Abstract
Human brain tumors such as glioblastomas are typically detected using conventional, nonquantitative magnetic resonance imaging (MRI) techniques, such as T2-weighted and contrast enhanced T1-weighted MRI. In this manuscript, we tested whether dynamic quantitative T1 mapping by MRI can localize orthotopic glioma tumors in an objective manner. Quantitative T1 mapping was performed by MRI over multiple time points using the conventional contrast agent Optimark. We compared signal differences to determine the gadolinium concentration in tissues over time. The T1 parametric maps made it easy to identify the regions of contrast enhancement and thus tumor location. Doubling the typical human dose of contrast agent resulted in a clearer demarcation of these tumors. Therefore, T1 mapping of brain tumors is gadolinium dose dependent and improves detection of tumors by MRI. The use of T1 maps provides a quantitative means to evaluate tumor detection by gadolinium-based contrast agents over time. This dynamic quantitative T1 mapping technique will also enable future quantitative evaluation of various targeted MRI contrast agents.
Introduction
Gliomas are the most common primary malignant brain tumor in adults [1]. High-grade glioma, or glioblastoma (GBM), is one of the most aggressive and lethal types of brain cancer. The standard treatment for GBM involves maximal safe surgical resection of the tumor mass [2]. More complete resection has been linked to improved survival [3]. Surgery is followed by radiation and adjuvant temozolomide chemotherapy, but despite this three-pronged treatment strategy, the median survival of a GBM patient is only 15 months [4]. One of the key features of GBMs is their dispersive, infiltrative nature, which makes visualization of the tumor difficult [5].
Magnetic resonance imaging (MRI) is essential in the diagnosis, surgical planning, and evaluation of treatment efficacy of GBM patients. Conventional MRI for brain tumor diagnosis employs T1-weighted, contrast-enhanced T1-weighted, and T2-weighted sequences, but these images are qualitative and require subjective interpretation. There is difficulty locating these tumors on MRI both before and after surgery due to their dispersal. These conventional MRI images generally show the location of the main tumor mass but suffer from poor tumor-to-background contrast resulting in imprecise location of the full extent of the tumor and its borders. MRI contrast agents, i.e., gadolinium chelates, improve the visibility of structures and pathologies, thereby increasing tumor-to-background contrast. Gadolinium-based MRI contrast agents provide contrast enhancement due to the nonspecific leakage of gadolinium out of abnormal vasculature into the extracellular space in the tumor [6]. Gadolinium-based MRI contrast agents shorten the T1 relaxation time and alter the T1 values differently in different tissues. This decrease in T1 relaxation time is directly proportional to the tissue concentration of contrast agent. Using the absolute T1 values in the brain before and after intravenous injection of contrast agent, a measurement of contrast agent concentration per voxel of the tumor can be calculated. T2-weighted/fluid-attenuated inversion-recovery (FLAIR) sequences have the ability to distinguish edema from tumor [6] and may also be useful in evaluating nonenhancing tumor regions [7]. Yet, the results of these various MRI sequences are highly variable in the clinic and are qualitative in nature [6].
T1 relaxation time mapping yields absolute T1 relaxation time values, thereby offering an objective quantitative, longitudinal evaluation of 1) tumor/tissue concentration of the MRI contrast agent at all imaging time points, 2) increased injected contrast agent, 3) invasive and noninvasive gliomas, and 4) therapeutic efficacy. The dynamic T1 mapping method enables quantitative assessments of gadolinium concentration in tissues and allows for the assessment of maximum contrast agent accumulation and clearance over time as opposed to subjective evaluation of changes in contrast enhancement on radiologic images. Importantly, through the use of T1 maps, radiologists will not have to rely on qualitative information to determine tumor location but will be able to quantitatively locate a tumor and its borders.
Measurable human disease in radiographic images of GBM is defined as a bidimensionally contrast-enhancing lesion with clearly defined margins by computed tomography or MRI scan, with 2 perpendicular diameters of at least 10 mm, visible on 2 or more axial slices that are preferably, at most, 5 mm apart with 0-mm skip [7]. We developed a series of xenograft models to study brain tumors in an orthotopic setting [8]. Depending upon the cell line and number of days postimplantation that the MRI was performed, these models have tumors that are of variable sizes of 1 to 4 mm. In these xenograft models, it is difficult to reliably detect any of these tumors with qualitative MRI as the various tumor models exhibit differential endogenous MRI properties (i.e., T1, T2, etc.) and contrast uptake, thereby limiting our ability to reliably study these glioma models in detail. Due to its quantitative nature, we hypothesize that dynamic mapping of T1 relaxation time during contrast agent injection can be used to objectively locate these xenograft brain tumors and to track gadolinium-based contrast uptake over time.
To test this hypothesis, we employed a T1 mapping sequence and generated quantitative T1 and gadolinium concentration maps using the gadolinium-based contrast agent Optimark (gadoversetamide) to enhance mouse orthotopic xenografts of human Gli36∆5 glioma cells. This orthotopic glioma model was generated by transplanting Gli36Δ5 cells that constitutively overexpress the vIII mutant forms of the EGFR gene, a mutation frequently found in GBM tumors [9], into the brains of athymic mice [8]. Using human Gli36Δ5 cells implanted in the mouse brain for 7 to 10 days, we created tumors that present as a highly vascularized mass of ~ 1.5 × 2 × 3.5 mm (or 10.5 mm3) in size. We observed the reliable detection of these orthotopic mouse brain tumors using quantitative T1 mapping techniques and a conventional clinical contrast agent at multiple dose levels on a 9.4-T preclinical MR scanner. To evaluate the quantitative nature of the T1 mapping technique, we compared tumor enhancement using two doses of Optimark. Doubling the dose of contrast agent in the mouse system resulted in a greater decrease in T1 relaxation time, allowing for clear demarcation of tumors in the maps.
These data demonstrate that T1 mapping of brain tumors is feasible, is gadolinium dose dependent, and provides reliable detection of tumors. By doubling the dose of the MRI contrast agent, we found that the tumor is further enhanced, facilitating the ability to detect smaller orthotopic mouse brain tumors. Evaluation of the T1 relaxation time maps as well as estimated tumor gadolinium concentrations calculated across animals over time demonstrates that these values are reproducible between different animals. Therefore, we conclude that T1 maps and gadolinium concentration maps provide a quantifiable and reproducible means of visualizing intracranial tumor enhancement over time. We suggest that the use of T1 mapping provides a quantitative means to evaluate tumors in the clinic to ultimately track tumor response to therapy.
Material and Methods
Orthotopic Xenograft Intracranial Tumors
NIH athymic nude female mice (NCr-nu/+, NCr-nu/nu) were bred in the Athymic Animal Core Facility and housed in the Case Center for Imaging Research at Case Western Reserve University according to institutional policies. The Institutional Animal Care and Use Committee approved all animal protocols.
The Gli36∆5 cells [10] were obtained from E.A. Chiocca and authenticated by Research Animal Diagnostic Laboratory at the University of Missouri (Columbia, MO) for interspecies and mycoplasma contamination by PCR analysis.
Gli36Δ5 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. Gli36∆5 cells were infected with lentivirus to express green fluorescent protein [11] 48 hours prior to harvesting. Cells were harvested for intracranial implantation by trypsinization and concentrated to 1 × 105 cells per microliter of PBS. Mice 6 to 7 weeks of age were anesthetized by intraperitoneal administration of 50 mg/kg ketamine/xylazine and fitted into a stereotaxic rodent frame (David Kopf Instruments, Tujunga, CA). A small incision was made to expose the bregma suture. A small (0.7 mm) burr hole was made 0.7 mm anterior and 2.0 mm lateral from bregma. Cells were slowly deposited at a rate of 1 μl/min for a total of 200,000 cells into the right striatum at a depth of − 3 mm from the dura using a 10-μl syringe (26-gauge needle; Hamilton Co, Reno, NV). The needle was slowly withdrawn, and the incision was closed with sutures. Mice were imaged as described below and then sacrificed 10 days after tumor implant. Brain tissue was collected for histological processing.
Magnetic Resonance Imaging
The MRI studies were performed using a Bruker Biospec 9.4-T preclinical MRI scanner (Bruker Corp., Billerica, MA) with a 35 mm inner diameter mouse radiofrequency coil. Mice bearing Gli36Δ5 brain tumors were scanned 7 to 10 days after tumor implantation using a clinically approved MRI contrast agent, Optimark (Mallinckrodt Pharmaceuticals, St. Louis, MO, FW 661.77).
Dynamic T1 Relaxation Time Mapping MRI Acquisition
Tumor-bearing mice were catheterized prior to the imaging study. In brief, polyurethane tubing (0.014″ ID × 0.033″ OD) (SAI Infusion Technologies, Lake Villa, IL) was connected to a 1-ml syringe and preloaded with the appropriate amount of Optimark diluted in PBS or saline. After mice were anesthetized with a 2% isoflurane-oxygen mixture in an induction chamber, tail veins were catheterized using a 26-gauge veterinary catheter and connected to the preloaded tubing described above. The animals were moved into the magnet in the prone position and kept under inhalation anesthesia with 1.5% isoflurane-oxygen via a nose cone. A respiratory sensor connected to a monitoring system (SA Instruments, Stony Brook, NY) was placed on the back of the animal to monitor rate and depth of respiration. Body temperature was maintained at 35 ± 2°C by blowing warm air into the magnet through a feedback control system.
T2-weighted images were first obtained for each mouse using a rapid acquisition with relaxation enhancement sequence [repetition time (TR)/echo time (TE) = 5000 milliseconds/40 milliseconds, resolution = 0.078 × 0.078 × 0.5 mm, field of view = 20 × 20 mm, and three signal averages] to select the imaging slice for the dynamic T1 relaxation time acquisition [12]. The quantitative T1 data were acquired using a snapshot gradient recalled echo (GRE) sequence with inversion recovery preparation described previously [10 inversion times (519, 1031, 1543, 2055, 2567, 3079, 3591, 4103, 4615, and 5127 milliseconds), GRE imaging readout TR/TE = 4.0 milliseconds/1.9 milliseconds, flip angle = 10°, resolution = 0.156 × 0.156 × 1 mm, field of view = 20 × 20 mm, and 10 signal averages] [13], [14]. The total acquisition for each T1 mapping scan was 2.5 minutes.
Three T1 mapping baseline images were acquired before injecting Optimark at a dose of 0.1 mmol Gd/kg. T1 maps were acquired every 5 minutes, alternating with T1-weighted scans, for a total of 60 minutes.
T1 Mapping
The T1 mapping data were then acquired using a slightly different sequence than that described above [10 inversion times (263, 775, 1287, 1799, 2311, 2823, 3335, 3847, 4359, and 4871 milliseconds), GRE imaging readout TR/TE = 4.0 milliseconds/1.3 milliseconds, flip angle = 10°, resolution = 0.234 × 0.234 with a slice thickness of 1.5 mm, field of view = 30 × 30 mm, 10 signal averages for a total scan time of 2.5 minutes]. As described above, Optimark at 0.1- or 0.2-mmol/kg was administered after acquiring three baseline scans (n = 9 for Optimark at 0.1-mmol/kg, n = 7 for Optimark at 0.2-mmol/kg).
Generation of T1 Relaxation and Gadolinium Concentration Maps
The MRI data were imported into MATLAB, and the quantitative T1 relaxation maps were generated from the T1 mapping acquisition using previously described methods based on monoexponential models [14]. Dynamic maps of gadolinium concentration maps were calculated from the dynamic T1 relaxation maps using the following formula:
where T1 is the measured T1, r1 is the relaxivity value measured at 37°C on the Bruker Biospec 9.4-T scanner, and [Gd] is the gadolinium concentration. Median filtering was performed in MATLAB on the normalized T1 relaxation and gadolinium concentration maps to remove salt and pepper noise.
Evaluation of Dynamic T1 Mapping Values and Gadolinium Concentration in Orthotopic Xenograft Mouse Glioma Models
Once the maps were generated, the mean T1 relaxation and gadolinium concentration values were determined by performing a region of interest (ROI) analysis. For each mouse, an ROI was drawn around the tumor using the parametric map generated at 5 minutes postinjection when tumor enhancement was greatest. The same ROI was then applied to all of the T1 maps, the average value in the ROI was calculated for each time point, and normalized T1 values were obtained by dividing each T1 value by the mean baseline T1 value. Gadolinium concentrations were calculated using measured T1 values and the relaxivity of Optimark.
Histology
Brain tissue was harvested and fixed in 4% formalin for approximately 72 hours. Tissues were paraffin embedded and sectioned at 7 to 10 μm in thickness. Hematoxylin and eosin (H&E) staining was performed to visualize the main tumor mass. Images were captured with a 20 × objective using a Leica SCN400 microscope. Full tissue images were viewed using the Leica SCN400 Image Viewer version 2.2 software. Images were viewed at a 20 × zoom using the Aperio ImageScope version 12.1 software.
Results
Tumor Preparation and Validation
To detect small mouse orthotopic brain tumors, we performed a contrast-enhanced T1 mapping study to directly evaluate tumor enhancement using a conventional contrast agent. High-resolution T2-weighted MR images were acquired of Gli36Δ5 glioma tumors in athymic mice 7 to 10 days postimplantation. Using T2-weighted images, we were able to localize the tumors and identify the largest tumor cross section on MRI. This cross section then served as the reference for the position of the T1 slice (Figure 1A). Despite our a priori knowledge of the implantation site of the noninvasive Gli36Δ5 tumors [8], tumors of this size were not routinely evident in the T1-weighted images (Figure 1, Figure 2).
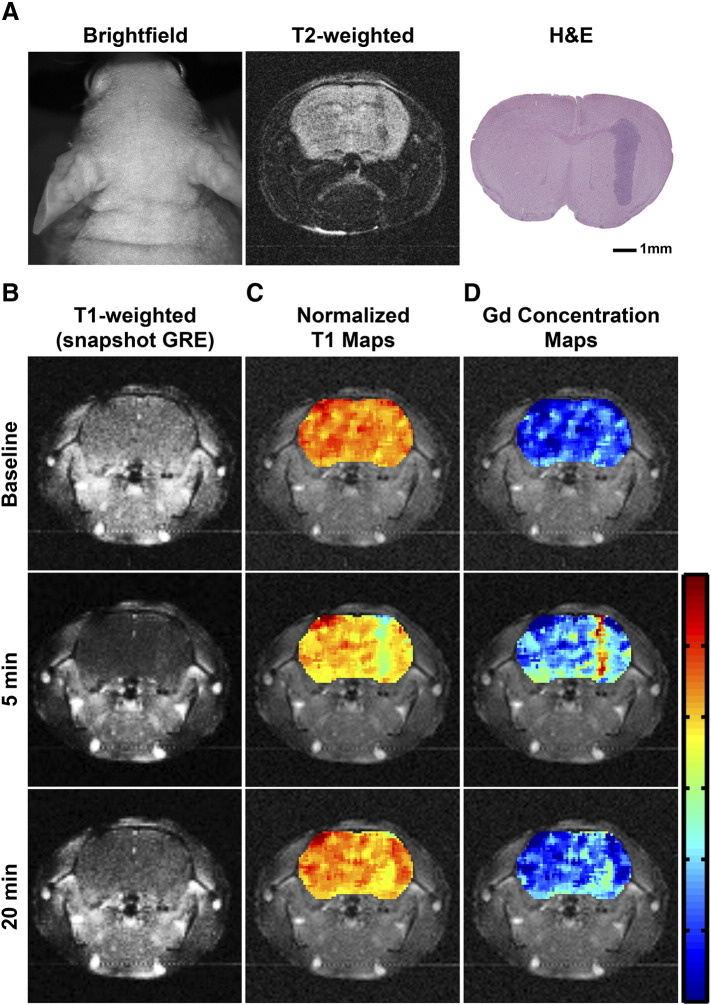
Figure 1.
Evaluation of tumor enhancement using quantitative T1 mapping. (A) Representative bright-field images of athymic mice bearing Gli36Δ5 glioma tumors. T2-weighted high-resolution images show the tumor mass and are co-registered with their corresponding T1-weighted images and normalized T1 maps. An H&E-stained histological section shows tumor size, shape, and location. Scale bar represents 1 mm. (B) T1-weighted, snapshot GRE acquired, images of an orthotopic Gli36Δ5 glioma tumor at baseline (before injection of 0.1-mmol/kg Optimark), 5 minutes following the injection (peak contrast), and 22.5 minutes postinjection (contrast agent clearance). (C) Normalized T1 maps of the tumor-bearing brains are overlaid onto T1-weighted images at baseline and at 5 and 22.5 minutes postinjection. The color-coded scale bar indicates normalized T1 map values with dark blue representing zero and red representing 1.2; the marks are in 0.2 increments. (D) Gadolinium concentration maps of the tumor-bearing brains are overlaid onto T1-weighted images at baseline and at 5 and 22.5 minutes postinjection. The color-coded scale bar indicates gadolinium concentration in mM with dark blue representing zero and red representing 0.06; the marks are in 0.01 increments.
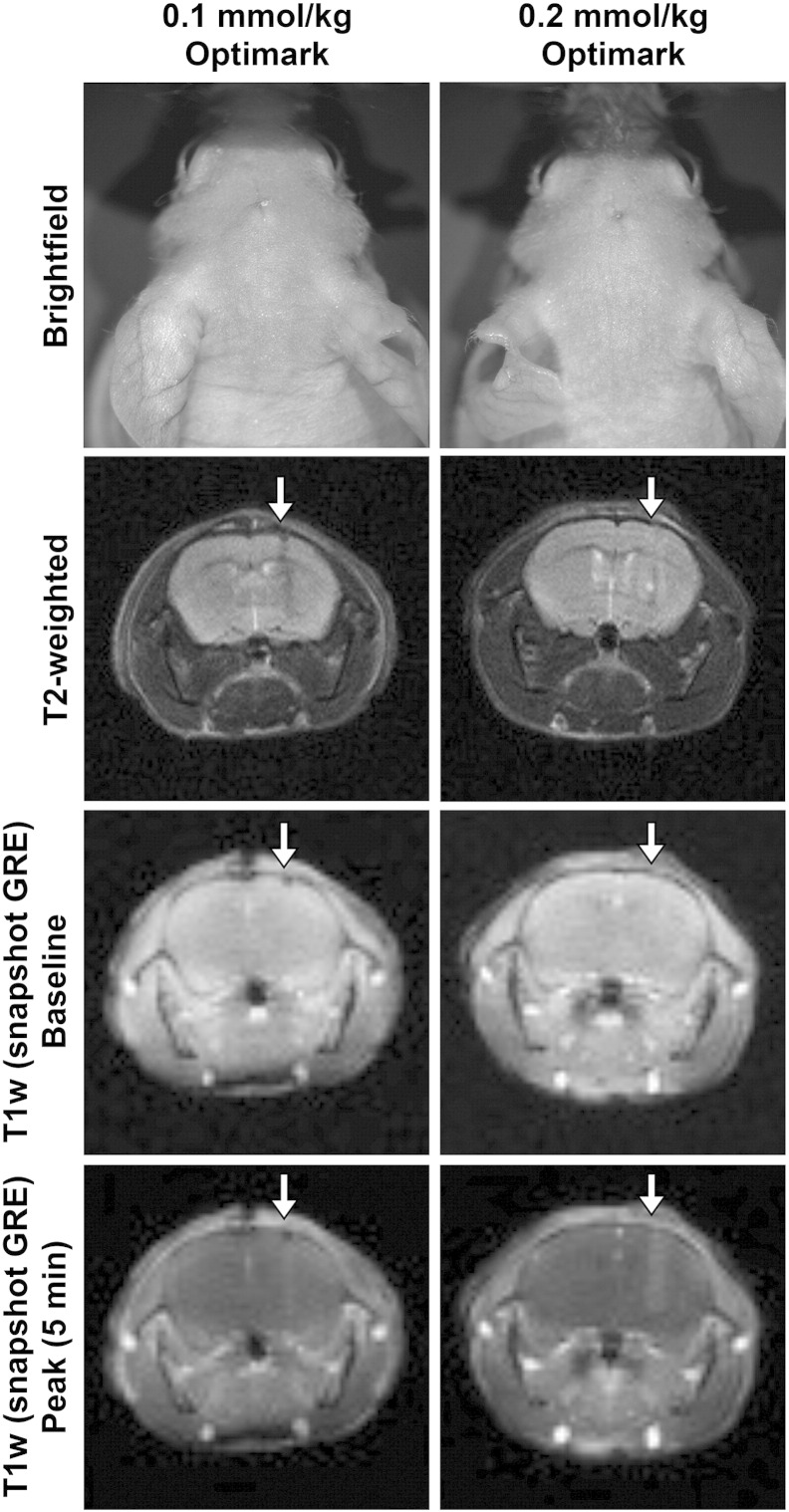
Figure 2.
The conventional imaging agent Optimark results in tumor enhancement of orthotopic glioma tumors in mice at 0.1- and 0.2-mmol/kg. Representative bright-field images of athymic mice bearing Gli36Δ5 glioma tumors that received 0.1- or 0.2-mmol/kg Optimark. T2-weighted high-resolution images show the tumor mass and are co-registered with their corresponding T1-weighted images below and quantitative maps in Figure 3, Figure 5. T1-weighted images of orthotopic Gli36Δ5 glioma tumors at baseline (before injection of contrast agents) and at time of maximum contrast (5 minutes) following intravenous administration are shown.
Evaluation of Orthotopic Tumor Using Quantitative T1 Mapping
We hypothesized that a contrast-enhanced quantitative T1 mapping technique would allow the generation of T1 parametric maps to enable visualization of contrast enhancement across the entire brain slice and thus allow us to detect these tumors. Quantitative T1 mapping is performed on a single MR slice. As a reference for the dimensions shown in the figures, the tumor size in this study was approximately 1.5 × 2 × 3.5 mm. The MR slice thickness is 1 to 1.5 mm. Therefore, the majority of the tumor is in 1 to 2 MR slices out of approximately 350 histological sections (7 μm thick) in 1 tumor.
T1 mapping sequences were acquired before and after administration of Optimark. Injection of the gadolinium-based contrast agent Optimark resulted in decreased T1 relaxation times, as expected (Figure 1). T1 mapping is a quantitative MRI method that yields absolute T1 relaxation values as opposed to the relative signal intensity values in conventional T1-weighted images. Using the measured T1 mapping values, quantitative, normalized T1 parametric maps were generated before and after contrast administration for the orthotopic brain tumor xenografts (Figure 1C). For clarity and to enable intermouse comparisons, the heat maps shown are normalized to the mean precontrast T1 value of whole brain. As shown in Figure 1C, the tumor is visible in these maps (represented by cyan; tumor average = 0.820) at 5 minutes and recovers towards baseline at 20 minutes after Optimark administration (tumor average = 0.902). Tissue concentration of gadolinium is proportional to the measured T1 mapping values and enables the generation of quantitative gadolinium concentration maps. The tumor is visible in the gadolinium concentration map (represented by yellow, orange, and red; tumor average = 0.013 mM) at 5 minutes after Optimark administration (Figure 1D). The 7 μm histological section shown in Figure 1A corroborates the general size and shape of the tumor observed. The normalized T1 and gadolinium concentration maps 5 minutes postcontrast represent the combined signal from an MR slice of 1.5 mm.
Quantitative, Contrast-Enhanced T1 Mapping
Quantitative T1 mapping and the generation of parametric maps enabled the detection of brain tumors in our mouse model of glioma (Figure 1). To evaluate the objective nature of T1 mapping methods, we hypothesized that administration of 0.1- and 0.2-mmol/kg Optimark would decrease T1 values in a dose-dependent manner. Representative brightfield, fluorescence, and MR images are shown in Figure 2. The T2-weighted scans do not clearly show the tumors, but an anatomical abnormality is apparent (Figure 2). The T1-weighted snapshot GRE images obtained with the T1 mapping sequence are shown at baseline and peak tumor enhancement postcontrast (5 minutes). Tumor masses were not apparent in the baseline T1 mapping images. Following injection of 0.1-mmol/kg Optimark, the tumor mass is not visible, as in Figure 1. However, there is an obvious change in overall signal in the brain indicating that delivery of contrast was successful (Figure 2). The 0.2-mmol/kg dose of Optimark resulted in a similar overall signal change in the brain along with tumor enhancement (Figure 2).
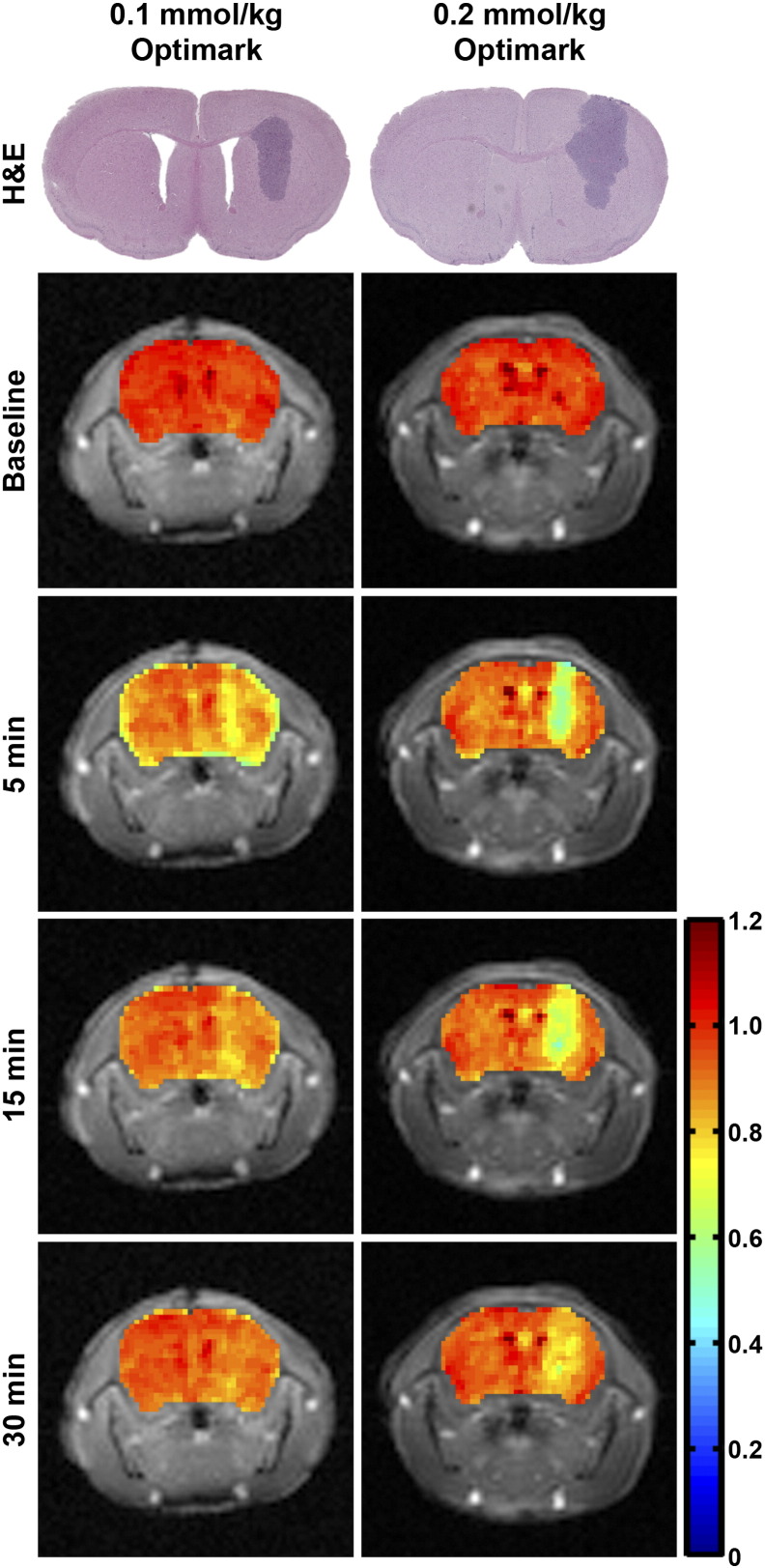
Quantitative parametric maps were generated to compare the tumor detection ability of the 0.1-versus 0.2-mmol/kg dose of Optimark. Representative normalized T1 maps are shown at baseline and at 5, 15, and 30 minutes postcontrast in Figure 3 for the 2 doses. Representative H&E-stained histological sections for each mouse are also shown in the first row of images of Figure 3 to verify tumor size and shape and to facilitate qualitative comparisons to the tumor enhancement observed in the normalized T1 maps. The 0.1-mmol/kg dose of Optimark resulted in a modest decrease in normalized T1 values (represented by yellow; tumor average = 0.798) at 5 minutes. At 15 minutes, the normalized T1 value exhibits recovery toward baseline due to Optimark clearance (tumor average = 0.899). By 30 minutes, there is no distinction between T1 value of normal brain and tumor with the 0.1-mmol/kg dose (tumor average = 0.935). The 0.2-mmol/kg dose of Optimark caused a larger decrease in normalized T1 values compared with 0.1-mmol/kg at 5 minutes (Figure 3, 0.2-mmol/kg dose; represented by green; tumor average = 0.646). By 15 minutes, the normalized T1 values begin to recover (tumor average = 0.690), and there is moderate enhancement observed beyond the initial tumor borders observed at 5 minutes (represented by yellow) that likely represents the full extent of the tumor within the 1.5 mm slice. At 30 minutes, a weak distinction remains between normalized T1 values of normal brain and tumor (tumor average = 0.823).
Figure 3.
The 0.1-mmol/kg and 0.2-mmol/kg doses of Optimark result in decreased normalized T1 relaxation times in orthotopic glioma tumors, with the 0.2-mmol/kg dose resulting in a greater decrease compared with 0.1-mmol/kg. Representative H&E-stained histological sections show tumor size, shape, and location. Normalized T1 maps of the tumor-bearing brains are overlaid onto T1-weighted images at baseline and at 5, 15, and 30 minutes postinjection. The color-coded scale bar indicates normalized T1 map values with dark blue representing the lowest and red representing the highest normalized T1 values. Maps represent the same slices as in Figure 2, Figure 5.
Comparison of mean normalized T1 values in the brain tumors of different animals demonstrates that both the 0.1- and 0.2-mmol/kg doses of Optimark exhibited peak enhancement in the tumors at 5 minutes and then steadily returned to baseline values over the next 30 minutes (Figure 4). The 0.2-mmol/kg dose of Optimark had a larger and statistically significant decrease (P < .00005 to P < .005 depending on the time point) in normalized T1 values at all time points compared with the 0.1-mmol/kg dose (Figure 4).
Figure 4.
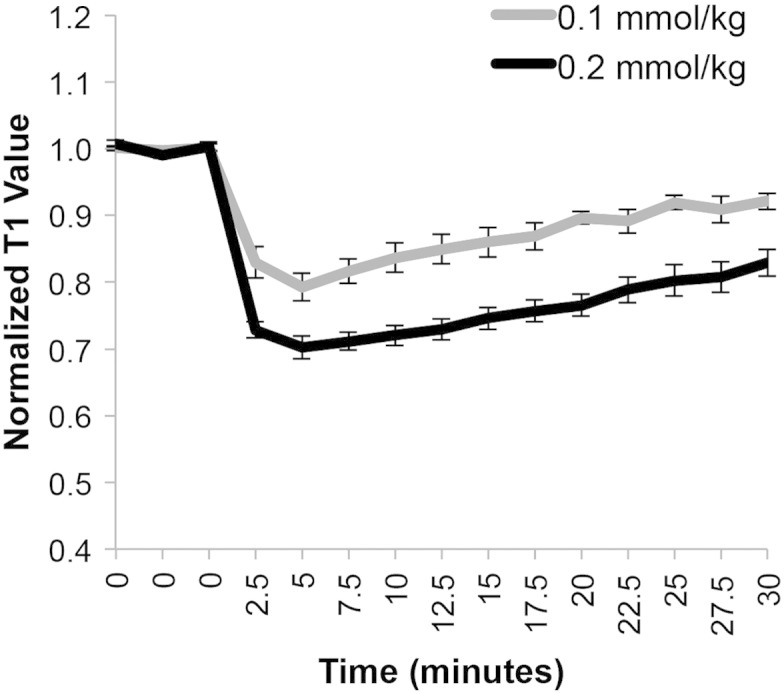
Mean normalized T1 values in the tumor before and after intravenous administration of 0.1- or 0.2-mmol/kg Optimark in cohorts of nu/nu athymic mice bearing orthotopic glioma tumors (n = 9 for 0.1-mmol/kg and n = 7 for 0.2-mmol/kg Optimark). Data plotted as means ± standard error. Mean tumor normalized T1 values at baseline and after contrast agent injection measured every 2.5 minutes for 30 minutes. Normalized T1 values are significantly different between the 0.1- and 0.2-mmol/kg doses (ranges from P < .00005 to P < .006 depending on the time point).
Quantitative Mapping of Gadolinium Concentrations
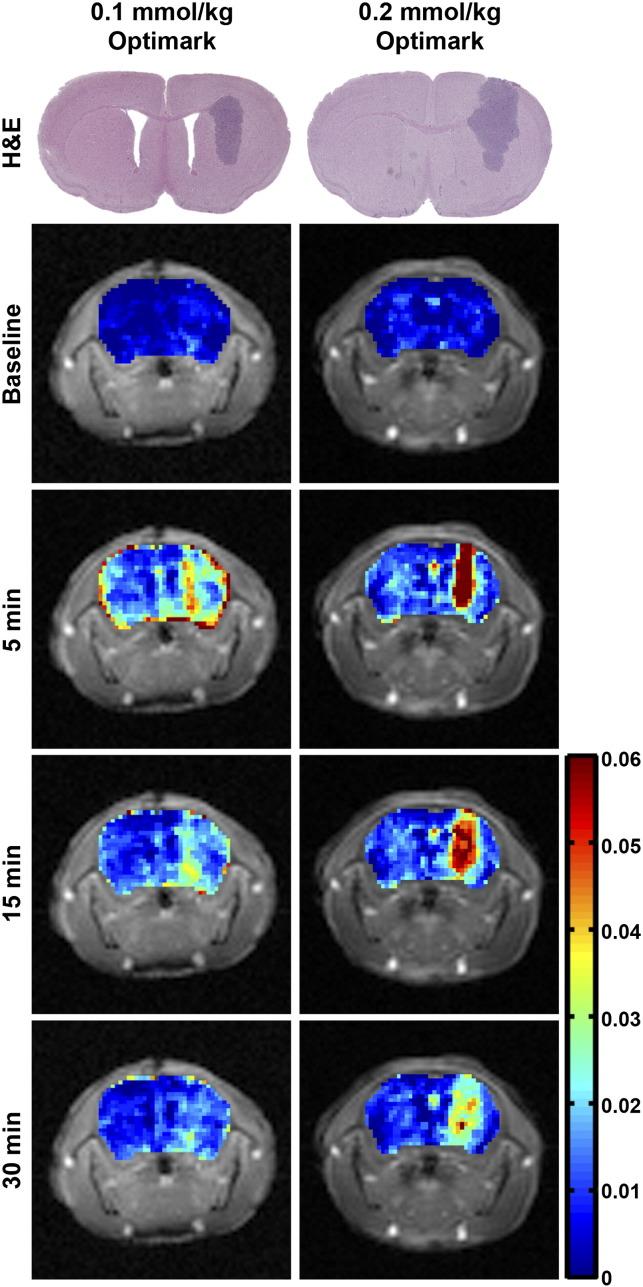
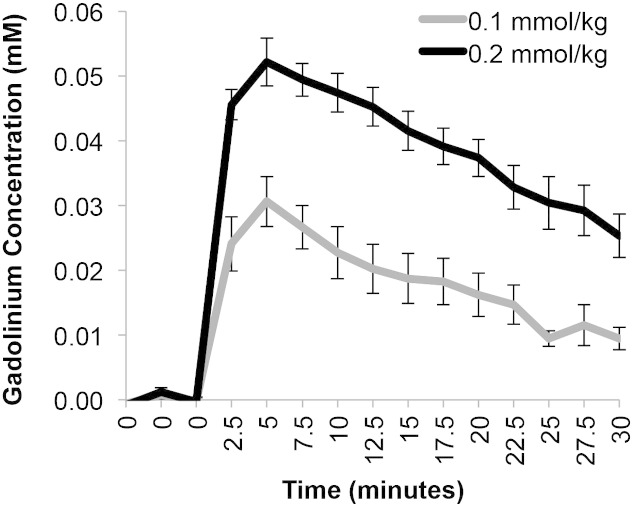
Gadolinium concentration maps can also evaluate contrast enhancement and hence tumor location using T1 mapping sequences. Representative gadolinium concentration maps are shown at baseline and at 5, 15, and 30 minutes postcontrast in Figure 5 for the 0.1- and 0.2-mmol/kg doses of Optimark. Representative H&E-stained histological sections are shown for comparison. The 0.1-mmol/kg dose of Optimark resulted in moderate increases in gadolinium concentration (represented by yellow and orange; tumor average = 0.030 mM) at 5 minutes. At 15 minutes, the gadolinium concentration in the 0.1-mmol/kg dose decreased toward baseline (represented by cyan; tumor average = 0.013 mM) and is virtually gone by 30 minutes (tumor average = 0.008 mM). The 0.2-mmol/kg dose of Optimark had higher concentrations of gadolinium compared with 0.1 mmol/kg at all time points. Peak gadolinium concentration is observed at 5 minutes (tumor average = 0.064 mM). Similar to the normalized T1 maps, gadolinium accumulates beyond the initial contrast area observed at 5 minutes while declining in intensity throughout the experiment. Interestingly, in the 0.2-mmol/kg dose at 30 minutes postcontrast, the tumor borders remain distinct (tumor average = 0.025 mM) in the gadolinium concentration maps.
Figure 5.
Gadolinium concentration maps of glioma-bearing mice injected with 0.1-mmol/kg and 0.2-mmol/kg doses of Optimark, with the 0.2-mmol/kg dose resulting in a greater concentration of gadolinium compared with 0.1-mmol/kg. Representative H&E-stained histological sections show tumor size, shape, and location. Gadolinium concentration maps of the tumor-bearing brains are overlaid onto T1-weighted images at baseline and at 5, 15, and 30 minutes postinjection. The color-coded scale bar indicates gadolinium concentration in mM with dark blue representing the lowest and red representing the highest gadolinium concentrations. Maps represent the same slices as in Figure 2, Figure 3.
Comparison of mean gadolinium concentration in different tumors over time confirms that peak contrast agent concentration is achieved at the 5-minute time point for both doses of Optimark and then steadily returns to baseline (Figure 6). Consistent with the normalized T1 values depicted in Figure 3, Figure 4, the 0.2-mmol/kg dose had significantly higher levels of gadolinium concentration compared with 0.1-mmol/kg at all time points (Figure 6).
Figure 6.
Mean gadolinium concentrations in the tumor before and after intravenous administration of 0.1- or 0.2-mmol/kg Optimark in cohorts of nu/nu athymic mice bearing orthotopic glioma tumors (n = 9 for 0.1-mmol/kg and n = 7 for 0.2-mmol/kg Optimark). Data plotted as means ± standard error. Mean tumor gadolinium concentration at baseline and after contrast agent injection measured every 2.5 minutes for 30 minutes. Gadolinium concentrations are significantly different between the 0.1- and 0.2-mmol/kg doses (ranges from P < .00005 to P < .005 depending on the time point).
Discussion
Novel MRI methods are being developed to detect brain tumors both in animal models and in patients [15], [16], [17], [18]. This study used orthotopic animal xenograft models that allow for the control of tumor size, location, genetics, and other parameters that permit standardization. Despite the standardization of the model, small tumors are still difficult to detect consistently. This is highly problematic because the assessment of treatment response and surgical planning are based on the accuracy of these qualitative radiographic findings.
Because clinical MRI protocols are not standardized across institutions and are not quantitative in nature, treatment response or failure can only be gauged when substantial changes in the tumor are observed. Conventional nonquantitative MRI protocols are susceptible to variations in signal intensity caused by radiofrequency coil sensitivity issues, magnetic field inhomogeneities, and many other sources of variation. Contrast-enhanced MRI is further complicated by surgery, treatment, corticosteroids, and the administered dose of the MRI contrast agent [7].
To circumvent the limitations of qualitative MRI imaging, we used a dynamic T1 mapping acquisition protocol and generated quantitative parametric maps. This study demonstrates that mapping of relaxation time and generation of T1 maps are highly sensitive for tumor detection. T1 relaxation time shortening in the tumor following administration of Optimark is evident in the color-coded T1 maps, with peak tumor enhancement occurring at 5 minutes. Correspondingly, color-coded gadolinium concentration maps, which are directly derived from the T1 maps, also show that the highest gadolinium concentrations occur at 5 minutes. A patient study evaluating the time course of brain lesion enhancement using Omniscan arrived at a similar conclusion; the optimal time to observe the effects of contrast agents in that study was 3.5 minutes postadministration [19].
Our study demonstrates that computer-generated maps are able to determine gadolinium concentration in tissue. These maps are sensitive enough to detect differences between 0.1- and 0.2-mmol/kg doses of Optimark. The 0.2-mmol/kg dose of Optimark afforded more complete tumor detection of these small glioma tumors than the standard clinical dose of 0.1-mmol/kg. Several studies in which administration of higher doses of contrast agent significantly improved the enhancement of intracranial tumors clinically [20], [21], [22], [23], [24] corroborate our findings. The T1 mapping sequence and quantitative maps provide an objective quantitative method for detection of small tumors and for the measurement of tumor enhancement over time.
Despite having a resolution of 1 mm, the minimum size of MR image-based detection of human brain tumors is considered to be a bidimensionally contrast-enhancing lesion with clearly defined margins of at least 10 mm visible in 2 or more axial slices 5 mm apart [7]. In this manuscript, we demonstrate reliable detection of brain tumors that are 1.5 × 2 × 3.5 mm using the quantitative T1 mapping techniques and a conventional clinical contrast agent on a 9.4-T preclinical scanner. Given that T1 mapping protocols can be easily translated to the clinic, quantitative MRI would be especially relevant for clinical trials to unambiguously define tumor response and/or progression.
The mechanism of action of these widely used gadolinium-based contrast agents is nonspecific. The coupling of quantitative T1 mapping with targeted molecular probes designed to detect tumor-specific markers could increase tumor enhancement without increasing the dose of gadolinium. For example, a peptide probe designed to target PTPμ fragments generated by GBM tumors was conjugated to a gadolinium chelate [25], [26]. This probe, called SBK2, is selectively retained in heterotopic glioma tumors compared with scrambled and Optimark controls using T1 mapping [26]. Detection of intracranial glioma tumors could be further improved upon by combining disease-specific molecular contrast agents and quantitative T1 mapping.
Conclusions
By using a controlled orthotopic model, we were able to demonstrate that T1 mapping is able to identify small tumors. We were able to quantitatively compare multiple time points in different animals using different contrast agent concentrations. Using T1 mapping, we found that doubling the contrast agent concentration has a significant impact on tumor enhancement. Given the importance of treatment comparisons for the development and approval of therapeutics, an objective, quantitative methodology for evaluating tumor size changes among the same patient over time and between patients is paramount. We demonstrate that, even with a conventional contrast agent, T1 mapping can produce significant image quality differences. Future studies will evaluate whether using this imaging strategy in concert with targeted molecular contrast agents can further improve MRI sensitivity.
Conflicts of Interest
None.
Acknowledgements
This study was funded by a grant from the National Cancer Institute of the National Institutes of Health (R01 CA179956). We thank Cathy Doller for help with histology, and the Visual Sciences Histology Core Facility is supported by a core grant from the National Eye Institute of the National Institutes of Health (EY11373). The authors thank Lan Lu and Ying Gao for providing the MATLAB code for T1 mapping analysis; Michael Howell and Steven Cady for increasing the efficiency and throughput of the T1 mapping analysis code; and Polly Phillips-Mason, Jason Vincent, and Ramamurthy Gopalakrishnan for technical expertise and experimental setup. We thank Sonya Craig for critical reading of the manuscript.
Footnotes
This study was funded by a grant from the National Cancer Institute of the National Institutes of Health (R01 CA179956). The Visual Sciences Histology Core Facility is supported by a core grant from the National Eye Institute of the National Institutes of Health (EY11373).
Contributor Information
Kelsey Herrmann, Email: kelsey.herrmann@case.edu.
Bernadette O. Erokwu, Email: bernadette.erokwu@case.edu.
Mette L. Johansen, Email: mette.johansen@case.edu.
James P. Basilion, Email: james.basilion@case.edu.
Vikas Gulani, Email: vikas.gulani@case.edu.
Mark A. Griswold, Email: mark.griswold@case.edu.
Chris A. Flask, Email: christopher.flask@case.edu.
Susann M. Brady-Kalnay, Email: susann.brady-kalnay@case.edu.
References
- 1.Kohler BA, Ward E, McCarthy BJ, Schymura MJ, Ries LA, Eheman C, Jemal A, Anderson RN, Ajani UA, Edwards BK. Annual report to the nation on the status of cancer, 1975-2007, featuring tumors of the brain and other nervous system. J Natl Cancer Inst. 2011;103:714–736. doi: 10.1093/jnci/djr077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 3.Sanai N, Polley MY, McDermott MW, Parsa AT, Berger MS. An extent of resection threshold for newly diagnosed glioblastomas. J Neurosurg. 2011;115:3–8. doi: 10.3171/2011.2.jns10998. [DOI] [PubMed] [Google Scholar]
- 4.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 5.Louis DN. Molecular pathology of malignant gliomas. Annu Rev Pathol. 2006;1:97–117. doi: 10.1146/annurev.pathol.1.110304.100043. [DOI] [PubMed] [Google Scholar]
- 6.Upadhyay N, Waldman AD. Conventional MRI evaluation of gliomas. Br J Radiol. 2011;84(Spec No 2):S107–S111. doi: 10.1259/bjr/65711810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wen PY, Macdonald DR, Reardon DA, Cloughesy TF, Sorensen AG, Galanis E, Degroot J, Wick W, Gilbert MR, Lassman AB. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol. 2010;28:1963–1972. doi: 10.1200/JCO.2009.26.3541. [DOI] [PubMed] [Google Scholar]
- 8.Burden-Gulley SM, Qutaish MQ, Sullivant KE, Lu H, Wang J, Craig SE, Basilion JP, Wilson DL, Brady-Kalnay SM. Novel cryo-imaging of the glioma tumor microenvironment reveals migration and dispersal pathways in vivid three-dimensional detail. Cancer Res. 2011;71:5932–5940. doi: 10.1158/0008-5472.CAN-11-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gan HK, Cvrljevic AN, Johns TG. The epidermal growth factor receptor variant III (EGFRvIII): where wild things are altered. FEBS J. 2013;280:5350–5370. doi: 10.1111/febs.12393. [DOI] [PubMed] [Google Scholar]
- 10.Tyminski E, Leroy S, Terada K, Finkelstein DM, Hyatt JL, Danks MK, Potter PM, Saeki Y, Chiocca EA. Brain tumor oncolysis with replication-conditional herpes simplex virus type 1 expressing the prodrug-activating genes, CYP2B1 and secreted human intestinal carboxylesterase, in combination with cyclophosphamide and irinotecan. Cancer Res. 2005;65:6850–6857. doi: 10.1158/0008-5472.CAN-05-0154. [DOI] [PubMed] [Google Scholar]
- 11.Tyagi M, Karn J. CBF-1 promotes transcriptional silencing during the establishment of HIV-1 latency. EMBO J. 2007;26:4985–4995. doi: 10.1038/sj.emboj.7601928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hennig J, Nauerth A, Friedburg H. RARE imaging: a fast imaging method for clinical MR. Magn Reson Med. 1986;3:823–833. doi: 10.1002/mrm.1910030602. [DOI] [PubMed] [Google Scholar]
- 13.Deichmann R. Haase A (1992) Quantification of T1 values by SNAPSHOT-FLASH NMR imaging. J Magn Reson. 1969;96:608–612. [Google Scholar]
- 14.Jakob PM, Hillenbrand CM, Wang T, Schultz G, Hahn D, Haase A. Rapid quantitative lung (1)H T(1) mapping. J Magn Reson Imaging. 2001;14:795–799. doi: 10.1002/jmri.10024. [DOI] [PubMed] [Google Scholar]
- 15.Sun Y, Schmidt NO, Schmidt K, Doshi S, Rubin JB, Mulkern RV, Carroll R, Ziu M, Erkmen K, Poussaint TY. Perfusion MRI of U87 brain tumors in a mouse model. Magn Reson Med. 2004;51:893–899. doi: 10.1002/mrm.20029. [DOI] [PubMed] [Google Scholar]
- 16.Coquery N, Francois O, Lemasson B, Debacker C, Farion R, Remy C, Barbier EL. Microvascular MRI and unsupervised clustering yields histology-resembling images in two rat models of glioma. J Cereb Blood Flow Metab. 2014;34:1354–1362. doi: 10.1038/jcbfm.2014.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sagiyama K, Mashimo T, Togao O, Vemireddy V, Hatanpaa KJ, Maher EA, Mickey BE, Pan E, Sherry AD, Bachoo RM. In vivo chemical exchange saturation transfer imaging allows early detection of a therapeutic response in glioblastoma. Proc Natl Acad Sci U S A. 2014;111:4542–4547. doi: 10.1073/pnas.1323855111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rose CJ, Mills SJ, O'Connor JP, Buonaccorsi GA, Roberts C, Watson Y, Cheung S, Zhao S, Whitcher B, Jackson A. Quantifying spatial heterogeneity in dynamic contrast-enhanced MRI parameter maps. Magn Reson Med. 2009;62:488–499. doi: 10.1002/mrm.22003. [DOI] [PubMed] [Google Scholar]
- 19.Akeson P, Nordstrom CH, Holtas S. Time-dependency in brain lesion enhancement with gadodiamide injection. Acta Radiol. 1997;38:19–24. doi: 10.1080/02841859709171236. [DOI] [PubMed] [Google Scholar]
- 20.Runge VM, Kirsch JE, Burke VJ, Price AC, Nelson KL, Thomas GS, Dean BL, Lee C. High-dose gadoteridol in MR imaging of intracranial neoplasms. J Magn Reson Imaging. 1992;2:9–18. doi: 10.1002/jmri.1880020103. [DOI] [PubMed] [Google Scholar]
- 21.Yousry I, Camelio S, Schmid UD, Horsfield MA, Wiesmann M, Bruckmann H, Yousry TA. Visualization of cranial nerves I-XII: value of 3D CISS and T2-weighted FSE sequences. Eur Radiol. 2000;10:1061–1067. doi: 10.1007/s003300000452. [DOI] [PubMed] [Google Scholar]
- 22.Yuh WT, Fisher DJ, Engelken JD, Greene GM, Sato Y, Ryals TJ, Crain MR, Ehrhardt JC. MR evaluation of CNS tumors: dose comparison study with gadopentetate dimeglumine and gadoteridol. Radiology. 1991;180:485–491. doi: 10.1148/radiology.180.2.2068317. [DOI] [PubMed] [Google Scholar]
- 23.Yuh WT, Fisher DJ, Runge VM, Atlas SW, Harms SE, Maravilla KR, Mayr NA, Mollman JE, Price AC. Phase III multicenter trial of high-dose gadoteridol in MR evaluation of brain metastases. AJNR Am J Neuroradiol. 1994;15:1037–1051. [PMC free article] [PubMed] [Google Scholar]
- 24.Yuh WT, Nguyen HD, Tali ET, Mayr NA, Fisher DJ, Atlas SW, Carvlin MC, Drayer BP, Pollei SR, Runge VM. Delineation of gliomas with various doses of MR contrast material. AJNR Am J Neuroradiol. 1994;15:983–989. [PMC free article] [PubMed] [Google Scholar]
- 25.Burden-Gulley SM, Zhou Z, Craig SE, Lu ZR, Brady-Kalnay SM. Molecular Magnetic Resonance Imaging of Tumors with a PTPmu Targeted Contrast Agent. Transl Oncol. 2013;6:329–337. doi: 10.1593/tlo.12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herrmann K, Johansen M, Craig S, Vincent J, Howell M, Gao Y, Lu L, Erokwu B, Agnes R, Lu Z-R. Molecular Imaging of Tumors Using a Quantitative T1 Mapping Technique via Magnetic Resonance Imaging. Diagnostics. 2015;5:318. doi: 10.3390/diagnostics5030318. [DOI] [PMC free article] [PubMed] [Google Scholar]