Abstract
Conformational states of the metastable drkN SH3 domain were characterized using single-molecule fluorescence techniques. Under nondenaturing conditions, two Förster resonance energy transfer (FRET) populations were observed that corresponded to a folded and an unfolded state. FRET-estimated radii of gyration and hydrodynamic radii estimated by fluorescence correlation spectroscopy of the two coexisting conformations are in agreement with previous ensemble x-ray scattering and NMR measurements. Surprisingly, when exposed to high concentrations of urea and GdmCl denaturants, the protein still exhibits two distinct FRET populations. The dominant conformation is expanded, showing a low FRET efficiency, consistent with the expected behavior of a random chain with excluded volume. However, approximately one-third of the drkN SH3 conformations showed high, nearly 100%, FRET efficiency, which is shown to correspond to denaturation-induced looped conformations that remain stable on a timescale of at least 100 μs. These loops may contain interconverting conformations that are more globally collapsed, hairpin-like, or circular, giving rise to the observed heterogeneous broadening of this population. Although the underlying mechanism of chain looping remains elusive, FRET experiments in formamide and dimethyl sulfoxide suggest that interactions between hydrophobic groups in the distal regions may play a significant role in the formation of the looped state.
Introduction
There is growing awareness that the biologically functional state of many proteins is not folded in a stable configuration, but instead contains one or more disordered regions. Although they lack a stable 3D fold, these intrinsically disordered proteins (IDPs) and regions generally exhibit fluctuating secondary and tertiary structures (1). IDPs are involved in cell signaling, molecular recognition, and transcriptional regulation, with a majority of oncogenic proteins having significant disordered regions (2, 3, 4). Many similarities exist between IDPs and the unfolded-state ensemble of globular proteins, as may be expected given that some IDPs undergo a disorder-to-order transition upon interaction with other proteins or upon posttranslational modification (4, 5). A more quantitative description of unfolded-state ensembles is much needed, not only from a protein-folding perspective, but also as a model or reference for IDPs. Since the 1960s, when Tanford showed that proteins in high concentrations of chemical denaturant obey a random-coil scaling law (6), the random-coil model has been widely used as the basis for understanding the physical properties of unfolded proteins. However, the validity of this model for describing the unfolded ensemble is limited, since significant residual structure was reported in several denatured proteins, such as apomyoglobin and staphylococcal nuclease (7), and local hydrophobic interactions restricting backbone motions have been shown to persist even under the harshest denaturing conditions (8).
Despite the biological importance of disordered/unfolded states of proteins, conventional experimental approaches for structural and thermodynamic characterization provide limited information and the nature of their transient structure remains elusive (9, 10). Computational methods have recently focused on the study of unfolded and disordered proteins (11, 12, 13), although the selection of the force field in the molecular simulations is critical and the most popular options seem to be biased toward describing the folded state. Recent computational studies point to an overestimation of the helical structure in small proteins and an underestimation of the size of denatured proteins compared to experimental findings (14). Hybrid computational-experimental approaches in which sets of conformers are selected to fit NMR and small-angle x-ray scattering (SAXS) experimental data have been developed (15), although powerful single-molecule fluorescence (SMF) data have yet to be incorporated to help provide a better evaluation and understanding of structural ensembles.
The N-terminal Src-homology 3 domain of Drosophila downstream of the receptor kinase (drkN SH3) is a 59-residue β-barrel consisting of five antiparallel β-strands (PDB: 2A36). It has a marginally stable structure, with the folded (Fexch) and unfolded (Uexch) states almost equally populated under nondenaturing conditions and interconverting at a slow exchange rate, ∼2 s−1 (16). NMR chemical shift, J-coupling, residual dipolar coupling, O2-induced paramagnetic shift, 15N relaxation, nuclear Overhauser effect (NOE), paramagnetic relaxation enhancement, and hydrodynamic radius (RH) measurements, as well as SAXS data, have been used to calculate structural ensembles for the Uexch (17). The data and the derived ensembles provide evidence for fluctuating structure in the Uexch, including significant nonnative α-helical secondary structure as well as both native-like and nonnative tertiary structure, in particular in the region of the central β-sheet of the folded state, which shows multiple contacts to the tryptophan at position 36. NMR and tryptophan fluorescence data point to significant differences between the unfolded state, Uexch, under nondenaturing conditions and the chemically denatured state and demonstrate that drkN SH3 is essentially fully unfolded in 2 M GdmCl (18, 19, 20).
SMF spectroscopy provides an elegant quantitative characterization of coexisting protein conformations (21), and it has become a powerful tool for studying the physical mechanism of protein folding (22, 23, 24). To describe and compare the conformations associated with the unfolded state and the chemically denatured state of the drkN SH3 domain, we performed SMF spectroscopy experiments such as fluorescence correlation spectroscopy (FCS) and single-molecule Förster resonance energy transfer (smFRET). The protein was found to preserve its conformational heterogeneity in the denatured state. Based on the single-molecule data, the hydrodynamic radius (RH), end-to-end distance (REE), and radius of gyration (RG) were estimated for different conformational states resolved for drkN SH3. The linear size and the topology of the denatured protein were interpreted using a coarse-grained, subensemble-based polymer model that accounts for excluded volume (25). The single-molecule results obtained in this study are compared with previous drkN SH3 ensemble data obtained by NMR and SAXS, and interpretations are proposed in terms of denaturant-induced intrachain interactions.
Materials and Methods
Materials
The fluorescent probes used for labeling the SH3 domain for SMF experiments were 5-carboxy-tetramethylrhodamine N-succinimidyl ester (TMR-NHS) (AnaSpec, Fremont, CA), Bodipy fluorescein (BFL) maleimide, Alexa 647 (A647) maleimide and Alexa 555 (A555) maleimide (ThermoFisher Scientific, Waltham, MA). Forty-basepair-long oligonucleotides labeled with Cy5 at the 5′ end and 6-FAM at the 3′ end were purchased from Integrated DNA Technologies (Coralville, IA). To prevent FRET between two conjugated dyes, the sequence chosen was Cy5-5′-TAA GCC TCG TCC TGC GTC GGA GCC CGT CTG CCA GCG GAAT-3′-6-FAM. A DNA sample was usually used as a detection volume calibration sample. Guanidinum chloride (GdmCl) (G9284, Sigma-Aldrich, St. Louis, MO) and urea (EM-9510, EMD Millipore, Billerica, MA) were used for protein denaturation. Immediately before use, urea was purified of hydrolysis products and ionic impurities as described previously (26). GdmCl and urea solutions were adjusted to pH 7.5 for all the experiments unless stated otherwise. Each GdmCl and urea concentration was also confirmed by measuring the solution viscosity by FCS. All other samples were diluted in Tris buffer (50 mM Tris and 150 mM NaCl, pH 7.5). Formamide (47671, Sigma-Aldrich), dimethyl sulfoxide (DMSO) (472301, Sigma-Aldrich), glycerol (EM-4750, EMD Millipore), sodium sulfate (238597, Sigma-Aldrich), and L-arginine (11009, Sigma-Aldrich) solutions were prepared in the Tris buffer.
Protein expression and purification
Detailed protein expression and purification procedures can be found in our previous articles (16, 22). Briefly, plasmid encoding the N-terminal SH3 domain of the drk protein (residues 1–59) with ampicillin resistance was transformed into BL21-CodonPlus (DE3)-RIL competent Escherichia coli cells. Bacterial cultures were grown overnight on a Lennox lysogeny broth (LB) (L7658, Sigma-Aldrich)-ampicillin agar plate in a 37°C incubator. A single colony was inoculated into LB medium with a 100 mg/L final concentration of ampicillin (AB0028, Bio Basic, Amherst, NY) and shaken in a 37°C incubator for ∼5 h until cell density reached an OD600 of 0.6. A 1 mM final concentration of isopropyl β-D-1-thiogalactopyranoside (IPTG) (IB0168, Bio Basic) was then added and the medium was transferred to a 16°C incubator for overnight growth up to a cell density having an OD600 of ∼1.3. Cells were lysed by sonication in a buffer containing 50 mM Tris-HCl, 2 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) (646547, Sigma-Aldrich), 1 mM EDTA (EDT111, BioShop Canada, Burlington, ON, Canada), and 2 mM benzamidine-HCl (BD0076, Bio Basic), pH 7.5. The drkN SH3 domain was purified on an ion-exchange column (17-1153-01 HiTrap Q HP, GE Healthcare, Little Chalfont, United Kingdom) with a linear gradient of NaCl (0–1 M) followed by a HiLoad Superdex 75 PG gel filtration column (28-9893-34, GE Healthcare) in 50 mM Tris, 150 mM NaCl, 2 mM TCEP, 1 mM EDTA, and 2 mM benzamidine-HCl, pH 7.5. The T22G, C2 (Cys inserted at N-terminus position 2) and the C2/C61/G62 (Cys inserted at N-terminus position 2, with Cys and Gly added at C-terminal positions 61 and 62) mutants were expressed and purified in a manner similar to that for the wild-type (WT) protein. The identity and purity of the three protein samples were assessed by mass spectrometry.
Fluorescence labeling
For FCS experiments, free amines in the WT and the T22G drkN SH3 proteins were labeled using TMR-NHS. The NHS-amine coupling reaction was performed in phosphate-buffered saline at pH 8.0 by adding the NHS-ester-activated fluorophores to a 50 μL solution of 200 μM protein at a dye/protein molar ratio of 1:2. This ratio was chosen to limit the fraction of multilabeled proteins. For dual-color FCS (dcFCS) control experiments, the SH3 C2 protein was singly labeled using BFL-maleimide and A647-maleimide. The samples were gently shaken for 3 h in the dark at room temperature. The excess dye was removed by size-exclusion chromatography using Sephadex G-25 gels (G2580, Sigma-Aldrich) in a BioLogic LP system (731-8300, Bio-Rad).
For smFRET experiments, the double-cysteine mutant (C2/C61/G62) was labeled in a site-specific manner by a pair of thiol-reactive dyes, i.e., BFL-maleimide (donor, neutral charge) and A647-maleimide (acceptor, −3 charge). To improve the solubility and labeling efficiency of BFL, 10 μL DMSO was added to a 50 μL solution of 100 μM protein in Tris buffer. TCEP was added to the protein at a 20-fold molar excess to reduce the disulfide bonds, and BFL was then added to the protein at a 5:1 molar ratio. Oxygen was removed by flushing the sample with argon gas in a desiccator for 5–10 min. The vial was capped tightly and shaken gently for 3 h at room temperature. Then, A647 was added to the protein at a 20-fold molar excess, and the solution was flushed with argon in a vacuum desiccator and kept at 4°C for 24 h in a sealed vial in the dark.
The free dyes were removed by size-exclusion chromatography. For some SMF experiments, a different donor dye was used: A555 (−4 charge) instead of BFL (neutral charge). To estimate the Förster radius and the intensity correction factors for smFRET analysis, donor-only and acceptor-only proteins were prepared using a similar protocol in which the protein was incubated for 3 h with only one species of thiol-reactive dye. Using these control samples, the fluorescence quantum yields of the dyes attached to drkN SH3 were found to be 0.55 for BFL, 0.39 for A555, and 0.37 for A647 in Tris buffer (pH 7.5); 0.38 for BFL, 0.40 for A555, and 0.36 for A647 in 6 M GdmCl; and 0.28 for BFL and 0.34 for A647 in 7 M urea. These represent ensemble-averaged and labeling-site-averaged values.
Sample preparation
All samples were diluted to concentrations of 1–20 nM and 20–100 pM, which are most suitable for dcFCS and smFRET burst experiments, respectively. For a typical experiment, a sample solution of ∼50–100 μL was dropped on the surface of plasma-cleaned coverslips. Nonspecific protein adsorption to the coverslip was prevented by adding 0.005% (v/v) Tween-20 (P2287, Sigma-Aldrich) to the solution, and bovine serum albumin (15260-037, ThermoFisher Scientific) was used to coat the clean coverslips. All experiments were performed at 20°C.
Instrumentation
Quantum yield and tryptophan fluorescence measurements were performed using a QuantaMaster PTI spectrofluorimeter (Photon Technology International, Birmingham, NJ) equipped with a red-sensitive photomultiplier tube (R928P, Hamamatsu, Hamamatsu City, Japan). Single-molecule measurements were performed on a custom-built multiparameter fluorescence microscope that has been described in detail elsewhere (27, 28). Laser excitation at 532 nm (LRS-0532, Laserglow Technologies, Toronto, ON, Canada) and 473 nm (04-01 series, Cobolt, Stockholm, Sweden) was used in the experiments and the fluorescence data were acquired using a single-photon-counting system (PicoHarp300, PicoQuant, Berlin, Germany).
In FCS measurements, the fluorescence from the sample was focused through a 50-μm pinhole and then divided into two channels by a nonpolarizing cube beamsplitter. Each beam was focused onto a separate single-photon avalanche diode (PD5CTC, MPD, Italy). A pseudo-autocorrelation curve with logarithmic time binning and 24 bins per temporal decade was calculated from these two signals using a photon-by-photon algorithm (27).
In smFRET measurements, a larger confocal pinhole was used (75 μm vs. 50 μm) and the beamsplitter cube was replaced by a dichroic mirror (FF560-Di01 or FF640-Di01, Semrock, Rochester, NY, USA), which splits the fluorescence into donor and acceptor dye contributions. In addition, a red-sensitive single-photon avalanche diode detector (COUNT-100, Laser Components, Bedford, NH) was used to increase the signal in the acceptor channel. The donor and acceptor signals were spectrally filtered using band-pass filters (Chroma, Bellows Falls, VT): BP530/50m and HQ690/70 for BFL-A647 and D575/70 and D705/80 for A555-A647. A custom-written LabView code was used to identify and analyze dual-color single-molecule fluorescence bursts, from which an smFRET histogram was constructed (29).
dcFCS measurements were performed on a different confocal microscope, on which the samples were excited with 488-nm (TECBL-488, World Star Tech, Toronto, ON, Canada) and 633-nm (TECRL-633, World Star Tech) lasers simultaneously. Fluorescence from blue (BFL or 6-FAM) and red (A647/Cy5) probes was passed through a 30-μm pinhole and a combination of long-pass and bandpass filters and focused on single-photon-counting avalanche photodiodes (SPCM-CD 3017, Perkin Elmer Optoelectronics, San Jose, CA). BLP01-488 (Semrock, Lake Forest, IL) and HQ 520/66 (Chroma, Bellows Falls, VT) filters were used in the BFL/6-FAM emission channel and BLP01-647 (Semrock) and HQ 685/80 (Chroma) were used in the A647/Cy5 emission channel. The signal output of the detectors was fed into a four-channel hardware correlator (Flex02-01D, Correlator.com, Bridgewater, NJ), which provided autocorrelation and cross-correlation curves.
FCS analysis
Under some simplifying assumptions, the fluorescence intensity correlation function for the free Brownian diffusion of a single molecular species with triplet-state blinking is given by (30)
| (1) |
Before each set of measurements, a dilute solution of a dye with a known diffusion coefficient was used to characterize the detection volume (27). Then, by fitting the measured correlation curve to the model described by Eq. 1, the local concentration, Neff, and the diffusion time, τd, were obtained and used to estimate the diffusion coefficient and then the hydrodynamic radius of the molecule via the Stokes-Einstein equation (27).
smFRET analysis
The FRET efficiency was calculated using the number of detected photons in the donor (ID) and acceptor (IA) channels in each single-molecule burst (31):
| (2) |
where γ is the ratio of the detection efficiencies (ζ) and the quantum yields (Φ), i.e., (0.72 for BFL-A647 and 0.91 for A555-A647 in Tris buffer, and 0.9 and 1.17 for BFL-A647 in 6 M GdmCl and 7 M urea, respectively). In addition, corrections were applied on both ID and IA to subtract the background and the spectral cross talk. An E value was estimated for each single-molecule intensity burst, and for each sample, ∼10,000 bursts were processed and a FRET histogram was produced.
The distance between the FRET probes, R, can be estimated from the measured energy transfer efficiency using the Förster equation:
| (3) |
The Förster radius, R0, was estimated to be 4.4 ± 0.2 nm for the BFL-A647 pair and 5.9 ± 0.2 nm for the A555-A647 pair. These estimates were made using the fluorophore spectra and quantum yields measured upon labeling drkN SH3. At each concentration of denaturant, the spectra and quantum yields of the donor and the acceptor were measured and used in the FRET calculations. For instance, at 6 M GdmCl, the estimated R0 values were 4.3 ± 0.2 nm for the BFL-A647 and 5.6 ± 0.2 nm for A555-A647.
Polymer model for inferring RG from smFRET data
The drkN SH3 domain with dyes and linkers was modeled as a coarse-grained self-avoiding walk (SAW) (25) with n = 66 residues, i.e., 60 residues separating the dyes plus six equivalent residues (three at each end) accounting for flexible dye linkers. Each amino acid residue is modeled by a single spherical bead with excluded volume defined by a hard core radius, Rhc = 0.4 nm. The SAW is sampled without energetic bias to populate conformations with equal a priori probabilities. From this unbiased full ensemble, an inferred subensemble is constructed by reweighting the conformations in accordance with a posteriori information from the smFRET histograms.
Briefly, this proceeds by seeking a distribution of end-to-end distances, , that minimizes the deviation where
| (4) |
To construct , conformations sampled in the unbiased full-ensemble SAW are sorted into subensembles conditioned on a narrow range of . The inferred is the one for which the deviation is minimized, and the uncertainty in is reported such that is in the range for which . The uncertainty in the mean experimental FRET efficiency, , was estimated to be ∼0.02 using the bootstrap technique by fitting the FRET efficiency histograms of 1000 replicate data sets generated by random sampling with replacement from the original set of fluorescence bursts. More details about the use of coarse-grained protein chains in the interpretation of smFRET data, and the comparison with conventional procedures that use either the Gaussian chain model or the mean-field Sanchez polymer theory can be found in (25).
The population-weighted root mean-squared (RMS) radius of gyration for a mixture of conformation clusters was calculated using the equation
| (5) |
where are the RMS RG values calculated or measured for each FRET subpopulation, and f1 and f2 = 1 − f1 are their respective fractions obtained by fitting the smFRET histograms. By adding Gaussian random noise to , and with standard deviation according to their respective uncertainties, 5000 in silico data sets were created. was calculated for each of the 5000 in silico data sets, and the standard deviation of these values was reported.
Results
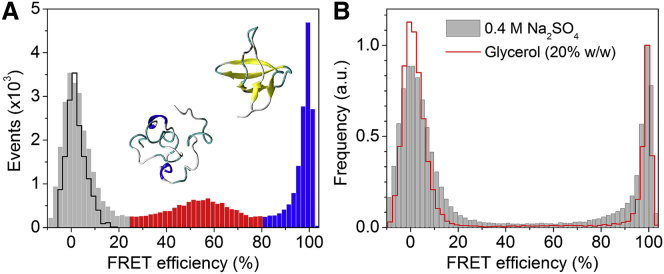
Measurements of the tryptophan fluorescence of WT and C2/C61/G62 mutant drkN SH3 samples in different denaturant concentrations showed that terminal mutations/insertions did not affect the denaturation midpoint (Fig. S1 in the Supporting Material). The distribution of protein end-to-end separations was measured by smFRET using the double-cysteine protein labeled with the BFL-A647 donor-acceptor dye pair. Fig. 1 shows the smFRET histograms obtained under nondenaturing conditions and in the presence of protein folding stabilizers.
Figure 1.
smFRET histograms of drkN SH3 end-labeled with BFL and A647. (A) Measurements in Tris buffer (pH 7.5). The blue and red parts of the histogram correspond to the folded (Fexch) and unfolded (Uexch) subpopulations, respectively; the black curve was obtained from a donor-only sample. The inset shows representative structures for the two states (17, 32). (B) smFRET histograms obtained in the presence of folding stabilizers: 0.4 M Na2SO4 (gray bars) and 20% (w/w) glycerol (red line). To see this figure in color, go online.
In Tris buffer (pH 7.5), the sample shows three distinct populations with average FRET efficiencies of 1%, 55%, and 99%, respectively (Fig. 1 A; see Table 2). We assign the second and third FRET peaks to the coexisting Uexch and Fexch, respectively, of drkN SH3 (18). Low or nearly-zero FRET peaks are often observed in single-molecule studies and are mostly attributed to donor-only or acceptor inactive or photobleached proteins. Alternatively, it would correspond to highly elongated conformations, in this case with end-to-end separations exceeding 90 Å. If the entire 60-residue drkN SH3 sequence was a single α-helix with no loops, the end-to-end distance would be 60 residues × 1.5 Å/residue = 90 Å. Since this situation is unrealistic, the zero-FRET peak will be discarded in the further analysis. For the folded state, a high FRET value yields an upper limit of REE ≈ 20 Å, in agreement with the three-dimensional structure, in which the N- and the C-termini are separated by ∼10 Å (32). Based on the area under the peaks, the population fractions of the folded and the unfolded states were estimated to be 0.53 and 0.47, respectively (see Table 2). The FRET fractional population fraction of 0.53 for Fexch at 20°C is in reasonable agreement with previous values measured by NMR, i.e., 0.50 at 30°C (33, 34) and 0.66 at 20°C (16).
Table 2.
Fitting Parameters Obtained for the smFRET Histogram Measured in Tris Buffer and in 6 M GdmCl
| Tris Buffer (pH 7.5) |
6 M GdmCl |
|||
|---|---|---|---|---|
| E (%) | Fraction | E (%) | Fraction | |
| Cluster 1 | 55.0 ± 19.6 | 0.53 ± 0.05 | 23.0 ± 16.4 | 0.68 ± 0.05 |
| Cluster 2′ | – | – | 91.3 ± 7.8 | 0.18 ± 0.03 |
| Cluster 2 | 99.1 ± 2.5 | 0.47 ± 0.05 | 99.6 ± 3.6 | 0.14 ± 0.03 |
To confirm the assignments of the FRET peaks, we performed experiments in solvents known to perturb the equilibrium between the folded and unfolded states of drkN SH3. For instance, in solutions containing sodium sulfate or glycerol, the FRET population around E = 55% is absent and the smFRET histogram contains only the 0% and 100% peaks (Fig. 1 B). Sodium sulfate is the strongest salt in the Hofmeister series of anions and it is known to stabilize the folded state of the drkN SH3 domain, and folded states in general, by promoting hydrophobic interactions (34). Glycerol is also known to stabilize proteins and prevent aggregation by shifting the folded protein ensemble toward more compact states and by destabilizing aggregation-prone partially unfolded intermediates (35).
Chemical denaturants lead to a heterogeneous set of conformations that include a high-FRET cluster
Chemical denaturants, such as GdmCl and urea, cause proteins to lose most of their secondary and tertiary structure and to approach a random-coil state (6, 36). smFRET studies of globular proteins such as RNase H (37), protein L (22), and the cold-shock protein CspTm (22, 38), and of IDPs such as the human prothymosin α (ProTα) (38), α-synuclein (39, 40), tau (40), troponin I (TnIc) (41), and yeast Sic1 (29) show the appearance of extended chain conformations in GdmCl, which become increasingly broader, more populated and more extended as the denaturant concentration increases. In view of these precedents, for the drkN SH3 domain, the population of the high-FRET peak was expected to gradually diminish in GdmCl, accompanied by an increase of the intermediate-FRET peak. This peak was also expected to gradually shift toward lower E values and become broader as [GdmCl] increased and the protein unfolded.
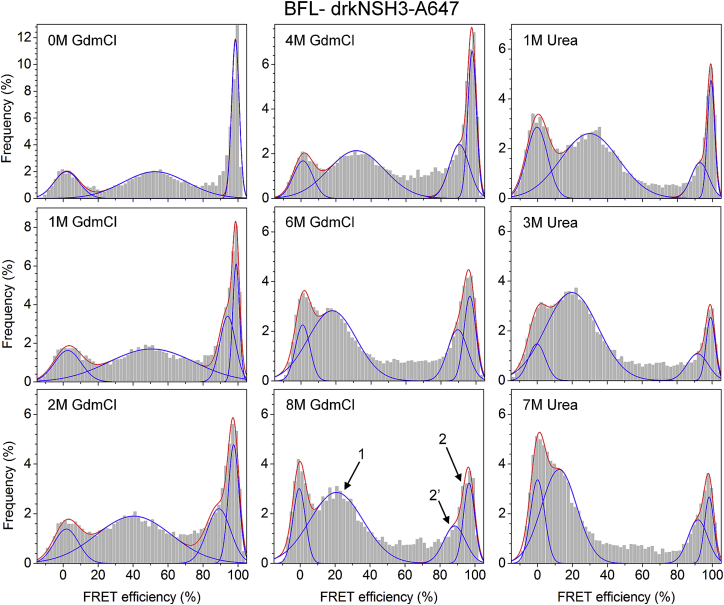
The smFRET data obtained for drkN SH3 under denaturing conditions are shown in Fig. 2. Quite surprisingly, for a marginally stable protein, the high-FRET (∼100%) population, although diminished, did not completely disappear even in the harshest denaturant conditions (8 M GdmCl and 7 M urea). The peaks of the smFRET histograms are typically approximated by Gaussian distributions and sometimes, near the limits of the FRET range, by beta distributions. However, the choices of fitting functions were found not to be critical for data interpretation (31). The smFRET histograms in this study were all decomposed into a minimal number of Gaussians.
Figure 2.
smFRET histograms measured at various concentrations of GdmCl and urea. Each histogram was constructed from a data set consisting of ∼10,000 intensity bursts and fitted to a sum of four Gaussians (three at no denaturant). The nonzero FRET peaks are denoted as 1, 2, and 2′, respectively, as labeled in the 8 M GdmCl histogram. The total area under the FRET peaks, excluding the zero-FRET peak (donor only and inactive and/or photobleached acceptor), was normalized to unity. To see this figure in color, go online.
The denaturant dependence of the smFRET histograms points to the presence of heterogeneous drkN SH3 conformations, with at least two distinct nonzero FRET clusters present at all denaturant concentrations. We define a FRET cluster as a population of protein conformations of similar end-to-end distance that gives rise to a distinct peak in the smFRET histogram data. The results of decomposing the two denaturation series into Gaussian peaks, including average FRET efficiency, width, and population fraction for each series, are given in Table 1.
Table 1.
FRET Efficiencies and Fractional Populations Obtained by Gaussian Decomposition of drkN SH3 smFRET Histograms in Denaturant
| [GdmCl] (M) | Cluster 1 | Cluster 2′ | Cluster 2 |
|---|---|---|---|
| 0 | 52.6 ± 21.5 (0.59) | – | 98.5 ± 2.5 (0.41) |
| 1 | 50.5 ± 28.6 (0.61) | 94.1 ± 5.2 (0.22) | 98.9 ± 2.3 (0.17) |
| 2 | 40.4 ± 25.3 (0.58) | 89.1 ± 7.9 (0.22) | 97.5 ± 3.6 (0.21) |
| 4 | 31.7 ± 18.8 (0.53) | 90.5 ± 6.7 (0.21) | 97.8 ± 2.9 (0.25) |
| 6 | 17.8 ± 16.2 (0.62) | 89.6 ± 6.8 (0.19) | 96.5 ± 4.0 (0.19) |
| 8 | 20.8 ± 17.9 (0.69) | 88.4 ± 7.3 (0.15) | 96.6 ± 3.8 (0.16) |
| [Urea] (M) | |||
| 1 | 29.7 ± 19.5 (0.72) | 92.8 ± 6.0 (0.12) | 99.1 ± 2.4 (0.16) |
| 3 | 19.8 ± 17.7 (0.80) | 91.7 ± 7.1 (0.10) | 99.1 ± 3.0 (0.10) |
| 7 | 12.3 ± 11.9 (0.69) | 91.7 ± 6.6 (0.18) | 98.2 ± 3.1 (0.13) |
Data are formatted as a percentage of the average FRET efficiency ± the half-width at half-maximum (HWHM). For each FRET peak (cluster), the fractional population is given in parentheses. See Fig. 2 for the histograms used to obtain the data.
As discussed above, in the absence of denaturant, two Gaussians are sufficient to describe the data, corresponding to the unfolded (cluster 1, E = 50–55%) and folded (cluster 2, E = 90–100%) subpopulations of this SH3 domain. When the denaturant concentration was increased, cluster 1 shifted to lower FRET values, with the center peak reaching 21% in GdmCl and 12% in urea, and it increased as a fraction of the total histogram. This dominant cluster was satisfactorily fitted by a single Gaussian for both denaturation agents at all measured concentrations. The broad distribution and the significant shift to lower FRET is consistent with the overall expansion of proteins upon denaturation, typically measured in smFRET studies (22, 29, 38). The remarkable difference compared to other proteins is that the unfolded fraction is already populated at zero denaturant due to unusual energetics, ΔG ≈ 1 kcal/mol (32). GdmCl and urea will diminish the transient secondary and tertiary structure in this cluster to a point where it may approach a random-coil state (see also below).
In the absence of denaturation agents, the high-FRET region of the histograms (E > 80%) is attributed to the folded state, Fexch, which is consistent with the close proximity of the N- and C-termini in the drkN SH3 crystal structure (PDB: 2A37). When denaturant was added, the average FRET efficiency of this population decreased overall and the peak broadened asymmetrically toward lower FRET (longer distance) values. To account for the asymmetric broadening, another Gaussian component, cluster 2′, was used for fitting. Cluster 2′ shifts from E = 94% in 1 M GdmCl to E = 88% in 8 M GdmCl and accounts for approximately half of the high-FRET population. A similar trend was observed in urea, although the peak shift was somewhat smaller than in GdmCl (Table 1).
Although fitting the high-FRET population at high denaturant by two Gaussians does not necessarily imply the existence of two sets of protein conformations that are significantly different, the procedure can nevertheless serve to quantify the asymmetric broadening of this cluster toward lower efficiency values. The total fraction of the high-FRET population decreases from 0.41 at zero denaturant to 0.31 at the highest denaturant concentrations, with a minimum of 0.2 in 3 M urea.
Overall, urea seems to be more effective than GdmCl at denaturing drkN SH3. This is not surprising, since urea is thought to be better than GdmCl at destabilizing β-sheets (42) and more efficiently solvates the protein backbone and side chains with few or no restrictions due to excluded-volume effects (43). Whereas urea accumulates in the first solvation shell, guanidinium displays a longer-range electrostatic effect that does not perturb the structure of the solvent close to the protein. On the other hand, the ionic nature of GdmCl leads to screening of all electrostatic interactions in a protein. Surprisingly, the high-FRET drkN SH3 population does not disappear completely in either GdmCl or urea, as typically seen in smFRET denaturation studies on other proteins (22, 38, 44).
Apart from the two major FRET clusters described above, the histograms show some population density at intermediate FRET values (E = 50–80%), particularly at GdmCl and urea concentrations of >3 M (Fig. 2). There is no distinguishable peak in this range, and this density may be caused by interconversion between the two major clusters (31, 45) or by photobleaching-induced “bridging” effects (45).
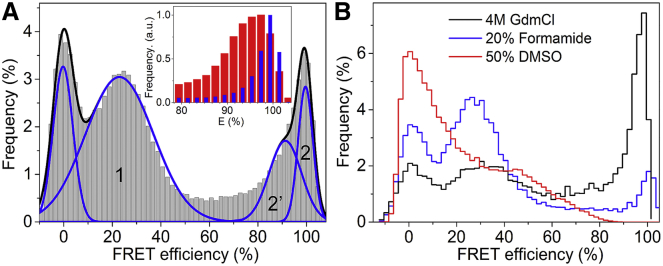
The different FRET clusters, corresponding to different subensembles of the drkN SH3 conformations, are better resolved in the smFRET histogram shown in Fig. 3 A. These data were acquired at 6 M GdmCl and were constructed from ∼100,000 intensity bursts. Utilizing a much larger number of single-molecule events allowed for finer binning along the FRET efficiency axis and provided a higher signal/noise ratio than the denaturation series data (Fig. 2). The histogram was fitted to a sum of three Gaussians, excluding the donor-only zero-FRET peak (Table 2).
Figure 3.
(A) smFRET histogram of drkN SH3 in 6 M GdmCl, constructed from ∼10,000 intensity bursts. The histogram was fitted to four Gaussians and normalized to the area excluding the zero-FRET peak. The parametric fitting results are listed in Table 2. The inset shows a comparison between the high-FRET peaks measured in Tris buffer (blue bars) and 6 M GdmCl (red bars). (B) FRET histograms of drkN SH3 in 20% (v/v) formamide (black), 4 M GdmCl (red), and 50% (v/v) DMSO (blue). DMSO data were acquired with a different donor dye, i.e., A555 instead of BFL. To see this figure in color, go online.
In the low-FRET region, a broad peak (cluster 1) covers nearly the entire range between E = 0% and E = 50%. With an average FRET efficiency of 23% and a fractional population of 0.68, this cluster is the predominant conformational subensemble at 6 M GdmCl. Cluster 1 exhibits a denaturant dependence similar to the chemically denatured states of other proteins (i.e., with peak broadening and a shift to lower FRET (overall expansion)), which are typically described by a random-coil polymer model.
About one-third of the total population shows an anomalously high FRET efficiency at this high denaturant concentration. This corresponds to a mixed population of conformations, all of which have the two ends of the chain in close proximity to each other, and it is described by a sum of two Gaussians of nearly equal area, which are centered at E = 91.3% (cluster 2′) and E = 99.6% (cluster 2). The high-FRET peak is broader and more asymmetric than the Fexch peak observed in Tris buffer, as its variance increased from ΔE ≈ 5% in Tris to ΔE ≈ 12% in 6 M GdmCl (Fig. 3 A, inset). Importantly, NMR and bulk fluorescence studies indicate that the drkN SH3 domain is nearly completely unfolded in 2 M GdmCl (19), thus ruling out the presence of folded-state conformers at high-denaturant concentrations.
Organic solvents are commonly used to change the native structure of macromolecules by disrupting the hydrophobic interactions between hydrophobic side chains of amino acids. To verify whether nonpolar hydrophobic interactions could cause the opposite ends of the protein to “stick” to each other in high denaturant concentrations, smFRET measurements of drkN SH3 in the presence of DMSO and formamide were performed.
Formamide and DMSO are widely used polar solvents that precipitate, crystallize, and denature proteins (46). In Fig. 3 B, the smFRET histograms measured in solutions containing either 4 M GdmCl, 20% formamide, or 50% DMSO are shown. Interestingly, the formamide diminished the high-FRET population nearly five times more efficiently than GdmCl. An even more pronounced change was recorded in DMSO, for which the high-FRET peak is absent. This suggests that groups at opposite ends of the chain are involved in interactions that are facilitated by guanidinium and urea, thus causing the persistence of a dynamic, high-FRET population of end-labeled drkN SH3 under denaturing conditions (see Discussion).
Effect of dye pairs on the high-FRET signal
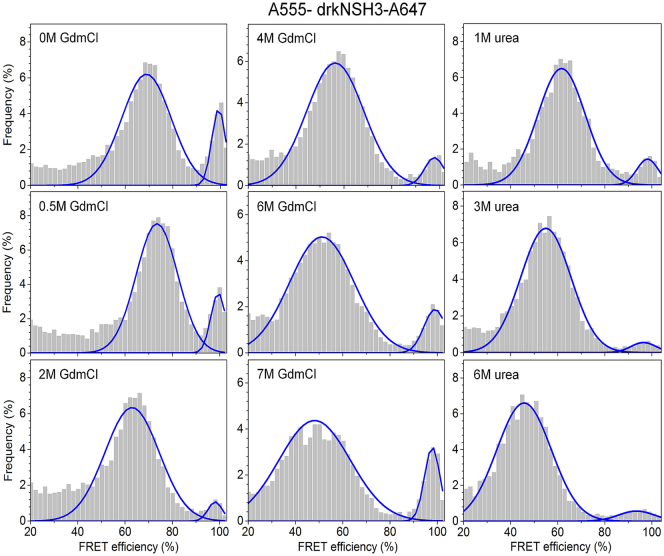
The significant fraction of drkN SH3 molecules with high FRET under denaturing conditions is highly unusual, and it could be due to “sticky” fluorescent labels promoting attractive interactions between the two ends of the protein. As a control, we replaced the donor dye, the neutral BFL, with the negatively charged dye, A555, and used the same acceptor dye, A647 (Fig. 4). According to the manufacturer, A555 and A647 are both negatively charged, with net charges of −4 and −3, respectively, and are therefore unlikely to cause attractive interactions between the two ends. On the contrary, this FRET pair exhibits repulsive electrostatics, and it could destabilize possible attractive intraprotein interactions.
Figure 4.
smFRET denaturation series of drkN SH3 measured with the A555/A647 donor-acceptor pair in GdmCl and urea solutions. Both dyes are negatively charged. To see this figure in color, go online.
Fig. 4 shows the smFRET histogram data obtained for drkN SH3 labeled with A555 and A647 at different GdmCl and urea concentrations (Fig. 4). The new dye pair has a larger Förster radius, R0 = 5.9 ± 0.2 nm, and consequently, the histogram peaks are shifted to higher FRET efficiencies compared to the data measured with the BFL-A647 pair. For instance, the peak centered at E = 23% in 6 M GdmCl shifts to a peak centered at E = 51% with the new donor-acceptor pair, and the two high-FRET subpopulations shift to the right and merge within the E ≈ 100% peak.
When adding denaturant, the mid-FRET peak corresponding to the unfolded state, Uexch, shifts slightly to the right (smaller REE) for [GdmCl] < 1 M, and then to the left (longer REE) for [GdmCl] > 1 M. The initial phase is consistent with overall chain compaction caused by the screening of electrostatic repulsion between the two ends of the protein (29). In the absence of denaturant, the fraction of the high-FRET folded state (∼0.2) is considerably smaller than the value measured with the other FRET pair (0.41; Table 1). This is likely the effect of electrostatic repulsion between the donor and acceptor dyes destabilizing the folded, Fexch conformation, and it highlights the importance of selecting fluorescent dyes for smFRET experiments that do not perturb the conformational equilibrium of the labeled protein. As the GdmCl concentration is increased, the high-FRET fractional population decreases sharply to ∼0.05 in 2 M GdmCl, followed by an increase to ∼0.15 in 7 M GdmCl. The electrostatic screening is absent in urea and the repulsion between the two dyes drives the high-FRET fractional population to a low value of ∼0.05, which remains roughly constant for urea concentrations >3 M.
Our smFRET data suggest that 1) the observed residual high-FRET population of the drkN SH3 protein in high denaturant concentration is unlikely to be caused primarily by an attractive interaction between fluorescent labels; and 2) GdmCl, and to a lesser extent urea, promote the formation of intra- or intermolecular contacts between residues near the N- and C-termini of drkN SH3, which are stable on the diffusion timescale, i.e., ∼100 μs. Due to the shallow energy landscape of unfolded, denatured, and other disordered states, their conformational ensembles can be highly sensitive to conditions and other factors. The smFRET data are thus consistent with interactions within the drkN SH3 domain synergizing with denaturants, and possibly dye interactions, to generate conformational preferences leading to high FRET.
The high-FRET cluster in denaturants is not attributable to aggregation
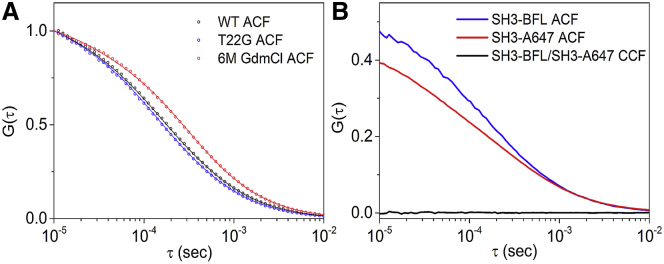
Although denaturants are generally known to inhibit the formation of protein aggregates (47), the persistence of a high-FRET population in denaturant could, in principle, be an effect of aggregation. To test this possibility, the hydrodynamic radius (RH) of the fully folded and the chemically denatured drkN SH3 was measured by FCS and the codiffusion fraction of singly labeled SH3-BFL and SH3-A647 was measured by dcFCS (Fig. 5, A and B). A single-point mutation of a threonine to a glycine (T22G) was shown to fully stabilize the folded state of the drkN SH3 domain (32). The T22G mutant was labeled with an amine-reactive TMR dye and the RH was measured by FCS. A value of RH,F = 14.4 Å was obtained (see Table 4), which is similar to the value obtained by NMR, RH,F = 15.6 Å (48), within the experimental error of both methods.
Figure 5.
(A) Autocorrelation-function data (symbols) and fitting according to Eq. 1 (solid curves) for WT SH3 in Tris buffer (pH 7.5) (black), T22G SH3 (fully folded) in Tris buffer (pH 7.5) (blue), and WT SH3 in 6 M GdmCl (red). (B) Autocorrelation-function and cross-correlation-function data were obtained for an equimolar mixture of SH3-C2-BFL and SH3-C2-A647 in a 6 M GdmCl solution. To see this figure in color, go online.
Table 4.
Hydrodynamic Radii, Radii of Gyration, and Shape Factors for Different Conformation States of drkN SH3
| Folded, Fexch (Native) | Unfolded, Uexch (Native) | Denatured, D (6 M GdmCl) | |
|---|---|---|---|
| RH (Å) | 14.4 ± 0.2 | 18.9 ± 0.3 | 17.8 ± 0.4 |
| RG (Å) | 12.0 ± 0.9 | 18.8 ± 1.0 | 20.6–21.9 |
| ρ = RG/RH | 0.83 | ∼1.0 | 1.16–1.23 |
The fitting of the FCS data of the WT protein under denaturing conditions (6 M GdmCl) required only one diffusive species. The obtained hydrodynamic radius is thus an average value for the entire denatured ensemble, namely corresponding to all the smFRET peaks combined. The viscosity of the 6 M GdmCl solution, η = 1.63 cP, which was determined by FCS using a free dye solution, was used to correct the diffusion parameters obtained for the denatured protein. After correction, the estimated average hydrodynamic radius of the entire denatured ensemble was RH,D = 17.8 ± 0.3 Å (see Table 4). The ratio between the hydrodynamic sizes of the denatured state and the folded state of drkN SH3 is RH,D/RH,F = 1.24 ± 0.03.
Since the average radius of folded proteins is roughly proportional to the cubic root of their molecular weight, dimerization will lead to an increase of RH by at least 25%. Larger aggregates will result in a more significant increase of the RH. If GdmCl or urea facilitate the aggregation of the drkN SH3 protein, the hydrodynamic radius is expected to increase with GdmCl (urea) concentration. However, FCS data measured between 2 M and 7 M GdmCl yielded virtually invariant RH values, between 17.7 and 17.9 Å (Table S1).
In addition, dcFCS measurements were performed to examine the possibility of protein aggregation during the labeling procedure. Single cysteine proteins (SH3-C2) were labeled with either BFL or A647 and subjected to the same sample conditions as during the dual labeling protocol. Fig. 5 B shows the autocorrelation and cross-correlation curves of the two spectral species in 6 M GdmCl. Similar data were measured in Tris buffer (50 mM Tris and 150 mM NaCl, pH 7.5) and in 50% v/v DMSO (Section S2 in the Supporting Material and Fig. S2). In all cases, the cross-correlation was virtually zero, indicating that there was no codiffusion of SH3-BFL and SH3-A647 and thus ruling out the presence of aggregates of drkN SH3 in high-denaturant conditions.
Discussion
The chemically denatured state of proteins is generally described by a random-coil model (36), and it is often used as a proxy for the unfolded-state ensemble. Accordingly, the high-FRET (small REE) population that corresponds to the folded state (Fexch) should vanish under strong denaturing conditions. However, a sizeable high-FRET fraction of drkN SH3 persisted in very high concentrations of denaturant, in contrast to the typical chemical denaturation behavior of proteins observed in smFRET studies (21, 22, 23, 49). Of potential relevance, NMR data point to the existence of significant transient structure in chemically denatured proteins (7, 9, 10), whereas computational studies emphasize the importance of nonnative and nonlocal interactions along the folding pathways (13, 50, 51). Note that the high-FRET peak in the presence of high [GdmCl] or [urea] is significantly broader than in Tris buffer (Fig. 3, inset), thus suggesting that it was not caused by the persistence of the folded state, Fexch, which gave rise to a narrow FRET peak at E = 98% (Fig. 1). Moreover, previous NMR studies showed that drkN SH3 is nearly completely denatured at 2 M GdmCl (18, 19). On the other hand, control dcFCS measurements performed at high denaturant concentrations ruled out protein oligomerization as a possible cause for this effect (Fig. 5 B). What, then, is the physical origin of the significantly populated (20–35%), unusually high-FRET (E > 90%) cluster observed for drkN SH3 in urea and GdmCl?
Molar-range concentrations of denaturants are typically “good solvents” for proteins (36), and denatured proteins are often described as freely jointed polymers (52). Thus, smFRET data on unfolded proteins are typically interpreted using homopolymer models that are either the Gaussian chain (38) or variants of the Sanchez model (53). Recently, our groups developed a new method, to our knowledge, to infer conformational properties of heterogeneous disordered proteins from smFRET data using a coarse-grained SAW polymer model with physical excluded volume (25). This subensemble SAW-based inference was applied to an IDP protein, Sic1 from yeast, which showed distinct coexisting smFRET subpopulations (29). Derived Sic1 conformational parameters, such as the hydrodynamic radius and the radius of gyration were consistent with those obtained in NMR and SAXS experiments. Here, we applied the same SAW framework to infer most probable RG values for each FRET cluster observed in the presence or in the absence of GdmCl (Table 3). Population-weighted RG averages were calculated according to Eq. 5 and compared to results of SAXS experiments. To that end, different assumptions regarding the conformations responsible for the high-FRET cluster in 6 M GdmCl were considered (Table 3).
Table 3.
RG of Different Conformations of drkN SH3 Estimated from the smFRET Data or from Previous Ensemble SAXS Data
| Fexcha | Uexcha | Denatured Random Coil | Denatured Looped | <RG>FRETb (Å) | <RG>SAXS | |
|---|---|---|---|---|---|---|
| Tris buffer (pH 7.5) | 12.0 ± 0.9c | 18.8 ± 1.0 | – | – | 15.5 ± 0.8 | 14.9 ± 0.5d |
| 6 M GdmCl | 12.0 ± 0.9c | – | 22.5 ± 1.0 | – | 19.7 ± 0.8 | 21.9 ± 0.5d |
| – | – | 22.5 ± 1.0 | 15.8 ± 0.2 | 20.6 ± 0.8 | ||
Fexch and Uexch are the folded and unfolded subpopulations under non-denaturing conditions.
The population-weighted average RG was calculated using Eq. 5 with the fractions obtained by fitting the smFRET histograms (Table 2).
The RG of the folded state was measured by SAXS (48) and adjusted for the presence of dye linkers (Section S5 in the Supporting Material).
Under nondenaturing conditions, drkN SH3 coexists in two different states, a folded (Fexch) and an unfolded (Uexch) state, as demonstrated here (Fig. 1) and in previous NMR studies (33). Choy et al used sodium sulfate to stabilize the folded state and measured RG,F = 11.9 ± 0.5 Å using SAXS (48). The presence of dye linkers at the two ends of the chain was found to be equivalent to adding between 4 and 16 residues (54), which results in the RG,F of the protein-dye construct being adjusted to 12.0 ± 0.9 Å.
Experimental studies and ENSEMBLE calculations were performed previously for the Uexch (55). This work primarily probes chemically denatured states. Chemical denaturants significantly affect the Uexch, based on previously published results (32), and it is also likely that chemical denaturants affect the folded state, but the current data do not offer strong additional insights into either of these states. Nevertheless, for the Uexch, the SAW model estimated a radius of gyration of RG,F = 18.9 ± 0.5 Å based on an average FRET efficiency of 55% and an average end-to-end separation of 43.0 ± 0.9 Å, which is very similar to the value obtained from the distribution of unfolded ENSEMBLE conformations (55) (Fig. S3). The FRET population-weighted average RG was estimated to be 15.6 ± 0.8 Å (Table 3), which is close to the SAXS value for the drkN SH3 ensemble, i.e., RG = 14.9 ± 0.5 Å (17). The small difference is mostly due to the presence of dyes and linkers in the fluorescence experiments, which are expected to lead to a small increase in the overall size of the chain. Another contributing factor is the relatively compact nature of the Uexch, which contains significant fluctuating structure and thus deviates from a random chain with excluded volume (see below). However, given the inherent limitations of the polymer model used for RG inference, the reasonable agreement between the smFRET and SAXS values is encouraging.
The shape factor, defined as the ratio between the radius of gyration and the hydrodynamic radius (ρ = RG/RH), is commonly used to define the average conformational shape of a polymer. The shape factor, ρF, of the drkN SH3 folded state was estimated to be 0.83 using the FCS-measured RH,F (Table 4) and 0.77 using the NMR-measured RH,F (48). Both values are similar to the theoretical shape factor predicted for globular proteins, i.e., ρ = (3/5)0.5 ≈ 0.775 (53, 56) and to experimental results for other proteins, e.g., ρ ≈ 0.75 for the HIV-1 capsid protein (57).
The shape factor of the native unfolded drkN SH3 state (Uexch) was estimated to be ∼1.0 (Table 4). Note that the RH,U is ∼30% larger than the RH,F, in excellent agreement with previous NMR data (48). As mentioned above, the FRET-inferred radius of gyration for the Uexch is slightly overestimated because this state is not a random chain. This implies that the actual value of ρU is lower, probably between 0.775 and 1.0, which is consistent with an expanded protein conformation with sizeable fluctuating structure.
The smFRET histograms at high denaturant concentrations do not show a single uniform peak (Fig. 2), which indicates that the denatured state ensemble is heterogeneous. To understand the nature of this heterogeneity, we applied the subensemble FRET inference based on the SAW polymer model to estimate the most probable RG for each FRET peak of end-labeled drkN SH3 at 6 M GdmCl (Fig. 3). Different premises about the nature of the protein conformations in the high-FRET cluster were used. As validation, the population-weighted average RG values, which were calculated based on the smFRET data (Table 2) and using Eq. 5, were compared to the SAXS-measured RG (Table 3). In addition, the subensemble RG values and the FCS-measured RH values were used to derive shape factors for the denatured ensemble and for the residual high-FRET state and thus help distinguish between different hypotheses (Table 4).
The predominant FRET population, i.e., cluster 1 in Fig. 3, was modeled as a random self-avoiding chain with 66 residues, i.e., 60 amino acid residues between the labeling positions and three equivalent residues at each end accounting for the dye linkers. By imposing that Esim matches Eexp = 23%, the radius of gyration of this random-coil subensemble was estimated to be RG,RC = 22.5 ± 1.0 Å (Table 3). The corresponding value obtained with the classical Gaussian chain model, which does not account for excluded volume, is much larger, i.e., 30.6 ± 1.0 Å. The expected value for a 66-residue chain based on the scaling relation for fully unfolded proteins (58) is 23.6 ± 4.3 Å, which is similar to the SAW smFRET inference. Moreover, since the Cα−Cα virtual bond length is ∼0.38 nm, the Gaussian-inferred RG value translates into an inferred Kuhn length of ∼6 residues, which is clearly unrealistic.
Since dcFCS controls ruled out the presence of aggregates in our single-molecule experiments, intermolecular FRET cannot cause the high-FRET peak at high denaturant (GdmCl and urea) concentrations. It is therefore reasonable to assume that this subensemble (clusters 2 and 2′ in Fig. 3) could arise from either a residual population of the folded state, Fexch, or a disordered looped state, L. The folded state is rather compact, i.e., RG,F = 12.0 Å, and using it in Eq. 5 to calculate the population-average radius of gyration of the denatured state, RG,D, yielded a value of 19.7 Å, which is >2 Å smaller than the SAXS value, 21.9 Å (48).
We next considered the alternate possibility, which is a looped (L) state in which the N- and C-termini are in close proximity. The simulations of the L state were performed with an n = 66 SAW polymer modified so that an attractive potential enforces two selected residues to be on average 5.7 Å apart. The “sticky” residues were chosen to be l residues away from each end of the chain. Using the simulated REE distribution , we estimated the average FRET efficiency, and compared it to the experimental values for peaks 2 and 2′ from Table 2. The % peak can be assigned to looped conformations with l ≈ 10, i.e., a total of ∼20 residues, that are left dangling at the two ends. The % peak can be assigned to a loop with l ≤ 5, i.e., a total of <10 residues are left dangling at the two ends (Section S4). Here, these simulated L states are used as putative subensembles that contribute to the compact portion of the drkN SH3 unfolded ensemble at 6 M GdmCl.
The RG values of both looped states were estimated using the SAW formalism and were found to be almost identical, i.e., RG,L = 15.8 ± 0.2 Å. Using this value in Eq. 5. we obtained a population-average RG,D value of 20.6 ± 0.8 Å for the denatured ensemble (Table 3). This RG value is closer, although not identical, to the SAXS value, which was in fact measured at a different GdmCl concentration (2 M instead of 6 M). This, corroborated by the observed peak broadening (Fig. 3 A, inset), suggests that a dynamic looped conformational subensemble most likely causes the appearance of the high-FRET peak at high urea and GdmCl concentrations.
The shape factor of the denatured drkN SH3 ensemble, ρD, was calculated using the FCS-measured RH. We obtained a ρD value between 1.16 and 1.23 (Table 4), depending on whether the FRET or the SAXS RG value was used. For a linear Gaussian chain, i.e., a polymer in a theta solvent, a value of was predicted by Tanford using the Zimm model (59). This is the theoretical limit for n → ∞, but for n = 66, our coarse-grained simulations gave ρ = 1.25 for a Gaussian chain and ρ = 1.2 for a SAW chain. Coincidentally, for a Gaussian ring polymer in a theta solvent, the shape factor is ρ = (60).
A similar shape factor, i.e., ρ ≈ 1.2, was estimated for the denatured IgG domain of protein L using SMF methods (61). In addition, ρ ≈ 1.06 was found for several denatured polypeptides based on NMR and SAXS measurements (56). This was interpreted as the proteins retaining some fluctuating structure even at high denaturant concentrations and thus being more compact than a random chain, or as an increase in RH due to binding of GdmCl, or to a change in the bound water layer. NOE and NMR chemical-shift experiments suggested that drkN SH3 is almost completely denatured at 2 M GdmCl (18, 19). It is thus plausible that the high-FRET drkN SH3 population observed in urea and GdmCl consists of solvated, disordered conformations, but which have some denaturant-mediated interactions between the N- and C-terminal regions.
The underlying mechanisms of protein destabilization by denaturants such as guanidinium salts and urea are not completely understood (42, 62). What type of interactions could possibly lead to two ends of the drkN SH3 domain coming close together in high-denaturant GdmCl and urea solution? Thirumalai and co-workers (43) used molecular dynamics simulations to estimate the interactions between hydrophobic and ionic solutes in aqueous GdmCl and urea solutions. They proposed that both denaturant solvents only mildly alter hydrophobic association, whereas they dramatically change electrostatic interactions by solvating the charged residues or by forming hydrogen bonds with the peptide backbone. Ramsden and co-workers (63) proposed that GdmCl and urea can tune the water structure to some extent to either increase or decrease the strength of the hydrophobic interactions. Godawat et al. (64) found evidence that small hydrophobic pairs are stabilized, whereas large hydrophobic pairs are destabilized, by adding the denaturant, and similar findings were reported recently by Khajehpour (65) based on fluorescence quenching experiments.
The high-FRET peak is absent in a 50% DMSO solution, suggesting that hydrophobic interactions play an important role in the formation of the denatured drkN SH3 looped state. Note that nearly half of the amino acids in the drkN SH3 sequence are nonpolar. Contacts involved in the loop formation were found to be located within the terminal 5–10 residues at each end and probably involve aromatic residues and residues with nonpolar side chains. Similarly, Chan and co-workers (50) reported that hydrophobic interactions in denatured Fyn SH3 lead to a short-lived transient looped folding intermediate. In the case of drkN SH3, the looped state seems to be more stable, since the high-FRET peak appears as a distinct feature in the smFRET histogram, implying that the loop opening-closing rate is slower than the average diffusion time of the protein through the detection volume (∼100 μs).
In a recent smFRET study, Muñoz and co-workers resolved two unfolded conformations of the chicken α-spectrin SH3 domain upon chemical denaturation (44). Their results showed that a significant fractional population remains in the high-FRET region at molar-range concentrations of urea and GdmCl. They assigned the denatured high-FRET population to the native folded conformation, which gradually expands and becomes increasingly disordered due to breaking of intramolecular hydrogen bonds. In other smFRET studies of protein folding, the folded-state high-FRET peak decreases gradually and disappears upon adding denaturation agents (22, 37, 38). In this connection, it is noteworthy that fluorescence decay analysis by Ittah and Haas suggested a prominent looped state in the early stage of the folding kinetics of the 58-residue bovine pancreatic trypsin inhibitor when nonlocal interactions bring the two chain ends of the protein into close proximity (66). This study is the first time, to our knowledge, that single-molecule experimental evidence of protein loop formation under denaturing conditions has been reported.
Even though the details of the mechanism of drkN SH3 loop formation are not yet clear, the complete suppression of the high-FRET peak in DMSO implies that nonpolar hydrophobic interactions have a major contribution. The underlying mechanism of looping and the interconversion rates between closed and open states could be further studied by single-molecule spectroscopy on various mutants targeting the end regions of the drkN SH3 sequence. Other osmolytes could be used to perturb intramolecular interactions to investigate the contribution of hydrogen bonds, nonpolar contacts, and salt bridges in the association of specific protein segments. It is worth pointing out that contacts between the termini are absent in the unfolded ensemble under nondenaturing conditions. The loops are formed in the presence of denaturants and stabilized by denaturants, thus suggesting the need for more caution when using chemical denaturation to access the unfolded ensemble. Our findings also point to a potential synergy between the denaturants, the intrinsic intraprotein interactions, and potential dye interactions. In addition, heterogeneous denatured states are consistent with the idea that the free-energy landscape of disordered/unfolded states is very malleable and the underlying conformational biases could be amplified by certain chemical denaturants.
Author Contributions
A.M., Z.Z., J.F.-K., and C.C.G. designed and guided the research; A.M., Z.Z., and A.B. performed the experiments; A.M., Z.Z., and G.G. analyzed the data; J.S. and H.S.C. performed the molecular dynamics simulations; H.L. expressed and purified protein constructs; A.M, Z.Z., J.F.-K., and C.C.G. wrote the manuscript.
Acknowledgments
Financial support for this work was provided by Natural Science and Engineering Research Council of Canada Discovery Grants (to C.C.G. and J.F.-K.) and Canadian Institutes of Health Research operating grants (to H.S.C. and J.F.-K.). Z.Z. was supported by the Canadian Institutes of Health Research Training Program in Protein Folding and Interaction Dynamics. We are grateful for the allotments of computational resources from SciNet of Compute Canada.
Editor: David Eliezer.
Footnotes
Amir Mazouchi and Zhenfu Zhang contributed equally to this work.
Supporting Materials and Methods, eight figures, and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30034-0.
Supporting Material
References
- 1.Habchi J., Tompa P., Uversky V.N. Introducing protein intrinsic disorder. Chem. Rev. 2014;114:6561–6588. doi: 10.1021/cr400514h. [DOI] [PubMed] [Google Scholar]
- 2.Dunker A.K., Silman I., Sussman J.L. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 2008;18:756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005;579:3346–3354. doi: 10.1016/j.febslet.2005.03.072. [DOI] [PubMed] [Google Scholar]
- 4.Forman-Kay J.D., Mittag T. From sequence and forces to structure, function, and evolution of intrinsically disordered proteins. Structure. 2013;21:1492–1499. doi: 10.1016/j.str.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bah A., Vernon R.M., Forman-Kay J.D. Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature. 2015;519:106–109. doi: 10.1038/nature13999. [DOI] [PubMed] [Google Scholar]
- 6.Tanford C. Protein denaturation. Adv. Protein Chem. 1968;23:121–282. doi: 10.1016/s0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]
- 7.Dyson H.J., Wright P.E. Unfolded proteins and protein folding studied by NMR. Chem. Rev. 2004;104:3607–3622. doi: 10.1021/cr030403s. [DOI] [PubMed] [Google Scholar]
- 8.Dyson H.J., Wright P.E. Insights into the structure and dynamics of unfolded proteins from nuclear magnetic resonance. Adv. Protein Chem. 2002;62:311–340. doi: 10.1016/s0065-3233(02)62012-1. [DOI] [PubMed] [Google Scholar]
- 9.Eliezer D. Characterizing residual structure in disordered protein states using nuclear magnetic resonance. In: Bai Y., Nussinov R., editors. Protein Folding Protocols. Humana; New York: 2006. pp. 49–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCarney E.R., Kohn J.E., Plaxco K.W. Is there or isn’t there? The case for (and against) residual structure in chemically denatured proteins. Crit. Rev. Biochem. Mol. Biol. 2005;40:181–189. doi: 10.1080/10409230591008143. [DOI] [PubMed] [Google Scholar]
- 11.Lindorff-Larsen K., Trbovic N., Shaw D.E. Structure and dynamics of an unfolded protein examined by molecular dynamics simulation. J. Am. Chem. Soc. 2012;134:3787–3791. doi: 10.1021/ja209931w. [DOI] [PubMed] [Google Scholar]
- 12.Rauscher S., Gapsys V., Grubmüller H. Structural ensembles of intrinsically disordered proteins depend strongly on force field: a comparison to experiment. J. Chem. Theory Comput. 2015;11:5513–5524. doi: 10.1021/acs.jctc.5b00736. [DOI] [PubMed] [Google Scholar]
- 13.Chen T., Song J., Chan H.S. Theoretical perspectives on nonnative interactions and intrinsic disorder in protein folding and binding. Curr. Opin. Struct. Biol. 2015;30:32–42. doi: 10.1016/j.sbi.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 14.Freddolino P.L., Park S., Schulten K. Force field bias in protein folding simulations. Biophys. J. 2009;96:3772–3780. doi: 10.1016/j.bpj.2009.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen M.R., Zweckstetter M., Blackledge M. Exploring free-energy landscapes of intrinsically disordered proteins at atomic resolution using NMR spectroscopy. Chem. Rev. 2014;114:6632–6660. doi: 10.1021/cr400688u. [DOI] [PubMed] [Google Scholar]
- 16.Tollinger M., Skrynnikov N.R., Kay L.E. Slow dynamics in folded and unfolded states of an SH3 domain. J. Am. Chem. Soc. 2001;123:11341–11352. doi: 10.1021/ja011300z. [DOI] [PubMed] [Google Scholar]
- 17.Marsh J.A., Forman-Kay J.D. Structure and disorder in an unfolded state under nondenaturing conditions from ensemble models consistent with a large number of experimental restraints. J. Mol. Biol. 2009;391:359–374. doi: 10.1016/j.jmb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Crowhurst K.A., Tollinger M., Forman-Kay J.D. Cooperative interactions and a non-native buried Trp in the unfolded state of an SH3 domain. J. Mol. Biol. 2002;322:163–178. doi: 10.1016/s0022-2836(02)00741-6. [DOI] [PubMed] [Google Scholar]
- 19.Zhang O., Forman-Kay J.D. NMR studies of unfolded states of an SH3 domain in aqueous solution and denaturing conditions. Biochemistry. 1997;36:3959–3970. doi: 10.1021/bi9627626. [DOI] [PubMed] [Google Scholar]
- 20.Farrow N.A., Zhang O., Kay L.E. Characterization of the backbone dynamics of folded and denatured states of an SH3 domain. Biochemistry. 1997;36:2390–2402. doi: 10.1021/bi962548h. [DOI] [PubMed] [Google Scholar]
- 21.Gambin Y., Schug A., Deniz A.A. Direct single-molecule observation of a protein living in two opposed native structures. Proc. Natl. Acad. Sci. USA. 2009;106:10153–10158. doi: 10.1073/pnas.0904461106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merchant K.A., Best R.B., Eaton W.A. Characterizing the unfolded states of proteins using single-molecule FRET spectroscopy and molecular simulations. Proc. Natl. Acad. Sci. USA. 2007;104:1528–1533. doi: 10.1073/pnas.0607097104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuler B., Eaton W.A. Protein folding studied by single-molecule FRET. Curr. Opin. Struct. Biol. 2008;18:16–26. doi: 10.1016/j.sbi.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferreon A.C., Moran C.R., Deniz A.A. Single-molecule fluorescence studies of intrinsically disordered proteins. Methods Enzymol. 2010;472:179–204. doi: 10.1016/S0076-6879(10)72010-3. [DOI] [PubMed] [Google Scholar]
- 25.Song J., Gomes G.-N., Chan H.S. An adequate account of excluded volume is necessary to infer compactness and asphericity of disordered proteins by Förster resonance energy transfer. J. Phys. Chem. B. 2015;119:15191–15202. doi: 10.1021/acs.jpcb.5b09133. [DOI] [PubMed] [Google Scholar]
- 26.Hempel J. An orientation to Edman chemistry. In: Howard G.C., Brown W.E., editors. Modern Protein Chemistry: Practical Aspects. CRC Press; Boca Raton, FL: 2001. pp. 103–122. [Google Scholar]
- 27.Mazouchi A., Liu B., Gradinaru C.C. On the performance of bioanalytical fluorescence correlation spectroscopy measurements in a multiparameter photon-counting microscope. Anal. Chim. Acta. 2011;688:61–69. doi: 10.1016/j.aca.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 28.Liu B., Mazouchi A., Gradinaru C.C. Trapping single molecules in liposomes: surface interactions and freeze-thaw effects. J. Phys. Chem. B. 2010;114:15191–15198. doi: 10.1021/jp104614d. [DOI] [PubMed] [Google Scholar]
- 29.Liu B., Chia D., Gradinaru C.C. The effect of intrachain electrostatic repulsion on conformational disorder and dynamics of the Sic1 protein. J. Phys. Chem. B. 2014;118:4088–4097. doi: 10.1021/jp500776v. [DOI] [PubMed] [Google Scholar]
- 30.Haustein E., Schwille P. Fluorescence correlation spectroscopy: novel variations of an established technique. Annu. Rev. Biophys. Biomol. Struct. 2007;36:151–169. doi: 10.1146/annurev.biophys.36.040306.132612. [DOI] [PubMed] [Google Scholar]
- 31.Gopich I.V., Szabo A. Single-Molecule Biophysics. John Wiley and Sons; New York: 2011. Theory of single-molecule FRET efficiency histograms; pp. 245–297. [Google Scholar]
- 32.Bezsonova I., Singer A., Forman-Kay J.D. Structural comparison of the unstable drkN SH3 domain and a stable mutant. Biochemistry. 2005;44:15550–15560. doi: 10.1021/bi0512795. [DOI] [PubMed] [Google Scholar]
- 33.Zhang O., Forman-Kay J.D. Structural characterization of folded and unfolded states of an SH3 domain in equilibrium in aqueous buffer. Biochemistry. 1995;34:6784–6794. doi: 10.1021/bi00020a025. [DOI] [PubMed] [Google Scholar]
- 34.Zhang O., Kay L.E., Forman-Kay J.D. Backbone 1H and 15N resonance assignments of the N-terminal SH3 domain of drk in folded and unfolded states using enhanced-sensitivity pulsed field gradient NMR techniques. J. Biomol. NMR. 1994;4:845–858. doi: 10.1007/BF00398413. [DOI] [PubMed] [Google Scholar]
- 35.Vagenende V., Yap M.G.S., Trout B.L. Mechanisms of protein stabilization and prevention of protein aggregation by glycerol. Biochemistry. 2009;48:11084–11096. doi: 10.1021/bi900649t. [DOI] [PubMed] [Google Scholar]
- 36.Linhananta A., Hadizadeh S., Plotkin S.S. An effective solvent theory connecting the underlying mechanisms of osmolytes and denaturants for protein stability. Biophys. J. 2011;100:459–468. doi: 10.1016/j.bpj.2010.11.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rieger R., Nienhaus G.U. A combined single-molecule FRET and tryptophan fluorescence study of RNase H folding under acidic conditions. Chem. Phys. 2012;396:3–9. [Google Scholar]
- 38.Müller-Späth S., Soranno A., Schuler B. From the Cover: Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc. Natl. Acad. Sci. USA. 2010;107:14609–14614. doi: 10.1073/pnas.1001743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferreon A.C.M., Gambin Y., Deniz A.A. Interplay of α-synuclein binding and conformational switching probed by single-molecule fluorescence. Proc. Natl. Acad. Sci. 2009;106:5645–5650. doi: 10.1073/pnas.0809232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nath A., Sammalkorpi M., Rhoades E. The conformational ensembles of α-synuclein and tau: combining single-molecule FRET and simulations. Biophys. J. 2012;103:1940–1949. doi: 10.1016/j.bpj.2012.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Metskas L.A., Rhoades E. Conformation and dynamics of the troponin I C-terminal domain: combining single-molecule and computational approaches for a disordered protein region. J. Am. Chem. Soc. 2015;137:11962–11969. doi: 10.1021/jacs.5b04471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Camilloni C., Rocco A.G., Tiana G. Urea and guanidinium chloride denature protein L in different ways in molecular dynamics simulations. Biophys. J. 2008;94:4654–4661. doi: 10.1529/biophysj.107.125799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Brien E.P., Dima R.I., Thirumalai D. Interactions between hydrophobic and ionic solutes in aqueous guanidinium chloride and urea solutions: lessons for protein denaturation mechanism. J. Am. Chem. Soc. 2007;129:7346–7353. doi: 10.1021/ja069232+. [DOI] [PubMed] [Google Scholar]
- 44.Campos L.A., Sadqi M., Muñoz V. Gradual disordering of the native state on a slow two-state folding protein monitored by single-molecule fluorescence spectroscopy and NMR. J. Phys. Chem. B. 2013;117:13120–13131. doi: 10.1021/jp403051k. [DOI] [PubMed] [Google Scholar]
- 45.Campos L.A., Liu J., Muñoz V. A photoprotection strategy for microsecond-resolution single-molecule fluorescence spectroscopy. Nat. Methods. 2011;8:143–146. doi: 10.1038/nmeth.1553. [DOI] [PubMed] [Google Scholar]
- 46.Jackson M., Mantsch H.H. Beware of proteins in DMSO. Biochim. Biophys. Acta. 1991;1078:231–235. doi: 10.1016/0167-4838(91)90563-f. [DOI] [PubMed] [Google Scholar]
- 47.Chen J., Liu Y., Su Z. Cooperative effects of urea and L-arginine on protein refolding. Protein Expr. Purif. 2009;66:82–90. doi: 10.1016/j.pep.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 48.Choy W.Y., Mulder F.A.A., Kay L.E. Distribution of molecular size within an unfolded state ensemble using small-angle x-ray scattering and pulse field gradient NMR techniques. J. Mol. Biol. 2002;316:101–112. doi: 10.1006/jmbi.2001.5328. [DOI] [PubMed] [Google Scholar]
- 49.Hoffmann A., Kane A., Schuler B. Mapping protein collapse with single-molecule fluorescence and kinetic synchrotron radiation circular dichroism spectroscopy. Proc. Natl. Acad. Sci. USA. 2007;104:105–110. doi: 10.1073/pnas.0604353104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zarrine-Afsar A., Wallin S., Chan H.S. Theoretical and experimental demonstration of the importance of specific nonnative interactions in protein folding. Proc. Natl. Acad. Sci. USA. 2008;105:9999–10004. doi: 10.1073/pnas.0801874105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Faísca P.F.N., Nunes A., Shakhnovich E.I. Non-native interactions play an effective role in protein folding dynamics. Protein Sci. 2010;19:2196–2209. doi: 10.1002/pro.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fleming, P. J., and G. D. Rose. 2008. Conformational properties of unfolded proteins. Protein Science Encyclopedia.
- 53.Ziv G., Thirumalai D., Haran G. Collapse transition in proteins. Phys. Chem. Chem. Phys. 2009;11:83–93. doi: 10.1039/b813961j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCarney E.R., Werner J.H., Plaxco K.W. Site-specific dimensions across a highly denatured protein; a single molecule study. J. Mol. Biol. 2005;352:672–682. doi: 10.1016/j.jmb.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 55.Krzeminski M., Marsh J.A., Forman-Kay J.D. Characterization of disordered proteins with ENSEMBLE. Bioinformatics. 2013;29:398–399. doi: 10.1093/bioinformatics/bts701. [DOI] [PubMed] [Google Scholar]
- 56.Wilkins D.K., Grimshaw S.B., Smith L.J. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry. 1999;38:16424–16431. doi: 10.1021/bi991765q. [DOI] [PubMed] [Google Scholar]
- 57.Ehrlich L.S., Liu T., Carter C.A. HIV-1 capsid protein forms spherical (immature-like) and tubular (mature-like) particles in vitro: structure switching by pH-induced conformational changes. Biophys. J. 2001;81:586–594. doi: 10.1016/S0006-3495(01)75725-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kohn J.E., Millett I.S., Plaxco K.W. Random-coil behavior and the dimensions of chemically unfolded proteins. Proc. Natl. Acad. Sci. USA. 2004;101:12491–12496. doi: 10.1073/pnas.0403643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Teraoka I. Wiley; New York: 2002. Polymer Solutions: An Introduction to Physical Properties. [Google Scholar]
- 60.Robertson R.M., Laib S., Smith D.E. Diffusion of isolated DNA molecules: dependence on length and topology. Proc. Natl. Acad. Sci. USA. 2006;103:7310–7314. doi: 10.1073/pnas.0601903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sherman E., Haran G. Coil-globule transition in the denatured state of a small protein. Proc. Natl. Acad. Sci. USA. 2006;103:11539–11543. doi: 10.1073/pnas.0601395103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lim W.K., Rösgen J., Englander S.W. Urea, but not guanidinium, destabilizes proteins by forming hydrogen bonds to the peptide group. Proc. Natl. Acad. Sci. USA. 2009;106:2595–2600. doi: 10.1073/pnas.0812588106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cacace M.G., Landau E.M., Ramsden J.J. The Hofmeister series: salt and solvent effects on interfacial phenomena. Q. Rev. Biophys. 1997;30:241–277. doi: 10.1017/s0033583597003363. [DOI] [PubMed] [Google Scholar]
- 64.Godawat R., Jamadagni S.N., Garde S. Unfolding of hydrophobic polymers in guanidinium chloride solutions. J. Phys. Chem. B. 2010;114:2246–2254. doi: 10.1021/jp906976q. [DOI] [PubMed] [Google Scholar]
- 65.Macdonald R.D., Khajehpour M. Effects of the protein denaturant guanidinium chloride on aqueous hydrophobic contact-pair interactions. Biophys. Chem. 2015;196:25–32. doi: 10.1016/j.bpc.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 66.Ittah V., Haas E. Nonlocal interactions stabilize long range loops in the initial folding intermediates of reduced bovine pancreatic trypsin inhibitor. Biochemistry. 1995;34:4493–4506. doi: 10.1021/bi00013a042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.