Cell karyotyping in patients with small lymphocytic lymphoma/chronic lymphocytic leukemia (SLL/CLL) is not easy to success, and small genomic lesions (<5 Mb) are not routinely detected by this method. It is likely that a complete genomic characterization of CLL requires a combination of fluorescence in situ hybridization (FISH), single nucleotide polymorphism (SNP) array profiling for comprehensive genome-wide analysis of acquired genomic copy number aberrations (aCNAs) and loss-of-heterozygosity (LOH) in dominant clones, and karyotyping for detection of balanced translocations, isochromosomes, and marker chromosomes. SNP array analysis can reveal chromothripsis, a phenomenon by which regions of the cancer genome are shattered and recombined to generate frequent oscillations between the lower and the higher DNA copy number states. This study provided cytogenetic findings in a CLL/SLL patient with v-myc avian myelocytomatosis viral oncogene homolog (C-MYC)-amplification by FISH, in which SNP arrays detected profound genomic upheaval due to chromothripsis that may lead to malignant transformation.
In June 2014, a 62-year-old woman was admitted to Center of Oncology and Hematology, First Affiliated Hospital of Guangzhou Medical University for bilateral neck mass, which showed enlargement progressively and were broad bean-sized. Peripheral blood studies were as follows: White blood cell count, 13.48 × 109/L (neutrophils 16.5%, lymphocytes 72.2%, monocytes 10.9%); lymphocytes count, 9.7 × 109/L; monocyte count, 1.5 × 109/L; eosinophils count, 0.03 × 109/L; erythrocyte count, 3.42 × 109/L; hemoglobin (Hb), 108 g/L; hematocrit, 33.2%; mean cell volume, 97.1 fL; and peripheral platelet count (PLT), 92 × 109/L. Microscopically, the structures of lymph nodes were completely damaged with lymph follicles depletion. The small tumors cells were diffuse distribution with scanty cytoplasm and spherical nucleus. Coarse chromatin, small nucleolus, and nuclear division could be seen in the nucleus of tumor cells. These small tumors cells were composed of cells positive for CD20, CD79a, CD5, CD23, CD43, and B-cell CLL/lymphoma 2. The bone marrow biopsy showed an obvious infiltrate of typical lymphocytes that were identical with positive immunophenotype of small tumors cells in lymph nodes. Based on these histological change and clinical history, the patient was diagnosed with SLL with bone marrow infiltration. Bone marrow aspiration showed a 91% infiltration of small lymphocytes. Based on these morphological findings, the patient was diagnosed with CLL according to the WHO classification. Positron emission tomography-computed tomography showed generalized lymphadenopathy, radioactive uptake and glucose metabolism increased in splenomegaly. The patient refused further treatment after examination.
An abnormal clonal population with numerical and structural abnormalities including 8q, 9q, 11p, 13q, 17 and 19p, monosomy 9, 11, 13, and X, and up to five marker chromosomes of unknown origin. The karyotypes were described according to International System for Chromosome Nomenclature (ISCN) 2009. Cytogenetic analysis of the bone marrow using conventional cytogenetic techniques showed that the ISCN karyogram was: 42–43, X, −X, −4, −8, −13, +mar1–mar5 [cp6]/46, XX[2] [Figure 1]. FISH analysis was performed using the following probes from Abbott Molecular (Des Plaines, Illinois 60018, USA): The Vysis C-MYC/CEP 8 Probes and TP53/CEP17 Probes, and showed loss of one copy of TP53 in 65 of 150 nuclei examined and more than three copies of 8q24 (C-MYC) in 20 of 150 nuclei [Figure 2a and 2b]. SNP-based array technique also found deletions in 2q, 5p, 7q, 8p, 9p, 9q 11p, 13q, 14q, 17p, 18p, 19p and gains in 8q and 17q. There were LOH in the regions of 7q31.33q32.1 arr (123,979,100–128,137,850 bp [hg19]: 4.16 Mb), 9p21.3q34.3 arr (24,095,293–141,018,648 bp [hg19]: 116.92 Mb), 11p15.5p12 arr (230,680–41,464,545 bp [hg19]: 41.23 Mb), 13q11q12.13 arr (19,436,286–26,030,997 [hg19]: 6.60 Mb), 13q12.3q34 arr (31,267,069–115,107,733 [hg19]: 83.84 Mb) and 18p11.32p11.21 arr (136,227–14,215,381 [hg19]: 14.08 Mb), all of chromosome was lost in X. arr (hg19) (8) cth and arr (hg19) (17) cth could be seen in the patient. There were some complex changes in 19p13.3p13.12 arr (260,911–14,947,397 [hg19] cth) [Figure 2c].
Figure 1.
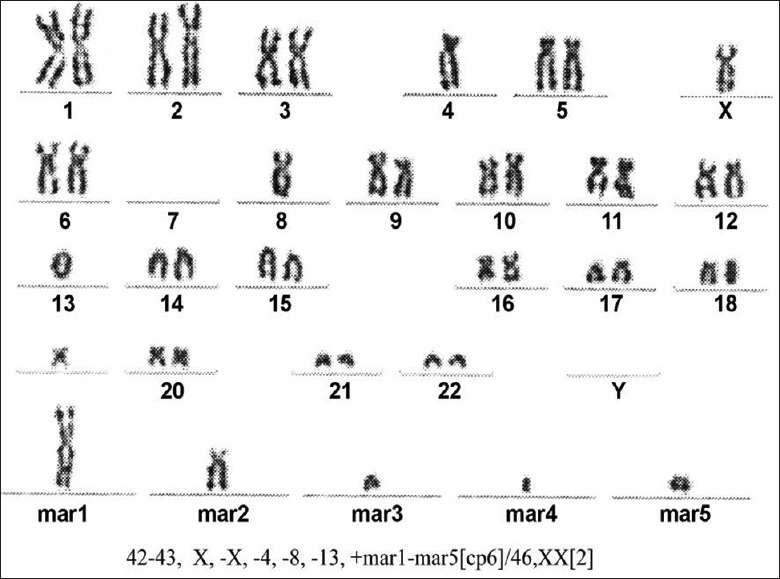
Cytogenetic studies of the bone marrow were performed using conventional cytogenetic techniques. The karyotypes were described according to the International System for Chromosome Nomenclature 2009.
Figure 2.
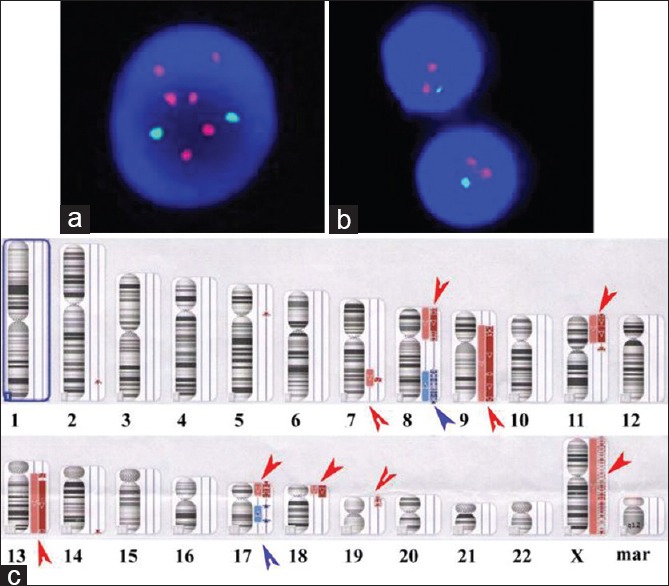
(a) Fluorescence in situ hybridization showing multiple copies the C-MYC gene (red color); (b) and a deletion of the p53 gene (green color); (c) Depiction of DNA copy number in the abnormal chromosomes identified in marrow samples from this chronic lymphocytic leukemia/small lymphocytic lymphoma patient. For each chromosome shown, the red arrows depict the deletions of DNA copy number, and blue arrows depict the amplification of DNA copy number. Multiple oscillations between two DNA copy number stated in chromosomes 8, 9, 11, 13, 17, 19, and X indicative of chromothripsis. Numerous discontinuous deletions are indicated by red arrows and discontinuous amplification in chromosome arms 8q and 17q are indicated by blue arrows. In addition to these focal deletions, larger single deletions could be seen in chromosome arms 2q, 5p, 7q, and 14q.
Chronic lymphocytic leukemia cell karyotyping is successful only in a minority of CLL cases without stimulation in history; the CLL/SLL cell karyotyping reported in our patient is one example. FISH probes have become a staple in the analysis of chromosomal aberrations in CLL/SLL, which circumvents the need for the mitotic stimulation. Although FISH analysis is now well-accepted in clinical utility of CLL/SLL and has wide applicability, the current standard clinical practice of identifying alterations with probes targeting is only 4–5 chromosomal sites.[1] SNP arrays permit high-resolution analysis of DNA copy number imbalances throughout the genome, and also allow for the detection of LOH and uncovers alterations overlooked by FISH analysis. Using SNP arrays, aberrations previously shown by FISH to have prognostic importance in CLL/SLL have been identified at the expected frequency.[2]
The sample from our patient exhibited a gain of 8q22.1q24.3 and 17q12q24.2, whereas the copy neutral LOH (cnLOH) at 7q31.33q32.1, 8p23.3p11.21, 9p21.3q34.3, 11p15.5p12, 13q12.3q34, 17p13.3.3p11.2, 18p11.32p11.21, 19p13.3p13.12 and X. The discontinuous regions of LOH in chromosomes 7, 8, 9, 11, 13, 17, 18, 19 and X were also separated by regions retaining heterozygosity. The sample in our case was collected from a 62-year-old woman with CLL/SLL, who had not previously received treatment, her subsequent clinical course showed rapid deterioration, and unfortunately, the patient refused further treatment, we could not assess whether the abnormalities seen in the pretreatment sample persisted in the relapsing cells or indeed showed further evolution. In this case, we found complex rearrangements involving chromosomes 8, 17 and 19p with the hallmarks of chromothripsis, the massive numbers of copy number changes with the lower and the higher copy number state on chromosome 8 were the typical appearances of chromothripsis. The regions with cnLOH in 9q, 11p and 13q were large (>30 Mb), which were usually acquired, not constitutional.
The concept of elevated genomic complexity (multiple aCNAs per CLL case) is reinforced as one of the strongest biological traits associated with aggressive CLL with short survival. The clinical course in the initial CLL patient reported by us showed rapid deterioration. The recurrent aCNAs in CLL were listed here in order of descending frequency: Del (13q14) (<50%), trisomy 12 (12%–18%), del11q (10%–15%), and del (17p) (7%–10%). Lai and Huang[3] reported that genomic aberrations in 77.9% of Chinese CLL patients; Chinese CLL patients have the similar frequencies of del (13q), trisomy 12 and del (11q), and a higher frequency of del (17p) when compared to those reported in the literatures. Del (17p) is associated with advanced stage and low levels of Hb and PLT. Some others CLL patients carrying a del (17p) clone show marked resistance against genotoxic chemotherapies that cannot be overcome by the addition of anti-CD20 antibodies in the context of state of the art chemo-immunotherapy and short survival.[4]
The frequent occurrence of del (13q14) in CLL suggested that it is a driver of CLL pathogenesis. Del (13q14) long enough to include the retinoblastoma 1 gene near 48 Mb (termed Type II lesions) indicate a CLL subset with del (13q14) that is clinically more aggressive and demonstrates shortened survival, especially after therapy.[5] In our case, the length of del (13q14) is more than 80 Mb, and the degree of (13q14) deletion may be related with the progression of the case. The deletion of 14q found in our patient was noteworthy, which is associated with a short disease-free survival. It was reported that these patients with del (14q) typically required more immediate therapy.[6] Amplifications of 8q24 focused on C-MYC in CLL had been reported, high expression levels of C-MYC genes were found to be closely associated with more aggressive forms of the disease requiring earlier therapy.[7]
In conclusion, this report illustrated the value of SNP array analysis for the detection of novel genomic alterations that may contribute to CLL/SLL onset or progression and the detection of complex alterations that would be overlooked with a standard FISH panel. With the exception of del (13q14), del (17p), del (14q) in the case, the identification of chromothripsis involving in chromosomes 8, 17, and 19p indicated that this phenomenon may be a recurrent change in this disease.
Footnotes
Edited by: Xin Chen
Source of Support: This study was supported by grants from Natural Science Foundation of China (No. 81100379 and No. 81302079), Science and Technology Planning Project of Guangdong Province, China (No. 2013B022000102), Medical Scientific Research Foundation of Guangdong Province, China (No. A2014292) and Key Clinical Disciplines of Guangdong Province (No. 20111219).
Conflict of Interest: None declared.
REFERENCES
- 1.Zenz T, Mertens D, Döhner H, Stilgenbauer S. Molecular diagnostics in chronic lymphocytic leukemia-pathogenetic and clinical implications. Leuk Lymphoma. 2008;49:864–73. doi: 10.1080/10428190701882955. [DOI] [PubMed] [Google Scholar]
- 2.Pfeifer D, Pantic M, Skatulla I, Rawluk J, Kreutz C, Martens UM, et al. Genome-wide analysis of DNA copy number changes and LOH in CLL using high-density SNP arrays. Blood. 2007;109:1202–10. doi: 10.1182/blood-2006-07-034256. [DOI] [PubMed] [Google Scholar]
- 3.Lai YY, Huang XJ. Cytogenetic characteristics of B cell chronic lymphocytic leukemia in 275 Chinese patients by fluorescence in situ hybridization: A multicenter study. Chin Med J. 2011;124:2417–22. [PubMed] [Google Scholar]
- 4.Hallek M, Fischer K, Fingerle-Rowson G, Fink AM, Busch R, Mayer J, et al. Addition of rituximab to fludarabine and cyclophosphamide in patients with chronic lymphocytic leukaemia: A randomised, open-label, phase 3 trial. Lancet. 2010;376:1164–74. doi: 10.1016/S0140-6736(10)61381-5. [DOI] [PubMed] [Google Scholar]
- 5.Mosca L, Fabris S, Lionetti M, Todoerti K, Agnelli L, Morabito F, et al. Integrative genomics analyses reveal molecularly distinct subgroups of B-cell chronic lymphocytic leukemia patients with 13q14 deletion. Clin Cancer Res. 2010;16:5641–53. doi: 10.1158/1078-0432.CCR-10-0151. [DOI] [PubMed] [Google Scholar]
- 6.Cosson A, Chapiro E, Belhouachi N, Cung HA, Keren B, Damm F, et al. 14q deletions are associated with trisomy 12, NOTCH1 mutations and unmutated IGHV genes in chronic lymphocytic leukemia and small lymphocytic lymphoma. Genes Chromosomes Cancer. 2014;53:657–66. doi: 10.1002/gcc.22176. [DOI] [PubMed] [Google Scholar]
- 7.Brown JR, Hanna M, Tesar B, Werner L, Pochet N, Asara JM, et al. Integrative genomic analysis implicates gain of PIK3CA at 3q26 and MYC at 8q24 in chronic lymphocytic leukemia. Clin Cancer Res. 2012;18:3791–802. doi: 10.1158/1078-0432.CCR-11-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]


