Abstract
Pir proteins are unique proteins with internal repeat sequences that are reported to be present in the cell wall of Saccharomyces cerevisiae. They are covalently attached to the cell wall and can be released by mild alkali treatment. In this study the biotinylated cell wall preparations from Candida albicans and S. cerevisiae were extracted by alkali and β-1,3 glucanase and analyzed in parallel. Among the four bands detected by streptavidin, two proteins were recognized by the antibody to the S. cerevisiae Pir protein Hsp150. The antibody also detected a high molecular mass protein secreted in the growth medium of C. albicans. Using S. cerevisiae HSP150/PIR2 gene as a probe, Southern and Northern hybridizations were performed with DNA and RNA of C. albicans. Hybridization with DNA digested with different restriction enzymes showed more than one hybridized fragment. An increased level of mRNA was found in heat shocked cells (37°C for 45 min compared to 25°C). Hybridization of ScHSP150 gene to mRNAs from cells grown in different media was also determined. Two transcripts of size approximately 3.5 kb and 2.0 kb were detected in mRNAs from cells grown in defined medium with glucose as carbon source or in the same medium supplemented with hemoglobin. The lower transcript of size 2.0 kb was absent in cells grown in medium with galactose as carbon source. A single band was also observed when cells were grown in rich medium. Together these results demonstrated the existence of β1,3 glucan linked proteins in C. albicans, which are related to Pir family proteins of S. cerevisiae.
Keywords: Candida albicans, Saccharomyces cerevisiae, Cell wall, PIR family protein
1. Introduction
Candida albicans is a commensal organism as well as one of the infectious agents commonly found in immunocompromised patients. The cell wall of C. albicans is considered to play a fundamental role in host interactions. The cell wall structure has been extensively studied in Saccharomyces cerevisiae, which is likely to be a model for C. albicans. The yeast cell wall is a complex structure whose major components are β-1,3 and β-1,6 glucan and a number of mannoproteins attached to the glucan network [1–3]. Non-covalently linked cell wall proteins (CWPs) are extractable with hot sodium dodecyl sulfate (SDS) [4,5]. The remaining proteins are extractable with β-1,3-glucanase or with mild alkali suggesting that they are covalently incorporated in the cell wall through a glucosidic linkage [5–7]. In S. cerevisiae two types of CWPs have been identified that are covalently linked to β-glucan namely glycosyl phosphatidyl inositol (GPI)-dependent CWPs and PIR (proteins with internal repeats) CWPs [5,8]. The GPI-CWPs are linked to the wall by β-1,6 glucan, which is coupled to β-1,3 glucan [8,9]. They can be released from cell wall by β-1,3 glucanase digestion. Some GPI-CWPs are resistant to β-1,3 glucanase since their β-1,6 glucan moiety is cross linked not only to β-1,3 glucan, but also to chitin [8,9]. Pir proteins are highly O-glycosylated, have one or more repeats but do not contain a GPI anchor [5,10]. They can be liberated from the cell wall by mild alkali extraction [5]. The Pir proteins have been immunolocalized to the cell wall, where they were found to confer resistance to plant antifungal protein, osmotin [11]. One of the Pir proteins, Hsp150, is regulated by heat as well as by nutrient stress. Using the HSP150 gene (PIR2) as well as antibody to its product from S. cerevisiae as probes we examined the members of CWPs in C. albicans. In this study, we report evidence that demonstrates the presence of Pir family proteins in the cell wall of C. albicans.
2. Materials and methods
2.1. Microbial strains and growth conditions
S. cerevisiae INVSC1(MATa/α his3Δ1 leu2 trp1-289 ura3-52) (Invitrogen, Carlsbad, CA, USA) and C. albicans NCPF 3153 were routinely grown in YPD (1% yeast extract, 2% bacto peptone, 2% glucose) with shaking at the selected temperatures. For specific experiments, cells were also cultured in 4×YNB (yeast nitrogen base with amino acids, Difco Laboratories, Detroit, MI, USA) with 500 mM galactose, 50 mM glucose or 50 mM glucose plus 0.1% (w/v) hemoglobin (Sigma Chemical Co., St. Louis, MO, USA).
2.2. Biotin labeling of CWPs
Yeast cells were harvested at mid-exponential phase and washed twice with 50 mM Tris–HCl, pH 7.4, and cells were resuspended in the same buffer containing SulfoNHS-LC biotin reagent (Pierce, Rockford, IL, USA) (1 mg ml−1) and incubated for 90 min on ice. Cells were washed repeatedly with Tris–HCl, pH 7.4 to remove the excess SulfoNHS-LC biotin. The biotin derivative that labels the CWP does not permeate the cell membrane [12]. The labeled cells were resuspended in Tris–HCl, pH 7.4 that contained 2 mM phenylmethylsulfonyl fluoride (PMSF) and broken by a homogenizer, with glass beads. Cells were chilled by liquid CO2 pulse during homogenization. The proportion of broken cells was determined by light microscopy. The slurry was centrifuged and the cell wall pellet was washed repeatedly with the above- mentioned buffer. Cell walls were treated with Laemmli SDS–mercaptoethanol buffer [13] for 10 min in a boiling water bath and washed 4–6 times with Tris–HCl buffer.
2.3. Extraction of biotinylated CWP, electrophoresis and blotting
Laemmli buffer treated cell walls were resuspended in Tris–HCl buffer, pH 7.4 and incubated with Quantazyme (Quantum Biotechnologies, Montreal, Que., Canada) 600 U g−1 wet weight of cell wall for 16 h at 37°C or with 30 mM NaOH at 4°C. Quantazyme is a recombinant β-1,3 glucanase which releases β-1,6 linked proteins [7]. The alkali treatment was stopped with acetic acid. Alkali or enzyme treated cell wall extracts were centrifuged and the clear supernatants containing CWP were used for further analysis. CWP were separated by SDS–PAGE using linear (4–20%) polyacrylamide gradient gels and transferred to nitrocellulose membranes. Membranes were blocked in 0.1% Nonidet P-40 (NP-40) in Tris buffered saline (TBS), pH 7.5 for 1 h at room temperature and then probed with ExtrAvidin-peroxidase (Sigma) diluted 1:10 000 in the same buffer with 0.1% bovine serum albumin (BSA) for 1 h at room temperature. After washing with 0.1% NP-40 in TBS four times for 5 min, the blots were developed with SuperSignal substrates for Western blotting (Pierce, Rockford, IL, USA). For Western analysis, the membranes were blocked in 4% milk in TBST (TBS+Tween-20, 0.05%) and then incubated for 1 h in TBST+1% BSA containing antibody to S. cerevisiae Hsp150, a generous gift of Dr. Marja Makarow (University of Helsinki, Finland), at a dilution of 1:1000. After three washes in TBST, the membranes were incubated for 1 h in TBST+1% BSA containing goat anti-rabbit IgG peroxidase at a dilution of 1:5000, washed with TBST and developed with solution containing H2O2 and 4-chloronaphthol.
2.4. Southern blot hybridization analysis
Genomic DNAs were prepared from C. albicans and S. cerevisiae by standard procedures [14]. C. albicans DNA was digested with different restriction enzymes and separated by 0.8% agarose gel electrophoresis, and transferred onto positive nylon membranes (Hybond N+, Amersham Pharmacia Biotech, Piscataway, NJ, USA) as described previously [15,16]. The coding region of the HSP150 gene of S. cerevisiae was amplified by PCR using gene specific synthetic oligonucleotides annealing to the stop and the start codon regions. ScHSP150 DNA was separated on agarose gel and purified before labeling with 32P (random priming kit, High Prime, Boehringer Mannheim Biochemicals, Indianapolis, IN, USA). Unincorporated nucleotides were removed by a desalting column (BioRad, Hercules, CA, USA). Filters were prehybridized for at least 2 h at 42°C in Hybrisol I solution containing 50% formamide (Intergen Company, Purchase, NY, USA). The 32P random primed DNA was added to the hybridization mixture and hybridized overnight at 42°C. Filters were washed twice (30 min each) in 2×SSC, 1% SDS at 50°C and once at room temperature for 30 min then exposed to X-ray films.
2.5. Northern blot analysis
For heat shock treatment, exponentially growing C. albicans yeast cells in YPD medium were transferred to fresh medium and subjected to heat shock at different temperatures for various time intervals. Total RNA of C. albicans was extracted by the hot acidic phenol method [17] from cells grown under different growth conditions. Using Oligo-dT cellulose (Amersco, Solon, OH, USA) the mRNAs were purified from total RNA. The RNAs were quantified by absorption (OD260) and separated by denaturing agarose electrophoresis. Following electrophoresis in formaldehyde containing 1% agarose gels, the RNAs were transferred to nylon membranes and hybridized with 32P labeled random primed HSP150 gene of S. cerevisiae. The actin (ACT1) gene from C. albicans was PCR amplified and used as a control to determine equivalent RNA loading.
3. Results and discussion
3.1. Analysis of CWPs of C. albicans in comparison with S. cerevisiae
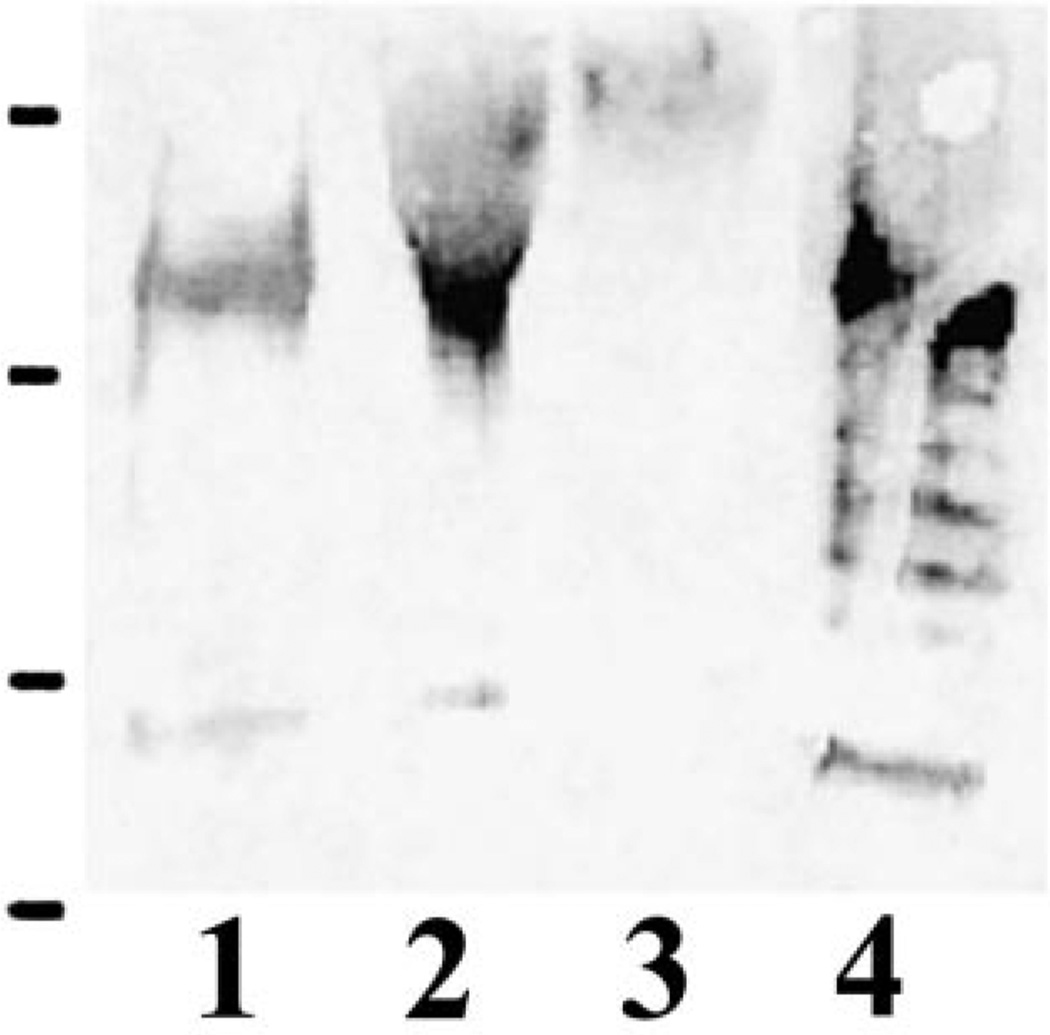
Recent studies [18,19] on C. albicans cell wall showed strong evidence for the existence of a family of GPI-CWPs. The cell wall of C. albicans contains several proteins that like the Pir CWPs in S. cerevisiae can be released by mild alkali [20] suggesting that also in C. albicans a Pir CWP→β-1,3 glucan-like complex might exist. Kapteyn and coworkers [7] showed the presence of proteins bound to β-1,3/β-1,6 glucan polymers in the cell walls of C. albicans and suggested that these polymers might be responsible for retaining CWPs. In the present study, the C. albicans and S. cerevisiae CWPs were labeled by NHS-LC biotin. The cell walls were then treated with SDS under reduced conditions to remove the non-covalently linked CWPs. The remaining proteins (most likely covalently linked) were extracted by Quantazyme or with 30 mM NaOH. Four bands corresponding to proteins sized approximately 150, 66, 54, and 45 kDa could be detected in NaOH and Quantazyme extracts of C. albicans by streptavidin (Fig. 1).
Fig. 1.
Analysis of biotinylated C. albicans CWPs. Biotinylated cell wall preparations were treated with hot SDS and subsequently digested either with Quantazyme (lane 1) or with 30 mM NaOH (lane 2). The extracts were separated by SDS–PAGE (5–20%), blotted and developed with ExtrAvidin-peroxidase conjugate. Protein molecular mass standards run in parallel (in kDa from the top 250, 150, 100, 75, 50, 37, 20, 15, and 10, Precision protein standards, Bio-Rad) are indicated on the left.
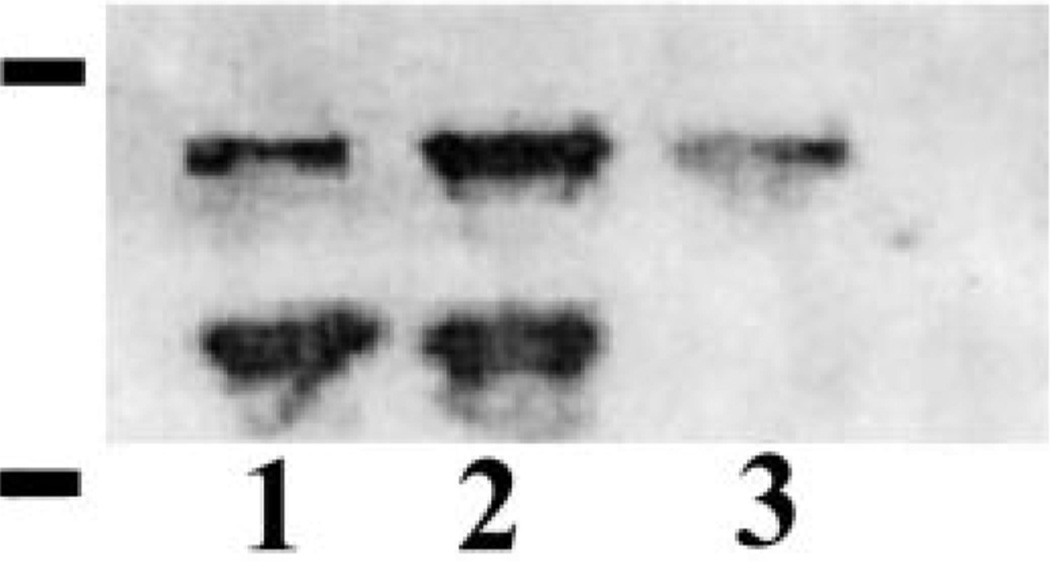
For Western blot analysis cell wall extractions by Quantazyme and NaOH treatment from C. albicans and S. cerevisiae and C. albicans culture medium were separated by SDS–PAGE and transferred to nitrocellulose membrane. Among the four protein bands identified by streptavidin in C. albicans, two were recognized by the antibody to S. cerevisiae Hsp150 (Fig. 2). They were the 150-kDa band that corresponds in size to Pir2/Hsp150 protein [5] of S. cerevisiae and the 66-kDa protein band. Since all the Pir family proteins are highly homologous the cross reactivity of ScHsp150 antibody to more than one Pir family protein is possible. In S. cerevisiae, Yun et al. [11] have reported the cross reactivity of Pir 3 polyclonal antibody with Pir1 and Pir2 proteins in which 75 and 79.5% of their amino acids were identical to Pir3 respectively. In the culture medium of C. albicans the antibody recognized a high molecular mass broad band. Among the Pir family proteins the Pir2/Hsp150 is known to be secreted into the growth medium. These observations supported that CWPs, which belong to the Pir family, are present in C. albicans and some of them are secreted into the medium.
Fig. 2.
Western blot analysis of C. albicans cell wall extracts and culture medium. The cell wall extracts of C. albicans obtained by NaOH (lane 1), Quantazyme (lane 2), culture medium of C. albicans (lane 3) and Quantazyme digest of S. cerevisiae were separated by SDS–PAGE (5–20%) and probed with ScHsp150 antibody. Protein molecular mass standards run in parallel (in kDa from the top 209, 124, 80, 49, and 34, Pre-stained SDS–PAGE standard, Bio-Rad) are indicated on the left.
3.2. Southern and Northern blot analyses
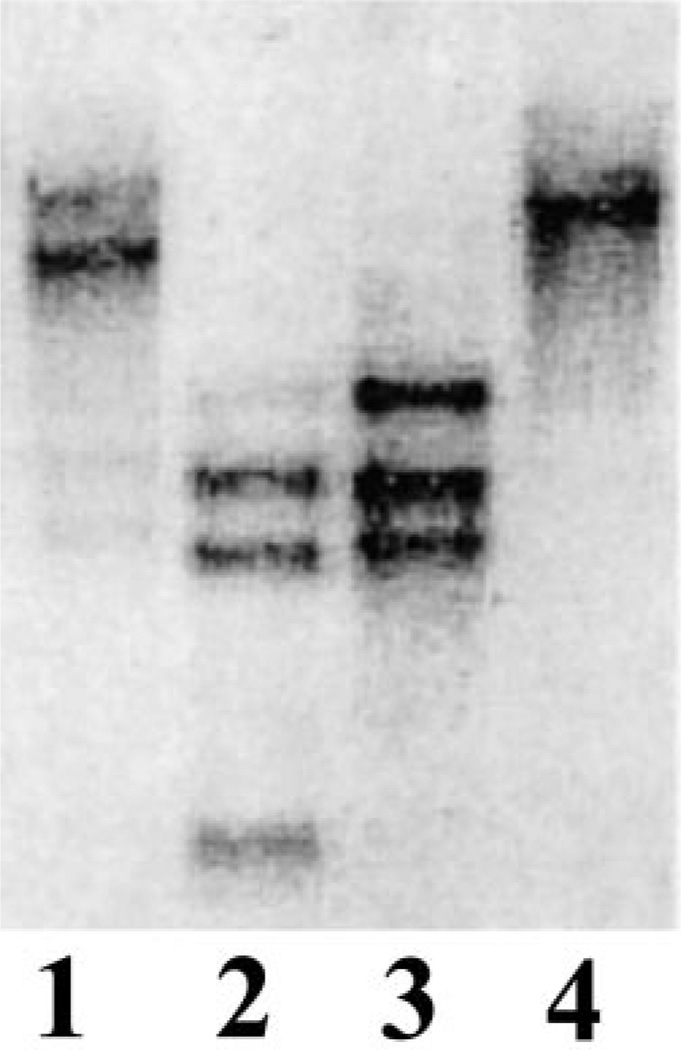
The Southern blot analysis in which genomic DNA of C. albicans was digested with various restriction endonucleases and probed with ScHSP150 DNA under high stringency condition showed more than one hybridized fragment (Fig. 3). Quantitative Northern analysis was performed on mRNA prepared from C. albicans yeast cells grown in different media using 32P labeled ScHSP150 DNA as a probe. Two transcripts of size approximately 3.5 and 2.0 kb were detected in the mRNA from cells grown in 4×YNB medium with glucose as a carbon source or in the same medium supplemented with 0.1% hemoglobin (Fig. 4). The smaller transcript (2 kb) was absent in the cells grown with galactose as a carbon source. Growth of C. albicans on galactose rather than glucose or in the presence of hemoglobin is reported to alter the profile of the CWPs [21,22]. A variety of nutritional conditions have been reported to affect the overall composition of the cell wall. In the absence of a suitable carbon source, S. cerevisiae cells activate the sporulation pathway accompanied by the up and down regulation of various CWP encoding genes [23]. The Southern and Northern analyses showed that there might be more than one gene related to the Pir family present in C. albicans and their expression may differ in different growth media. In this case an alteration in the number of transcripts present in C. albicans mRNA was observed when the organism was grown on different carbon source.
Fig. 3.
Southern analysis of genomic DNA from C. albicans. Membrane was probed with 32P labeled ScHSP150 DNA at 42°C in a hybridization solution containing 50% formamide. Enzymes used to digest genomic DNAs were HindIII (lane 1), HincII (lane 2), EcoRI (lane 3), and BamHI (lane 4).
Fig. 4.
Northern analysis of mRNA from C. albicans cells. Yeast cells grown in 4×YNB with 50 mM glucose (lane 1), with 50 mM glucose+0.1% hemoglobin (lane 2), or with 500 mM galactose (lane 3). 32P labeled ScHSP150 DNA was used as probe at 42°C in a hybridization solution containing 50% formamide. RNA standard run in parallel is indicated on the left (from the top, 4.4 and 2.37 kb).
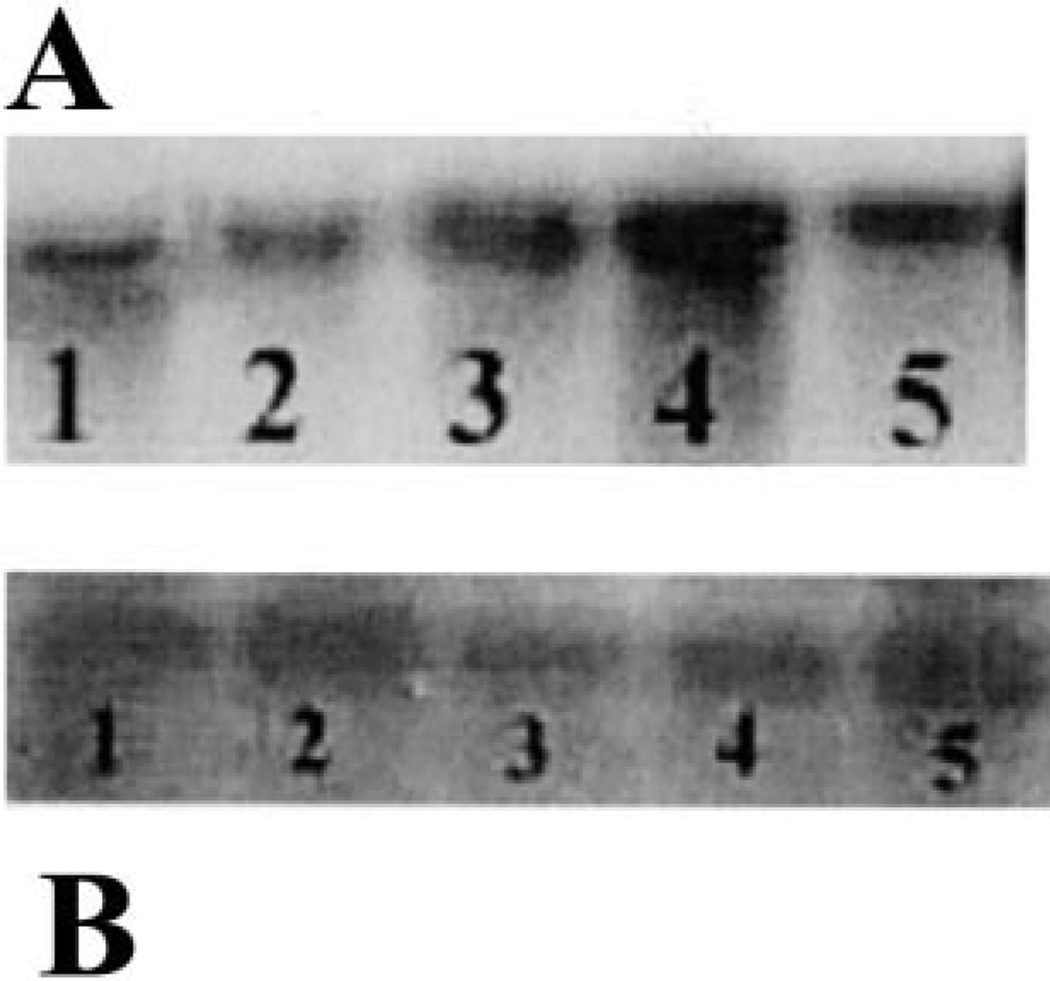
Among all the Pir family proteins reported in S. cerevisiae, the Hsp150 is the only secretory protein that is heat shock inducible [24,25]. In this study, we observed the reactivity of ScHsp150 antibody with the secretory protein of higher molecular mass in the growth medium of C. albicans. Hence, we decided to monitor the RNA transcription level in the heat shocked cells of C. albicans using 32P labeled ScHSP150 DNA as a probe. The total RNA was prepared from C. albicans yeast cells grown in YPD at 24°C that were heat shocked at 37°C and 45°C for 30 or 45 min. A single band of size around 3 kb was observed after hybridization and the smaller transcript is absent here also as in the mRNA from galactose supplemented cells. The RNA transcript level was increased in cells heat shocked at 37°C for 45 min compared to all the other conditions as well as in control at 24°C (Fig. 5).
Fig. 5.
The expression of the homologue of the ScHSP150 gene in C. albicans cells exposed to temperature upshift. A: Total RNA was extracted from cells treated at 24°C before (lane 5) and after the shift of the temperature to 37°C for 30 min (lane 3), 45 min (lane 4) and 45°C for 30 min (lane 1) and 45 min (lane 2). Probe and hybridization conditions were the same as in Fig. 4. B: Actin message after hybridization of the same blot probed with the actin gene (ACT1) of C. albicans.
In summary, we have analyzed the CWPs of C. albicans in several ways to assess the presence of Pir family related proteins. The protein analysis using ScHsp150 antibody confirmed their presence in the cell wall as well as in the growth medium. Southern hybridization showed the existence of Pir family genes in C. albicans and the Northern hybridization results suggested that one of them might be related to the heat shock inducible HSP150 gene of S. cerevisiae. Future studies may be focused on isolation and characterization of these genes and their specific role in the human pathogen C. albicans.
After the submission of this manuscript, Kapteyn et. al. [26], using Western analysis, reported that at least two Pir2 homologues were differentially expressed in the cell wall of C. albicans.
Acknowledgments
We thank Dr. Marja Makarow for kindly giving us the antibody of Saccharomyces cerevisiae Pir protein Hsp150.
References
- 1.Klis FM. Review: cell wall assembly in yeast. Yeast. 1994;10:851–869. doi: 10.1002/yea.320100702. [DOI] [PubMed] [Google Scholar]
- 2.Kollar R, Reinhold BB, Petrakova E, Yeh HJC, Ashwell G, Draonova J. Architecture of the yeast cell wall: β-1,6 glucan interconnects mannoproteins, β-1,3 glucan and chitin. J. Biol. Chem. 1997;272:17762–17775. doi: 10.1074/jbc.272.28.17762. [DOI] [PubMed] [Google Scholar]
- 3.Orlean P. Biogenesis of yeast wall and surface components. In: Pringle JR, Broach JR, Jones EW, editors. Molecular and Cellular Biology of the Yeast Saccharomyces. Vol. 3, Cell Cycle and Cell Biology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 229–362. [Google Scholar]
- 4.Valentin E, Herrero W, Pastor JFI, Sentandreu R. Solubilization and analysis of mannoprotein molecules from the cell wall of Saccharomyces cerevisiae . J. Gen. Microbiol. 1984;130:1419–1428. [Google Scholar]
- 5.Mrsa V, Seidl T, Gentzsch M, Tanner W. Specific labeling of cell wall proteins by biotinylation. Identification of four covalently linked O-mannosylated proteins of Saccharomyces cerevisiae . Yeast. 1997;13:1145–1154. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1145::AID-YEA163>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 6.Van Rinsum J, Klis FM, Van Den Ende H. Cell wall glucomannoproteins of S. cerevisiae mnn9. Yeast. 1991;7:717–726. doi: 10.1002/yea.320070707. [DOI] [PubMed] [Google Scholar]
- 7.Kapteyn JC, Montijn RC, Dijkgraf GJP, Van Den Ende H, Klis FM. Covalent association of β-1,3 glucan with β-1,6 glycosylated mannoproteins in cell walls of C. albicans . J. Bacteriol. 1995;177:3788–3792. doi: 10.1128/jb.177.13.3788-3792.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kapteyn JC, Montijn RC, Vink R, de la Cruz J, Llobell A, Douwes JE, Shimoi H, Lipke PN, Klis FM. Retention of Saccharomyces cerevisiae cell wall proteins through a phosphodiester-linked β-1,3/β1-6 glucan heteropolymer. Glycobiology. 1996;6:337–345. doi: 10.1093/glycob/6.3.337. [DOI] [PubMed] [Google Scholar]
- 9.Kapteyn JC, Van Egmond P, Sievi E, Van Den Ende H, Makarow M, Klis FM. The contribution of the O-glycosylated protein Pir2/Hsp150 to the construction of the yeast cell wall in wild type cells and β-1-6 glucan deficient mutants. Mol. Microbiol. 1999;31:1835–1844. doi: 10.1046/j.1365-2958.1999.01320.x. [DOI] [PubMed] [Google Scholar]
- 10.Toh-E A, Yasunaga S, Nisogi K, Oguchi T, Matsui Y. Three yeast genes PIR1, PIR2 and PIR3 containing internal random repeats, are related to each other, and PIR1 and PIR2 are required for heat tolerance to heat shock. Yeast. 1993;9:481–494. doi: 10.1002/yea.320090504. [DOI] [PubMed] [Google Scholar]
- 11.Yun DJ, Zhoa Y, Pardo JM, Narasimhan ML, Dasz B, Lee H, Abad LR, D'Urzo MP, Hasegawa PM, Bressan RA. Stress proteins on the yeast cell surface determine resistance to osmotin, a plant antifungal protein. Proc. Natl. Acad. Sci. USA. 1997;94:7082–7087. doi: 10.1073/pnas.94.13.7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole SR, Ashman LK, Ev PL. Biotinylation: an alternative to radio iodination for the identification of cell surface antigens in immunoprecipitates. Mol. Immunol. 1987;24:699–705. doi: 10.1016/0161-5890(87)90051-4. [DOI] [PubMed] [Google Scholar]
- 13.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 14.Treco DD. In: Current Protocols in Molecular Biology, Section IV: Preparation of Yeast DNA, RNA, and Proteins; Unit 13.1: Preparation of Yeast DNA. Ausubel FM, et al., editors. New York: Green/Wiley; 1989. [Google Scholar]
- 15.Glover DM. DNA Cloning: A Practical Approach. Oxford: IRL Press; 1985. [Google Scholar]
- 16.Perbal B. A Practical Guide to Molecular Cloning. New York: Wiley-Interscience; 1988. [Google Scholar]
- 17.Wise JA. Preparation and analysis of low molecular mass RNAs and small ribonucleoproteins. Methods Enzymol. 1991;194:405–415. doi: 10.1016/0076-6879(91)94031-7. [DOI] [PubMed] [Google Scholar]
- 18.Hoyer LL, Payne TL, Hecht JE. Identification of C. albicans ALS2 and ALS4 and localization of Als proteins to the fungal cell surface. J. Bacteriol. 1998;180:5334–5343. doi: 10.1128/jb.180.20.5334-5343.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoyer LL, Payne TL, Bell M, Myers AM, Scherer S. C. albicans ALS3 and insights into the nature of the ALS gene family. Curr. Genet. 1998;33:451–459. doi: 10.1007/s002940050359. [DOI] [PubMed] [Google Scholar]
- 20.Mormeneo S, Marcilla A, Iranzo M, Sentandreu R. Structural mannoproteins released by β-elimination from C. albicans cell walls. FEMS Microbiol. Lett. 1994;123:131–136. doi: 10.1111/j.1574-6968.1994.tb07212.x. [DOI] [PubMed] [Google Scholar]
- 21.Yan S, Rodrigues RG, Cahn-Hidalgo D, Walsh TJ, Roberts DD. Hemoglobin induces binding of several extracellular matrix proteins to Candida albicans. Identification of a common receptor for fibronectin, fibrinogen, and laminin. J. Biol. Chem. 1998;273:5638–5644. doi: 10.1074/jbc.273.10.5638. [DOI] [PubMed] [Google Scholar]
- 22.McCourtie J, Douglas L. Relationship between cell surface composition of Candida albicans and adherence to acrylic after growth on different carbon sources. Infect. Immun. 1981;32:1234–1241. doi: 10.1128/iai.32.3.1234-1241.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional programme of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 24.Russo P, Kalkkinen N, Sareneva H, Paakkola J, Makarow M. A heat shock gene from Saccharomyces cerevisiae encoding a secretory glycoprotein. Proc. Natl. Acad. Sci. USA. 1992;89:3671–3675. doi: 10.1073/pnas.89.9.3671. Correction 89, 8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russo P, Simonen M, Uimari A, Teesalu T, Makarow M. Dual regulation of heat and nutrient stress of the yeast HSP150 gene encoding a secretory glycoprotein. Mol. Gen. Genet. 1993;239:273–280. doi: 10.1007/BF00281628. [DOI] [PubMed] [Google Scholar]
- 26.Kapteyn JC, Hoyer LL, Hecht JE, Muller WH, Andel A, Verkleij AJ, Makarow M, Van Den Ende H, Klis FM. The cell wall architecture of Candida albicans wild-type cells and cell wall-defective mutants. Mol. Microbiol. 2000;35:601–611. doi: 10.1046/j.1365-2958.2000.01729.x. [DOI] [PubMed] [Google Scholar]