 and Liu et al.44 reported closed-chamber microfluidic devices utilizing hydrodynamic focusing for focal delivery of soluble reagents to cells. Chen and Lee used a hydrodynamically focused microfluidic device to investigate gap junctions by delivering calcein AM to target cells and tracking its transport into neighboring unexposed cells.45 While these devices enabled insightful experiments, their utility was limited to short-term experiments on the order of minutes.
In this paper, we describe a microfluidic device that produces stable, reproducible hydrodynamically focused streams in an “open-chamber” design that can be sustained for long-term operation on the order of hours. The open-chamber design allows for the device to be accessible and cells to be maintained at high viability using conventional cell culture protocols. We demonstrate the advantage of the open-chamber architecture in enabling long-term cell cultures of myoblasts and differentiation into myotubes over 7–10 days. Moreover, we coupled the open microfluidic system with a poly(dimethylsiloxane) (PDMS) surface modification technique for long-term attachment and growth of C2C12 myoblasts and a trapping technique for primary muscle fibers. We show localized delivery of soluble molecules to cells at microscale resolution.
II. MATERIALS AND METHODS
A. Device overview
The microfluidic device operates on the principle of hydrodynamic focusing, which involves laminar flow with outer flanking streams that hydrodynamically “squeeze” a central focal stream.42,46,47 Using appropriate flow rates, the effect of lateral diffusion of the soluble species can be minimized,46 maintaining a steady state focal stream for long periods of time. Furthermore, by tuning the relative flow rates of the flanking and focal streams, the width of the focal stream can be controlled.42 In our device (Figure 1), the focal stream is ejected into an open cell culture reservoir and applied to focal regions on the apical surface of cells micropatterned orthogonal to the flow direction. Since the microjets of the two flanking channels and focal channel are aligned in only the x dimension (Figure 4(a)), we expect the hydrodynamic focusing effect to dominate in the XY (top-view) and XZ planes but be diminished in the YZ (side cross-sectional) plane, thereby allowing for transport of molecules in the z-direction down into the trench. The configuration of this device is in contrast to that of a flow cytometer nozzle, where the central stream is focused with an outer sheath stream that surrounds it, creating a 3D hydrodynamic focusing effect. In this paper, we demonstrate this localized transport phenomenon both experimentally and computationally.
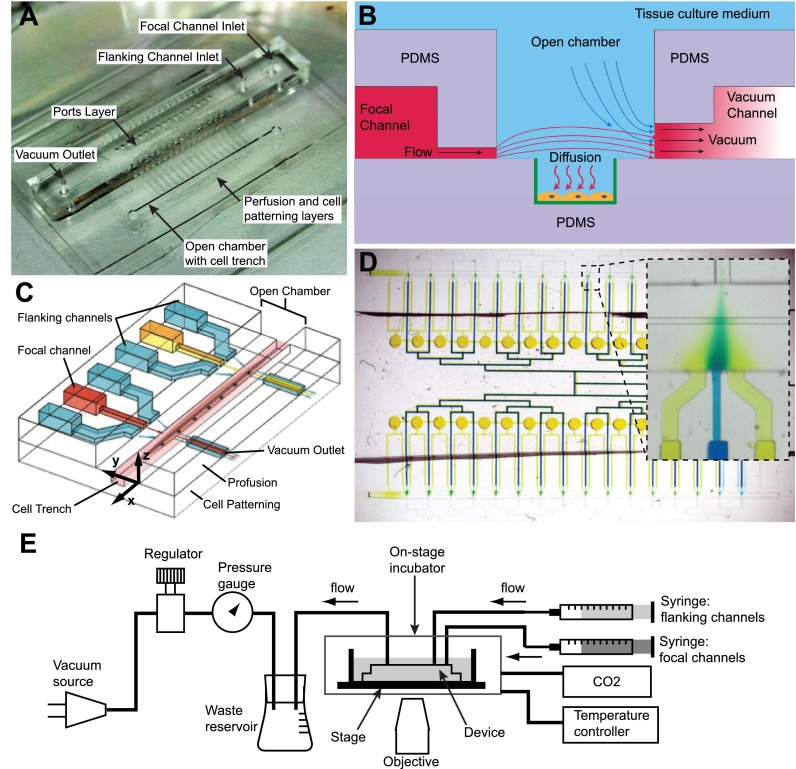
FIG. 1.
Schematics and operation of the microfluidic device. (a) Photograph of the device shows that the focal and flanking channel inlets and the vacuum outlet are introduced into the chip via a central ports layer. The focal and flanking microjets are on a middle perfusion layer, while the cell trench lies within the open chamber. (b) A cross-sectional side view (YZ plane) of the device through the channels: there is continuous flow driven by positive pressure from syringe pumps and negative pressure from vacuum channels. The molecule of interest diffuses to cells, which are patterned within trenches modified with epoxysilane/poly-lysine coating (green). (c) A 3D schematic shows two units of focal and flanking channels apposed by vacuum aspiration channels. The flanking channels squeeze the stream from the central focal channel by hydrodynamic focusing to deliver factors to cells (e.g., myotubes and myofibers) in trenches, which lie in a chamber open to the bulk cell culture medium. (d) The device consists of 32 units of focal and flanking channels, as represented by the blue and yellow dyes in the focal and flanking channels, respectively. The inset shows one unit of focal and flanking channels, showing hydrodynamic focusing. (e) Diagram of fluidic connections. For cell experiments, the device was enclosed within an on-stage incubator with temperature control and CO2 source. Panels (b) and (c) reproduced with permission from Cheng et al., in Proceedings of The 15th International Conference on Miniaturized Systems for Chemistry and Life Sciences, Seattle, 2–6 October 2011, pp. 356–358. Copyright 2011 Chemical and Biological Microsystems Society, CBMS.67
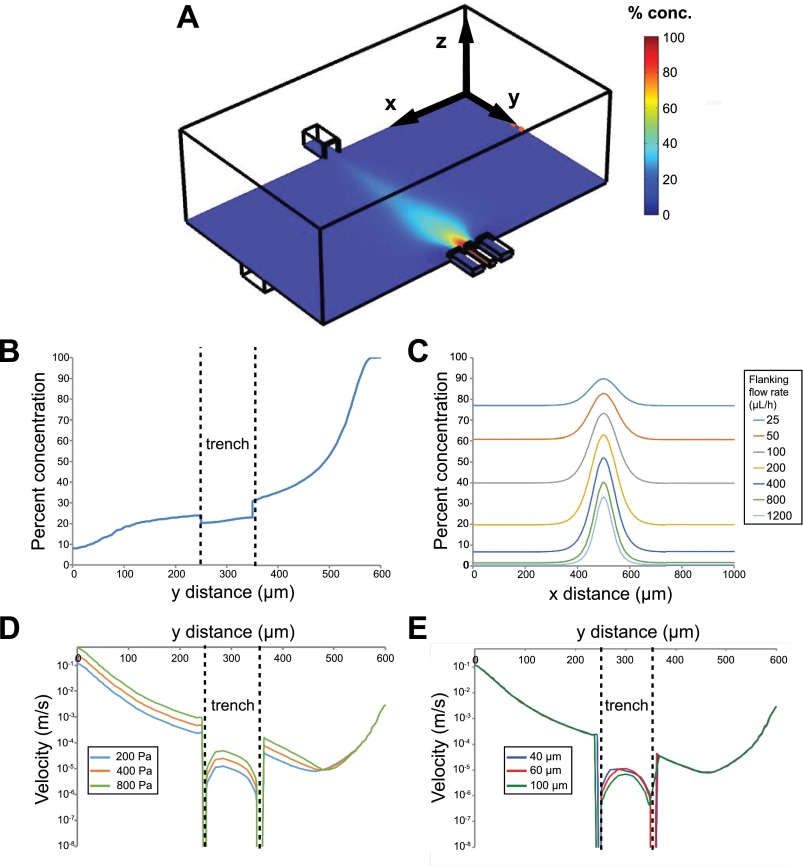
FIG. 4.
Computational simulations of local concentrations and velocities. (a) An isometric view of one microjet unit, showing focal and flanking channels on the bottom right and vacuum channel on the top left, and a color map of local concentrations (arbitrary 0%–100%) at the plane of the bottom of the microjets. (b) Concentration profile in the YZ cross-section of the device shows that there is ∼22% concentration at 10 μm above the trench floor. Note that the y distance is from the vacuum outlet to the focal inlet. (c) Simulated linescans at a constant focal channel flow rate of 100 μl/h, and 7 varying flanking channel flow rates ranging from 25 μl/h to 1200 μl/h. (d) Predicted flow velocity profile along y (z = 0) and at 10 μm above the trench floor for various vacuum channel pressures. (e) Predicted flow velocity profile along y (z = 0) and at 10 μm above the trench floor for various trench depths.
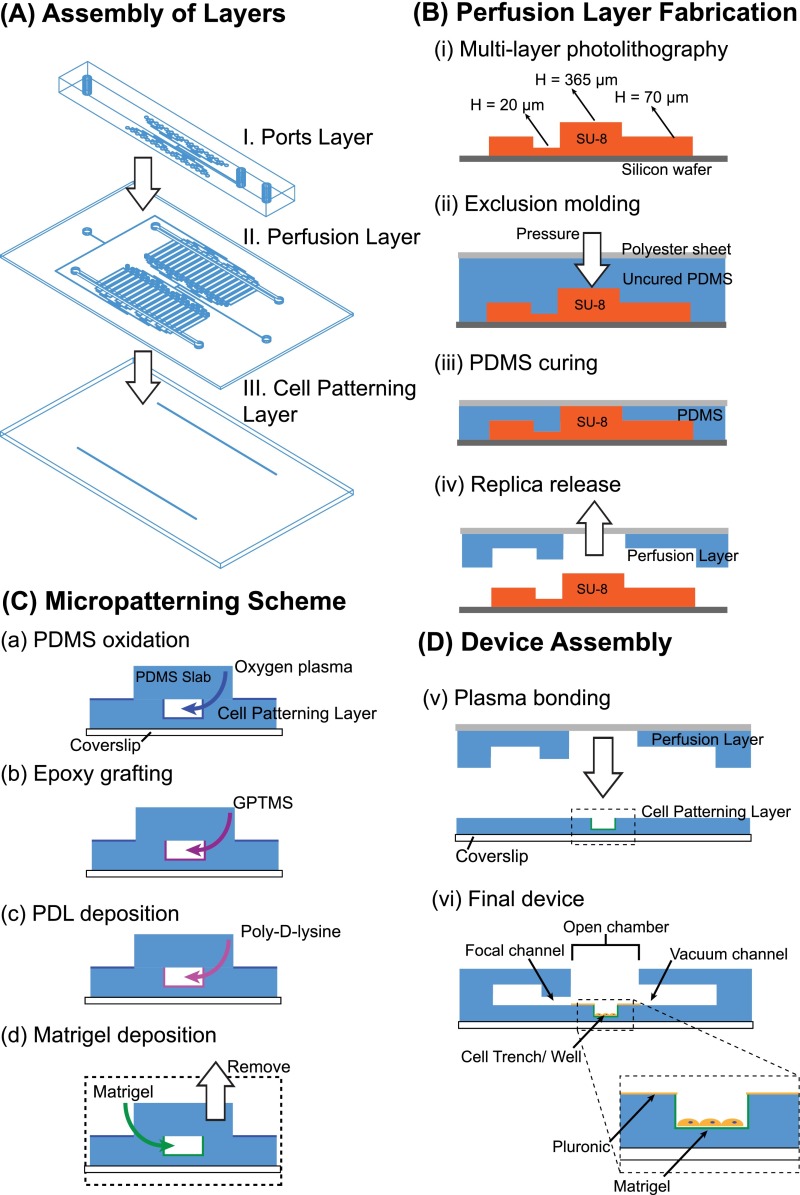
The device consists of three layers of PDMS bonded to a custom-made glass-bottom petri dish. The three layers from bottom to top are: Layer I, the cell patterning layer, Layer II, the perfusion layer, and Layer III, the ports layer (Figures 1(a) and 2(A)). For myotube focal experiments, the cell patterning layer consists of two micro-scale trenches that serve to contain and promote aligned micropatterns of myotubes; for myofiber focal experiments, the cell patterning layer consists of two rows of microtraps for trapping myofibers. The depressed trenches and microtraps serve to shield the cells from flow-induced shear stresses (Figure 1(b)).48 The perfusion layer consists of one focal channel, adjacent to which are two flanking channels which drive flow into an open chamber containing the cells, and apposed on the other side of the open chamber by a vacuum channel which aspirates the flow (Figures 1(b) and 1(c)). We define the focal and flanking channel openings as “microjets,” since the cross-sectional areas of these openings are much smaller than the rest of the connected fluidic channels: 20 μm × 20 μm for the focal channel and 50 μm × 50 μm for the flanking channel.22,35 The soluble species diffuses to the cells on the bottom of the trench or microtrap (Figure 1(b)). Finally, the ports layer provides for off-chip connections for the focal and flanking channel system inlets and an outlet for a vacuum source. The binary network of channels feeding the focal and flanking microjets is connected into two planes by a system of vertical passages (vias) and converges only at the microjets opening at the perfusion layer. We scaled up the device into configurations for large-scale experiments: 32 and 64 sets of focal and flanking channels for the myotubes and myofibers devices, respectively. The device was fabricated using a combination of: (1) photolithography to define a series of master molds and (2) soft lithography to form the PDMS structures (Figure 2). For detailed fabrication methods, please refer to the supplementary material.66 Notably, an established method from our group, termed “exclusion molding,”49 was used to make the open chamber, vias, and inlet holes in PDMS for the perfusion layer (Layer II).
FIG. 2.
Fabrication of the microfluidic device. (A) The device consists of three layers: (I) the cell patterning layer containing either cell trenches or myofiber wells, (II) the perfusion layer containing the focal and flanking microjets, their associated channels, and vacuum channels, and (III) the ports layer that contains the off-chip connections. (B) (i) A master mold for the perfusion layer is fabricated using multi-layer photolithography. (ii)-(iv) Exclusion molding, where PDMS is pressed down to the highest SU-8 features with a polyester sheet backing forms the open chamber and vias. (C) Micropatterning: (a) insides of the cell trench are selectively oxidized by oxygen plasma, (b) an epoxysilane GPTMS is grafted to oxidized PDMS, (c) Polylysine (PDL) covalent coated onto PDMS to promote, and (d) protein (Matrigel) adsorption. (D-v) The perfusion layer is plasma bonded to the cell trench layer. After bonding, the background areas are backfilled with Pluronic F127 to prevent cell attachment. (vi) Cells can be easily top loaded into the open chamber in the completed device.
B. Device operation and characterization of focal stream
The device setup is shown in Figure 1(e). Flow was driven simultaneously by both a “push” positive pressure source (two syringe pumps) and a “pull” negative pressure source (vacuum). Syringes ranging from 3 ml to 25 ml carrying the focal and flanking solutions (BD Falcon #309604) were mounted on separate syringe pumps (Kent Scientific, Chemyx Fusion 200 Syringe Pump) and connected to the ports layer of the device via stainless steel 19 gauge blunt needle connectors using Cole-Parmer silicone tubing (0.020 in. inner diameter (ID), 0.092 in. outer diameter (OD), #95609–18) for dye-based experiments or Sanipure silicone tubing (1/32 in. ID, 3/32 in. OD, Cole Parmer #96132–00) for cell-based experiments. Syringe pump flow rates between 100 μl/h and 3000 μl/h were used to drive flow. The average channel velocities and local velocities within the open-chamber are also reported in the finite element method (FEM) simulations section. Low adsorption Sanipure silicone tubing was used to minimize protein adsorption and loss. House vacuum was also connected to the vacuum outlet using the same connector type. A vacuum regulator (Airtrol) was used to control the vacuum strength, which was measured by a vacuum gauge (Ashcroft). The vacuum was set to a pressure (1 psi) that is low enough to minimize shear stress on cells yet capable of generating focal streams. For experiments on delivery of soluble reagents to live cells and myofibers, devices were imaged in an on-stage incubator, with a temperature controller and 5% carbon dioxide source.
In pilot studies on device operation, blue and yellow food coloring dyes (FD&C Yellow #5—tartrazine and FD&C Blue #1—erioglaucine) were delivered through the focal and flanking channels, respectively, and observed under a stereomicroscope. To measure the effectiveness of focal soluble factor delivery to specific regions, we used fluorescently labeled bovine serum albumin (BSA): Alexa Fluor 594 BSA conjugate (7.6 μM, #A13101, Life Technologies) to quantify the stream width. BSA was used as a model protein to simulate the transport of large molecules of interest, e.g., proteins. For quantification of long-term stream stability, we used 10 kDa Dextran–Alexa Fluor 594 conjugate at 10 μM (#D-22913, Life Technologies). Here, Dextran was chosen due to its low surface adsorption, reducing the likelihood of fluorescence accumulation and signal saturation during long timelapse acquisitions. Operation conditions, including focal and flanking flow rates of 100 μl/h and 400 μl/h, respectively, and a negative pressure of 1 psi, were kept constant in stream stability and cell staining experiments.
Fluid flow in the devices was measured using a widefield epifluorescence microscope: Nikon Eclipse Ti-E model with Plan Fluor 4×/0.13 NA, Plan Fluor 10×/0.30 NA, and Plan Fluor extra long working distance (ELWD) 20×/0.45 NA objectives (Nikon Instruments, Melville, NY) with a 12-bit cooled CCD camera (ORCA-ER, Hamamatsu, Japan) with an automated stage. Image analysis was performed using Fiji (ImageJ) software and curve fitting and statistics done with MATLAB. To characterize the stream width, we took fluorescence images of the same field of view of each of the 32 channels under varying flow conditions. A 1344-pixel wide linescan was taken in the open chamber along a line equidistant from the focal channel and the vacuum channel. The full width at half maximum (FWHM) fluorescence intensity was then calculated to represent the stream width.
C. Finite element modeling
The 3D simulation of the open-chamber hydrodynamic focusing device was generated using the FEM software COMSOL Multiphysics (Burlington, MA). The laminar flow module was used to define the pressure-driven flow. The transport of diluted species module was used to define the concentration inlets of focal and flanking channels with BSA (M.W. = 60 kDa) as the diffusing species (diffusion coefficient, D = 4 × 10−8 m2 s−1) to simulate concentration profiles at the open-chamber region.
D. Spatially selective PDMS surface modification for cell micropatterning
To selectively render the insides of the PDMS cell trenches hydrophilic and permissive while keeping the outside regions hydrophobic and non-permissive to cells, a PDMS slab (2.1 cm × 2.1 cm) spanning the length of the trenches was placed in conformal contact, covering the open trenches and turning them into microchannels. Oxygen plasma was then applied (60 W, 670 mTorr, and 60 s) to generate hydroxyl groups on the inner surfaces of the PDMS trenches (Figure S1 (Ref. 66)). 50 μl of (3-glycidoxypropyl) trimethoxysilane (GPTMS, 1% v/v in H2O, Sigma Aldrich #440167) was then drawn into the trenches by vacuum, incubated at room temperature for 20 min, and washed three times with sterile H2O. Subsequently, 50 μl of a positively charged polyamino acid, 100 μg/ml poly-ornithine (Molecular weight 30 000–70 000, Sigma Aldrich #P4957, St. Louis, MO), was flowed through the covered trenches, incubated for 1 h at room temperature, and washed three times by flowing sterile H2O through the trenches. The basement membrane extract Matrigel (BD Biosciences #356234, San Jose, CA), diluted 1:50 in Dulbecco's Modified Eagle's Medium (DMEM, Gibco, Invitrogen, #11995), was infused into the trenches, allowed to polymerize at 37 °C for 30 min, and dried for 5 h or overnight within the trenches. After Matrigel coating, the fluidic part of the device, Layers II and III were aligned and plasma bonded. Surprisingly, these bonding steps did not negatively affect the Matrigel coating, as discussed in Section III D. After bonding, PDMS was allowed to recover hydrophobically50 such that the non-permissive regions outside of the cell trenches would adsorb Pluronic. The device was bathed in Pluronic F127 (0.2% w/v in phosphate buffered saline (PBS), Invitrogen, #P6866), [(poly(ethylene oxide) (PEO))100–(poly(propylene oxide) (PPO)70)–(PEO)100] for 1 h. To eliminate bubbles from microchannels, the device was de-gassed in a vacuum desiccator for 1 h prior to cell seeding and operation. For cell culture experiments, the device was naturally de-bubbled for the course of 7 days by the high CO2 environment of the cell culture incubator.
E. Myoblast cell seeding and culture
Myotubes were derived from the mouse myogenic cell line C2C12 (American Type Cell Culture #CRL-1772, Manassas, VA). Myoblasts were expanded on gelatin-coated (Sigma-Aldrich, #1393) petri dishes in DMEM supplemented with 20% fetal bovine serum (FBS, PAA Laboratories) and 1% penicillin/streptomycin in a 5% CO2 incubator at 37 °C. Cells were passaged at 80% confluence to prevent differentiation and were used before passage 10. Cells were seeded as a 400 μl droplet (4.0 × 105 cells/ml) on top of the trenches and allowed to settle by gravity and attach for 25 min in the cell culture incubator. Cells that did not attach were rinsed away with three washes of media. Cells were allowed to grow for 24 h or longer until confluence inside the trenches, after which media were changed to low serum differentiation media (DM): DMEM supplemented with 2% horse serum (Sigma-Aldrich, #H1138) and 1% antibiotic/antimycotic, and replenished every 2 days. C2C12 myoblasts were allowed to fuse into mature myotubes for 7–10 days after switching to DM.
F. Myofiber harvest and microfluidic device adaptation
Muscle fibers from mouse hindlimbs were harvested and cultured using a previously described method.51 All animal procedures were approved by the Institutional Animal Care and Use Committee of the University of Washington and were performed in accordance with guidelines of the National Institutes of Health and the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). Adult C57BL6 mice, 2 months–1 year old (Jackson Laboratories), were euthanized by cervical dislocation and the flexor digitorum brevis (FDB) muscles were immediately dissected. The myofibers were dissociated from the muscle in 0.2% collagenase (Sigma-Aldrich, #C0130) solution in DMEM for 2–3 h in a cell culture incubator at 37 °C. Using a sterilized, scored, and flame-polished Pasteur pipette, the collagenase-digested muscle was triturated, liberating myofibers. The myofibers and their associated satellite cells were further purified from fibroblasts and other mononuclear cells by passing the digested muscle solution through a series of horse serum-coated glass centrifuge tubes three times (Corex, 10 ml glass centrifuge tube, Cole Parmer) and collecting the myofibers from the bottom of the column. Individual myofibers could be manually picked using a P-20 micropipette. Under a stereomicroscope, the myofibers were suspended in solution above the microtraps using a P-10 micropipette and then allowed to settle by gravity into microtraps. For myofibers, the microfluidic device design was altered to contain 64 focal channels with 128 flanking channels. Rectangular microtraps for trapping myofibers had dimensions of L × W × H = 400 μm × 80 μm × 110 μm or 360 μm × 60 μm × 110 μm.
G. Focal delivery of soluble factors to myoblasts and myofibers
Calcein AM (5 μM, #C3100MP, Life Technologies) was applied onto myoblasts for 30 min using focal and flanking flow rates of 200 μl/h and imaged in the green fluorescent protein (GFP) channel. Hoechst 33342 and Hoechst 33258, both at 5 μg/ml (#H1399, #H3569, Life Technologies) in PBS were used to label myoblast and myofiber nuclei, respectively. For image analysis of Hoechst 33258 labeling of myofiber nuclei, the individual nuclei were segmented using Fiji (ImageJ) software by user-defined thresholding and marking the nuclei as regions of interest (ROIs). The normalized mean intensities of the nuclei were plotted against their positions along the x-axis of microtrap (x-axis).
III. RESULTS AND DISCUSSION
A. Characterization of flow focusing
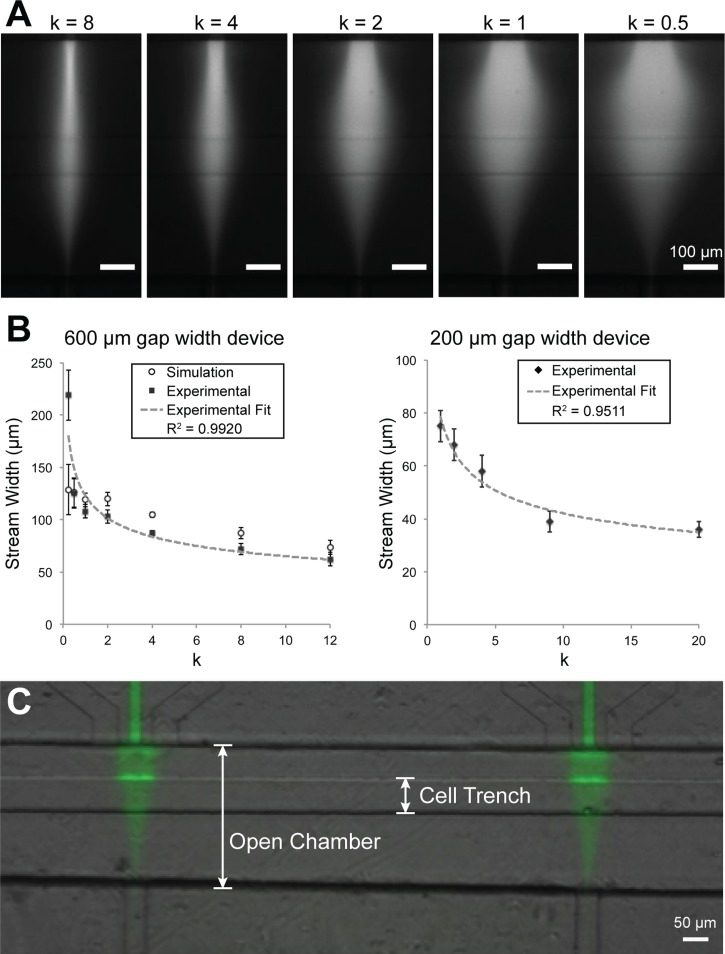
We characterized the ability of the device to control steady state focal streams in the open chamber (Figure 3(a)). We showed that stream width, measured by the linescan FWHM of continuously flowing BSA-Alexa Fluor 594, can be controlled by varying the ratio k of flanking and focal flow rates. We tested this phenomenon in two device configurations: open-chambers with 600 μm and 200 μm separation distances between the focal and vacuum channel openings. In the 600 μm gap device, we were able to vary the stream width ranging from 71 μm at k = 12 to 187 μm at k = 0.25. In the 200 μm gap device, we obtained stream widths ranging from 36 μm at k = 20 to 75 μm at k = 1. In Figure 3(b), the plot of k versus focal stream width shows an inverse relationship, which is consistent with theoretical models that describe the relationship as an inverse power function.52 In both device formats (600 μm and 200 μm), our experimental results showed a strong inverse power function relationship between k and the stream width (R2 = 0.9920 and 0.9511, respectively).
FIG. 3.
Characterization of hydrodynamic focusing control of microfluidic device. (a) The width of the focal stream (BSA-Alexa Fluor 594) can be controlled by adjusting the ratio of flanking and focal rates (k). (b) In 600 μm and 200 μm gap width devices, plot of stream width (FWHM) versus k, showing an inverse relationship. Black squares and diamonds represent the experimental and open circles represent simulated stream widths and have a strong fit with an inverse function (R2 values). Simulation and experimental results also show strong correlation (r = 0.970). (c) After focused flow and washing, a localized pattern of BSA-Alexa Fluor 488 deposited on the trench floor.
We further characterized the transport of a large molecular weight species, BSA-Alexa Fluor 488, to the trench floor. After 1 h of focused flow and washing with PBS, a fluorescence pattern similar to the active flow pattern remained on the PDMS substrate, indicating that fluorescently labeled BSA was locally transported and adsorbed onto the substrate (Figure 3(c)). This result is notable because it indicates that BSA deposited locally onto the regions of the cell trench beneath the focal stream, suggesting that cells would locally receive the soluble signal. In Figure 3(c), the bright signal on the trench walls is likely out-of-focus signal from accumulation of BSA on the trench walls, in part due to the presence of a dead zone where BSA signal accumulated over time.
B. FEM simulations of local concentrations and velocities
FEM simulations of the velocities, local concentrations, and stream widths (FWHM) of BSA under the same flow conditions as the experimental section within the 600 μm gap device were computed (Figure 4). The simulated stream widths (Figure 4(c)) were found to be strongly correlated with experimental stream widths, with a Pearson correlation coefficient r = 0.970 (Figure 3(b)). One outlier was found at extremely low flanking flow rates of 25 μl/h (k = 0.25), likely due to higher dominance of diffusion. We note that the simulated local BSA concentration under 200 Pa at 10 μm above the trench floor directly under the focal stream was 22 ± 1% of the inlet concentration, which is an approximation for the local cell fluid microenvironment (Figure 4(b)). We also calculated the flow conditions in which lateral diffusion of molecules becomes significant. For reference, under syringe pump focal/flanking flow rates of 100 μl/h and 2000 μl/h, the average channel velocity at the opening of the microjet is 0.00217 and 0.00350 m/s for the focal and flanking channels. Notably, at low flanking flow rates, there is considerable lateral molecular diffusion, resulting in much higher baseline concentrations outside of the focal stream (Figure 4(c)). Our calculations indicated that the flanking flow rate should be at least 4 times the focal flow rate (k > 4) to minimize diffusion of the species into the culture bath.
In our microfluidic system, it is important to consider the effect of flow-induced shear stress on cultured cells. Shear stress from microfluidic flows may be deleterious to cells, introduce migration bias, and generally alter the underlying biology of interest.29,53 We designed a micro trench for cells to be shielded from flow-induced shear. Due to the difficulty of experimentally measuring shear stresses directly on cells, we performed simulations of local velocities in our COMSOL model. Figures 4(d) and 4(e) show local flow velocities with 3 different outlet pressures and 3 different trench depths at 10 μm above the bottom of the trench. As expected, higher vacuum pressures lead to higher velocities, while deeper trenches can better shield cells from high flow velocities and shear stresses, as seen in the drop in local velocities within the trench. We compared the magnitudes of simulated shear stresses in our system versus values known to adversely affect cortical neurons, some of the most sensitive cell types to shear stress. Corresponding with the velocities shown in Figures 4(d) and 4(e), the average shear stress on the cells at 10 μm above the floor of the trench is 3.26 × 10−3 dyn/cm2, which is ∼20 times lower than the shear stress that leads to flow-induced retraction of neuron growth cones (0.07 dyn/cm2),30 and ∼200 times lower than the threshold shear stress ∼0.7 dyn/cm2 needed to maintain healthy cortical neurons.22 Therefore, we conclude that the micro trenches effectively protect cells from flow-induced shear stress, and the shear stress remains relatively insensitive to outlet flow velocity.
C. Characterization of focal stream stability and reproducibility
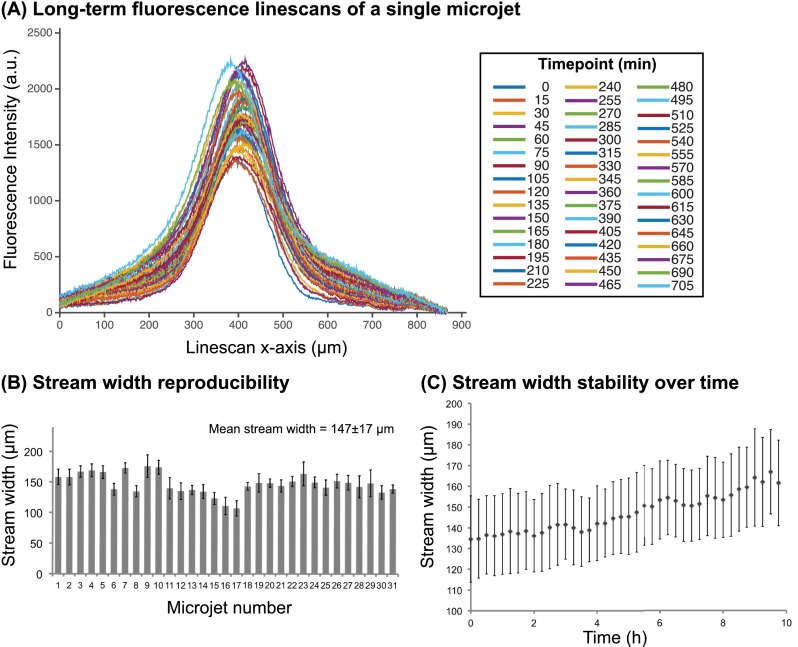
One key feature of this microfluidic system is the ability to generate multiple focal streams, enabling parallel acquisition of multiple data points. To determine focal stream reproducibility between microjets, we quantified the position and stream widths of fluorescent Dextran in 31 of the total 32 microjets averaged over a timespan of 10 h. One microjet's field of view was obstructed by a particle, so it was discarded from the analysis. We characterized two parameters of focal stream stability: (1) the stream width, as defined by the full width at half maximum (FWHM) of the fluorescence linescan, and (2) the stream position, as defined by the x-displacement of the fluorescence linescan maximum with respect to its initial position. The stream's x-displacement was defined as the absolute value of the x-coordinate of the linescan maximum at each timepoint subtracted by the x-coordinate of the maximum at the initial timepoint. For reference, an x-displacement of zero represents a perfectly invariant stream from the initial measurement.
Figure 5(a) shows the variability of linescans of a single representative microjet taken at 49 timepoints over 10 h of device operation. Here, all timepoint images were background-corrected such that the baseline of all linescans was equal. We observed that the linescan FWHM only increased 7.9% from 138 μm at the initial timepoint to 149 μm at 9.75 h. Moreover, the mean x-displacement was 9 μm, which was only 6.1% of the FWHM, indicating the consistent positioning of the focal stream. In Figure 5(b), we show the stream widths of the 31 microjets averaged over 10 h. The average stream width of all the microjets was 147 ± 17 μm. Therefore, under the same flow conditions, the device can generate multiple focal streams with less than 12% variation. The variation in stream widths between microjets may be attributed to channel clogging due to fabrication imperfections, particulates, or air bubbles. Over 10 h of operation, the cumulative average stream width of all microjets changed from 135 ± 21 μm to 162 ± 21 μm, a 20% increase (Figure 5(c)), whereas the average shift (x-displacement) was 9 ± 2 μm (Figure S2 (Ref. 66)). The stream width increase over time may be due to a decrease in negative pressure, possibly attributed to a drop in the hydrostatic pressure head from depletion of the culture medium bath or possibly from particulates clogging the vacuum channels. Furthermore, surface accumulation of Dextran onto PDMS may have contributed to increased fluorescence intensity, effectively broadening the stream width. Together, these results showed that the focal streams generated in this open-chamber microfluidic device have low variability between multiple microjets, enabling future high-throughput designs, and were stable over long-term operation timescales.
FIG. 5.
Characterization of focal stream reproducibility and long-term stability. (a) Linescans of a focal stream of Dextran 10 kDa - Alexa Fluor 594 at 49 timepoints taken in 15 min intervals over a 10-h operation period. (b) The time-averaged stream widths of 31 microjets in a single device. Error bars represent standard deviations of stream width over the 10 h timelapse. (c) The average width of the 31 focal streams as a function of time. Error bars represent standard deviations of stream width between 31 microjet units in a device.
D. Micropatterning of myoblasts and myotubes in trenches
We found that native PDMS, due to its hydrophobicity, did not support protein (i.e., Matrigel) coatings for long-term (∼2 week) cell viability (data not shown). Furthermore, even after oxygen plasma treatment, PDMS undergoes fast hydrophobic recovery within hours and also did not support long-term cell viability.50,54 To address this problem, we developed a method to selectively covalently modify the PDMS surface inside the trenches with a positively charge molecule to support Matrigel adsorption. We utilized a covalent grafting approach using an epoxy-silane, (3-glycidoxypropyl) trimethoxysilane (GPTMS), to modify PDMS surfaces.55,56 The inside of the trench was selectively rendered hydrophilic by plasma oxidation (Figure 2(C)). The silane groups on GPTMS covalently bond with hydroxyl groups on PDMS, creating a surface of epoxide groups. The amine groups on poly-D-lysine or poly-ornithine react with the epoxide groups from GPTMS, forming a covalently bound layer of poly-D-lysine or poly-ornithine inside the trench surface. The positively charged layer of poly-D-lysine/poly-ornithine serves to improve protein adsorption and preserves protein biological activity,57 facilitating Matrigel coating within the trench. Conversely, in the zones outside of the trench, which were protected from plasma oxidation by the PDMS slab, we expected Pluronic F127, a triblock copolymer consisting of a central hydrophobic poly(propylene oxide) (PPO) block, flanked by two blocks of poly(ethylene oxide) (PEO), to adsorb to the untreated hydrophobic PDMS substrate and repel cell attachment.58,59 Pluronic forms a “brushlike” molecular conformation, in which the hydrophobic PPO blocks adsorb to the hydrophobic PDMS substrate and the hydrophilic PEO blocks face the solution, resisting protein adsorption by repulsive osmotic force, thereby minimizing cell adhesion.59,60
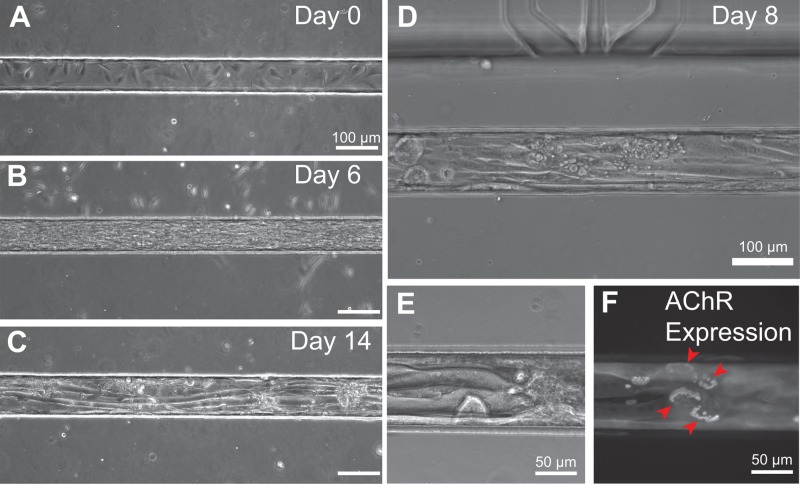
We found that our micropatterning scheme supported long-term micropatterned cultures of myotubes within trenches (Figure 6). Within 1 h of cell seeding, C2C12 myoblasts attached inside the Matrigel-coated trenches, while a lower density of cells also adhered to the zones outside of the trenches. While bathed in growth media, myoblasts proliferated and spread within the trenches. When the cells became 80% confluent at Days 1–3, media were changed to low-serum differentiation media and myoblasts fused into myotubes, aligning along the axis of the trenches (Figures 6(a)–6(d)). Mature myotubes formed within 5–7 days after media change, both inside the trenches and on flat substrate controls. Mature myotubes could be maintained for several days thereafter, i.e., for at least 14 days after initial seeding. Moreover, we found that narrower trench dimensions of 15–20 μm width and 40 μm height allowed confinement of single myotubes, allowing for single cell manipulation (Figure S3 (Ref. 66)). Meanwhile, during the first few days of culture, the cells outside of the trenches became rounded, died, and detached from the substrate, suggesting that the Pluronic coating was sufficiently cell-repellant.
FIG. 6.
Long term cell culture and myoblast differentiation into myotubes. (a) Day 0: One hour after cell seeding, C2C12 myoblasts were adherent and spread inside the Matrigel-coated PDMS trench. Few cells attached on the outside of the trench, showing the effectiveness of the Pluronic F127 cell-repellant coating. Cells grew confluent and filled the trench by Day 3, during which the growth media were changed to differentiation media containing low serum (not shown). (b) At Day 6, 3 days after change to differentiation media, myoblasts fused and formed elongated myotubes. (c) Day 14: Mature myotubes are shown within a 100 μm wide trench. Note that cells outside of the trench have died and detached. (d) On-chip differentiation of C2C12-derived myotubes at Day 9 showed their elongated, mature phenotype. (e) Elongated mature myotubes at Day 8 within a 100 μm wide trench, and (f) expression of mature acetylcholine receptor “pretzel” patterns labeled by fluorescent bungarotoxin.
The microscale geometry of the open-chamber (gap widths of 600 μm or 200 μm) necessitated that the protein micropatterning step be done prior to bonding of the cell patterning layer and the perfusion layer. Therefore, there was concern that the bonding of these two layers, which would expose the Matrigel-coated trenches to oxygen plasma, would cause denaturation of the protein coating. Surprisingly, we found no problems with cell adhesion and long-term viability after this step. Matrigel, a mouse sarcoma extract rich in extracellular matrix components, provided a cell culture substrate that mimics the skeletal muscle basement membrane. We observed robust long-term myotube maturation, growth, and expression of laminin-induced acetylcholine receptor cluster “pretzels” within the devices (Figure 6(f)). These mature, elaborate AChR cluster pretzels, which form independently in the absence of motor neuron signaling, indicated that laminin, which constitutes 60% of Matrigel, was biologically active.61 We hypothesize that there is a sufficiently thick Matrigel coating that remains active even if a superficial layer of Matrigel gets stripped or denatured by plasma treatment. Together, we demonstrated on-chip cell viability, micropatterning, and differentiation of muscle cells within the open architecture.
E. Focal delivery of soluble factors to micropatterned myoblasts
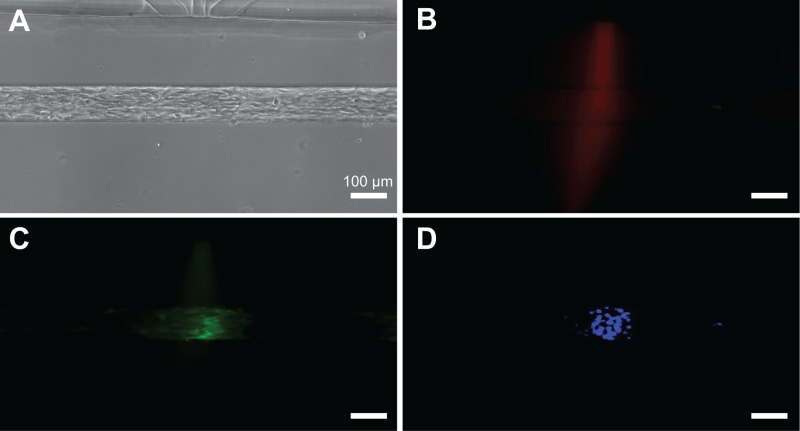
We tested the focal delivery of a cytoplasmic live cell stain, a calcein AM, and a nucleic acid stain, Hoechst 33342 to micropatterned myoblasts (Figure 7). In this experiment, C2C12 myoblasts were seeded and confined inside 100 μm wide trenches and allowed to grow for 24 h of cell seeding. For the focal delivery experiment, calcein AM was used to track cells, Hoechst 33342 was used to track nuclei, and BSA-Alexa Fluor 594 was used to track the profile of the stream. A mixture of the three molecules was introduced into the focal channels along with media in the flanking channels. Within 10 min of onset of flow, we observed localized calcein staining of cell cytoplasm and localized Hoechst staining of nuclei. We observed that calcein AM staining of cell cytoplasms and Hoechst 33342 staining of cell nuclei were largely spatially confined to the areas beneath the focal streams as indicated by the fluorescent BSA tracer. Localized staining was maintained for up to 4 h of observation. Due to the effectiveness of the Pluronic coating, non-specific cell attachment and cellular debris were not a problem as devices operated with good efficiency: about 80% microjets showed expected focal staining in these experiments.
FIG. 7.
Focal delivery of soluble reagents to micropatterned cells. (a) Micropatterned C2C12 myoblasts in a trench (100 μm wide, 110 μm deep). (b) Fluorescent BSA-Alexa Fluor 594 was used to track the stream profile. Focused staining of cell cytoplasm with calcein AM (c) and nuclei with Hoechst 33342 (d).
F. Focal delivery of soluble factors to trapped myofibers
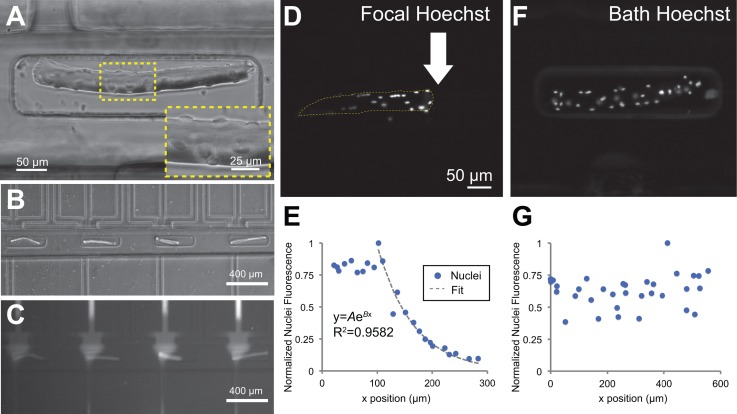
The open-chamber architecture of the microfluidic device allowed simple top-loading of cells, which enabled microfluidic manipulation of alternative cell culture models. In an alternative design, the cell patterning layer featured an array of 64 microtraps to immobilize myofibers, which are used as a model system for understanding the growth, differentiation, and migration of satellite cells within their native microenvironment.51,62–65 FDB myofibers isolated from mouse hindlimbs were viable and stained positively for myosin heavy chain 48 h after dissection and immobilization in the microfluidic device. Myofibers showed striations, multiple cell nuclei, and satellite cells on their surfaces (Figure 8(a)). The microtraps were offset during the plasma bonding process such that one end of the trap was aligned with the focal channel and the other end was distal to it. This configuration allowed application of a focal stream on one end of the myofiber, effectively generating a gradient along the length of the myofiber (Figures 8(b) and 8(c)).
FIG. 8.
Focal delivery of Hoechst DNA stain to nuclei in trapped myofibers. (a) Brightfield image of a FDB myofiber inside a microtrap. Inset shows the muscle fiber striations and myonuclei. (b) The traps were offset from the focal channel opening, generating in situ gradients within single myofibers shown by flow of BSA-Alexa Fluor 594 (c). (d) and (e) A focal stream of Hoechst 33258 live nuclear stain was applied to one side of a myofiber, selectively staining myonuclei and surface-lying satellite cell nuclei on the right side of the myofiber. (f) and (g) Negative control: bath application of Hoechst nuclear stain resulted in uniform nuclei staining.
As a proof-of-concept experiment to show localized soluble factor delivery, we applied focal streams of Hoechst DNA stain to locally label nuclei in myofibers. A solution of Hoechst was introduced into the focal channel, while media were flowed through the flanking channels. Within 20 min after initial flow onset, we observed a gradient pattern of Hoechst 33258 staining of myonuclei and satellite cell nuclei along the lengths of the myofibers (Figures 8(d) and 8(e)). In contrast, nuclei on myofibers simply bathed in a solution of Hoechst 33258 were homogeneously stained (Figures 8(f) and 8(g)). The gradient of nuclei staining intensity indicated that there was a gradient of local concentrations of the soluble factor, Hoechst, along the myofiber length. The nuclei staining intensities followed an exponential profile from the source immediately underneath the focal stream (near the white arrow in Figure 8(d)) to the sink away from the focal stream (left of the white arrow in Figure 8(d)). The normalized intensity values were fitted to an exponential function of the form , where A and B were constants with values of A = 4.503 ans B = −0.01529, to a good coefficient of determination, R2 = 0.9582. Note that the points within the first 100 μm were discarded because there were a few nuclei in that zone that reached maximum saturation intensities. From n = 5 myofibers that were focally targeted with Hoechst dye, we observed a distance dependence of myonuclei staining fluorescence intensity with respect to the source focal channel (Figure S4 (Ref. 66)). One limitation of the myofiber system was that, unlike the myoblast/myotube system, there was no chemical surface modification to confine cells only in trenches. The myofiber system was based on physical trapping of relatively large and non-adherent myofibers. We observed that cellular and tissue debris (e.g., fibroblasts and dead myofibers) from the myofiber isolation process at times clogged the vacuum channels, altering the flow and stream widths, and reducing the number of functional channels. Improvements in the purification protocol of myofibers should mitigate this problem. Overall, our finding suggests that the open-chamber flow-focused microfluidic device can generate soluble gradients along the length of live primary myofibers, with potential applications in investigating satellite cell migration and differentiation within their native microenvironment.
IV. CONCLUSIONS
We have developed a microfluidic platform for the focal delivery of soluble factors to micropatterned cells that operates within an open architecture. With the open chamber, cells can be seeded, cultured, and differentiated with the straightforward ease of use of a conventional petri dish. The device operates on the principle of hydrodynamic focusing, with the ability to generate focused streams (36–187 μm wide) in the open space. Cells and muscle fibers can be confined within trench or trap geometries, respectively, that allow focal stimulation and provide physiological mimicry. The PDMS trenches can be selectively micropatterned using an epoxy-silane surface modification strategy that facilitates long-term cell culture and differentiation on-chip. FEM simulations of local concentration correlated strongly with experimental results, and local velocity simulations further showed that the trenches shielded cells from high shear stresses while still maintaining ∼22% of inlet concentration within the open chamber. We demonstrated focal delivery of calcein AM and Hoechst nuclear stain to cells. Finally, we showed generation of in situ gradients of Hoechst nuclear stain on live myofibers. We envision that our open-chamber flow-focusing microfluidic concept will be useful for cell manipulation applications in muscle and stem cell biology.
ACKNOWLEDGMENTS
This work was kindly supported by the National Institutes of Health/National Institute of Biomedical Imaging and Bioengineering (Grant No. R01 EB007526-01). We thank Philip Lindstedt and Ross Boitano for their help with device fabrication and image analysis, and Rainer Ng and Lisa Horowitz for donating mice and providing training on myofiber dissections.
References
- 1. Yarmush M. L. and King K. R., Annu. Rev. Biomed. Eng. 11(1), 235 (2009). 10.1146/annurev.bioeng.10.061807.160502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Young E. W. K. and Beebe D. J., Chem. Soc. Rev. 39(3), 1036 (2010). 10.1039/b909900j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li N., Tourovskaia A., and Folch A., Crit. Rev. Biomed. Eng. 31(5–6), 423 (2003). 10.1615/CritRevBiomedEng.v31.i56.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Folch A. and Toner M., Annu. Rev. Biomed. Eng. 2, 227 (2000). 10.1146/annurev.bioeng.2.1.227 [DOI] [PubMed] [Google Scholar]
- 5. Chen C. S., Science 276(5317), 1425 (1997). 10.1126/science.276.5317.1425 [DOI] [PubMed] [Google Scholar]
- 6. Paguirigan A. L. and Beebe D. J., BioEssays 30(9), 811 (2008). 10.1002/bies.20804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Keenan T. M. and Folch A., Lab Chip 8(1), 34 (2008). 10.1039/B711887B [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Healy K. E., Nat. Mater. 8(9), 700 (2009). 10.1038/nmat2524 [DOI] [PubMed] [Google Scholar]
- 9. Tavana H., Mosadegh B., and Takayama S., Adv. Mater. 22(24), 2628 (2010). 10.1002/adma.200904271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kaigala G. V., Lovchik R. D., and Delamarche E., Angew. Chem., Int. Ed. 51(45), 11224 (2012). 10.1002/anie.201201798 [DOI] [PubMed] [Google Scholar]
- 11. Dittrich P. S. and Manz A., Nat. Rev. Drug Discovery 5(3), 210 (2006). 10.1038/nrd1985 [DOI] [PubMed] [Google Scholar]
- 12. Scott A., Weir K., Easton C., Huynh W., Moody W. J., and Folch A., Lab Chip 13(4), 527 (2013). 10.1039/C2LC40826K [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Croushore C. A. and Sweedler J. V., Lab Chip 13(9), 1666 (2013). 10.1039/c3lc41334a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chang T. C., Mikheev A. M., Huynh W., Monnat J., Raymond J., Rostomily R. C., and Folch A., Lab Chip 14, 4540 (2014). 10.1039/C4LC00642A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Klauke N., Smith G., and Cooper J. M., Anal. Chem. 81(15), 6390 (2009). 10.1021/ac9008429 [DOI] [PubMed] [Google Scholar]
- 16. Tourovskaia A., Kosar T. F., and Folch A., Biophys. J. 90(6), 2192 (2006). 10.1529/biophysj.105.074864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tourovskaia A., Li N., and Folch A., Biophys. J. 95(6), 3009 (2008). 10.1529/biophysj.107.128173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beebe D. J., Mensing G. A., and Walker G. M., Annu. Rev. Biomed. Eng. 4(1), 261 (2002). 10.1146/annurev.bioeng.4.112601.125916 [DOI] [PubMed] [Google Scholar]
- 19. Squires T. M. and Quake S. R., Rev. Mod. Phys. 77(3), 977 (2005). 10.1103/RevModPhys.77.977 [DOI] [Google Scholar]
- 20. Takayama S., Ostuni E., LeDuc P., Naruse K., Ingber D. E., and Whitesides G. M., Chem. Biol. 10(2), 123 (2003). 10.1016/S1074-5521(03)00019-X [DOI] [PubMed] [Google Scholar]
- 21. Takayama S., Ostuni E., LeDuc P., Naruse K., Ingber D. E., and Whitesides G. M., Nature 411(6841), 1016 (2001). 10.1038/35082637 [DOI] [PubMed] [Google Scholar]
- 22. Bhattacharjee N., Li N., Keenan T. M., and Folch A., Integr. Biol. 2(11–12), 669 (2010). 10.1039/c0ib00038h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Walker G. M., Zeringue H. C., and Beebe D. J., Lab Chip 4(2), 91 (2004). 10.1039/b311214d [DOI] [PubMed] [Google Scholar]
- 24. Tourovskaia A., Figueroa-Masot X., and Folch A., Nat. Protoc. 1(3), 1092 (2006). 10.1038/nprot.2006.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Su X., Theberge A. B., January C. T., and Beebe D. J., Anal. Chem. 85(3), 1562 (2013). 10.1021/ac3027228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paguirigan A. L. and Beebe D. J., Integr. Biol. 1(2), 182 (2009). 10.1039/b814565b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meyvantsson I. and Beebe D. J., Annu. Rev. Anal. Chem. 1(1), 423 (2008). 10.1146/annurev.anchem.1.031207.113042 [DOI] [PubMed] [Google Scholar]
- 28. Yin H., Zhang X., Pattrick N., Klauke N., Cordingley H. C., Haswell S. J., and Cooper J. M., Anal. Chem. 79(18), 7139 (2007). 10.1021/ac071146k [DOI] [PubMed] [Google Scholar]
- 29. Garanich J. S., Am. J. Physiol.: Heart Circ. Physiol. 288(5), H2244 (2005). 10.1152/ajpheart.00428.2003 [DOI] [PubMed] [Google Scholar]
- 30. Joanne Wang C., Li X., Lin B., Shim S., Ming G.-l., and Levchenko A., Lab Chip 8(2), 227 (2008). 10.1039/b713945d [DOI] [PubMed] [Google Scholar]
- 31. Cate D. M., Sip C. G., and Folch A., Biomicrofluidics 4(4), 044105 (2010). 10.1063/1.3490784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sip C. G., Bhattacharjee N., and Folch A., Lab Chip 14(2), 302 (2014). 10.1039/C3LC51052B [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lovchik R. D., Bianco F., Tonna N., Ruiz A., Matteoli M., and Delamarche E., Anal. Chem. 82(9), 3936 (2010). 10.1021/ac100771r [DOI] [PubMed] [Google Scholar]
- 34. Jowhar D., Wright G., Samson P. C., Wikswo J. P., and Janetopoulos C., Integr. Biol. 2(11–12), 648 (2010). 10.1039/c0ib00110d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Keenan T. M., Hsu C.-H., and Folch A., Appl. Phys. Lett. 89(11), 114103 (2006). 10.1063/1.2345914 [DOI] [Google Scholar]
- 36. Juncker D., Schmid H., and Delamarche E., Nat. Mater. 4(8), 622 (2005). 10.1038/nmat1435 [DOI] [PubMed] [Google Scholar]
- 37. Qasaimeh M. A., Ricoult S. G., and Juncker D., Lab Chip 13(1), 40 (2013). 10.1039/C2LC40898H [DOI] [PubMed] [Google Scholar]
- 38. Kaigala G. V., Lovchik R. D., Drechsler U., and Delamarche E., Langmuir 27(9), 5686 (2011). 10.1021/la2003639 [DOI] [PubMed] [Google Scholar]
- 39. Cosson S., Allazetta S., and Lutolf M. P., Lab Chip 13(11), 2099 (2013). 10.1039/c3lc50219h [DOI] [PubMed] [Google Scholar]
- 40. Regenberg B., Kruhne U., Beyer M., Pedersen L. H., Simon M., Thomas O. R. T., Nielsen J., and Ahl T., Lab Chip 4(6), 654 (2004). 10.1039/B409141H [DOI] [PubMed] [Google Scholar]
- 41. Chabert M. and Viovy J.-L., Proc. Natl. Acad. Sci. U. S. A. 105(9), 3191 (2008). 10.1073/pnas.0708321105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Knight J. B., Vishwanath A., Brody J. P., and Austin R. H., Phys. Rev. Lett. 80(17), 3863 (1998). 10.1103/PhysRevLett.80.3863 [DOI] [Google Scholar]
- 43. Wang F., Wang H., Wang J., Wang H.-Y., Rummel P. L., Garimella S. V., and Lu C., Biotechnol. Bioeng. 100(1), 150 (2008). 10.1002/bit.21737 [DOI] [PubMed] [Google Scholar]
- 44. Liu Y., Butler W. B., and Pappas D., Anal. Chim. Acta 743, 125 (2012). 10.1016/j.aca.2012.06.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen S. and Lee L. P., Integr. Biol. 2(2–3), 130 (2010). 10.1039/b919392h [DOI] [PubMed] [Google Scholar]
- 46. Golden J. P., Justin G. A., Nasir M., and Ligler F. S., Anal. Bioanal. Chem. 402(1), 325 (2012). 10.1007/s00216-011-5415-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jacobson S. C. and Ramsey J. M., Anal. Chem. 69(16), 3212 (1997). 10.1021/ac961093z [DOI] [Google Scholar]
- 48. Manbachi A., Shrivastava S., Cioffi M., Chung B. G., Moretti M., Demirci U., Yliperttula M., and Khademhosseini A., Lab Chip 8(5), 747 (2008). 10.1039/b718212k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hsu C.-H., Chen C., and Folch A., Lab Chip 4(5), 420 (2004). 10.1039/B404956J [DOI] [PubMed] [Google Scholar]
- 50. Donzel C., Geissler M., Bernard A., Wolf H., Michel B., Hilborn J., and Delamarche E., Adv. Mater. 13(15), 1164 (2001). [DOI] [Google Scholar]
- 51. Shefer G. and Yablonka-Reuveni Z., Methods Mol. Biol. 290, 281 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stiles T., Fallon R., Vestad T., Oakey J., Marr D. W. M., Squier J., and Jimenez R., Microfluid. Nanofluid. 1(3), 280 (2005). 10.1007/s10404-005-0033-z [DOI] [Google Scholar]
- 53. Walker G. M., Sai J., Richmond A., Stremler M., Chung C. Y., and Wikswo J. P., Lab Chip 5(6), 611 (2005). 10.1039/b417245k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhou J., Ellis A. V., and Voelcker N. H., Electrophoresis 31(1), 2 (2010). 10.1002/elps.200900475 [DOI] [PubMed] [Google Scholar]
- 55. Nam Y., Branch D. W., and Wheeler B. C., Biosens. Bioelectron. 22(5), 589 (2006). 10.1016/j.bios.2006.01.027 [DOI] [PubMed] [Google Scholar]
- 56. Moorcroft M. J., Nucleic Acids Res. 33(8), e75 (2005). 10.1093/nar/gni075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tourovskaia A., Figueroa-Masot X., and Folch A., Lab Chip 5(1), 14 (2005). 10.1039/b405719h [DOI] [PubMed] [Google Scholar]
- 58. Tan J. L., Liu W., Nelson C. M., Raghavan S., and Chen C. S., Tissue Eng. 10(5–6), 865 (2004). 10.1089/1076327041348365 [DOI] [PubMed] [Google Scholar]
- 59. Nejadnik M. R., Olsson A. L. J., Sharma P. K., van der Mei H. C., Norde W., and Busscher H. J., Langmuir 25(11), 6245 (2009). 10.1021/la9001169 [DOI] [PubMed] [Google Scholar]
- 60. Tenstad E., Tourovskaia A., Folch A., Myklebost O., and Rian E., Lab Chip 10(11), 1401 (2010). 10.1039/b926738g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kummer T. T., J. Cell Biol. 164(7), 1077 (2004). 10.1083/jcb.200401115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gilbert P. M., Corbel S., Doyonnas R., Havenstrite K., Magnusson K. E. G., and Blau H. M., Integr. Biol. 4(4), 360 (2012). 10.1039/c2ib00148a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dhawan J. and Rando T. A., Trends Cell Biol. 15(12), 666 (2005). 10.1016/j.tcb.2005.10.007 [DOI] [PubMed] [Google Scholar]
- 64. Siegel A. L., Atchison K., Fisher K. E., Davis G. E., and Cornelison D. D. W., Stem Cells 27(10), 2527 (2009). 10.1002/stem.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hawke T. J. and Garry D. J., J. Appl. Physiol. 91(2), 534–551 (2001). [DOI] [PubMed] [Google Scholar]
- 66.See supplementary material at http://dx.doi.org/10.1063/1.4946801E-BIOMGB-10-034602 for fabrication and device assembly methods, micropatterning, and myofiber gradient data, including Figures S1–S4.
- 67. Cheng J., Chang T., Bhattacharjee N., and Folch A., in Proceedings of The 15th International Conference on Miniaturized Systems for Chemistry and Life Sciences, Seattle, 2–6 October 2011, pp. 356–358. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1063/1.4946801E-BIOMGB-10-034602 for fabrication and device assembly methods, micropatterning, and myofiber gradient data, including Figures S1–S4.