Abstract
BACKGROUND: The PI3K/AKT/mTOR pathway alterations have been shown to play significant roles in the development, progression, and metastatic spread of breast cancer. Furthermore, they have been implicated in the process of drug resistance, especially endocrinal therapies. In this study, we aimed to define the correlation between the PI3K mutations and the expression of the phosphorylated forms of different downstream molecules in women with estrogen receptor (ER)–positive, human epidermal growth factor receptor 2–negative (luminal) early breast cancer treated at Cairo university hospitals. METHODS: Next-generation sequencing was used to detect mutations in the PIK3CA hotspots (in exons 9 and 20). Immunohistochemistry was performed on tissue microarray blocks prepared from samples of 35 Egyptian luminal breast cancer patients in the pathology department of Centre Léon Bérard (CLB). The intensity and the percentage of stained tumor cells were integrated to define high versus low biomarker expression. The cytoplasmic and nuclear stainings were graded separately. Patients were followed for a median of 4.7 years (2.1 to 6.9 years). Correlation was done between PI3K mutations and the immunohistochemistry expression of pAKT, LKB1, p4EBP1, and pS6 ribosomal protein (pS6RP) with the clinicopathologic features and disease free survival (DFS) of the patients. RESULTS: Median age at diagnosis was 51.3 years (range, 25 to 82 years). Tumors were larger than 20 mm in 79.2% of the cases, whereas 57.9% had axillary lymph node deposits. Only 12.3% of the patients had SBR grade I tumors, 50.8% had grade II, and 36.8% had grade III. ERs were negative in 6 patients (17%) after pathology review. Thirty-two cases were assessable for LKB1 and pAKT, 33 for p4EBP1 and pS6RP, and 24 for PI3K mutations. Nuclear LKB1, cytoplasmic LKB1, nuclear pAKT, cytoplasmic pAKT, nuclear p4EBP1, and cytoplasmic pS6RP expression was high in 65.6%, 62.5%, 62.5%, 68.8%, 42.4%, and 57.6%, respectively. PIK3CA mutations were found in 7 patients (29.2%). PI3K mutations were correlated with nuclear localization of pAKT (i.e., decreased cytoplasmic pAKT, P = .04; and increased nuclear pAKT, P = .10). There was a tendency toward an inverse correlation between PI3K mutations and the expression of pS6RP (P = .10) and p4EBP1 (P = .19). Nuclear LKB1 expression was a marker of good prognosis. It was associated with smaller tumors (P = .05), more ER (P = .08) and progesteron receptor (PgR) positivity (P = .002). In the Kaplan Meier (KM) model, patients with high nuclear LKB1 had longer DFS (hazard ratio = 0.36; 95% confidence interval, 0.15-1.10; P = .08). Nuclear pAKT high expression also carried a tendency toward longer DFS (hazard ratio = 0.51; 95% confidence interval, 0.11-1.16; P = .13). The expression of p4EBP1, pS6RP, and the PI3K mutational status did not show any prognostic significance in our cohort. CONCLUSION: Among the studied biomarkers, only nuclear expression of LKB1 and pAKT tended to predict better survival in breast cancer patients. PI3K mutation was correlated with the expression of nuclear pAKT but not pS6RP or p4EBP1.
Introduction
The PI3K/AKT/mTOR pathway is a key regulator of cell growth, proliferation, and survival in normal as well as cancer cells. Deregulation of PI3K signaling may occur through mutation in PIK3CA gene (which codes for ligand-independent PI3K catalytic subunit), overexpression of upstream receptors (such as EGFR, Her2, and IGF-R1), or loss of function of its negative regulators (PTEN, TSC, and LKB1). LKB1 (also known as serine-threonine kinase 11 or STK11) has been described to negatively control mTOR signaling derived by the intracellular energy sensing via activation of TSC 1/2. Activated PI3K is responsible for AKT phosphorylation with its subsequent translocation to the plasma membrane, where it can activate many downstream signaling networks [1], [2], [3].
Of particular interest among the Akt targets is its downstream effect on mTOR kinase. The later integrates signals from nutrients, energy status, and extracellular growth factors to regulate many cellular processes, including cell cycle progression, angiogenesis, ribosome biogenesis, and metabolism. Most of the mTOR-mediated cellular processes are executed via phosphorylation of its two downstream effectors: 4E-BP1 and S6 kinase [2]. It is of note that ongoing studies are testing specific inhibitors of S6 kinases such as saquinavir-NO in vitro and in vivo for treatment of different cancers and autoimmune conditions with promising results [4], [5].
The importance of abnormal PI3K/AKT/mTOR signaling has been demonstrated to occur in several diseases including different cancers, HIV, and HCV infections in addition to autoimmunity [6], [7]. This has highlighted the relevance of this pathway as a possible therapeutic target for these diseases. In particular, several clinical trials have been initiated to address the role of mTOR inhibitors, whether alone or in combination across different tumor types [8], [9], [10].
In breast cancer, extensive preclinical evidence has implicated PI3K/AKT/mTOR pathway deregulation in acquired resistance to endocrine therapy, which gave a molecular rational to the combined inhibition of estrogen receptor (ER) and mTOR pathways in these patients [11]. In two randomized clinical trials (TAMRAD and BOLERO2), hormone receptor (HR)–positive/human epidermal growth factor receptor 2 (HER2)–negative metastatic breast cancer patients treated with of the mTOR inhibitor everolimus in combination with endocrinal therapy exhibited a significant increase in progression-free survival (PFS) and clinical benefit compared with patients treated with endocrinal therapy alone [12], [13].
Importantly, future treatment optimization of these drugs will certainly require assessing multiple biomarkers related to the activation state of the different components of the PI3K/AKT/m-TOR pathway with validation of these markers on different cohorts of patients.
In the current study, we aimed to assess the prevalance of PI3K/AKT/m-TOR pathway effectors dysregulation, namely, pS6 ribosomal protein (pS6RP), LKB1, pAKT, and p4E-BP1 expression and PIK3CA mutations, in a group of Egyptian women with early HR +/HER2 − breast cancer. Biomarker analysis was done in collaboration with the pathology laboratory in Centre Léon Bérard (CLB), which has served as a central laboratory for the TAMRAD study. Furthermore, we tried to correlate these biomarkers with the known clinicopathologic factors and relapse events of the studied patients.
Patients and Methods
Patient Population
Between January 2007 and December 2011, we screened 400 consecutive female patients with operable primary breast cancer who underwent radical surgery and presented for postoperative management at the Department of Clinical Oncology, Cairo University. For eligibility, female patients older than 18 years at diagnosis should have a documented pathological proof of primary breast cancer and should have had their primary curative surgery (either modified radical mastectomy or breast saving surgery) and all adjuvant treatment and follow-up at our institution. The patients should have luminal breast cancer subtype, positive for ER +/− PgR (> 10%) and negative for HER2 overexpression. All patients received adjuvant endocrine therapy, either tamoxifen if premenopausal or aromatase inhibitors if postmenopausal, according to the local guidelines.
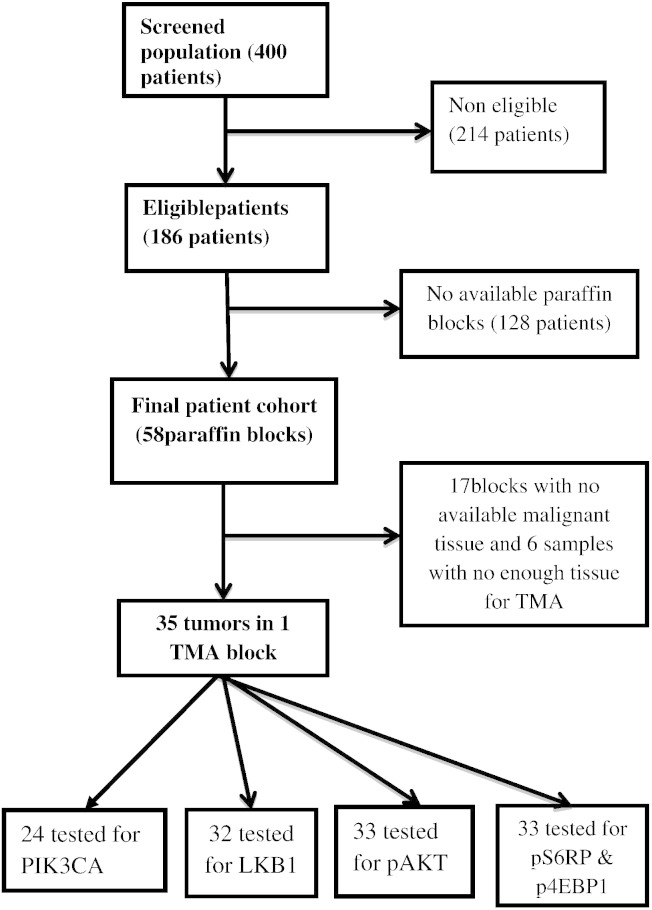
Among the screened 400 patients, 186 patients were found eligible (as per the previously mentioned criteria) for the study, and 214 cases were excluded. Paraffin blocks of tumor tissue were only available for 58 patients. Among those, not enough malignant tissue could be found in the paraffin blocks of 23 cases, and only 35 cases contained enough tumor tissue for tissue microarray (TMA) construction and biomarker testing in the pathology laboratory of CLB (Figure 1).
Figure 1.
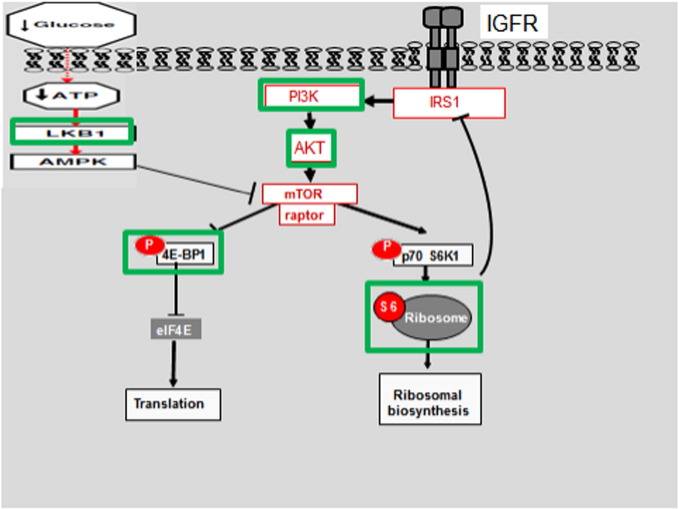
Different molecules involved in the mTOR activation cascade. Studied molecules (marked with green rectangles) are PI3K mutations, pAKT, LKB1, pS6 ribosomal protein, and p4E-BP. Adapted with modifications from Azim et al [3].
Among those 35 samples (and because of further tissue loss during sectioning of the TMA block), 32 cases were assessable for pAKT and LKB1, and 33 cases were assessable for pS6RP and p4E-BP1. PI3K hotspot mutations were assessed in 29 patients (see later). A flowchart of the whole population and subsets tested for different biomarkers is shown in Figure 2. The database was locked to further events on 30 September 2014, and all patients who were alive and/or disease free on that date were censored.
Figure 2.
A flowchart of the whole population and subsets tested for different biomarkers.
The study population was followed up for a median of 36 months (range, 15.6 to 74 months).
Patients underwent either modified radical mastectomy or breast-conserving surgery. Lymph node (LN) invasion was assessed by axillary level I and II LN dissection, and the number of LNs harboring metastasis was determined based on histologic examination. No cases had sentinel LN sampling. Tumor size was defined as the maximum tumor diameter measured on the tumor specimens at the time of surgery. ERs and progesterone receptors (PgRs) were detected by immunohistochemistry (IHC), and tumors were considered positive if they have nuclear staining in 10% or more of the tumor cells (or score of 3 or more by Allred's method). HER2 expression was determined using IHC, and tumors were considered positive if they had 3 + staining (intense staining in 10% or more cancer cells) by IHC or 2 + staining with HER2 amplification detected by fluorescence in situ hybridization.
The data exported from the patients' files for analysis included age, histologic subtype, maximum tumor size, number of LNs involved, Scarf Bloom Richardson (SBR) grade, ER, PgR expression, HER2 overexpression, date of diagnosis, date of relapse, and date of death or last follow-up. Tumor samples and clinical data were obtained under Institutional Review Board approval. This study is reported according to the “Reporting Recommendations for Tumor Marker Prognostic Studies” criteria [14].
Immunohistochemical Analysis
A 4-μm–thick section was obtained from each of the selected paraffin blocks chosen for the study. Two regions containing the highest percentage of malignant cells were selected, one for the TMA and the other for DNA extraction and PIK3CA sequencing. Breast tumor cores were inserted as triplicates (three cores for each patient) using a 600-μm needle in one TMA block. The block containing invasive carcinomas was sectioned at a thickness of 4 μm. After deparaffinization and rehydration, endogenous peroxidases were blocked by incubating the slides in 5% hydrogen peroxide in sterile water. Heat-induced antigen retrieval was performed for all the antibodies in CC1 standard on Discovery automate (Roche Diagnostic, Meylan, France) for 60 minutes.
The slides were then incubated in Discovery automate for 60 minutes with the following antibodies diluted in Dako Real Antibody Diluent (Dako, Trappes, France): anti-LKB1 (rabbit polyclonal antibody from Abcam [Cambridge, UK]) at 1/50, anti-pAKT (rabbit monoclonal antibody from Abcam) at 1/25, anti-pS6RP (rabbit monoclonal antibody 91B2 from Cell Signaling [Danvers, MA]) at 1/100, and anti-p4EBP1 antibody (rabbit polyclonal from Cell Signaling) at 1/200. Primary bound antibodies were revealed using Rb OmniMap Kit (Roche) for LKB1, pS6RP, and p4EBP1 antibodies or Rb UltraMap Kit (Roche) for pAKT antibody and then DAB ChromoMap Kit (Roche). Sections were counterstained with hematoxylin and Bluing (Roche).
Blinded to the clinical data, the biomarkers’ expression was evaluated by two observers who assessed both the percentage and the intensity of nuclear and cytoplasmic staining for LKB1 and pAKT and of nuclear staining for pS6RP and p4EBP1 in the infiltrative carcinomatous cells only. For scoring purposes, the highest intensity of staining in malignant cells was classified into four levels (0: no staining, 1: weak staining, 2: moderate staining, 3: strong staining), and the percentage of stained cells was also classified into four levels (0: no stained cells, 1: staining in less than one third of the malignant cells, 2: staining in one to two thirds of the malignant cells, 3: staining in more than two thirds of the malignant cells). Then both intensity and the percentage scores were added to conclude a single score (from 0 to 6) in a manner similar to the Allred score for ER and PR staining [15]. In cases where both a nuclear staining and cytoplasmic staining were observed, then two different scores were obtained independently, one for the nuclear and one for the cytoplasmic staining.
For the purpose of correlation and survival analysis, tumors were considered to have low expression of nuclear or cytoplasmic LKB1 if they had a score of 0 and high expression if they had a score of 1 or more. For nuclear or cytoplasmic pAKT and for pS6RP, expression was considered low with a score of 0 to 1 and high with a score of 2 or more. For p4E-BP1, expression was considered low with a score of 0 to 2 and high with a score of 3 or more.
Gene Sequencing
Paraffin blocks were scraped in a selected area containing more than 50% of tumor cells, and DNA was extracted. Twenty nanograms of DNA was used for the Ion Torrent library preparation of exons 9 and 20 of PIK3CA gene (NM_006218.2) following the manufacturer’s protocol for the Ion AmpliSeq Libray Kit 2.0 (Life Technologies). The size distribution of the DNA amplicons was analyzed on the 2200 TapeSation (Agilent) using the high-sensitivity kit (Agilent). Template preparation, emulsion PCR, and ion sphere particle enrichment was performed using the One Touch 2 kit (Life Technologies) according to manufacturer’s instructions. The ion sphere particles were loaded onto a 318 chip (Life Technologies) and sequenced using an Ion PGM 200 v2 sequencing kit (Life Technologies) on the Ion Torrent PGM for 500 cycles. The raw signal data were analyzed using NextGENe Software Suite v3.4.2 (Softgenetics; NGS). The pipeline includes quality score assignment, alignment to human genome 19 reference, mapping quality QC, coverage analysis, and variant calling.
Statistical Analysis
The correlation between LKB1, pAKT, pS6RP, and p4EBP1 expression and clinicopathologic characteristics was determined using Fisher’s exact test for categorical variables and Student's t test for numerical variables. Relapse-free survival (RFS) was defined as the time from the date of diagnosis of breast cancer to the date of any cancer recurrence (local, distant, or contralateral) or death. Overall survival was defined as the time from the date of diagnosis of breast cancer to the date of death.
The database was locked at the same time point for all patients corresponding to 30 August 2014, and no events were recorded after that point.
Survival curves and mean and median RFS (if reached), in addition to 60-month RFS and overall survival rates (with 95% confidence intervals [CIs]), were derived from Kaplan-Meier estimates, and the curves were compared using log-rank test [16]. Hazard ratios and 95% CIs were calculated using Cox regression model [17]. All statistical tests were two-sided, and in view of the small size of our sample and the exploratory nature of our analysis, the P value was considered statistically significant if less than 0.1. The statistical analyses were performed using SPSS 17.0 statistics package (SPSS Inc., Chicago, IL).
Results
Clinicopathological Characteristics
For the 58 patients eligible for the first stage of the study, the median follow-up interval was 4.7 years (range, 2.1 to 6.9 years). Median age at diagnosis was 51.3 years (range, 25 to 82 years). The majority of the patients (79.2%) had tumors larger than 2 cm in maximum dimension, and 57.9% had axillary LN metastasis. Twenty patients (35.1%) had four or more positive LN. Invasion by a malignant deposit less than 2 mm (N1mic) was not considered positive in this analysis. Most of the patients (81%) had invasive ductal carcinomas or mixed subtypes. Only 12.3% of the patients had SBR grade I tumors, 50.9% had grade II tumors, and 36.8% had grade III tumors.
Estrogen receptor was expressed in all the patient population at the central laboratory in Egypt, whereas at CLB laboratory, 6 patients (10.5%) were found to lack expression of ER. Progesterone receptor was expressed in 77.6% of the patients' samples. HER2 was found to be expressed in four patients (10%) after revision at CLB laboratory. As a result of absence of validated and standardized technique for Ki-67 testing at the time of starting our work, it was not tested in this study.
Forty-one patients (70.7%) received adjuvant chemotherapy, 15 of them received anthracyclin-based therapy, 23 of them received sequential anthracyclins and taxanes, whereas 3 patients had a taxane-only chemotherapy. Fifty-seven (98.3%) patients received adjuvant hormonal treatment, 33 of them received tamoxifen only, whereas 24 patients had sequential tamoxifen and aromatase inhibitors. Table S1 shows the clinicopathological characteristics of the whole patient cohort.
Patterns of Expression of mTOR Pathway Markers
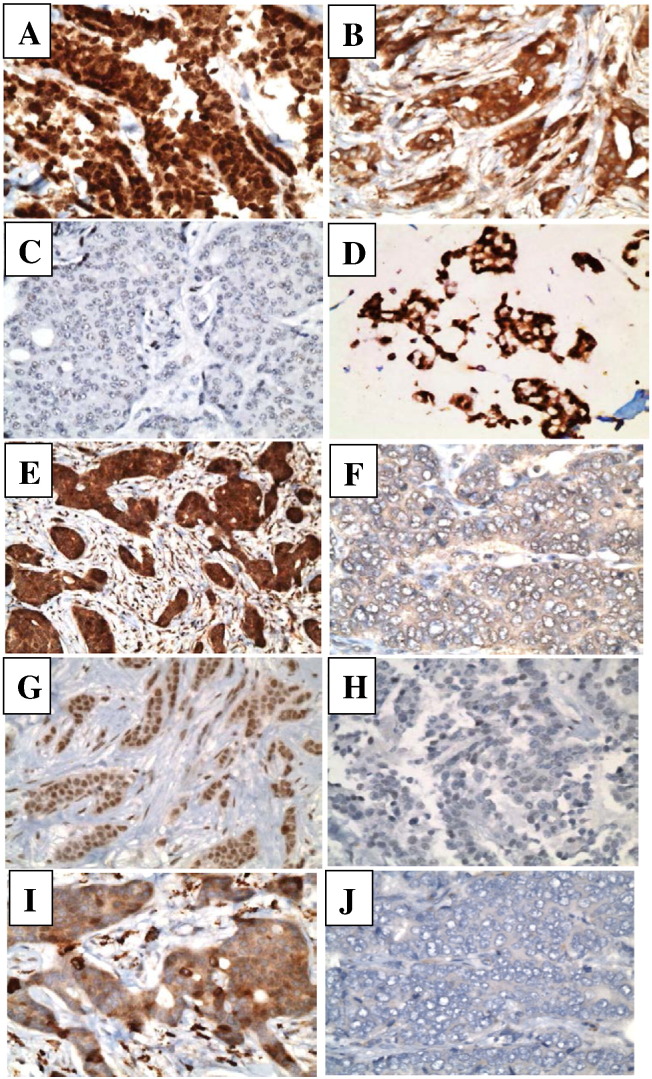
Representative images of microscopic pictures showing tumor cells with positive and negative staining for all markers are shown in Figure 3. Nuclear LKB1 expression was high in 21 cases (65.6%) (Figure 3A), whereas 11 cases (34.4%) had low expression (Figure 2C). Cytoplasmic LKB1 expression was high in 20 cases (62.5%) (Figure 3B), whereas 12 cases (37.5%) showed low expression.
Figure 3.
Representative images of microscopic pictures showing tumor cells with high expression of nuclear LKB1 (A), cytoplasmic LKB1 (B) and low expression of LKB1 (C). Tumor cells with high expression of nuclear pAKT (D), cytoplasmic pAKT (E), and low expression of pAKT (F). Tumor cells with high (G) and low (H) expression of p4E-BP1, and tumor cells with high (I) and low (J) expression of pS6RP.
Nuclear pAKT was high in 20 cases (62.5%) (Figure 3D) and low in 12 cases (37.5%), whereas cytoplasmic pAKT was high in 22 cases (68.8%) (Figure 3E) and low in 10 cases (31.2%) (Figure 3F). Phospho-4EBP1 expression was high in 14 patients (42.4%) (Figure 3G) and low in 19 patients (57.6%) (Figure 3H). Phospho-S6 ribosomal protein expression was high in 19 patients (57.6%) (Figure 3I) and low in 14 patients (42.2%) (Figure 3J).
By NGS and among the 24 patients' samples tested for PIK3CA hotspot mutations, 2 mutations were detected in exon 9 (P539R and E542K) and 5 mutations were detected in exon 20 (3 mutations in H1047L, 1 in G1049R, and 1 in R1049G), with a total of 7 patients who had mutated PIK3CA (29.2%). Four of the patients' samples could not be sequenced for the target exons despite two experiments for sequencing. Table 1 shows the expression distribution of the different biomarkers.
Table 1.
Biomarker Distribution in the Final Patient Cohort (58 Patients)
| Biomarker | Number | Percent | |
|---|---|---|---|
| PI3K hotspot mutations | Wild | 17 | 70.2% |
| Mutant | 7 | 29.2% | |
| Missing | 34 | ||
| Nuclear LKB1 | Low | 11 | 34.4% |
| High | 21 | 65.6% | |
| Missing | 26 | ||
| Cytoplasmic LKB1 | Low | 12 | 37.5% |
| High | 20 | 62.5% | |
| Missing | 26 | ||
| Nuclear pAKT | Low | 12 | 37.5% |
| High | 20 | 62.5% | |
| Missing | 26 | ||
| Cytoplasmic pAKT | Low | 10 | 31.2% |
| High | 22 | 68.8% | |
| Missing | 26 | ||
| P4EBP1 | Low | 19 | 57.6% |
| High | 14 | 42.4% | |
| Missing | 25 | ||
| pS6RP | Low | 14 | 42.2% |
| High | 19 | 57.6% | |
| Missing | 25 | ||
PI3K Hotspot Mutations Were Associated with Nuclear pAKT Localization but Not with Downstream Activation Markers
The tumors with PIK3CA hotspot mutations had more association with nuclear localization of pAKT, i.e., increased incidence of high nuclear pAKT expression (85.7% vs 53.3%; P = .10) and decreased incidence of high cytoplasmic pAKT expression (28.6% vs 73.3%; P = .04) compared with PIK3CA nonmutant tumors.
Moreover, there was a tendency toward negative correlation between PIK3CA mutations and the higher expression of the downstream substrates of mTOR. For example, in PIK3CA mutant tumors, pS6RP expression was high in 28.6% of the cases versus 62.5% in PIK3CA wild-type tumors (P = .10). Also, p4E-BP1 showed similar tendency with high expression in only 14.3% of PIK3CA mutant versus 37.5% in PIK3CA wild-type tumors (P = .19).
No correlation was found between PI3K mutational status and the expression or the subcellular localization of LKB1. A summary of correlations between the PI3K mutations by NGS and the protein expression by IHC of pAKT, pS6RP, LKB1, and p4E-BP1is shown in Table 2.
Table 2.
Correlation between PI3K Hotspot Mutations (by NGS) and Biomarker Distribution (by IHC)
| Marker | PI3K Wild No. (%) | PI3K Mutant No. (%) | P Value⁎ | |
|---|---|---|---|---|
| Nuclear LKB1 | Low | 6 (40%) | 3 (42.9%) | .89 |
| High | 9 (60%) | 4 (57.1%) | ||
| Cytopl. LKB1 | Low | 6 (40%) | 3 (42.9%) | .89 |
| High | 9 (60%) | 4 (57.1%) | ||
| Nuclear pAKT | Low | 7 (46.7%) | 1 (14.3%) | .10 |
| High | 8 (53.3%) | 6 (85.7%) | ||
| Cytopl. pAKT | Low | 4 (26.7%) | 5 (71.4%) | .04 |
| high | 11 (73.3%) | 2 (28.6%) | ||
| pS6RP | Low | 6 (37.5%) | 5 (71.4%) | .10 |
| High | 10 (62.5%) | 2 (28.6%) | ||
| P4E-BP1 | Low | 10 (62.5%) | 6 (85.7%) | .19 |
| High | 6 (37.5%) | 1 (14.3%) | ||
Correlations by Fisher's exact test.
Correlations between Expression of the Different Biomarkers Downstream of the PI3K
High nuclear and cytoplasmic LKB1 expression showed tendency for correlation with the expression of the phosphorylated forms of the downstream mTOR substrates: pS6RP and p4E-BP1. For example, in tumors with high pS6RP levels, 78.9% highly expressed LKB1, whereas only 21.1% had low LKB1 expression (P = .05).
Also, the higher the pAKT was expressed in both the cytoplasm and the nuclei, the higher was the expression of the downstream molecules. Among the 12 tumors with low nuclear pAKT expression, only 5 tumors had high pS6RP levels versus 14 of 20 tumors with high nuclear pAKT expression (P = .11). Similarly, among the 12 tumors with low pAKT levels, only 2 patients had high p4E-BP1 expression versus 12 tumors (with high p4EBP1) in the high–pAKT expression subgroup (P = .01).
Moreover, the expressions of both pS6RP and p4E-BP1 were correlated together, i.e., expression of one marker predicted the expression of the other (P = .03). Table 3 summarizes the correlations between the different biomarkers and their statistical significance.
Table 3.
Correlations between Expression of pS6RP, p4EBP1, pAKT, and LKB1
| Marker | pS6RP Low No. (%) | pS6RP High No. (%) | P Value⁎ | p4E-BP1 Low No. (%) | p4E-BP1 High No. (%) | P Value⁎ | |
|---|---|---|---|---|---|---|---|
| Nuclear pAKT | Low | 7 (53.8%) | 5 (26.3%) | P = .11 | 10 (55.6%) | 2 (14.3%) | P= .01 |
| High | 6 (46.2%) | 14 (73.7%) | 8 (44.4%) | 12 (85.7%) | |||
| Cytopl. pAKT | Low | 8 (61.5%) | 2 (10.5%) | P< .01 | 8 (44.4%) | 2 (14.3%) | P= .06 |
| High | 5 (38.5%) | 17 (89.5%) | 10 (55.6%) | 12 (85.7%) | |||
| Nuclear LKB1 | Low | 7 (53.8%) | 4 (21.1%) | P= .05 | 8 (44.4%) | 3 (21.4%) | P = .17 |
| High | 6 (46.2%) | 15 (78.9%) | 10 (55.6%) | 11 (78.6%) | |||
| Cytopl. LKB1 | Low | 8 (61.5%) | 4 (21.1%) | P= .02 | 9 (50.0%) | 3 (21.4%) | P= .09 |
| high | 5 (38.5%) | 15 (78.9%) | 9 (50.0%) | 11 (78.6%) | |||
| P4E-BP1 | Low | 11 (78.6%) | 8 (42.1%) | P= .03 | |||
| High | 3 (21.4%) | 11 (57.9%) | |||||
Correlations tested by Fisher's exact test.
Nuclear LKB1 Association with Good Prognosis
Patients with tumors that highly express LKB1 in their nuclei were more likely to be younger than 50 years (P = .06) and premenopausal (P = .06). Nuclear LKB1 expression was associated with features of good prognosis such as smaller tumor sizes (P = .05) and more ER positivity (P = .08), PgR positivity (P = .002), and HER2 negativity (P = .06).
Cytoplasmic LKB1, on the other hand, was not associated with age or with ER, PgR, or HER2 status, but its expression was associated with smaller tumors (P = .03). Table S2 shows a summary of the correlations between nuclear and cytoplasmic expression of LKB1 with the different clinicopathologic factors.
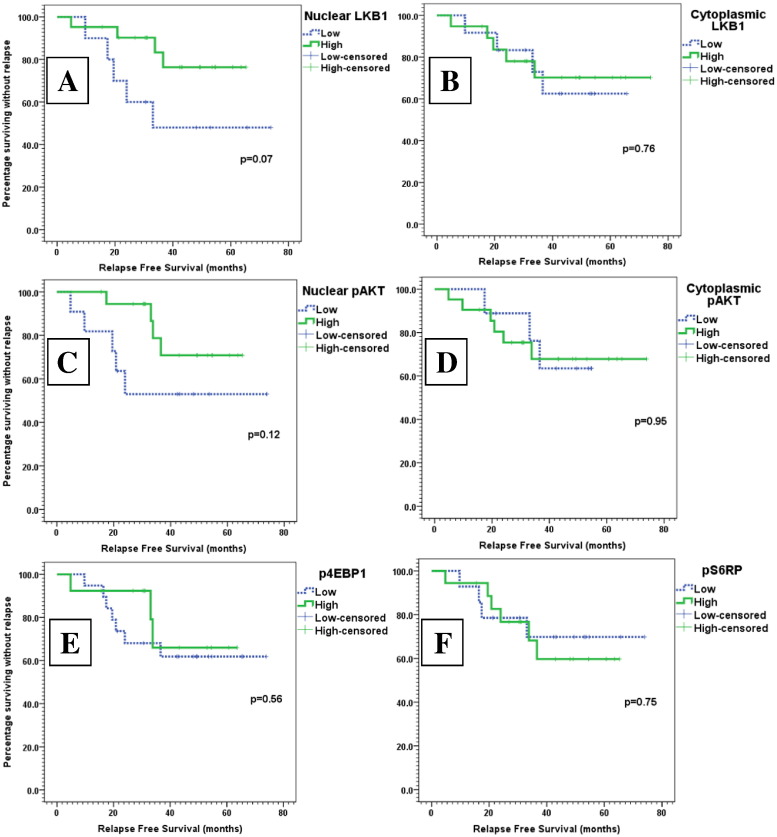
In the Kaplan-Meier model, the 60-month RFS rate of patients with high expression of nuclear LKB1 was 76.3% versus 48% in patients with low expression (P = .07). In the Cox univariate model, nuclear LKB1 expression was associated with decreased risk of recurrence or death (hazard ratio = 0.36; 95% CI, 0.15 to 1.10; P = .08).
However, expression of cytoplasmic LKB1 was not predictive of the outcome of breast cancer (hazard ratio = 0.81; 95% CI, 0.22 to 3.03; P = .76). The Kaplan-Meier curves for high versus low expression of nuclear and cytoplasmic LKB1 are shown in Figure 4, A and B.
Figure 4.
Kaplan-Meier curves of RFS (months) across different levels of biomarker expression: high (green) versus low (dotted blue) expression of nuclear and cytoplasmic LKB1, nuclear and cytoplasmic pAKT, p-4E-BP1, and p-S6RP.
Nuclear pAKT Had a Prognostic Significance
Patients with tumors that highly express nuclear pAKT tended to have more ER and PgR positivity (P = .10 and .15, respectively), whereas cytoplasmic expression of pAKT was associated with younger age and more premenopausal women (P = .11) and did not show any correlation with the different histopathologic factors. Table S3 shows a summary of the correlations between nuclear and cytoplasmic expression of pAKT with the different clinicopathologic factors.
In the Kaplan-Meier model, the 60-month RFS rate of patients with high expression of nuclear pAKT was 70.8% versus 53% in patients with low expression (P = .12). In the Cox univariate model, nuclear pAKT expression tended to decrease risk of recurrence or death (hazard ratio = 0.51; 95% CI, 0.11 to 1.16; P = .13).
However, expression of cytoplasmic pAKT did not predict the outcome of breast cancer (hazard ratio = 1.11; 95% CI, 0.28 to 4.45; P = .88). Kaplan-Meier curves for high versus low expression of nuclear and cytoplasmic pAKT are shown in Figure 4, C and D.
Impact of Downstream Markers Expression
Tumors with high expression of p4E-BP1 and pS6RP were more likely to occur in patients younger than 50 years (P = .01 and .07, respectively) and in premenopausal women (P = .01 and .07, respectively). Such high expression of p4E-BP1 and pS6RP did not consistently correlate with the tumor stage or with the biological characteristics of breast cancer such as HR status, HER2 status, and SBR grade. A summary of these correlations is shown in Table S4.
Regarding the RFS analysis, neither p4E-BP1 nor pS6RP was a significant prognostic factor in patients with luminal breast cancer. For p4E-BP1 high expression, HR for relapse or death was 0.61 (95% CI, 0.16 to 2.37; P = .48), and for pS6RP high expression, HR for relapse or death was 1.15 (95% CI, 0.32 to 4.07; P = .83). Figure 4, E and F, shows the Kaplan-Meier curves for RFS of high versus low expression of p4E-BP1 and pS6RP.
Discussion
Endocrine therapy is the cornerstone treatment in luminal breast cancers. Nevertheless, more than 50% of HR-positive patients will escape hormone sensitivity in the adjuvant setting, with subsequent relapse of their disease, representing a major therapeutic challenge in such patients. Extensive evidence has implicated PI3K/AKT/mTOR axis aberrations in the process of secondary endocrinal resistance, which gave a strong rational for the development of several PI3K pathways inhibitors in clinical trials. Our study was designed in 2011, influenced by the impressive results of the mTOR inhibitor everolimus in prolonging PFS in patients with HR +/HER − metastatic breast cancer in the context of aromatase inhibitors resistance. On the other hand, pivotal preclinical studies have strongly suggested that earlier intervention with combined endocrine and PI3K-directed therapies could limit escape from endocrine therapy (ET) during the adjuvant phase of the disease, a hypothesis that is now being tested in at least two ongoing phase 3 studies. Of note, the toxicity from this combination is significant, and many patients treated with this approach during the metastatic settings were reported to decline treatment as a result of adverse events. Therefore, it is of prime importance to explore specific tumor biomarkers that can a priori identify those patients who might benefit from such effective, albeit toxic, treatment, particularly in the adjuvant setting. Accordingly, in our study, we tried to characterize some molecular changes associated with PIK3CA mutation status and their prognostic relevance in a cohort of ER +/HER2 − Egyptian patients with early breast cancer treated with adjuvant endocrine therapy.
Our study was designed in 2011, influenced by the promising success of mTOR inhibitors in improving the outcome of metastatic breast cancer patients, particularly in the aspect of overcoming resistance to hormonal therapies. The different studies showing high prevalence of the PI3K/AKT/mTOR axis aberrations in that population, in addition to the success of the mTOR inhibitor everolimus in the prolongation of PFS in ER-positive, HER2-negative advanced breast cancer, offered an open opportunity to explore the potential role of the different components of the PI3K/AKT/mTOR in the development of hormone resistance in breast cancer. Such resistance in the adjuvant setting should manifest by shorter RFS.
In the current report, we detected PIK3CA hotspot mutations in exons 9 and 20 in 29.2% of the studied population. This is concordant with what has been reported by others [18], [19], [20] including a single study on a relatively similar ethnicity from Saudia Arabia, which detected PIK3CA mutations in 32% in a cohort of 50 cases of luminal breast cancer [21]. Of notice, a higher incidence of PIK3CA mutations is consistently reported among patients with strong ER and PR expression status compared with other breast cancer phenotypes. In a recent study by the Cancer Genome Atlas Network, PIK3CA mutations were detected in 45% and 29% of luminal A and luminal B subtypes, respectively [22].
Prognostically speaking, it has been originally suggested that PIK3CA mutations could be responsible for the oncogenic deregulation in the poor prognosis luminal breast cancers, and consequently, these patients would be most suitable for treatment by PI3K/m-TOR pathway inhibitor.
In our analysis, no prognostic impact of the PIK3CA mutational status was found in our small cohort of luminal breast cancers. Other larger studies described conflicting results on the prognostic value of PIK3CA mutational status in this patient population [18], [20], [23]. The largest, and probably the most reliable, data set regarding the prognostic significance of PIK3CA mutations in luminal breast cancer evolved from a study of 4540 patients with luminal breast cancers included in the TEAM trial. In this study, PIK3CA mutations (found in approximately 40% of the cases) were associated with a significantly better 5-year distant RFS (hazard ratio, 0.76; 95% CI, 0.63 to 0.91; P = .003). However, after adjustment for other clinical and biological parameters, such favorable prognostic impact did not remain significant in a multivariate analysis (hazard ratio, 0.92; 95% CI, 0.75 to 1.12; P = .4012) [24]. We think that the TEAM results in addition to other smaller studies including ours would probably close the debate on the prognostic value of PI3K mutation, which may display a limited favorable prognostic impact, but it cannot be considered a strong independent predictor of outcome in the context of hormone receptor (HR) positive early breast cancer (EBC). Indeed, PIK3CA mutations in EBC may serve as a marker of highly hormone-dependent, relatively indolent tumors in which ER and PI3K signaling coexists but with a dominant effect of ER transcription. Of note, PIK3CA mutations in endometrial cancer were also reported to be associated with a less aggressive phenotype.
In the metastatic settting, a strong preclinical evidence on the involvment of PI3K/mTOR pathway activation in the development of acquired (secondary) resistance to hormonal therapy has been well acknowledged. In the clinic, this was clearly evident by the therapeutic benefit achieved by the addition of everolimus to hormonal therapy in two randomized trials [12], [13]. However, the predictive role of PIK3CA mutation on the likelihood to benefit from mTOR inhibitors could not be demonstrated in either trial [25], [26]. More interiguingly, Loi et al., in a cohort of 58 HR +/HER2 − patients, have shown that patients with gene signature associated to PIK3CA wild-type—and not PIK3CA mutation—were those who made a significant therapeutic benefit from everolimus plus letrozole compared with letrozole alone when given during the neoadjuvant setting [27]. Taken together, although high hopes were expected from the presence of PIK3CA mutations to serve as a prognostic and/or predictive biomarker in HR +/HER − breast cancers, the current evidence does not indicate a clinically relevant role of PIK3CA mutations in this cohort of patients. This stands in clear contradistinction with the situation in HER-positive metastatic breast cancer where PIK3CA mutations have been shown to be associated with a significantly worse prognosis and less response to trastuzumab treatment.
An important result detected in our work is the correlation between PI3K mutations and the nuclear localization of pAKT which might indicate its activation. However, this was not correlated with higher expression of downstream effectors of the mTOR pathway. On the contrary, PIK3CA mutated cases tended to have lower expression of pS6RP and p4EBP1 compared with patients with wild-type PIK3CA. Importantly, an extensive study, including 1800 HR + breast cancer patients, trying to link PIK3CA mutation with mTOR pathway activation (as detected by gene signature) could not demostrate a positive correlation between the two events. Paradoxically, PIK3CA mutations in this study were associated with a low mTORC1 signaling gene signature [28]. This underscores the importance of other alternative pathways responsible for mTOR activation and the potential role of complex feedback loops regulated signaling of this pathway.
Regarding the pAKT expression, we found different patterns of expression of nuclear and cytoplasmic pAKT. No correlation was found between the nuclear and cytoplasmic expression, denoting complete independence of both stainings. We might hence conclude that this molecule has different actions depending on its subcellular localization. Only nuclear expression of pAKT carried tendency to predict a longer RFS that did not reach statistical significance because of the small sample size. This finding is opposite to what is expected from the role of pAKT in tumor progression and growth from preclinical studies and several other retrospective analyses [29], [30], [31], [32]. However, in the same line with our work, a study conducted by Badve et al. in 377 breast cancer patients found a better outcome in patients with nuclear localization of pAKT in ER/PgR-positive patients [33].
In our work (as with pAKT), high nuclear LKB1 expression showed a tendency to predict better DFS and was associated with markers of good prognosis such as smaller tumor size, and ER and PgR positivity. Such associations were not found with the cytoplasmic expression. These findings are concordant with what was observed by Bouchekioua-Bouzaghou and colleagues about the differential prognostic impact of the subcellular localization of LKB1. In their study, they described a better outcome with nuclear LKB1 and a poorer survival with cytoplasmic LKB1 expression [34].
A growing interest in LKB1 expression has increased after the translational study of TAMRAD showed that patients with low cytoplasmic LKB1 were more likely to benefit from the addition of everolimus, and suggested an inverse correlation between cytoplasmic LKB1 and nuclear p4EBP1. However, in a larger series of breast cancers (n = 154), the same group demonstrated a positive correlation between both markers, which seems to contradict the former results [34]. In fact, no statistical analysis has been done for such correlation in the TAMRAD study which aimed at discovering predictive markers for everolimus response.
We have also studied the expression of the downstream markers of mTOR activation: pS6RP and p4E-BP1. Both biomarkers’ expressions were correlated, i.e., pS6RP and p4E-BP1 had a high probability of mutual expression. Neither the high expression of p4E-BP1 nor that of pS6RP had an impact on the prognosis of breast cancer patients.
Despite the unclear prognostic impact of the upstream biomarkers, the downstream molecules (particularly S6K and p4E-Bp1) predicted poorer outcome of breast cancer in most of the studied series. In a Swedish study done on more than 700 patients from the Stockholm tamoxifen studies, high mRNA levels of S6K2 and 4E-BP1 (which also had strong tendency toward mutual expression) were associated with poor prognostic factors such as higher grade and larger tumors and were independent predictors of poor survival. In the same study, higher 4E-BP1 and p4E-BP1 expression by IHC predicted distant recurrences and breast cancer mortality [35]. Such notion (of poor prognostic impact of the downstream markers) is in line with other retrospective studies [36], [37], [38]. Such impact was not observed in our sample of Egyptian patients probably as a result of small sample size.
Although our work was comprehensive in terms of the spectrum of biomakers analyzed, their subcellular localization, and their prognostic impact, still it is obviously limited by its retrospective nature, the use of TMA sections that bear only cores of the whole tumor, and the heterogeneity of our patient cohort regarding the adjuvant treatment received that may bias the results. In our opinion, the biggest limitation is the small size of our sample that makes drawing solid conclusions very difficult. A larger prospective biomarker-oriented study powered to account for the various prognostic variables in a multivariate analysis is needed to further clarify the missing pieces of the PI3K/AKT/mTOR story.
Finally, this study is throwing lights on the technical difficulties to conduct this kind of retrospective-prospective studies in developing countries, where the absence of standardized tumor tissue banks is still a major problem. The importance of collaborative translational studies like the one we did in our work will certainly support the idea of having a high-quality research on different ethnic groups where breast cancer patients may exhibit a molecularly different profile.
The extreme complexity of the mTOR pathway activation mechanisms, the multiplicity and redundancy of the interactions between its components, and the presence of various feedback loops that control mTOR activation are all responsible for the inconsistent results of similar studies, and that is why none of those biomarkers were adopted into clinical practice. What we know so far is that suppression of the mTOR pathway can improve the outcome in a subgroup of luminal breast cancer patients, and this means that the mTOR pathway activation is responsible for tumor progression in such subgroup. What we also know (from the translational data of the BOLERO2 and TAMRAD studies) is that a consistent PIK3CA mutational status is not predictive of benefit from the mTOR inhibitors.
In conclusion, our work supports the idea of the presence of at least two pathways for mTOR activation: AKT dependent and AKT independent. PIK3CA hotspot mutations in exons 9 and 20 are mostly associated with AKT activation but have little correlation with the downstream markers such as p4E-BP1 and pS6RP. AKT-independent mTOR activation, which is associated with wild-type PIK3CA and high expression of the downstream markers, seems to define a subgroup of tumors that might be deriving the most benefit from rapamycin-like mTOR inhibitors. Our findings need further confirmation in larger validation cohort and longer follow-up to test for the effects of other prognostic variables.
Disclosures
None of the authors has any conflict of interests to disclose.
Acknowledgments
The authors acknowledge the support done by Novartis Oncology Egypt team in funding resources for biomarker testing. We also acknowledge the efforts of Amelie Colombe in samples preparation and of J. Perrossier and V. Attignon in mutation testing.
Footnotes
Funding: This work was in part funded by a research grant from Novartis Oncology Egypt.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.tranon.2016.01.001.
Contributor Information
Hamdy A. Azim, Email: Azimonc@cairocure.com.
Loay Kassem, Email: Loay.kassem@cairocure.com.
Appendix A. Supplementary data
Table S1. Clinicopathological Characteristics in the Final Patient Cohort (58 Patients)
Table S2: Correlation Between Nuclear and Cytoplasmic LKB1 Expression and the Clinicopathologic Factors
Table S3: Correlation Between Nuclear and Cytoplasmic paKT Expression With The Clinicopathologic Factors
Table S4: Correlation Between pS6RP, p4EBP1 Expression With The Clinicopathologic Factors
References
- 1.Katso R, Okkenhaug K, Ahmadi K, White S, Timms J, Waterfield MD. Cellular function of phosphoinositide 3-kinases: implications for development, homeostasis, and cancer. Annu Rev Cell Dev Biol. 2001;17:615–675. doi: 10.1146/annurev.cellbio.17.1.615. [DOI] [PubMed] [Google Scholar]
- 2.Yip CK, Murata K, Walz T, Sabatini DM, Kang SA. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol Cell. 2010;38(5):768–774. doi: 10.1016/j.molcel.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azim H, Azim HA, Jr., Escudier B. Targeting mTOR in cancer: renal cell is just a beginning. Target Oncol. 2010;5(4):269–280. doi: 10.1007/s11523-010-0141-x. [DOI] [PubMed] [Google Scholar]
- 4.Maksimovic-Ivanic D, Mojic M, Bulatovic M, Radojkovic M, Kuzmanovic M, Ristic S, Stosic-Grujicic S, Miljkovic D, Cavalli E, Libra M. The NO-modified HIV protease inhibitor as a valuable drug for hematological malignancies: role of p70S6K. Leuk Res. 2015;39(10):1088–1095. doi: 10.1016/j.leukres.2015.06.013. [DOI] [PubMed] [Google Scholar]
- 5.Petkovic F, Blazevski J, Momcilovic M, Timotijevic G, Zocca MB, Mijatovic S, Maksimović-Ivanić D, Mangano K, Fagone P, Stošić-Grujičić S. Saquinavir-NO inhibits S6 kinase activity, impairs secretion of the encephalytogenic cytokines interleukin-17 and interferon-gamma and ameliorates experimental autoimmune encephalomyelitis. J Neuroimmunol. 2013;259(1-2):55–65. doi: 10.1016/j.jneuroim.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 6.Nicoletti F, Fagone P, Meroni P, McCubrey J, Bendtzen K. mTOR as a multifunctional therapeutic target in HIV infection. Drug Discov Today. 2011;16(15-16):715–721. doi: 10.1016/j.drudis.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 7.Donia M, Mangano K, Amoroso A, Mazzarino MC, Imbesi R, Castrogiovanni P, Coco M, Meroni P, Nicoletti F. Treatment with rapamycin ameliorates clinical and histological signs of protracted relapsing experimental allergic encephalomyelitis in Dark Agouti rats and induces expansion of peripheral CD4 + CD25 + Foxp3 + regulatory T cells. J Autoimmun. 2009;33(2):135–140. doi: 10.1016/j.jaut.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Baselga J, Semiglazov V, van Dam P, Manikhas A, Bellet M, Mayordomo J, Campone M, Kubista E, Greil R, Bianchi G. Phase II randomized study of neoadjuvant everolimus plus letrozole compared with placebo plus letrozole in patients with estrogen receptor–positive breast cancer. J Clin Oncol. 2009;27(16):2630–2637. doi: 10.1200/JCO.2008.18.8391. [DOI] [PubMed] [Google Scholar]
- 9.Yao JC, Shah MH, Ito T, Bohas CL, Wolin EM, Van Cutsem E, Hobday TJ, Okusaka T, Capdevila J, de Vries EG. Everolimus for advanced pancreatic neuroendocrine tumors. N Engl J Med. 2011;364(6):514–523. doi: 10.1056/NEJMoa1009290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ray-Coquard I, Favier L, Weber B, Roemer-Becuwe C, Bougnoux P, Fabbro M, Floquet A, Joly F, Plantade A, Paraiso D. Everolimus as second-or third-line treatment of advanced endometrial cancer: ENDORAD, a phase II trial of GINECO. Br J Cancer. 2013;108(9):1771–1777. doi: 10.1038/bjc.2013.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steelman LS, Navolanic P, Chappell WH, Abrams SL, Wong EW, Martelli AM, Cocco L, Stivala F, Libra M, Nicoletti F. Involvement of Akt and mTOR in chemotherapeutic- and hormonal-based drug resistance and response to radiation in breast cancer cells. Cell Cycle. 2011;10(17):3003–3015. doi: 10.4161/cc.10.17.17119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bachelot T, Bourgier C, Cropet C, Ray-Coquard I, Ferrero JM, Freyer G, Abadie-Lacourtoisie S, Eymard JC, Debled M, Spaëth D. Randomized phase II trial of everolimus in combination with tamoxifen in patients with hormone receptor-positive, human epidermal growth factor receptor 2–negative metastatic breast cancer with prior exposure to aromatase inhibitors: a GINECO study. J Clin Oncol. 2012;30(22):2718–2724. doi: 10.1200/JCO.2011.39.0708. [DOI] [PubMed] [Google Scholar]
- 13.Baselga J, Campone M, Piccart M, Burris HA, III, Rugo HS, Sahmoud T, Noguchi S, Gnant M, Pritchard KI, Lebrun F. Everolimus in postmenopausal hormone-receptor–positive advanced breast cancer. N Engl J Med. 2012;366(6):520–529. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. REporting recommendations for tumor MARKer prognostic studies (REMARK) Breast Cancer Res Treat. 2006;100(2):229–235. doi: 10.1007/s10549-006-9242-8. [DOI] [PubMed] [Google Scholar]
- 15.Allred DC, Bustamante MA, Daniel CO, Gaskill HV, Cruz AB., Jr. Immunocytochemical analysis of estrogen receptors in human breast carcinomas. Evaluation of 130 cases and review of the literature regarding concordance with biochemical assay and clinical relevance. Arch Surg. 1990;125(1):107–113. doi: 10.1001/archsurg.1990.01410130113018. [DOI] [PubMed] [Google Scholar]
- 16.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53(282):457–481. [Google Scholar]
- 17.Cox DR. Regression models and life-tables. J R Stat Soc B. 1972;34(2):187–220. [Google Scholar]
- 18.Abramson VG, Cooper Lloyd M, Ballinger T, Sanders ME, Du L, Lai D, Su Z, Mayer I, Levy M, LaFrance DR. Characterization of breast cancers with PI3K mutations in an academic practice setting using SNaPshot profiling. Breast Cancer Res Treat. 2014;145(2):389–399. doi: 10.1007/s10549-014-2945-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dupont Jensen J, Laenkholm AV, Knoop A, Ewertz M, Bandaru R, Liu W, Hackl W, Barrett JC, Gardner H. PIK3CA mutations may be discordant between primary and corresponding metastatic disease in breast cancer. Clin Cancer Res. 2011;17(4):667–677. doi: 10.1158/1078-0432.CCR-10-1133. [DOI] [PubMed] [Google Scholar]
- 20.Arsenic R, Lehmann A, Budczies J, Koch I, Prinzler J, Kleine-Tebbe A, Schewe C, Loibl S, Dietel M, Denkert C. Analysis of PIK3CA mutations in breast cancer subtypes. Appl Immunohistochem Mol Morphol. 2014;22(1):50–56. doi: 10.1097/pdm.0b013e318297afea. [DOI] [PubMed] [Google Scholar]
- 21.Karakas B, Colak D, Kaya N, Ghebeh H, Al-Qasem A, Hendrayani F, Toulimat M, Al-Tweigeri T, Park BH, Aboussekhra A. Prevalence of PIK3CA mutations and the SNP rs17849079 in Arab breast cancer patients. Cancer Biol Ther. 2013;14(10):888–896. doi: 10.4161/cbt.25945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancer-Genome-Atlas-Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mangone FR, Bobrovnitchaia IG, Salaorni S, Manuli E, Nagai MA. PIK3CA exon 20 mutations are associated with poor prognosis in breast cancer patients. Clinics (Sao Paulo) 2012;67(11):1285–1290. doi: 10.6061/clinics/2012(11)11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sabine VS, Crozier C, Brookes CL, Drake C, Piper T, van de Velde CJ, Hasenburg A, Kieback DG, Markopoulos C, Dirix L. Mutational analysis of PI3K/AKT signaling pathway in tamoxifen exemestane adjuvant multinational pathology study. J Clin Oncol. 2014;32(27):2951–2958. doi: 10.1200/JCO.2013.53.8272. [DOI] [PubMed] [Google Scholar]
- 25.Hortobagyi G, Piccart-Gebhart M, Rugo H, Burris H, Campone M, Noguchi S. Correlation of molecular alterations with efficacy of everolimus in hormone receptor–positive, HER2-negative advanced breast cancer: results from BOLERO-2. J Clin Oncol. 2013;31(Suppl. 15):509. [Google Scholar]
- 26.Treilleux I, Arnedos M, Cropet C, Wang Q, Ferrero JM, Abadie-Lacourtoisie S, Levy C, Legouffe E, Lortholary A, Pujade-Lauraine E. Translational studies within the TAMRAD randomized GINECO trial: evidence for mTORC1 activation marker as a predictive factor for everolimus efficacy in advanced breast cancer. Ann Oncol. 2015;26(1):120–125. doi: 10.1093/annonc/mdu497. [DOI] [PubMed] [Google Scholar]
- 27.Loi S, Michiels S, Baselga J, Bartlett JM, Singhal SK, Sabine VS, Sims AH, Sahmoud T, Dixon JM, Piccart MJ. PIK3CA genotype and a PIK3CA mutation-related gene signature and response to everolimus and letrozole in estrogen receptor positive breast cancer. PLoS One. 2013;8(1):e53292. doi: 10.1371/journal.pone.0053292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loi S, Haibe-Kains B, Majjaj S, Lallemand F, Durbecq V, Larsimont D, Gonzalez-Angulo AM, Pusztai L, Symmans WF, Bardelli A. PIK3CA mutations associated with gene signature of low mTORC1 signaling and better outcomes in estrogen receptor–positive breast cancer. Proc Natl Acad Sci U S A. 2010;107(22):10208–10213. doi: 10.1073/pnas.0907011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vestey SB, Sen C, Calder CJ, Perks CM, Pignatelli M, Winters ZE. Activated Akt expression in breast cancer: correlation with p53, Hdm2 and patient outcome. Eur J Cancer. 2005;41(7):1017–1025. doi: 10.1016/j.ejca.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 30.Perez-Tenorio G, Stal O. Activation of AKT/PKB in breast cancer predicts a worse outcome among endocrine treated patients. Br J Cancer. 2002;86(4):540–545. doi: 10.1038/sj.bjc.6600126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tokunaga E, Kimura Y, Oki E, Ueda N, Futatsugi M, Mashino K, Yamamoto M, Ikebe M, Kakeji Y, Baba H. Akt is frequently activated in HER2/neu-positive breast cancers and associated with poor prognosis among hormone-treated patients. Int J Cancer. 2006;118(2):284–289. doi: 10.1002/ijc.21358. [DOI] [PubMed] [Google Scholar]
- 32.Benesch C, Schneider C, Voelker HU, Kapp M, Caffier H, Krockenberger M, Dietl J, Kammerer U, Schmidt M. The clinicopathological and prognostic relevance of pyruvate kinase M2 and pAkt expression in breast cancer. Anticancer Res. 2010;30(5):1689–1694. [PubMed] [Google Scholar]
- 33.Badve S, Collins NR, Bhat-Nakshatri P, Turbin D, Leung S, Thorat M, Dunn SE, Geistlinger TR, Carroll JS, Brown M. Subcellular localization of activated AKT in estrogen receptor– and progesterone receptor–expressing breast cancers: potential clinical implications. Am J Pathol. 2010;176(5):2139–2149. doi: 10.2353/ajpath.2010.090477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bouchekioua-Bouzaghou K, Poulard C, Rambaud J, Lavergne E, Hussein N, Billaud M, Bachelot T, Chabaud S, Mader S, Dayan G. LKB1 when associated with methylatedERalpha is a marker of bad prognosis in breast cancer. Int J Cancer. 2014;135(6):1307–1318. doi: 10.1002/ijc.28781. [DOI] [PubMed] [Google Scholar]
- 35.Karlsson E, Perez-Tenorio G, Amin R, Bostner J, Skoog L, Fornander T, Sgroi DC, Nordenskjöld B, Hallbeck AL, Stål O. The mTOR effectors 4EBP1 and S6K2 are frequently coexpressed, and associated with a poor prognosis and endocrine resistance in breast cancer: a retrospective study including patients from the randomised Stockholm tamoxifen trials. Breast Cancer Res. 2013;15(5):R96. doi: 10.1186/bcr3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perez-Tenorio G, Karlsson E, Waltersson MA, Olsson B, Holmlund B, Nordenskjold B, Fornander T, Skoog L, Stål O. Clinical potential of the mTOR targets S6K1 and S6K2 in breast cancer. Breast Cancer Res Treat. 2011;128(3):713–723. doi: 10.1007/s10549-010-1058-x. [DOI] [PubMed] [Google Scholar]
- 37.Armengol G, Rojo F, Castellvi J, Iglesias C, Cuatrecasas M, Pons B, Baselga J, Ramón y Cajal S. 4E-binding protein 1: a key molecular "funnel factor" in human cancer with clinical implications. Cancer Res. 2007;67(16):7551–7555. doi: 10.1158/0008-5472.CAN-07-0881. [DOI] [PubMed] [Google Scholar]
- 38.Rojo F, Najera L, Lirola J, Jimenez J, Guzman M, Sabadell MD, Baselga J, Ramón y Cajal S. 4E-binding protein 1, a cell signaling hallmark in breast cancer that correlates with pathologic grade and prognosis. Clin Cancer Res. 2007;13(1):81–89. doi: 10.1158/1078-0432.CCR-06-1560. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Clinicopathological Characteristics in the Final Patient Cohort (58 Patients)
Table S2: Correlation Between Nuclear and Cytoplasmic LKB1 Expression and the Clinicopathologic Factors
Table S3: Correlation Between Nuclear and Cytoplasmic paKT Expression With The Clinicopathologic Factors
Table S4: Correlation Between pS6RP, p4EBP1 Expression With The Clinicopathologic Factors