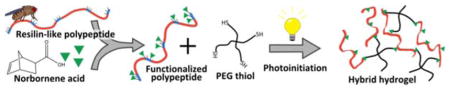
Abstract
The efficient and biocompatible cross-linking of recombinant polypeptide hydrogels offers opportunities to develop these materials for applications in tissue regeneration. Accordingly, a range of chemical strategies have been developed for cross-linking these scaffolds, although few photocross-linking approaches for recombinant polypeptides have been reported. Here, we capitalize on the novel mechanical properties of the insect protein, resilin, and the versatility of click reactions to introduce a chemically modified resilin-like polypeptide (RLP) that is capable of photoinitiated cross-linking via a thiol-ene step polymerization. Lysine residues present in the RLP were functionalized with norbornene acid and the resulting RLPN conjugates confirmed via 1H-NMR spectroscopy. RLPNs were subsequently cross-linked with multi-arm PEG thiols in the presence of a photoinitiator and UV light to form elastic hybrid hydrogels as confirmed via oscillatory rheology. Further, the crosslinking and resulting RLP-PEG networks demonstrated cytocompatibility with human mesenchymal stem cells in both 2D cell-adhesion and 3D photoencapsulation studies.
Keywords: biomaterials, protein, hydrogels, thiol-ene, photocross-linking
Graphical Abstract
Efficient photoinitiated cross-linking via thiol-ene step polymerization is conferred to hybrid hydrogels composed of poly(ethylene glycol) star macromers and a recombinant elastomeric protein polymer based upon resilin, an insect protein The hybrid hydrogels form elastic polymer networks and demonstrate cytocompatibility with human mesenchymal stem cells making them potential candidates as tissue engineering scaffolds for mechanically-demanding applications.
1. Introduction
Over the last few decades, photopolymerization has become an important technique utilized in the development of materials for biomedical applications.[1–5] In tissue engineering applications, photo-initiated reactions have been employed both to cross-link macromolecules into hydrogel networks[2, 3, 6–10] and to pattern materials with enhanced functionality.[11–17] The use of visible or UV light to initiate polymerization or cross-linking reactions has significant advantages, including very fast cure rates under physiological conditions, generation of very little heat, the ability to be performed using aqueous precursors that are generally biocompatible,[2] and the spatiotemporal control over cross-linking, a feature which provides a more direct means of manipulating the material properties of the hydrogel[2, 6, 13, 14, 16, 18] than afforded by conventional chemical strategies.[19, 20] In addition, light intensity, exposure time, and/or location of the irradiation[6, 21] may be manipulated so that a single precursor composition may be cross-linked into hydrogels with various mechanical stiffness.[22] Additionally, precursors remain stable after mixing and may be easily positioned prior to irradiation;[6] hydrogels can be formed into a variety of complex shapes that conform and adhere to tissue,[2] as well as patterned to create precise two and three dimensional geometries.[11–17, 23, 24] More recently, photodegradation strategies have afforded means to manipulate the mechanical and biochemical properties of hydrogels post-gelation.[25–28]
Although synthetic polymers, such as PEG, have been the main macromolecular precursors employed for photocross-linkable hydrogels, well-defined biosynthetic protein polymers offer an interesting alternative for imparting specific biofunctionality and desirable mechanical properties to photocross-linked tissue engineering scaffolds.[29–33] Implementing sequences from elastomeric structural proteins, including those from the mammalian protein elastin and the insect protein resilin, has demonstrated tremendous utility in the developing materials with novel mechanical properties.[29, 31, 32, 34–42] For example, hydrogels composed of resilin-like polypeptides (RLPs) demonstrate the same outstanding extensibility and resilience that is characteristic of the natural protein[31, 36, 37] and enables its high-frequency performance in insect organs.[34, 36] Despite the advantages of employing such mechanically interesting polypeptides in hydrogels, there has been limited exploration of photoreactive chemistries for their cross-linking.[11, 34, 37–39, 43, 44] Most reports have focused instead on creating photocross-linkable materials via the acrylation/methacrylation of naturally-derived proteins (e.g. gelatin, fibrinogen).[45–61] We thus sought to establish the feasibility of producing RLP-based hydrogels that could be cross-linked via efficient photocross-linking strategies while maintaining their excellent mechanical properties and cytocompatibility.[31, 32, 35, 36, 62]
Thiol-ene reactions have been an important part of polymer chemistry for the better part of a century,[63–67] but more recently have been applied to formation of PEG-based hydrogels for tissue engineering and drug delivery applications.[6, 68–73] Traditional photoreactive chemistries, such as the acrylate/methacrylate systems mentioned above, are sensitive to oxygen inhibition[64] and are driven by radical chain-growth polymerization that results in networks with heterogeneous cross-links composed of poly(meth)acrylate chains of varying size.[6, 28] In contrast, thiol-ene chemistry is less sensitive to oxygen inhibition and proceeds via step-growth mechanisms that form cross-links between complementary reactive groups which results in more homogeneous networks suggested to possess superior mechanical properties to those of chain-growth networks.[6, 74] Fairbanks et al. introduced PEG-peptide hydrogels that are cross-linked via a photoinitiated thiol-norbornene step polymerization;[6] norbornene serves as an efficient ‘ene’ moiety due to its high reactivity and selectivity for thiol-ene click reactions.[64, 75] A number of hydrogels successfully utilizing the thiol-norbornene chemistry have been reported since its introduction;[23, 68–70, 72, 76–83] an in-depth review of these hydrogels has been reported by Lin et al.[18] Of particular relevance for the development of these photochemical methods for biological applications, has recently been demonstrated that thiol-norbornene chemistry may be more cytocompatible than radical chain growth mechanisms due to the relatively fast gelation kinetics, which limits cell exposure to damaging radical species.[80]
Here we introduce a thiol-ene photocross-linkable recombinant polypeptide-PEG hybrid hydrogel comprising a chemically modified RLP and a multiarm star PEG macromer. RLPs were functionalized with norbornene acid through simple amide bond coupling chemistry with lysine residues contained in the RLP sequence; functionality was determined via 1H NMR spectroscopy as well as a TNBSA assay that is specific for primary amines. Oscillatory rheology was utilized to investigate the gelation and resulting mechanical properties of these hybrid hydrogels while cyto-compatibility was confirmed through cell adhesion studies as well as the encapsulation and long-term culture of human mesenchymal stem cells.
2. Experimental Section
2.1. Materials
All chemicals or reagents were purchased from Sigma-Aldrich (St. Louis, MO) or Fisher Scientific (Waltham, MA) and used as received unless otherwise noted. Ni-NTA agarose used for protein purification was purchased from Qiagen (Valencia, CA) or Thermo Scientific (Rockford, IL). Water for buffers or media was deionized and filtered using either a ThermoFisher Barnstead NANOpure Diamond water purifier or a Purelab Classic (Siemens, Munich, Germany). The thiol-terminated 4-arm star PEG (10 kDa) was purchased from JenKem Technology USA (Plano, TX). The photoinitiator, lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP), was synthesized as described previously.[22]
2.2. Expression and Purification of RLPs
The design and cloning of the RLP and RLPRGD protein has been previously reported elsewhere.[31, 35] Expression was performed using the QIAexpress® expression system (QIAGEN, Valencia, CA). Briefly, E. coli M15[pREP4] was transformed with a pQE-80L plasmid containing the RLP or RLPRGD gene. To express the RLP proteins, overnight cultures of transformed E. coli were used to inoculate six Fernbach flasks containing 500 or 750 mL of 2×YT medium containing 100 μg/mL ampicillin. To express the polypeptide these cultures were induced at an O.D. of approximately 0.6 by adding isopropyl β-D-thiogalactopyranoside (IPTG) at a 1 mM final concentration and were then cultured for four hours at 37 °C. Cells were pelleted via centrifugation and frozen at −20 °C. The polypeptide was purified under native conditions according to the manufacturer’s guidelines for Ni-NTA affinity chromatography.[84] The eluate containing the RLP or RLPRGD product was dialyzed in a 20:1 volumetric ratio of dialysis buffer to protein solution once against 1 M NaCl and then against ddH2O (5×). The dialyzed solutions were subsequently frozen and lyophilized to a dry powder. The purity of the polypeptide was monitored via gel electrophoresis (SDS-PAGE),[85] and the composition confirmed via amino acid analysis as has been previously reported (see Table S1).[31]
2.3. Functionalization and Characterization of RLPN
The RLP proteins were functionalized with norbornene via modification of regularly positioned lysine residues on the polypeptide chain (see Figure 1A/B, Figure S1A/B, Figure S2). Simple amide bond coupling chemistry was employed to conjugate exo-5-norbornenecarboxylic acid (Sigma Aldrich) to the amine side chain of the lysine residues. First, the RLP proteins were dissolved using heat and sonication into dimethyl sulfoxide (DMSO) at a 5 mg/mL concentration; drop-wise addition of triethylamine (TEA) was performed until the protein dissolved. The molar ratio of TEA to lysine for any given reaction was never greater than 10:1. The norbornene acid was separately dissolved in DMSO with TEA added in a 1:1 ratio. To initiate the reaction, the coupling reagent, (2-(7-Aza-1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate) (HATU) was dissolved in DMSO, combined with the norbornene/DMSO/TEA mixture and promptly added to the RLP/DMSO/TEA solution.
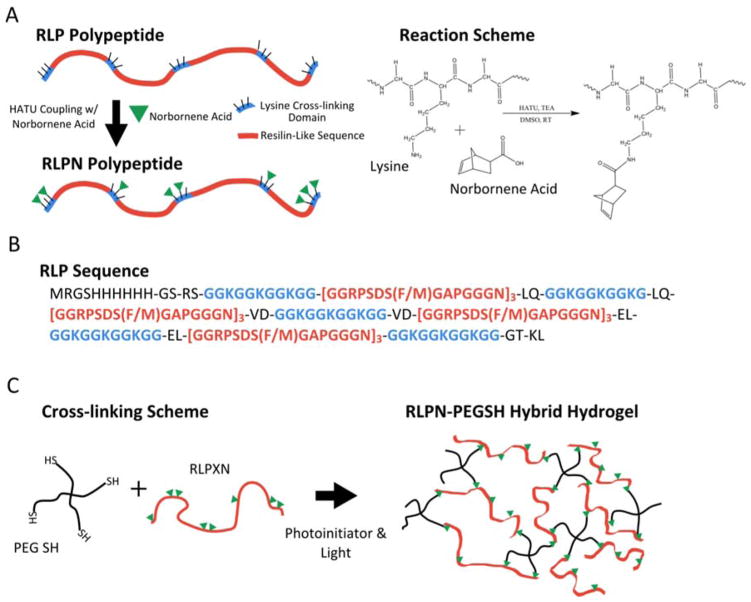
Figure 1.
Schemes illustrating the norbornene functionalization of RLP, the RLP sequence and the formation of hybrid hydrogels. (A) Lysine residues occurring in five ‘cross-linking bundles’ along the polypeptide chain can be functionalized with photoreactive norbornene acid through simple amide bond coupling. (B) The amino acid sequence of RLP; parentheses indicate repeated sequences and (F/M) indicates the presence of either a phenylalanine or methionine residue. For each group of three resilin repeats, there were two phenylalanine residues and one methionine residue encoded in the gene sequence; the RLP had a total of eight phenylalanine and four methionine residues. (C) Scheme illustrating the cross-linking reaction between 4-arm star PEG thiol macromers and RLPN to form hybrid hydrogels.
The reaction was incubated at room temperature for approximately 2 hours. The ratio of norbornene acid to lysine was varied depending upon the desired functionality of the conjugate. However, the ratio of HATU to norbornene was kept at a 1:1 stoichiometry to avoid side reactions. The reaction was terminated with the addition of an equal volume of water containing 100 μM ammonium hydroxide. This solution was promptly dialyzed at room temperature (Snakeskin™, 10 kDa, Thermo Scientific) against ddH2O for two hours in a 20:1 volumetric ratio of buffer to dialysate; this removed most of the DMSO. In later stages of dialysis, precipitation was prevented by adding a minute concentration (< 10 μM) of sodium hydroxide to the ddH2O dialysis solution. However, in the final step of dialysis, the protein was always returned to pure water. Finally, the RLPN (or RLPRGDN) solution was filtered, frozen in liquid N2, and lyophilized.
The functionality of the RLP proteins was characterized via 1H NMR. Samples were prepared at a ~5 mg/mL concentration of functionalized protein in D2O with approximately 0.2% NaOD (Cambridge Isotope Laboratories, Tewksbury, MA) and were analyzed using either an AVIII 600 MHz NMR spectrometer equipped with a 5 mm Bruker SMART probe or an AV400 NMR spectrometer equipped with a cryogenic QNP probe (Bruker, Billerica, MA) courtesy of Dr. Shi Bai (Department of Chemistry and Biochemistry, University of Delaware, Newark, DE). Water suppression was employed in the analysis. Eight phenylalanine residues per RLP molecule were used as a reference for the quantification of norbornene functionality. The integration of the aromatic protons of phenylalanine (δH 7.15–7.35 (m, 5H)) was compared to the integration of the olefin protons of the norbornene (δH 6.00–6.15 (d, 2H)). A modified TNBSA assay (Life Technologies, Carlsbad, CA) for primary amines confirmed the functionality of the RLPRGD conjugate (see Supporting Information, Figure S2).
2.4. Cross-linking and Oscillatory Rheology of RLPN-PEGSH Hydrogels
Characterization of gelation and the mechanical properties of the hydrogels was conducted using an AR-G2 Rheometer (TA Instruments, New Castle, DE) equipped with an OmniCure® S1000 UV lamp assembly for photocuring applications; a stainless steel 8 mm diameter parallel plate geometry was used for these experiments. RLPN and PEGSH were dissolved separately in PBS buffer (pH 7.4) that included the LAP photoinitiator at a 0.067 wt% (w/v) concentration (2.3 mM). Different weight percentages of precursor (i.e., total solids of both the RLP and PEG) were analyzed, but the cross-linking stoichiometry of norbornene to thiol was kept constant at a 1:1 ratio. Precursor solutions were briefly mixed on a vortex mixer and deposited on to a quartz crystal that would guide UV light and serve as the rheometer stage during the experiment. Unless otherwise noted, the solutions were cured using 365 nm light at an intensity of 5 mW/cm2. Oscillatory time sweeps were conducted at 1% strain (in the linear viscoelastic regime) and 1 Hz while frequency sweeps from 0.01 – 10 Hz were analyzed at 1% strain. For all gels analyzed, the composition of the RLPN-PEGSH hydrogels was approximately 50% protein and 50% PEG. The elastic moduli are reported as a simple mean of three experiments and the error is reported as the standard deviation.
2.5. 3D Photoencapsulation and Viability of hMSCs
The viability of bone-derived human mesenchymal stems encapsulated within RLPN-PEG hydrogels was analyzed via confocal microscopy. The hMSCs (Lonza, Basel, Switzerland) were subcultured according to the manufacturer’s specifications at 37 °C, 5% CO2, and using the BulletKitTM hMSC growth medium provided by Lonza. The cells were kept below 80% confluency and utilized between passage three and ten. The RLPN-PEGSH hydrogels used for the encapsulation experiments were 6 wt% (w/v) (total solids) and had a final volume of 50 μL. First, the LAP initiator was dissolved in PBS buffer at a 0.084% (w/v) concentration to compensate for the lack of photoinitiator in the cell media. RLPN and PEGSH were then dissolved separately in this PBS buffer; usually, a small amount (< 1 μL) of 2 N NaOH was needed to help solubilize the protein in the 50 μL sample. Next, the hMSCs were lifted via treatment with tryspin/EDTA (Lonza), counted using a hemocytometer, and separated into 10 μL aliquots of 20,000 cells. The precursors were vortex mixed and then triturated with the cells before being deposited onto a glass-bottom dish (MatTek Corporation, Ashland, MA), which was then placed under 365 nm UV light (B-100 High Intensity Long Wave Length Lamp, UltraViolet Products, Upland, CA) at ~5 mW/cm2 for 30 seconds. Immediately following, 3 mL of growth media was added to the dishes and the gels containing encapsulated cells were placed in an incubator (37 °C, 5% CO2); lifting the gels encouraged diffusion to the bottom edge and improved cell viability.
Live/Dead® stain (Life Technologies) was utilized to determine the viability of the hMSCs over a 28 day time period. Prior to imaging, hydrogels were placed in PBS buffer containing 2 μM Calcein AM and 4 μM ethidium homodimer-1 for 30 minutes. The cell-gels were then imaged via laser scanning confocal microscopy on a Zeiss LSM 510 NLO multiphoton microscope (Carl Zeiss, Inc., Thornwood, NY) courtesy of the Delaware Biotechnology Institute (Newark, DE) Bioimaging Center. Z-stack images were acquired from several sample gels for a given time point. Representative maximum intensity projections are reported.
2.6. 2D Cell Culture and Adhesion of hMSCs
The adhesion of hMSCs to the RLPN-PEG hydrogels with and without integrin binding domains (RGD) was assessed using two dimensional cell-seeding studies. The hMSCs were subcultured according to protocols stated above. RLPN-PEGSH or RLPRGDN-PEGSH hydrogels were prepared at 6 wt% (w/v) (total solids) and had a final volume of 30 μL; LAP initiator was dissolved into PBS buffer at a 0.067 wt% (w/v) concentration and the precursors were dissolved separately into the buffer containing initiator. The precursors were mixed and then deposited onto a glass-bottom dish that was then placed under 365 nm UV light at ~5 mW/cm2 for 45 seconds. Gels were prepared under sterile conditions in a biosafety cabinet to prevent contamination and sterilized using UV light (254 nm) for a period of 30 minutes. Following lifting and counting per the protocols stated above, the hMSCs were seeded at a 3500 cells/cm2 density to the gel surface in approximately 20 μL of cell media and incubated for 15 minutes at 37 °C; gels were then immersed in 3 mL of cell media. Cell attachment and spreading was analyzed the following day via the use of a commercially available F-actin staining kit (Alexa Fluor® 568 Phalloidin) from Life Technologies. The manufacturer’s instructions were followed: briefly, the cells were fixed in a 4% paraformaldehyde solution, permeabilized using 0.1% Triton X-100 and stained using the Alexa Fluor-568 conjugated phalloidin. Syto13, prepared at 1:1000 ratio in PBS, was used to stain for cell nuclei. The cell-seeded gels were then imaged via laser scanning confocal microscopy on a Zeiss 5 LIVE DUO Highspeed/Spectral confocal microscope (Carl Zeiss, Inc., Thornwood, NY) courtesy of the Delaware Biotechnology Institute (Newark, DE) Bioimaging Center. Z-stack images were acquired from several sample gels for a given time point. Representative maximum intensity projections are reported.
3. Results and Discussion
3.1. Design, Expression, and Purification of RLP
The design, expression, and purification of RLPs has been previously reported by Li, et al.[31] As illustrated in Figure 1A/B, the RLP protein comprises 12 repeats of a resilin-like sequence based upon the consensus sequence (GGRPSDSYGAPGGGN) derived from the first exon of the Drosophila melanogaster gene, CG15920.[86] Fifteen lysine residues are intermittently distributed throughout each RLP molecule in five distinct ‘cross-linking domains’ composed of three lysine residues separated by two glycine spacers (GGKGGKGGKGG); the lysine residues enabled the functionalization of the RLP through amide bond coupling of norbornene acid to the N-alkyl amine side chain of the lysine. Two versions of RLP were investigated in this report: the first (RLP) contained no bioactive domains while the second (RLPRGD), contained the well-established integrin binding domain, RGDSP (see Figure S1A/B). The RLPs, when functionalized with norbornene, are referred to as RLPN or RLPRGDN; both were capable of cross-linking with thiol-functionalized PEG macromers to form hydrogels as illustrated in Figure 1C.
The expression of RLP and RLPRGD was performed as previously described;[31, 35, 84] the high purity of the RLP polypeptide product was indicated by acrylamide gel electrophoresis (see Figure S3) and confirmed via amino acid compositional analysis (Table S1). The yield for expressions varied between 10–30 mg/L of culture and between 1.90–5.03 mg/g of cell paste, consistent with previous reports.[31]
3.2. Functionalization of RLP
To equip the RLP proteins with photoreactive groups, norbornene acid was conjugated to the polypeptide via simple amide bond coupling of the acid to the lysine side chains (see Figure 1A). Modification of the RLP was selected as the means to enable thiol-ene cross-linking, rather than use of a thiolated RLP and PEG-norbornene, owing our interests in exploring a range of cross-linking approaches for these RLP constructs; the lysine-rich domains of the RLP can be modified with a variety of different cross-linkable groups to facilitate these comparisons. Following purification via dialysis, the norbornene functionality of the RLP was determined utilizing the downfield region of the 1H NMR spectrum owing to the complexity of the upfield region (see Figure S4). By comparing the integration of the phenylalanine benzyl ring protons (δH 7.15–7.35 (m, 5H)) with that of the norbornene alkene protons (δH 6.00–6.15 (d, 2H)), the extent of functionalization of the RLPN was determined.
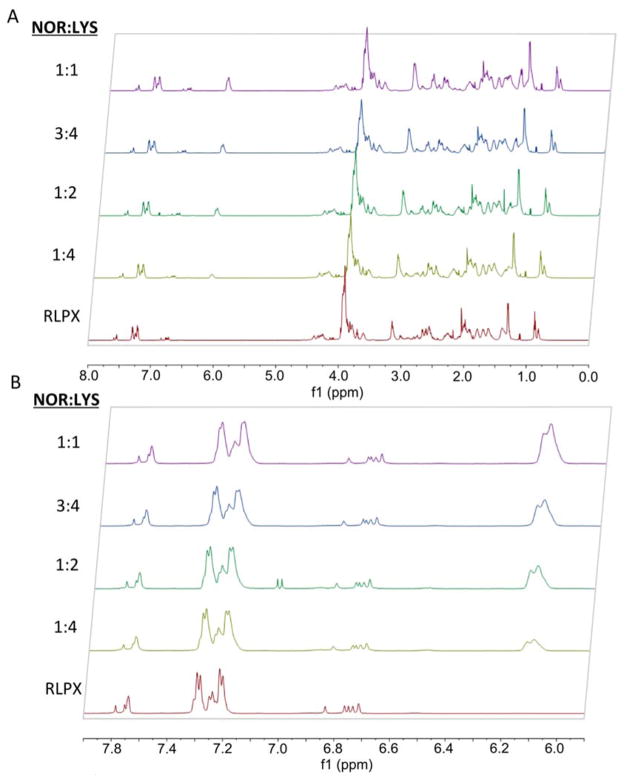
The norbornene functionality of RLPN could be modulated through variations in reaction stoichiometry. Figure 2A depicts the entire 1H NMR spectra of an unfunctionalized RLP polypeptide (red) as well as RLPNs prepared using four different norbornene to lysine reaction stoichiometries: 1:1 (purple), 3:4 (blue), 1:2 (green) and 1:4 (yellow). The expansion of the 6.0–8.0 ppm region shown in Figure 2B clearly presents the decreasing intensity of the norbornene alkene proton peaks; the functionality of RLPN calculated from NMR indicated that, depending on the reaction stoichiometry, approximately 3 to 13 of the 15 lysine residues of the RLP were modified with norbornene (Table 1). Given that the RLPNs of the highest functionality were only sparingly soluble and precipitated when mixed with PEG thiol, RLPNs with approximately 10 norbornenes were employed in subsequent rheology and cell culture experiments. The high reactivity of the norbornenes in the thiol-ene reaction rendered the modified RLPs reactive for efficient photocross-linking even at the lower functionalities, which motivated our use of the norbornene in these modifications. Figure S5 and Figure S6 present the NMR spectra of a preparation of RLPN and RLPRGDN, respectively; RLPN was equipped with approximately 9 norbornenes (ca. 60% conversion of lysines to norbornenes) while the RLPRGDN was functionalized with approximately 11 norbornenes (ca. 73% conversion). Batch preparations of the norbornene-functionalized RLPs could be achieved in as high as 80% yield.
Figure 2.
1H NMR spectra of RLPN with different functionality as a function of stoichiometric ratio. By modulating the stoichiometric ratio of norbornene to lysine, RLPN with different functionality could be attained as reflected by the corresponding increase in peak size at 6.1 ppm with increasing stoichiometric ratio of norbornene to lysine. A full spectrum is presented for the 1:1 (purple), 3:4 (blue), 1:2 (green) and 1:4 (yellow) stoichiometries in panel (A) and an expansion of the 6.0–8.0 ppm region in panel (B). The bottom (red) spectrum represents an unfunctionalized RLP control.
Table 1.
Reaction stiochiometries and functionalities of RLPN
| Feed Ratio (Nor:Lys)a | # of Norbornene 1Hb | Norbornene Functionality | % Lysine Reactedc |
|---|---|---|---|
|
|
|||
| 1 : 1 | 25.8 | 12.9 | 85.9 |
| 0.75 : 1 | 18.2 | 9.1 | 60.9 |
| 0.50 : 1 | 14.8 | 7.4 | 49.3 |
| 0.25 : 1 | 7.7 | 3.3 | 22.0 |
Ratio of norbornene acid to lysine in the reaction.
Number of norbornene protons calculated from NMR integration.
Norbornene functionality divided by the 15 available lysine residues in the RLP
3.3. Oscillatory Rheology of Photopolymerized RLPN-PEG Hydrogels
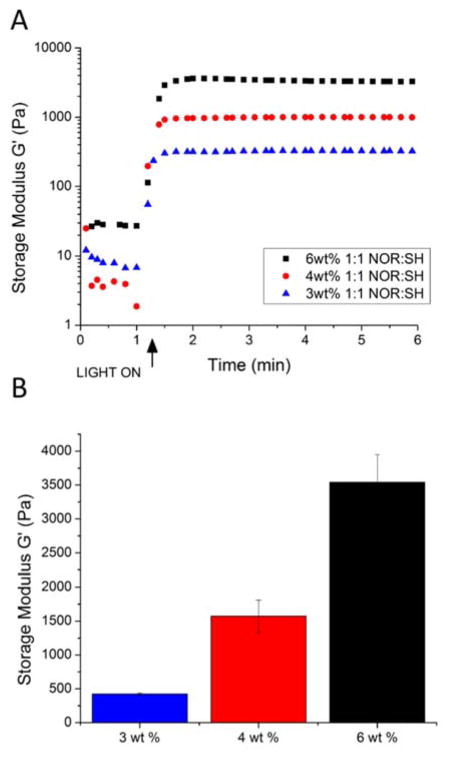
The RLPN-PEGSH hybrid hydrogels were formed via a thiol-ene step polymerization between the norbornenes of the RLPN and thiols of the PEG macromer. Of the many potential photoinitiators, LAP was chosen due to its demonstrated water solubility and its robust reactivity when exposed to light at 365 nm.[22] To analyze the gelation and mechanical properties of the RLPN-PEGSH hydrogels, oscillatory rheology was employed with in situ photopolymerization of the precursors conducted via a photocuring accessory on the rheometer. Precursors of RLPN and PEG thiol were prepared at 3, 4 and 6 wt% (w/v) concentrations while maintaining a 1:1 ratio of thiol to norbornene; the resulting data are presented in Figure 3. The oscillatory rheology time sweeps presented in Figure 3A illustrate the rapid gelation of the precursor solutions when cross-linked at a 5 mW/cm2 irradiation intensity; the storage modulus of each formulation stabilized approximately 30–45 seconds after the start of irradiation. Figure 3B illustrates that the storage moduli of the RLPN-PEGSH hydrogels could be varied, in a range relevant to the engineering of soft tissues, by altering the precursor concentration. 3 wt% gels exhibited storage moduli of approximately 400 Pa; 4 wt% gels exhibited storage moduli of approximately 1500 Pa; and 6 wt% gels exhibited storage moduli of approximately 3500 Pa, commensurate with the elastic moduli of tissues including brain, kidney, liver, thyroid and lung.[87] In comparison, RLP-only hydrogels, cross-linked via tri(hydroxyl methyl) phosphines, exhibited shear moduli between 1.2 and 20 kPa; however, these hydrogels required concentrations greater than 10 wt% RLP in order to reach these moduli.[31, 36] The photocross-linked hybrid hydrogels reported here form elastic networks at far lower concentrations, which conserves the RLP protein and suggests more robust network formation. All formulations of the RLPN-PEG hydrogels exhibited frequency-independent behavior consistent with that expected for networks with permanent cross-links (Figure S7). Further, RLPN-PEGSH gels tested in cyclic tension experiments demonstrated very low hysteresis (with resilience calculated to be approximately 90% resilience at 60% strain, see Figure S8), which is in excellent agreement with the resilience values of other RLP hydrogels cross-linked via alternative chemistries, as well as with the reported properties of the native resilin protein.[31, 36, 37] Together these observations suggest that the RLPs can undergo a variety of chemical modifications and cross-linking approaches and still maintain mechanical behavior similar to that of the native protein. In addition, PEG hydrogels cross-linked using the same chemistry and similar precursor concentrations were found to have shear moduli ranging from 300 to 6000 Pa,[6, 68] consistent with the mechanical properties of the RLPN-PEGSH hydrogels. These results reflect the efficiency of the thiol-ene chemistry as examples of other hydrogels polymerized via traditional radical polymerization require greater precursor concentrations to match the elasticity of the RLPN-PEGSH hydrogels. [51, 80]
Figure 3.
Effect of weight percent on the mechanical properties of RLPN-PEGSH hydrogels. A representative oscillatory rheometry time sweep conducted at using 1% strain at 1 Hz was used to investigate the gelation (A). A summary of the average storage moduli for hydrogels with different polymer concentrations is also presented (B). The simple mean of at least three samples for a given concentration was used for the summary; the error represents the standard deviation.
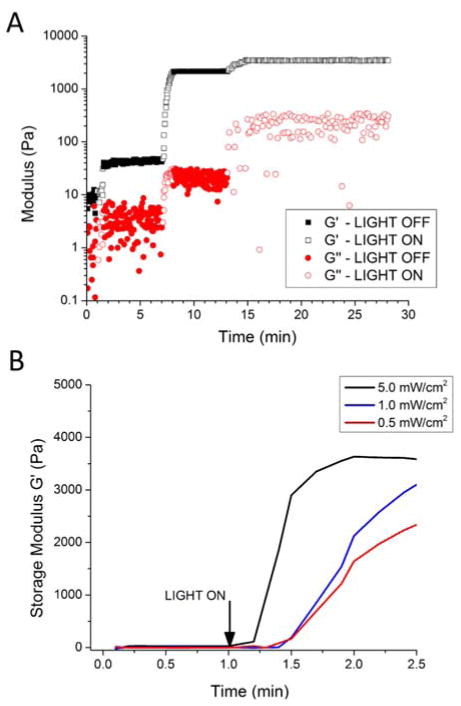
To confirm the temporal control over cross-linking of these gels, precursor solutions were irradiated at low intensity in a step-wise manner and monitored via oscillatory rheology as depicted in Figure 4A. Over three separate periods of irradiation, the stiffness of the hydrogel was increased, from approximately 45 Pa to 2.2 kPa to 3.5 kPa, as indicated by the open symbols. By switching off the UV light, the cross-linking reaction and the corresponding increase in storage modulus was halted as indicated by the closed symbols in Figure 4A. Fairbanks et al, demonstrated similar control over hydrogel mechanical properties through step-wise irradiation of solutions of PEG-norbornene and bis-thiol peptides.[6] Additionally, the intensity of irradiation could be utilized to control the cross-linking rate of the RLPN-PEGSH precursors. As depicted in Figure 4B, at the highest irradiation intensity, cross-linked hydrogels were formed within 15 seconds, while at the lower intensities, cross-linked networks were formed after 30 seconds, with the storage moduli of the hydrogels increasing more gradually. Taken together, these data illustrate the control over cross-linking afforded by the photocross-linking of the RLPN-PEGSH system; a single precursor composition may be cross-linked into hydrogels with a variety of different moduli simply by controlling exposure to light or the irradiation intensity.
Figure 4.
The effect of irradiation on RLPN/PEGSH solutions (6 wt% (w/v); 1:1, norbornene to thiol) is demonstrated via oscillatory rheology. (A) RLP-PEGSH precursors were irradiated (open symbols): at 1.5 min at an intensity of 1 mW/cm2 for 30 seconds and cross-linked to approximately 45 Pa (G′, storage modulus); at 7 min at an intensity of 1 mW/cm2 for 60 seconds and cross-linked to approximately 2.2 kPa; and, at 13 min at a 0.1 mW/cm2 intensity for 10 minutes that cross-linked to a final stiffness of approximately 3.5 kPa. In the intervening periods (closed symbols) the samples were not exposed to UV irradiation and the modulus did not change. (B) The effect of irradiation intensity on gelation is depicted. Solutions exposed to the higher irradiation intensity, 5 mW/cm2, cross-linked within 15 seconds and quickly reached a plateau. Solutions exposed to the lower irradiation intensities, 1 mW/cm2 and 0.5 mW/cm2, required approximately 30 seconds to form networks and the rate of increase in the storage moduli was much slower.
3.4. Photoencapsulation and Spreading Analysis of hMSCs
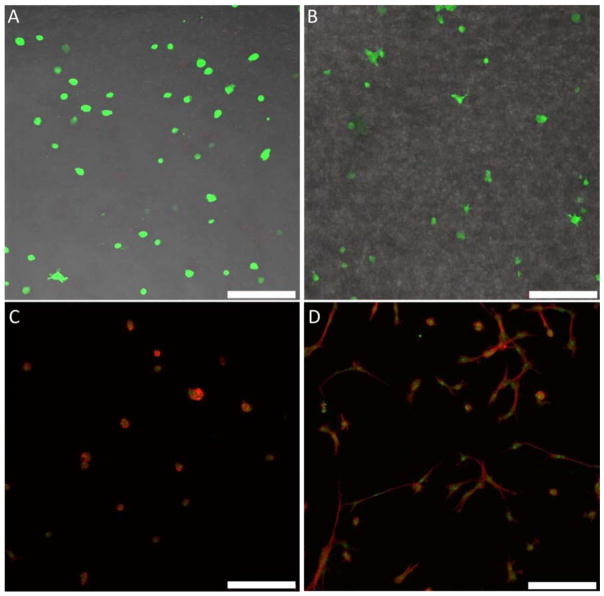
The cytocompatibility of the RLPN-PEGSH and RLPRGDN-PEGSH hydrogels was demonstrated through the photoencapsulation and 3D culture of human mesenchymal stem cells. As displayed by the representative composite Live/Dead™ confocal images for the RLPN-PEGSH hydrogels presented in Figure 5A/B, encapsulated hMSCs remained viable through the experimental period and out to 28 days of culture. In fact, nearly universal viability was observed within the hydrogels at day 0, day 14 and day 28 as depicted by representative confocal microscopy (see Figure S9); viable cells (green) comprised approximately 99% of the entire population of encapsulated cells. This outstanding cytocompatibility of the RLP-PEG hydrogels is in agreement with that observed for other hydrogels utilizing thiol-ene reactions of thiol with norbornene.[6, 23, 69, 80, 82] The fast kinetics of the thiol-ene chemistry and consequent rapid gelation of the networks likely limits cell exposure to cytotoxic UV irradiation and radical species.[23, 80] The highly viable hMSCs extended a few small processes into the matrix but mainly remained small and rounded, which is consistent with our previous studies of 3D cell encapsulation in homogeneous RLP-based matrices.[31]
Figure 5.
Representative images of composite (‘live’, ‘dead’, and transmitted channels) confocal z-stack maximum intensity projections for 3D cultures of encapsulated hMSCs in RLP-PEG hydrogels at day 0 (A) and day 28 (B). Representative images of cell spreading/adhesion analysis for the 2D cultures of seeded hMCs; composite confocal images of actin-stained (red) and nuclei-stained (green) cells for RLPN-PEGSH (control) gels (C) and RLPRGDN-PEGSH (adhesive) gels (D) one day following hMSC seeding. Images are 10× magnification and Z-stacks were between 100 to 300 μm in depth. (Scale bar represents 200 μm).
Two-dimensional hMSC studies were thus undertaken to confirm that cells could interact with cell adhesion domains in the RLP-PEG hydrogels; Figure 5C/D depicts the confocal microscopy results of actin and nuclei-stained hMSCs a day following their initial seeding on RLP-PEG gels. A substantial increase in spreading and adhesion of the hMSCs was observed on the RLPRGDN-PEGSH gels, while the cells on the RLPN-PEGSH gels remained largely rounded. The hydrogels incorporating RGD (at an approximate 1.1 mM concentration) clearly demonstrated that hMSCs adopted a more natural spindle-like phenotype, confirming the specific interaction of RGD with the seeded cells as expected and in agreement with previously explored RLP-RGD hydrogels[31] and with many other examples of RGD-mediated cell adhesion.[88, 89] Interestingly, hMSCs encapsulated in RLPN-PEGSH networks with tethered RGD peptide conjugates were unable to spread (unpublished observation), in contrast with the behavior of cells encapsulated in our other reported RLP-PEG hydrogel networks;[32] this is likely due to the lack of degradability of the RLPN-PEGSH hydrogels (which do not carry enzymatically degradable domains) and/or due to potential differences in microstructure. Nevertheless, the superb cytocompatibility of the photo-crosslinked RLPN-PEGSH networks illustrates the promise of recombinant resilin-PEG hybrid hydrogels for use as mechanically robust, chemically versatile, cell-compatible scaffolds.
4. Conclusions
Recombinant resilin-like polypeptides can be modified readily with photoreactive molecules to facilitate the use of photocross-linking methods in the production of hybrid hydrogels containing a PEG macromer. Various degrees of functionalization can be achieved simply by modulating the ratio of norbornene acid to lysine residue. The resulting RLPN were successfully cross-linked into hybrid hydrogels via thiol-ene reactions with a thiol-terminated PEG and using the photoinitiator LAP. Oscillatory rheology confirmed that gels quickly form within 45 seconds when irradiated with UV light and that the stiffness of the hydrogels could be modulated through precursor concentration and light exposure; the final mechanical properties were consistent with intended applications in soft tissue engineering. Finally, the cytocompatibility and cell adhesive nature of the materials was demonstrated through the viable 3D encapsulation and 2D spreading in cultures of hMSCs, suggesting the potential of photochemical approaches for the production of RLP-based hydrogels.
Supplementary Material
Acknowledgments
This publication was made possible by the Delaware COBRE program, supported by a grant from the National Institute of General Medical Sciences (NIGMS, 1 P30 GM110758-01) from the National Institutes of Health, and also by the National Institute on Deafness and Other Communication Disorders (RO1 DC011377A) and the National Heart Lung and Blood Institute (RO1 HL108110). The authors would like to thank Dr. Kenneth C. Koehler for the synthesis of lithium phenyl-2,4,6-trimethylbenzoylphosphinate photoinitiator used in these studies, Dr. Shi Bai (Department of Chemistry and Biochemistry) for his assistance with the 1H NMR, and Michael Moore (Delaware Biotechnology Institute) for his assistance with the confocal microscopy.
Footnotes
Supporting Information is available online from the Wiley Online Library or from the author.
Contributor Information
Christopher L. McGann, Department of Materials Science and Engineering, University of Delaware, Newark, Delaware 19716, United States
Rebekah E. Dumm, Department of Chemistry and Biochemistry, University of Delaware, Newark, Delaware 19716, United States
Anna K. Jurusik, Department of Materials Science and Engineering, University of Delaware, Newark, Delaware 19716, United States
Ishnoor Sidhu, Department of Biological Sciences, University of Delaware, Newark, Delaware 19716, United States.
Kristi L. Kiick, Email: kiick@udel.edu, Department of Materials Science and Engineering, University of Delaware, Newark, Delaware 19716, United States. Department of Biomedical Engineering, University of Delaware, Newark, Delaware 19716, United States. Delaware Biotechnology Institute, 15 Innovation Way, Newark, Delaware 19711, United States
References
- 1.Baroli B. J Chem Technol Biotechnol. 2006;81:491. [Google Scholar]
- 2.Nguyen KT, West JL. Biomaterials. 2002;23:4307. doi: 10.1016/s0142-9612(02)00175-8. [DOI] [PubMed] [Google Scholar]
- 3.Ifkovits JL, Burdick JA. Tissue Eng. 2007;13:2369. doi: 10.1089/ten.2007.0093. [DOI] [PubMed] [Google Scholar]
- 4.Cramer NB, Stansbury JW, Bowman CN. J Dent Res. 2011;90:402. doi: 10.1177/0022034510381263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fisher JP, Dean D, Engel PS, Mikos AG. Annual Review of Materials Research. 2001;31:171. [Google Scholar]
- 6.Fairbanks BD, Schwartz MP, Halevi AE, Nuttelman CR, Bowman CN, Anseth KS. Adv Mater. 2009;21:5005. doi: 10.1002/adma.200901808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu J. Biomaterials. 2010;31:4639. doi: 10.1016/j.biomaterials.2010.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burdick JA, Anseth KS. Biomaterials. 2002;23:4315. doi: 10.1016/s0142-9612(02)00176-x. [DOI] [PubMed] [Google Scholar]
- 9.Drury JL, Mooney DJ. Biomaterials. 2003;24:4337. doi: 10.1016/s0142-9612(03)00340-5. [DOI] [PubMed] [Google Scholar]
- 10.Bryant SJ, Nuttelman CR, Anseth KS. Journal of Biomaterials Science-Polymer Edition. 2000;11:439. doi: 10.1163/156856200743805. [DOI] [PubMed] [Google Scholar]
- 11.Carrico IS, Maskarinec SA, Heilshorn SC, Mock ML, Liu JC, Nowatzki PJ, Franck C, Ravichandran G, Tirrell DA. J Am Chem Soc. 2007;129:4874. doi: 10.1021/ja070200b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Culver JC, Hoffmann JC, Poché RA, Slater JH, West JL, Dickinson ME. Adv Mater. 2012;24:2344. doi: 10.1002/adma.201200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeForest CA, Polizzotti BD, Anseth KS. Nat Mater. 2009;8:659. doi: 10.1038/nmat2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn MS, Miller JS, West JL. Adv Mater. 2006;18:2679. [Google Scholar]
- 15.Lee SH, Moon JJ, West JL. Biomaterials. 2008;29:2962. doi: 10.1016/j.biomaterials.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nemir S, Hayenga HN, West JL. Biotechnol Bioeng. 2010;105:636. doi: 10.1002/bit.22574. [DOI] [PubMed] [Google Scholar]
- 17.Polizzotti BD, Fairbanks BD, Anseth KS. Biomacromolecules. 2008;9:1084. doi: 10.1021/bm7012636. [DOI] [PubMed] [Google Scholar]
- 18.Lin C-C, Ki CS, Shih H. J Appl Polym Sci. 2015;132:n/a. doi: 10.1002/app.41563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anseth KS, Bowman CN, Brannon-Peppas L. Biomaterials. 1996;17:1647. doi: 10.1016/0142-9612(96)87644-7. [DOI] [PubMed] [Google Scholar]
- 20.Slaughter BV, Khurshid SS, Fisher OZ, Khademhosseini A, Peppas NA. Adv Mater. 2009;21:3307. doi: 10.1002/adma.200802106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anseth KS, Wang CM, Bowman CN. Polymer. 1994;35:3243. [Google Scholar]
- 22.Fairbanks BD, Schwartz MP, Bowman CN, Anseth KS. Biomaterials. 2009;30:6702. doi: 10.1016/j.biomaterials.2009.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gramlich WM, Kim IL, Burdick JA. Biomaterials. 2013;34:9803. doi: 10.1016/j.biomaterials.2013.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adzima BJ, Tao Y, Kloxin CJ, DeForest CA, Anseth KS, Bowman CN. Nat Chem. 2011;3:256. doi: 10.1038/nchem.980. [DOI] [PubMed] [Google Scholar]
- 25.Griffin DR, Kasko AM. J Am Chem Soc. 2012;134:13103. doi: 10.1021/ja305280w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kloxin AM, Benton JA, Anseth KS. Biomaterials. 2010;31:1. doi: 10.1016/j.biomaterials.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Science. 2009;324:59. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kloxin AM, Kloxin CJ, Bowman CN, Anseth KS. Adv Mater. 2010;22:3484. doi: 10.1002/adma.200904179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Charati MB, Ifkovits JL, Burdick JA, Linhardt JG, Kiick KL. Soft Matter. 2009;5:3412. doi: 10.1039/b910980c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiick KL. Polymer Reviews. 2007;47:1. [Google Scholar]
- 31.Li L, Tong Z, Jia X, Kiick KL. Soft Matter. 2013;9:665. doi: 10.1039/C2SM26812D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGann CL, Levenson EA, Kiick KL. Macromol Chem Phys. 2013;214:203. doi: 10.1002/macp.201200412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Straley KS, Heilshorn SC. Soft Matter. 2009;5:114. [Google Scholar]
- 34.Elvin CM, Carr AG, Huson MG, Maxwell JM, Pearson RD, Vuocolo T, Liyou NE, Wong DCC, Merritt DJ, Dixon NE. Nature. 2005;437:999. doi: 10.1038/nature04085. [DOI] [PubMed] [Google Scholar]
- 35.Li L, Kiick K. Frontiers in Chemistry. 2014;2 doi: 10.3389/fchem.2014.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li L, Teller S, Clifton RJ, Jia X, Kiick KL. Biomacromolecules. 2011;12:2302. doi: 10.1021/bm200373p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lyons RE, Nairn KM, Huson MG, Kim M, Dumsday G, Elvin CM. Biomacromolecules. 2009;10:3009. doi: 10.1021/bm900601h. [DOI] [PubMed] [Google Scholar]
- 38.Nagapudi K, Brinkman WT, Leisen JE, Huang L, McMillan RA, Apkarian RP, Conticello VP, Chaikof EL. Macromolecules. 2002;35:1730. [Google Scholar]
- 39.Raphel J, Parisi-Amon A, Heilshorn SC. J Mater Chem. 2012;22:19429. doi: 10.1039/C2JM31768K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Betre H, Ong SR, Guilak F, Chilkoti A, Fermor B, Setton LA. Biomaterials. 2006;27:91. doi: 10.1016/j.biomaterials.2005.05.071. [DOI] [PubMed] [Google Scholar]
- 41.Nettles DL, Chilkoti A, Setton LA. Adv Drug Del Rev. 2010;62:1479. doi: 10.1016/j.addr.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Werkmeister JA, Ramshaw JAM. Biomed Mater. 2012;7:29. doi: 10.1088/1748-6041/7/1/012002. [DOI] [PubMed] [Google Scholar]
- 43.Halstenberg S, Panitch A, Rizzi S, Hall H, Hubbell JA. Biomacromolecules. 2002;3:710. doi: 10.1021/bm015629o. [DOI] [PubMed] [Google Scholar]
- 44.Lv S, Dudek DM, Cao Y, Balamurali MM, Gosline J, Li H. Nature. 2010;465:69. doi: 10.1038/nature09024. [DOI] [PubMed] [Google Scholar]
- 45.Xiao W, He J, Nichol JW, Wang L, Hutson CB, Wang B, Du Y, Fan H, Khademhosseini A. Acta Biomater. 2011;7:2384. doi: 10.1016/j.actbio.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang H, Zhou L, Liao J, Tan Y, Ouyang K, Ning C, Ni G, Tan G. J Mater Sci Mater Med. 2014;25:2173. doi: 10.1007/s10856-014-5261-x. [DOI] [PubMed] [Google Scholar]
- 47.Daniele MA, Adams AA, Naciri J, North SH, Ligler FS. Biomaterials. 2014;35:1845. doi: 10.1016/j.biomaterials.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 48.Shin H, Olsen BD, Khademhosseini A. Biomaterials. 2012;33:3143. doi: 10.1016/j.biomaterials.2011.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hutson CB, Nichol JW, Aubin H, Bae H, Yamanlar S, Al-Haque S, Koshy ST, Khademhosseini A. Tissue Engineering Part A. 2011;17:1713. doi: 10.1089/ten.tea.2010.0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nichol JW, Koshy ST, Bae H, Hwang CM, Yamanlar S, Khademhosseini A. Biomaterials. 2010;31:5536. doi: 10.1016/j.biomaterials.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Almany L, Seliktar D. Biomaterials. 2005;26:2467. doi: 10.1016/j.biomaterials.2004.06.047. [DOI] [PubMed] [Google Scholar]
- 52.Elvin CM, Vuocolo T, Brownlee AG, Sando L, Huson MG, Liyou NE, Stockwell PR, Lyons RE, Kim M, Edwards GA. Biomaterials. 2010;31:8323. doi: 10.1016/j.biomaterials.2010.07.032. [DOI] [PubMed] [Google Scholar]
- 53.Lee BH, Tin SPH, Chaw SY, Cao Y, Xia Y, Steele TWJ, Seliktar D, Bianco-Peled H, Venkatraman SS. J Biomater Sci Polym Ed. 2013;25:394. doi: 10.1080/09205063.2013.862401. [DOI] [PubMed] [Google Scholar]
- 54.Van den Bulcke AI, Bogdanov B, De Rooze N, Schacht EH, Cornelissen M, Berghmans H. Biomacromolecules. 2000;1:31. doi: 10.1021/bm990017d. [DOI] [PubMed] [Google Scholar]
- 55.Benton JA, DeForest CA, Vivekanandan V, Anseth KS. Tissue Engineering Part A. 2009;15:3221. doi: 10.1089/ten.tea.2008.0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peyton SR, Kim PD, Ghajar CM, Seliktar D, Putnam AJ. Biomaterials. 2008;29:2597. doi: 10.1016/j.biomaterials.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dikovsky D, Bianco-Peled H, Seliktar D. Biomaterials. 2006;27:1496. doi: 10.1016/j.biomaterials.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 58.Gonen-Wadmany M, Oss-Ronen L, Seliktar D. Biomaterials. 2007;28:3876. doi: 10.1016/j.biomaterials.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 59.Shapira-Schweitzer K, Seliktar D. Acta Biomater. 2007;3:33. doi: 10.1016/j.actbio.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 60.Aubin H, Nichol JW, Hutson CB, Bae H, Sieminski AL, Cropek DM, Akhyari P, Khademhosseini A. Biomaterials. 2010;31:6941. doi: 10.1016/j.biomaterials.2010.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen YC, Lin RZ, Qi H, Yang Y, Bae H, Melero-Martin JM, Khademhosseini A. Adv Funct Mater. 2012;22:2027. doi: 10.1002/adfm.201101662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Renner JN, Cherry KM, Su RSC, Liu JC. Biomacromolecules. 2012;13:3678. doi: 10.1021/bm301129b. [DOI] [PubMed] [Google Scholar]
- 63.Lowe AB, Bowman CN. Thiol-X Chemistries in Polymer and Materials Science. Royal Society of Chemistry; 2013. [Google Scholar]
- 64.Hoyle CE, Bowman CN. Angew Chem Int Ed. 2010;49:1540. doi: 10.1002/anie.200903924. [DOI] [PubMed] [Google Scholar]
- 65.Cramer NB, Bowman CN. J Polym Sci, Part A: Polym Chem. 2001;39:3311. [Google Scholar]
- 66.Hoyle CE, Lowe AB, Bowman CN. Chem Soc Rev. 2010;39:1355. doi: 10.1039/b901979k. [DOI] [PubMed] [Google Scholar]
- 67.Kloxin CJ, Scott TF, Bowman CN. Macromolecules. 2009;42:2551. doi: 10.1021/ma802771b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aimetti AA, Machen AJ, Anseth KS. Biomaterials. 2009;30:6048. doi: 10.1016/j.biomaterials.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anderson SB, Lin CC, Kuntzler DV, Anseth KS. Biomaterials. 2011;32:3564. doi: 10.1016/j.biomaterials.2011.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Benton JA, Fairbanks BD, Anseth KS. Biomaterials. 2009;30:6593. doi: 10.1016/j.biomaterials.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ki CS, Lin TY, Korc M, Lin CC. Biomaterials. 2014;35:9668. doi: 10.1016/j.biomaterials.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shih H, Mirmira RG, Lin CC. J Mater Chem B. 2015;3:170. doi: 10.1039/C4TB01593B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lin TY, Ki CS, Lin CC. Biomaterials. 2014;35:6898. doi: 10.1016/j.biomaterials.2014.04.118. [DOI] [PubMed] [Google Scholar]
- 74.Malkoch M, Vestberg R, Gupta N, Mespouille L, Dubois P, Mason AF, Hedrick JL, Liao Q, Frank CW, Kingsbury K, Hawker CJ. Chem Commun. 2006:2774. doi: 10.1039/b603438a. [DOI] [PubMed] [Google Scholar]
- 75.Hoyle CE, Lee TY, Roper T. J Polym Sci, Part A: Polym Chem. 2004;42:5301. [Google Scholar]
- 76.McCall JD, Anseth KS. Biomacromolecules. 2012;13:2410. doi: 10.1021/bm300671s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kyburz KA, Anseth KS. Acta Biomater. 2013;9:6381. doi: 10.1016/j.actbio.2013.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gould ST, Darling NJ, Anseth KS. Acta Biomater. 2012;8:3201. doi: 10.1016/j.actbio.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McKinnon DD, Kloxin AM, Anseth KS. Biomaterials Science. 2013;1:460. doi: 10.1039/c3bm00166k. [DOI] [PubMed] [Google Scholar]
- 80.Lin CC, Raza A, Shih H. Biomaterials. 2011;32:9685. doi: 10.1016/j.biomaterials.2011.08.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mnoz Z, Shih H, Lin CC. Biomaterials Science. 2014;2:1063. doi: 10.1039/c4bm00070f. [DOI] [PubMed] [Google Scholar]
- 82.Raza A, Lin CC. Macromol Biosci. 2013;13:1048. doi: 10.1002/mabi.201300044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shih H, Fraser AK, Lin CC. ACS applied materials & interfaces. 2013;5:1673. doi: 10.1021/am302690t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.The QIAexpressionist. 5. QIAGEN, Inc; 2003. [Google Scholar]
- 85.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Short Protocols in Molecular Biology. Wiley; 2002. [Google Scholar]
- 86.Ardell DH, Andersen SO. Insect Biochem Mol Biol. 2001;31:965. doi: 10.1016/s0965-1748(01)00044-3. [DOI] [PubMed] [Google Scholar]
- 87.Levental I, Georges PC, Janmey PA. Soft Matter. 2007;3:299. doi: 10.1039/b610522j. [DOI] [PubMed] [Google Scholar]
- 88.Hersel U, Dahmen C, Kessler H. Biomaterials. 2003;24:4385. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- 89.Hern DL, Hubbell JA. J Biomed Mater Res. 1998;39:266. doi: 10.1002/(sici)1097-4636(199802)39:2<266::aid-jbm14>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.