Abstract
Lower motor neuron (LMN) degeneration occurs in several diseases that affect patients from neonates to elderly and can either be genetically transmitted or occur sporadically. Among diseases involving LMN degeneration, spinal muscular atrophy (SMA) and spinal bulbar muscular atrophy (Kennedy’s disease, SBMA) are pure genetic diseases linked to loss of the SMN gene (SMA) or expansion of a polyglutamine tract in the androgen receptor gene (SBMA) while amyotrophic lateral sclerosis (ALS) can either be of genetic origin or occur sporadically. In this review, our aim is to put forward the hypothesis that muscle fiber atrophy and weakness might not be a simple collateral damage of LMN degeneration, but instead that muscle fibers may be the site of crucial pathogenic events in these diseases. In SMA, the SMN gene was shown to be required for muscle structure and strength as well as for neuromuscular junction formation, and a subset of SMA patients develop myopathic pathology. In SBMA, the occurence of myopathic histopathology in patients and animal models, along with neuromuscular phenotype of animal models expressing the androgen receptor in muscle only has lead to the proposal that SBMA may indeed be a muscle disease. Lastly, in ALS, at least part of the phenotype might be explained by pathogenic events occuring in skeletal muscle. Apart from its potential pathogenic role, skeletal muscle pathophysiological events might be a target for treatments and/or be a preferential route for targeting motor neurons.
1. Introduction : lower motor neuron degeneration affects first neuromuscular junctions
Voluntary locomotion is governed by motor units, composed of lower motor neurons (LMNs) (alpha-motor neurons) that innervate skeletal muscle fibers and control their contraction. Myelinated axons of the LMNs contact muscle fibers on a discrete region of the fiber, highly speciallized, called the neuromuscular junction (NMJ). A number of pathologies target this critical region. First, myasthenic syndromes affect the efficacy of synaptic transmission through either immunological or genetic processes. In these pathologies, the absolute number of NMJs remain roughly the same but their efficacy to trigger a muscle action potential in response to motor neuron stimulation is decreased leading to fatigable muscle weakness. NMJs are also affected during lower motor neuron degeneration. In this review, we will focus on pathologies involving LMN degeneration, but sparing sensory innervation. This group of diseases affects patients from neonates to elderly people and can either be genetically transmitted or occur sporadically in a family. The table 1 summarizes the clinical features of the diseases we will focus on in this review, namely spinal muscular atrophy (SMA)[1], spinal bulbar muscular atrophy (Kennedy’s disease, SBMA) [2] and amyotrophic lateral sclerosis (ALS)[3].
Table 1.
clinical presentation of diseases with LMN degeneration disease onset Cause LMN involvement
| disease | onset | Cause | LMN involvement | UMN involvement | Myopathic features |
|---|---|---|---|---|---|
| SMA | |||||
| type I | birth | Loss of SMN1 | + | − | + |
| type II | Before 18 mo | Loss of SMN1 | + | − | + |
| type III | After 18 mo | Loss of SMN1 | + | − | ++ |
| SBMA | |||||
| SBMA | Adult-onset | Expansion of the polyglutamine tract of AR | + | − | +++ |
| ALS | |||||
| sALS | Adult-onset | Unknown (sporadic) | + | + | Occasionally reported |
| fALS | Adult onset | Genetic (multiple loci : sod1, tdp-43, fus, vapb…) | + | + | Occasionally reported |
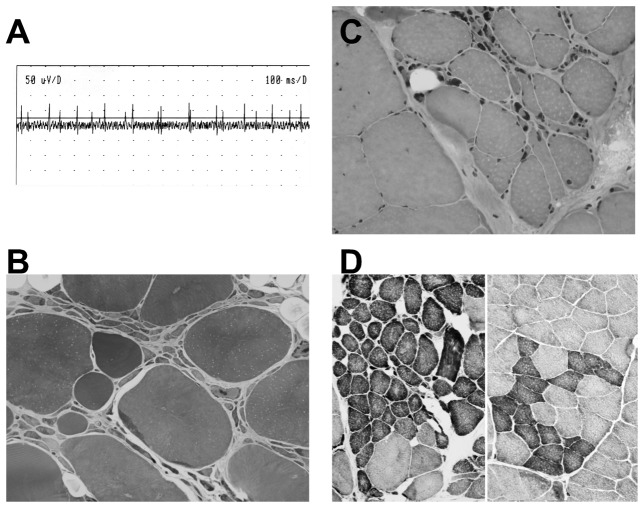
During LMN degeneration, NMJs are progressively destroyed, leading to fully denervated muscle fibers and neurogenic figures in histopathology. The figure 1 presents typical histopathological and electrophysiological results obtained in patients afflicted with a LMN degeneration. One is able to observe that muscle fibers of these patients display spontaneous denervation activities as judged in electromyography (figure 1A). Histopathology shows the occurence of small grouped, angulated fibers (figure 1B–D). It is thought that reinnervation of muscle fibers by the same motor neuron leads to these grouped fibers. NMJ destruction during LMN degeneration is associated with the degeneration of lower motor neuron cell bodies, hence their name and a widely accepted paradigm was that LMN death was the cause of NMJ destruction in these pathologies. Indeed, muscle denervation by nerve transection leads to roughly similar histopathological figures, suggesting that in LMN degeneration, the same cause (disruption of the nerve to muscle communication) leads to the same consequence (muscle denervation). However, recent histopathological evidences suggest that grouped atrophic fibers are highly homogenous in type in polyneuropathies, but heterogenous in LMN degeneration [4]. The reason for this dyscrepancy is unknown, but might indeed reflect profound differences in the mechanisms of denervation/reinnervation between purely denervating diseases and diseases with LMN degeneration.
Figure 1. electrophysiological and muscle pathological features of LMN degeneration in skeletal muscle.
A. Electrophysiological (EMG) recording of a patient with ALS demonstrating fibrillation potentials at rest, i.e. spontaneous denervation-related muscle activity.
B–D. Muscle biopsy of a patient with ALS. All the features associated with neurogenic disorders, i.e. grouping of atrophic fibers (B,C), predominance of one type of fiber (D), and presence of angulated fibers (D), are observed.
(B: semi-thin section, X 250; C: H–E staining, X 200; D: NADH-TR staining, X 100).
In this review, we would like to put forward the hypothesis that muscle fibers might not be the collateral damages of LMN degeneration, but rather, one of their murderers. For this, we will first describe how muscle is involved in the patterning and development of fully functional NMJs and how muscle participates in the pathology of ALS, SBMA and SMA. We will also give insights into how drugs might target muscle pathophysiological events or target motor neurons but be delivered through muscles to treat LMN degeneration.
2. Muscle and motor neuron : a peer to peer dialog to establish neuromuscular junctions
The establishment of neuromuscular synapses is a highly controlled developmental event. Motor axons, emerging from the embryonic spinal cord, contact newly formed myofibers around E13. It was long known that motor neurons and muscles are interdependant for their development. Indeed, during development, motor neurons die if deprived of their target muscle [5–8]. Conversely, muscle development is arrested in the absence of motor neurons. A neurocentric view of the development of NMJs long prevailed and postulated that this process was mainly driven by motor neurons, with muscle fibers being passive players of NMJ development. Indeed, motor neurons secrete a very active isoform of the glycoprotein agrin (neural agrin), that is sufficient to cluster nicotinic acetylcholine receptors on the myofiber [9, 10] through a complex receptor pathway involving the tyrosine kinase Musk and a co-receptor LRP4 [11, 12]. In this model, the presence of agrin secreted by the nerve is sufficient to cluster AchR and form NMJs. However, in the absence of nerves, clusters of nicotinic acetylcholine receptors still form in the correct, central, region of the developping muscle [13, 14]. Moreover, diaphragm muscles from E14 embryos cultured in the presence of recombinant agrin as well as muscles from transgenic mice overexpressing a miniaturised form of agrin in the whole muscle fiber retain the formation of AchR clusters in their central region [15]. Conversely, the transient muscle overexpression of a constitutively active form of ErbB2 widened the zone of AchR transcription and of the region where NMJs formed [16]. Altogether, these data show that the site where NMJs form in skeletal muscle is pre-patterned by muscle cells, independantly of nerve derived cues such as agrin. In the current model, NMJs thus form through a dialog between muscle and motor neurons, muscle driving the zone where NMJs will form and neurons strengthening the newly formed contacts by the secretion of agrin.
This pathway of NMJ development has been largely studied in the context of congenital myasthenic syndromes in which mutations in Musk [17], or more recently in agrin [18] were found. Up to now, there is no documented association between this pathway and LMN degeneration. However, it is striking to note that a mutation in Musk, when introduced in knockin mice, not only leads to the full phenotypic spectrum of myasthenic syndrome, but also to denervation of endplates and molecular features of denervation [19]. In line with this, the recently described mutation in agrin in a patient with congenital myastheniav [18] leads to not only to post-synaptic defects but also to pre-synaptic pathology suggesting that the Agrin/Musk pathway might also be involved in the maintenance of NMJs in adults and, thus potentially in LMN degeneration.
3. Muscle pathogenic role in spinal muscular atrophy
3-1). spinal muscular atrophy
Childhood spinal muscular atrophy (hereafter called spinal muscular atrophy, SMA) is a child-onset motor neuron disease involving mutations in the survival of motor neuron gene [20]. SMA patients are divided in three clinical groups according to the severity of their disease [1]. Type I SMA is characterized by severe, generalized muscle weakness and hypotonia at birth or within the first 6 months, with death usually occuring within the first 2 years. Type II children are able to sit, although they cannot stand or walk unaided, and they survive beyond 2 years. In type III SMA patients have proximal muscle weakness, starting after the age of 18 months.
In the human genome, the SMN gene is duplicated with a highly homologous copy called SMN2. Both SMN and SMN2 genes are expressed and only five nucleotides are different between both genes. We showed that even the promoter sequences and activities of these two genes were strikingly similar [21]. Importantly, only SMN1 deletions cause SMA, while up to 5% of individuals are lacking the SMN2 gene. The subtle difference in nucleotide sequence between SMN1 and 2 has no effect on the encoded open reading frame but profound effects on SMN2 splicing. Indeed, one of the nucleotides divergent between SMN1 and SMN2 creates a new splicing site in SMN2 mRNA by skipping exon 7. Thus, the SMN1 gene produces exclusively full length fully functional transcripts, while the transcripts derived from SMN2 lack exon 7. Most importantly, the amount of SMN2 protein are strikingly invertly correlated with the clinical severity of disease [22, 23], suggesting that SMN2 is a modifying gene in SMA. Thus, SMA is caused by SMN1 mutation and the severity of the disease is linked to the potential compensation by SMN2 protein products. That SMA is caused by the lack of full-length SMN proteins is largely substantiated by the evidence that ablating specifically the exon 7 in motor neurons lead to motor neuron degeneration in mice [24–26].
3-2). a housekeeping function for SMN protein ?
What is the function of SMN, lost in SMA, that might lead to motor neuron degeneration ? SMN forms a large multiprotein complex with at least 7 other proteins (called gemins) both in the cytoplasm and in the nucleus where it is concentrated in a structure called gems (for “gemini of coiled bodies”), associated with Cajal bodies [27]. This complex including SMN and gemins appears crucial for the biogenesis of small nuclear ribonucleoprotein particles (snRNPs) [28–30] that are involved in the splicing of pre-mRNA. Consistent with a key role in RNA metabolism, the ablation of exon 7 of SMN in either muscle or neuron leads to strong up-regulation of a number of genes involved in pre-mRNA splicing, ribosomal RNA processing, or RNA decay [31]. Furthermore, a recent study used exon microarray in SMA mice and observed widely distributed splicing defects in numerous mRNAs [32]. Interestingly, while splicing defects were prominently observed in all the tissues studied, the genes that were abnormaly spliced were different between tissues, suggesting that the selectivity of SMA for the neuromuscular system is explained by tissue specific alterations in mRNA splicing.
3-3). SMN is required for muscle function
The SMN protein is ubiquitously expressed and the tissue specific ablation of SMN leads to drastic alterations in the physiology of the targeted tisue [24–26, 33]. A number of studies have convincingly shown that SMN expression was required for muscle structure and function. In vitro, Shafey and collaborators demonstrated that SMN knockdown in C2C12 myoblasts lead to decreased myoblast proliferation and impaired myotube fusion [34]. Consistently, in vivo, exon 7 ablation of the SMN gene in mouse skeletal muscle leads to a massive muscle dystrophy and death of the animals [24] and RNAi knockdown of SMN in drosophila muscles is letal in drosophila in a much more severe way than neuronal knockdown [35]. These studies were all based on the knock-down or knock-out of the SMN gene, a situation much more drastic of what occurs in real pathological situations. Their relevance for the human pathology is however strengthened by the work of Arnold and collaborators [36] that had previously observed decreased proliferation and impaired fusion of type I SMA myoblasts. More recently, Martinez-Hernández and collaborators showed that myotubes of fetuses affected with type I SMA were smaller and with abnormal arrangements suggesting delayed developmental maturation [37]. Importantly, about a quarter of SMA type 3 are reported to have a dystrophic phenotype with high serum creatine kinase (CK) levels and “myopathic” histopathology [38]. Thus, SMN is required for normal muscle structure and development either in experimental situations or in SMA patients.
The function of SMN in muscle is likely to be related to sarcomeric structure. In drosophila bearing an hypomorphic allele of SMN, Rajendra and collaborators in 2007 observed a severe disorganization of muscle filaments associated with decreased actin expression [39]. Indeed, SMN hypomorph displayed a similar phenotype than flies with ablation of one muscle isoform of actin. These authors suggested a function for SMN in sarcomere formation since both endogenous and transgenic SMN localized to the sarcomeric region of fly muscle. Such a function was further suggested by the work of Walker and collaborators [40]. Indeed, not only SMN, but also its associated proteins localize to the sarcomeric Z-disc in both cardiac and skeletal myofibrils of the mouse. This localization of the SMN complex appeared independent of its role in pre-mRNA splicing since snRNPs themselves were not found in the sarcomere. SMA mutant muscles exhibit numerous morphological defects, including vacuoles and altered Z-disc spacing and an overall loss of sarcomeric uniformity and alignment. Whether this phenotype is primarily myopathic or if it is a secondary consequence of denervation is not known but these results establish that myofibrils from SMA type I mice display defects that are consistent with those observed in other myopathies. Furthermore, these observations are consistent with the occurence of congenital heart defects in SMA. This function of SMN in sarcomere formation might be related to the postulated role of SMN in beta-actin mRNA translation [41]. In all, the data available point to a key function for SMN in muscle development and function.
3-4). SMA is a developmental pathology of NMJs
Recent evidence suggest that the SMA pathology begins at NMJs. Indeed, Kariya and collaborators used severe- and mild-SMN2 expressing mouse models of SMA as well as material from human patients to understand the initial stages of neurodegeneration in the human disease [42]. In these studies, the earliest defects appear at the NMJ. Indeed, lack of SMN protein lead to the failure of post-natal development of the NMJ. In particular, NMJs of SMA mice showed impaired maturation of acetylcholine receptor (AChR) clusters into ‘pretzels’ that were reflected in functional deficits at the NMJ characterized by intermittent neurotransmission failures. Similar results were also reported by Kong and collaborators which observed immaturity of NMJs in SMA mice as assessed for instance by persistance of embryonic gene expression, reduced post-synaptic apparatus and electrophysiological abnormalities [43]. Importantly, the pre-synaptic pathology of severe SMA mice is dissociated from the post-synaptic pathology, suggesting that both events might be independent [44]. These data are indeed consistent with earlier studies showing reduced expression of AchR genes in SMA myocytes [36]. Altogether, SMA should now be viewed as a developmental NMJ synaptopathy
3-5). SMA as a global neuromuscular pathology
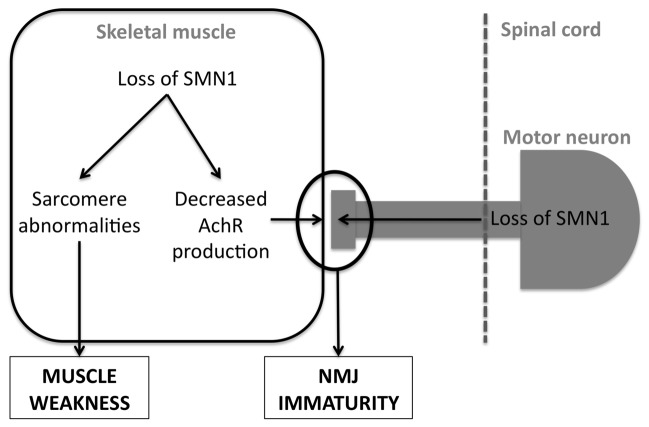
Given that SMN has a crucial function in muscle, and that SMA pathology is initiated at the neuromuscular synapse, one could hypothesize that SMA is indeed a muscle disease spreading to the motor neuron. Indeed, muscle knockdown of SMN leads to a more drastic phenotype than neuronal knockdown in Drosophila [35]. This is however not true in all animal models, since SMN ablation is partially rescued by neuronal but not muscle transgenic expression in the nematode [45]. Furthermore, overexpression of SMN in skeletal muscles did not allow the rescue of the phenotype of severe SMA mice, while a more widespread overexpression including high levels in neurons and low levels in muscle strongly alleviated the phenotype of these mice [46]. However, it should be kept in mind that the transgenic overexpression of SMN2 in these animals might be sufficient on its own to fully rescue the muscle phenotype. In this situation, one should not expect an even more increased survival of the mice after increasing SMN expression. In any case, the bunch of litterature currently available strongly suggests that SMA is a global neuromuscular pathology, involving pathological events in muscle and neurons, and potentially a number of other cell types (figure 2).
Figure 2. potential mechanisms involving skeletal muscle in SMA.
Loss of SMN1 leads to two consequences in skeletal muscle. First, SMN1 loss leads to abnormalities in sarcomere structure, which are a likely cause of muscle weakness. Second, SMN1 loss decreases the potential of muscle to produce mature AchR subunits, leading to an abnormal development of NMJs. SMN1 loss in motor neurons has also profound effects on NMJ development. See text (section 3) for further details.
4. Muscle pathogenic role in Kennedy’s disease
4-1). Kennedy’s disease
Spinal and bulbar muscular atrophy (SBMA), or Kennedy’s disease, is a purely genetic LMN degeneration caused by expansion of a CAG repeat in the first exon of the androgen receptor (AR) gene, leading to an AR protein with an expanded polyglutamine (polyQ) tract [47]. Like in other triplet repeat disorder, including Huntington’s disease, the disease develops only in individuals bearing an expansion of the polyQ tract over a certain threshold (>36 Gln) and the length of the polyQ is correlated with a number of clinical parameters [48, 49]. SBMA is an X-linked, gender specific disease since only male carriers of a pathogenic mutation are affected. The phenotype of females carrying a pathogenic allele is absent or very mild even if homozygous for an expanded CAG repeat [50, 51]. SBMA patients develop proximal muscle weakness, fasciculations, and atrophy, along with lower motor neuron degeneration in the brainstem and spinal cord [2, 52]. A subset of patients also display additional features, associated with androgen dysfunction such as androgen insensitivity, oligozoospermia or azoospermia, testicular atrophy, feminized skin changes, and gynecomastia [53, 54].
Different animal models of SBMA have been generated in the recent years. In particular, expansions of CAG repeats have been introduced in knock-in mice [55–57] or in transgenic mice [58–61]. For instance, male mice with a 113 polyglutamine tract in the endogenous AR developped an SBMA-like neuromuscular pathology, with early muscle pathology but late spinal cord disease and relatively spared motor neurons. In this mouse model, there was prominent early death of males caused by obstruction of the urinary tract [55]. Interestingly, affected males display several signs of androgen insensitivity, as commonly observed in SBMA.
The physiological function of the mutant protein in SBMA is extremely well documented. The AR is a transcription factor, whose transcriptional activity is activated by testosterone or dihydrotestosterone [62]. After ligand activation, AR drives the expression of its target genes, including key factors for muscle growth, male reproductive function and male secondary sexual phenotypes [63]. The lack of functional AR protein leads to testicular feminization and complete infertility in both mice and humans [64, 65]. In skeletal muscle, AR is especially required for the maintenance in muscle mass and fiber type [66] as well as muscle strength in males [67].
Studies in animal models indicate that hormonal ligands are crucial for the development of SBMA. First, only male transgenic mice develop an SBMA-like phenotype. Second, castration of transgenic males alleviates the phenotype while testosterone-treated females develop the pathology [58, 60, 61]. This is reminiscent of the absence of phenotype of women carriers of SBMA mutations [50, 51]. Consistently, a phase 2 clinical trial has recently suggested that androgen deprivation by leuprorelin acetate may be beneficial to patients [68] Thus, SBMA is due to a toxic gain of function in mutant AR unmasked by the presence of the ligand.
4-2). Myopathic pathology in SBMA
Studies in both patients and mice have shown that, apart from lower motor neuron degeneration, SBMA patients display myopathic features. In particular, SBMA patients showed CK elevation in muscle biopsies [69]. Recently, we described a family with early onset and rapidly progressive SBMA mimicking muscle dystrophy [70]. Three out of seven patients in this family displayed increased CK levels indicating rhabdomyolysis, suggesting that the occurence of such muscle abnormalities is indeed very common in SBMA. It should however be noted that CK elevation is a poor indicator of myopathy since it is subject to large variations and increased in a number of conditions. Apart from plasma CK levels more elevated than expected in denervative diseases, SBMA patients also often show myopathic abnormalities in muscle biopsies with centrally nucleated fibers or other myopathic features [69] and such abnormalities do not appear to be the sole consequence of denervation. In the same line, Yu and collaborators have shown that muscle pathology long preceeds spinal cord pathology in an animal model of SBMA. Furthermore, these animals display mixed features of myopathy and denervation and showed prominent and lethal myotonic discharges [55]. Thus, while SBMA was considered as a pure motor neuron disorder, recent research reevaluated this notion and showed that SBMA is a mixed condition involving not only neurogenic denervation, but also myopathic features.
4-3). A myogenic origin for KD related motor neuron degeneration?
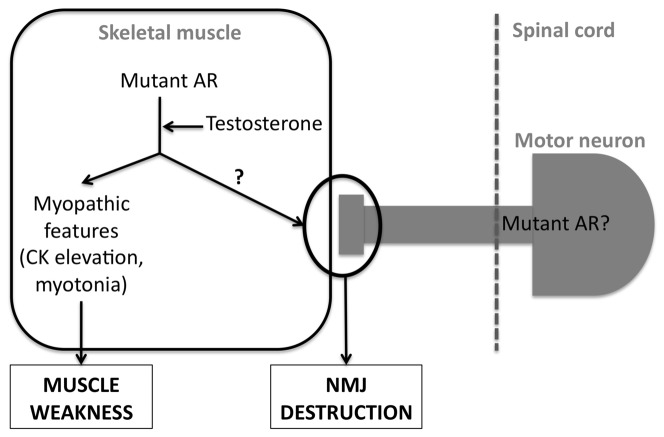
These findings prompted to test the hypothesis that LMN pathology of SBMA patients indeed resulted from a myogenic pathology. This idea is further supported by the crucial role of AR in the skeletal muscle [63–65] and by the intriguing observation that AR accumulates at NMJs [71]. Indeed, the pathology of animal models of SBMA is primarily muscular, with both neurogenic and myogenic pathology occuring long before motor neuron degeneration is observed [55]. Indeed, in most SBMA animal models, there is no detectable motor neuron degeneration [55, 58, 59]. Most importantly, the muscle specific overexpression of a non-expanded AR leads to a SBMA-like phenotype. In this model, the phenotype was androgen dependent, with a pathology strictly affecting males but alleviated by castration [72]. Importantly, mice present with a pathology not restricted to muscle (muscle necrosis) but also show axonal loss and denervation related changes in gene expression but not loss of MN cell bodies [72]. Last, the pathology did not affect females except when treated with testosterone, and the cessation of testosterone treatment in females allows complete recovery of the phenotype [73, 74]. These studies thus provide the convincing proof of principle that SBMA might be a LMN degeneration of muscle origin [74] (figure 3). Further work is needed to delineate the mechanisms linking muscle AR and NMJ denervation, as well as the pathogenic role of polyglutamine expansion.
Figure 3. potential mechanisms involving skeletal muscle in SBMA.
The mutant AR toxicity is unmasked by testosterone. This leads to myopathic features, such as myotonic discharges and elevation of blood CK, contributing to muscle weakness and letality. On the other hand, mutant AR toxicity (or wild type AR overexpression) dismantles NMJs through yet unknown mechanisms. A pathogenic function of mutant AR in motor neurons has also been documented. See text (section 4) for further details.
5. Muscle pathogenic role in amyotrophic lateral sclerosis
5-1). Amyotrophic lateral sclerosis
Amyotrophic lateral sclerosis (ALS) is the most frequent LMN degeneration of adult onset. ALS worldwide incidence is estimated to be of 1–3 new cases per 100 000 individuals, which ranks ALS as the most frequent neurodegenerative disease after Alzheimer’s and Parkinson’s diseases [3]. Like SMA and SBMA, ALS presents with a progressive paralysis affecting first either limb muscles (spinal onset) or cranial muscles (bulbar onset). A difference between ALS and other diseases with LMN degeneration is that LMNs are not the only neuronal cell type to be affected. Indeed, in parallel to LMN involvement, upper motor neurons (corticospinal motor neurons) also degenerate. Most of the ALS patients die within two to five years after the diagnosis but the disease is heterogeneous in its duration and clinical presentation. No current treatment is able to stop the disease process. Riluzole remains the only FDA approved drug and increases the survival of the patients by a few months.
Most ALS cases are not associated with a family history and are, hence, termed sporadic; the remainders (20%) are of genetic origin, generally transmitted with an autosomal dominant inheritance. Sporadic and familial ALS are clinically indistinguishable. Indeed, even in a single affected family, the clinical presentation of the patients may vary, strongly suggesting that genetic and/or environmental cues are of pathological importance [75–77]. Several genes including angiogenin [78], vapb [79], dynactin [80, 81], and more recently tdp43 [82 86], fus [87, 88]and fig4 [89] have been genetically linked to familial forms of ALS, but how these specific mutations lead to ALS is currently unknown [90]. The recent description of a mouse model overexpressing mutant TDP-43 will hopefully lead to new insights in the field [91]. On the contrary, the mechanisms underlying ALS linked to mutations in the sod1 gene, the first and major gene linked to familial ALS in 1993, have been extensively studied [75–77]. It is interesting to note that variations in the SMN1 and 2 genes involved in SMA have been reported to be a risk factor in ALS also [92–97].
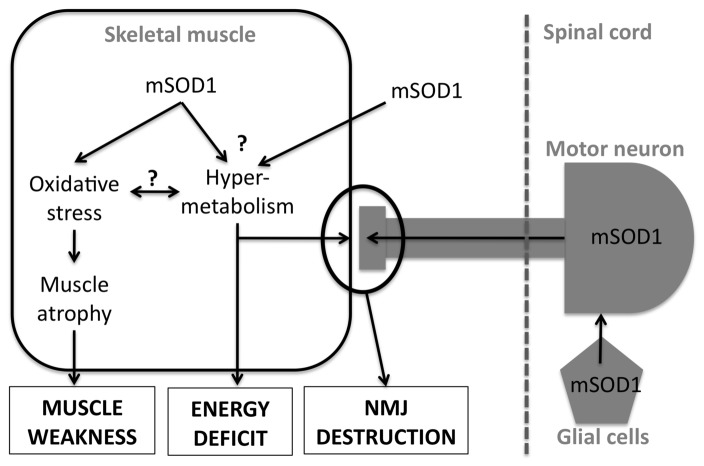
Figure 4. potential mechanisms involving skeletal muscle in SOD1-linked ALS.
Mutant SOD1 expression in skeletal muscle leads to oxidative stress, muscle atrophy and weakness. mSOD1 mice skeletal muscles are also hypermetabolic but whether mSOD1 expression in muscle or in other cell types is responsible of this phenotype is unknown. Muscle hypermetabolism is sufficient to drive NMJ destruction and systemic energy deficit. mSOD1 expression in both motor neurons and glial cells is also involved in the overall ALS phenotype of mSOD1 mice. See text (section 5) for further details.
5-2). Myopathic features in ALS
Myopathic features have been rarely described in ALS and are certainly much less frequent than in SMA or SBMA. However, it was reported that a large number of ALS muscle biopsies displayed myopathic features [98, 99] and moderately elevated CK levels [100]. The extent of these findings is however very limited since similar changes in magnitude are observed upon denervation. Most interestingly, a number of case report studies suggest that even typical ALS patients might initially display myopathic features. For instance, a patient bearing a SOD1 mutation displayed CK elevation before EMG abnormalities [101]. Moreover, ALS patients might present ragged-red muscle fibers, indicative of mitochondrial pathology in their muscles [102]. It is noteworthy that mitochondrial alterations in skeletal muscle might contribute to the pathology [103]. However, this interpretation is complicated by the fact that detectable mitochondrial dysfunction in muscle is a rather late event [104, 105] that could indeed be the result of muscle denervation. Thus, ALS is also associated with myopathic changes, although milder than in SMA or SBMA and mitochondrial defects in muscle might be associated in at least a subset of ALS cases.
5-3). NMJ destruction is the critical pathogenic event in SOD1-linked ALS
Studies in ALS during the last ten years have been focused on the elucidation of SOD1-linked ALS, through the use of transgenic mice overexpressing mutant SOD1 (mSOD1 mice) isoforms. Between 3 and 12 months of age, mSOD1 mice develop muscle weakness linked to muscle denervation and both upper and lower motor neuron degeneration. The first event in this disease process is the destruction of the NMJ, more specifically of the postsynaptic apparatus, followed by axonal degeneration and late-onset degeneration of motor neuron cell body [106, 107]. In years 2000, the accepted paradigm was that neuronal expression of mSOD1 was responsible for motor neuron degeneration, leading to muscle denervation and paralysis. Thus, most research efforts sought to understand the mechanisms leading from mutant SOD1 expression in motor neurons to cell death. Indeed, apoptosis was shown to be the main mechanism of motor neuron death in ALS and multiple signaling pathways are now known to contribute in vivo to the degeneration of motor neuron cell body [108]. However, none of these pathways did seem to contribute to overall survival of mSOD1 mice and all preclinical trials based on interference with cell death pathways only marginally affected mSOD1 mice lifespan Most importantly, clinical trials based on those results, including the recent minocycline trial [109], not only did not improve patient outcome, but in some instances even worsen it.
On top of this, we and others have shown that even a complete rescue of motor neuron cell bodies does not cure mSOD1 mice. For instance, gene ablation of bax, a key player in motor neuron apoptosis, completely rescued mSOD1 mice motor neurons from apoptosis, while only modestly delaying muscle denervation and animal death [110]. Along the same line, sodium valproate, a drug inhibiting epigenetic chromatin remodeling during apoptosis, or an inhibitor of p38MAPK, a protein kinase involved in initiating cell death, are able to rescue the cell bodies but have no effects on muscle denervation and animal lifespan [111, 112]. Last, ablating mSOD1 from motor neurons, while delaying onset modestly, did not cure the pathology [113]. Thus, the primary pathogenic event, determining the survival of the animal, is not motor neuron death itself, but rather the loss of motor neuron/muscle contacts. Hence, preserving motor neuron cell bodies is therapeutically not sufficient since the rescued motor neurons are unable to recreate destroyed NMJs. In this model, motor neuron degeneration represents a late, secondary consequence of synaptic destruction
5-4). Is muscle mutant SOD1 expression sufficient to trigger ALS ?
In SBMA, recent work suggested that the muscle overexpression of AR, even not bearing an expanded polyglutamine tract could lead to the pathology [72]. Could mSOD1 expression in muscle also lead to such a concept in ALS ? Indeed, the transgenic overexpression of mSOD1 was sufficient to induce severe muscle atrophy associated with significant reduction in muscle strength, sarcomere disorganization, significant changes in mitochondria morphology and disposition, and disorganization of the sarcotubular system. The authors involved several signalling pathways, including autophagy and oxidative stress in the deleterious effects of muscle mSOD1 expression. However, while muscle-restricted mSOD1 expression promote spinal cord astrocytosis and inflammation, no motor neuron loss was observed. Thus, in this study, muscle mSOD1 expression was not sufficient to support full blown ALS [114]. A very recent study confirmed and significantly extended this initial work. Wong and Martin created transgenic mice overexpressing either wild type or two mutations in the SOD1 cDNA under the control of the skeletal muscle actin promoter [115]. These authors, in a manner consistent with Dobrowolny and collaborators, found important clues for this overexpression triggering localized oxidative stress and subsequent muscle atrophy. Most interestingly, they found NMJ denervation and animals developped paresis and motor neuron degeneration. In this study, muscle SOD1 expression, either wild type or mutant was sufficient to trigger the full ALS phenotype. It is intriguing to note that there was no difference between wild type and mutant SOD1 overexpression, reminiscent of the effects of wild type AR overexpression in muscle [72].
Intriguingly, the knock-down of mSOD1 in muscle did not appear to be sufficient to slow down the pathology [116, 117]. This suggests either that muscle mSOD1 expression is not key in the pathology, or that even the small remaining amounts of mSOD1 in knocked down muscles were sufficient to lead to their toxic effects, or that the knock down occured too late in animal lifespan to yield its protective potential. In all, muscle appears to be one of the sites of mSOD1 toxicity, although it remains elusive whether NMJ destruction is primarily driven by muscle mSOD1 expression.
5-5). Muscle energy metabolism abnormalities as a cause of NMJ destruction
What could be the mechanisms underlying muscle toxicity to motor neurons in ALS ? We have shown that the neurite outgrowth inhibitor Nogo-A is massively expressed in the skeletal muscle of mSOD1 mice and ALS patients [118–120]. Importantly, Nogo-A ablation increased mSOD1 mice lifespan while its overexpression in muscle fibers lead to NMJ shrinkage [119]. These findings provide the proof of principle that alterations in muscle gene expression are able to modulate the disease process in mSOD1 mice.
From a more general point of view, our work suggests that abnormalities in muscle energy metabolism might the direct cause. We and others have observed that muscles of mSOD1 mice display decreased cellular levels of ATP [121, 122], along with increased expression of mitochondrial uncoupling proteins and of markers of both lipid and carbohydrate use [123, 124]. Indeed, as previously mentioned, mitochondrial dysfunction occured in muscles of mSOD1 mice, and, to a lesser extent, in muscles of ALS patients [103]. These findings suggested that muscles of ALS patients and mSOD1 mice show an exacerbation of their energy metabolism. To determine whether this was sufficient to lead to motorneuron degeneration, we studied the neuromuscular system of mice overexpressing the mitochondrial uncoupling protein 1 in their muscles, as a model of muscle restricted hypermetabolism. These animals displayed age-dependent deterioration of the NMJ that correlated with progressive signs of denervation and a mild late-onset motor neuron pathology [125]. Furthermore, NMJ regeneration and functional recovery were profoundly delayed following injury of the sciatic nerve and crossing these mice with mSOD1 mice exacerbated ALS-like pathology [125]. Thus, a muscle restricted mitochondrial defect is sufficient to generate motor neuron degeneration.
5-5). ALS as a systemic pathology
Muscle hypermetabolism of mSOD1 mice has broad consequences on the overall energetic physiology of these animals, and provides clues for a potential therapeutic strategy. Indeed, mSOD1 mice show body weight deficit as compared to wild types due to an increase in the basal metabolic rate, as a reflect of muscle hypermetabolism [123]. Furthermore, energy metabolism, especially lipid metabolism, was strikingly altered in these animals [126]. We sought to determine whether correcting energy deficit of mSOD1 mice could delay their pathology and fed these mice with a diet enriched in saturated fats. High fat fed mSOD1 mice lived longer and showed reduced muscle denervation associated with improved motor neuron survival, a finding recently confirmed by Mattson’s group in another mSOD1 strain [123, 127].
ALS patients also show abnormalities in their systemic energy homeostasis. Couratier’s group has found an increased energy expenditure in these patients, similar to what observed in mSOD1 mice [128–132]. Most importantly, ALS patients show increased blood lipid levels and hyperlipemia was associated with increased survival [133]and better respiratory capacity in these patients. However, the translation from mSOD1 mice to ALS patients is not straightforward since patients show an important trend to become insulin resistant [134]. In these conditions, the increase in energy intake, in the form of an high fat diet, might precipitate insulin resistance and worsen patient outcome. Potential therapeutic strategies based on nutrition should take this trend to insulin resistance into account.
In all, skeletal muscle is involved in ALS through an increase in its metabolic rate, precipitating systemic defects in energy homeostasis, such as leanness. Muscle might also directly influence NMJ stability through energy metabolism defects and yet unknown signalling pathways (figure 4).
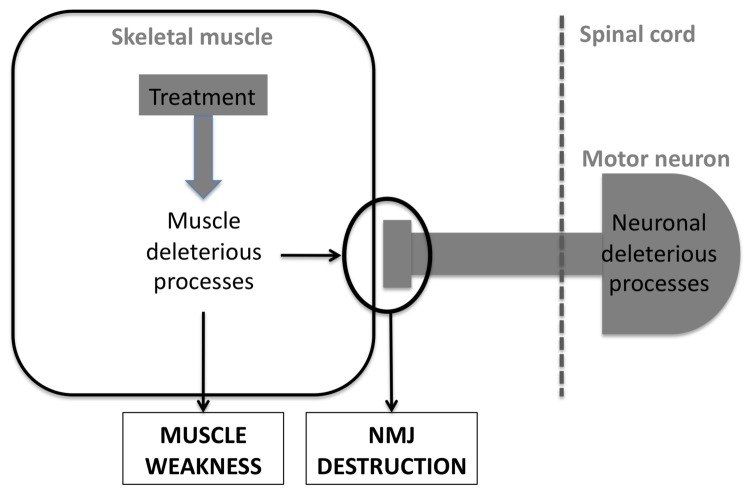
6. Muscle as a drug target in lower motor neuron diseases
The previous results in either SMA, SBMA or ALS point to muscle being an active contributor to these pathologies. Based on these studies, a number of therapeutic strategies have been proposed to target muscle pathology (figure 5).
Figure 5. targeting skeletal muscle in LMN degeneration.
A potential therapeutic treatment for LMN degeneration might target deleterious processes occuring in muscle itself.
In SMA mice, treatment with follistatin, an inhibitor of myostatin [135] that increases muscle mass, was reported to lessen disease severity in SMA mice [136] but not in ALS mice [116]. In SMA mice, but not in ALS mice, follistatin-treated mice performed better than their vehicle-treated littermates. The most widely documented factor that could be beneficial in LMN degeneration through a muscle action is IGF1. In 2002, Antonio Musaro and colleagues generated a transgenic mouse line overexpressing an isoform of IGF1 (termed mIGF1) that is retained locally in skeletal muscle and does not systematically diffuse [137]. Crossing these mice with either mSOD1 mice [138] or SBMA mice [139] potently increased the lifespan of both models. In mSOD1 mice, mIGF1 expression delayed the disease, enhanced survival, stabilized NMJs and had also distant protective effects by decreasing astrocytosis [138]. In SBMA mice, mIGF1 increased AR phosphorylation, promoted the degradation of aggregated AR, rescued the muscle phenotype, increased motor neuron counts and potently increased the lifespan of SBMA mice [139]. These data thus suggest that a pharmacological intervention solely targeted at muscles is able to provide global protection to motor neurons. Indeed, it should be noted that the protection mediated by exercise [140–142], high fat feeding [123, 127], creatine [143] or carnitine [144] in animal models of LMN degeneration might well be due to an action of these treatments on skeletal muscle rather than on motor neurons. Such an indirect effect is suggested by the profound effects of exercise on genes involved in NMJ maintenance [145].
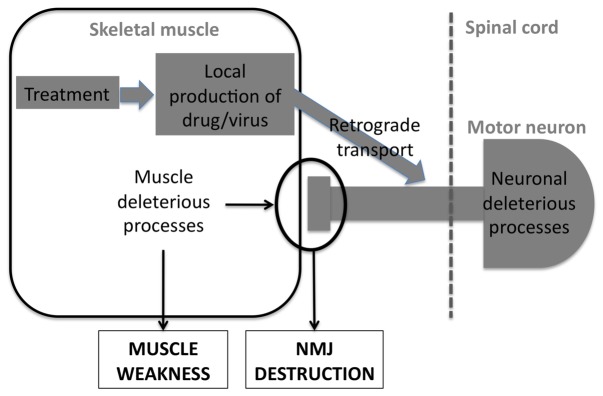
7. Muscle a a delivery route to cure motor neurons in lower motor neuron diseases
While skeletal muscle is involved in at least some of the key pathogenic events in LMN degeneration, one could also consider to use muscle as a way to target motor neurons (figure 6). For instance, local production of neurotrophic factors by the skeletal muscle might sustain neuronal survival and enhance muscle reinnervation. Such an example was provided by the study of Li and collaborators [146] showing that muscle production of GDNF, but not astrocyte derived-GDNF delayed disease onset and slowed down disease progression of mSOD1 mice.
Figure 6. skeletal muscle as a delivery route in LMN degeneration.
Skeletal muscle is also a privilegied route to deliver drugs targeting motor neurons through retrograde transport.
In the same line, retrograde transport of a therapeutic gene through muscle injections of a virus could provide protection to motor neurons. Two different examples provide the proof of concept for such a strategy. First, delivery of IGF1 through AAV vectors in skeletal muscles leads to protection of motor neurons and delayed disease onset. This protective effect might be due to retrograde transport of viral particles in the axon, leading to IGF1 production in motor neurons and autocrine protection of these cells. Alternatively, and as suggested by A. Musaro and colleagues, IGF1 has profound local effects on skeletal muscle that might account for its potential for therapeutics in ALS [138]. A second example of a therapeutic strategy aiming at delivering a neurotrophic factor though skeletal muscle is VEGF. M. Azzouz and collaborators used a lentiviral vector to provide VEGF directly to motor neurons through retrograde transport. They observed that a single injection of a VEGF-expressing lentiviral vector into various muscles delayed onset and slowed progression of in mSOD1 mice [147]. Interestingly, either viral delivery of VEGF or IGF1 slowed disease progression even when treatment was initiated at the onset of disease, suggesting that these strategies might be relevant in ALS patients.
Viral delivery might prove useful not only for providing a neuroprotective factor, but also to restore the causative deficient gene. Indeed, multiple muscular injections of a lentiviral vector expressing SMN restored SMN expression in motor neurons and delayed the pathology of severe SMA mice [148]. Conversely, similar viral strategies might be used to silence the toxic gene when gain of function of the mutant protein is shown to lead to the pathogenic effects. In this line, targeting of SOD1 mutations through siRNA has been achieved by Ralph and collaborators in mSOD1 mice [149] through muscle injections of viral vectors retrogradely transported to motor neurons. This mode of delivery achieved what remains probably the most impressive protective effect in mSOD1 mice. Thus, muscle might not only be a tissue to target, but also a route to deliver drugs or treatments.
8. Conclusion
As a conclusion, when considering all the data discussed above, one may hypothesize that LMN degeneration may be “synaptopathies” rather than « motor neuron diseases ». As such, one should focus on designing treatments to strengthen and stabilize the remaining NMJs and/or to stimulate the generation of newly formed NMJs. To our knowledge, no treatment has been specifically designed to target NMJs in motor neuron diseases until now. It may worth to consider that a drug or treatment targeting the common final pathway of diseases with LMN degeneration i.e. the NMJ, would be equally beneficial for ALS, SMA and SBMA patients.
Lastly, it should be kept in mind that while skeletal muscle is a much more important actor in LMN degeneration than previously anticipated, these affections target motor neurons and involve other cell types, i.e. astrocytes, schwann cells and microglia [75, 150–157]. An efficient therapy should take all these cellular actors into account.
References
- 1.Lunn MR, Wang CH. Spinal muscular atrophy. Lancet. 2008;371:2120–2133. doi: 10.1016/S0140-6736(08)60921-6. [DOI] [PubMed] [Google Scholar]
- 2.Katsuno M, Adachi H, Waza M, Banno H, Suzuki K, Tanaka F, Doyu M, Sobue G. Pathogenesis, animal models and therapeutics in spinal and bulbar muscular atrophy (sbma) Exp Neurol. 2006;200:8–18. doi: 10.1016/j.expneurol.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 3.Brooks BR, Sanjak M, Belden D, Juhasz-Poscine K, Waclawik A. Natural history of amyotrophic lateral sclerosis. In: Brown RHJ, Meininger V, Swash M, editors. Amyotrophic lateral sclerosis. London: Dunitz; 2000. pp. 31–58. [PubMed] [Google Scholar]
- 4.Baloh RH, Rakowicz W, Gardner R, Pestronk A. Frequent atrophic groups with mixed-type myofibers is distinctive to motor neuron syndromes. Muscle Nerve. 2007;36:107–110. doi: 10.1002/mus.20755. [DOI] [PubMed] [Google Scholar]
- 5.Henderson CE, Bloch-Gallego E, Camu W, Gouin A, Lemeulle C, Mettling C. Motoneuron survival factors: Biological roles and therapeutic potential. Neuromuscul Disord. 1993;3:455–458. doi: 10.1016/0960-8966(93)90096-3. [DOI] [PubMed] [Google Scholar]
- 6.Mettling C, Camu W, Henderson CE. Embryonic wing and leg motoneurons have intrinsically different survival properties. Development. 1993;118:1149–1156. doi: 10.1242/dev.118.4.1149. [DOI] [PubMed] [Google Scholar]
- 7.Henderson CE, Camu W, Mettling C, Gouin A, Poulsen K, Karihaloo M, Rullamas J, Evans T, McMahon SB, Armanini MP, et al. Neurotrophins promote motor neuron survival and are present in embryonic limb bud. Nature. 1993;363:266–270. doi: 10.1038/363266a0. [DOI] [PubMed] [Google Scholar]
- 8.Henderson CE, Huchet M, Changeux JP. Denervation increases a neurite-promoting activity in extracts of skeletal muscle. Nature. 1983;302:609–611. doi: 10.1038/302609a0. [DOI] [PubMed] [Google Scholar]
- 9.Madhavan R, Peng HB. Molecular regulation of postsynaptic differentiation at the neuromuscular junction. IUBMB Life. 2005;57:719–730. doi: 10.1080/15216540500338739. [DOI] [PubMed] [Google Scholar]
- 10.Ruegg MA, Bixby JL. Agrin orchestrates synaptic differentiation at the vertebrate neuromuscular junction. Trends Neurosci. 1998;21:22–27. doi: 10.1016/s0166-2236(97)01154-5. [DOI] [PubMed] [Google Scholar]
- 11.Zhang B, Luo S, Wang Q, Suzuki T, Xiong WC, Mei L. Lrp4 serves as a coreceptor of agrin. Neuron. 2008;60:285–297. doi: 10.1016/j.neuron.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim N, Stiegler AL, Cameron TO, Hallock PT, Gomez AM, Huang JH, Hubbard SR, Dustin ML, Burden SJ. Lrp4 is a receptor for agrin and forms a complex with musk. Cell. 2008;135:334–342. doi: 10.1016/j.cell.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin W, Burgess RW, Dominguez B, Pfaff SL, Sanes JR, Lee KF. Distinct roles of nerve and muscle in postsynaptic differentiation of the neuromuscular synapse. Nature. 2001;410:1057–1064. doi: 10.1038/35074025. [DOI] [PubMed] [Google Scholar]
- 14.Yang X, Arber S, William C, Li L, Tanabe Y, Jessell TM, Birchmeier C, Burden SJ. Patterning of muscle acetylcholine receptor gene expression in the absence of motor innervation. Neuron. 2001;30:399–410. doi: 10.1016/s0896-6273(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 15.Lin S, Landmann L, Ruegg MA, Brenner HR. The role of nerve- versus muscle-derived factors in mammalian neuromuscular junction formation. J Neurosci. 2008;28:3333–3340. doi: 10.1523/JNEUROSCI.5590-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vock VM, Ponomareva ON, Rimer M. Evidence for muscle-dependent neuromuscular synaptic site determination in mammals. J Neurosci. 2008;28:3123–3130. doi: 10.1523/JNEUROSCI.5080-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chevessier F, Faraut B, Ravel-Chapuis A, Richard P, Gaudon K, Bauche S, Prioleau C, Herbst R, Goillot E, Ioos C, Azulay JP, Attarian S, Leroy JP, Fournier E, Legay C, Schaeffer L, Koenig J, Fardeau M, Eymard B, Pouget J, Hantai D. Towards the molecular elucidation of congenital myasthenic syndromes: Identification of mutations in musk. Acta Myol. 2005;24:55–59. [PubMed] [Google Scholar]
- 18.Huze C, Bauche S, Richard P, Chevessier F, Goillot E, Gaudon K, Ben Ammar A, Chaboud A, Grosjean I, Lecuyer HA, Bernard V, Rouche A, Alexandri N, Kuntzer T, Fardeau M, Fournier E, Brancaccio A, Ruegg MA, Koenig J, Eymard B, Schaeffer L, Hantai D. Identification of an agrin mutation that causes congenital myasthenia and affects synapse function. Am J Hum Genet. 2009;85:155–167. doi: 10.1016/j.ajhg.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chevessier F, Girard E, Molgo J, Bartling S, Koenig J, Hantai D, Witzemann V. A mouse model for congenital myasthenic syndrome due to musk mutations reveals defects in structure and function of neuromuscular junctions. Hum Mol Genet. 2008;17:3577–3595. doi: 10.1093/hmg/ddn251. [DOI] [PubMed] [Google Scholar]
- 20.Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 21.Echaniz-Laguna A, Miniou P, Bartholdi D, Melki J. The promoters of the survival motor neuron gene (smn) and its copy (smnc) share common regulatory elements. Am J Hum Genet. 1999;64:1365–1370. doi: 10.1086/302372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lefebvre S, Burglen L, Frezal J, Munnich A, Melki J. The role of the smn gene in proximal spinal muscular atrophy. Hum Mol Genet. 1998;7:1531–1536. doi: 10.1093/hmg/7.10.1531. [DOI] [PubMed] [Google Scholar]
- 23.Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A, Dreyfuss G, Melki J. Correlation between severity and smn protein level in spinal muscular atrophy. Nat Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 24.Cifuentes-Diaz C, Frugier T, Tiziano FD, Lacene E, Roblot N, Joshi V, Moreau MH, Melki J. Deletion of murine smn exon 7 directed to skeletal muscle leads to severe muscular dystrophy. J Cell Biol. 2001;152:1107–1114. doi: 10.1083/jcb.152.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cifuentes-Diaz C, Nicole S, Velasco ME, Borra-Cebrian C, Panozzo C, Frugier T, Millet G, Roblot N, Joshi V, Melki J. Neurofilament accumulation at the motor endplate and lack of axonal sprouting in a spinal muscular atrophy mouse model. Hum Mol Genet. 2002;11:1439–1447. doi: 10.1093/hmg/11.12.1439. [DOI] [PubMed] [Google Scholar]
- 26.Frugier T, Tiziano FD, Cifuentes-Diaz C, Miniou P, Roblot N, Dierich A, Le Meur M, Melki J. Nuclear targeting defect of smn lacking the c-terminus in a mouse model of spinal muscular atrophy. Hum Mol Genet. 2000;9:849–858. doi: 10.1093/hmg/9.5.849. [DOI] [PubMed] [Google Scholar]
- 27.Gubitz AK, Feng W, Dreyfuss G. The smn complex. Exp Cell Res. 2004;296:51–56. doi: 10.1016/j.yexcr.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 28.Fischer U, Liu Q, Dreyfuss G. The smn-sip1 complex has an essential role in spliceosomal snrnp biogenesis. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 29.Liu Q, Fischer U, Wang F, Dreyfuss G. The spinal muscular atrophy disease gene product, smn, and its associated protein sip1 are in a complex with spliceosomal snrnp proteins. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- 30.Meister G, Hannus S, Plottner O, Baars T, Hartmann E, Fakan S, Laggerbauer B, Fischer U. Smnrp is an essential pre-mrna splicing factor required for the formation of the mature spliceosome. EMBO J. 2001;20:2304–2314. doi: 10.1093/emboj/20.9.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olaso R, Joshi V, Fernandez J, Roblot N, Courageot S, Bonnefont JP, Melki J. Activation of rna metabolism-related genes in mouse but not human tissues deficient in smn. Physiol Genomics. 2006;24:97–104. doi: 10.1152/physiolgenomics.00134.2005. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G. Smn deficiency causes tissue-specific perturbations in the repertoire of snrnas and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vitte JM, Davoult B, Roblot N, Mayer M, Joshi V, Courageot S, Tronche F, Vadrot J, Moreau MH, Kemeny F, Melki J. Deletion of murine smn exon 7 directed to liver leads to severe defect of liver development associated with iron overload. Am J Pathol. 2004;165:1731–1741. doi: 10.1016/S0002-9440(10)63428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shafey D, Cote PD, Kothary R. Hypomorphic smn knockdown c2c12 myoblasts reveal intrinsic defects in myoblast fusion and myotube morphology. Exp Cell Res. 2005;311:49–61. doi: 10.1016/j.yexcr.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 35.Chang HC, Dimlich DN, Yokokura T, Mukherjee A, Kankel MW, Sen A, Sridhar V, Fulga TA, Hart AC, Van Vactor D, Artavanis-Tsakonas S. Modeling spinal muscular atrophy in drosophila. PLoS One. 2008;3:e3209. doi: 10.1371/journal.pone.0003209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arnold AS, Gueye M, Guettier-Sigrist S, Courdier-Fruh I, Coupin G, Poindron P, Gies JP. Reduced expression of nicotinic achrs in myotubes from spinal muscular atrophy i patients. Lab Invest. 2004;84:1271–1278. doi: 10.1038/labinvest.3700163. [DOI] [PubMed] [Google Scholar]
- 37.Martinez-Hernandez R, Soler-Botija C, Also E, Alias L, Caselles L, Gich I, Bernal S, Tizzano EF. The developmental pattern of myotubes in spinal muscular atrophy indicates prenatal delay of muscle maturation. J Neuropathol Exp Neurol. 2009;68:474–481. doi: 10.1097/NEN.0b013e3181a10ea1. [DOI] [PubMed] [Google Scholar]
- 38.Muqit MM, Moss J, Sewry C, Lane RJ. Phenotypic variability in siblings with type iii spinal muscular atrophy. J Neurol Neurosurg Psychiatry. 2004;75:1762–1764. doi: 10.1136/jnnp.2003.018614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rajendra TK, Gonsalvez GB, Walker MP, Shpargel KB, Salz HK, Matera AG. A drosophila melanogaster model of spinal muscular atrophy reveals a function for smn in striated muscle. J Cell Biol. 2007;176:831–841. doi: 10.1083/jcb.200610053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walker MP, Rajendra TK, Saieva L, Fuentes JL, Pellizzoni L, Matera AG. Smn complex localizes to the sarcomeric z-disc and is a proteolytic target of calpain. Hum Mol Genet. 2008;17:3399–3410. doi: 10.1093/hmg/ddn234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rossoll W, Jablonka S, Andreassi C, Kroning AK, Karle K, Monani UR, Sendtner M. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mrna in growth cones of motoneurons. J Cell Biol. 2003;163:801–812. doi: 10.1083/jcb.200304128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kariya S, Park GH, Maeno-Hikichi Y, Leykekhman O, Lutz C, Arkovitz MS, Landmesser LT, Monani UR. Reduced smn protein impairs maturation of the neuromuscular junctions in mouse models of spinal muscular atrophy. Hum Mol Genet. 2008;17:2552–2569. doi: 10.1093/hmg/ddn156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kong L, Wang X, Choe DW, Polley M, Burnett BG, Bosch-Marce M, Griffin JW, Rich MM, Sumner CJ. Impaired synaptic vesicle release and immaturity of neuromuscular junctions in spinal muscular atrophy mice. J Neurosci. 2009;29:842–851. doi: 10.1523/JNEUROSCI.4434-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murray LM, Comley LH, Thomson D, Parkinson N, Talbot K, Gillingwater TH. Selective vulnerability of motor neurons and dissociation of pre- and post-synaptic pathology at the neuromuscular junction in mouse models of spinal muscular atrophy. Hum Mol Genet. 2008;17:949–962. doi: 10.1093/hmg/ddm367. [DOI] [PubMed] [Google Scholar]
- 45.Briese M, Esmaeili B, Fraboulet S, Burt EC, Christodoulou S, Towers PR, Davies KE, Sattelle DB. Deletion of smn-1, the caenorhabditis elegans ortholog of the spinal muscular atrophy gene, results in locomotor dysfunction and reduced lifespan. Hum Mol Genet. 2009;18:97–104. doi: 10.1093/hmg/ddn320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gavrilina TO, McGovern VL, Workman E, Crawford TO, Gogliotti RG, DiDonato CJ, Monani UR, Morris GE, Burghes AH. Neuronal smn expression corrects spinal muscular atrophy in severe sma mice while muscle-specific smn expression has no phenotypic effect. Hum Mol Genet. 2008;17:1063–1075. doi: 10.1093/hmg/ddm379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in x-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 48.Suzuki K, Katsuno M, Banno H, Takeuchi Y, Atsuta N, Ito M, Watanabe H, Yamashita F, Hori N, Nakamura T, Hirayama M, Tanaka F, Sobue G. Cag repeat size correlates to electrophysiological motor and sensory phenotypes in sbma. Brain. 2008;131:229–239. doi: 10.1093/brain/awm289. [DOI] [PubMed] [Google Scholar]
- 49.Rhodes LE, Freeman BK, Auh S, Kokkinis AD, La Pean A, Chen C, Lehky TJ, Shrader JA, Levy EW, Harris-Love M, Di Prospero NA, Fischbeck KH. Clinical features of spinal and bulbar muscular atrophy. Brain. 2009 doi: 10.1093/brain/awp258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmidt BJ, Greenberg CR, Allingham-Hawkins DJ, Spriggs EL. Expression of x-linked bulbospinal muscular atrophy (kennedy disease) in two homozygous women. Neurology. 2002;59:770–772. doi: 10.1212/wnl.59.5.770. [DOI] [PubMed] [Google Scholar]
- 51.Sobue G, Doyu M, Kachi T, Yasuda T, Mukai E, Kumagai T, Mitsuma T. Subclinical phenotypic expressions in heterozygous females of x-linked recessive bulbospinal neuronopathy. J Neurol Sci. 1993;117:74–78. doi: 10.1016/0022-510x(93)90157-t. [DOI] [PubMed] [Google Scholar]
- 52.Atsuta N, Watanabe H, Ito M, Banno H, Suzuki K, Katsuno M, Tanaka F, Tamakoshi A, Sobue G. Natural history of spinal and bulbar muscular atrophy (sbma): A study of 223 japanese patients. Brain. 2006;129:1446–1455. doi: 10.1093/brain/awl096. [DOI] [PubMed] [Google Scholar]
- 53.Arbizu T, Santamaria J, Gomez JM, Quilez A, Serra JP. A family with adult spinal and bulbar muscular atrophy, x-linked inheritance and associated testicular failure. J Neurol Sci. 1983;59:371–382. doi: 10.1016/0022-510x(83)90022-9. [DOI] [PubMed] [Google Scholar]
- 54.Casella R, Maduro MR, Lipshultz LI, Lamb DJ. Significance of the polyglutamine tract polymorphism in the androgen receptor. Urology. 2001;58:651–656. doi: 10.1016/s0090-4295(01)01401-7. [DOI] [PubMed] [Google Scholar]
- 55.Yu Z, Dadgar N, Albertelli M, Gruis K, Jordan C, Robins DM, Lieberman AP. Androgen-dependent pathology demonstrates myopathic contribution to the kennedy disease phenotype in a mouse knock-in model. J Clin Invest. 2006;116:2663–2672. doi: 10.1172/JCI28773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu Z, Dadgar N, Albertelli M, Scheller A, Albin RL, Robins DM, Lieberman AP. Abnormalities of germ cell maturation and sertoli cell cytoskeleton in androgen receptor 113 cag knock-in mice reveal toxic effects of the mutant protein. Am J Pathol. 2006;168:195–204. doi: 10.2353/ajpath.2006.050619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Albertelli MA, Scheller A, Brogley M, Robins DM. Replacing the mouse androgen receptor with human alleles demonstrates glutamine tract length-dependent effects on physiology and tumorigenesis in mice. Mol Endocrinol. 2006;20:1248–1260. doi: 10.1210/me.2006-0021. [DOI] [PubMed] [Google Scholar]
- 58.Chevalier-Larsen ES, O’Brien CJ, Wang H, Jenkins SC, Holder L, Lieberman AP, Merry DE. Castration restores function and neurofilament alterations of aged symptomatic males in a transgenic mouse model of spinal and bulbar muscular atrophy. J Neurosci. 2004;24:4778–4786. doi: 10.1523/JNEUROSCI.0808-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Katsuno M, Adachi H, Kume A, Li M, Nakagomi Y, Niwa H, Sang C, Kobayashi Y, Doyu M, Sobue G. Testosterone reduction prevents phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Neuron. 2002;35:843–854. doi: 10.1016/s0896-6273(02)00834-6. [DOI] [PubMed] [Google Scholar]
- 60.Katsuno M, Adachi H, Inukai A, Sobue G. Transgenic mouse models of spinal and bulbar muscular atrophy (sbma) Cytogenet Genome Res. 2003;100:243–251. doi: 10.1159/000072860. [DOI] [PubMed] [Google Scholar]
- 61.Katsuno M, Adachi H, Doyu M, Minamiyama M, Sang C, Kobayashi Y, Inukai A, Sobue G. Leuprorelin rescues polyglutamine-dependent phenotypes in a transgenic mouse model of spinal and bulbar muscular atrophy. Nat Med. 2003;9:768–773. doi: 10.1038/nm878. [DOI] [PubMed] [Google Scholar]
- 62.Lee HJ, Chang C. Recent advances in androgen receptor action. Cell Mol Life Sci. 2003;60:1613–1622. doi: 10.1007/s00018-003-2309-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mooradian AD, Morley JE, Korenman SG. Biological actions of androgens. Endocr Rev. 1987;8:1–28. doi: 10.1210/edrv-8-1-1. [DOI] [PubMed] [Google Scholar]
- 64.McPhaul MJ. Androgen receptor mutations and androgen insensitivity. Mol Cell Endocrinol. 2002;198:61–67. doi: 10.1016/s0303-7207(02)00369-6. [DOI] [PubMed] [Google Scholar]
- 65.McPhaul MJ. Molecular defects of the androgen receptor. Recent Prog Horm Res. 2002;57:181–194. doi: 10.1210/rp.57.1.181. [DOI] [PubMed] [Google Scholar]
- 66.Ophoff J, Van Proeyen K, Callewaert F, De Gendt K, De Bock K, Vanden Bosch A, Verhoeven G, Hespel P, Vanderschueren D. Androgen signaling in myocytes contributes to the maintenance of muscle mass and fiber type regulation but not to muscle strength or fatigue. Endocrinology. 2009;150:3558–3566. doi: 10.1210/en.2008-1509. [DOI] [PubMed] [Google Scholar]
- 67.MacLean HE, Chiu WS, Notini AJ, Axell AM, Davey RA, McManus JF, Ma C, Plant DR, Lynch GS, Zajac JD. Impaired skeletal muscle development and function in male, but not female, genomic androgen receptor knockout mice. FASEB J. 2008;22:2676–2689. doi: 10.1096/fj.08-105726. [DOI] [PubMed] [Google Scholar]
- 68.Banno H, Katsuno M, Suzuki K, Takeuchi Y, Kawashima M, Suga N, Takamori M, Ito M, Nakamura T, Matsuo K, Yamada S, Oki Y, Adachi H, Minamiyama M, Waza M, Atsuta N, Watanabe H, Fujimoto Y, Nakashima T, Tanaka F, Doyu M, Sobue G. Phase 2 trial of leuprorelin in patients with spinal and bulbar muscular atrophy. Ann Neurol. 2009;65:140–150. doi: 10.1002/ana.21540. [DOI] [PubMed] [Google Scholar]
- 69.Soraru G, D’Ascenzo C, Polo A, Palmieri A, Baggio L, Vergani L, Gellera C, Moretto G, Pegoraro E, Angelini C. Spinal and bulbar muscular atrophy: Skeletal muscle pathology in male patients and heterozygous females. J Neurol Sci. 2008;264:100–105. doi: 10.1016/j.jns.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 70.Echaniz-Laguna A, Rousso E, Anheim M, Cossee M, Tranchant C. A family with early-onset and rapidly progressive x-linked spinal and bulbar muscular atrophy. Neurology. 2005;64:1458–1460. doi: 10.1212/01.WNL.0000158617.41819.F3. [DOI] [PubMed] [Google Scholar]
- 71.Monks DA, O’Bryant EL, Jordan CL. Androgen receptor immunoreactivity in skeletal muscle: Enrichment at the neuromuscular junction. J Comp Neurol. 2004;473:59–72. doi: 10.1002/cne.20088. [DOI] [PubMed] [Google Scholar]
- 72.Monks DA, Johansen JA, Mo K, Rao P, Eagleson B, Yu Z, Lieberman AP, Breedlove SM, Jordan CL. Overexpression of wild-type androgen receptor in muscle recapitulates polyglutamine disease. Proc Natl Acad Sci U S A. 2007;104:18259–18264. doi: 10.1073/pnas.0705501104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Johansen JA, Yu Z, Mo K, Monks DA, Lieberman AP, Breedlove SM, Jordan CL. Recovery of function in a myogenic mouse model of spinal bulbar muscular atrophy. Neurobiol Dis. 2009;34:113–120. doi: 10.1016/j.nbd.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jordan CL, Lieberman AP. Spinal and bulbar muscular atrophy: A motoneuron or muscle disease? Curr Opin Pharmacol. 2008;8:752–758. doi: 10.1016/j.coph.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boillee S, Vande Velde C, Cleveland DW. Als: A disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 76.Gonzalez de Aguilar JL, Echaniz-Laguna A, Fergani A, Rene F, Meininger V, Loeffler JP, Dupuis L. Amyotrophic lateral sclerosis: All roads lead to rome. J Neurochem. 2007 doi: 10.1111/j.1471-4159.2006.04408.x. [DOI] [PubMed] [Google Scholar]
- 77.Dupuis L, Loeffler JP. Neuromuscular junction destruction during amyotrophic lateral sclerosis: Insights from transgenic models. Curr Opin Pharmacol. 2009;9:341–346. doi: 10.1016/j.coph.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 78.Greenway MJ, Andersen PM, Russ C, Ennis S, Cashman S, Donaghy C, Patterson V, Swingler R, Kieran D, Prehn J, Morrison KE, Green A, Acharya KR, Brown RH, Jr, Hardiman O. Ang mutations segregate with familial and ‘sporadic’ amyotrophic lateral sclerosis. Nat Genet. 2006;38:411–413. doi: 10.1038/ng1742. [DOI] [PubMed] [Google Scholar]
- 79.Nishimura AL, Mitne-Neto M, Silva HC, Richieri-Costa A, Middleton S, Cascio D, Kok F, Oliveira JR, Gillingwater T, Webb J, Skehel P, Zatz M. A mutation in the vesicle-trafficking protein vapb causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am J Hum Genet. 2004;75:822–831. doi: 10.1086/425287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Puls I, Jonnakuty C, LaMonte BH, Holzbaur EL, Tokito M, Mann E, Floeter MK, Bidus K, Drayna D, Oh SJ, Brown RH, Jr, Ludlow CL, Fischbeck KH. Mutant dynactin in motor neuron disease. Nat Genet. 2003;33:455–456. doi: 10.1038/ng1123. [DOI] [PubMed] [Google Scholar]
- 81.Munch C, Sedlmeier R, Meyer T, Homberg V, Sperfeld AD, Kurt A, Prudlo J, Peraus G, Hanemann CO, Stumm G, Ludolph AC. Point mutations of the p150 subunit of dynactin (dctn1) gene in als. Neurology. 2004;63:724–726. doi: 10.1212/01.wnl.0000134608.83927.b1. [DOI] [PubMed] [Google Scholar]
- 82.Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, Bouchard JP, Lacomblez L, Pochigaeva K, Salachas F, Pradat PF, Camu W, Meininger V, Dupre N, Rouleau GA. Tardbp mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 83.Kuhnlein P, Sperfeld AD, Vanmassenhove B, Van Deerlin V, Lee VM, Trojanowski JQ, Kretzschmar HA, Ludolph AC, Neumann M. Two german kindreds with familial amyotrophic lateral sclerosis due to tardbp mutations. Arch Neurol. 2008;65:1185–1189. doi: 10.1001/archneur.65.9.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rutherford NJ, Zhang YJ, Baker M, Gass JM, Finch NA, Xu YF, Stewart H, Kelley BJ, Kuntz K, Crook RJ, Sreedharan J, Vance C, Sorenson E, Lippa C, Bigio EH, Geschwind DH, Knopman DS, Mitsumoto H, Petersen RC, Cashman NR, Hutton M, Shaw CE, Boylan KB, Boeve B, Graff-Radford NR, Wszolek ZK, Caselli RJ, Dickson DW, Mackenzie IR, Petrucelli L, Rademakers R. Novel mutations in tardbp (tdp-43) in patients with familial amyotrophic lateral sclerosis. PLoS Genet. 2008;4:e1000193. doi: 10.1371/journal.pgen.1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, Baralle F, de Belleroche J, Mitchell JD, Leigh PN, Al-Chalabi A, Miller CC, Nicholson G, Shaw CE. Tdp-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yokoseki A, Shiga A, Tan CF, Tagawa A, Kaneko H, Koyama A, Eguchi H, Tsujino A, Ikeuchi T, Kakita A, Okamoto K, Nishizawa M, Takahashi H, Onodera O. Tdp-43 mutation in familial amyotrophic lateral sclerosis. Ann Neurol. 2008;63:538–542. doi: 10.1002/ana.21392. [DOI] [PubMed] [Google Scholar]
- 87.Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis P, Rouleau GA, Hosler BA, Cortelli P, de Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance MA, Yan J, Ticozzi N, Siddique T, McKenna-Yasek D, Sapp PC, Horvitz HR, Landers JE, Brown RH., Jr Mutations in the fus/tls gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 88.Ticozzi N, Silani V, Leclerc AL, Keagle P, Gellera C, Ratti A, Taroni F, Kwiatkowski TJ, Jr, McKenna-Yasek DM, Sapp PC, Brown RH, Jr, Landers JE. Analysis of fus gene mutation in familial amyotrophic lateral sclerosis within an italian cohort. Neurology. 2009 doi: 10.1212/WNL.0b013e3181bbff05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chow CY, Landers JE, Bergren SK, Sapp PC, Grant AE, Jones JM, Everett L, Lenk GM, McKenna-Yasek DM, Weisman LS, Figlewicz D, Brown RH, Meisler MH. Deleterious variants of fig4, a phosphoinositide phosphatase, in patients with als. Am J Hum Genet. 2009;84:85–88. doi: 10.1016/j.ajhg.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lagier-Tourenne C, Cleveland DW. Rethinking als: The fus about tdp-43. Cell. 2009;136:1001–1004. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wegorzewska I, Bell S, Cairns NJ, Miller TM, Baloh RH. Tdp-43 mutant transgenic mice develop features of als and frontotemporal lobar degeneration. Proc Natl Acad Sci U S A. 2009;106:18809–18814. doi: 10.1073/pnas.0908767106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Corcia P, Camu W, Praline J, Gordon PH, Vourch P, Andres C. The importance of the smn genes in the genetics of sporadic als. Amyotroph Lateral Scler. 2009;10:436–440. doi: 10.3109/17482960902759162. [DOI] [PubMed] [Google Scholar]
- 93.Corcia P, Camu W, Halimi JM, Vourc’h P, Antar C, Vedrine S, Giraudeau B, de Toffol B, Andres CR. Smn1 gene, but not smn2, is a risk factor for sporadic als. Neurology. 2006;67:1147–1150. doi: 10.1212/01.wnl.0000233830.85206.1e. [DOI] [PubMed] [Google Scholar]
- 94.Corcia P, Mayeux-Portas V, Khoris J, de Toffol B, Autret A, Muh JP, Camu W, Andres C. Abnormal smn1 gene copy number is a susceptibility factor for amyotrophic lateral sclerosis. Ann Neurol. 2002;51:243–246. doi: 10.1002/ana.10104. [DOI] [PubMed] [Google Scholar]
- 95.Veldink JH, Kalmijn S, Van der Hout AH, Lemmink HH, Groeneveld GJ, Lummen C, Scheffer H, Wokke JH, Van den Berg LH. Smn genotypes producing less smn protein increase susceptibility to and severity of sporadic als. Neurology. 2005;65:820–825. doi: 10.1212/01.wnl.0000174472.03292.dd. [DOI] [PubMed] [Google Scholar]
- 96.Veldink JH, van den Berg LH, Cobben JM, Stulp RP, De Jong JM, Vogels OJ, Baas F, Wokke JH, Scheffer H. Homozygous deletion of the survival motor neuron 2 gene is a prognostic factor in sporadic als. Neurology. 2001;56:749–752. doi: 10.1212/wnl.56.6.749. [DOI] [PubMed] [Google Scholar]
- 97.Echaniz-Laguna A, Guiraud-Chaumeil C, Tranchant C, Reeber A, Melki J, Warter JM. Homozygous exon 7 deletion of the smn centromeric gene (smn2): A potential susceptibility factor for adult-onset lower motor neuron disease. J Neurol. 2002;249:290–293. doi: 10.1007/s004150200007. [DOI] [PubMed] [Google Scholar]
- 98.Iwasaki Y, Sugimoto H, Ikeda K, Takamiya K, Shiojima T, Kinoshita M. Muscle morphometry in amyotrophic lateral sclerosis. Int J Neurosci. 1991;58:165–170. doi: 10.3109/00207459108985432. [DOI] [PubMed] [Google Scholar]
- 99.Achari AN, Anderson MS. Myopathic changes in amyotrophic lateral sclerosis. Pathologic analysis of muscle biopsy changes in 111 cases. Neurology. 1974;24:477–481. doi: 10.1212/wnl.24.5.477. [DOI] [PubMed] [Google Scholar]
- 100.Amrit AN, Anderson MS. Serum creatine phosphokinase in amyotrophic lateral sclerosis. Correlation with sex, duration, and skeletal muscle biopsy. Neurology. 1974;24:834–837. doi: 10.1212/wnl.24.9.834. [DOI] [PubMed] [Google Scholar]
- 101.Stewart HG, Mackenzie IR, Eisen A, Brannstrom T, Marklund SL, Andersen PM. Clinicopathological phenotype of als with a novel g72c sod1 gene mutation mimicking a myopathy. Muscle Nerve. 2006;33:701–706. doi: 10.1002/mus.20495. [DOI] [PubMed] [Google Scholar]
- 102.Hirano M, Angelini C, Montagna P, Hays AP, Tanji K, Mitsumoto H, Gordon PH, Naini AB, DiMauro S, Rowland LP. Amyotrophic lateral sclerosis with ragged-red fibers. Arch Neurol. 2008;65:403–406. doi: 10.1001/archneurol.2007.65. [DOI] [PubMed] [Google Scholar]
- 103.Dupuis L, Gonzalez de Aguilar JL, Oudart H, de Tapia M, Barbeito L, Loeffler JP. Mitochondria in amyotrophic lateral sclerosis: A trigger and a target. Neurodegener Dis. 2004;1:245–254. doi: 10.1159/000085063. [DOI] [PubMed] [Google Scholar]
- 104.Echaniz-Laguna A, Zoll J, Ponsot E, N’Guessan B, Tranchant C, Loeffler JP, Lampert E. Muscular mitochondrial function in amyotrophic lateral sclerosis is progressively altered as the disease develops: A temporal study in man. Exp Neurol. 2006;198:25–30. doi: 10.1016/j.expneurol.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 105.Echaniz-Laguna A, Zoll J, Ribera F, Tranchant C, Warter JM, Lonsdorfer J, Lampert E. Mitochondrial respiratory chain function in skeletal muscle of als patients. Ann Neurol. 2002;52:623–627. doi: 10.1002/ana.10357. [DOI] [PubMed] [Google Scholar]
- 106.Fischer LR, Culver DG, Tennant P, Davis AA, Wang M, Castellano-Sanchez A, Khan J, Polak MA, Glass JD. Amyotrophic lateral sclerosis is a distal axonopathy: Evidence in mice and man. Exp Neurol. 2004;185:232–240. doi: 10.1016/j.expneurol.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 107.Pun S, Santos AF, Saxena S, Xu L, Caroni P. Selective vulnerability and pruning of phasic motoneuron axons in motoneuron disease alleviated by cntf. Nat Neurosci. 2006;9:408–419. doi: 10.1038/nn1653. [DOI] [PubMed] [Google Scholar]
- 108.Guegan C, Przedborski S. Programmed cell death in amyotrophic lateral sclerosis. J Clin Invest. 2003;111:153–161. doi: 10.1172/JCI17610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gordon PH, Moore DH, Miller RG, Florence JM, Verheijde JL, Doorish C, Hilton JF, Spitalny GM, MacArthur RB, Mitsumoto H, Neville HE, Boylan K, Mozaffar T, Belsh JM, Ravits J, Bedlack RS, Graves MC, McCluskey LF, Barohn RJ, Tandan R. Efficacy of minocycline in patients with amyotrophic lateral sclerosis: A phase iii randomised trial. Lancet Neurol. 2007;6:1045–1053. doi: 10.1016/S1474-4422(07)70270-3. [DOI] [PubMed] [Google Scholar]
- 110.Gould TW, Buss RR, Vinsant S, Prevette D, Sun W, Knudson CM, Milligan CE, Oppenheim RW. Complete dissociation of motor neuron death from motor dysfunction by bax deletion in a mouse model of als. J Neurosci. 2006;26:8774–8786. doi: 10.1523/JNEUROSCI.2315-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rouaux C, Panteleeva I, Rene F, Gonzalez de Aguilar JL, Echaniz-Laguna A, Dupuis L, Menger Y, Boutillier AL, Loeffler JP. Sodium valproate exerts neuroprotective effects in vivo through creb-binding protein-dependent mechanisms but does not improve survival in an amyotrophic lateral sclerosis mouse model. J Neurosci. 2007;27:5535–5545. doi: 10.1523/JNEUROSCI.1139-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dewil M, dela Cruz VF, Van Den Bosch L, Robberecht W. Inhibition of p38 mitogen activated protein kinase activation and mutant sod1(g93a)-induced motor neuron death. Neurobiol Dis. 2007;26:332–341. doi: 10.1016/j.nbd.2006.12.023. [DOI] [PubMed] [Google Scholar]
- 113.Boillee S, Yamanaka K, Lobsiger CS, Copeland NG, Jenkins NA, Kassiotis G, Kollias G, Cleveland DW. Onset and progression in inherited als determined by motor neurons and microglia. Science. 2006;312:1389–1392. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- 114.Dobrowolny G, Aucello M, Rizzuto E, Beccafico S, Mammucari C, Bonconpagni S, Belia S, Wannenes F, Nicoletti C, Del Prete Z, Rosenthal N, Molinaro M, Protasi F, Fano G, Sandri M, Musaro A. Skeletal muscle is a primary target of sod1g93a-mediated toxicity. Cell Metab. 2008;8:425–436. doi: 10.1016/j.cmet.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 115.Wong M, Martin LJ. Skeletal muscle-restricted expression of human sod1 causes motor neuron degeneration in transgenic mice. Hum Mol Genet. doi: 10.1093/hmg/ddq106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Miller TM, Kim SH, Yamanaka K, Hester M, Umapathi P, Arnson H, Rizo L, Mendell JR, Gage FH, Cleveland DW, Kaspar BK. Gene transfer demonstrates that muscle is not a primary target for non-cell-autonomous toxicity in familial amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2006;103:19546–19551. doi: 10.1073/pnas.0609411103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Towne C, Raoul C, Schneider BL, Aebischer P. Systemic aav6 delivery mediating rna interference against sod1: Neuromuscular transduction does not alter disease progression in fals mice. Mol Ther. 2008;16:1018–1025. doi: 10.1038/mt.2008.73. [DOI] [PubMed] [Google Scholar]
- 118.Dupuis L, Gonzalez de Aguilar JL, di Scala F, Rene F, de Tapia M, Pradat PF, Lacomblez L, Seihlan D, Prinjha R, Walsh FS, Meininger V, Loeffler JP. Nogo provides a molecular marker for diagnosis of amyotrophic lateral sclerosis. Neurobiol Dis. 2002;10:358–365. doi: 10.1006/nbdi.2002.0522. [DOI] [PubMed] [Google Scholar]
- 119.Jokic N, Gonzalez de Aguilar JL, Dimou L, Lin S, Fergani A, Ruegg MA, Schwab ME, Dupuis L, Loeffler JP. The neurite outgrowth inhibitor nogo-a promotes denervation in an amyotrophic lateral sclerosis model. EMBO Rep. 2006;7:1162–1167. doi: 10.1038/sj.embor.7400826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jokic N, Gonzalez de Aguilar JL, Pradat PF, Dupuis L, Echaniz-Laguna A, Muller A, Dubourg O, Seilhean D, Hauw JJ, Loeffler JP, Meininger V. Nogo expression in muscle correlates with amyotrophic lateral sclerosis severity. Ann Neurol. 2005;57:553–556. doi: 10.1002/ana.20420. [DOI] [PubMed] [Google Scholar]
- 121.Derave W, Van Den Bosch L, Lemmens G, Eijnde BO, Robberecht W, Hespel P. Skeletal muscle properties in a transgenic mouse model for amyotrophic lateral sclerosis: Effects of creatine treatment. Neurobiol Dis. 2003;13:264–272. doi: 10.1016/s0969-9961(03)00041-x. [DOI] [PubMed] [Google Scholar]
- 122.Dupuis L, di Scala F, Rene F, de Tapia M, Oudart H, Pradat PF, Meininger V, Loeffler JP. Up-regulation of mitochondrial uncoupling protein 3 reveals an early muscular metabolic defect in amyotrophic lateral sclerosis. Faseb J. 2003;17:2091–2093. doi: 10.1096/fj.02-1182fje. [DOI] [PubMed] [Google Scholar]
- 123.Dupuis L, Oudart H, Rene F, Gonzalez de Aguilar JL, Loeffler JP. Evidence for defective energy homeostasis in amyotrophic lateral sclerosis: Benefit of a high-energy diet in a transgenic mouse model. Proc Natl Acad Sci U S A. 2004;101:11159–11164. doi: 10.1073/pnas.0402026101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gonzalez de Aguilar JL, Niederhauser-Wiederkehr C, Halter B, De Tapia M, Di Scala F, Demougin P, Dupuis L, Primig M, Meininger V, Loeffler JP. Gene profiling of skeletal muscle in an amyotrophic lateral sclerosis mouse model. Physiol Genomics. 2008;32:207–218. doi: 10.1152/physiolgenomics.00017.2007. [DOI] [PubMed] [Google Scholar]
- 125.Dupuis L, Gonzalez de Aguilar JL, Echaniz-Laguna A, Eschbach J, Rene F, Oudart H, Halter B, Huze C, Schaeffer L, Bouillaud F, Loeffler JP. Muscle mitochondrial uncoupling dismantles neuromuscular junction and triggers distal degeneration of motor neurons. PLoS One. 2009;4:e5390. doi: 10.1371/journal.pone.0005390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fergani A, Oudart H, Gonzalez De Aguilar JL, Fricker B, Rene F, Hocquette JF, Meininger V, Dupuis L, Loeffler JP. Increased peripheral lipid clearance in an animal model of amyotrophic lateral sclerosis. J Lipid Res. 2007;48:1571–1580. doi: 10.1194/jlr.M700017-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mattson MP, Cutler RG, Camandola S. Energy intake and amyotrophic lateral sclerosis. Neuromolecular Med. 2007;9:17–20. doi: 10.1385/nmm:9:1:17. [DOI] [PubMed] [Google Scholar]
- 128.Desport JC, Preux PM, Truong TC, Vallat JM, Sautereau D, Couratier P. Nutritional status is a prognostic factor for survival in als patients. Neurology. 1999;53:1059–1063. doi: 10.1212/wnl.53.5.1059. [DOI] [PubMed] [Google Scholar]
- 129.Desport JC, Preux PM, Truong CT, Courat L, Vallat JM, Couratier P. Nutritional assessment and survival in als patients. Amyotroph Lateral Scler Other Motor Neuron Disord. 2000;1:91–96. doi: 10.1080/14660820050515386. [DOI] [PubMed] [Google Scholar]
- 130.Desport JC, Preux PM, Magy L, Boirie Y, Vallat JM, Beaufrere B, Couratier P. Factors correlated with hypermetabolism in patients with amyotrophic lateral sclerosis. Am J Clin Nutr. 2001;74:328–334. doi: 10.1093/ajcn/74.3.328. [DOI] [PubMed] [Google Scholar]
- 131.Desport JC, Torny F, Lacoste M, Preux PM, Couratier P. Hypermetabolism in als: Correlations with clinical and paraclinical parameters. Neurodegenerative Dis. 2005;2:202–207. doi: 10.1159/000089626. [DOI] [PubMed] [Google Scholar]
- 132.Funalot B, Desport JC, Sturtz F, Camu W, Couratier P. High metabolic level in patients with familial amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2008:1–5. doi: 10.1080/17482960802295192. [DOI] [PubMed] [Google Scholar]
- 133.Dupuis L, Corcia P, Fergani A, Gonzalez De Aguilar JL, Bonnefont-Rousselot D, Bittar R, Seilhean D, Hauw JJ, Lacomblez L, Loeffler JP, Meininger V. Dyslipidemia is a protective factor in amyotrophic lateral sclerosis. Neurology. 2008;70:1004–1009. doi: 10.1212/01.wnl.0000285080.70324.27. [DOI] [PubMed] [Google Scholar]
- 134.Pradat PF, Bruneteau G, Gordon PH, Dupuis L, Bonnefont-Rousselot D, Simon D, Salachas F, Corcia P, Frochot V, Lacorte JM, Jardel C, Coussieu C, Forestier NL, Lacomblez L, Loeffler JP, Meininger V. Impaired glucose tolerance in patients with amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2009:1–6. doi: 10.3109/17482960902822960. [DOI] [PubMed] [Google Scholar]
- 135.Tsuchida K, Nakatani M, Hitachi K, Uezumi A, Sunada Y, Ageta H, Inokuchi K. Activin signaling as an emerging target for therapeutic interventions. Cell Commun Signal. 2009;7:15. doi: 10.1186/1478-811X-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rose FF, Jr, Mattis VB, Rindt H, Lorson CL. Delivery of recombinant follistatin lessens disease severity in a mouse model of spinal muscular atrophy. Hum Mol Genet. 2009;18:997–1005. doi: 10.1093/hmg/ddn426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Musaro A, McCullagh K, Paul A, Houghton L, Dobrowolny G, Molinaro M, Barton ER, Sweeney HL, Rosenthal N. Localized igf-1 transgene expression sustains hypertrophy and regeneration in senescent skeletal muscle. Nat Genet. 2001;27:195–200. doi: 10.1038/84839. [DOI] [PubMed] [Google Scholar]
- 138.Dobrowolny G, Giacinti C, Pelosi L, Nicoletti C, Winn N, Barberi L, Molinaro M, Rosenthal N, Musaro A. Muscle expression of a local igf-1 isoform protects motor neurons in an als mouse model. J Cell Biol. 2005;168:193–199. doi: 10.1083/jcb.200407021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Palazzolo I, Stack C, Kong L, Musaro A, Adachi H, Katsuno M, Sobue G, Taylor JP, Sumner CJ, Fischbeck KH, Pennuto M. Overexpression of igf-1 in muscle attenuates disease in a mouse model of spinal and bulbar muscular atrophy. Neuron. 2009;63:316–328. doi: 10.1016/j.neuron.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Deforges S, Branchu J, Biondi O, Clement G, Pariset C, Lecolle S, Lopes P, Vidal PP, Chanoine C, Charbonnier F. Motor neuron survival is promoted by specific exercise in a mouse model of amyotrophic-lateral-sclerosis. J Physiol. 2009 doi: 10.1113/jphysiol.2009.169748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Grondard C, Biondi O, Armand AS, Lecolle S, Della Gaspera B, Pariset C, Li H, Gallien CL, Vidal PP, Chanoine C, Charbonnier F. Regular exercise prolongs survival in a type 2 spinal muscular atrophy model mouse. J Neurosci. 2005;25:7615–7622. doi: 10.1523/JNEUROSCI.1245-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kirkinezos IG, Hernandez D, Bradley WG, Moraes CT. Regular exercise is beneficial to a mouse model of amyotrophic lateral sclerosis. Ann Neurol. 2003;53:804–807. doi: 10.1002/ana.10597. [DOI] [PubMed] [Google Scholar]
- 143.Klivenyi P, Ferrante RJ, Matthews RT, Bogdanov MB, Klein AM, Andreassen OA, Mueller G, Wermer M, Kaddurah-Daouk R, Beal MF. Neuroprotective effects of creatine in a transgenic animal model of amyotrophic lateral sclerosis. Nat Med. 1999;5:347–350. doi: 10.1038/6568. [DOI] [PubMed] [Google Scholar]
- 144.Kira Y, Nishikawa M, Ochi A, Sato E, Inoue M. L-carnitine suppresses the onset of neuromuscular degeneration and increases the life span of mice with familial amyotrophic lateral sclerosis. Brain Res. 2006;1070:206–214. doi: 10.1016/j.brainres.2005.11.052. [DOI] [PubMed] [Google Scholar]
- 145.Ferraiuolo L, De Bono JP, Heath PR, Holden H, Kasher P, Channon KM, Kirby J, Shaw PJ. Transcriptional response of the neuromuscular system to exercise training and potential implications for als. J Neurochem. 2009;109:1714–1724. doi: 10.1111/j.1471-4159.2009.06080.x. [DOI] [PubMed] [Google Scholar]
- 146.Li W, Brakefield D, Pan Y, Hunter D, Myckatyn TM, Parsadanian A. Muscle-derived but not centrally derived transgene gdnf is neuroprotective in g93a-sod1 mouse model of als. Exp Neurol. 2007;203:457–471. doi: 10.1016/j.expneurol.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 147.Azzouz M, Ralph GS, Storkebaum E, Walmsley LE, Mitrophanous KA, Kingsman SM, Carmeliet P, Mazarakis ND. Vegf delivery with retrogradely transported lentivector prolongs survival in a mouse als model. Nature. 2004;429:413–417. doi: 10.1038/nature02544. [DOI] [PubMed] [Google Scholar]
- 148.Azzouz M, Le T, Ralph GS, Walmsley L, Monani UR, Lee DC, Wilkes F, Mitrophanous KA, Kingsman SM, Burghes AH, Mazarakis ND. Lentivector-mediated smn replacement in a mouse model of spinal muscular atrophy. J Clin Invest. 2004;114:1726–1731. doi: 10.1172/JCI22922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ralph GS, Radcliffe PA, Day DM, Carthy JM, Leroux MA, Lee DC, Wong LF, Bilsland LG, Greensmith L, Kingsman SM, Mitrophanous KA, Mazarakis ND, Azzouz M. Silencing mutant sod1 using rnai protects against neurodegeneration and extends survival in an als model. Nat Med. 2005;11:429–433. doi: 10.1038/nm1205. [DOI] [PubMed] [Google Scholar]
- 150.Barbeito LH, Pehar M, Cassina P, Vargas MR, Peluffo H, Viera L, Estevez AG, Beckman JS. A role for astrocytes in motor neuron loss in amyotrophic lateral sclerosis. Brain Res Brain Res Rev. 2004;47:263–274. doi: 10.1016/j.brainresrev.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 151.Pehar M, Cassina P, Vargas MR, Castellanos R, Viera L, Beckman JS, Estevez AG, Barbeito L. Astrocytic production of nerve growth factor in motor neuron apoptosis: Implications for amyotrophic lateral sclerosis. J Neurochem. 2004;89:464–473. doi: 10.1111/j.1471-4159.2004.02357.x. [DOI] [PubMed] [Google Scholar]
- 152.Vargas MR, Pehar M, Cassina P, Beckman JS, Barbeito L. Increased glutathione biosynthesis by nrf2 activation in astrocytes prevents p75ntr-dependent motor neuron apoptosis. J Neurochem. 2006;97:687–696. doi: 10.1111/j.1471-4159.2006.03742.x. [DOI] [PubMed] [Google Scholar]
- 153.Cassina P, Cassina A, Pehar M, Castellanos R, Gandelman M, de Leon A, Robinson KM, Mason RP, Beckman JS, Barbeito L, Radi R. Mitochondrial dysfunction in sod1g93a-bearing astrocytes promotes motor neuron degeneration: Prevention by mitochondrial-targeted antioxidants. J Neurosci. 2008;28:4115–4122. doi: 10.1523/JNEUROSCI.5308-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Olivera S, Fernandez A, Latini A, Rosillo JC, Casanova G, Wajner M, Cassina P, Barbeito L. Astrocytic proliferation and mitochondrial dysfunction induced by accumulated glutaric acidemia i (gai) metabolites: Possible implications for gai pathogenesis. Neurobiol Dis. 2008;32:528–534. doi: 10.1016/j.nbd.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 155.Vargas MR, Pehar M, Diaz-Amarilla PJ, Beckman JS, Barbeito L. Transcriptional profile of primary astrocytes expressing als-linked mutant sod1. J Neurosci Res. 2008;86:3515–3525. doi: 10.1002/jnr.21797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yamanaka K, Chun SJ, Boillee S, Fujimori-Tonou N, Yamashita H, Gutmann DH, Takahashi R, Misawa H, Cleveland DW. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat Neurosci. 2008;11:251–253. doi: 10.1038/nn2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lobsiger CS, Boillee S, McAlonis-Downes M, Khan AM, Feltri ML, Yamanaka K, Cleveland DW. Schwann cells expressing dismutase active mutant sod1 unexpectedly slow disease progression in als mice. Proc Natl Acad Sci U S A. 2009;106:4465–4470. doi: 10.1073/pnas.0813339106. [DOI] [PMC free article] [PubMed] [Google Scholar]