Abstract
Background
Determining whether methicillin-resistant Staphylococcus aureus (MRSA) is a true causative pathogen or reflective of colonization when MRSA is cultured from the respiratory tract remains important in treating patients with pneumonia.
Methods
We evaluated the bacterial microbiota in bronchoalveolar lavage fluid (BALF) using the clone library method with a 16S ribosomal RNA (rRNA) gene analysis in 42 patients from a pneumonia registry who had MRSA cultured from their sputum or BALF samples. Patients were divided into two groups: those treated with (Group A) or without (Group B) anti-MRSA agents, and their clinical features were compared.
Results
Among 248 patients with pneumonia, 42 patients who had MRSA cultured from the respiratory tract were analyzed (Group A: 13 patients, Group B: 29 patients). No clones of S. aureus were detected in the BALF of 20 out of 42 patients. Twenty-eight of 29 patients in Group B showed favorable clinical outcomes, indicating that these patients had non-MRSA pneumonia. Using a microflora analysis of the BALF, the S. aureus phylotype was predominant in 5 of 28 (17.9 %) patients among the detected bacterial phylotypes, but a minor population (the percentage of clones ≤ 10 %) in 19 (67.9 %) of 28 patients. A statistical analysis revealed no positive relationship between the percentage of clones of the S. aureus phylotype and risk factors of MRSA pneumonia.
Conclusions
The molecular method using BALF specimens suggests that conventional cultivation method results may mislead true causative pathogens, especially in patients with MRSA pneumonia. Further studies are necessary to elucidate these clinically important issues.
Keywords: 16S rRNA gene; Methicillin-resistant Staphylococcus aureus, MRSA; Clone library; Contamination; Pneumonia; Bronchoalveolar lavage, BALF; Pneumonia
Background
Patients with nosocomial pneumonia caused by methicillin-resistant Staphylococcus aureus (MRSA) have been increasing over the past half century. Approximately 20–40 % of all hospital-acquired pneumonia (HAP), ventilator-associated pneumonia (VAP) [1, 2] and the number of MRSA pneumonia patients is increasing in step with aging of the population [3]. Several guidelines [4, 5], including Japanese guidelines for nursing and healthcare-associated pneumonia (NHCAP) [6] and HAP [7, 8], suggest the use of anti-MRSA antimicrobials in pneumonia patients when the risks of MRSA are suggested. However, there have been only a few clinical studies that describe the pathogenicity of MRSA in bacterial pneumonia and accurate diagnostic methods for evaluating MRSA pneumonia [9–11]. Generally, the diagnostic criteria of respiratory infection caused by MRSA are positive results of a quantitative culture of MRSA over 106 colony forming units (CFU)/ml in sputum samples, 104 CFU/ml in lower respiratory specimens and/or phagocytosis of S. aureus by polymorphonuclear neutrophils [4]. However, it is occasionally difficult to differentiate whether the detected MRSA is a true causative pathogen of pneumonia or only reflective of colonization when MRSA is cultured from the lower respiratory tract samples. Physicians should carefully consider whether or not cultured MRSA is actually causative in each case because many patients fulfill these criteria and improve without anti-MRSA agents in real-world clinical settings. Differentiation of MRSA as a cause of pneumonia or merely colonization remains an important clinical issue and is of a particular interest in clinical settings.
We hypothesized that the percentage of S. aureus clones in bronchoalveolar lavage fluid (BALF) directly obtained from the affected lesions of pneumonia identified by chest CT might be helpful to distinguish true MRSA pneumonia from colonization of MRSA. In the present study, we used the data from the pneumonia registry, which included 16S ribosomal RNA gene analyses of BALF, and patients with pneumonia in whom MRSA was cultured from the respiratory samples were enrolled. Then we divided these patients into two groups: Group A included MRSA pneumonia patients treated with anti-MRSA agents and Group B were patients with MRSA cultured from respiratory samples but who improved without anti-MRSA treatment, and the clinical features of these two groups were compared.
Methods
Patients
Among 248 Japanese patients with community-acquired pneumonia (CAP), healthcare-associated pneumonia (HCAP) and HAP at the University of Occupational and Environmental Health, Japan and referred hospitals between April 2010 and January 2015, 42 patients with positive cultures for MRSA from respiratory specimens (i.e., sputum, endobronchial aspirates and BALF) were enrolled (Fig. 1). This cohort included patients in previous studies of CAP [12] and HCAP [13]. Patients who had MRSA positively cultured from respiratory specimens were divided into two groups: Group A consisted of patients that had been treated with anti-MRSA agents, and Group B included patients that had been treated without anti-MRSA agents, and the clinical features of these two groups were compared. This study was approved by the Human and Animal Ethics Review Committee of the University of Occupational and Environmental Health, Japan (No.09-118), and all patients provided their written informed consent.
Fig. 1.
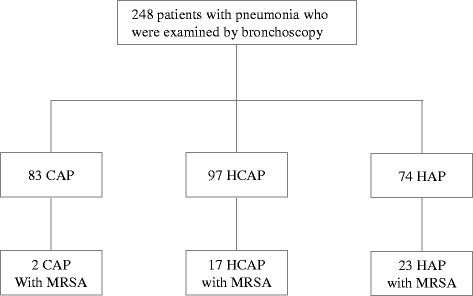
A flow chart of the study participants. CAP: community-acquired pneumonia, HCAP: healthcare-associated pneumonia, HAP: hospital-acquired pneumonia, MRSA: methicillin-resistant Staphylococcus aureus, BALF: bronchoalveolar lavage fluid
Definitions
The diagnosis of pneumonia was made by the fulfillment of the following three criteria: (1) at least one of the following clinical symptoms (a fever ≥ 37 °C, cough, purulent sputum, moist rales, pleural pain, dyspnea, or tachypnea); (2) new infiltrates on a chest X-ray or computed tomography (CT); and (3) at least one sign of systemic inflammation (a white blood cell (WBC) count > 10,000/mm3 or < 4,500/mm3 or an increased C-reactive protein (CRP) level). The definitions of CAP, HCAP and HAP were made according to the Infectious Diseases Society of America (IDSA)/American Thoracic Society (ATS) guidelines. Briefly, CAP was defined as pneumonia acquired in the community with no risk factors for HCAP. HCAP was defined as pneumonia acquired in the community with one or more of the following risk factors: (a) hospitalization for 2 days in the preceding 90 days; (b) residence in a nursing home or extended care facility; (c) home infusion therapy (including antibiotics); (d) long-term dialysis (including hemodialysis and peritoneal dialysis) within 30 days of entering the study; and (e) home wound care. HAP was defined as pneumonia acquired in the hospital ≥ 48 h after admission [4, 14]. The criteria for aspiration risk factors, such as neurologic dysphagia, anatomical abnormalities of the upper aerodigestive tract and poor oral hygiene defined by Marik et al. [15] were used, and patients with gastroesophageal disorders (including disruption of the gastroesophageal junction) were included in this study.
Data and sample collection
The laboratory findings and radiological information on chest X-rays and/or CT were collected. BALF specimens using 40 ml of sterile saline were obtained from lung lesions of pneumonia, as previously described [12, 13].
Evaluation of the clinical efficacy
The clinical efficacies of the antimicrobials were evaluated by an improvement in the clinical symptoms, laboratory and chest radiography findings, which fulfill the definitions proposed by the Japan Society of Chemotherapy [16]. The treatment medication for pneumonia was considered to be clinically “effective” when more than three of the following criteria were satisfied: (1) improvement or complete resolution of the clinical symptoms, (2) improvement in the body temperature to ≤ 37 °C, (3) chest radiography score of ≤ 70 % of the previous value, (4) WBC count ≤ 9,000/mm3 and (5) CRP level ≤ 30 % of the previous value. When the efficacy criteria were not satisfied for any reason, the case was considered to be “ineffective.” Physicians followed the guidelines for CAP, HCAP and HAP to use anti-MRSA agents as the first antimicrobial treatment. As the present study was a retrospective cohort study, there were no strict criteria that regulate an intervention of anti-MRSA therapy as an additive antibiotic treatment, but clinical response to antibiotics was firstly evaluated three days after the start of antimicrobial treatment, physicians decided to add anti-MRSA agents when the clinical response to antimicrobials were ineffective with positive culture results for MRSA.
Criteria for the identification of bacterial isolates
Microbiological evaluation using cultivation methods
Cultivation of BALF and sputum samples was performed as previously described [12, 13]. The samples were inoculated onto the appropriate Vitek apparatus with or without the associated API identification strip using the Vitek 2 apparatus (bioMerieux), and bacterial identification was confirmed according to an identification percentage of more than or equal to 90 %. When the percentage was less than 90 %, subsequent bacterial identification was performed using API. Each macroscopically recognized bacterial colony containing normal bacterial flora was recorded as normal flora. All S. aureus isolates were identified according to a morphologic analysis of the bacterial colony, Gram staining and catalase and coagulase tests. MRSA was identified if the minimum inhibitory concentration of oxacillin was ≥ 4 μg/ml.
Total cell count, cell lysis efficiency analysis and bacterial identification using the molecular method
Using BALF specimens, the total bacterial cell count and cell lysis efficiency were evaluated using epifluorescent microscopy, and DNA extraction, amplification of the 16S rRNA gene using polymerase chain reaction (PCR), clone library construction and a sequencing analysis were performed, as previously described [12, 13, 17–20]. Detected DNA sequences were then compared with those of the type strains using the basic local alignment search tool (BLAST) algorithm, as described previously. The 16S rRNA gene sequences of type strains were obtained from the DNA Data Bank of Japan (http://www.ddbj.nig.ac.jp/) and the Ribosomal Database Project II (http://rdp.cme.msu.edu/) [12, 13, 17–20].
Evaluation of the radiologic findings
Chest X-rays or CT performed within 48 h of the onset of pneumonia were analyzed and evaluated by two experienced respirologists without any clinical information. Radiological findings were sorted into four different patterns as follows: “lobar pneumonia pattern” that showed an air-space consolidation limited to one lobe or one segment, “aspiration pneumonia pattern” that showed pulmonary infiltrations in the bilateral lower lobes, “pulmonary abscess pattern” where the infiltrations accompanied the cavity and “others” that included any other remaining patterns of pneumonia, such as bronchopneumonia, according to previous reports [21, 22].
Statistical analysis
The baseline characteristics were summarized using descriptive statistics. Continuous variables were compared using the Mann–Whitney U-test and Student’s t-tests, while categorical variables were compared using Fisher’s exact test or the chi-square test, as appropriate. The SPSS software package (version 19) was used for the statistical analysis, and a P <0.05 was considered statistically significant.
Results
Clinical characteristics and laboratory findings of the participants
The clinical characteristics and laboratory findings of 42 patients from whom MRSA was detected are shown in Table 1. Thirteen (31.0 %) and 29 (69.0 %) patients had been treated with (Group A) and without (Group B) anti-MRSA antimicrobials. Between Groups A and B, the rate of patients with malignancy was significantly higher in Group B (37.9 %, 11/29) than in Group A (7.7 %, 1/13) (p = 0.016) (Table 1). In these 42 patients, the risk factors of MRSA, such as the use of corticosteroid or immunosuppressants (14.3 %), nasogastric tube feeding or percutaneous gastrostomy tube feeding (7.1 %), antibiotics use within 90 days (28.6 %), detection of MRSA within 90 days (23.1 %), were observed, however, these findings were not significantly different between the groups (Table 1). In addition, patients with aspiration risks were observed in 54.8 % of the total cohort. Group B included significantly more HCAP patients than Group A (P = 0.015), however, the radiological findings were not significantly different between Groups A and B (Table 1).
Table 1.
The clinical characteristics and laboratory findings of 42 patients treated with or without anti-MRSA drugs in this study
| Clinical variates | Group A | Group B | P-value |
|---|---|---|---|
| Treated with anti-MRSA drugs | Treated without anti-MRSA drugs | ||
| (n = 13) | (n = 29) | ||
| Age ± SD (years) | 74.8 ± 10.1 | 74.6 ± 17.5 | 0.764 |
| Male: Female | 10:3 | 19:10 | 0.458 |
| Underlying diseases** | |||
| None | 2 (15.4 %) | 4 (13.8 %) | 0.898 |
| Malignancies | 1 (7.7 %) | 11 (37.9 %) | 0.016 |
| Cerebrovascular disorders | 5 (38.5 %) | 10 (34.5 %) | 0.813 |
| Chronic pulmonary diseases | 4 (30.8 %) | 5 (17.2 %) | 0.382 |
| COPD | 2 (15.4 %) | 2 (6.9 %) | 0.469 |
| Bronchiectasis/NTM | 1 (7.7 %) | 5 (17.2 %) | 0.370 |
| Interstitial lung diseases | 1 (7.7 %) | 1 (3.4 %) | 0.621 |
| Diabetes mellitus | 4 (30.8 %) | 3 (10.3 %) | 0.178 |
| Dementia | 4 (30.8 %) | 6 (20.7 %) | 0.519 |
| Heart diseases | 4 (30.8 %) | 5 (17.2 %) | 0.382 |
| Hepatic diseases | 1 (7.7 %) | 1 (3.4 %) | 0.621 |
| Renal diseases | 1 (7.7 %) | 4 (13.8 %) | 0.550 |
| Hematology/Collagen-vascular diseases | 1 (7.7 %) | 4 (13.8 %) | 0.550 |
| ECOG performance status 3-4 | 75.0 % (9/12) | 60.0 % (15/25) | 0.371 |
| Use of Glucocorticoid**/Immunosuppresant | 1 (7.7 %) | 5 (17.2 %) | 0.37 |
| Use of gastric tube | 1 (7.7 %) | 2 (6.9 %) | 0.931 |
| Histories/Risks of aspiration | 8 (61.5 %) | 15 (51.7 %) | 0.568 |
| Antibiotic therapy in the preceding 90 days | 5 (38.5 %) | 7 (24.1 %) | 0.387 |
| History of MRSA detection in the preceding 90 days | 4 (30.8 %) | 5 (17.2 %) | 0.382 |
| Type of pneumonia | |||
| CAP | 0 (0.0 %) | 2 (6.9 %) | 0.161 |
| HCAP | 2 (15.4 %) | 15 (51.7 %) | 0.015 |
| HAP | 11 (84.6 %) | 12 (41.3 %) | 0.099 |
| Radiologic findings of Chest CT* | |||
| Consolidation | 7 (63.6 %) | 12 (41.4 %) | 0.474 |
| Bronchopneumonia | 1 (9.1 %) | 9 (31.0 %)) | 0.052 |
| Complicated with cavitation/abscess formation | 0 (0.0 %) | 0 (0.0 %) | – |
| Complicated with atelectasis | 0 (0.0 %) | 3 (10.3 %) | 0.083 |
| Centrilobular nodules (DPB-like) | 0 (0.0 %) | 0 (0.0 %) | – |
| Diffuse alveolar shadow (ARDS-like) | 2 (18.2 %) | 0 (0.0 %) | 0.165 |
| Bronchiectasis | 1 (9.1 %) | 6 (20.7 %) | 0.239 |
| Parapneumonic pleural effusion | 5 (45.5 %) | 3 (10.3 %) | 0.082 |
SD standard deviation, COPD chronic obstractive pulmonary disease, NTM nontuberculous mycobacterial infection, MRSA methicillin-resistant Staphylococcus aureus, ECOG Eastern Cooperative Oncology Group, CAP community-acquired pneumonia, HCAP helathcare-associated pneumonia, HAP hospital-acquired pneumonia, CT computed tomography, DPB diffuse pulmonary bronchiolitis, ARDS acute respiratory distress syndrome
*includes diplicates, **corresponds to prednisolone 5 mg daily or greater
Comparison between bacterial cultivation and the clone library method of 16S rRNA gene sequencing analysis
Table 2 shows the comparison of the results of the bacterial culture and bacterial floral analysis using the 16S rRNA gene with the clinical course of the patients. Cultivation results demonstrated that MRSA were isolated in all 19 patients in whom sputum culture was performed, and 37 of 41 (90.5 %) patients excluding No. 33 (BALF culture was not analyzed) showed positive culture results of MRSA using BALF samples. The molecular method detected the S. aureus phylotype in 22 patients, whereas 20 patients showed no S. aureus phylotypes in the BALF samples (Table 2). In Group A, S. aureus was detected in 69.2 % (9 of 13, Cases 1–9) of the patients by both cultivation methods and the molecular method, and the molecular method demonstrated that the S. aureus phylotype was predominant in 38.5 % (5 of 13, Cases 1–5) of the BALF specimens (Table 2). In 8 of 13 (61.5 %) Group A patients where S. aureus was not the predominant phylotype (Cases 6–13), S. aureus comprised a minor population (percentage of clones ≤ 10 %). In these 8 patients (Cases 6–13), no discrepancies were observed between the most occupied bacterial phylotypes by the molecular method and the clinical outcomes after antibiotic therapy. All of the patients with poor clinical outcomes in Group A (5 of 13, Cases 1, 4, 6, 10, 11) had obvious complicated poor prognostic factors, such as asphyxiation due to tracheobronchial secretion, exacerbation of heart failure and aspergillosis.
Table 2.
Comparison of Bacteria Between Conventional Cultivation Methods and 16S rDNA Sequencing Analysis in the Bacterial Infection Group
| Age | Pneumonia type | Sputum | BALF | Prior antibiotics | Treatments | Clinical outcome | |||
|---|---|---|---|---|---|---|---|---|---|
| Culture | Culture | Clone library analysis | |||||||
| Predominant phylotype, % | S. aureus | ||||||||
| GROUP A | |||||||||
| 1 | 72 | HAP | not analyzed | MRSA | Staphylococcus aureus 97.5 % | 97.5 % | ABK | VCM | ineffective |
| 2 | 70 | VAP | not analyzed | MRSA | Staphylococcus aureus 91.8 % | 91.8 % | None | TEIC | effective |
| 3 | 78 | HAP | MRSA | MRSA | Staphylococcus aureus 91.0 % | 91.0 % | None | ABK | effective |
| 4 | 77 | HCAP | MRSA | MRSA | Staphylococcus aureus 53.1 % | 51.8 % | IPM/CS MINO | BIPM, PZFX + CLDM, CZOP + ABK | ineffective |
| 5 | 65 | HAP | not analyzed | MRSA | Staphylococcus aureus 50.0 % | 50.0 % | MEPM | MEPM+VCM | effective |
| 6 | 76 | HAP | not analyzed | MRSA | Corynebacterium simulans 41.9 % | 8.1 % | None | VCM | ineffective |
| 7 | 61 | HAP | S. pneumoniae MRSA | MRSA | Haemophilus influenzae 35.3 % | 3.5 % | MEPM | MEPM + VCM | effective |
| 8 | 87 | HAP | MRSA | MRSA | Corynebacterium spp. 97.8 % | 2.2 % | SBT/ABPC | TEIC | effective |
| 9 | 66 | HAP | not analyzed | H. influenzae, MRSA | Haemophilus influenzae 84.0 % | 1.1 % | None | IPM/CS + VCM SBT/ABPC | effective |
| 10 | 82 | VAP | not analyzed | MRSA P. aeruginosa | Pseudomonas aeruginosa 94.6 % | 0.0 % | IPM/CS | PZFX+VCM | ineffective |
| 11 | 61 | HCAP | not analyzed | MRSA, Aspergillus fumigatus | Streptococcus spp. 90.7 % | 0.0 % | DRPM | DPPM PZFX + CLDM/L-AMB + VCM/TAZ/PIPC | ineffective |
| 12 | 91 | HAP | MRSA | MRSA | Streptococcus oralis 58.5 % | 0.0 % | MEPM | LZD | effective |
| 13 | 87 | HAP | not analyzed | MRSA, Neisseria | Neisseria perflava 95.5 % | 0.0 % | TEIC | TEIC, AMK | effective |
| GROUP B | |||||||||
| 14 | 81 | HAP | not analyzed | MRSA | Staphylococcus aureus 100 % | 100.0 % | None | DRPM | effective |
| 15 | 21 | VAP | not analyzed | MRSA | Staphylococcus aureus 88.6 % | 88.6 % | None | MEPM | effective |
| 16 | 73 | CAP | not analyzed | MRSA | Staphylococcus aureus 60.8 % | 60.8 % | None | GRX | effective |
| 17 | 76 | HCAP | MRSA P. aeruginosa | MSSA | Staphylococcus aureus 57.1 % | 57.1 % | None | TAZ/PIPC | ineffective |
| 18 | 81 | HCAP | MRSA P. aeruginosa | no growth | Staphylococcus aureus 55.4 % | 55.4 % | None | CZOP + CLDM | effective |
| 19 | 62 | CAP | MRSA | MRSA | Staphylococcus aureus 48.7 % | 48.7 % | FQ | CPFX | effective |
| 20 | 80 | HAP | MRSA | MRSA | Streptococcus intermedius 56.5 % | 40.6 % | None | DRPM | effective |
| 21 | 76 | HAP | MRSA | MRSA | Corynebacterium spp. 25.3 % | 18.4 % | TAZ/PIPC | SBT/ABPC | effective |
| 22 | 22 | HAP | not analyzed | MRSA, A. baumannii | Neisseria elongata 81.2 % | 15.1 % | TEIC | IPM/CS | effective |
| 23 | 81 | HCAP | MRSA | MRSA | Streptococcus spp. 46.7 % | 12.0 % | None | LVFX | effective |
| 24 | 85 | HAP | not analyzed | MRSA | Streptococcus oralis/mitis 37.3 % | 8.0 % | CPFX | LVFX | effective |
| 25 | 83 | HCAP | MRSA, E. coli | MRSA, E. coli | Moraxella catarrhalis 69.2 % | 7.7 % | None | TAZ/PIPC | effective |
| 26 | 76 | HAP | not analyzed | K. pneumoniae, MRSA, Proteus mirabilis | Corynebacterium simulans 58.4 % | 1.3 % | None | TAZ/PIPC | effective |
| 27 | 85 | HAP | MRSA | MRSA | Fusobacterium nucleatum 55.7 % | 0.0 % | None | LVFX | effective |
| 28 | 61 | HCAP | not analyzed | MRSA, B. cepacia, F. mortiferum | Rothia spp. 45.2 % | 0.0 % | None | MEPM | effective |
| 29 | 45 | HAP | not analyzed | S. maltophilia, MRSA | Enterococcus hirae 25.8 % | 0.0 % | None | MEPM | effective |
| 30 | 74 | HCAP | P. aeruginosa MRSA | P. aeruginosa, MRSA | Streptococcus salivarius 43.0 % | 0.0 % | None | MEPM | effective |
| 31 | 80 | HCAP | not analyzed | MRSA | Streptococcus spp. 98.9 % | 0.0 % | Unknown | LVFX | effective |
| 32 | 80 | HCAP | not analyzed | MRSA | Streptococcus spp. 97.4 % | 0.0 % | Unknown | LVFX | effective |
| 33 | 98 | HCAP | MRSA | not analyzed | Streptococcus spp. 78.8 % | 0.0 % | Unknown | TAZ/PIPC | effective |
| 34 | 80 | HCAP | MRSA | oral bacteria | Neisseria spp. 55.0 % | 0.0 % | LVFX | MEPM | effective |
| 35 | 82 | HCAP | not analyzed | P. aeruginosa, MRSA, Streptococcus | Streptococcus oralis/mitis 70.7 % | 0.0 % | Unknown | SBT/ABPC | effective |
| 36 | 64 | HCAP | not analyzed | P. aeruginosa, MRSA | Pseudomonas aeruginosa 97.4 % | 0.0 % | None | TAZ/PIPC + LVFX | effective |
| 37 | 86 | HCAP | MRSA | K. pneumoniae, MRSA | Lactobacillus spp. 51.1 % | 0.0 % | None | TAZ/PIPC | effective |
| 38 | 80 | HCAP | E. coli, MRSA | oral bacteria | Streptococcus spp. 45.2 % | 0.0 % | None | TAZ/PIPC | effective |
| 39 | 93 | HCAP | not analyzed | MRSA, oral bacteria | Corynebacterium spp. 94.3 % | 0.0 % | None | LVFX | effective |
| 40 | 81 | HAP | not analyzed | MRSA, E. coli | Haemophilus influenzae 34.5 % | 0.0 % | None | TAZ/PIPC | effective |
| 41 | 73 | HAP | not analyzed | Enterobacter cloacae, MRSA | Enterobacter asburiae 70.0 % | 0.0 % | None | DRPM | effective |
| 42 | 74 | HAP | MRSA | Corynebacterium, MRSA | Corynebacterium simulans 98.9 % | 0.0 % | TAZ/PIPC | TAZ/PIPC | effective |
Abbreviations: CAP community-acquired pneumonia, healthcare-associated pneumonia, HAP hospital-acquired pneumonia, VAP ventilator-associated pneumonia, MRSA methicillin-resistant Staphylococcus aureus, BALF bronchoalveolar lavage fluid, ABK arbekacin, VCM vancomycin, TEIC teicoplanin, LZD linezolid, IPM/CS imipenem/cilastatin, MEPM meropenem, DRPM doripenem, BIPM biapenem, CZOP cefozopran, SBT/ABPC sulbactam/ampicillin, TAZ/PIPC tazobactam/piperacillin, CPFX ciprofloxacin, LVFX levofloxacin, GRNX garenoxacin, MINO minomycin, CLDM clindamycin, L-AMB liposomal amphotericin B, NA not applicable
In Group B, 96.5 % of patients (28 of 29, Cases 14–16, 18–42) showed good clinical outcomes without anti-MRSA antimicrobials; one patient (Case 17) died because of asphyxiation due to tracheobronchial secretion. The S. aureus phylotype was a minor population (percentage of clones ≤10 %) or undetectable (0 %) in 10.7 % (3 of 28, Cases 24–26) and 57.1 % (16 of 28, Cases 27–42), respectively, of the patients in Group B who had good clinical outcomes. In addition, 5 of these 28 patients (Cases 14–18) showed that the S. aureus phylotype was the most detected phylotype, including one patient (Case 14) with 100 % of the percentage of clones of S. aureus phylotype.
Correlation of the percentage of clones of S. aureus phylotype and risk factors of MRSA pneumonia
Table 3 shows the relationship between the percentage of clones of the S. aureus phylotype and the risk factors of MRSA pneumonia among all 42 patients. Gastrostomy or nasogastric tube feeding was significantly negatively correlated with the percentage of clones of S. aureus phylotype using the molecular method (Table 3).
Table 3.
Comparison of Bacteria Between Conventional Cultivation Methods and 16S rDNA Sequencing Analysis in the Bacterial Infection Group
| Risk factors (Positive/Negative)** | The percentage of clones of MRSA phylotype (%) | ||
|---|---|---|---|
| Positive | Negative | P-value | |
| Use of Glucocorticoid**/Immunosuppressant (6/36) | 40.2 ± 41.4 | 19.0 ± 28.3 | 0.305 |
| Histories/Risks of aspiration (23/19) | 18.7 ± 33.2 | 25.2 ± 29.2 | 0.422 |
| Antibiotic therapy in the preceding 90 days (12/30) | 30.1 ± 40.7 | 17.5 ± 25.7 | 0.391 |
| History of pathogens detection in the preceding 90 days* (9/30) | 38.2 ± 35.1 | 15.4 ± 28.9 | 0.105 |
*Unknown data in three cases, **“Positive” and “Negative” indicate the number of patients with or without each risk factor, respectively.
Abbreviations: MRSA methicillin-resistant Staphylococcus aureus
Discussion
We analyzed the cultivation results and bacterial phylotypes according to the molecular method using BALF samples in patients with MRSA cultured from respiratory samples, and interestingly, no clones of S. aureus were detected in the BALF samples in 47.6 % (20/42) of these patients. Most of the patients (n = 28 of 29; 96.5 %) treated without anti-MRSA antimicrobials (Group B) showed favorable clinical outcomes despite the cultivation of MRSA, and these 28 patients were suspected to have non-MRSA pneumonia; the cultured MRSA from the respiratory samples might have been due to colonization in the respiratory tract. In addition, the S. aureus phylotype was only a minor population (the percentage of clones ≤ 10 %) in 67.9 % (19/28) of these 28 patients in Group B according to the molecular method, which was compatible with their clinical courses. Several previous reports have described that MRSA is occasionally a non-causative pathogen of pneumonia in some patients [23–26] even when MRSA is cultured from sputum samples, and our results suggest that even when MRSA was cultured using samples obtained from the lower respiratory tract, MRSA was clinically considered not to be a causative agent in more than two-thirds of these patients.
A similar report by Nagaoka et al. showed that approximately half (51.4 %, 36/70) of the patients were considered to have true MRSA pneumonia when hospital-acquired MRSA pneumonia was defined according to the positive responses and/or clinical demand of anti-MRSA agents with a positive culture of MRSA and detection of clustered Gram-positive cocci within polymorphonuclear cells in the respiratory samples, such as BALF or transthoracic aspiration [11]. Moreover, at least 66.7 % (28/42) of the patients were possibly considered to have MRSA colonization, and MRSA was considered to be a causative pathogen in 33.3 % (14/42) of the patients in this study. These data suggest that it remains clinically controversial whether or not MRSA is a true causative pathogen of pneumonia, even in patients with MRSA cultured from the lower respiratory samples, and the ratio of true MRSA pneumonia in these patients might be lower than previously believed.
Five of 28 (17.9 %) patients in Group B who showed good clinical outcomes without anti-MRSA agents demonstrated that the S. aureus phylotype was predominant among the detected bacterial phylotypes in the samples, which may be inconsistent with the colonization of MRSA. The molecular method we used could not evaluate drug resistance, and a differentiation between MRSA and methicillin-susceptible S. aureus (MSSA) was not possible in this retrospective study. Spontaneous remission of MRSA pneumonia is another potential explanation. In addition, there are presently no criteria to differentiate causative pathogens using the ratio of bacterial phylotypes in the samples, thus careful discretion is necessary to interpret these data, and further studies are needed to elucidate this issue.
Several guidelines [4, 6–8] and clinical trials [11] have described the risk factors of MRSA pneumonia. According to a report by Nagaoka et al. [11] that used a multiple regression analysis for the risk factors of MRSA, a past history of head and neck, esophageal or stomach surgery (odds ratio (OR) 8.63), radiological findings of other than lobar pneumonia (OR 10.2), severity of pneumonia with the Pneumonia Patient Outcomes Research Team (PORT) score 5 (OR 5.23), more than 106 CFU/ml of MRSA using a quantitative culture (OR 12.8), and a single cultivation of MRSA (OR 19.9) were significantly correlated with MRSA pneumonia. More HCAP patients were observed in Group B than in Group A, however, no other factors were significantly different between Groups A and B in this study. When considering the percentage of clones of the S. aureus phylotype in BALF samples according to each different risk factor in this study, only the “use of gastric tube feeding” was inversely correlated with the percentage of clones of S. aureus (Table 3), suggesting that such condition may be a clue to avoid an abuse of anti-MRSA agents.
The analysis using the 16S rRNA gene can detect only bacterial DNA, and does not equally indicate that the detected bacterial phylotype causes bacterial infection. Therefore, we have been investigating and validating this molecular method in several diseases to compare this molecular method with the results of cultivation methods in several settings [12, 13]. Further investigations for validating this method should be performed.
Study limitation
There are several limitations associated with the present study. First, the universal primers we used for the molecular analysis could not amplify all of the bacterial 16S rRNA genes. The primers we used cover approximately 92 % of the registered bacterial species in the Ribosomal Database Project II database, however, the remaining undetectable bacteria with these primers included no causative pathogens that have been reported in humans [19]. Second, approximately 100 clones per each clone library were analyzed, meaning that bacterial 16S rRNA gene sequences present at less than 1 % of each sample may not be detectable using this method. Third, this study was retrospective, and anti-MRSA agents were administered with no particular criteria. Fourth, a quantitative culture and evaluation of neutrophil phagocytosis of the organisms were not performed. Fifth, study population was relatively small and elderly patients were mostly included in HCAP or HAP patients, and only two CAP patients were included. Further investigations should be considered to elucidate the data in younger population.
Conclusion
We evaluated the clinical course and the ratios of bacterial phylotypes in BALF specimens using the clone library method and conventional cultivation methods in patients with MRSA detected by cultivation from respiratory samples. The results of this study demonstrated that these patients were heterogeneous, and approximately two-thirds of these patients might be considered to have MRSA colonization or non-MRSA pneumonia. In addition, the results of the cultivation-independent molecular method we used indicated that the detection of MRSA by cultivation methods may not correctly reflect the pathogenicity of MRSA in patients with pneumonia. Further prospective studies are necessary to elucidate the pathogenicity of MRSA in pneumonia.
Ethics approval and consent to participate
This study was approved by the Human and Animal Ethics Review Committee of the University of Occupational and Environmental Health, Japan (No.09-118), and all patients provided their written informed consent.
Availability of data and materials
We declare that the data supporting the conclusions of this article are fully described in the article.
Acknowledgments
We thank Drs. Chiharu Yoshii, Hideto Obata, Yukiko Kawanami, Yugo Yoshida, Takashi Kido, Takeshi Orihashi, Chinatsu Nishida, Naoyuki Inoue, Takaaki Ogoshi, Susumu Tokuyama and Keishi Oda for collecting the samples and Yoshiko Yamazaki and Kumiko Matsuyama for their technical assistance.
Abbreviations
- ATS
American Thoracic Society
- BALF
bronchoalveolar lavage fluid
- BLAST
the basic local alignment search tool
- CAP
community-acquired pneumonia
- CFU
colony forming units
- CT
computed tomography
- HAP
hospital-acquired pneumonia
- HCAP
healthcare-associated pneumonia
- IDSA
Infectious Diseases Society of America
- MRSA
methicillin-resistant Staphylococcus aureus
- MSSA
methicillin-susceptible S. aureus
- NHCAP
nursing and healthcare-associated pneumonia
- OR
odds ratio
- PCR
polymerase chain reaction
- PORT
Pneumonia Patient Outcomes Research Team
- VAP
ventilator-associated pneumonia
- WBC
white blood cell
Footnotes
Competing interests
The authors declare that there are no competing interests.
Authors’ contributions
Conceived and designed the experiments: TK, SN, HM, KYatera, KF, HI, HT, KYamasaki. Performed the experiments: TK, SN, KYatera, KF, KA, KYamasaki, KN. Analyzed the data: SN, TK, KYatera, KF, KYamasaki. Contributed reagents/materials/analysis tools: TK, SN, HM, K. Yatera, KF, HI, HT, KYamasaki. Wrote the paper: TK, KYatera, SN, KYamasaki, KF, HM. All authors read and approved the final manuscript.
Contributor Information
Toshinori Kawanami, Email: namihei@med.uoeh-u.ac.jp.
Kazuhiro Yatera, Phone: +81-93-691-7453, Email: yatera@med.uoeh-u.ac.jp.
Kei Yamasaki, Email: yamasaki@med.uoeh-u.ac.jp.
Shingo Noguchi, Email: sn0920@med.uoeh-u.ac.jp.
Kazumasa Fukuda, Email: kfukuda@med.uoeh-u.ac.jp.
Kentarou Akata, Email: kentarouakata@med.uoeh-u.ac.jp.
Keisuke Naito, Email: k-naito@med.uoeh-u.ac.jp.
Takashi Kido, Email: t-kido@med.uoeh-u.ac.jp.
Hiroshi Ishimoto, Email: h-ishimoto@med.uoeh-u.ac.jp.
Hatsumi Taniguchi, Email: hatsumi@med.uoeh-u.ac.jp.
Hiroshi Mukae, Email: hmukae@med.uoeh-u.ac.jp.
References
- 1.Kollef MH, Shorr A, Tabak YP, Gupta V, Liu LZ, Johanners RS. Epidemiology and outcomes of health-care-associated pneumonia. Chest. 2005;128:3854–62. doi: 10.1378/chest.128.6.3854. [DOI] [PubMed] [Google Scholar]
- 2.Rubinstein E, Kollef MH, Nathwani D. Pneumonia caused by methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 2008;46(Sppl 5):S378–85. doi: 10.1086/533594. [DOI] [PubMed] [Google Scholar]
- 3.Ministry of Health, Labour and Welfare of Japan. Summary of Vital Statistics; Trends in leading causes of death. Website. Available: http://www.mhlw.go.jp/english/database/db-hw/populate/index.html. Accessed 17 December 2014.
- 4.Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am J Respir Crit Care Med. 2005;171:388–416. [DOI] [PubMed]
- 5.Masterton RG, Galloway A, French G, Street M, Armstrong J, Brown E, et al. Guidelines for the management of hospital-acquired pneumonia in the UK: report of the working party on hospital-acquired pneumonia of the British Society for Antimicrobial Chemotherapy. J Antimicrob Chemother. 2008;62(1):5–34. doi: 10.1093/jac/dkn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohno S, Imamura Y, Shindo Y, Seki M, Ishida T, Teramoto S, et al. Clinical practice guidelines for nursing- and healthcare-associated pneumonia (NHCAP) [complete translation] Respir Investig. 2013;51:103–26. doi: 10.1016/j.resinv.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Japanese Respiratory Society Establishment of new severity ratings based on analysis of hospital-acquired pneumonia. Respirology. 2009;14(Suppl 2):S4–9. doi: 10.1111/j.1440-1843.2009.01571.x. [DOI] [PubMed] [Google Scholar]
- 8.Seki M, Watanabe A, Mikasa K, Kadota J, Kohno S. Revision of the severity rating and classification of hospital-acquired pneumonia in the Japanese Respiratory Society guidelines. Respirology. 2008;13:880–5. doi: 10.1111/j.1440-1843.2008.01348.x. [DOI] [PubMed] [Google Scholar]
- 9.González C, Rubio M, Romero-Vivas J, González M, Picazo JJ. Bacteremic pneumonia due to Staphylococcus aureus: A comparison of disease caused by methicillin-resistant and methicillin-susceptible organisms. Clin Infect Dis. 1999;29:1171–7. doi: 10.1086/313440. [DOI] [PubMed] [Google Scholar]
- 10.Rello J, Sole-Violan J, Sa-Borges M, Garnacho-Montero J, Muñoz E, Sirgo G, et al. Pneumonia caused by oxacillin-resistant Staphylococcus aureus treated with glycopeptides. Crit Care Med. 2005;33:1983–7. doi: 10.1097/01.CCM.0000178180.61305.1D. [DOI] [PubMed] [Google Scholar]
- 11.Nagaoka K, Yanagihara K, Harada Y, Yamada K, Migiyama Y, Morinaga Y, et al. Predictors of the pathogenicity of methicillin-resistant Staphylococcus aureus nosocomial pneumonia. Respirology. 2014;19:556–62. doi: 10.1111/resp.12288. [DOI] [PubMed] [Google Scholar]
- 12.Yamasaki K, Kawanami T, Yatera K, Fukuda K, Noguchi S, Nagata S, et al. Significance of anaerobes and oral bacteria in community-acquired pneumonia. PLoS One. 2013;8:e63103. doi: 10.1371/journal.pone.0063103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noguchi S, Mukae H, Kawanami T, Yamasaki K, Fukuda K, Akata K, et al. Bacteriological Assessment of Healthcare-associated Pneumonia using a Clone Library Analysis. PLoS One. 2015;10:e0124697. doi: 10.1371/journal.pone.0124697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mandell LA, Wunderink RG, Anzueto A, Bartlett JG, Campbell GD, Dean NC, et al. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis. 2007;44(Suppl 2):S27–72. doi: 10.1086/511159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marik PE. Aspiration pneumonitis and aspiration pneumonia. N Engl J Med. 2001;344:665–71. doi: 10.1056/NEJM200103013440908. [DOI] [PubMed] [Google Scholar]
- 16.Saito A, Miki F, Oizumi K, Rikitomi N, Watanabe A, Koga H, et al. Clinical evaluation methods for new antimicrobial agents to treat respiratory infections: Report of the Committee for the Respiratory System, Japan Society of Chemotherapy. J Infect Chemother. 1999;5:110–23. doi: 10.1007/s101560050020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawanami T, Fukuda K, Yatera K, Kido M, Mukae H, Taniguchi H. A higher significance of anaerobes: the clone library analysis of bacterial pleurisy. Chest. 2011;139:600–8. doi: 10.1378/chest.10-0460. [DOI] [PubMed] [Google Scholar]
- 18.Morotomi N, Fukuda K, Nakano M, et al. Evaluation of intestinal microbiotas of healthy Japanese adults and effect of antibiotics using the 16S ribosomal RNA gene based clone library method. Biol Pharm Bull. 2011;34(7):1011–20. doi: 10.1248/bpb.34.1011. [DOI] [PubMed] [Google Scholar]
- 19.Aoki R, Fukuda K, Ogawa M, Ikeno T, Kondo H, Tawara A, et al. Identification of causative pathogens in eyes with bacterial conjunctivitis by bacterial cell count and microbiota analysis. Ophthalmology. 2013;120:668–76. doi: 10.1016/j.ophtha.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 20.Akiyama T, Miyamoto H, Fukuda K, Sano N, Katagiri N, Shobuike T, et al. Development of a novel PCR method to comprehensively analyze salivary bacterial flora and its application to patients with odontogenic infections. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2010;109:669–76. doi: 10.1016/j.tripleo.2009.10.045. [DOI] [PubMed] [Google Scholar]
- 21.Vilar J, Domingo ML, Soto C, Cogollos J. Radiology of bacterial pneumonia. Eur J Radiol. 2004;51:102–13. doi: 10.1016/j.ejrad.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 22.Hota B, Lyles R, Rim J, Popovich KJ, Rise T, Aroutcheva A, et al. Predictors of clinical virulence in community-onset methicillin-resistant Staphylococcus aureus infections: the importance of USA300 and pneumonia. Clin lnfect Dis. 2011;53:757–65. doi: 10.1093/cid/cir472. [DOI] [PubMed] [Google Scholar]
- 23.Ewig S, Welte T, Chartre J, Torres A. Rethinking the concepts of community-acquired pneumonia and healthcare-associated pneumonia. Lancet Infect Dis. 2010;10:279–87. doi: 10.1016/S1473-3099(10)70032-3. [DOI] [PubMed] [Google Scholar]
- 24.Shorr AF, Zilberberg MD, Reichley R, Kan J, Hoban A, Hoffman J, et al. Validation of a clinical score for assessing the risk of resistant pathogen in patients with pneumonia presenting to the emergency department. Clin Infect Dis. 2012;54:193–8. doi: 10.1093/cid/cir813. [DOI] [PubMed] [Google Scholar]
- 25.Oshitani Y, Nagai H, Matsui H, Aoshima M. Reevaluation of the Japanese guideline for healthcare-associated pneumonia in a medium-sized community hospital in Japan. J Infect Chemother. 2013;19:579–87. doi: 10.1007/s10156-012-0517-1. [DOI] [PubMed] [Google Scholar]
- 26.Sakaguchi M, Shime N, Fujita N, Fujiki S, Hashimoto S. Current problems in the diagnosis and treatment of hospital-acquired methicillin-resistant Staphylococcus aureus pneumonia. J Anesth. 2008;22:125–30. doi: 10.1007/s00540-007-0600-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We declare that the data supporting the conclusions of this article are fully described in the article.


