Abstract
Regulatory protein-protein interactions are ubiquitous in biology, and small molecule protein-protein interaction inhibitors are an important focus in drug discovery. Remarkably little attention has been given to the opposite strategy – stabilization of protein-protein interactions, despite the fact that several well-known therapeutics act through this mechanism. From a structural perspective, we consider representative examples of small molecules that induce or stabilize the association of protein domains to inhibit, or alter, signaling for nuclear hormone, GTPase, kinase, phosphatase, and ubiquitin ligase pathways. These SPLINTS (small-molecule protein ligand interface stabilizers) drive interactions that are in some cases physiologically relevant, and in others entirely adventitious. The diverse structural mechanisms employed suggest approaches for a broader and systematic search for such compounds in drug discovery.
Introduction
Biological assemblies frequently encompass a large number of subunits, many of which only transiently associate. In this review, we highlight representative examples of small molecules that stabilize regulatory interactions in signal transduction through nuclear hormone receptors, GTPases, kinases, phosphatases, and ubiquitin ligases. We refer broadly to these compounds as SPLINTS, or small-molecule protein ligand interface stabilizers, and we focus on selected examples that exhibit diverse structural and mechanistic features in these signaling pathways. We have not attempted to be comprehensive in our analysis; excellent recent reviews cover a panoply of additional examples of agents that both stabilize and disrupt protein interactions[1-3]. We consider well-known natural products such as cyclosporin and rapamycin, which induce adventitious binding of immunophilins to target proteins, as well as synthetic small molecules such as GNF2, which was developed to stabilize autoinhibitory interactions within the Bcr-Abl oncoprotein. Some SPLINTS are endogenous regulators or mimics thereof, while others exploit binding sites with no known physiologic ligands. Structurally, these examples exhibit diverse mechanisms. In some cases, they act as “molecular glue”, contributing contacts to both protein or domain partners to stabilize an interface. In other examples, SPLINTS do not directly participate in the interface, but instead act to remodel a protein surface to induce an interaction-competent conformation. Finally, we discuss the broader implications of these examples for drug discovery.
Nuclear hormones and remodeling of transcription factors
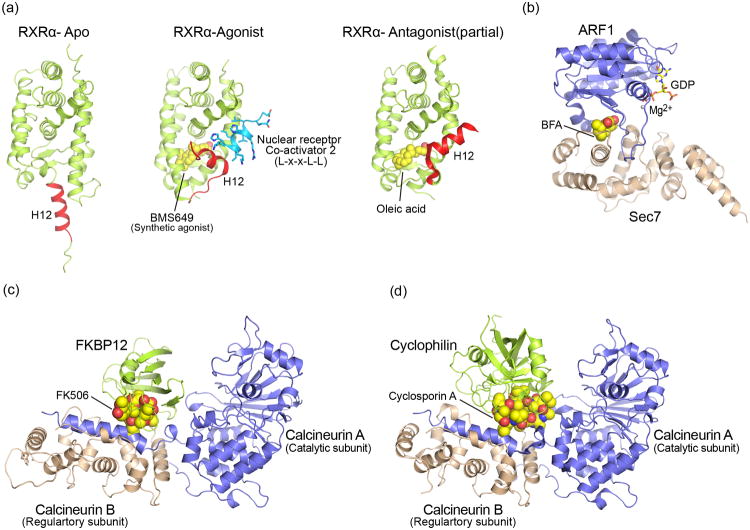
Nuclear hormone receptors function as ligand-dependent transcription factors[4] that are subject to regulation by steroid hormones and other metabolites. The large family of nuclear hormone receptors are comprised of two folded domains, a highly conserved two Zinc-finger DNA binding module, and a ligand binding, dimerization and transactivation domain, known as the AF-2 (ligand dependent activation function 2) domain (Figure 1a). On structural grounds, agonist/antagonist binding induces a conformational change that modulates the ability of the receptor to recruit co-receptors (Figure 1a) [5-7]. Agonist binding to the interior of the AF2 domain directly influences the conformation of an α -helix (H12 in the retinoic acid receptor), and results in productive co-receptor binding. In the productive conformation, this helix facilitates binding of the L-x-x-L-L motif of the co-receptor, while in the ligand-free, or antagonist bound form, co-receptor binding is disfavored.
Figure 1.
(a) Structural comparison among three different conformational state of nuclear receptor ligand binding domain. Activation domain is represented red helix in Apo (left, PDB ID: 1LBD), Agonist (middle, PDB ID: 1FBY) and Antagonist (Right, PDB ID: 2DKF). (b) Structure of Arf-Sec7 in complex with Brefeldin A (BFA). BFA is represented by space filling model (PDB ID: 1S9D). (c),(d) Structural comparison between FKBP-FK506-calcineurin (PDB ID:1TCO) and Cyclophilin-CyclosporinA-calcineurin(PDB ID:1M63), respectively.
Trapping of a GTPase/GEF complex by the fungal metabolite Brefeldin A
The Sec7 family of proteins act as guanine nucleotide exchange factors (GEFs) for ARF GTPases. Transient complex formation between ARF1-GDP and Sec7 occurs as part of the GTPase cycle. Brefeldin A (BFA) is a fungal metabolite that selectively binds the complex of ARF-GDP and Sec7 and prevents guanine nucleotide release[8]. By inhibiting the Golgi-associated guanine nucleotide exchange for Arf1, BFA interferes with the assembly of COPI coats on the Golgi and impairs vesicular transport between endoplasmic reticulum and Golgi[9-11]. BFA does not interact with isolated ARF1-GDP or Sec7. Rather, it binds only to a site formed in the complex, where it stabilizes the interaction between ARF1-GDP and Sec7 and enforces a conformation that does not permit GDP release from ARF1 (Figure 1b). Brefeldin A is a planar lactone macrocycle, decorated with keto- and hydroxyl-groups. Crystal structures of ternary ARF1-BFA-Sec7 complexes reveal that BFA is fully buried at the interface of Sec7 and Arf1-GDP, where it contacts both proteins[11,12]. The predominant interactions between BFA and Arf1 involve the switch 1 and 2 loops of the Arf1 GTPase. A hydrophobic cage that is largely composed of Arf1 aromatic residues encircles the macrocyle on three sides. The BFA hydrophilic groups are bound in this hydrophobic environment together with buried water molecules. Consistent with biochemical studies, the overall Sec7/Arf1 surface area in the absence/presence of BFA is extensive, such that the natural product stabilizes a distinct Arf1 conformation, rather than driving complex association per se.
Cyclosporin A and FK506, de novo complex formation
Cyclosporin A (CsA) and FK506 are natural microbial products with immunosuppressant activity[13]. CsA is a non-ribosomal cyclic peptide, while FK506 is a macrolide lactone ring. CsA and FK506 bind to their cellular receptors cyclophilin A (CypA) and FKBP12, respectively. Both CypA and FKBP12 belong to a family of small cis-trans peptidyl-prolyl isomerases termed immunophilins. While CsA and FK506 are strong inhibitors of their cognate isomerase, the immunosuppressant activity stems from ligand induced binding of the CsA-cyclophilin or FK506-FKBP12 complex to calcineurin (CaN), a Ser/Thr phosphatase important for T-cell activation (also referred to as Protein phosphatase 2B). The CaN heterodimer consists of subunit A (CaNA) and subunit B (CaNB). CaNA comprises a globular catalytic domain (residues 14 - 342), an extended helical domain involved in CaNB binding (residues 343 – 373) followed by a largely unstructured c-terminal region (374 – 521). The structure of the FKBP12-FK506-CaN complex (Figure 1c) reveals that FK506 and surrounding residues of FKBP12 contribute to extensive interactions with both CaN subunits[14,15]. The CyPA-CsA complex (Figure 1d) binds to the same interface of the CaNA-CaNB dimer[16,17]. Both, CyPA-CsA and FK506-FKBP12, share a common set of signature residues on CaN. Despite the large number of shared residues, distinct contacts for CyPA-CsA or FK506-FKBP12 exist, with differences in the detailed hydrogen bonding networks resulting in distinct binding modes. The ability of calcineurin to accommodate two structurally diverse immunophilin-drug complexes with distinct binding modes, may be indicative of an interface evolved to recognize a multitude of different substrates.
Kinase inhibition via stabilization of domain interfaces, by chance and by design
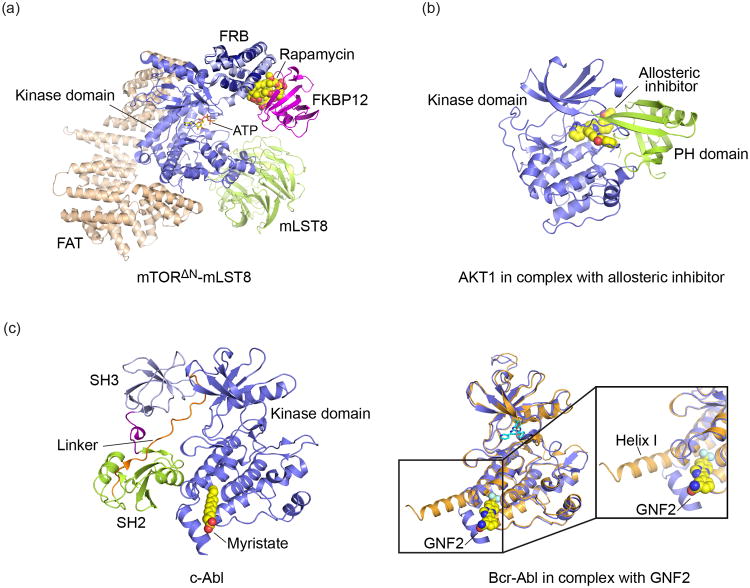
Interestingly, the immunophilin FKBP12 can also be hijacked as an inhibitor of the protein kinase mTOR. Binding of FKBP12 to mTOR is induced not by FK506, but by the structurally-related macrolide rapamycin[18-20]. mTOR (mammalian target of rapamycin) is a member of the PI3K-related protein kinase family. The mTOR kinase is the catalytic engine of two distinct multiprotein complexes termed mTORC1 and mTORC2. These regulators control cellular processes including protein and lipid synthesis, energy metabolism, and autophagy in response to a variety of inputs including amino acid levels, stress and growth factor signaling[21]. The mTORC1 complex, for example, controls protein synthesis by phosphorylating translational regulators 4E-BP1 and S6 kinase 1 (S6K1). Rapamycin binds FKBP12 in a manner similar to FK506, but the composite surface of the rapamycin/FKBP12 complex creates a recognition interface for mTOR, rather that calcineurin. The rapamycin/FKPBP12 complex binds the FRB region of mTOR, a small helical bundle that extends from the N-terminal lobe of the kinase domain. The structure of a ternary FRB/rapamycin/FKBP12 complex revealed extensive interactions between rapamycin and both FKBP12 and the mTOR FRB domain, and also significant contacts between the two proteins, including hydrogen bond interactions on either side of the sandwiched rapamycin molecule[22]. This interaction interferes with normal mTOR function by altering access of protein substrates to the kinase active site[23]. While no structure is available for an intact mTOR kinase inhibited by rapamycin and FKBP12, superposition of the ternary complex described above on the recently elucidated structure of a partial mTORC complex reveals that the induced interaction of FKBP12 with the FRB domain constricts access to the kinase catalytic cleft[23] (Figure 2a). In addition, the highly conserved FRB surface that is blocked by FKBP12 is important for recruitment of a key substrate of the mTORC1 complex, S6K1 [23].
Figure 2.
Small molecules that inhibit kinases by inducing or stabilizing domain interactions. (a) Rapamycin recruits FKBP12 to mTOR, thereby restricting access of substrates to the kinase. The crystal structure of a partial mTOR complex revealed the structure and interactions of the mTOR FAT, kinase and FRB domains, and of the mLST8 subunit (shown in tan, blue and green, respectively, drawn from PDB ID 4JSP). Modeling of the interaction of rapamycin and FKBP12 with mTOR, based on superposition of the structure of an FRB:rapamycin:FKBP12 ternary complex (PDB ID 1FAP), showed how this induced protein interaction occupies a substrate binding surface on the FRB domain and constricts access to the kinase active site (shown with nucleotide substrate analog ATPγS bound). (b) Structure of human AKT in complex with allosteric inhibitor compound VIII (PDB ID: 3O96). The inhibitor stabilizes the interaction of the PH domain with the kinase domain, rendering it inactive due dismantling of key elements of the active site. (c) Allosteric Inhibition of Bcr-Abl by restoration of autoinhibitory interactions of the SH2 domain. In autoinhibited cAbl (PDB ID: 1OPK), insertion of the N-terminal myristoyl group (represented by myristate in this structure) into a pocket on the kinase domain promotes docking of the SH2 and SH3 domains onto the kinase domain. These interactions lock the kinase in an inactive conformation. In the oncogenic Bcr-Abl fusion protein, this myristoyl group is not present, but docking of the SH2 domain can be restored by small molecules such as GNF2, which mimic the action of myristate. Superposition of the Abl kinase domain determined in the presence of absence of GNF2 (right panel, blue and tan ribbons respectively) showed that the compound induces the “kinked” conformation of helix I required for the inhibitory SH2 domain interaction (drawn from PDB ID codes 2F4J and 3K5V).
Recruitment of FKBP12 by rapamycin as an inhibitor of mTOR is apparently adventitious; FKBP12 has no known physiologic role in mTOR signaling. However, kinases are typically regulated by inter- or intra- molecular protein interactions, and small molecules that stabilize such regulatory interactions have also been developed. One illustrative example is the serine/threonine kinase AKT1 (also known as protein kinase B, PKB). AKT1 is one of three AKT isoforms[24], and is maintained in an autoinhibited state by intramolecular interaction of its N-terminal pleckstrin-homology (PH) domain with the kinase domain[25]. AKT1 is activated in the course of PI3-kinase signaling by binding of its PH domain to phospholipids, including PI[3,4,5]P3, the product of PI3-kinase[26]. Phosphoinositide binding results in recruitment of AKT to the membrane, and concomitantly, in release of the autoinhibitory interaction of the PH domain with the kinase domain. Release of the PH domain interaction also promotes activating phosphorylation of AKT1 by upstream kinases including PDK1 and the mTORC2 complex. While most AKT inhibitors (and the vast majority of all kinase inhibitors) bind in the ATP-site and exert their effect by blocking binding of this substrate, allosteric inhibitors that stabilize the inhibitory interaction of the PH domain have also been developed[27]. A breakthrough structural study of one such agent, inhibitor VIII, revealed that the compound binds at the interface between the PH and kinase domains, interacting extensively with both and effectively “cementing” the autoinhibitory contacts of the PH domain[28] (Figure 2b). The interactions of the PH domain disrupt the ATP-site, and also displace and disorder the C-helix, which plays an important role in catalysis in AKT and other kinases. A recent a co-crystal structure with a chemically unrelated allosteric AKT inhibitor revealed a closely similar binding site and AKT conformation[29], supporting the hypothesis that these agents are further stabilizing a physiologically relevant regulatory interaction between the kinase and PH domains.
Such intramolecular, autoinhibitory interactions are a recurring theme in kinase regulation. Diverse non-receptor tyrosine kinases are regulated by intramolecular contacts of their modular targeting domains. Src, Abl, and Tec family kinases all contain an SH3-SH2-kinase module in which contacts of the SH3 and SH2 domains with the “back” of the kinase domain stabilize an autoinhibited conformation of the kinase domain[30-33]. In Abl, this constellation of inhibitory interactions is promoted by insertion of an N-terminal myristoyl group into a pocket on the C-lobe of the kinase domain. Insertion of this lipid group remodels a helix (Helix-I) on the surface of the C-lobe, creating the conformation required for the inhibitory SH2 domain interaction [32,34]. A crystal structure of myristate-bound Abl is shown in Figure 2c. In Bcr-Abl, the oncogenic fusion protein produced by a 9:22 chromosomal translocation, loss of this lipid modification and other inhibitory contacts in the Abl N-terminus renders the kinase constitutively active and leads to chronic myelogenous leukemia (CML). A cell-based screen for inhibitors of Bcr-Abl led to the discovery of compounds GNF-2 and GNF-5, which inhibit the oncoprotein by binding the vacant myristate pocket[35-37]. A series of elegant mechanistic studies including crystallography, NMR and hydrogen deuterium exchange mass spectrometry have shown that like the myristoyl modification, these compounds inhibit the kinase by remodeling Helix-I in the C-lobe so as to restore docking of the SH2 domain[36,38] [39] (Figure 2c, right panel). Interestingly, variants of GNF-2 that retain the ability to bind the myristate pocket but fail to remodel Helix-I act as agonists rather than antagonists of the kinase[40].
The plant hormone auxin, a molecular glue for a ubiquitin ligase
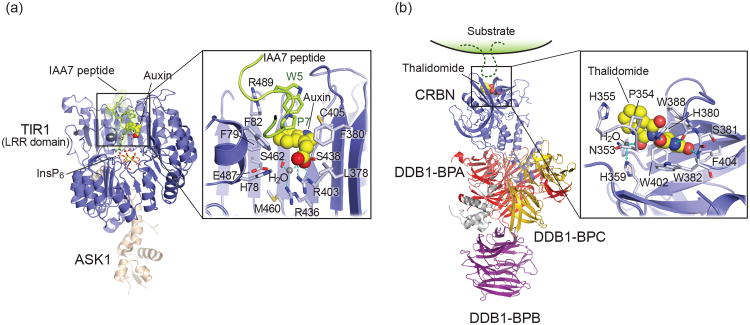
The phytohormone auxin (indole-3-acetic acid) is an essential regulator of plant development[41-44]. The Cullin-RING ubiquitin ligase SCFTIR1 serves as the auxin receptor, which ubiquitinates the Aux/IAA transcriptional repressors in response to auxin[45]. Structural studies[46] established the TIR1 subunit of the SCFTIR1 complex as the hormone receptor. TIR1 is a canonical F-box protein connected through its F-box domain to an ASK1-CUL1-RBX1 ligase module[47]. Following the three-helix F-box domain, TIR1 comprises a leucine-rich-repeat (LRR) domain with 18 LRR motifs, which binds inositol hexakiphosphate (InsP6) at the center of the LRR solenoid adjacent to the auxin (Figure 3a). Auxin is accommodated in a hydrophobic pocket on the top surface of the LRR domain, which simultaneously provides the majority of interactions to bind a peptide derived from the auxin-responsive protein IAA7. The IAA7 peptide stacks directly on top of the auxin molecule, utilizing a hydrophobic motif conserved across Aux/IAA variants by which it fully engulfs auxin. Additional hydrophobic interactions are provided by extended loops of the LRR motifs adjacent to the pocket. Neither binding of auxin, nor Aux/IAA, results in significant structural rearrangements of TIR1. Auxin hence serves as “molecular glue”, which by filling the gap at the bottom of the Aux/IAA binding pocket increases the available hydrophobic surface area for Aux/IAA binding to strengthen the interaction.
Figure 3.
(a) Structure of TIR1-ASK1 complex with auxin and IAA7 degron peptide (PDB ID: 2P1Q). A detailed view of the TIR1-auxin-IAA7 interface is shown in the inset, with key residues labeled. (b) Structure of DDB1-CRBN E3 ligase in complex with thalidomide (PDB ID: 4CI1). The predicted site of substrate recognition is indicated as schematically. Inset: Close up of the Thalidomide binding pocket in the CRBN-CTD with key residues depicted as sticks.
Thalidomide and its IMiD derivatives
Thalidomide and its second-generation derivatives lenalidomide and pomalidomide, collectively known as IMiDs, are low molecular weight drugs commonly used in the treatment of hematological malignancies[48,49]. The thalidomide efficacy target is CRBN, the substrate receptor of a Cullin-RING ubiquitin ligase (CRL4CRBN) complex[50]. Thalidomide and its derivatives prevent the CRBN receptor from engaging an endogenous substrate[51,52]. IMiD binding to CRBN further induces recruitment and degradation of the Ikaros/Aiolos transcription factors and Casein Kinase 1 alpha (Ck1 α) [53-56]. These factors likely represent neo-substrates, which are exclusively degraded by CRL4CRBN in the presence of the drug. Their degradation depends on the exposed C4, C5 and C6 positions of the phthalimide ring. While the structural basis of Ikaros/Aiolos and Ck1 α recruitment to CRBN induced by the drug is unclear, thalidomide and its derivatives likely function as molecular glue, similar to auxin, whose binding will most likely involve surrounding residues of CRBN.
Concluding Remarks
As the examples above illustrate, small molecules that act by stabilizing protein interfaces are not rare. By nature, their mechanisms of action are highly idiosyncratic, a property that recommends them for further exploitation in drug discovery. In contrast to active-site directed inhibitors, for which refining specificity for a particular member of a large enzyme family can be a major challenge (kinases or serine proteases, for example), SPLINTS can exhibit intrinsically greater specificity by exploiting regulatory interactions that are unique to a particular target of interest.
For target classes in which endogenous small molecules act as regulators of protein interactions (nuclear hormone receptors, for example), development of pharmaceutical antagonists and agonists is obviously a long-established paradigm. By contrast, the seemingly random recruitment of cyclophilins to calcineurin and mTOR has quite understandably not inspired broad efforts to reproduce this mechanism with alternate targets. However, the “accidental” discovery of stabilizers of interdomain interactions in proteins such as AKT and Bcr-Abl and of the E3-ligase altering agents like thalidomide does argue that compounds with such unique mechanisms of action can be more systematically discovered and exploited with greater awareness of the “splinting” mechanism and attention to assay design. Many protein classes of pharmaceutical interest are enzymes regulated by inter- or intramolecular protein interactions or are part of larger regulated complexes. A few examples include diverse protein kinases (BRAF, Tec, Jak, and Src-family kinases), phosphatases (PP2A, SHP2), ubiquitin ligases, and epigenetic modifiers (histone acetyl transferases or histone methyl transferases such as EZH2). A focus on the discovery of active site-directed inhibitors of these targets often leads to screening of truncated proteins containing only the isolated catalytic domain or subunit. Developing assays in which full-length proteins are used, where regulatory elements remain intact and inhibited states are kinetically accessible, is expected to lead to the discovery of more compounds that exploit these autoregulatory contacts. In principle, finding small-molecules that contribute a small amount of binding energy to stabilize an inhibitory interface should be much more tractable than finding those with sufficient binding energy to disrupt activating or targeting interactions.
Highlights.
SPLINTS are small-molecules that stabilize/remodel protein-protein interactions
They mimic physiological regulators, or confer de novo regulatory function
SPLINT compounds offer novel possibilities for drug discovery
Acknowledgments
MJE and EP are supported in part by grants GM110352 and CA116020 from the National Institutes of Health. NHT is supported in part by a senior ERC grant (ERC-2014-ADG 666068) and the Novartis Research Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Eric S. Fischer, Email: eric_fischer@dfci.harvard.edu.
Michael J. Eck, Email: eck@crystal.harvard.edu.
Nicolas H. Thomä, Email: nicolas.thoma@fmi.ch.
References
- 1**.Milroy LG, Grossmann TN, Hennig S, Brunsveld L, Ottmann C. Modulators of protein-protein interactions. Chem Rev. 2014;114:4695–4748. doi: 10.1021/cr400698c. Comprehensive review of current methods and technologies available to the discovery of small molecule PPI modulators. Reviews broad examples of low molecular weight PPI stabilizers. [DOI] [PubMed] [Google Scholar]
- 2.Fischer G, Rossmann M, Hyvonen M. Alternative modulation of protein-protein interactions by small molecules. Curr Opin Biotechnol. 2015;35:78–85. doi: 10.1016/j.copbio.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bier D, Thiel P, Briels J, Ottmann C. Stabilization of Protein-Protein Interactions in chemical biology and drug discovery. Prog Biophys Mol Biol. 2015;119:10–19. doi: 10.1016/j.pbiomolbio.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Bourguet W, Germain P, Gronemeyer H. Nuclear receptor ligand-binding domains: three-dimensional structures, molecular interactions and pharmacological implications. Trends Pharmacol Sci. 2000;21:381–388. doi: 10.1016/s0165-6147(00)01548-0. [DOI] [PubMed] [Google Scholar]
- 5**.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-alpha. Nature. 1995;375:377–382. doi: 10.1038/375377a0. First structure of retinoid-X receptor (RXR) ligand-binding domain revealing its overall architecture and proposing the presence of a retionic acid binding pocket. [DOI] [PubMed] [Google Scholar]
- 6.Egea PF, Mitschler A, Rochel N, Ruff M, Chambon P, Moras D. Crystal structure of the human RXRalpha ligand-binding domain bound to its natural ligand: 9-cis retinoic acid. EMBO J. 2000;19:2592–2601. doi: 10.1093/emboj/19.11.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lippert WP, Burschka C, Gotz K, Kaupp M, Ivanova D, Gaudon C, Sato Y, Antony P, Rochel N, Moras D, et al. Silicon analogues of the RXR-selective retinoid agonist SR11237 (BMS649): chemistry and biology. ChemMedChem. 2009;4:1143–1152. doi: 10.1002/cmdc.200900090. [DOI] [PubMed] [Google Scholar]
- 8.Chardin P, McCormick F. Brefeldin A: the advantage of being uncompetitive. Cell. 1999;97:153–155. doi: 10.1016/s0092-8674(00)80724-2. [DOI] [PubMed] [Google Scholar]
- 9.Donaldson JG, Finazzi D, Klausner RD. Brefeldin A inhibits Golgi membrane-catalysed exchange of guanine nucleotide onto ARF protein. Nature. 1992;360:350–352. doi: 10.1038/360350a0. [DOI] [PubMed] [Google Scholar]
- 10.Helms JB, Rothman JE. Inhibition by brefeldin A of a Golgi membrane enzyme that catalyses exchange of guanine nucleotide bound to ARF. Nature. 1992;360:352–354. doi: 10.1038/360352a0. [DOI] [PubMed] [Google Scholar]
- 11**.Renault L, Guibert B, Cherfils J. Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor. Nature. 2003;426:525–530. doi: 10.1038/nature02197. Structural characterization of nucleotide dissociation from Arf1/GEF complexes and their transisition states. Transition stabilized through bound Brefeldin A or mutations. [DOI] [PubMed] [Google Scholar]
- 12**.Mossessova E, Corpina RA, Goldberg J. Crystal structure of ARF1*Sec7 complexed with Brefeldin A and its implications for the guanine nucleotide exchange mechanism. Mol Cell. 2003;12:1403–1411. doi: 10.1016/s1097-2765(03)00475-1. Structural characterization of Brefeldin A binding to the ARF1/GDP/Sec7 complex. Detailed description of Brefeldin A mechanism of action. [DOI] [PubMed] [Google Scholar]
- 13.Schreiber SL, Crabtree GR. The mechanism of action of cyclosporin A and FK506. Immunol Today. 1992;13:136–142. doi: 10.1016/0167-5699(92)90111-J. [DOI] [PubMed] [Google Scholar]
- 14**.Kissinger CR, Parge HE, Knighton DR, Lewis CT, Pelletier LA, Tempczyk A, Kalish VJ, Tucker KD, Showalter RE, Moomaw EW, et al. Crystal structures of human calcineurin and the human FKBP12-FK506-calcineurin complex. Nature. 1995;378:641–644. doi: 10.1038/378641a0. Structure of a FKBP12-FK506-Calcineurin complex revealing the molecular mechanism of action for FK506. [DOI] [PubMed] [Google Scholar]
- 15**.Griffith JP, Kim JL, Kim EE, Sintchak MD, Thomson JA, Fitzgibbon MJ, Fleming MA, Caron PR, Hsiao K, Navia MA. X-ray structure of calcineurin inhibited by the immunophilin-immunosuppressant FKBP12-FK506 complex. Cell. 1995;82:507–522. doi: 10.1016/0092-8674(95)90439-5. Structure of a FKBP12-FK506-Calcineurin complex revealing the molecular mechanism of action for FK506. [DOI] [PubMed] [Google Scholar]
- 16.Huai Q, Kim HY, Liu Y, Zhao Y, Mondragon A, Liu JO, Ke H. Crystal structure of calcineurin-cyclophilin-cyclosporin shows common but distinct recognition of immunophilin-drug complexes. Proc Natl Acad Sci U S A. 2002;99:12037–12042. doi: 10.1073/pnas.192206699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin L, Harrison SC. Crystal structure of human calcineurin complexed with cyclosporin A and human cyclophilin. Proc Natl Acad Sci U S A. 2002;99:13522–13526. doi: 10.1073/pnas.212504399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18*.Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. Discovery of mTOR as FRAP (FKBP-rapamycin-associated protein) [DOI] [PubMed] [Google Scholar]
- 19*.Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. Discovery of mTOR as RAFT1 (Rapamycin and FKBP12 target 1) [DOI] [PubMed] [Google Scholar]
- 20.Sabers CJ, Martin MM, Brunn GJ, Williams JM, Dumont FJ, Wiederrecht G, Abraham RT. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J Biol Chem. 1995;270:815–822. doi: 10.1074/jbc.270.2.815. [DOI] [PubMed] [Google Scholar]
- 21.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22**.Choi J, Chen J, Schreiber SL, Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. Structure of FKBP12-Rapamycin bound to the FRB domain of mTOR. [DOI] [PubMed] [Google Scholar]
- 23**.Yang H, Rudge DG, Koos JD, Vaidialingam B, Yang HJ, Pavletich NP. mTOR kinase structure, mechanism and regulation. Nature. 2013;497:217–223. doi: 10.1038/nature12122. Structure of a mTOR/mLST8 complex revealing the mTOR kinase architecture, mechanism and regulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanada M, Feng J, Hemmings BA. Structure, regulation and function of PKB/AKT--a major therapeutic target. Biochim Biophys Acta. 2004;1697:3–16. doi: 10.1016/j.bbapap.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 25.Calleja V, Alcor D, Laguerre M, Park J, Vojnovic B, Hemmings BA, Downward J, Parker PJ, Larijani B. Intramolecular and intermolecular interactions of protein kinase B define its activation in vivo. PLoS Biol. 2007;5:e95. doi: 10.1371/journal.pbio.0050095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calleja V, Laguerre M, Parker PJ, Larijani B. Role of a novel PH-kinase domain interface in PKB/Akt regulation: structural mechanism for allosteric inhibition. PLoS Biol. 2009;7:e17. doi: 10.1371/journal.pbio.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindsley CW. The Akt/PKB family of protein kinases: a review of small molecule inhibitors and progress towards target validation: a 2009 update. Curr Top Med Chem. 2010;10:458–477. doi: 10.2174/156802610790980602. [DOI] [PubMed] [Google Scholar]
- 28**.Wu WI, Voegtli WC, Sturgis HL, Dizon FP, Vigers GP, Brandhuber BJ. Crystal structure of human AKT1 with an allosteric inhibitor reveals a new mode of kinase inhibition. PLoS One. 2010;5:e12913. doi: 10.1371/journal.pone.0012913. This work provided a structural basis for understanding regulation of AKT kinase activity by its PH domain, and furhter showed how this interaction can be stabilized by a small-molecule inhibitor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashwell MA, Lapierre JM, Brassard C, Bresciano K, Bull C, Cornell-Kennon S, Eathiraj S, France DS, Hall T, Hill J, et al. Discovery and optimization of a series of 3-(3-phenyl-3H-imidazo[4,5-b]pyridin-2-yl)pyridin-2-amines: orally bioavailable, selective, and potent ATP-independent Akt inhibitors. J Med Chem. 2012;55:5291–5310. doi: 10.1021/jm300276x. [DOI] [PubMed] [Google Scholar]
- 30.Xu W, Harrison SC, Eck MJ. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 31.Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 32**.Nagar B, Hantschel O, Young MA, Scheffzek K, Veach D, Bornmann W, Clarkson B, Superti-Furga G, Kuriyan J. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell. 2003;112:859–871. doi: 10.1016/s0092-8674(03)00194-6. This paper, together with Ref. 34 below, provided a structural understanding of regulation of c-Abl by its SH3 and SH2 domains and myristoyl-modified N-terminus. [DOI] [PubMed] [Google Scholar]
- 33**.Wang Q, Vogan EM, Nocka LM, Rosen CE, Zorn JA, Harrison SC, Kuriyan J. Autoinhibition of Bruton's tyrosine kinase (Btk) and activation by soluble inositol hexakisphosphate. Elife. 2015;4 doi: 10.7554/eLife.06074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34**.Hantschel O, Nagar B, Guettler S, Kretzschmar J, Dorey K, Kuriyan J, Superti-Furga G. A myristoyl/phosphotyrosine switch regulates c-Abl. Cell. 2003;112:845–857. doi: 10.1016/s0092-8674(03)00191-0. An elegant biochemical dissection of the regulation of Abl by its myristoylated N-terminal “cap” region. [DOI] [PubMed] [Google Scholar]
- 35.Adrian FJ, Ding Q, Sim T, Velentza A, Sloan C, Liu Y, Zhang G, Hur W, Ding S, Manley P, et al. Allosteric inhibitors of Bcr-abl-dependent cell proliferation. Nat Chem Biol. 2006;2:95–102. doi: 10.1038/nchembio760. [DOI] [PubMed] [Google Scholar]
- 36**.Zhang J, Adrian FJ, Jahnke W, Cowan-Jacob SW, Li AG, Iacob RE, Sim T, Powers J, Dierks C, Sun F, et al. Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors. Nature. 2010;463:501–506. doi: 10.1038/nature08675. This work showed how an allosteric inhibitor of Bcr-Abl that targets its myristate binding pocket can combine with active-site directed inhibitors to inhibit drug-resistant variants of BCR-Abl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gray NS, Fabbro D. Discovery of allosteric BCR-ABL inhibitors from phenotypic screen to clinical candidate. Methods Enzymol. 2014;548:173–188. doi: 10.1016/B978-0-12-397918-6.00007-0. [DOI] [PubMed] [Google Scholar]
- 38.Iacob RE, Zhang J, Gray NS, Engen JR. Allosteric interactions between the myristate- and ATP-site of the Abl kinase. PLoS One. 2011;6:e15929. doi: 10.1371/journal.pone.0015929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39*.Skora L, Mestan J, Fabbro D, Jahnke W, Grzesiek S. NMR reveals the allosteric opening and closing of Abelson tyrosine kinase by ATP-site and myristoyl pocket inhibitors. Proc Natl Acad Sci U S A. 2013;110:E4437–4445. doi: 10.1073/pnas.1314712110. An insightful NMR-based analysis of conformational switching in Abl and its modulation by both allosteric myristoyl pocket binders and ATP competitive inhibitors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jahnke W, Grotzfeld RM, Pelle X, Strauss A, Fendrich G, Cowan-Jacob SW, Cotesta S, Fabbro D, Furet P, Mestan J, et al. Binding or bending: distinction of allosteric Abl kinase agonists from antagonists by an NMR-based conformational assay. J Am Chem Soc. 2010;132:7043–7048. doi: 10.1021/ja101837n. [DOI] [PubMed] [Google Scholar]
- 41.Muday GK, DeLong A. Polar auxin transport: controlling where and how much. Trends Plant Sci. 2001;6:535–542. doi: 10.1016/s1360-1385(01)02101-x. [DOI] [PubMed] [Google Scholar]
- 42.Calderon-Villalobos LI, Tan X, Zheng N, Estelle M. Auxin perception--structural insights. Cold Spring Harb Perspect Biol. 2010;2:a005546. doi: 10.1101/cshperspect.a005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jurgens G, Estelle M. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 44.Leyser O. Auxin distribution and plant pattern formation: how many angels can dance on the point of PIN? Cell. 2005;121:819–822. doi: 10.1016/j.cell.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 45*.Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature. 2001;414:271–276. doi: 10.1038/35104500. This work uncovers that auxin targets AUX/IAA proteins for SCFTIR1 mediated degradation. First example of a small molecule/phytohormone to target ubiquitin ligase and direct its activity towards a target protein. [DOI] [PubMed] [Google Scholar]
- 46**.Tan X, Calderon-Villalobos LIA, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. Structural basis for auxin mediated AUX/IAA binding to the TIR substrate receptor of the SCFTIR1 ubiquitin ligase. [DOI] [PubMed] [Google Scholar]
- 47.Zimmerman ES, Schulman BA, Zheng N. Structural assembly of cullin-RING ubiquitin ligase complexes. Current opinion in structural biology. 2010 doi: 10.1016/j.sbi.2010.08.010. 10.1016/j.sbi.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bartlett JB, Dredge K, Dalgleish AG. The evolution of thalidomide and its IMiD derivatives as anticancer agents. Nature reviews Cancer. 2004;4:314–322. doi: 10.1038/nrc1323. [DOI] [PubMed] [Google Scholar]
- 49.Singhal S, Mehta J, Desikan R, Ayers D, Roberson P, Eddlemon P, Munshi N, Anaissie E, Wilson C, Dhodapkar M, et al. Antitumor activity of thalidomide in refractory multiple myeloma. The New England journal of medicine. 1999;341:1565–1571. doi: 10.1056/NEJM199911183412102. [DOI] [PubMed] [Google Scholar]
- 50*.Ito T, Ando H, Suzuki T, Ogura T, Hotta K, Imamura Y, Yamaguchi Y, Handa H. Identification of a primary target of thalidomide teratogenicity. Science (New York, NY) 2010;327:1345–1350. doi: 10.1126/science.1177319. Through chemical-proteomics experiments the authors identified Cereblon (CRBN) as the cellular target of thalidomide. [DOI] [PubMed] [Google Scholar]
- 51**.Fischer ES, Böhm K, Lydeard JR, Yang H, Stadler MB, Cavadini S, Nagel J, Serluca F, Acker V, Lingaraju GM, et al. Nature. 2014. Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. 10.1038/nature13527. First structure of the anti-cancer therapeutics thalidomide, lenalidomide and pomalidomide (IMiDs) bound to their cellular receptor DDB1-CRBN. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52**.Chamberlain PP, Lopez-Girona A, Miller K, Carmel G, Pagarigan B, Chie-Leon B, Rychak E, Corral LG, Ren YJ, Wang M, et al. Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs. Nat Struct Mol Biol. 2014;21:803–809. doi: 10.1038/nsmb.2874. Structure of lenalidomide bound to human DDB1-CRBN and thalidomide and pomalidomide bound to the mouse CRBN compound binding domain. Provided first apo-structure of mouse CRBN compound binding domain. [DOI] [PubMed] [Google Scholar]
- 53*.Lu G, Middleton RE, Sun H, Naniong M, Ott CJ, Mitsiades CS, Wong KK, Bradner JE, Kaelin WG., Jr The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science. 2014;343:305–309. doi: 10.1126/science.1244917. Discovery of IKZF1/3 transcription factors as IMiD dependent neo-substrates of the CRL4CRBN ubiquitin ligase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54*.Kronke J, Udeshi ND, Narla A, Grauman P, Hurst SN, McConkey M, Svinkina T, Heckl D, Comer E, Li X, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–305. doi: 10.1126/science.1244851. Discovery of IKZF1/3 transcription factors as IMiD dependent neo-substrates of the CRL4CRBN ubiquitin ligase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gandhi AK, Kang J, Havens CG, Conklin T, Ning Y, Wu L, Ito T, Ando H, Waldman MF, Thakurta A, et al. Immunomodulatory agents lenalidomide and pomalidomide co-stimulate T cells by inducing degradation of T cell repressors Ikaros and Aiolos via modulation of the E3 ubiquitin ligase complex CRL4(CRBN.) Br J Haematol. 2014;164:811–821. doi: 10.1111/bjh.12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56*.Kronke J, Fink EC, Hollenbach PW, MacBeth KJ, Hurst SN, Udeshi ND, Chamberlain PP, Mani DR, Man HW, Gandhi AK, et al. Lenalidomide induces ubiquitination and degradation of CK1alpha in del(5q) MDS. Nature. 2015;523:183–188. doi: 10.1038/nature14610. Discovery of CSNK1A1 as a lenalidomide dependent neo-substrates of the CRL4CRBN ubiquitin ligase important for lenalidomide efficacy in the treatmtent of 5q-MDS. [DOI] [PMC free article] [PubMed] [Google Scholar]