Abstract
The atypical E3 ubiquitin ligase RNF31 is highly expressed in human breast cancer, the most frequent neoplastic lethality among women. Here, RNF31 depletion in breast cancer cells in combination with global gene expression profiling revealed p53 (TP53) signaling as a potential RNF31 target. Interestingly, RNF31 decreased p53 stability, whereas depletion of RNF31 in breast cancer cells caused cell cycle arrest and cisplatin-induced apoptosis in a p53-dependent manner. Furthermore, RNF31 associated with the p53/MDM2 complex and facilitated p53 polyubiquitination and degradation by stabilizing MDM2, suggesting a molecular mechanism by which RNF31 regulates cell death. Analysis of publically available clinical data sets displayed a negative correlation between RNF31 and p53 target genes, including IGFBP3 and BTG1, consistent with RNF31 regulating p53 function in vivo as well. Together, our findings suggest RNF31 as a potential therapeutic target to restore p53 function in breast cancer.
Introduction
Breast cancer is one of the most common cancers worldwide and the most frequent neoplastic lethality among women.1 Chemotherapy is frequently used in patients resistant to endocrine therapy and in patients presenting with cancer that is negative for the expression of estrogen receptors (ERs), progesterone receptors and HER2, the so-called triple-negative breast cancer. A particular challenge is breast cancer resistance to chemotherapy causing refractory disease.2 Thus, it is important to further characterize signaling pathways in breast cancer with the ultimate goal to identify novel therapeutic strategies.
The atypical E3 ubiquitin ligase RNF31 (alias HOIP and ZIBRA), belonging to the RING-between ring-RING (RBR) protein family of E3 ubiquitin ligases,3 was initially cloned from breast cancer cells based on its elevated mRNA expression.4 We previously showed that RNF31 mRNA expression is higher in human breast cancer compared with that in adjacent tissues.5
The tumor suppressor protein p53 (TP53), discovered 30 years ago,6 induces genes promoting cell cycle arrest and apoptosis, including CDKN1A, BTG2 and BAX.7 The induced proteins can either inhibit cell cycle progression by prohibiting cyclin-dependent kinases or promote apoptosis via the mitochondria pathway.8 Mutant p53 is frequently observed in cancer and is often associated with a loss of function to induce cell cycle arrest and apoptosis.9 The frequency of p53 mutations in breast cancer is ~20%.10, 11
The p53-mediated pathways are activated by genotoxic compounds, such as the chemotherapeutic compound cisplatin, causing cell cycle arrest and cell death. If p53 is absent or mutated, the execution of apoptosis is drastically reduced. Consistent with this, the p53 status is a response marker to chemotherapy.12 Activation of p53 function is also being explored as a therapeutic target in cancers.13, 14
The p53 protein is the substrate of several E3 ubiquitin ligases, including MDM2, p300 (EP300), COP1 and CHIP.15, 16, 17, 18 MDM2 is considered the major regulator of p53 and, on interaction, MDM2 induces p53 polyubiquitination and degradation.15 Furthermore, as a short-lived protein, the MDM2 protein level is tightly controlled. A number of E3 ubiquitin ligases regulate the MDM2/p53 axis by stabilizing the MDM2 protein, rather than directly modifying p53 stability.19, 20
Using an unbiased approach of global gene expression profiling after RNF31 depletion, we reveal RNF31 as a novel regulator of the MDM2/p53 complex in breast cancer cells. We show that RNF31 by stabilizing MDM2 promotes p53 degradation, leading to suppression of p53 target genes, ultimately facilitating breast cancer cell proliferation and inhibition of cisplatin-induced apoptosis.
With this effect on a critical regulator, RNF31 should be explored as a therapeutic target in breast cancer.
Results
RNF31 depletion increases the expression of p53 target genes in breast cancer cells
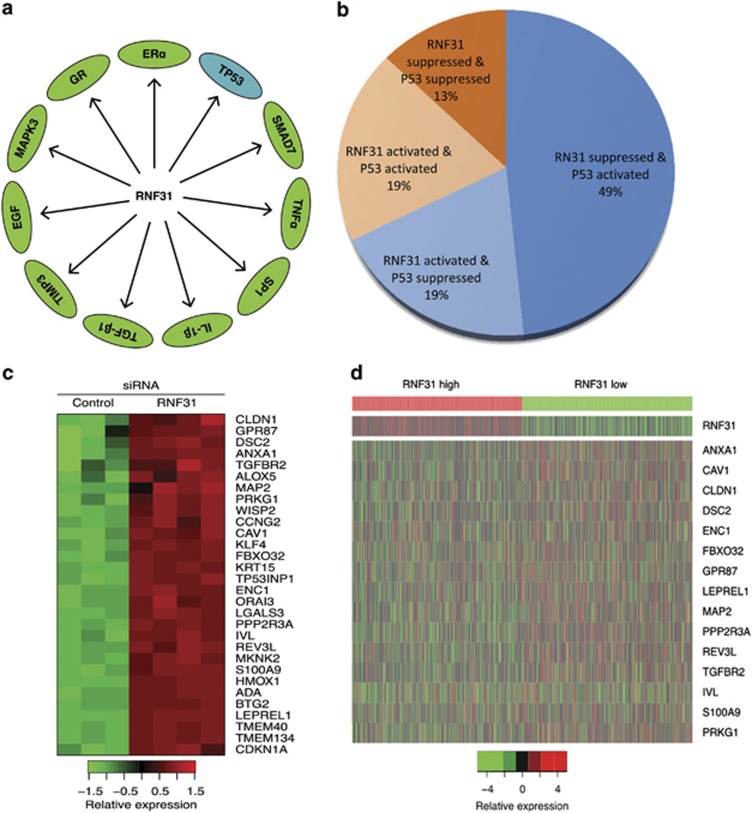
To approach the function of RNF31 in breast cancer cells in an unbiased way, we analyzed changes in previously generated global gene expression profiles following RNF31 depletion in MCF-7 breast cancer cells.5 Interestingly, pathway analysis revealed that RNF31 depletion is associated with changes in specific signaling pathways, including p53 signaling (Figure 1a). By overlapping published profiles of p53 target genes in MCF-7 cells21 with our derived gene expression profiles after RNF31 depletion in the same cell line, 62 genes that are subject to regulation by both p53 and RNF31 were identified, supporting common signaling pathways for RNF31 and p53 (Supplementary Table S2). Figure 1b shows that the majority (68%) of these genes were oppositely regulated by RNF31 and p53, suggesting that RNF31 inhibits p53 signaling (Figure 1b; blue segments). Among these genes, 30 genes are p53 positively regulated genes that also display increased expression by RNF31 depletion (Figure 1c). Analysis of data in The Cancer Genome Atlas clinical sample database shows that 15 out of these 30 p53-activated genes (50%) are negatively and significantly correlated with RNF31 expression, thus supporting RNF31 inhibition of p53 signaling in vivo (Supplementary Table S3 and Figure 1d).
Figure 1.
RNF31 depletion increases the expression of p53 target genes in breast cancer cells. (a) Schematic graph illustrates significantly changed signaling by RNF31 depletion in MCF-7 cells. Signal pathway-enrichment analysis was used to derive the related pathways, using P<0.001 and fold change>2 as cutoff to derive regulated genes and P<0.001 to define significantly enriched pathways using Pathway Studio (Elsevier). (b) Pie graph shows the relative fractions of genes regulated by p53 and RNF31 depletion. p53-activated/RNF31-suppressed genes together with p53-suppressed/RNF31-activated genes account for 68% of the total genes. (c) The heatmap graph shows the p53-activated/RNF31-suppressed genes in MCF-7 cells. (d) The heatmap graph shows that p53-activated genes are negatively correlated with RNF31 expression in clinical breast cancer samples from The Cancer Genome Atlas.
RNF31 depletion induces G1-phase cell cycle arrest and apoptosis in a p53-dependent manner
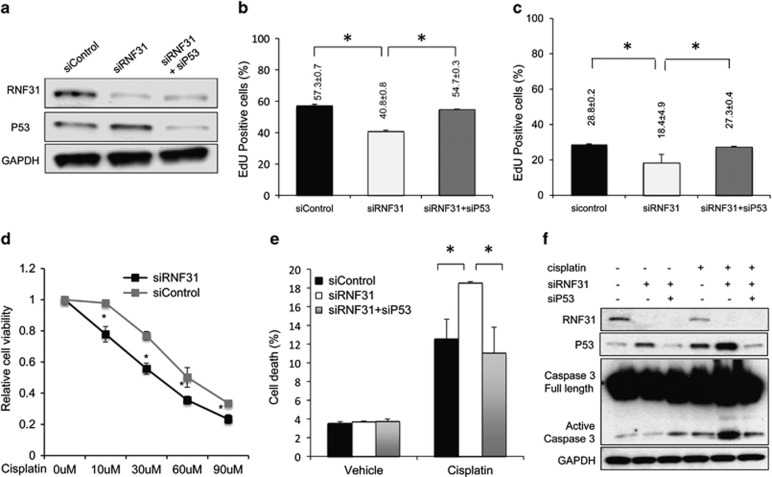
Consistent with the observation that RNF31 depletion induces p53 signaling in MCF-7 cells, RNF31 depletion induced p53 levels in this cell line (Figure 2a). To investigate the role of RNF31 in cell proliferation, in particular in relation to p53 signaling, we depleted RNF31 or both RNF31 and p53 in MCF-7 cells. RNF31 depletion reduced the number of cells in S phase, an effect that was reversed on depletion of p53 (Table 1 and Supplementary Figure S1A). In order to test the effect of RNF31 in an additional cell line and to employ an alternative assay (ethynly-deoxyuridine (EdU) incorporation), RNF31 or both RNF31 and p53 were depleted in both MCF-7 cells and ZR-75-1 cells. RNF31 depletion decreased EdU incorporation in both cells lines (Figures 2b and c, and Supplementary Figures S1B and C), whereas an additional depletion of p53 restored the EdU incorporation. Next, we investigated whether RNF31 had an impact on cisplatin sensitivity. As shown in Figure 2d and Supplementary Figure S2A, siRNF31 increased cisplatin sensitivity in ZR-75-1 and MDA-MB-175 cells, as measured by cell viability at increasing concentrations of cisplatin. MCF-7 cells are relatively resistant to cisplatin-induced apoptosis due to the lack of caspase-3 expression, the main effector of apoptosis, and were therefore not explored in these studies.22 The potential effect of RNF31 depletion for cisplatin-induced apoptosis was explored in ZR-75-1 cells. By propidium iodide and annexin V double staining, we found that siRNF31 increased cisplatin-induced apoptosis, which in turn was restored by depletion of p53 (Figure 2e and Supplementary Figure S2B). Furthermore, Figure 2f shows that RNF31 depletion increased activated caspase-3 in the presence of cisplatin, whereas depletion of p53 in addition to RNF31 depletion restored activated caspase-3 to the level observed without RNF31 depletion. Altogether, these data suggest that RNF31 modifies cell proliferation and cisplatin-induced apoptosis through the p53 pathway.
Figure 2.
RNF31 depletion induces G1 cell cycle arrest and cisplatin-induced apoptosis in a p53-dependent manner. (a) The efficiency of RNF31 and p53 knockdown in MCF-7 cells. RNF31 and p53 protein levels were determined by western blot analysis. Glyceraldehydes 3-phophate dehydrogenase (GAPDH) was used as internal control. A representative blot of three independent experiments is shown. (b) RNF31 knockdown decreases cell proliferation in MCF-7 cells as determined by EdU incorporation, which can be rescued by p53 depletion. MCF-7 cells were treated with siControl, siRNF31 or siRNF31 and siP53 for 48 h. EdU was added at a concentration of 10 μM and incubated for 1 h. The cells were subject to fluorescence-activated cell sorting (FACS) analysis. All values are mean±s.d. (n=3, *P<0.05, Student's t-test). (c) RNF31 knockdown decreases cell proliferation in ZR-75-1 cells as determined by EdU incorporation, which can be rescued by p53 depletion. ZR-75-1 cells were treated with siControl, siRNF31 or siRNF31 and siP53 for 48 h. EdU was added at a concentration of 10 μM and incubated for 1 h. The cells were subject to FACS analysis. All values are mean±s.d. (n=3, *P<0.05, Student's t-test). (d) RNF31 depletion reduces cell viability after cisplatin treatment in ZR-75-1 cells. ZR-75-1 cells were transfected with siRNF31 or siControl. After 48 h, cells were treated with indicated concentrations of cisplatin for 24 h. *P<0.05 for siRNF31 versus siControl. All values are mean±s.d. (n=3, Student's t-test). (e) RNF31 knockdown sensitizes cells to cisplatin-induced apoptosis, an effect which is rescued by p53 depletion. ZR-75-1 cells were transfected with siControl, siRNF31 or the combination siRNF31 and siP53. Forty-eight hours after transfection, cells were treated with 10 μM cisplatin or vehicle for 24 h. The percentage of apoptotic cells was determined by FACS. Experiments were done in triplicates. All values are mean±s.d. (n=3, *P<0.05, Student's t-test). (f) RNF31 knockdown facilitates cisplatin-induced caspase-3 activation, an effect which is rescued by p53 depletion. ZR-75-1 cells were transfected with siControl, siRNF31 or the combination siRNF31 and siP53. After 48 h, cells were treated with 10 μM cisplatin or vehicle for 24 h. p53, RNF31, full-length and activated caspase-3 levels were determined by western blot analysis. GAPDH was used as internal control.
Table 1. Cell cycle comparison among siControl, siRNF31 and siRNF31+sip53-treated MCF-7 cells.
| G1-phase | S-phase | G2–M phase | |
|---|---|---|---|
| siControl | 66.4±0.6 | 10.8±0.4 | 23.2±0.3 |
| siRNF31 | 76.7±0.6* | 7.5±0.3* | 15.9±0.6 |
| siRNF31+sip53 | 66.5±0.8# | 15.6±0.4# | 18.3±0.7 |
*P<0.05 for siControl versus siRNF31; #P<0.05 for siRNF31 versus siRNF31+sip53.
RNF31 knockdown increases p53 protein levels and its target gene expression in breast cancer cells
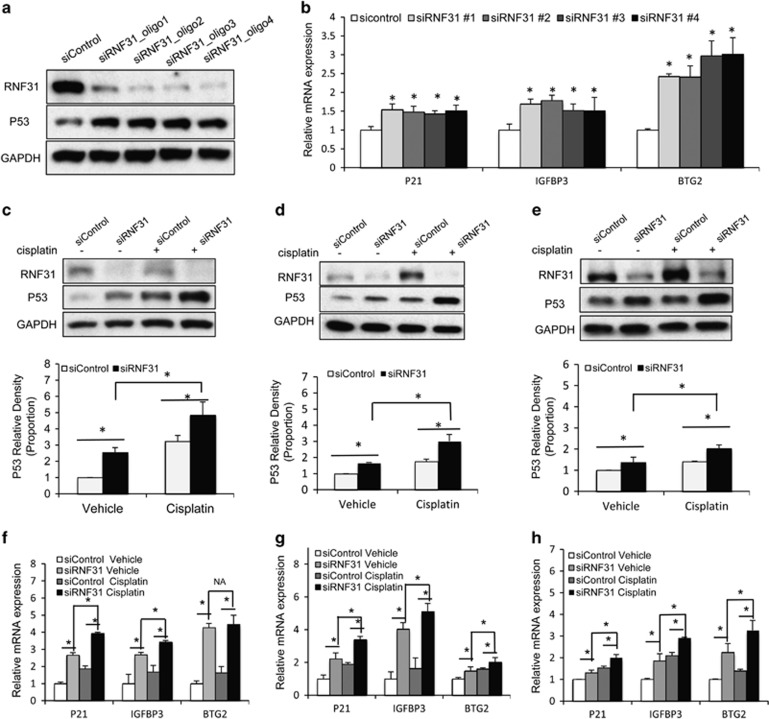
Increased p53 levels in MCF-7 cells were observed using the four individual small interfering RNAs (siRNAs) constituting a SMART pool (Figure 3a). Furthermore, in the same cells, increased expression of the p53 target genes, p21 (CDKN1A), IGFBP3 and BTG2, were observed using the same four individual siRNAs (Figure 3b).
Figure 3.
RNF31 depletion increases p53 protein levels and expression of p53 target genes. (a) RNF31 depletion increases p53 protein levels using four different siRNA oligos. MCF-7 cells were transfected with siRNF31 or siControl. After 72 h, p53 and RNF31 levels were determined by western blot analysis. Glyceraldehydes 3-phophate dehydrogenase (GAPDH) was used as internal control. (b) RNF31 depletion increases p53 target genes using four different siRNA oligos. MCF-7 cells were transfected with siRNF31 or siControl. After 72 h RNA was prepared and the expression of the endogenous p53 target genes, P21, IGFBP3 and BTG2, were determined by quantitative PCR (qPCR). Shown are the results from three experiments. n=3, *P<0.05 for siRNF31 versus siControl. (c–e) RNF31 depletion increases p53 protein levels in multiple cell lines. (c) MCF-7, (d) ZR-751 or (e) MDA-MB-175 cells were transfected with siRNF31 or siControl. After 48 h, cells were treated with 10 μM cisplatin or vehicle. p53 and RNF31 levels were determined by western blot analysis. GAPDH was used as the internal control. Each experiment was repeated for three times. The relative p53 gray density was measured by ImageJ. n=3, *P<0.05 for siRNF31 versus siControl. (f–h) RNF31 depletion increases the expression of endogenous p53 target genes. (f) MCF-7, (g) ZR-75-1 or (h) MDA-MB-175 cells were transfected with siRNF31 or siControl. After 72 h cells were treated with 10 μM cisplatin or vehicle for 6 h and RNA was prepared. The expression levels of the endogenous p53 target genes, P21, IGFBP3 and BTG2, were determined by qPCR. Shown are the results from triplicate experiments. n=3, *P<0.05 for siRNF31 versus siControl.
RNF31 depletion increased p53 levels in three distinct breast cancer cell lines that all express wt p53 (MCF-7, ZR-75-1 and MDA-MB-175;23 Figures 3c–e). Furthermore, cisplatin-induced p53 levels were further enhanced by RNF31 depletion in all these three cell lines (Figures 3c–e). In addition, RNF31 depletion caused increased expression of the p53-target genes p21, IGFBP3 and BTG2 in all the three breast cancer cell lines (Figures 3f–h). Furthermore, cisplatin-induction of these genes was further enhanced by RNF31 depletion (Figures 3f–h).
RNF31 regulates p53 protein stability
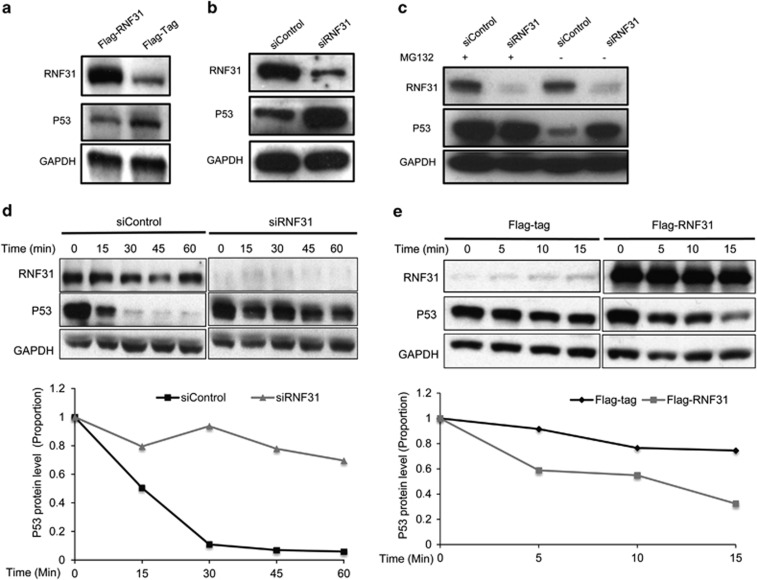
In line with the increased p53 levels on RNF31 depletion, overexpression of RNF31 decreased p53 protein levels (Figure 4a). p53 protein levels were increased within 24 h of RNF31 knockdown (Figure 4b), at a time point when p53 mRNA is not yet increased (Supplementary Figure S4A), suggesting that RNF31 directly regulates p53 protein levels. Furthermore, when cells were treated with the proteasome inhibitor MG132, there was no further increase of p53 in RNF31-depleted cells (Figure 4c). Finally, RNF31 depletion significantly increased the half-life of endogenous p53 (Figure 4d), whereas overexpression of RNF31 increases p53 degradation in MCF-7 cells (Figure 4e). Together, these data suggest that RNF31 regulates the stability and subsequent protein levels of p53.
Figure 4.
RNF31 regulates p53 protein stability. (a) Overexpression of RNF31 decreases endogenous p53 protein levels in MCF-7 cells. MCF-7 cells were transfected with plasmids expressing Flag-tagged RNF31 or the Flag tag alone. After 48 h, whole-protein extracts were prepared and the levels of RNF31, p53 and the internal control Glyceraldehydes 3-phophate dehydrogenase (GAPDH) were determined by western blot analysis. (b) RNF31 depletion increases p53 protein levels. MCF-7 cells were transfected with siRNF31 or siControl. Cells were collected after 24 h. p53 and RNF31 levels were determined by western blot analysis. GAPDH was used as internal control. (c) RNF31 depletion does not further increase the stability of p53 in the presence of the proteasome inhibitor MG132. MCF-7 cells were transfected with siRNF31 or siControl. After 48 h, cells were treated with 10 μM MG132 or vehicle. Cells were collected 6 h after treatment and whole-protein extracts were prepared. The levels of RNF31, p53 and the internal control GAPDH were determined by western blot analysis. (d) Depletion of RNF31 increases p53 protein stability. MCF-7 cells were transfected with siRNF31 or siControl. After 48 h, cells were treated with protein biosynthesis inhibitor (100 μM cycloheximide) for different times before whole-protein extraction. The levels of RNF31, p53 and the internal control GAPDH were determined by western blot analysis. ImageJ was used to quantify the p53 band density, followed by a normalization of the p53 level, with the level at time point zero set as 1. (e) Overexpression of RNF31 decreases p53 protein stability. MCF-7 cells were transfected with siRNF31 or siControl. After 48 h, cells were treated with protein biosynthesis inhibitor (100 μM cycloheximide) for different times before whole-protein extraction. The levels of RNF31, p53 and the internal control GAPDH were determined by western blot analysis. ImageJ was used to quantify the p53 band density, followed by a normalization of the p53 level, with the level at time point zero set as 1.
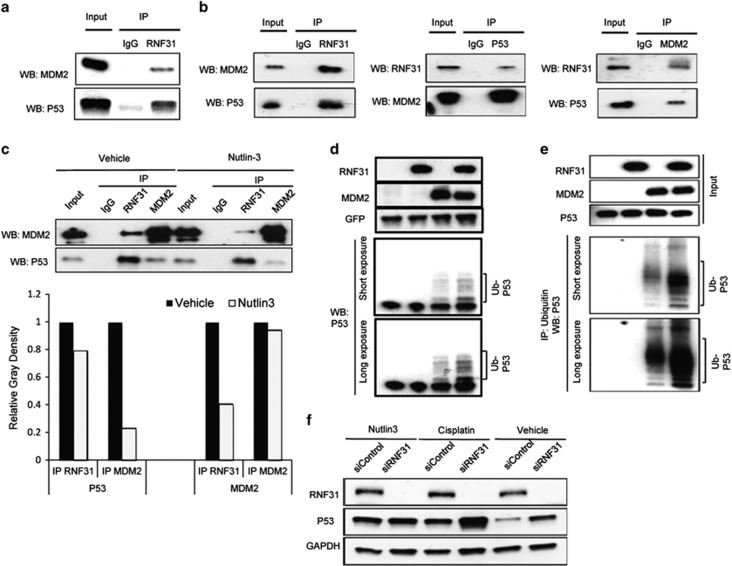
RNF31 associates with the p53/MDM2 complex and increases p53 polyubiquitination in an MDM2-dependent manner
Further support for a functional cooperation of RNF31 and p53 was obtained by co-immunoprecipitation. RNF31 associated with the p53/MDM2 complex in MCF-7 cells (Figure 5a). Owing to a limitation of available antibodies, we needed to further explore interactions between p53, MDM2 and RNF31 using proteins with appropriate Tags for antibody detection and choose to overexpress proteins in HEK293 cells. However, it was also important to confirm interactions between p53, MDM2 and RNF31 in this system, as the known regulation of MDM2 mRNA by p53 could be a confounder when we study the effects on p53 protein stability in a system where MDM2 is derived from the endogenous gene. Association of RNF31 with the p53/MDM2 complex was confirmed in HEK293 cells after the introduction of RNF31, p53 and MDM2 (Figure 5b). Figure 5c shows that RNF31 still interacts with p53 in the presence of Nutlin-3 (compare lanes 7 and 3), which blocks the interaction between p53 and MDM2 (compare lanes 8 and 4), whereas the RNF31 association with MDM2 was partially blocked by Nutlin-3. This suggests that RNF31 associates with p53 independent of the p53-MDM2 interaction, whereas RNF31's association with MDM2 at least partially depends on the p53-MDM2 interaction. HEK293 cells were used to investigate the function of RNF31 on the p53/MDM2 complex. As shown in Figures 5d and e, MDM2-mediated p53-polyubiquitination was increased by RNF31. Interestingly, in the presence of Nutlin-3, RNF31 depletion did not increase p53 protein levels or the expression of its target genes (Figure 5f and Supplementary Figure S4B). Thus, MDM2 may have an important role in mediating the RNF31-mediated effect on p53 protein stability.
Figure 5.
RNF31 associates with p53/MDM2 complex and increases p53 polyubiquitination and degradation in an MDM2-dependent manner. (a) Co-immunoprecipitation (Co-IP) assays reveal associations between endogenous RNF31 and the p53/MDM2 complex in MCF-7 cells. IgG was used as control. (b) RNF31 associates with the p53/MDM2 complex in HEK293 cells. Cells were seeded in 10-cm dishes. After 24 h, cells were transfected with 2 μg myc-MDM2 plasmid, 2 μg p53 plasmid and 2 μg Flag-RNF31 plasmid. After another 24 h, cells were collected. Co-IP was performed using antibodies as indicated. (c) Nutlin-3 does not affect the association between RNF31 and p53. MCF-7 cells were treated with vehicle or Nutlin-3 for 2 h. Co-IP was performed using antibodies as indicated. IgG was used as negative control for IP. IP of MDM2 was used as control for Nutlin-3 function. ImageJ was applied to quantify p53 and MDM2 band densities. (d) RNF31 facilitates p53 polyubiquitination. HEK293 cells were transfected with 0.5 μg each of the Flag-RNF31, myc-MDM2 and p53. Green fluorescent protein (GFP) was used as the transfection control. After 24 h, MG132 (10 μM) was added. Four hours later, whole-cell extracts were prepared for western blot analysis. (e) RNF31 facilitates interaction between p53 and ubiquitin. HEK293 cells were transfected with 0.5 μg each of the Flag-RNF31, myc-MDM2 and p53. After 24 h, MG132 (10 μM) was added. Four hours later, whole-cell extracts were prepared and thereafter subjected to immunoprecipitation using ubiquitin antibody. Western blot analysis using p53 antibody was used to detect ubiquitinated p53 forms. The predicted molecular weight of polyubiquitinated p53 is indicated. (f) The effect of RNF31 on p53 stability is dependent on interaction between p53 and MDM2. MCF-7 cells were transfected with siRNF31 or siControl and treated with vehicle, 10 μM cisplatin or 10 μM nutlin-3 for 24 h. p53 and RNF31 levels were determined by western blot analysis. Glyceraldehydes 3-phophate dehydrogenase (GAPDH) was used as an internal control.
RNF31 stabilizes MDM2 by inhibiting MDM2 polyubquitination and proteasome-mediated degradation
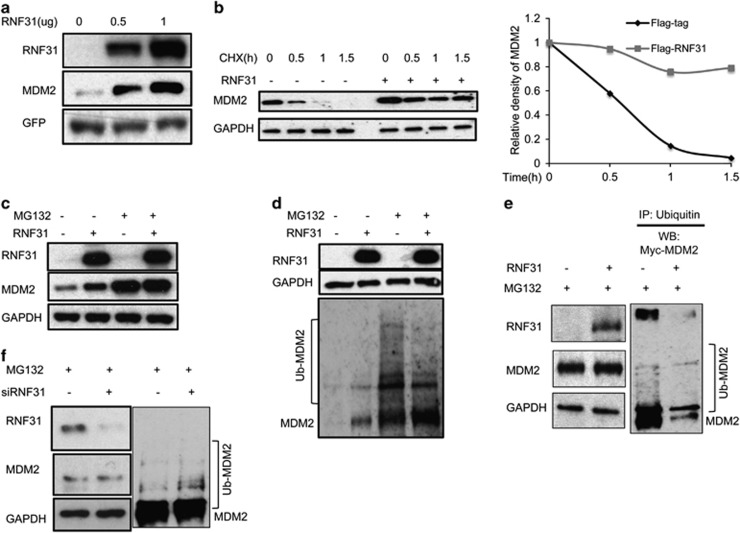
To examine how MDM2 mediates RNF31 regulation of p53 stability, we determined the effects of RNF31 on MDM2. RNF31 overexpression increased MDM2 protein levels in HEK293 cells (Figure 6a). Furthermore, RNF31 overexpression stabilized the MDM2 protein (Figure 6b). In the presence of the proteasome inhibitor (MG132), RNF31 did not further increase MDM2 protein levels (Figure 6c) and we observed a decreased polyubiquitination of MDM2 in the presence of RNF31 (Figures 6d and e). Importantly, as shown in Figure 6f, RNF31 depletion increased endogenous MDM2 polyubiquitination in MCF-7 cells. In summary, these data suggest that RNF31 stabilizes the MDM2 protein, thereby increasing p53 polyubiquitination resulting in decreased p53 protein stability.
Figure 6.
RNF31 stabilizes MDM2 and inhibits proteasome-mediated MDM2 degradation. (a) RNF31 increases MDM2 protein levels. HEK293 cells were transfected with 0.2 μg enhanced green fluorescent protein (EGFP) plasmid, 0.5 μg myc-MDM2 plasmid and the indicated amounts of Flag-RNF31 plasmid. After 24 h, whole-cell extracts were prepared for western blot analysis. (b) RNF31 increases MDM2 half-life in HEK293 cells. HEK293 cells were transfected with 0.5 μg myc-MDM2 plasmid and 0.5 μg Flag-tag or Flag-RNF31 plasmids. After 24 h, cells were treated with 100 μM cycloheximide/vehicle for indicated times. Cell lysates were prepared for western blot analysis. (c) RNF31 inhibits proteasome-mediated MDM2 degradation. HEK293 cells were transfected with 0.5 μg MYC-MDM2 plasmid and 0.5 μg Flag-tag/Flag-RNF31 plasmid. After 24 h, cells were treated with 10 μM MG132/vehicle. Cell lysis was prepared for western blot analysis. (d) RNF31 inhibits MDM2 polyubiquitination in HEK293 cells. HEK293 cells were transfected with 0.5 μg myc-MDM2 plasmid and 0.5 μg Flag-tag or Flag-RNF31 plasmids. After 24 h, cells were treated with 10 μM MG132/vehicle for 4 h. Cell lysates were prepared for western blot analysis. MDM2 was measured by MYC antibody. The expected ubiquitin bands are indicated. (e) Co-IP assay shows that RNF31 inhibits MDM2 polyubiquitination in HEK293 cells. HEK293 cells were transfected with 0.5 μg MYC-MDM2 plasmid and 0.5 μg Flag-tag or Flag-RNF31 plasmids. After 24 h, cells were treated with 10 μM MG132 or vehicle for 4 h. Ubiquitin-binding beads were used to enrich the ubiquitin-ligated MDM2. MYC antibody was used to detect the ubiquitinated and unmodified MDM2. (f) RNF31 depletion facilitates MDM2 polyubiquitination. MCF-7 cells were transfected with siRNF31 or siControl. After 48 h, cells were treated with MG132 for 4 h. MDM2 antibody was used to detect MDM2. The possible ubiquitin-ligated MDM2 bands are indicated.
Discussion
In this report we demonstrate that the RBR E3 ubiquitin ligase RNF31 associates with the MDM2/p53 complex. We propose a model where RNF31 stabilizes MDM2, resulting in increased p53 ubiquitinylation and subsequent degradation. Furthermore, RNF31 depletion induces the p53 signaling pathway, cell cycle arrest and facilitates cisplatin-induced apoptosis, making it an interesting therapeutic target.
In this study we describe the effects of RNF31 on p53 levels, cellular proliferation and apoptosis under non-stressed and stressed (cisplatin induced) conditions (Figures 2e and f). We show that RNF31 depletion was associated with increased p53 protein levels under both non-stressed and stressed conditions (Figures 3c–e). Furthermore, RNF31 depletion was associated with decreased cell proliferation under both non-stressed and stressed conditions (Figures 2b and c, and Table 1). On the contrary, RNF31 depletion was only associated with increased apoptosis under stressed conditions (Figures 2e and f). This is consistent with our observation that overexpression of p53 did not increase apoptosis under non-stressed conditions (Supplementary Figure S3). This is in line with the described diverse effects of p53 on cell proliferation and apoptosis.24, 25
RNF31 was initially cloned from breast cancer cells based on its elevated mRNA expression and shown to display higher expression in breast tumors compared with adjacent tissues.7 Based on this information, we hypothesize that deregulated expression of RNF31 may contribute to breast cancer development and/or growth. We used an unbiased approach to identify signaling pathways that are regulated by RNF31 in breast cancer cells. Interestingly, this approach identified p53 signaling as a potential target of RNF31. RING finger protein family members harbor E3 ubiquitin ligase functions and some of them have been demonstrated to modify p53 protein levels and p53 signaling.19, 20, 21, 22, 24 However, the effects could be quite different by different E3 ubiquitin ligases. For example, BRCA1 (RNF53) facilitates p53 signaling and suppress carcinogenesis in breast cancer cells,26 whereas MDM2 can suppress p53 signaling and promote breast cancer progression.19, 20 RNF31 is similar to BRCA1 in stabilizing a protein, in this case MDM2. On the other hand, RNF31 is distinct from BRCA1 in that it suppresses p53 signaling and facilitates breast cancer cell progression. Future studies need to address the relative importance of, and potential synergy between, different members of E3 ubiquitin ligases in relation to modifying p53 signaling in cancer.
The RBR family includes several atypical E3 ligases, such as RNF31, RBCK1 and Parkin (PARK2).3, 27 These proteins mainly localize to the cytoplasm, but harbor the ability to translocate to the nucleus.28 Furthermore, RBR proteins have been shown to associate with transcriptional factors and regulate the corresponding pathways.23, 28 Importantly, RBR proteins can ubiquitinate several proteins, thereby modifying their downstream signal transduction pathways. However, the functions of RBR proteins need further exploration.
As one of the RBR family members, RNF31 regulates nuclear factor-κB, ERα and c-Jun NH(2)-terminal kinase pathways in cancers.5, 29, 30, 31 RNF31 also mediates protein stabilization by ubiquitination in these pathways distinct from the classical mechanism of polyubiquitination-mediated protein degradation. Consistent with this, we demonstrate that RNF31 stabilizes MDM2, which in turn regulates p53 levels. The details of how RNF31 interacts with MDM2 need further investigation. For example, it remains to be determined whether RNF31 directly interacts with p53 and/or MDM2 by analyzing potential interactions between purified proteins.
In this study, we examined the role of RNF31 on p53 signaling in three distinct breast cancer cell lines, MCF-7, ZR-75-1 and MDA-MB-175, which are all ERα positive and express wild-type p53. We previously reported that RNF31 facilitates ERα signaling and proliferation in breast cancer cells.5 We demonstrated a non-genomic mechanism by which RNF31 could stabilize the ERα (ERα/ESR1) and enhance its signaling.5 In addition, ERα has been shown to suppress the function of p53 in relation to gene regulation.32 Therefore, it cannot be excluded that RNF31 exerts its effect on p53 via ERα signaling. The relative importance of the effect of RNF31 on ERα and p53 signaling in relation to cell proliferation needs to be addressed.
Functional p53 is required for chemotherapy-induced cell death in cancer.25, 33 Modulation of p53 protein levels is one approach to increase p53 signaling and downstream cell cycle arrest and apoptosis. Treatment with the compound Nutlin-3, which blocks the interaction between MDM2 and p53, and thereby stabilizes p53, causes cell cycle arrest and apoptosis.34, 35 Consistently, Nutlin-3 was effective in several cancers when combined with chemotherapy.36, 37, 38 We found that RNF31 depletion could arrest the cell cycle and enhance cisplatin-induced cell death, providing insight into the molecular mechanism for the regulation of p53 signaling in breast cancer cells. As a novel target to affect p53 signaling, inhibition of RNF31 activity may be a potential therapy to enhance the effect of chemotherapy.
Materials and methods
Cell culture
The original MCF-7 cells were obtained from the Michigan Cancer Foundation (Detroit, MI, USA). The MDA-MB-175 cells and HEK293 cells come from ATCC cell bank. The ZR-75-1 cells developed from ATCC (LGC Standards GmbH, Wesel, Germany) cell bank and were kindly provided by Dr Bergh Jonas. MCF-7 and HEK293 cells were cultured in DMEM (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Invitrogen) at 37 °C in a humidified atmosphere of 5% CO2 in air. ZR-75-1 and MDA-MB-175 cells were cultured in RPMI 1640 (Invitrogen) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Cisplatin was purchased from Santa Cruz (Heidelberg, Germany). Cells were periodically tested for mycoplasma contamination and were not used for more than 3 months after being thawed.
Plasmids
RNF31 (pcDNA-MYC-RNF31 and pcDNA-Flag-RNF31) constructs were previously described.28 p53 and MYC-MDM2 plasmids were obtained from the Addgene Company (Cambridge, MA, USA) and were previously described.39
siRNA transfection
Cells were transfected with 100 nM siRNA. RNF31 siRNAs correspond to the siGENOME SMARTpool RNF31 (Dharmacon, Epsom, UK; D-0021419). p53 siRNA and control siRNA are Stealth Select siRNA (Invitrogen). INTERFERin transfection reagent (Polyplus Transfection, Illkirch, France) was used according to the manufacturer's protocol.
RNA extraction and quantitative PCR analysis
RNeasy kits were used to extract total RNA (Qiagen, Stockholm, Sweden). After that 1 μg RNA was subject to reverse transcription. Next, quantitative PCR was performed as previously described.27 36B4 was used as the internal control.5 Primer sequences for quantitative PCR are provided in Supplementary Table S1.
Western blotting
Cells were lysed with NP-40 lysis buffer. After that, 10 μg protein lysis was subject to immunobloting. Anti-GAPDH (ab8425), anti-RNF31 (ab46322) and anti-caspase-3 (ab32351) antibodies were from Abcam (Cambridge, UK). Anti-Flag (F9291) and anti-GFP (G1544) antibodies were from Sigma-Aldrich (St Louis, MO, USA). Anti-myc (9E10), anti-ubiquitin (FL76) and anti-p53 (DO1 and FL-393) antibodies were from Santa Cruz.
Quantification of cell viability
ZR-75-1 and MDA-MB-175 cells were transfected with siRNF31 or siControl in 24-well plates. After 24 h, the cells were seeded into 96-well plates with 104 cells in each well. Next, cells were treated with the indicated concentration of cisplatin. The cell numbers were determined using the WST-1 cell proliferation reagent as previously described.23
Flow cytometry
For propidium iodide staining, 106 MCF-7 cells were seeded in 10-cm dishes. After 24 h, cells were transfected with siRNF31, sip53 or siControl. After an additional 24 h, cells were fixed in 70% ethanol for 30 min and stained by propidium iodide. For EdU-labeled DNA stain, cells were transfected with siRNF31, sip53 and siControl. After 24 h, 10 μM EdU was added into each plate for the last 60 min. The BD LSR II flow cytometer (BD Bioscience, Franklin Lakes, NJ, USA) was used to measure the flow of fluorescence intensity. For apoptosis assays, ZR-75-1 cells were seeded in 10-cm dishes and transfected with siRNF31, sip53 or siControl. After 24 h, cells were treated with vehicle or 10 μM cisplatin. After another 24 h, cells were stained by propidium iodide and annexin V. The BD LSR II flow was used to measure the flow fluorescence intensity.
Co-immunoprecipitation
Co-immunoprecipitation was performed essentially as previously described.40 First, 100 μg cell lysates were pre-cleared with rabbit IgG for 2 h and subsequently incubated overnight with RNF31 rabbit antibody (ab46322), whereas rabbit IgG was used as the negative control. The bound proteins were analyzed by western blot with mouse MDM2 and p53 antibody.
For overexpression experiment, HEK293 cells were transfected with 10 μg Flag-RNF31, myc-MDM2 and p53 plasmids in 10-cm dish. Cell lysates were pre-cleared with rabbit IgG for 2 h and subsequently incubated overnight with Flag rabbit antibody/myc rabbit antibody/p53 rabbit antibody, whereas rabbit IgG was used as the negative control. The bound proteins were analyzed by western blotting with mouse antibodies.
Protein stability assays
MCF-7 cells were transfected with 100 nM siRNF31 or siControl. Twenty-four hours post transfection, cells were treated with cycloheximide (100 μM) or MG132 (10 μM), or MCF-7 cells were transfected with 10 μg Flag-RNF31 or Flag vector, and 48 h post transfection cells were treated with cycloheximide (100 μM) for indicated time points. Samples were analyzed by western blotting for p53 degradation.
Analysis of protein ubiquitination
HEK293 cells (107 cells) were transfected with 4 μg pCMV-myc-MDM2 together with 4 μg pCDNA3-Flag-RNF31. Forty-eight hours post transfection, cells were treated with 10 μM MG132 or ethanol for 2 h and thereafter lysed. Modified and unmodified p53, respectively, were detected by western blot analysis.
Ubiquitination immunoprecipitation assay
HEK293 cells (107 cells) were transfected with 1 μg pcDNA3-p53 together with 1 μg pCMV-myc-MDM2 and 1 μg pCDNA3-Flag-RNF31. After 24 h, cells were treated with MG132 for 2 h after which total protein was extracted and pre-cleared by 20 μl protein A slurry and 1 μg rabbit IgG for 2 h at 4 °C. The supernatant was collected and immunoprecipitated with 20 μl ubiquitin beads (CalBiochem, Darmstadt, Germany; 662200). Western blotting with anti-p53 (DO1) antibody was performed to detect ubiquitinated p53.
Sub-network enrichment of differentially expressed genes
Analysis was performed for differentially expressed genes (P<0.001 and fold change >2) using microarray data previously published5 and the Pathway Studio software (Elsevier, Inc., Rockville, MD, USA). P-values indicating overrepresentation using the Pathway Studio Gene Ontology analysis were calculated with Fisher's exact test and considered significant when P<0.001.
Analysis of gene expression in publicly available data sets
Analysis of gene expression in 1106 breast cancer samples from The Cancer Genome Atlas was carried out in the statistical environment R.41 Presented correlations are Pearson's product–moment correlations. Samples were separated into high/low expression of RNF31 by setting a cutoff at the median value.
Statistics
Student's t-test, Pearson's correlation coefficient and Cox regression analysis were used for comparisons. A P-value of<0.05 was considered to be significant.
Acknowledgments
We thank members of the KDW, SS and ET group members for sharing materials and for valuable discussions. We thank Marie Franzen for assisting with manuscript submission and Lars-Arne Haldosen for critical comments. This study was supported by scholarships from the China Scholarship Council, a PhD student grant (KID) from the Karolinska Institutet, funding from the Center for Innovative Medicine at Karolinska Institutet, Marie Curie Actions FP7-PEOPLE-2011-COFUND [GROWTH 291795] via the VINNOVA programme Mobility for Growth, the Swedish Cancer Society, the Swedish Research Council and the Karolinska Institutet.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin 2014; 64: 52–62. [DOI] [PubMed] [Google Scholar]
- Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst 2008; 100: 672–679. [DOI] [PubMed] [Google Scholar]
- Smit JJ, Monteferrario D, Noordermeer SM, van Dijk WJ, van der Reijden BA, Sixma TK. The E3 ligase HOIP specifies linear ubiquitin chain assembly through its RING-IBR-RING domain and the unique LDD extension. Embo J 2012; 31: 3833–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson HG, Harris JW, Lin L, Brody JP. Identification of the protein Zibra, its genomic organization, regulation, and expression in breast cancer cells. Exp Cell Res 2004; 295: 448–459. [DOI] [PubMed] [Google Scholar]
- Zhu J, Zhao C, Kharman-Biz A, Zhuang T, Jonsson P, Liang N et al. The atypical ubiquitin ligase RNF31 stabilizes estrogen receptor alpha and modulates estrogen-stimulated breast cancer cell proliferation. Oncogene 2014; 33: 4340–4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DP. Cancer. p53, guardian of the genome. Nature 1992; 358: 15–16. [DOI] [PubMed] [Google Scholar]
- Huang B, Vassilev LT. Reduced transcriptional activity in the p53 pathway of senescent cells revealed by the MDM2 antagonist nutlin-3. Aging (Albany NY) 2009; 1: 845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M et al. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 2004; 303: 1010–1014. [DOI] [PubMed] [Google Scholar]
- van Oijen MG, Slootweg PJ. Gain-of-function mutations in the tumor suppressor gene p53. Clin Cancer Res 2000; 6: 2138–2145. [PubMed] [Google Scholar]
- Uji K, Naoi Y, Kagara N, Shimoda M, Shimomura A, Maruyama N et al. Significance of TP53 mutations determined by next-generation "deep" sequencing in prognosis of estrogen receptor-positive breast cancer. Cancer Lett 2014; 342: 19–26. [DOI] [PubMed] [Google Scholar]
- Williams C, Norberg T, Ahmadian A, Ponten F, Bergh J, Inganas M et al. Assessment of sequence-based p53 gene analysis in human breast cancer: messenger RNA in comparison with genomic DNA targets. Clin Chem 1998; 44: 455–462. [PubMed] [Google Scholar]
- Di Leo A, Tanner M, Desmedt C, Paesmans M, Cardoso F, Durbecq V et al. p-53 gene mutations as a predictive marker in a population of advanced breast cancer patients randomly treated with doxorubicin or docetaxel in the context of a phase III clinical trial. Ann Oncol 2007; 18: 997–1003. [DOI] [PubMed] [Google Scholar]
- Van Maerken T, Ferdinande L, Taildeman J, Lambertz I, Yigit N, Vercruysse L et al. Antitumor activity of the selective MDM2 antagonist nutlin-3 against chemoresistant neuroblastoma with wild-type p53. J Natl Cancer Inst 2009; 101: 1562–1574. [DOI] [PubMed] [Google Scholar]
- Selivanova G, Iotsova V, Okan I, Fritsche M, Strom M, Groner B et al. Restoration of the growth suppression function of mutant p53 by a synthetic peptide derived from the p53 C-terminal domain. Nat Med 1997; 3: 632–638. [DOI] [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett 1997; 420: 25–27. [DOI] [PubMed] [Google Scholar]
- Grossman SR, Deato ME, Brignone C, Chan HM, Kung AL, Tagami H et al. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science 2003; 300: 342–344. [DOI] [PubMed] [Google Scholar]
- Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 2004; 429: 86–92. [DOI] [PubMed] [Google Scholar]
- Esser C, Scheffner M, Hohfeld J. The chaperone-associated ubiquitin ligase CHIP is able to target p53 for proteasomal degradation. J Biol Chem 2005; 280: 27443–27448. [DOI] [PubMed] [Google Scholar]
- Wen W, Peng C, Kim MO, Ho Jeong C, Zhu F, Yao K et al. Knockdown of RNF2 induces apoptosis by regulating MDM2 and p53 stability. Oncogene 2014; 33: 421–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie J, Xie P, Liu L, Xing G, Chang Z, Yin Y et al. Smad ubiquitylation regulatory factor 1/2 (Smurf1/2) promotes p53 degradation by stabilizing the E3 ligase MDM2. J Biol Chem 2010; 285: 22818–22830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikulenkov F, Spinnler C, Li H, Tonelli C, Shi Y, Turunen M et al. Insights into p53 transcriptional function via genome-wide chromatin occupancy and gene expression analysis. Cell Death Differ 2012; 19: 1992–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicke RU, Sprengart ML, Wati MR, Porter AG. Caspase-3 is required for DNA fragmentation and morphological changes associated with apoptosis. J Biol Chem 1998; 273: 9357–9360. [DOI] [PubMed] [Google Scholar]
- Gustafsson N, Zhao C, Gustafsson JA, Dahlman-Wright K. RBCK1 drives breast cancer cell proliferation by promoting transcription of estrogen receptor alpha and cyclin B1. Cancer Res 2010; 70: 1265–1274. [DOI] [PubMed] [Google Scholar]
- Reinhardt HC, Schumacher B. The p53 network: cellular and systemic DNA damage responses in aging and cancer. Trends Genet 2012; 28: 128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes DM, Camplejohn RS. P53, apoptosis, and breast cancer. J Mammary Gland Biol Neoplasia 1996; 1: 163–175. [DOI] [PubMed] [Google Scholar]
- Zhang H, Somasundaram K, Peng Y, Tian H, Bi D, Weber BL et al. BRCA1 physically associates with p53 and stimulates its transcriptional activity. Oncogene 1998; 16: 1713–1721. [DOI] [PubMed] [Google Scholar]
- Tatematsu K, Yoshimoto N, Okajima T, Tanizawa K, Kuroda S. Identification of ubiquitin ligase activity of RBCK1 and its inhibition by splice variant RBCK2 and protein kinase Cbeta. J Biol Chem 2008; 283: 11575–11585. [DOI] [PubMed] [Google Scholar]
- Ehrlund A, Anthonisen EH, Gustafsson N, Venteclef N, Robertson Remen K, Damdimopoulos AE et al. E3 ubiquitin ligase RNF31 cooperates with DAX-1 in transcriptional repression of steroidogenesis. Mol Cell Biol 2009; 29: 2230–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay C, Carroll E, Ibrahim AF, Garg A, Inman GJ, Hay RT et al. E3 ubiquitin ligase HOIP attenuates apoptotic cell death induced by cisplatin. Cancer Res 2014; 74: 2246–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumati P, Dikic I. Germline polymorphisms in RNF31 regulate linear ubiquitination and oncogenic signaling. Cancer Discov 2014; 4: 394–396. [DOI] [PubMed] [Google Scholar]
- Yang Y, Schmitz R, Mitala J, Whiting A, Xiao W, Ceribelli M et al. Essential role of the linear ubiquitin chain assembly complex in lymphoma revealed by rare germline polymorphisms. Cancer Discov 2014; 4: 480–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayeed A, Konduri SD, Liu W, Bansal S, Li F, Das GM. Estrogen receptor alpha inhibits p53-mediated transcriptional repression: implications for the regulation of apoptosis. Cancer Res 2007; 67: 7746–7755. [DOI] [PubMed] [Google Scholar]
- Weller M. Predicting response to cancer chemotherapy: the role of p53. Cell Tissue Res 1998; 292: 435–445. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004; 303: 844–848. [DOI] [PubMed] [Google Scholar]
- Carvajal D, Tovar C, Yang H, Vu BT, Heimbrook DC, Vassilev LT. Activation of p53 by MDM2 antagonists can protect proliferating cells from mitotic inhibitors. Cancer Res 2005; 65: 1918–1924. [DOI] [PubMed] [Google Scholar]
- Jiang M, Pabla N, Murphy RF, Yang TX, Yin XM, Degenhardt K et al. Nutlin-3 protects kidney cells during cisplatin therapy by suppressing Bax/Bak activation. J Biol Chem 2007; 282: 2636–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirce SK, Findley HW. The MDM2 antagonist nutlin-3 sensitizes p53-null neuroblastoma cells to doxorubicin via E2F1 and TAp73. Int J Oncol 2009; 34: 1395–1402. [PubMed] [Google Scholar]
- Roberts AM, Watson IR, Evans AJ, Foster DA, Irwin MS, Ohh M. Suppression of Hypoxia-inducible factor 2 alpha restores p53 activity via Hdm2 and reverses chemoresistance of renal carcinoma cells. Cancer Res 2009; 69: 9056–9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA et al. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol 2003; 23: 8902–8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Matthews J, Tujague M, Wan J, Strom A, Toresson G et al. Estrogen receptor beta2 negatively regulates the transactivation of estrogen receptor alpha in human breast cancer cells. Cancer Res 2007; 67: 3955–3962. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012; 490: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.