Summary
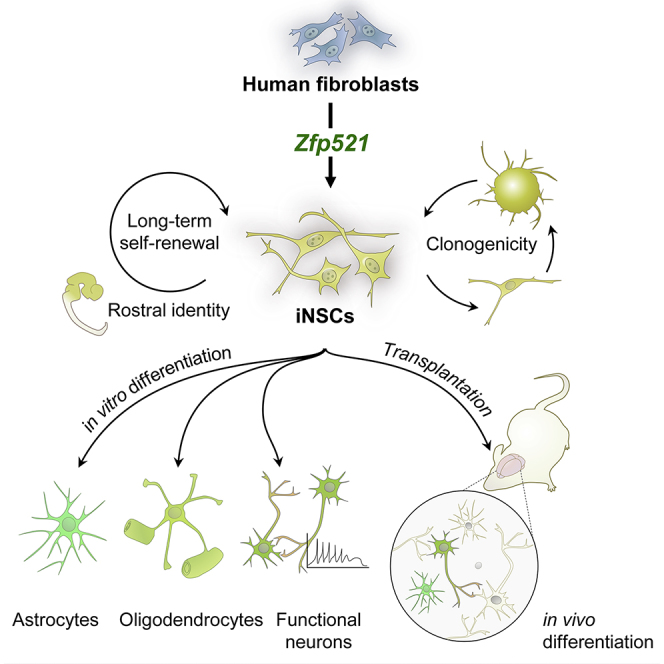
Direct conversion of somatic cells into neural stem cells (NSCs) by defined factors holds great promise for mechanistic studies, drug screening, and potential cell therapies for different neurodegenerative diseases. Here, we report that a single zinc-finger transcription factor, Zfp521, is sufficient for direct conversion of human fibroblasts into long-term self-renewable and multipotent NSCs. In vitro, Zfp521-induced NSCs maintained their characteristics in the absence of exogenous factor expression and exhibited morphological, molecular, developmental, and functional properties that were similar to control NSCs. In addition, the single-seeded induced NSCs were able to form NSC colonies with efficiency comparable with control NSCs and expressed NSC markers. The converted cells were capable of surviving, migrating, and attaining neural phenotypes after transplantation into neonatal mouse and adult rat brains, without forming tumors. Moreover, the Zfp521-induced NSCs predominantly expressed rostral genes. Our results suggest a facilitated approach for establishing human NSCs through Zfp521-driven conversion of fibroblasts.
Graphical Abstract
Highlights
-
•
ZFP521 can directly convert human fetal, neonatal, and adult fibroblasts into NSCs
-
•
iNSCs exhibit NSC properties, i.e., markers, long-term self-renewal, and tripotency
-
•
ZFP521-iNSCs predominantly express rostral markers
-
•
Single-seeded ZFP521-iNSCs are clonogenically comparable with control NSCs
Baharvand, Saric, and colleagues show that a single transcription factor Zfp521 is capable of converting murine and human fibroblasts into long-term self-renewing neural stem cells that are clonogenic and possess morphological, functional and molecular characteristics similar to their endogenous counterparts. These cells represent a valuable alternative source of cells for developmental studies, toxicology, and regenerative therapies of the nervous system.
Introduction
Differentiated cells can be reprogrammed to become induced pluripotent stem cells (iPSCs) by exogenous supplementation of defined factors (Takahashi and Yamanaka, 2006). The iPSCs provide an interminable source of a broad range of differentiated cells for applications such as in vitro disease modeling, drug development, toxicity testing, and cell-replacement therapies. Mature neurons and neural stem cells (NSCs) are among the most clinically useful cells that can be produced from pluripotent stem cells (PSCs) (Nemati et al., 2011). However, their clinical utility has been hampered by the tumorigenic potential elicited by the residual PSCs in the differentiated cell population, the lengthy and inefficient differentiation process (Hu et al., 2010), and genomic instability (Weissbein et al., 2014). In the process of trans-differentiation, one mature somatic cell type can be converted into another functional mature or progenitor cell type without undergoing an intermediate pluripotent state by using a variety of inducers, such as transcription factors, epigenetic modifiers, and microRNAs (Moradi et al., 2014, Pournasr et al., 2011). Mature neurons have been successfully trans-differentiated from several cell sources (Ambasudhan et al., 2011, Ladewig et al., 2012, Marro et al., 2011, Vierbuchen et al., 2010). However, their inability to proliferate and survive for long periods of time in culture conditions limits their use. An alternative approach is to convert somatic cells into NSCs, which are expandable in vitro and have the potential to differentiate into major neural cell types, such as neurons, oligodendrocytes, and astrocytes.
Ectopic expression of several combinations of genes via lentiviruses with or without small molecules have been used to produce induced NSCs (iNSCs) from somatic cells (Cassady et al., 2014, Han et al., 2012a, Lujan et al., 2012, Ring et al., 2012, Thier et al., 2012, Wang et al., 2013). Most of these NSC-induction cocktails depend on the use of potentially tumorigenic pluripotency-associated factors in reprogramming or a multi-factor strategy that increases the intricacy of the approach. Hence, the use of a single reprogramming factor for generation of iNSCs may represent a more controllable, easier, and safer approach.
Here, we demonstrate that iNSCs could be generated from human fibroblasts by ectopic expression of a single neurogenic factor, zinc-finger protein 521 (Zfp521). Our data indicate that Zfp521 alone is sufficient for conversion of fibroblasts into iNSCs, which may serve as an alternative and more accessible source of cells for neural cell-replacement therapies as well as in vitro disease modeling, toxicity testing, and drug development.
Results
Reprogramming Potential of Different Combinations of Neurogenic Transcription Factors
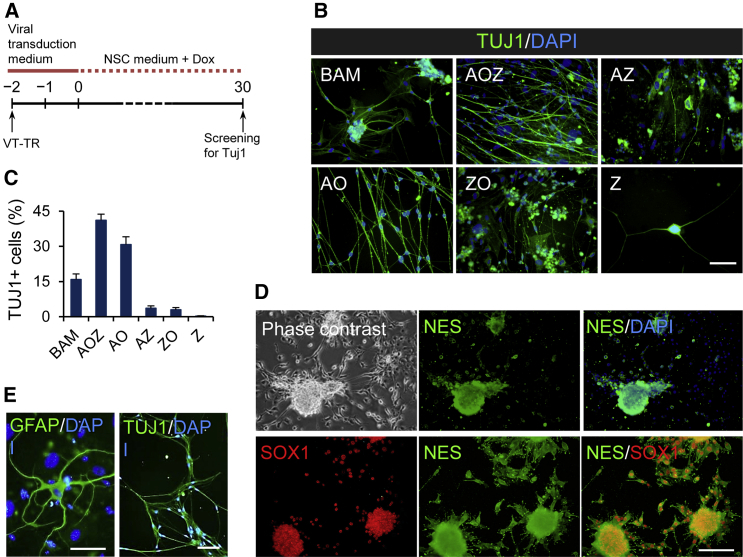
We initially used murine 3T3 fibroblasts to screen for a novel reprogramming cocktail that can efficiently derive neuronal-like cells. Cells were transfected with inducible lentiviruses in six combinations of neurogenic transcription factors encoding Math3, Ngn2, Oct6, Zfp521, Ascl1, Myt1l, and Brn2. These neural lineage-instructive transcription factors were selected based on their key roles in normal neurogenesis (Kamiya et al., 2011, Son et al., 2011). The previously reported cocktail Brn2-Ascl1-Myt1l (BAM) (Vierbuchen et al., 2010) served as the positive control. For controlled ectopic expression of the transgenes, we used lentiviruses that were inducible with doxycycline (Dox). First, we confirmed that Tuj1 or other neuronal markers were not expressed in native 3T3 fibroblasts (Figure S1A) or in fibroblasts that were cultured in neural induction medium without Dox after transduction with Zfp521-expressing lentivirus or in cells that were transduced with empty vector and cultured in neural induction medium on day 30 after exposure to Dox (Figure S1B).
Detection of the neuronal marker Tuj1 by using immunofluorescence on day 30 after transduction with Zfp521-expressing lentivirus and exposure to Dox was also used as readout for successful neuronal reprogramming (Figure 1A). With this approach, we found that, in addition to the BAM group, five gene combinations (Ascl1-Oct6-Zfp521 [AOZ]; Ascl1-Oct6 [AO]; Ascl1-Zfp521 [AZ]; Zfp521-Oct6 [ZO]; and Zfp521 alone [Z]) yielded Tuj1-positive cells with different efficiencies (Figure 1B). To estimate the conversion efficiency on day 30 of reprogramming, we determined the frequency of Tuj1-positive cells relative to the number of initially seeded 3T3 cells in three independent experiments (Vierbuchen et al., 2010). The efficiencies ranged from 0.2% ± 0.1% (Z group) to 30% ± 3.3% (AO group), and 40% ± 2.5 (AOZ group), which was even higher than in the BAM group (16% ± 2.4%; Figure 1C). This result indicates that our gene cocktails could successfully induce the neuronal phenotype in cultured fibroblasts. Unexpectedly, in the Z group, several cell spheroids emerged that were morphologically similar to spheres typically formed by wild-type NSCs, and expressed the NSC markers Nes (Nestin) and Sox1 (Figure 1D). These spheroids could also be differentiated into Tuj1- and Gfap-positive cells (Figure 1E). Therefore, Zfp521 seemed to be capable of directly converting murine fibroblasts into NSC-like cells.
Figure 1.
Reprogramming Potential of Different Combinations of Neurogenic Transcription Factors
(A) Schematic procedure for reprogramming of mouse 3T3 fibroblasts to neuronal cells by screening diverse neurogenic factors.
(B) Immunofluorescent staining for TUJ1 protein on day 30 after Dox treatment.
(C) The efficiency of neuronal reprogramming of 3T3 fibroblasts by different gene cocktails calculated 30 days after infection as the number of TUJ1-positive cells relative to the number of starting cells initially seeded. Data are shown as means ± SD of three independent experiments.
(D) Phase-contrast image of NSC-like colony emerged in Zfp521 (Z) treatment group and immunofluorescent double-staining of these NSC-like spheres for NES (Nestin) and SOX1 on day 30 after Dox treatment.
(E) Neurons (TUJ1) and astrocytes (GFAP) differentiated from NSC-like spheres that were induced by Zfp521.
Nuclei in (B), (D), and (E) were counterstained with DAPI. Scale bars represent 100 μm in (B) and (E), and 200 μm in (D). See also Figure S1.
Generation of Stably Expandable iNSCs from Human Neonatal Fibroblasts
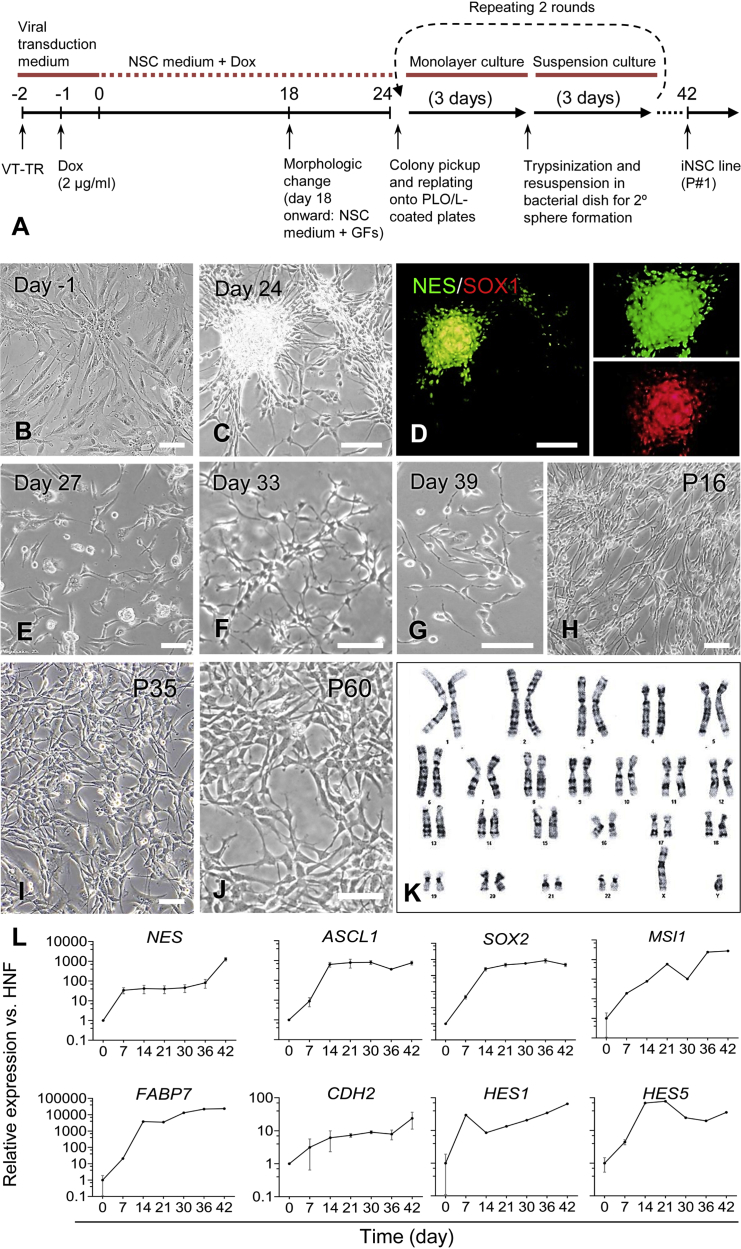
In order to further test the hypothesis that Zfp521 might act as a factor capable of reprogramming somatic cells into iNSCs, we switched to human neonatal (foreskin) fibroblasts (HNFs) as the starting cell type. Like murine fibroblasts, HNFs did not express any of the tested neural markers in the absence (Figure S2A) or the presence of a transgene when cultured in neural medium with or without Dox (Figure S2B). As per the procedure summarized in Figure 2A, overexpression of Zfp521 in HNFs (Figure 2B) led to the formation of NSC-like colonies (Figure 2C) that expressed the NSC markers NES and SOX1 (Figure 2D). The efficiency of fibroblast reprogramming into these colonies (referred to as primary neurospheres) was 0.4% ± 0.01%, as assessed in 12 independent biological replicates by the number of NES- and SOX2-positive primary neurospheres relative to the number of starting fibroblasts initially seeded (Ring et al., 2012). These NSC-like colonies were picked up manually, dissociated into single cells and plated onto laminin/poly-L-ornithine-coated dishes. At 60%–70% confluency, the cells were trypsinized and cultured in a non-adherent dish to allow for the formation of secondary neurospheres. This procedure (trypsinization and re-plating onto tissue culture dishes followed by re-suspension into bacterial dishes) was repeated two additional times to increase the purity of iNSCs. iNSCs generated by this method exhibited high morphological homogeneity (Figures 2E–2I) and could be stably expanded for more than 60 passages (Figure 2J) without acquiring karyotypic abnormalities (Figure 2K).
Figure 2.
Generation of iNSCs from Human Neonatal Foreskin Fibroblasts
(A) Schematic of the experimental procedure used for iNSC generation by overexpression of Zfp521. VT-TR stands for viral co-transduction of Zfp521-and Tet Repressor-expressing constructs.
(B) Phase-contrast image of HNFs on day −1.
(C) Phase-contrast image of an NSC-like cell colony on day 24 of reprogramming.
(D) Immunofluorescent staining of the NSC-like colony shown in (C) for NES (Nestin) and SOX1.
(E) Distinct bipolar morphology of reprogrammed cells following trypsinization and plating of primary neurospheres on day 27 of reprogramming.
(F) NSC-like cells on day 33 after initiation of reprogramming.
(G) NSC-like cells on day 39 after initiation of reprogramming.
(H–J) Established iNSCs at passages 16 (H), 35 (I), and 60 (J).
(K) A representative example of a diploid karyogram of iNSCs at passage 60. At least 50 metaphase spreads were screened from which ten metaphase spreads were assessed for chromosomal re-arrangements.
(L) Temporal expression patterns of selected neural lineage genes over the course of reprogramming. qRT-PCR was performed at different time points after induction of Zfp521 expression. The fibroblasts were treated with Dox for 24 days, the minimum time needed for stable acquisition of iNSC identity. Data are shown as means ± SD of three independent experiments.
Scale bars represent 100 μm in (B), (C), (D), (F), (G), and (J), and 50 μm in (E), (H), and (I). See also Figure S2.
In order to determine the shortest time period of Zfp521 expression required to achieve stable acquisition of iNSC identity, the HNFs were treated with Dox for 3, 6, 9, 12, 18, 24, and 30 days in a 42-day reprogramming procedure, as illustrated in Figure 2A. Although NSC-like colonies emerged in the 6-, 9-, 12-, and 18-day treatment groups, reprogrammed cells derived from these colonies could only be maintained for up to two passages (data not shown), which indicates an incomplete reprogramming and/or inability of the reprogrammed cells to establish a self-sustaining, NSC-like gene regulatory circuitry. At least 24 days of Dox treatment was required for generation of stable iNSC colonies from HNFs, which was preceded by a significant upsurge in the relative expression of the neural markers NES, ASCL1, SOX2, MSI1 (Musashi), FABP7 (BLBP), CDH2 (N-Cadherin), HES1, and HES5, after Dox-induced Zfp521 expression (Figure 2L).
Neural Stem Cell Identity of Established HNF-iNSCs
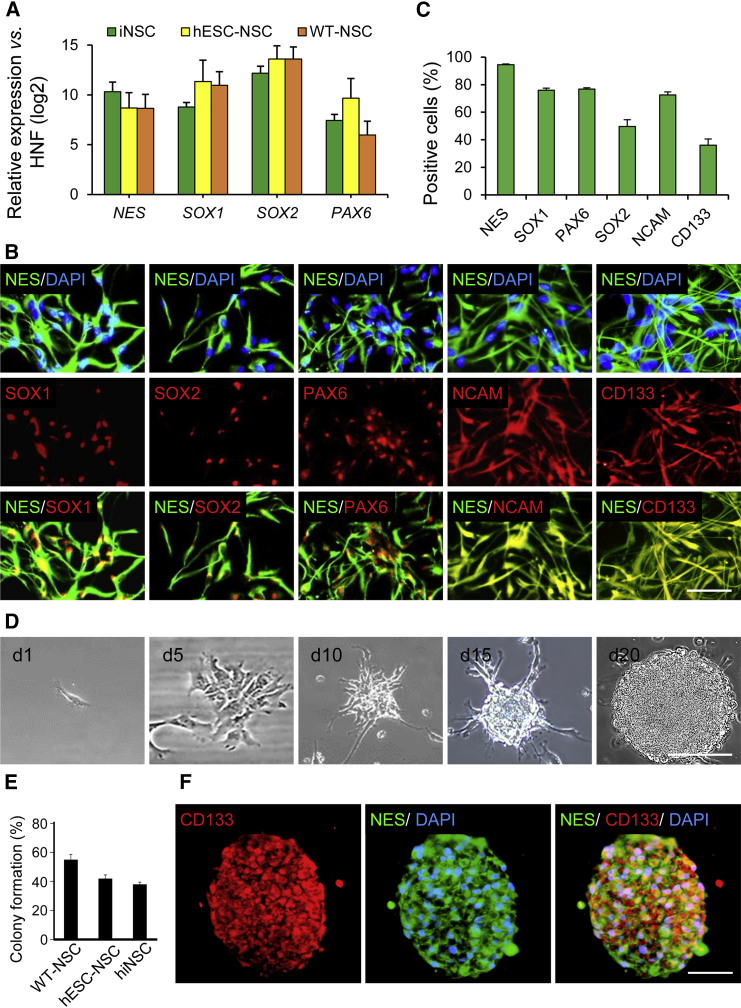
In order to further confirm the neural identity of Zfp521-iNSCs derived from HNFs, we probed for the relative transcript levels of NES, SOX1, SOX2, and PAX6 mRNAs by qRT-PCR. These analyses confirmed that they are expressed in iNSCs at levels comparable with control wild-type NSCs (WT-NSCs) that were derived from human fetal brain (18–21 week fetus) or from human embryonic stem cells (hESC-NSCs) (Figure 3A). In addition, immunofluorescence analyses showed that HNF-iNSCs express the key NSC-associated markers NES, SOX1, SOX2, PAX6, NCAM, and CD133 (Figure 3B). Flow cytometric quantification revealed that most HNF-iNSCs expressed NES (94.4% ± 0.5%), SOX1 (75.9% ± 1.5%), PAX6 (76.7% ± 0.9%), and NCAM (72.5% ± 2.2%) while lesser fraction of cells were positive for SOX2 (49.6% ± 4.9%) and a common stem cell marker, CD133 (36.1% ± 4.4%) (n = 3; Figure 3C). To evaluate the self-renewal capacity of HNF-iNSCs, we performed the clonality assay as a stringent test of NSC identity and seeded single iNSCs into the individual wells of a laminin/poly-L-ornithine-coated 96-well plate (one cell/well). This analysis showed that single iNSCs were able to proliferate and form NSC-like colonies (Figure 3D) with an efficiency comparable with control NSCs (Figure 3E), and that these clonally derived iNSCs maintained the expression of NES and CD133 (Figure 3F).
Figure 3.
Characterization of HNF-Derived iNSCs
(A) qRT-PCR analysis of indicated NSC markers in HNF-derived iNSCs and control NSCs derived from hESCs (hESC-NSC) and human fetal brain (WT-NSC). Data were normalized to the expression level of GAPDH and presented relative to the expression of each indicated gene in HNFs. Data are shown as means ± SD of three independent experiments.
(B) Immunofluorescent staining of HNF-iNSCs for NES, SOX1, SOX2, PAX6, NCAM, and CD133. Data are representative stainings of three independent experiments that gave similar results.
(C) Flow cytometry analysis of indicated NSC protein markers in iNSCs. Data are given as means ± SD of three independent experiments.
(D) Analysis of the clonogenicity of a single HNF-derived iNSC. Representative bright field images show a time course of colony formation on days 1, 5, 10, 15, and 20 after single-cell plating.
(E) Colony formation efficiency of iNSCs plated at a density of one cell/well in 96-well plates. After 2 hr, all wells were scored for the presence of single cells (64% for WT-NSC, 50% for hESC-NSC, and 56% for iNSC). Wells containing single cells were assessed on day 20 for the presence of a colony larger than 50 μm in diameter. Results are shown as means ± SD of three independent experiments.
(F) Expression of CD133 and NES in clonally derived iNSC spheres.
Scale bars represent 50 μm in (B), (D), and (F). See also Figure S3.
Zfp521-iNSCs Can Also Be Established from Human Fetal and Adult Fibroblasts
In addition to HNFs, Zfp521 could successfully reprogram human fetal fibroblasts (HFFs, 18 weeks fetus) (Figure S3Ai) to iNSCs, with an efficiency of 0.7% ± 0.02% (n = 12 independent biological replicates). After 24 days of Dox treatment, HFF-iNSCs formed primary neurospheres (Figure S3Aii) that expressed NES and SOX1 (Figure S3Aiii) and gave rise to a cell line with characteristics similar to WT-NSCs in terms of morphology (Figure S3Bi), stable maintenance in culture (Figure S3Bii), NSC marker expression (Figure S3C), and retention of a stable karyotype following serial passaging (Figure S3D).
We also sought to determine whether adult human dermal fibroblasts (HDFs) could be converted into iNSCs with the same factor. We observed that Zfp521 alone was not sufficient to reprogram HDFs (derived from a 40-year-old man) into iNSCs. To overcome the HDF reprogramming barrier to iNSC state, we utilized a small-molecule cocktail consisting of valproic acid, CHIR099021, and SB431542 under hypoxic conditions, which was shown by Cheng et al. (2014) to be sufficient for generation of iNSCs from mouse embryonic and neonatal fibroblasts and human urinary cells. As per our protocol, small-molecule treatment alone was not sufficient to generate iNSCs from HDFs. However, when this chemical cocktail was combined with Zfp521, according to the scheme in Figure S3E, the HDFs (Figure S3Fi) were successfully reprogrammed to assume iNSC identity (Figure S3Fii). The HDF-iNSCs could be serially passaged to establish a self-renewing cell line (Figure S3Fiii) and expressed NSC markers NES, SOX1, SOX2, and PAX6 at levels comparable with hESC-NSCs and WT-NSCs (Figure S3G). In addition, immunofluorescence analyses demonstrated that HDF-iNSCs expressed NES, SOX1, SOX2, and PAX6 (Figure S3H). The efficiency of HDF reprogramming with this protocol was 0.03% ± 0.01% (n = 12 independent biological replicates). Taken together, these data demonstrate that the transcription factor Zfp521 is capable of converting human fetal, neonatal, and adult fibroblasts, when co-treated with a particular combination of small molecules under hypoxic conditions, into stably expandable iNSCs.
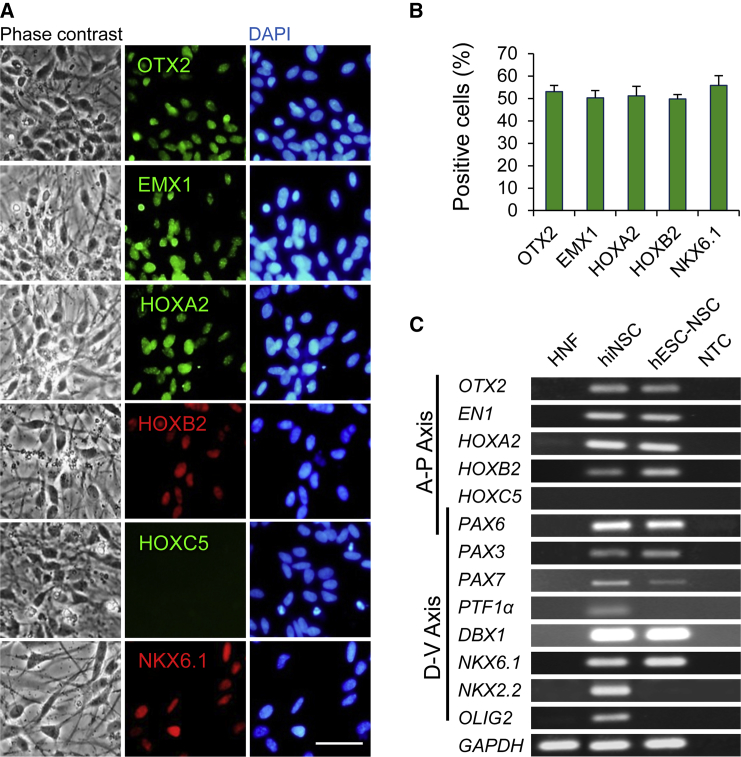
Regional Specificity of iNSCs
To determine the regional identity of Zfp521-iNSCs, the expression of region-specific markers known to be expressed along the anterior-posterior and dorsal-ventral axes of the neural tube was determined in iNSCs derived from HNFs. Immunofluorescence analyses showed that the forebrain markers OTX2 and EMX1 are highly expressed in more than 50% of iNSCs (Figures 4A and 4B). Another forebrain marker, namely PAX6, was detected at transcript (Figure 4C) and protein levels (Figure 3B). The midbrain marker Engrailed 1 (EN1) was also observed to be expressed in iNSCs, as shown by RT-PCR results (Figure 4C). In addition, key markers associated with the hindbrain (HOXA2 and HOXB2) were upregulated in iNSCs at both the protein (Figures 4A and 4B) and mRNA levels (Figure 4C). However, the spinal cord marker HOXC5 was not detected either at the transcript level or at the protein level (Figures 4A and 4C). The transcripts of dorsally expressed markers PAX3, PAX7, and PTF1a and the ventrally expressed markers DBX1, NKX6.1, NKX2.2, and OLIG2 were all detected in iNSCs (Figure 4C). The expression of NKX6.1 in iNSCs was also confirmed by immunocytochemistry (Figures 4A and 4B). With the exception of PTF1α, NKX2.2, and OLIG2 transcripts, all other analyzed regional-specific transcripts were also detected at similar levels in hESC-derived NSCs (Figure 4C). In contrast, the mesodermal and endodermal markers BRACHURY and SOX17 were undetectable in iNSCs (data not shown). Collectively, these results suggest that, similar to their brain-derived counterparts (Kamiya et al., 2011), Zfp521-iNSCs have a rostral identity (Figure S4).
Figure 4.
Regional Identity of iNSCs
(A) Immunofluorescent staining for indicated region-associated proteins in HNF-iNSCs. Scale bar represents 50 μm.
(B) Quantification of the regional identity data of iNSCs as determined via immunostaining for OTX2, EMX1, HOXA2, HOXB2, HOXC5, and NKX6.1. Data are given as means ± SD in three independent experiments.
(C) RT-PCR analysis of region-associated genes in iNSCs along anterior-posterior (A-P) and dorsal-ventral (D-V) axes.
See also Figure S4.
Global Gene Expression in Human iNSCs
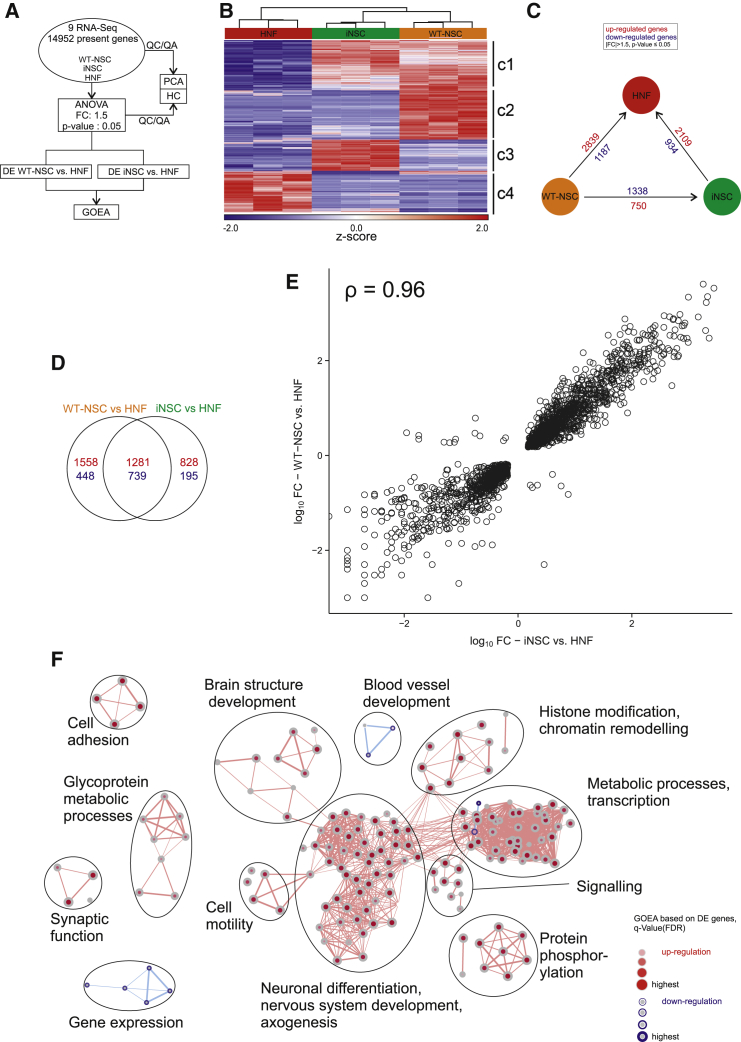
To assess the overall gene expression changes between HNFs, HNF-iNSCs, and native WT-NSCs, we generated transcriptomic data by RNA-seq and assessed the cellular differences through bioinformatics analyses (Figure 5A). These analyses revealed a high degree of similarity between iNSCs and WT-NSCs and that both these cell types strongly differed from HNFs. This is supported by the visualization of the hierarchical clustering of the 4,587 variable genes (Figure 5B). For the majority of variable genes, the expression profiles of iNSCs and WT-NSCs strongly corresponded to each other and differed from HNFs (gene clusters 1 and 4 in Figure 5B). Nevertheless, a fraction of variable genes were differentially expressed between iNSCs and WT-NSCs with gene cluster 2 being more prominently expressed in WT-NSCs and gene cluster 3 in iNSCs (Figure 5B). A statistical one-way ANOVA model further revealed that the number of significantly differentially expressed (DE) genes was higher between WT-NSCs versus HNFs and between iNSCs versus HNFs than between WT-NSCs versus iNSCs (Figure 5C). In all comparisons, most of the DE genes were shown to be upregulated (2,109 in iNSC versus HNF, 2,839 in WT-NSC versus HNF, and 1,338 in WT-NSC versus iNSC comparisons) while a lower number of genes were downregulated (934 in iNSC versus HNF, 1,187 in WT-NSC versus HNF, and 750 in WT-NSC versus iNSC comparisons). The comparison of DE genes between the control HNFs and iNSCs or WT-NSCs shows that the largest fractions of genes are equally regulated in both NSC types (Figure 5E). The FC-FC plot indicates that these similarly regulated genes also exhibit similar fold changes in comparison with control HNFs (Figure 5D).
Figure 5.
Global Gene Expression Profiling of HNFs, HNF-Derived iNSCs, and hESC-Derived NSCs
(A) Flowchart depicting the workflow for this figure and Figure S5.
(B) Hierarchical clustering showing the z-transformed expression values of the 4,587 present and variable genes.
The four gene clusters are indicated as c1, c2, c3 and c4.
(C) Differentially expressed (DE) genes (|FC| > 1.5, p ≤ 0.05) between WT-NSCs and HNFs, iNSCs and HNFs, and WT-NSCs and iNSCs.
(D) FC-FC plot of intersection from (E), log10 scaled.
(E) Venn diagram showing overlap of DE genes that were upregulated (red) or downregulated (blue) in WT-NSCs and iNSCs versus control HNFs.
(F) Network visualization of Gene Ontology Enrichment Analysis (GOEA) based on DE genes from quadrant I (right, top) and III (left, bottom) in (D). Enriched GO terms based on upregulated genes are depicted as red nodes, enriched GO terms based on downregulated genes as blue nodes, where color intensity and size represent the corresponding enrichment p value (q-value) adjusted for the false discovery rate. Overlap of genes between nodes is indicated by edge thickness. QC, quality control; QA, quality assurance; DE, differential expression.
See also Figure S5.
Gene Ontology Enrichment Analysis
To test that the biological processes are similarly regulated in WT-NSCs and iNSCs, we performed a Gene Ontology Enrichment Analysis (GOEA), followed by network visualization (Figures 5F, S5A, and S5B). Focusing the analyses first on those genes similarly DE between HNFs versus WT-NSCs and HNFs versus iNSCs (i.e., overlapping genes in the Venn diagram in Figure 5E), we found that the largest cluster of GO terms is associated with neuronal differentiation, nervous system development, and axogenesis (Figure 5F). Further clusters were associated with the GO terms brain structure development and synaptic function. One cluster was associated with GO terms histone modification and chromatin remodeling, indicating increased plasticity of NSCs compared with HNFs. When focusing on genes that were not similarly DE in WT-NSC versus HNF and iNSC versus HNF comparisons, the largest clusters of GO terms were still related to nervous system development and neurogenesis (Figures S5A and S5B), indicating that genes upregulated in iNSCs compared with WT-NSCs and genes upregulated in WT-NSCs compared with iNSCs are associated with similar biological functions. In addition, the GOEA network of specific DE genes in an iNSC versus HNF comparison (Figure S5B) contains a cluster that is associated with embryonic morphogenesis, embryonic development, and neural tube patterning indicating that iNSCs may represent a stem cell population at an earlier developmental stage than WT-NSCs.
Human iNSCs Are Multipotent In Vitro
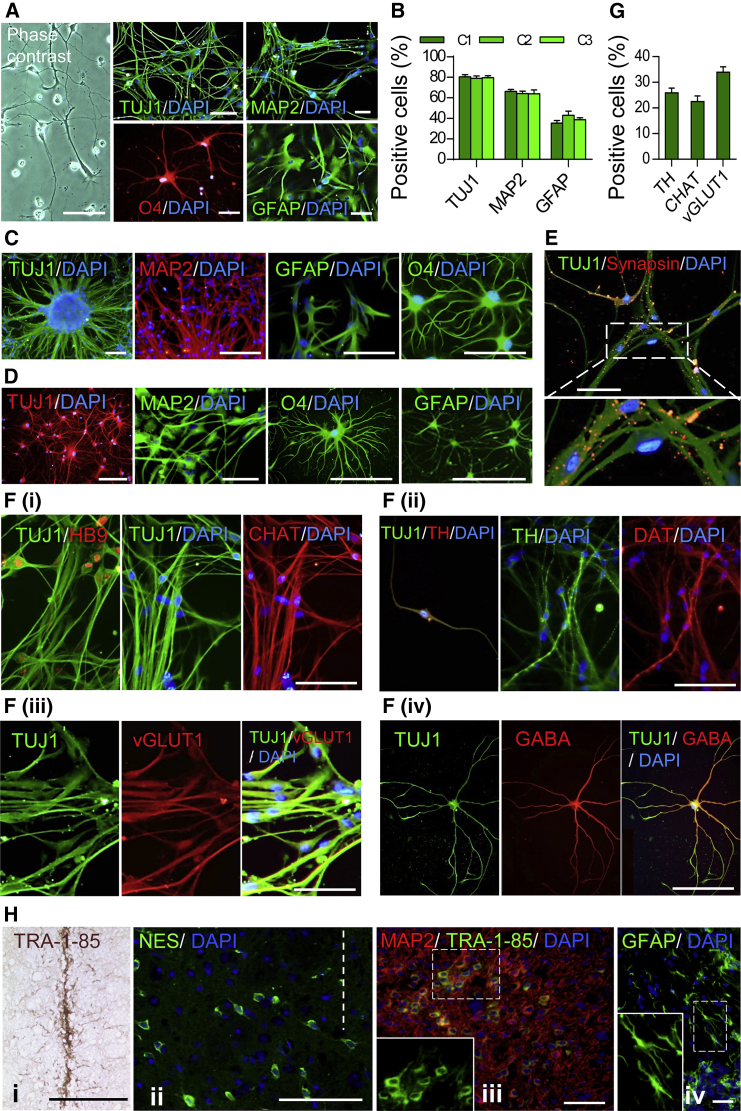
The developmental potential of Zfp521-iNSCs was evaluated by assessing their ability to generate the three main neural cell types: neurons, astrocytes, and oligodendrocytes. Under permissive differentiation conditions, iNSCs derived from HNFs, HFFs, and HDFs differentiated into TUJ1- and MAP2-positive neurons, GFAP-positive astrocytes, and O4-positive oligodendrocytes, all with characteristic morphological properties (Figures 6A, 6B, S6A, and S6B). Of note, the in vitro developmental potential of HNF-iNSCs was preserved in clonally derived HNF-iNSCs (Figures 6B and 6C) and even after 60 passages (Figure 6D). The iNSC-derived neurons exhibited punctate distribution of Synapsin protein, suggestive of the formation of synaptic structures (Figure 6E). These data demonstrate that Zfp521-iNSCs retain their phenotypic stability and multipotency following long-term expansion and single-cell subcloning.
Figure 6.
Differentiation Potential of HNF-iNSCs In Vitro and In Vivo
Figures (A–G) and (H) show the in vitro and in vivo data, respectively.
(A) Representative images of in vitro differentiation of iNSCs into TUJ1-and MAP2-positive neurons, O4-positive oligodendrocytes and glial fibrillary acidic protein (GFAP)-positive astrocytes.
(B) Quantification of the efficiency of iNSC differentiation as determined by immunostaining for TUJ1, MAP2, and GFAP. Experiments were performed with three different single-cell-derived iNSC clones (C1–C3). Data are shown as means ± SD of three biological replicates.
(C) Representative images of trilineage differentiation in clonally derived HNF-iNSCs.
(D) Representative images of differentiation of late passage (P60) HNF-iNSCs into neurons, oligodendrocytes, and astrocytes.
(E) A representative image for synapse formation between HNF-iNSC-derived neurons using antibodies specific for TUJ1 and Synapsin 6 weeks after spontaneous differentiation.
(F) Representative images for directed differentiation of HNF-iNSCs into (i) CHAT- and HB9-positive cholinergic neurons, (ii) TH- and DAT-positive dopaminergic neurons, (iii) vGLUT1-positive glutamatergic neurons, and (iv) GABA-positive GABAergic neurons.
(G) Quantification of the efficiency of iNSC differentiation as determined by immunostaining for TH, CHAT, and vGLUT1. Experiments were performed with one clone of iNSCs (C1). Data are shown as means ± SD of three biological replicates.
(H) Survival and differentiation potential of HNF-iNSCs in vivo. The iNSCs were transplanted into the striatum of adult nude rats (i, ii) or the cortices of newborn mice (iii, iv).
(i) Specific labeling against human marker TRA-1-85 visualized by DAB revealed the presence of the transplanted cells at the site of transplantation. (ii) Three weeks post-implantation, the cells migrated into the brain tissue at close proximity to the initial implantation site (dotted line) and stained positive for NES using antibodies specific for human protein. Scale bar represents 100 μm. (iii) Four weeks post-implantation, specific labeling against human TRA-1-85 (green) revealed the expression of neuronal marker MAP2 (red) in cells transplanted into the cortex of the neonatal mouse brain. Specific labeling against human GFAP shows astrocyte differentiation potential of human HNF-iNSC. Oligodendrocytes could not be detected by staining with antibody against human PDGFRα at this time point.
Scale bars represent 50 μm in (A), (C), and (F) and 100 μm in (D), (E), and (H). See also Figure S6.
Next, we sought to determine the type and frequency of the neuronal subtypes derived from HNF-iNSCs (Figures 6F and 6G). The iNSCs differentiated into cholinergic neurons as determined by the expression of choline acetyltransferase (CHAT, 22.5% ± 2.2%; Figures 6Fi and 6G) and HB9 as well as to dopaminergic neurons as judged by the expression of the dopaminergic neuron markers, dopamine transporter (DAT), and tyrosine hydroxylase (TH, 25.8% ± 1.9%; Figures 6Fii and 6G). In addition, 33.9% ± 2.1% of iNSC-derived neurons exhibited a glutamatergic phenotype, as shown by the expression of vesicular glutamate transporter 1 (vGLUT1), which is involved in the packaging of glutamate into the synaptic vesicles (Figures 6Fiii and 6G). We also detected a smaller number of GABAergic neurons, as demonstrated by immunofluorescence (Figure 6Fiv). These data indicate that diverse neuronal subtypes could be derived in vitro from Zfp521-iNSCs.
In Vivo Survival and Developmental Potential of iNSCs
In order to determine the capacity of iNSCs to survive after transplantation into adult brain tissue, we injected HNF-iNSCs into the striatum of adult nude rats. Analysis of tissue sections 1 week post-transplantation showed implanted cells that expressed human marker TRA-1-85 and were tightly packed at the site of implantation (Figure 6Hi). Evaluation at 3 weeks post-transplantation revealed that transplanted iNSCs survived and started migrating into the brain tissue in close proximity of the implantation site. These cells retained the expression of NES, suggestive of retention of neural-precursor characteristics of the surviving and migrating human iNSCs (Figure 6Hii). Differentiation into mature neural cell lineages was not observed at this early time interval. The in vivo developmental potential of iNSCs was further assessed by transplanting HNF-iNSCs into the cortices of newborn mice. Analysis of tissue sections 4 weeks post-transplantation revealed that the transplanted iNSCs survived and integrated into the brain tissue and expressed MAP2 and GFAP (Figures 6Hiii and 6Hiv). The human origin of transplanted cells for MAP2 and GFAP expression was confirmed by staining with human-specific TRA-1-85 and human-GFAP-specific antibodies, respectively. These data indicate that Zfp521-iNSCs possess the capability to survive and integrate into adult and neonatal brain tissue with the latter also being capable of supporting their differentiation into mature neural cells.
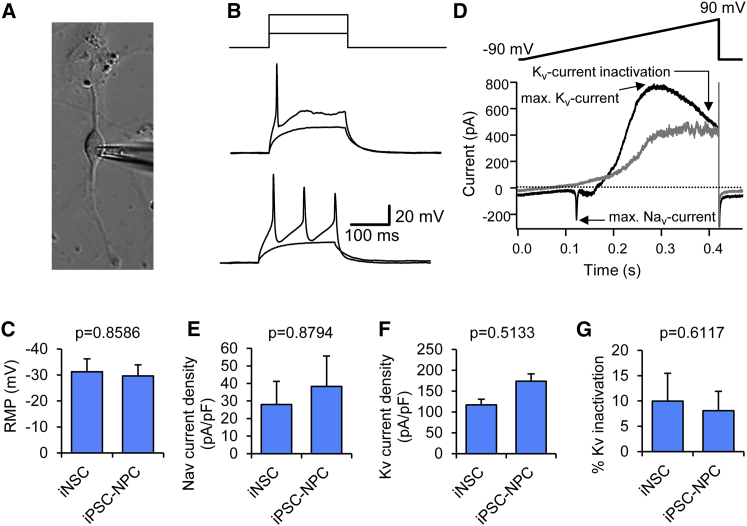
Electrophysiological Properties of Neurons Derived from iNSCs
In order to functionally characterize iNSC-derived neurons, electrophysiological properties of neurons derived from HNF-iNSCs were assessed in patch-clamp experiments and compared with those derived from human iPSC-NSCs. In both experimental groups, electrically active cells were observed. In the iNSC group, 32 cells were electrically active and 31% of them showed action potentials (APs) with a mature shape, including quickly rising upstrokes, an overshoot to positive potentials and an after hyperpolarization (Figures 7A and 7B). In comparison, APs were recorded in 63% of 19 patch-clamped, electrically active iPSC-NSC-derived neurons. The resting membrane potential was similar between both groups (about −30 mV) (Figure 7C). To investigate the firing properties of neurons in more detail, cells were held at −70 mV. Under this condition, two firing patterns were observed: repetitive firing (50.0% of the iPSC-NSC-derived neurons with APs and 40.0% of iNSC-derived neurons with APs) and phasic firing, which is characterized by a single AP, even if double the current needed to reach threshold is injected (Figure 6B). Thus, both NSC populations produced functionally active neurons.
Figure 7.
Electrophysiological Characteristics of HNF-iNSC-Derived Neurons
(A) Bright field image of a recorded HNF-iNSC-derived neuron.
(B) Representative current clamp recordings of a phasic and tonically firing iNSC-derived neuron. Upper panel: current injection protocol (23 pA and 53 pA for middle panel, 20 pA and 50 pA for recordings shown in lower panel). Cells were held at −70 mV.
(C) Resting membrane potential (RMP) of cells directly after break does not differ between iNSC-derived neurons (n = 10) and iPSC-derived neurons (n = 12). Only cells that displayed an action potential were analyzed. Data are shown as means ± SD.
(D) Voltage ramp and example current recording of an electrically active neuron (black line) and an inactive cell (gray line).
(E and F) Sodium current density was determined as maximal inward current (E), and potassium current density determined as maximal outward current (F) during a voltage ramp divided by cell capacitance as a measure for cell size; see (D) for protocol. Data are shown as means ± SD. n = 6 (iNSC-neurons) and 11 (iPSC-NSC-neurons).
(G) Percent of Kv inactivation is calculated as outward current at the end of the ramp divided by maximal outward current. Prominent inactivation indicates the expression of the delayed rectifier potassium channels. Data are shown as means ± SD.
In voltage-clamp mode, we investigated the currents evoked by ramp stimulation. In electrically active cells, the maximal fast inward current was carried by voltage-gated sodium channels (Nav), which were activated at negative potentials (black line in Figure 7D; compared with the current response of an electrically inactive cell [gray line]). Nav current density was calculated as maximal inward current divided by cell capacitance (a measure of the cell size) and revealed comparable values for iNSC- and iPSC-NSC-derived neurons (Figure 7E). Voltage-gated potassium currents (Kv) are recorded as outward currents at positive potentials, which often showed some inactivation on more positive voltages (Figure 7D). Kv current density was comparable for both investigated neuron types (Figure 7F) as well as their inactivating component (Figure 7G), which may hint at the expression of delayed rectifier potassium channels. Thus, neurons in both NSC populations showed similar functional maturity, AP properties, and Kv and Nav expression profiles.
Discussion
In this study, we demonstrate that iNSCs can be successfully generated from human fetal, neonatal, and adult fibroblasts by ectopic expression of a single Dox-inducible, lentivirally encoded neurogenic transcription factor Zfp521. These iNSCs display morphological characteristics of endogenous NSCs, are clonogenic, exhibit rostral regional specificity, and maintain their self-renewal ability and tripotency over prolonged passaging with retention of a stable karyotype following serial passaging. Therefore, Zfp521 seems to be sufficient to trigger a self-sustaining gene regulatory network that confers a rostral NSC identity to fetal, neonatal, and adult human fibroblasts (in the presence of a specific cocktail of small molecules), which is concordant with its reported permissive and rostralizing role in differentiation of neuroectodermal cells from undifferentiated ESCs and epiblast-stage embryos (Kamiya et al., 2011). Analysis of a global iNSC transcriptome confirmed the acquisition of neural gene expression profiles and activation of biological processes associated with neuronal differentiation and development in Zfp521-iNSCs. Although many of these genes were also expressed at similar levels in endogenous human NSCs isolated from fetal brain, these two cell types differed in the expression of a subset of genes that were differentially expressed between each NSC type and HNFs. However, GO analysis of these DE genes revealed that they still belonged to functional categories related to nervous system development and neurogenesis, supporting the conclusion that differences in expression profiles observed between iNSCs and WT-NSCs are more of a quantitative than of a qualitative nature and may reflect technical variance rather than biological differences.
We further show that Zfp521-iNSCs lines established from fetal, neonatal, and adult fibroblasts have the capacity to differentiate in vitro into the three main cell types: neurons, astrocytes and oligodendrocytes. This trilineage differentiation potential was also retained in three clonally derived iNSC lines that were tested, which further confirms that these cells are bona fide self-renewing multipotent stem cells. Functional analyses revealed that neurons derived from Zfp521-iNSCs exhibit resting membrane potentials, express sodium and potassium currents, and fire APs with characteristics indistinguishable from human iPSC-NSC-derived neurons that were used as controls and human PSC-derived neurons described by others (Pre et al., 2014, Song et al., 2013, Telias et al., 2014).
iNSCs transplanted into the neonatal mouse brain integrated into brain tissue and retained the capability to differentiate into neurons and astrocytes after 4 weeks. When transplanted into the adult rat brain, iNSCs survived and started migrating into the neighboring brain tissue. Nonetheless, in contrast to cells transplanted into neonatal brain, they retained the characteristics of neural precursors 3 weeks after transplantation. This result is in agreement with published data, which showed that a much longer time interval of several months is required for formation of mature neurons from transplanted human iNSCs and PSC-derived NSCs (Hemmer et al., 2014, Koch et al., 2009).
Several gene combinations have been successfully used for reprogramming human somatic cells to an NSC-like state (Cassady et al., 2014, Corti et al., 2012, Han et al., 2012a, Lu et al., 2013, Ring et al., 2012, Thier et al., 2012). These strategies, however, mostly rely on the use of multiple pluripotency-associated factors for reprogramming and the resultant cells have been characterized inadequately or may have limited self-renewal capacity. Our approach bypasses these limitations, as it is based on a single reprogramming factor, which provides a safer tool to generate iNSCs applicable for potential cell-based therapies and a unique platform for unraveling the mechanisms of iNSC reprogramming. Moreover, Zfp521-iNSCs could be stably propagated for more than 60 passages and were confirmed to be clonogenic. Furthermore, in the present study, we showed that Zfp521 enables the direct conversion of fetal and neonatal fibroblasts into stable iNSC lines, however NSCs from adult human fibroblasts can be derived by combinatorial treatment of Zfp521 and a cocktail of small molecules to overcome restrictions on neural fate conversion. Recently, it was also reported that inhibition of the let-7 microRNA, or expression of its target HMGA2, facilitates SOX2-mediated direct reprogramming of human adult fibroblasts and blood cells into iNSCs (Yu et al., 2015). Moreover, ectopic expression of OCT4 in the presence of SB431542, LDN-193189, Noggin, and CHIR99021 cocktail resulted in the generation of iNSCs from human neonatal and adult peripheral blood progenitors (Lee et al., 2015).
Zfp521 is a transcriptional co-repressor protein, the expression of which seems to be restricted to immature/multipotent cells of the body, including mesenchymal stem cells and hematopoietic stem cells (Bond et al., 2008, Han et al., 2012b). The highest expression levels of Zfp521 in the nervous system have been observed in the cerebellum, NSCs, and striatonigral neurons (Lobo et al., 2006). The importance of Zfp521 for neural development is demonstrated by the finding that NSCs/neuroectoderm cells cannot be generated from ESCs/epiblast when Zfp521 is abrogated (Kamiya et al., 2011). In addition, although homozygous Zfp521-deficient embryos apparently develop normally and are phenotypically indistinguishable from normal littermates at the time of birth, they exhibit behavioral abnormalities, lose weight over time, and are unable to survive for more than 10 weeks post-partum (Ohkubo et al., 2014) indicating the indispensable roles for Zfp521 in post-natal neurogenesis and development. Zfp521 exhibits a conserved amino acid and protein domain composition across diverse animal species (>90% identity), which suggests indispensable and universal roles for Zfp521 in organismal development (Shen et al., 2011). Notably, in this study, mouse Zfp521 has been used for conversion of human cells. Thus, it will be worthwhile to determine the mechanism underlying the regulatory role of Zfp521 during reprogramming in future studies.
Experimental Procedures
Generation of iNSCs
All animal-use procedures were in strict accordance with the approval of the Royan Institutional Review Board and Institutional Ethical Committee (approval ID J/90/1397) and the permission of the LANUV NRW (84-02.04.2012.A227) according to the guidelines of the Deutsche Tierschutzgesetz and the Versuchstierordnung. Written informed consent was provided for all donors.
Fibroblasts were transduced with lentiviral vectors expressing mouse Zfp521 (Life Technologies) and cultured for the first 2 days in fibroblast medium, as described in more detail in the Supplemental Information. Two days after viral infection, cells were switched into the neural medium, which included DMEM/Ham's F12 (DMEM/F12; Invitrogen) supplemented with 10% knockout serum replacement (KOSR), 1% non-essential amino acids (NEAA), 2 mM L-glutamine, ITS (1 mg/ml insulin, 0.55 mg/ml transferrin, 0.67 mg/ml selenium), 1% N2, 0.05% B27, penicillin/streptomycin (all Invitrogen), and 1.6 g/l glucose (Sigma-Aldrich). Dox (2 μg/ml) was added to the culture medium from this stage through day 24 to induce Zfp521 expression. The medium was changed every second day. Epidermal growth factor (EGF, 20 ng/ml; Royan Institute) and basic fibroblast growth factor (bFGF, 20 ng/ml; Royan Institute) were added to the neural medium from day 18 (when changes in the morphology of the cells were evident) until day 24. On day 24, NSC-like colonies emerged. Of these, a fraction detached from the plate and was released into the culture medium while the others remained adherent. Both cell populations, designated as adherent and non-adherent primary neurospheres (10–15 spheres), were collected under the microscope, transferred into a dish and trypsinized into single cells. Cells were subsequently seeded onto tissue culture dishes that were coated with 0.001% poly-L-ornithine and 1 μg/ml laminin (Sigma-Aldrich) for 1 hr at 37°C and were cultured in the neural medium supplemented with EGF and bFGF in the absence of Dox. Two to three days later, each dish that reached 60%–70% confluence was trypsinized, and the cells were transferred as a single-cell suspension into a 6-cm non-adherent dish to form secondary neurospheres. The procedure of plating onto tissue culture dishes followed by plating into bacterial dishes was repeated two additional times in order to obtain a pure population of induced neurosphere-forming NSC-like cells (iNSCs). The resultant neurospheres were trypsinized and plated onto laminin/poly-L-ornithine-coated 6-cm tissue culture dishes in neural medium containing EGF and bFGF. The cells at this stage were considered to be in passage 1. The iNSCs were then passaged after reaching a density of 80%–90% once every 3–4 days at a split ratio of 1:2. The medium was changed every other day. For reprogramming HDFs, we used a small-molecule cocktail consisting of valproic acid (0.5 mM; Sigma-Aldrich), CHIR099021 (3 μM; Stemgent), and SB431542 (1 μM; Sigma-Aldrich) under hypoxic conditions (5% O2) (Cheng et al., 2014) along with induced overexpression of Zfp521 as described above.
Statistical Analyses
Values are expressed as means ± SD. ANOVA or the t test was used to assess differences between means. A p value of <0.05 was considered statistically significant.
Author Contributions
E.S. and H.B. conceived this study. E.S. designed and performed most experiments. H.B. provided financial support, designed and analyzed experiments, discussed the results, wrote the paper, and approved the manuscript. S.M. wrote the paper. S.N. performed and analyzed colony-forming assays and RT-PCRs and wrote the paper. M.B. analyzed RT-PCRs and wrote the paper. L.S. performed in vivo transplantation and its analysis. H.G. analyzed karyotying. N.Z.M. differentiated iNSCs into neurons for electrophysiological studies, prepared and validated samples for RNA sequencing. T.Š. designed experiments for electrophysiological and transcriptional analyses, analyzed the data, discussed the results, and wrote the paper. K.H., P.G. and J.L.S. performed and analyzed RNA-seq experiments and wrote the corresponding manuscript sections. D.S. and A.L. designed and performed electrophysiology experiments, and A.L. wrote the corresponding manuscript sections. M.M. and F.H. carried out the tissue processing, histology, immunohistochemistry, and microscopic evaluation, and M.M. wrote the corresponding manuscript sections. J.H. provided financial support and approved the manuscript.
Acknowledgments
We thank the members of the Department of Stem Cells and Developmental Biology for their discussions. We are grateful to Najmeh Sadat Masoudi for karyotyping, Fazel Samani for assistance with flow cytometry, and Mostafa Najar-Asl for assistance with immunohistochemistry. This work was supported by a grant provided by the Iranian Council of Stem Cell Research and Technology, the Iran National Science Foundation (INSF), and Iran Science Elites Federation to H.B. and grants provided by Köln Fortune Program and Deutsche Forschungsgemeinschaft (DFG, grant number SA 1382/7-1) to T.Š. D.S. was funded by the IZKF N3 grant (to B. Winner, FAU Erlangen-Nürnberg).
Published: March 24, 2016
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes Supplemental Experimental Procedures, six figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.02.013.
Contributor Information
Tomo Saric, Email: tomo.saric@uni-koeln.de.
Hossein Baharvand, Email: baharvand@royaninstitute.org.
Accession Numbers
The accession number for the RNA-seq data reported in this paper is GEO: GSE64882.
Supplemental Information
References
- Ambasudhan R., Talantova M., Coleman R., Yuan X., Zhu S., Lipton S.A., Ding S. Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell. 2011;9:113–118. doi: 10.1016/j.stem.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond H.M., Mesuraca M., Amodio N., Mega T., Agosti V., Fanello D., Pelaggi D., Bullinger L., Grieco M., Moore M.A. Early hematopoietic zinc finger protein-zinc finger protein 521: a candidate regulator of diverse immature cells. Int. J. Biochem. Cell Biol. 2008;40:848–854. doi: 10.1016/j.biocel.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Cassady J.P., D'Alessio A.C., Sarkar S., Dani V.S., Fan Z.P., Ganz K., Roessler R., Sur M., Young R.A., Jaenisch R. Direct lineage conversion of adult mouse liver cells and B lymphocytes to neural stem cells. Stem Cell Rep. 2014;3:948–956. doi: 10.1016/j.stemcr.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Hu W., Qiu B., Zhao J., Yu Y., Guan W., Wang M., Yang W., Pei G. Generation of neural progenitor cells by chemical cocktails and hypoxia. Cell Res. 2014;24:665–679. doi: 10.1038/cr.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti S., Nizzardo M., Simone C., Falcone M., Donadoni C., Salani S., Rizzo F., Nardini M., Riboldi G., Magri F. Direct reprogramming of human astrocytes into neural stem cells and neurons. Exp. Cell Res. 2012;318:1528–1541. doi: 10.1016/j.yexcr.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D.W., Tapia N., Hermann A., Hemmer K., Hoing S., Arauzo-Bravo M.J., Zaehres H., Wu G., Frank S., Moritz S. Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell Stem Cell. 2012;10:465–472. doi: 10.1016/j.stem.2012.02.021. [DOI] [PubMed] [Google Scholar]
- Han R., Kan Q., Sun Y., Wang S., Zhang G., Peng T., Jia Y. MiR-9 promotes the neural differentiation of mouse bone marrow mesenchymal stem cells via targeting zinc finger protein 521. Neurosci. Lett. 2012;515:147–152. doi: 10.1016/j.neulet.2012.03.032. [DOI] [PubMed] [Google Scholar]
- Hemmer K., Zhang M., van Wullen T., Sakalem M., Tapia N., Baumuratov A., Kaltschmidt C., Kaltschmidt B., Scholer H.R., Zhang W. Induced neural stem cells achieve long-term survival and functional integration in the adult mouse brain. Stem Cell Rep. 2014;3:423–431. doi: 10.1016/j.stemcr.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B.Y., Weick J.P., Yu J., Ma L.X., Zhang X.Q., Thomson J.A., Zhang S.C. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc. Natl. Acad. Sci. USA. 2010;107:4335–4340. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya D., Banno S., Sasai N., Ohgushi M., Inomata H., Watanabe K., Kawada M., Yakura R., Kiyonari H., Nakao K. Intrinsic transition of embryonic stem-cell differentiation into neural progenitors. Nature. 2011;470:503–509. doi: 10.1038/nature09726. [DOI] [PubMed] [Google Scholar]
- Koch P., Opitz T., Steinbeck J.A., Ladewig J., Brustle O. A rosette-type, self-renewing human ES cell-derived neural stem cell with potential for in vitro instruction and synaptic integration. Proc. Natl. Acad. Sci. USA. 2009;106:3225–3230. doi: 10.1073/pnas.0808387106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladewig J., Mertens J., Kesavan J., Doerr J., Poppe D., Glaue F., Herms S., Wernet P., Kogler G., Muller F.J. Small molecules enable highly efficient neuronal conversion of human fibroblasts. Nat. Methods. 2012;9:575–578. doi: 10.1038/nmeth.1972. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Mitchell R.R., McNicol J.D., Shapovalova Z., Laronde S., Tanasijevic B., Milsom C., Casado F., Fiebig-Comyn A., Collins T.J. Single transcription factor conversion of human blood fate to NPCs with CNS and PNS developmental capacity. Cell Rep. 2015;11:1367–1376. doi: 10.1016/j.celrep.2015.04.056. [DOI] [PubMed] [Google Scholar]
- Lobo M.K., Karsten S.L., Gray M., Geschwind D.H., Yang X.W. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat. Neurosci. 2006;9:443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- Lu J., Liu H., Huang C.T., Chen H., Du Z., Liu Y., Sherafat M.A., Zhang S.C. Generation of integration-free and region-specific neural progenitors from primate fibroblasts. Cell Rep. 2013;3:1580–1591. doi: 10.1016/j.celrep.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan E., Chanda S., Ahlenius H., Sudhof T.C., Wernig M. Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc. Natl. Acad. Sci. USA. 2012;109:2527–2532. doi: 10.1073/pnas.1121003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marro S., Pang Z.P., Yang N., Tsai M.C., Qu K., Chang H.Y., Sudhof T.C., Wernig M. Direct lineage conversion of terminally differentiated hepatocytes to functional neurons. Cell Stem Cell. 2011;9:374–382. doi: 10.1016/j.stem.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradi S., Asgari S., Baharvand H. Concise review: harmonies played by MicroRNAs in cell fate reprogramming. Stem Cells. 2014;32:3–15. doi: 10.1002/stem.1576. [DOI] [PubMed] [Google Scholar]
- Nemati S., Hatami M., Kiani S., Hemmesi K., Gourabi H., Masoudi N., Alaei S., Baharvand H. Long-term self-renewable feeder-free human induced pluripotent stem cell-derived neural progenitors. Stem Cells Dev. 2011;20:503–514. doi: 10.1089/scd.2010.0143. [DOI] [PubMed] [Google Scholar]
- Ohkubo N., Matsubara E., Yamanouchi J., Akazawa R., Aoto M., Suzuki Y., Sakai I., Abe T., Kiyonari H., Matsuda S. Abnormal behaviors and developmental disorder of hippocampus in zinc finger protein 521 (ZFP521) mutant mice. PLoS One. 2014;9:e92848. doi: 10.1371/journal.pone.0092848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pournasr B., Khaloughi K., Salekdeh G.H., Totonchi M., Shahbazi E., Baharvand H. Concise review: alchemy of biology: generating desired cell types from abundant and accessible cells. Stem Cells. 2011;29:1933–1941. doi: 10.1002/stem.760. [DOI] [PubMed] [Google Scholar]
- Pre D., Nestor M.W., Sproul A.A., Jacob S., Koppensteiner P., Chinchalongporn V., Zimmer M., Yamamoto A., Noggle S.A., Arancio O. A time course analysis of the electrophysiological properties of neurons differentiated from human induced pluripotent stem cells (iPSCs) PLoS One. 2014;9:e103418. doi: 10.1371/journal.pone.0103418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring K.L., Tong L.M., Balestra M.E., Javier R., Andrews-Zwilling Y., Li G., Walker D., Zhang W.R., Kreitzer A.C., Huang Y. Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell. 2012;11:100–109. doi: 10.1016/j.stem.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S., Pu J., Lang B., McCaig C.D. A zinc finger protein Zfp521 directs neural differentiation and beyond. Stem Cell Res. Ther. 2011;2:20. doi: 10.1186/scrt61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son E.Y., Ichida J.K., Wainger B.J., Toma J.S., Rafuse V.F., Woolf C.J., Eggan K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M., Mohamad O., Chen D., Yu S.P. Coordinated development of voltage-gated Na+ and K+ currents regulates functional maturation of forebrain neurons derived from human induced pluripotent stem cells. Stem Cells Dev. 2013;22:1551–1563. doi: 10.1089/scd.2012.0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Telias M., Segal M., Ben-Yosef D. Electrical maturation of neurons derived from human embryonic stem cells. F1000Res. 2014;3:196. doi: 10.12688/f1000research.4943.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thier M., Worsdorfer P., Lakes Y.B., Gorris R., Herms S., Opitz T., Seiferling D., Quandel T., Hoffmann P., Nothen M.M. Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell. 2012;10:473–479. doi: 10.1016/j.stem.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Vierbuchen T., Ostermeier A., Pang Z.P., Kokubu Y., Sudhof T.C., Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Huang W., Su H., Xue Y., Su Z., Liao B., Wang H., Bao X., Qin D., He J. Generation of integration-free neural progenitor cells from cells in human urine. Nat. Methods. 2013;10:84–89. doi: 10.1038/nmeth.2283. [DOI] [PubMed] [Google Scholar]
- Weissbein U., Ben-David U., Benvenisty N. Virtual karyotyping reveals greater chromosomal stability in neural cells derived by transdifferentiation than those from stem cells. Cell Stem Cell. 2014;15:687–691. doi: 10.1016/j.stem.2014.10.018. [DOI] [PubMed] [Google Scholar]
- Yu K.R., Shin J.H., Kim J.J., Koog M.G., Lee J.Y., Choi S.W., Kim H.S., Seo Y., Lee S., Shin T.H. Rapid and efficient direct conversion of human adult somatic cells into neural stem cells by HMGA2/let-7b. Cell Rep. 2015 doi: 10.1016/j.celrep.2014.12.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.