Abstract
OBJECTIVES: Recurrence of hepatocellular carcinoma can arise from the primary tumor (“early recurrence”) or de novo from tumor formation in a cirrhotic environment (“late recurrence”). We aimed to develop one simple gene expression score applicable in both the tumor and the surrounding liver that can predict the recurrence risk. METHODS: We determined differentially expressed genes in a cell model of cancer aggressiveness. These genes were first validated in three large published data sets of hepatocellular carcinoma from which we developed a seven-gene risk score. RESULTS: The gene score was applied on two independent large patient cohorts. In the first cohort, with only tumor data available, it could predict the recurrence risk at 3 years after resection (68 ± 10% vs 35 ± 7%, P = .03). In the second cohort, when applied on the tumor, this gene score predicted early recurrence (62 ± 5% vs 37 ± 4%, P < .001), and when applied on the surrounding liver tissue, the same genes also correlated with late recurrence. Four patient classes with each different time patterns and rates of recurrence could be identified based on combining tumor and liver scores. In a multivariate Cox regression analysis, our gene score remained significantly associated with recurrence, independent from other important cofactors such as disease stage (P = .007). CONCLUSIONS: We developed a Global Risk Score that is able to simultaneously predict the risk of early recurrence when applied on the tumor itself, as well as the risk of late recurrence when applied on the surrounding liver tissue.
Introduction
Hepatocellular carcinoma (HCC) is the sixth most prevalent cancer and the third most frequent cause of cancer-related death. Treatments with curative intent, such as resection, are feasible at an early stage. Still, even after complete resection, patients remain at a high risk for disease recurrence, either due to early recurrence of the initial tumor or due to the formation of new lesions (leading to late recurrence) [1]. The latter is driven by the malignant potential of the remnant liver because the majority of patients with HCC share a history of liver cirrhosis.
Current decision making on HCC is based on a combination of factors regarding the status of the liver (synthesis capacity, cirrhosis) and characteristics of the tumor (size, vascular invasion, distant metastasis) [2], [3], [4]. In early stages, liver transplantation has the clearest benefit. However, due to the organ shortage, resection and radiofrequency ablation are alternatives [5]. Different prognostic indicators have been identified including liver function [6], [7], extent of cirrhosis and α-fetoprotein levels [8], and morphological criteria (vascular invasion) [9], [10]. There has also been extensive research on gene expression signatures in HCC that can objectively predict patient survival or disease recurrence. However, none of these signatures [11], [12], [13], [14], [15], [16], [17] are able to stratify patients on both rate and timing of disease recurrence. In the current study, we present a novel translational approach of gene expression signature training using microarray data derived from a human sorafenib-resistant hepatoma cell line, an in vitro model for hepatocyte dedifferentiation and tumor aggressiveness. By combining the transcriptome of this model with five large patient data sets submitted at the Gene Expression Omnibus (GEO), we developed a simple combination model based on gene expression that can be applied to tumor and surrounding liver to stratify patients into low and high risk for early and late recurrence of HCC.
Materials and Methods
Cell Culture and Development of Sorafenib Resistance
Full details on the development of a sorafenib-resistant cell line were previously published [18]. Briefly, HepG2 human hepatoblastoma cells (HB-8065-ATCC, Rockville, MD) were incubated with increasing doses of sorafenib (Bayer HealthCare, Leverkusen, Germany) over several months, resulting in a cell line resistant to sorafenib (HepG2S1).
Microarray
Whole transcriptome analysis of HepG2 and HepG2S1 cells (both in triplicate) was performed using the Affymetrix Human Gene 1.0 ST Array. Microarray data were analyzed with the Limma package from Bioconductor (http://www.bioconductor.org) [19]. Differentially expressed genes were assessed using a moderated t test. The resulting P values were corrected for multiple testing with Benjamini-Hochberg [20]. For selecting differentially expressed genes, a cutoff of 2log fold change >+1 or <−1 and a corrected P < .05 was applied.
Gene Set Enrichment Analysis
To explore the features of the differentially expressed genes in the in vitro model, gene set enrichment analysis (GSEA) was performed testing their significance in all gene sets of the Molecular Signature Database v5.0 [21]. Gene sets smaller than 15 or larger than 500 genes were excluded from the analysis. Gene sets with a family wise error rate P value < .05 were considered significantly enriched.
Gene Score Training
To determine the clinical relevance of the in vitro model, we compared the microarray data of the cell culture experiment with published data sets of patients with HCC. Three data sets submitted at the GEO containing 640 suitable samples of liver and/or tumor tissue were considered (GSE9843, GSE25097, GSE40873) (Table 1).
Table 1.
Characteristics of the Three Data Sets Used for Training of the GRS
| GEO Accession | GSE9843 | GSE25097 | GSE40873 |
|---|---|---|---|
| Responsible author | Chiang | Zhang | Kudo |
| Related publication | [22] | [23] | [24] |
| Microarray platform | Affymetrix Human Genome U133 Plus 2.0 Array | Rosetta/Merck Human RSTA Affymetrix 1.0 | Affymetrix Human Genome U133 Plus 2.0 Array |
| Preprocessing | RMA | RMA | RMA |
| Tumor or liver samples | Tumor | Both | Liver |
| Outcome parameter for training | BCLC C vs 0 & A & B | Tumor vs adjacent nontumor | Multicentric occurrence vs no multicentric occurrence |
| Number of positive outcomes | 8 | 268 | 17 |
| Number of negative outcomes | 72 | 243 | 32 |
RMA, robust multiarray; BCLC, Barcelona Clinic Liver Cancer.
More detailed information can be found at http://www.ncbi.nlm.nih.gov/geo/.
For each data set, a training outcome parameter was selected (Table 1). Using a global test described by Goeman et al. [25], we evaluated if a gene differentially expressed in the in vitro experiment correlated with the outcome parameter of each data set. Only genes with a Goeman Z-score of < 3 (which implies 3 standard deviations or 99.7% confidence of coefficient) in all 3 data sets and with the same direction of differential expression in vitro as well as in vivo were withheld. Their expression values are added or subtracted (depending on the direction of expression) to form a risk score (Global Risk Score, GRS).
Gene Score Validation
Two HCC data sets with abundant phenotypical information were found appropriate for validation purpose. Expression values of six out of seven score genes were available. Characteristics of the validation data sets are summarized in Table 2.
Table 2.
Characteristics of the Independent Data Sets Used for Validation of the GRS
| GEO Accession | GSE1898-GSE4024 | GSE14520 |
|---|---|---|
| Abbreviation used | LEC (Laboratory of Experimental Carcinogenesis) | NCI (National Cancer Institute) |
| Related publication | [11] | [12] |
| Microarray platform | NCI/ATC Hs-OperonV2 | Affymetrix Human Genome U133A 2.0 Array |
| Number of GRS genes available | 6/7 | 6/7 |
| Number of patients | 67 | 231 |
| Gender (male/female/NA) | 46/21/0 | 201/27/3 |
| Age in years [median (min-max)] | 59 (26-85) | 50 (21-77) |
| Cirrhosis (yes/no/NA) | 41/26/0 | 211/17/3 |
| Tumor size (< 5 cm/> 5cm/NA) | 28/21/7 | 145/82/4 |
| TNM stage (I/II/III/NA) | 3/9/27/28 | 93/75/38/25 |
| BCLC stage (0/A/B/C/NA) | NA | 20/144/22/26/19 |
| aFP levels (< 300 ng/ml/> 300 ng/ml/NA) | 36/25/6 | 120/104/7 |
| Median follow-up in months (min-max) | 65 (0.1-169) | 51.9 (1.8-67.4) |
NA, not available; aFP, alpha-fetoprotein.
More detailed information can be found at http://www.ncbi.nlm.nih.gov/geo/.
Statistics
To assess the performance of the GRS in the data sets, receiver operating characteristic (ROC) curves were used. Survival analysis was performed with Kaplan-Meier curves and log-rank test. Cutoff values to define high and low GRS values were calculated using the value with the highest Youden index and differed for tumor and liver. Univariate association with survival was determined using Cox regression. The significant variables with P values < .05 were included in a multivariable Cox regression. Statistics were performed using SPSS package 22 (IBM).
Results
Characteristics of the In Vitro Model
As previously described [18], the phenotype of the HepG2S1 cells consisted of marked epithelial-to-mesenchymal transition with augmented motility and invasiveness. Between the parental HepG2 cell line and its derived resistant lineage HepG2S1, 3545 probes representing 3201 genes were differentially expressed. Microarray data are available at the GEO (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE62813.
Two hundred three gene sets were significantly enriched using GSEA (Supplementary Table 1). Of note, the gene set most significant enriched in resistant cells pointed to a marked loss of hepatocyte-associated genes, indicating dedifferentiation of the resistant cell line compared with the nonresistant parental cell line. This was further illustrated by the loss of pathways involved in liver functions including blood coagulation, steroid hormone synthesis, and drug metabolism (Supplementary Figure 1, A–D). Resistant cells showed loss of expression of genes included in the good survival signature proposed by Lee [11] and changes concordant with poor prognostic HCC such as the proliferation subclass [22] or the G3 subtype described by Boyault et al. [26] (Supplementary Figure 1, E–G). These findings encouraged the use of the HepG2/HepG2S1 model as a model of hepatocyte dedifferentiation and tumor aggressiveness and therefore formed the starting point for training the GRS.
Training of the GRS
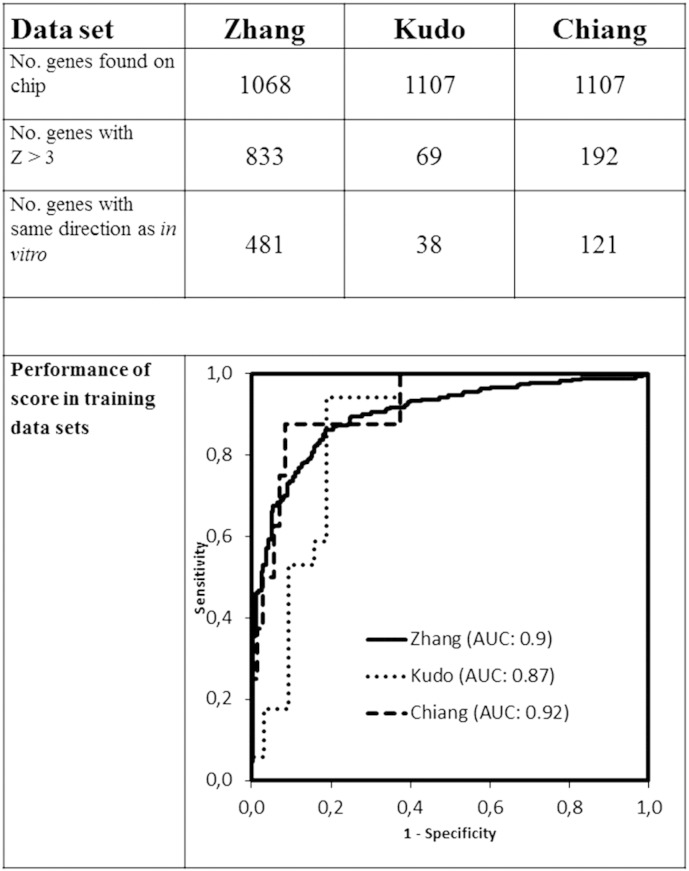
About a third of the differentially expressed genes of our in vitro model were annotated on the microarray chips of the HCC training data sets (Figure 1 and Table 1). The Goeman test identified a variable number of genes from each data set highly correlated with the outcome parameter. The number of genes was further downsized by looking at concordant direction of expression between all data sets and the in vitro condition. Finally, seven genes met all criteria for inclusion in the gene score. The GRS is therefore defined as the sum of expression values of COL4A2, OXCT1, and LRRC16A and subtraction of the expression values of F11, GCKR, ATP11C, and PCSK6 (Table 3). ROC curves confirmed the good performance of the GRS to predict the outcome parameter in the training data sets (Figure 1).
Figure 1.
Training of the GRS. Performance was tested using an ROC curve in the Zhang (tumor versus nontumoral tissue), Kudo (multicentric occurrence versus no multicentric occurrence), and Chiang (BCLC C versus 0-B) data sets (Table 1).
Table 3.
Overview of the Seven Genes Included in the GRS and Their Main Function
| Official Gene Symbol | Direction of Expression | Gene Name | Processes Involved |
|---|---|---|---|
| COL4A2 | ▲ | Collagen, type IV, Alpha 2 | Extracellular matrix structural constituent |
| OXCT1 | ▲ | 3-Oxoacid CoA transferase 1 | Cell metabolism |
| LRRC16A | ▲ | Leucine rich repeat containing 16A | Protein complex binding |
| F11 | ▼ | Coagulation factor XI | Blood coagulation |
| GCKR | ▼ | Glucokinase regulator | Cell metabolism |
| ATP11C | ▼ | ATPase, class VI, type 11C | Phospholipid transport |
| PCSK6 | ▼ | Proprotein convertase subtilisin/kexin type 6 | Protein and peptide processing |
GRS = COL4A2 + OXCT1 + LRRC16A - F11 - GCKR - ATP11C - PCSK6.
The GRS assessed in Tumor Tissue
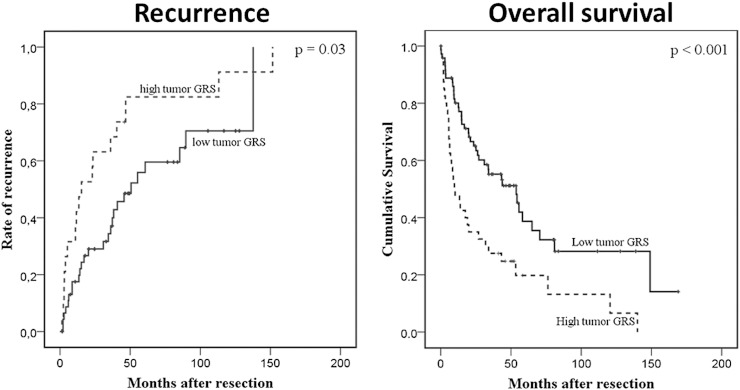
After determining an optimal tumor cutoff based on ROC analysis and Youden index, the GRS assessed in tumor tissue could adequately stratify patients into high and low recurrence risk groups in both data sets. Estimated recurrence rates 3 years after surgery were 68 ± 10% veresus 35 ± 7% (P = .03) in the Laboratory of Experimental Carcinogenesis (LEC) data set and 62 ± 5% versus 37 ± 4% (P < .001) in the National Cancer Institute (NCI) data set (Figure 2, Figure 3). In addition, patients with a low GRS had better mean overall survival rates compared with patients with high GRS in the LEC data set {67.2 [95% confidence interval (CI) 50-85.5] vs 33.2 [95% CI 18.3-48] months} as well as NCI data set [54.3 (95% CI 50.6-58) vs 40.7 (95% CI 35-46) months] (P < .001 for both comparisons) (Figure 2, Figure 3).
Figure 2.
Recurrence rates and overall survival in the LEC cohort [11]. Low (solid line) versus high (dashed line) GRS patients were assessed in tumor tissue.
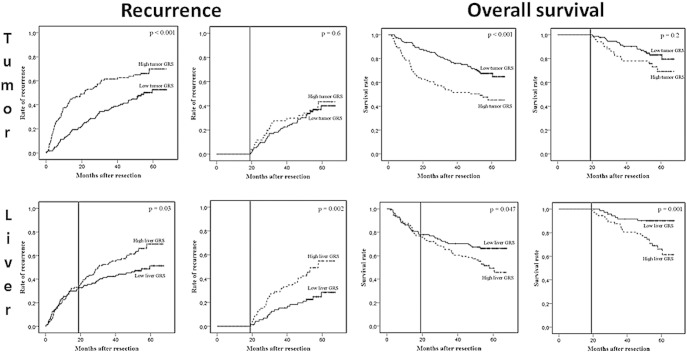
Figure 3.
Recurrence rates and overall survival in the LCI cohort [12]. Low (solid line) versus high (dashed line) GRS patients were assessed in tumor (first row) and liver (second row) tissue. Recurrence and death before 19 months were censored in the second and fourth column, respectively.
The GRS Assessed in the Surrounding Liver Tissue
The GRS was assessed on the surrounding nontumoral liver tissue of 228 patients in the NCI data set. Using an optimal cutoff specific for liver tissue, the recurrence curves of patients with low and high GRS started to separate at about 19 months (P = .03) (Figure 3).
When we excluded all patients with recurrence before 19 months, the GRS when assessed in tumor tissue could not distinguish patients with high versus low recurrence risk (Figure 3). In contrast, excluding these early recurrences enhanced the prognostic power of the GRS when applied on the surrounding liver.
Combining Tumor and Liver Gene Expression Data
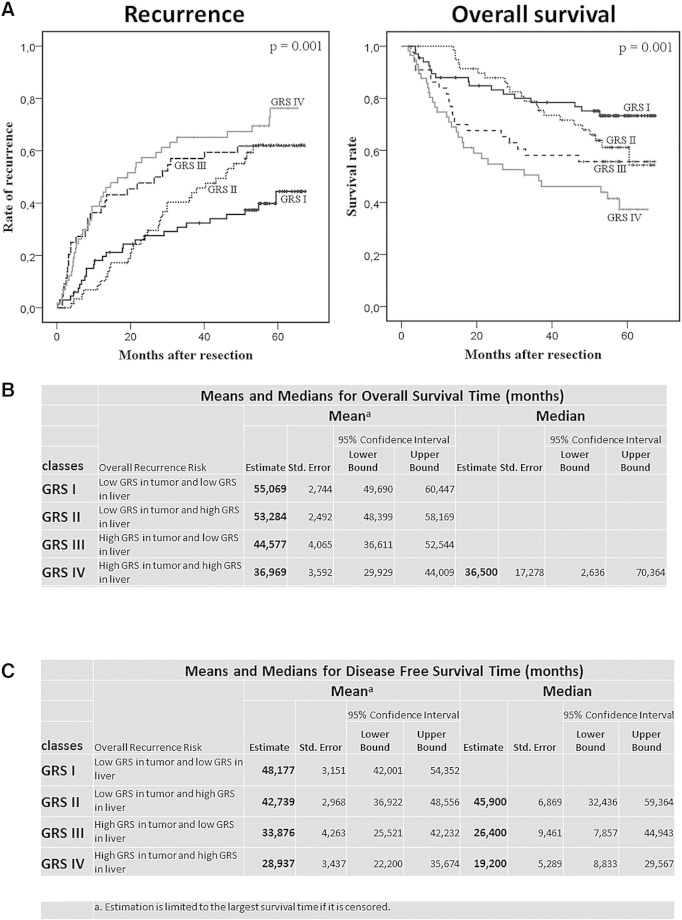
By combining the GRS of both tumor and liver tissue, we identified four classes of patients associated with different early and late recurrence risks (GRS class I-IV, overall P value for recurrence < .001) (Figure 4). Patients in GRS I, with low GRS both in liver and tumor tissue, had a 3-year recurrence rate of 32% ± 6% compared with 65% ± 6% in patients in GRS IV (high GRS in both tissues). Patients with low GRS in tumor and high GRS in liver (GRS II) had similar recurrence risk as high–tumor GRS/low–liver GRS patients (GRS III). However, GRS II and III differed with regard to median time to recurrence: 46 months (95% CI 32-59) versus 26 months (95% CI 8-45), respectively (P = .03 by Breslow test).
Figure 4.
Combining tumor and liver gene expression data. (A) Recurrence rates and overall survival in the LCI cohort in the four GRS classes (GRS I = low GRS in tumor and liver; GRS II = low GRS in tumor, high in liver; GRS III = high GRS in tumor, low in liver; GRS IV = high GRS in tumor and liver). (B) Mean overall survival time for different GRS classes. (C) Mean disease-free survival time for different GRS classes.
In a multivariate analysis, GRS class remained significantly associated with disease-free survival and overall survival independent from Barcelona Clinic Liver Cancer (BCLC) class and other established prognostic factors (Table 4 and Supplementary Table 2).
Table 4.
Multivariate Cox Regression Analysis on Selected Clinical Variables in the NCI Data Set Based on Significance in Univariate Analysis (see Supplementary Table 2)
| Disease-Free Survival |
Overall Survival |
|||||||
|---|---|---|---|---|---|---|---|---|
| P Value | Hazard Ratio | 95% CI |
P Value | Hazard Ratio | 95% CI |
|||
| Lower | Upper | Lower | Upper | |||||
| Gender (female vs male) | .083 | .544 | .273 | 1.082 | ||||
| Size largest nodule (> 5 cm vs < 5 cm) | .715 | .900 | .513 | 1.580 | ||||
| Multinodular (no vs yes) | .145 | 1.785 | .819 | 3.888 | ||||
| Cirrhosis (yes vs no) | .060 | 3.941 | .945 | 16.434 | ||||
| BCLC | .000 | .000 | ||||||
| BCLC (A vs 0) | .045 | 2.381 | 1.021 | 5.552 | .035 | 4.648 | 1.113 | 19.403 |
| BCLC (B vs 0) | .002 | 4.564 | 1.749 | 11.915 | .002 | 14.850 | 2.597 | 84.908 |
| BCLC (C vs 0) | .000 | 6.744 | 2.640 | 17.224 | .000 | 23.956 | 4.850 | 118.331 |
| aFP (> 300 ng/ml vs < 300 ng/ml) | .410 | 1.228 | .754 | 2.001 | ||||
| GRS group | .007 | .024 | ||||||
| GRS II vs I | .002 | 2.458 | 1.380 | 4.377 | .025 | 2.315 | 1.112 | 4.820 |
| GRS III vs I | .033 | 1.810 | 1.048 | 3.127 | .086 | 1.848 | .916 | 3.730 |
| GRS IV vs I | .002 | 2.372 | 1.378 | 4.083 | .003 | 2.870 | 1.446 | 5.696 |
GRS groups: GRS I = low GRS in tumor and liver; GRS II = low GRS in tumor, high in liver; GRS III = high GRS in tumor, low in liver; GRS IV = high GRS in tumor and liver.
Discussion
The fact that the recurrence rate after surgery comes in two waves each with different etiologies—early recurrence caused by recurrence of the resected primary tumor versus late recurrence caused by de novo tumor formation in the cirrhotic environment—is a unique feature of HCC [1]. We developed a GRS based on gene expression data of a limited number of genes that, when assessed in both tumor and the surrounding liver tissue, could predict the risk of early and late recurrence in patients with HCC treated with resection. Identifying this risk at the time of surgery is crucial in considering all further treatment options and their optimal timing, including liver transplantation [3], [4], [5].
It is now generally accepted that, to make an adequate estimate of the recurrence risk after resection of HCC, information on the resected tumor as well as the surrounding liver tissue needs to be combined [15]. Our approach, in which we developed one gene expression risk score (GRS) that predicts both early and late recurrence by combining this information on the tumor and liver tissue, has never been performed before. Previous gene expression signatures have focused on either tumor or liver tissue [11], [12], [13], [14], [15], [16], [17], [26]. With a well-validated five-gene score applied on tumor tissue, disease recurrence and survival after resection could be predicted independently of the established clinical and pathologic markers [16]. In another study, the authors could not predict prognosis-based gene expression of the tumor [13]. This is due to the limited number of early recurrence events (n = 6) in the training data set of 80 early-stage tumor samples. Interestingly, gene expression of the noncancerous surrounding liver tissue did predict survival. The authors developed a 186-gene signature, which was validated to predict recurrence after HCC resection as well as in hepatitis C cirrhosis without HCC [13], [17]. Similarly, a model of hepatic injury was used to develop another (233) gene signature that predicted late recurrence [14]. Although the latter two studies both predicted late recurrence of HCC and both used noncancerous liver tissue, their gene scores show remarkable little overlap (only four genes) despite containing a high number of genes. Our seven-gene GRS has one gene in common with both signatures (Supplementary Figure 2). To be noted, the majority of expression studies all point to a subgroup within the molecular classification of HCC that has a bad prognosis. And although the individual studies came up with different sets of genes to predict this subgroup, when applied to the same 287 HCC patients, there is a great concordance of the poor-outcome signatures [15]. This concordance is also indicated by GSEA with the HepG2S1 signature used to develop the GSR (Supplementary Figure 1, E–G).
The occurrence of de novo tumors is named late recurrence, although, theoretically, it can take place at any time after resection of HCC. Most authors have adopted a threshold of 2 years to distinguish early from late recurrence based on differences in risk factors and recurrence rates [27]. In this study, the GRS, when applied on the noncancerous surrounding liver tissue, predicted recurrence starting at about 19 months after resection. Similarly, when we excluded patients with recurrence before 19 months, the GRS in tumor tissue could not distinguish patients with high versus low risk. These results demonstrate that, from this time point on, the formation of new lesions starts to outnumber the incidence of recurrence of the primary lesion. The primary tumor is thus noninformative for the risk of de novo lesions. Gene expression data of both the primary tumor as well as the surrounding liver are needed to accurately predict the risk of recurrence. Therefore, our seven-gene GRS for both tumor and liver can guide treatment options and timing. But how could such a gene signature be implemented? The limited number of genes in the GSR allows the use of simple cost-effective platforms, such as RT-qPCR, for future validation and application. It will be possible to determine the expression in the tumor and in the surrounding tissue as well as in a set of noncancerous livers and subsequently calculate the GSR score and compare it with the reference value.
This study has several potential limitations. We aimed to improve treatment stratification by developing a gene score based on gene expression differences between HepG2 cells made resistant to sorafenib and their parental lineage. We are aware of the fact that none of the patients included in this study have been exposed to sorafenib. However, as HepG2S1 cells lose their hepatocyte differentiation, undergo epithelial-to-mesenchymal transition, and obtain invasive capacities, this model reflects an aggressive disease behavior and malignant potential rather than merely a drug resistance profile [18]. This is supported by the GSEA analysis where the resistant cells show strong overlap with the poor prognosis subclasses of Lee [11], Chiang [22], and Boyault [26] (see Supplementary Figures 1, E–G). The final, independent validation in two additional data sets justifies our approach. Second, by using one in vitro model, we might have lost important prognostic genes early in the development process. However, true downsizing of the number of genes was done using patient samples leading to a prognostic score. This approach was similar to what was done successfully in the past in the context of hypoxia in HCC [28] and colon cancer [29]. Using a cell line also has advantages; the results are more pure, which is reflected in the small set of genes necessary for the GRS as compared with other published studies that used patients’ tissue and that resulted sometimes in gene scores of 180 to 250 genes to classify. Third, one might argue that the differences between the four GRS classes are only modest, questioning its relevance in clinical decision making. As more than 70% of patients will show tumor recurrence within 5 years after surgery [27], it becomes difficult to acquire larger between-group differences with a prognostic factor. Indeed, in such setting, timing of recurrence becomes more of interest as the disease will almost certainly recur. Timing is addressed by the GRS as GRS III patients have only half median disease-free survival time compared with GRS II patients (Figure 4C). Finally, before the GRS can find its way to the clinic, further validation needs to determine a platform-specific cutoff and reveal its role in the context of other established clinical and pathological prognostic markers.
Conclusion
We developed and validated a gene score based on an in vitro model of aggressive tumor biology and hepatocyte dedifferentiation. The obtained score shows unique features, as it can be assessed in tumor as well as in liver tissue to predict early and late recurrence, respectively. Together, a global estimate of tumor recurrence by the GRS can help to stratify treatment options in an individual patient, and the particular small set of genes could facilitate its clinical use.
Funding
This work was supported by GOA/10/09 MaNet, IWT: TBM Rectal Cancer, iMinds SBO, VLK Stichting E. van der Schueren: rectal cancer,Federal Government Cancer Plan 2012-2015KPC-29-023 (prostate). J. Dekervel was supported by the Flemish League against Cancer (VLK).
Conflict of Interest Statement
None declared.
Acknowledgements
The authors thank Prof. David Cassiman and Prof. Schalk van der Merwe for their critical review of the manuscript.
Footnotes
This work was supported by GOA/10/09 MaNet, IWT: TBM Rectal Cancer, iMinds SBO, VLK Stichting E. van der Schueren: rectal cancer,Federal Government Cancer Plan 2012-2015KPC-29-023 (prostate). J. Dekervel was supported by the Flemish League against Cancer (VLK).
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.tranon.2016.02.003.
Appendix A. Supplementary data
Supplementary materials.
References
- 1.Poon RT, Fan ST, Ng IO, Wong J. Different risk factors and prognosis for early and late intrahepatic recurrence after resection of hepatocellular carcinoma. Cancer. 2000;89:500–507. [PubMed] [Google Scholar]
- 2.Llovet JM, Di Bisceglie AM, Bruix J, Kramer BS, Lencioni R, Zhu AX, Sherman M, Schwartz M, Lotze M, Talwalkar J, Gores GJ, Panel of Experts in HCC-Design Clinical Trials Design and endpoints of clinical trials in hepatocellular carcinoma. J Natl Cancer Inst. 2008;100(10):698–711. doi: 10.1093/jnci/djn134. [ http://jnci.oxfordjournals.org/content/100/10/698.long] [DOI] [PubMed] [Google Scholar]
- 3.Verslype C, Van Cutsem E, Dicato M, Arber N, Berlin JD, Cunningham D, De Gramont A, Diaz-Rubio E, Ducreux M, Gruenberger T. The management of hepatocellular carcinoma. Current expert opinion and recommendations derived from the 10th World Congress on Gastrointestinal Cancer, Barcelona, 2008. Ann Oncol. 2009;20(Suppl 7):vii1–vii6. doi: 10.1093/annonc/mdp281. [ http://annonc.oxfordjournals.org/content/20/suppl_7/vii1.long] [DOI] [PubMed] [Google Scholar]
- 4.Bruix J, Han KH, Gores G, Llovet JM, Mazzaferro V. Liver cancer: approaching a personalized care. J Hepatol. 2015;62(1 Suppl):S144–S156. doi: 10.1016/j.jhep.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roayaie S, Jibara G, Tabrizian P, Park JW, Yang J, Yan L, Schwartz M, Han G, Izzo F, Chen M. The role of hepatic resection in the treatment of hepatocellular cancer. Hepatology. 2015;62(2):440–451. doi: 10.1002/hep.27745. [ http://onlinelibrary.wiley.com/doi/10.1002/hep.27745/full] [DOI] [PubMed] [Google Scholar]
- 6.Zhang JP, Wang HB, Lin YH, Xu J, Wang J, Wang K, Liu WL. Lactate dehydrogenase is an important prognostic indicator for hepatocellular carcinoma after partial hepatectomy. Transl Oncol. 2015;8(6):497–503. doi: 10.1016/j.tranon.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Ruan DY, Yi HM, Wang GY, Yang Y, Jiang N. A three-factor preoperative scoring model predicts risk of recurrence after liver resection or transplantation in hepatocellular carcinoma patients with preserved liver function. Hepatobiliary Pancreat Dis Int. 2015;14(5):477–484. doi: 10.1016/s1499-3872(15)60412-x. [ http://www.hbpdint.com/EN/Y2015/V14/I5/477] [DOI] [PubMed] [Google Scholar]
- 8.Yang SL, Liu LP, Sun YF, Yang XR, Fan J, Ren JW, Chen GG, Lai PB. Distinguished prognosis after hepatectomy of HBV-related hepatocellular carcinoma with or without cirrhosis: a long-term follow-up analysis. J Gastroenterol. 2015 doi: 10.1007/s00535-015-1146-0. [[Epub ahead of print] PubMed PMID: 26607653. http://link.springer.com/article/10.1007%2Fs00535-015-1146-0] [DOI] [PubMed] [Google Scholar]
- 9.Shindoh J, Makuuchi M, Matsuyama Y, Mise Y, Arita J, Sakamoto Y, Hasegawa K, Kokudo N. Complete removal of the tumor-bearing portal territory decreases local tumor recurrence and improves disease-specific survival of patients with hepatocellular carcinoma. J Hepatol. 2015 doi: 10.1016/j.jhep.2015.10.015. [[Epub ahead of print] PubMed PMID: 26505120] [DOI] [PubMed] [Google Scholar]
- 10.Hou YF, Wei YG, Yang JY, Wen TF, Xu MQ, Yan LN, Li B, Chen KF. Microvascular invasion patterns affect survival in hepatocellular carcinoma patients after second hepatectomy. J Surg Res. 2016;200(1):82–90. doi: 10.1016/j.jss.2015.06.069. [DOI] [PubMed] [Google Scholar]
- 11.Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, Durnez A, Demetris AJ, Thorgeirsson SS. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 12.Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoshida Y, Villanueva A, Kobayashi M, Peix J, Chiang DY, Camargo A, Gupta S, Moore J, Wrobel MJ, Lerner J. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359:1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim JH, Sohn BH, Lee HS, Kim SB, Yoo JE, Park YY, Jeong W, Lee SS, Park ES, Kaseb A. Genomic predictors for recurrence patterns of hepatocellular carcinoma: model derivation and validation. PLoS Med. 2014;11 doi: 10.1371/journal.pmed.1001770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villanueva A, Hoshida Y, Battiston C, Tovar V, Sia D, Alsinet C, Cornella H, Liberzon A, Kobayashi M, Kumada H. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology. 2011;140:1501–1512. doi: 10.1053/j.gastro.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nault JC, De Reyniès A, Villanueva A, Calderaro J, Rebouissou S, Couchy G, Decaens T, Franco D, Imbeaud S, Rousseau F. A hepatocellular carcinoma 5-gene score associated with survival of patients after liver resection. Gastroenterology. 2013;145:176–187. doi: 10.1053/j.gastro.2013.03.051. [DOI] [PubMed] [Google Scholar]
- 17.Hoshida Y, Villanueva A, Sangiovanni A, Sole M, Hur C, Andersson KL, Chung RT, Gould J, Kojima K, Gupta S. Prognostic gene expression signature for patients with hepatitis C-related early-stage cirrhosis. Gastroenterology. 2013;144:1024–1030. doi: 10.1053/j.gastro.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Malenstein H, Dekervel J, Verslype C, Van Cutsem E, Windmolders P, Nevens F, van Pelt J. Long-term exposure to sorafenib of liver cancer cells induces resistance with epithelial-to-mesenchymal transition, increased invasion and risk of rebound growth. Cancer Lett. 2013;329:74–83. doi: 10.1016/j.canlet.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 19.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 21.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiang DY, Villanueva A, Hoshida Y, Peix J, Newell P, Minguez B, LeBlanc AC, Donovan DJ, Thung SN, Solé M. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68:6779–6788. doi: 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sung WK, Zheng H, Li S, Chen R, Liu X, Li Y, Lee NP, Lee WH, Ariyaratne PN, Tennakoon C. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet. 2012;44:765–769. doi: 10.1038/ng.2295. [ http://www.nature.com/ng/journal/v44/n7/fp/ng.2295.html] [DOI] [PubMed] [Google Scholar]
- 24.Kudo A, Mogushi K, Takayama T, Matsumura S, Ban D, Irie T, Ochiai T, Nakamura N, Tanaka H, Anzai N. Mitochondrial metabolism in the noncancerous liver determine the occurrence of hepatocellular carcinoma: a prospective study. J Gastroenterol. 2014;49:502–510. doi: 10.1007/s00535-013-0791-4. [ http://link.springer.com/article/10.1007%2Fs00535-013-0791-4] [DOI] [PubMed] [Google Scholar]
- 25.Goeman JJ, van de Geer SA, de Kort F, van Houwelingen HC. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;20:93–99. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- 26.Boyault S, Rickman DS, de Reyniès A, Balabaud C, Rebouissou S, Jeannot E, Hérault A, Saric J, Belghiti J, Franco D. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 27.Imamura H, Matsuyama Y, Tanaka E, Ohkubo T, Hasegawa K, Miyagawa S, Sugawara Y, Minagawa M, Takayama T, Kawasaki S. Risk factors contributing to early and late phase intrahepatic recurrence of hepatocellular carcinoma after hepatectomy. J Hepatol. 2003;38:200–207. doi: 10.1016/s0168-8278(02)00360-4. [DOI] [PubMed] [Google Scholar]
- 28.van Malenstein H, Gevaert O, Libbrecht L, Daemen A, Allemeersch J, Nevens F, Van Cutsem E, Cassiman D, De Moor B, Verslype C. A seven-gene set associated with chronic hypoxia of prognostic importance in hepatocellular carcinoma. Clin Cancer Res. 2010;16(16):4278–4288. doi: 10.1158/1078-0432.CCR-09-3274. [ http://clincancerres.aacrjournals.org/content/16/16/4278.full.pdf+html] [DOI] [PubMed] [Google Scholar]
- 29.Dekervel J, Hompes D, van Malenstein H, Popovic D, Sagaert X, De Moor B, Van Cutsem E, D'Hoore A, Verslype C, van Pelt J. Hypoxia-driven gene expression is an independent prognostic factor in stage II and III colon cancer patients. Clin Cancer Res. 2014;20(8):2159–2168. doi: 10.1158/1078-0432.CCR-13-2958. [ http://clincancerres.aacrjournals.org/content/20/8/2159.full.pdf+html] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials.