Abstract
In an ongoing investigation of 20-sulfonylamidine derivatives (9, YQL-9a) of camptothecin (1) as potential anticancer agents directly and selectively inhibiting topoisomerase (Topo) I, the sulfonylamidine pharmacophore was held constant, and a camptothecin derivatives with various substitution patterns were synthesized. The new compounds were evaluated for antiproliferative activity against three human tumor cell lines, A-549, KB, and multidrug resistant (MDR) KB subline (KBvin). Several analogues showed comparable or superior antiproliferative activity compared to the clinically prescribed 1 and irinotecan (3). Significantly, the 20-sulfonylamidine derivatives exhibited comparable cytotoxicity against KBvin, while 1 and 3 were less active against this cell line. Among them, compound 15c displayed much better cytotoxic activity than the controls 1, 3, and 9. Novel key structural features related to the antiproliferative activities were identified by structure-activity relationship (SAR) analysis. In a molecular docking model, compounds 9 and 15c interacted with Topo I-DNA through a different binding mode from 1 and 3. The sulfonylamidine side chains of 9 and 15c could likely form direct hydrogen bonds with Topo I, while hydrophobic interaction with Topo I and π-π stacking with double strand DNA were also confirmed as binding driving forces. The results from docking models were consistent with the SAR conclusions. The introduction of bulky substituents at the 20-position contributed to the altered binding mode of the compound by allowing them to form new interactions with Topo I residues. The information obtained in this study will be helpful for the design of new derivatives of 1 with most promising anticancer activity.
Keywords: Camptothecin, cytotoxic activity, molecular docking, sulfonylamidine, synthesis
Graphical Abstract

CPT (green), 9 (magenta), and 15c (blue) in the binding site of DNA-Topo-I.
1. Introduction
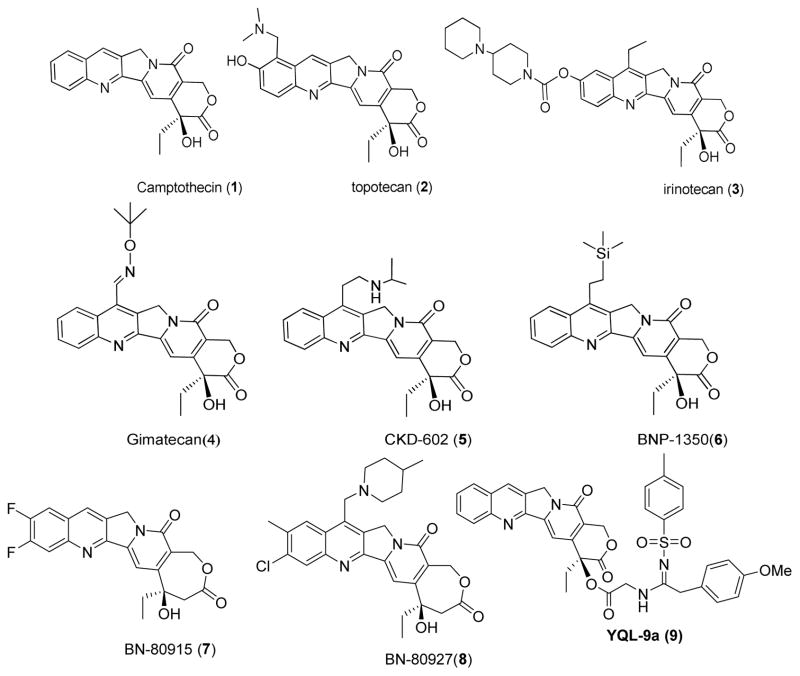
Camptothecin (1, Figure 1) is a cytotoxic alkaloid isolated from the Chinese tree Camptotheca acuminate [1–3]. Its semisynthetic analogues such as topotecan (2) and irinotecan (3) are the only current topoisomerase I (Topo I) inhibitors approved by the US Food and Drug Administration (FDA) for the treatment of various forms of cancer, while several derivatives, such as gimatecan (4), CKD-602 (5), and BNP-1350 (6), are in various stages of preclinical or clinical development [4–6]. Despite these compounds’ clinical successes, derivatives of 1 still suffer from poor solubility, cleavable-complex reversibility, and dose-limiting toxicity. In addition, the E-ring lactone exists in equilibrium with its ring-opened, hydroxyacid form in vivo [7,8]. While the latter form retains some potency, it also possesses high affinity for human serum albumin. Due to such pharmacokinetic problems, several approaches are being explored to improve the antitumor efficiency and anticancer therapeutic profiles of the 1-family. Such approaches include prodrugs (conjugates and polymer bound camptothecins), new formulations (liposomes or microparticulate carriers), and synthetic lipophilic camptothecins [9,10]. Also, newly emerging homocamptothecin (hCPT) derivatives, BN-80915 (7, diflomotecan) and BN-80927 (8), with a stabilized 7-membered hydroxylactone ring are currently undergoing clinical trials [11,12]. Most of these strategies aim to maintain the closed-lactone form in the plasma compartment. Additionally, because a free 20-hydroxy group favors lactone ring-opening by forming intra-molecular hydrogen bonds (H-bonds), acylation of the 20-hydroxy group should disfavor ring opening [13]. Accordingly, our own results [14,15], as well as those of others with 20(S)-O-acyl esters [16,17], 20(S)-O-carbonate linked tripeptide conjugates [18], and 20(S)-O-linked glycoconjugates [19], have supported the importance of various esters at the 20-position of derivatives of 1 for potent cytotoxic activity. Esterification of the 20-hydroxy group also enhances plasma stability and augments in vivo antitumor activity compared with unmodified 1. In continuing these efforts, we recently reported that a series of 20-sulfonylamidine camptothecin derivatives displayed potent antitumor activity with significantly different drug-resistance profiles from those of 1 [20]. Among them, 9 was more active than 3 against the growth of A549, DU-145, KB, and KBvin with IC50 values of 0.031, 0.050, 0.14 and 0.026 μM, respectively. Mechanistically, 9 induced significant DNA damage by selectively inhibiting Topo I and activating the ATM/Chk related DNA damage-response pathway. In mouse xenograft models, 9 demonstrated significant activity without overt adverse effects at 5 and 10 mg/kg, with two and three mice, respectively, among groups of eight undergoing complete regression. Notably, 9 at 300 mg/kg (i.p.) showed no overt acute toxicity in contrast to 1 (LD50 56.2 mg/kg, i.p.) and 3 (LD50 177.5 mg/33 kg, i.p.) [20]. Thus, 9 is attractive as a potential candidate for anticancer chemotherapy and the modification with sulfonylamidine-substituted side chains may overcome some limitations of 1. The present report intends to explore the structure-activity relationship (SAR) correlation, concentrating on structural variations of the substituents on the camptothecin pharmacophore, as well as to identify further more promising anticancer drug candidates.
Fig.1.
Structures of camptothecin derivatives
2. Results and discussion
2.1. Chemistry
The synthetic routes to target compounds are outlined in Scheme 1. Briefly, the 20-hydroxy groups of variously substituted derivatives of 1 (I-1) were converted to N-Boc protected amino esters (10) using a combination of DIPC (N,N′-diisopropyl carbodiimide) and DMAP (4-dimethylaminopyridine). Removal of the N-Boc group of 10 with TFA in CH2Cl2 (1:1) formed the TFA salts 11. Subsequently, these key precursors were successfully reacted with sulfonyl azides and alkynes in a Cu-catalyzed three-component reaction to produce the corresponding target compounds 12 in moderate yields. Under optimized conditions established in our prior study [20], a wide range of sulfonyl azide components, including aliphatic, aryl, and heterocyclic types, were all efficiently coupled to furnish the corresponding amidines. Moreover, various types of alkynes were likewise incorporated with almost the same efficiency. The coupling reaction has a wide substrate scope, a high tolerance to various functional groups, and very mild reaction conditions. The reaction proceeds through a ketenimine intermediate, which is generated in situ from the triazole cycloadduct upon release of N2 gas [21]. All newly synthesized compounds were purified by column chromatography and their structures were confirmed by 1H-NMR, 13C-NMR, ESI-MS, and elemental analysis.
Scheme 1.
General synthetic procedure for target compounds
2.2. Cytotoxicity
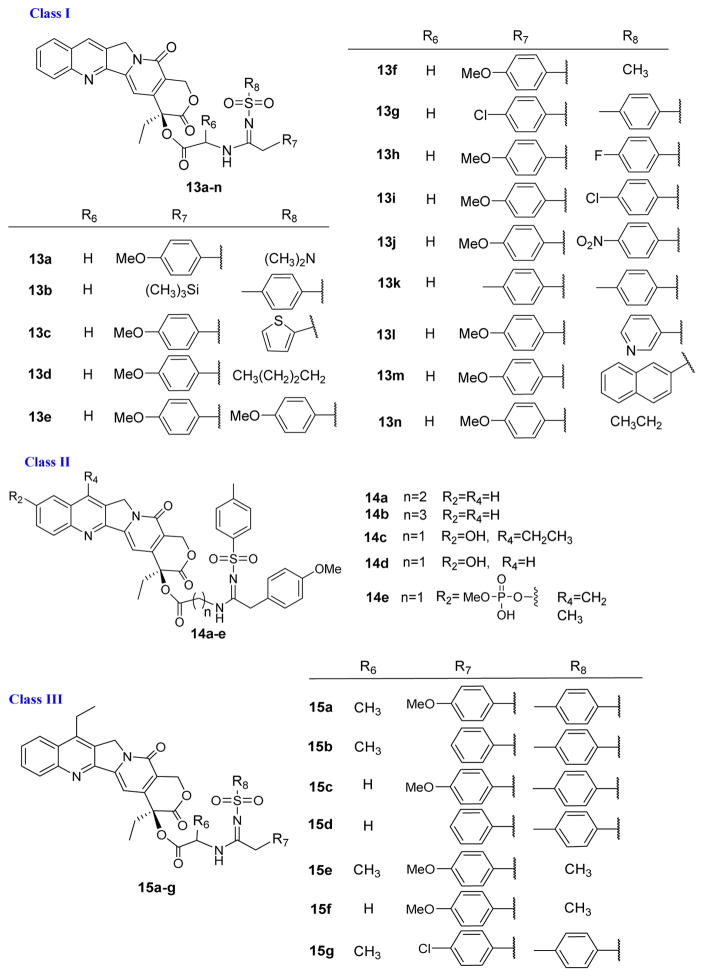
The newly synthesized compounds (Class I 13a–13n, Class II 14a–14e, Class III 15a–15g and Class IV 16a–16h) were evaluated for in vitro cytotoxic activity against three human tumor cell lines, KB (nasopharyngeal carcinoma), A-549 (lung carcinoma) and KBvin (MDR KB subline), by employing a sulforhodamine B colorimetric assay [22]. Compounds 1, 3 and 9 were used as reference drugs and the screening results are shown in Table 1.
Table 1.
In vitro cytotoxicity of compounds against three tumor cell lines.
| Entry | IC50 (μM)
|
||
|---|---|---|---|
| A-549 | KB | KBvin | |
| 13a | 0.0408 ± 0.0228 | 0.0665 ± 0.0293 | 0.0453 ± 0.0018 |
| 13b | 0.0302 ± 0.0229 | 0.0578 ± 0.0274 | 0.0399 ± 0.0170 |
| 13c | 0.0375 ± 0.0130 | 0.0550 ± 0.0209 | 0.0325 ± 0.0025 |
| 13d | 0.0349 ± 0.0253 | 0.0542 ± 0.0158 | 0.0435 ± 0.0158 |
| 13e | 0.0319 ± 0.0144 | 0.0516 ± 0.0259 | 0.0222 ± 0.0149 |
| 13f | 0.0540 ± 0.0179 | 0.1048 ± 0.0211 | 0.0635 ± 0.0246 |
| 13g | 0.0549 ± 0.0122 | 0.1000 ± 0.0167 | 0.0648 ± 0.0392 |
| 13h | 0.0141 ± 0.0033 | 0.0169 ± 0.0030 | 0.0225 ± 0.0051 |
| 13i | 0.0165 ± 0.0016 | 0.0730 ± 0.0111 | 0.0248 ± 0.0090 |
| 13j | 0.0149 ± 0.0010 | 0.0149 ± 0.0024 | 0.0149 ± 0.0019 |
| 13k | 0.0174 ± 0.0020 | 0.0739 ± 0.0114 | 0.0275 ± 0.0082 |
| 13l | 0.0765 ± 0.0065 | 0.0938 ± 0.0030 | 0.1226 ± 0.0134 |
| 13m | 0.0162 ± 0.0030 | 0.0687 ± 0.0056 | 0.0256 ± 0.0108 |
| 13n | 0.0481 ± 0.0232 | 0.0642 ± 0.0288 | 0.0786 ± 0.0287 |
| 14a | 0.0889 ± 0.0047 | 0.6219 ± 0.0496 | 0.5267 ± 0.0093 |
| 14b | 0.4478 ± 0.0185 | 0.8299 ± 0.0521 | 0.9550 ± 0.0339 |
| 14c | 0.0218 ± 0.0034 | 0.0045 ± 0.0014 | 0.0282 ± 0.0111 |
| 14d | 0.0640 ± 0.0009 | 0.0631 ± 0.0046 | 0.1010 ± 0.0184 |
| 14e | 0.3445 ± 0.0129 | 0.1414 ± 0.0064 | 0.3048 ± 0.0201 |
| 15a | 0.0160 ± 0.0051 | 0.0842 ± 0.0187 | 0.0227 ± 0.0031 |
| 15b | 0.0102 ± 0.0014 | 0.0836 ± 0.0279 | 0.0195 ± 0.0041 |
| 15c | 0.0068 ± 0.0001 | 0.0094 ± 0.0015 | 0.0101 ± 0.0026 |
| 15d | 0.0072 ± 0.0009 | 0.0895 ± 0.0344 | 0.0185 ± 0.0086 |
| 15e | 0.0610 ± 0.0083 | 0.0952 ± 0.0178 | 0.0982 ± 0.0326 |
| 15f | 0.0198 ± 0.0078 | 0.0167 ± 0.0001 | 0.0258 ± 0.0080 |
| 15g | 0.0129 ± 0.0034 | 0.0133 ± 0.0030 | 0.0160 ± 0.0042 |
| 16a | 0.0869 ± 0.0005 | 0.1087 ± 0.0196 | 0.1508 ± 0.0366 |
| 16b | 0.1399 ± 0.0051 | 0.2326 ± 0.0253 | 0.1592 ± 0.0312 |
| 16c | 8.2696 ± 0.1356 | 9.0460 ± 0.6230 | 9.1334 ± 0.4951 |
| 16d | 6.2726 ± 0.5033 | 7.0354 ± 0.8968 | 4.5833 ± 0.1234 |
| 16e | 13.9377 ± 0.4889 | 11.2886 ± 0.4375 | 10.7481 ± 0.9274 |
| 16f | 7.3051 ± 0.0591 | 7.5645 ± 0.0671 | 6.8826 ± 0.2515 |
| 16g | 0.0836 ± 0.0100 | 0.2315 ± 0.0194 | 0.2628 ± 0.0373 |
| 16h | 0.6877 ± 0.0035 | 0.9372 ± 0.0105 | 0.9469 ± 0.0721 |
| 9 (YQL-9a) | 0.0314 ± 0.0035 | 0.142 ± 0.0178 | 0.0263 ± 0.0130 |
| 1 (Camptothecin) | 0.016 ± 0.0005 | 0.037 ± 0.0031 | 0.12 ± 0.0091 |
| 3 (Irinotecan) | 9.480 ± 0.106 | 9.828 ± 0.481 | >20 |
As shown in Table 1, all synthesized compounds were more potent than 3 in the cytotoxicity assays. Significantly, the new compounds exhibited comparable cytotoxicity against the parental KB cell line and MDR KB subline KBvin, while 3 was inactive against KBvin. These results implied that the introduction of a sulfonylamidine group at C20 might combat the tumor MDR phenotype caused by P-glycoprotein overexpression. The cytotoxic profiles of class-I derivatives (13a–13n, Figure 2,) suggested that cytotoxic potency was dual-controlled by both the R6 and NR7R8 groups in the sulfonylamidine side chain.
Fig. 2.
Chemical structures of target compounds
Compared with 9, the higher IC50 values of 14a and 14b clearly demonstrated that the length of the C20 amino acid linker between the 1-skeleton and the sulfonylamidine-side chain also influenced the cytotoxic activity. In general, a compound with a CH2 linker (i.e., 9) showed better activity than compounds with a CH2CH2 or CH2CH2CH2 linker (i.e., 14a and 14b). This effect may be caused by interference with the H-bond between the C20 oxygen and Topo I (Supporting Figure S2). The H-bond’s stability could be affected by a longer linker in the sulfonylamidene-side chain. Class-II compounds containing hydrophilic tails (14c–14e) were designed for improved water-solubility. From Table 1, these three new derivatives showed superior cytotoxic activity to 3. In addition, phosphate 14e was less potent than 14c, suggesting that this common prodrug modification is not a suitable moiety for the design of a 9-derived prodrug.
With IC50 values ranging from 0.0068 to 0.0982 μM, all seven Class-III 7-ethyl-camptothecin-derived compounds (15a–15g) exhibited significant in vitro cytotoxic activity against the three tested tumor cell lines, indicating that the introduction of an ethyl group at C4 position contributed to improved cytotoxicity. Compared with 9, 15c was the most potent compound against the three tested tumor cell lines. Interestingly, 15c also showed greater cytotoxic activity against KBvin (IC50 0.0101 μM) compared with 9 and 3 (IC50 0.0263 and >20 μM, respectively).
In addition, class-IV compounds with modified 1-skeletons (16a–h) displayed a broad range of potency from comparable to much weaker cell growth inhibition compared with 9. The results demonstrated that the carboxyamide in ring-D can be replaced by a thioamide (16a and 16b). Placing a nitro group at C-9 (16c) or C-12 (16e) of ring-A dramatically decreased antiproliferative activity, suggesting that this group may interfere with the compound’s association with Topo-I. Modification of ring-C at C5 with a methoxy group (16f) also disrupted antiproliferative activity. These SAR studies will be further discussed below in light of the molecular docking studies.
2.3. Molecular Docking Study
The docking study was carried out using Autodock 4.2 [23] software with Lamarckian Genetic Algorithm [24]. The X-ray crystallographic structure of Topo I complexed with compound 1 was obtained from the RCSB Protein Data Bank (http://www.pdb.org/pdb/home/home.do) with PDB code 1T8I. The details about molecular docking process are described in the experimental section. All figures displaying the docking results were obtained using scientific software PyMol 0.99 [25].
To validate the reliability of the docking method, the ligand 1 was initially re-docked to the binding site of the protein-DNA complex. The top ranked pose with binding free energy −10.47 kcal/mol is shown in Supporting Figures S1 and S2. As can be seen from two figures, the docked pose of compound 1 agreed strongly with the status in X ray crystallographic structure, except for a flipped ethyl group. Compound 1 formed the strong π-π stacking interactions with DNA by stacking between thymine (T) 10 and guanine (G) 11 of one chain as well as cytosine (C) 112 and adenine (A) 113 of the other chain, forming a sandwich structure (Supporting Figure S2). At the same time, two hydrogen bonds (H-bonds) contributed to the ligand-protein interaction: one H-bond between the hydroxy group of 1 and the Asp533 side chain of Topo I and a second H-bond between the nitrogen-1 of 1 and the Arg354 guanidine group of Topo I. The interactions between the ligand and the bases of DNA as well as the protein residues of Topo promote the stable binding of 1 in the protein-DNA pocket and then interfere with the normal function of Topo I.
As Compound 2 is an extensively used anticancer drug, it was also docked into the binding site of DNA-Topo I for comparative purposes. The binding mode of compound 2 is displayed in Supporting Figure S3. Compounds 1 and 2 bound similarly in the cavity with their rigid rings inserting into the cleaved gap of DNA and forming H-bonds interactions with Topo I. The computed binding free energy for 2 was −11.02 kcal/mol, slightly less than that for compound 1.
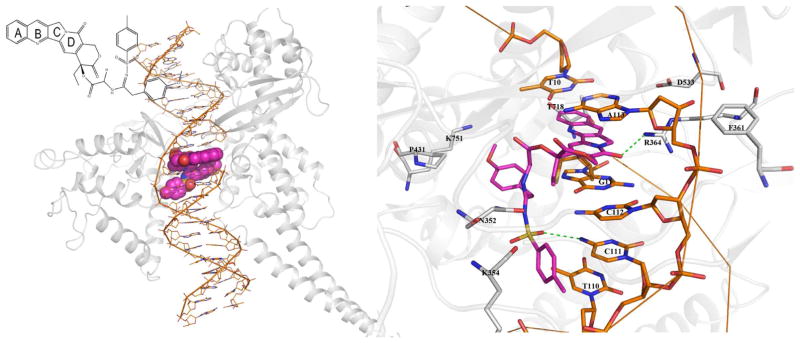
Because compound 15c exhibited the highest activity among our newly synthesized compounds, 15c might interact with Topo I more efficiently than 1. Thus, we analyzed the binding mode of our prior lead 9 as well as new 15c. The binding modes of all three compounds are shown in Fig. 3. The rigid rings of 9 and 15c overlaid well with each other, but were oriented oppositely to those of 1, whose lactone ring protruded into a DNA cleft rather than laterally along the DNA strand (Figure 3). The disparity might be attributed to the increased volume of 9 and 15, as well as the opportunity for additional interactions occurring at the C20 side chains. In spite of this difference, all three compounds inserted into the DNA cavity and stacked between planes formed by the same DNA bases via π-π stacking and hydrophobic interactions, which played critical roles in the ligands’ binding.
Figure 3.
Compounds 1, 9, and 15c in the binding site of DNA-Topo-I. Topo I is shown as a grey ribbon diagram, double strand DNA is displayed in orange, and the ligands are indicated by sticks with carbon atoms in different colors (1 in green, 9 in magenta, 15c in blue).
Compound 9 bound to DNA-Topo I with a binding free energy of −11.56 kcal/mol, From the detailed binding mode in Figures 4 and 5, the A, B, C and D rings stacked between the bases T10, G11, A113, and C112, forming π-π stacking and hydrophobic interactions. Additionally, the flexible 20-substitutent bound to the lateral side of the DNA strand and formed an H-bond with C111. Furthermore, the interactions with Topo I residues Lys751, Lys354, P431, Phe361, Asp533 and Thr718 also contributed to formation of the ternary complex, supporting 9 as a potential anticancer candidate molecule.
Figure 4.
The binding mode of compound 9 (spheres on the left and sticks on the right with carbon atoms colored magenta). The atoms from double strand DNA bases are shown in orange, and the residues from Topo I are shown in grey ribbon diagrams as well as grey sticks. The direct H-bonds formed between 9 and DNA bases or Topo I residues are indicated by green dashed lines.
Figure 5.
The interactions between compound 9 and DNA. Double strand DNA is shown by spheres.
Compound 15c lay in the binding pocket with five rings stacked between the planes formed by A113 and G11 and the flexible C20 substituent flanked to make H-bond contact with Lys436 using the methoxy oxygen and form π-π stacking interaction with G11 using the methylphenyl group on the sulfonylamidine (Figure 6). Another H-bond was also formed between residue Asn352 of Topo I and the lactone carbonyl oxygen of 15c. The binding free energy of 15c is −11.56 kcal/mol, identical to that of 9. Due to the flexibility, the bulky substituent in 9 and 15c adopted different conformations and made interactions with different residues. In spite of the discrepancy, the two compounds bound to the DNA-Topo I complex with high affinity laying the foundation that they would be potent candidates in anticancer treatment.
Figure 6.
The binding mode of compound 15c. The protein residues represented by sticks with carbons in white; the bases from DNA involved in the interaction network are shown as orange sticks.
Compounds 9 and 15c bound to Topo I adopting a pose that was similar to 1 with the five rings inserted into the slot formed by DNA base pairs, while the lactone rings of 9 and 15c protruded in the opposite orientation to accommodate the 20-substituent at the interface of DNA and Topo I. Consequently, the direct H-bond interaction between the ligand and Asp533, which was observed in the 1- or 3-complex, disappeared when 9 and 15c bound to Topo I. In spite of losing the interaction with Asp533, the 20-substituents of 9 and 15c were able to form new interactions with other residues, such as Lys436 or Asn352. In addition, the two synthetic compounds also formed new hydrophobic interactions with the protein residues.
In conclusion, the studied molecules could bind to DNA-Topo I complex with high affinity and the main driving forces were π-π stacking, direct hydrogen bonding, and hydrophobic interactions.
3. Conclusion
In summary, new 20(S)-sulfonylamidine derivatives of 1 have been synthesized and evaluated for cytotoxic activity. Most of the synthesized compounds exhibited potent in vitro cytotoxic activity against the tested tumor cell lines, including MDR cancer cell lines. Preliminary SAR correlations were proposed based on the cytotoxic activity results. Furthermore, a molecular docking analysis indicated that 9 and 15c have lower binding free energy than 1 and 2. By comparing their binding modes, we found that the four compounds had some common interaction features, including strong π-π stacking with DNA as well as direct hydrogen bonds and hydrophobic interactions with Topo I. However, to accommodate the 20-substituent at the interface of DNA and Topo I, the lactones of 9 and 15c protruded in opposite orientations to those of 1 and 2. Furthermore, the introduction of a bulky substituent at 20-position allowed the ligands to form new interactions with Topo I residues. These results enhanced our understanding of the interaction mechanism between these derivatives and Topo I and provided useful information for further structural modification. Additional work is underway regarding pre-clinical evaluation of 15c.
4. Experimental section
4.1. Chemistry
Reagents were purchased from commercial sources and were used as received. All reagents and solvents were of reagent grade or purified according to standard methods before use. Analytical thin-layer chromatography (TLC) and preparative thin-layer chromatography (PTLC) were performed with silica gel plates using silica gel 60 GF254 (Qingdao Haiyang Chemical Co., Ltd.). Melting points were determined on a Kofler apparatus and are uncorrected. IR spectra were measured on a Nicolet 380 FT-IR spectrometer on neat samples placed between KBr plates. Mass spectra were recorded on a Bruker Daltonics APEXII49e spectrometer with ESI ionization source. 1H and 13C NMR spectra were recorded at 400 MHz and 100 MHz on a Bruker AM-400 spectrometer using TMS as reference (Bruker Company, USA). Camptothecin was isolated from the Chinese medicinal plant C. acuminate, and was purified before being used. The intermediate camptothecin-20-esters of N-Boc-amino acid derivatives 10 and their TFA salts 11 were synthesized according to our previous procedures [14].
4.2. General synthetic procedures for target compounds
Triethylamine (1.2 mmol) was added slowly to a suspension of the various camptothecin amino acids TFA salts 11 (0.5 mmol) in CH2Cl2 (35 mL), and this mixture was stirred for 10 min when a clear solution was obtained. Under an N2 atmosphere, alkynes (0.5 mmol), sulfonyl azide (0.6 mmol), and CuI (0.05 mmol) was added into this reaction mixture at room temperature. After the reaction was completed, as monitored by TLC, the reaction mixture was diluted by adding CH2Cl2 (4 mL) and aqueous NH4Cl solution (6 mL). The mixture was stirred for an additional 30 min and two layers were separated. The aqueous layer was extracted with CH2Cl2 (3 mL × 3). The combined organic layers were dried over MgSO4, filtered, and concentrated in vacuo. The crude residue was purified by flash column chromatograph with an appropriate eluting solvent system.
4.2.1. Compound 13a
Yield 43%; m.p. 150–152 °C; 1H NMR (CDCl3, 400 MHz) δ: 8.40 (s, 1H, C7-H), 8.19 (d, 1H, J=8.4Hz, C9-H), 7.95 (d, 1H, J=8.0Hz, C12-H), 7.85 (t, 1H, J=7.2Hz, C11-H), 7.69 (t, 1H, J=7.60Hz, C10-H), 7.14 (m, 3H, C14-H, p-CH3OPh-H), 6.83 (d, 2H, J=8.0Hz, p-CH3OPh-H), 5.52 (ABq, 2H, J=17.6Hz, C17-H), 5.26 (s, 2H, C5-H), 4.10–4.33 (m, 4H, C23-H, C30-H), 3.72 (s, 3H, p-CH3OPh), 2.77 (s, 6H, -N(CH3)2), 2.13–2.28 (m, 2H, C18-H), 0.96 (t, 3H, J=7.2Hz, C19-H); 13C NMR (100 MHz, CDCl3) δ: 167.6, 166.9, 159.3, 157.1, 152.0, 148.7, 146.5, 144.8, 131.3, 131.2, 130.9, 129.4, 128.2, 124.3, 120.2, 114.8, 114.7, 95.6, 76.7, 67.2, 55.2, 50.0, 42.8, 39.1, 38.9, 38.4, 31.8, 7.5; MS-ESI m/z: 682.2 [M+Na]+.
4.2.2. Compound 13b
Yield 45%; m.p. 147–149 °C; 1H NMR (CDCl3, 400 MHz) δ: 8.40 (s, 1H, C7-H), 8.24 (d, 1H, J=8.4Hz, C9-H), 7.94 (d, 1H, J=8.4Hz, C12-H), 7.85 (t, 1H, J=7.6Hz, C11-H), 7.78 (d, 2H, J=8.0Hz, Ts-H), 7.68 (t, 1H, J=7.6Hz, C10-H), 7.20 (m, 3H, C14-H, Ts-H), 5.55 (ABq, 2H, C17-H), 5.30 (s, 2H, C5-H), 4.35 (m, 2H, C23-H), 2.44 (s, 2H, C30-H), 2.41 (s, 3H, Ts-CH3), 2.13–2.38 (m, 2H, C18-H), 0.98 (t, 3H, J=7.2Hz, C19-H), 0.13 (m, 9H, -Si(CH3)3); 13C NMR (100 MHz, CDCl3) δ: 168.6, 168.3, 167.0, 157.3, 152.0, 148.8, 146.6, 145.1, 141.9, 140.8, 131.2, 131.1, 130.6, 129.7, 129.0, 128.3, 128.1, 126.3, 126.1, 120.0, 95.8, 76.7, 67.1, 50.0, 43.2, 31.6, 25.8, 21.4, 7.5; MS-ESI m/z: 673.3 [M+H]+.
4.2.3. Compound 13c
Yield 57%; m.p.141–143 °C; 1H NMR (CDCl3, 400 MHz) δ: 8.40 (s, 1H, C7-H), 8.24 (d, 1H, J=8.4 Hz, C9-H), 7.94 (d, 1H, J=8.0Hz, C12-H), 7.85 (t, 1H, J=7.6Hz, C11-H), 7.68 (t, 1H, J=7.6Hz, C10-H), 7.62 (s, 2H, thiophene-H), 7.45 (d, 1H, J=4.8Hz, thiophene-H), 7.14 (s, 1H, C14-H), 7.11 (d, 2H, J=8.4Hz, p-CH3OPh-H), 6.84 (d, 2H, J =8.0 Hz, p-CH3OPh-H), 5.52 (ABq, 2H, J=17.2Hz, C17-H), 5.27 (s, 2H, C5-H), 4.14–4.43 (m, 4H, C23-H, 30-H), 3.75 (s, 3H, p-CH3OPh), 2.09–2.27 (m, 2H, C18-H), 0.94 (t, 3H, J=7.2Hz, C19-H); 13C NMR (100 MHz, CDCl3) δ: 167.5, 166.8, 159.4, 157.2, 152.0, 148.7, 146.6, 144.8, 131.2, 130.7, 130.5, 130.4, 129.6, 128.3, 128.1, 126.7, 123.6, 120.0, 114.9, 95.7, 76.7, 67.1, 55.1, 50.0, 43.3, 38.4, 31.7, 7.4; MS-ESI m/z: 721.1 [M+Na]+.
4.2.4. Compound 13d
Yield 45%; m.p.154–157 °C; 1H NMR (CDCl3, 400 MHz) δ: 8.41 (s, 1H, C7-H), 8.23 (d, 1H, J=8.8Hz, C9-H), 7.95 (d, 1H, J=8.0Hz, C12-H), 7.85 (t, 1H, J=7.6Hz, C11-H), 7.69 (t, 1H, J=7.6Hz, C10-H), 7.14 (m, 3H, C14-H, p-CH3OPh-H), 6.85 (d, 2H, J=8.0Hz, p-CH3OPh-H), 5.53 (ABq, 2H, J=17.2Hz, C17-H), 5.29 (s, 2H, C5-H), 4.13–4.29 (m, 4H, C23-H, C30-H), 3.75 (s, 3H, p-CH3OPh), 3.06–3.16 (m,2H, -CH2CH2CH2CH3), 2.10–2.19 (m, 2H, C18-H), 1.80–1.89 (m, 2H, -CH2CH2CH2CH3), 1.39–1.45 (m, 2H, -CH2CH2CH2CH3), 0.90–0.97 (m, 6H, -CH2CH2CH2CH3, C19-H); 13C NMR (100 MHz, CDCl3) δ: 167.6, 167.0, 166.8, 159.2, 157.1, 152.0, 148.7, 146.5, 144.9, 131.2, 130.7, 129.5, 128.4, 128.1, 124.1, 120.1, 114.7, 95.5, 76.7, 67.1, 55.1, 54.7, 50.0, 43.0, 38.6, 31.7, 25.6, 21.5, 13.6, 7.4; MS-ESI m/z: 695.2 [M+Na]+.
4.2.5. Compound 13e
Yield 46%; m.p. °C; 1H NMR (CDCl3, 400 MHz) δ: 8.40 (s, 1H, C7-H), 8.24 (d, 1H, J=8.8Hz, C9-H), 7.94 (d, 1H, J=8.4Hz, C12-H), 7.86 (m, 3H, -SO2PhOCH3, C11-H), 7.68 (t, 1H, J=7.2Hz, C10-H), 7.09 (m, 3H, C14-H, -SO2PhOCH3), 6.89 (d, 2H, J=8.8Hz, p-CH3OPh-H), 6.83 (d, 2H, J=8.4Hz, p-CH3OPh-H), 5.52 (ABq, 2H, J=17.2Hz, C17-H), 5.28 (s, 2H, C5-H), 4.12–4.37 (m, 4H, C23-H, C30-H), 3.81 (s, 3H, -SO2PhOCH3), 3.75 (s, 3H, p-CH3OPh), 2.05–2.25 (m, 2H, C18-H), 0.94 (t, 3H, J=7.6Hz, C19-H); 13C NMR (100 MHz, CDCl3) δ: 167.6, 167.0, 166.8, 162.1, 159.3, 157.1, 152.0, 148.7, 146.5, 144.9, 135.1, 131.2, 130.7, 129.6, 128.4, 128.1, 123.9, 119.9, 114.6, 113.6, 95.7, 76.7, 67.1, 55.4, 55.1, 50.0, 43.2, 38.3, 31.7,7.4; MS-ESI m/z: 723.2 [M+H]+.
4.2.6. Compound 13f
Yield 60%; m.p. 138–140 °C; 1H NMR (DMSO-d6, 400 MHz) δ: 8.68–8.72 (s, 1H, NH), 8.31 (s, 1H, C7-H), 8.13 (d, 2H, J=8.6 Hz, C12-H,C9-H), 7.86 (t, 1H, J=6.9Hz, C11-H), 7.71 (t, 1H, J=7.9Hz, C10-H), 7.28 (d, 2H, J=8.6Hz, p-CH3OPh-H), 7.08 (s, 1H, C14-H), 6.83 (d, 2H, J=8.7Hz, p-CH3OPh-H), 5.50 (s, 2H, C17-H), 5.29 (s, 2H, C5-H), 4.30 (dd, 1H, J=6.0, 18.0Hz, C23-H), 4.14 (dd, 1H, J=5.8, 18.0Hz, C23-H), 4.01 (d, 1H, J=14.7Hz, C30-H), 3.67 (s, 3H, p-CH3OPh), 2.84 (s, 3H, Ms-CH3), 2.11–2.16 (m, 2H, C18-H), 0.89 (t, 3H, J=7.3Hz, C19-H); 13C NMR (100 MHz, DMSO-d6) δ: 167.8, 166.9, 166.6, 158.2, 156.5, 152.2, 147.8, 146.0, 145.1, 130.3, 129.7, 128.8, 128.5, 127.9, 127.7, 126.6, 118.8, 113.8, 94.8, 76.6, 66.2, 55.0, 50.2, 43.2, 42.4, 37.1, 30.3, 7.5; MS-ESI m/z: 731.2 [M+H]+.
4.2.7. Compound 13g
Yield 57%; m.p. 148–150 °C; 1H NMR (DMSO-d6, 400 MHz) δ: 9.14 (s, 1H, NH), 8.68 (s, 1H, C7-H), 8.12 (d, 2H, J=8.8Hz, C9-H, C12-H), 7.84 (t, 1H, J=6.9Hz, C11-H), 7.70 (t, 1H, J=8.1Hz, C10-H), 7.46 (d, 2H, J=8.2Hz, Ts-H), 7.24 (s, 4H, p-ClPh-H, Ts-H), 7.11 (s, 1H, C14-H), 6.99 (d, 2H, J=8.0Hz, p-ClPh-H), 5.51 (s, 2H, C17-H), 5.27 (s, 2H, C5-H), 4.39 (dd, 1H, J=5.8, 18.0Hz, C23-H), 4.28 (dd, 1H, J=6.0, 18.0Hz, C23-H), 4.06 (s, 2H, C30-H), 2.19 (s, 3H, Ms-CH3), 2.12 (q, 2H, J=6.9Hz, C18-H), 0.84 (t, 3H, J=7.3Hz, C19-H); 13C NMR (100 MHz, DMSO-d6) δ: 167.8, 166.9, 166.3, 156.5, 152.2, 147.9, 146.1, 144.6, 141.4, 140.6, 133.7, 131.5, 130.7, 130.3, 129.6, 128.7, 128.5, 128.2, 127.9, 127.7, 125.6, 119.1, 95.0, 76.6, 66.4, 50.2, 42.4, 37.1, 30.4, 20.8, 7.4; MS-ESI m/z: 733.2 [M+Na]+.
4.2.8. Compound 13h
Yield 53%; m.p.149–151 °C; 1H NMR (CDCl3, 400 MHz) δ: 8.41 (s, 1H, C7-H), 8.22 (d, 1H, J=8.5 Hz, C9-H), 7.94–7.98 (m, 3H, p-FPh-H, C12-H), 7.86 (t, 1H, J=7.2Hz, C11-H), 7.69 (t, 1H, J=7.3Hz, C10-H), 7.08–7.12 (m, 5H, p-CH3OPh-H, C14, p-FPh-H), 6.84(d, 2H, J=8.6Hz, p-CH3OPh-H), 5.52 (ABq, 2H, J=17.3Hz, C17-H), 5.28 (s, 2H, C5-H), 4.16–4.33 (m, 4H, C23-H, 30-H), 3.75 (s, 3H, p-CH3OPh), 2.08–2.26 (m, 2H, C18-H), 0.94 (t, 3H, J=7.4Hz, C19-H); 13C NMR (100 MHz, DMSO-d6) δ: 167.7, 167.4, 166.9, 163.3, 158.1, 156.5, 152.2, 147.8, 146.1, 144.6, 139.9, 131.6, 130.3, 130.0, 129.6, 128.8, 128.5, 127.9, 127.6, 126.1, 119.1, 115.3, 115.1, 113.7, 104.5, 95.0, 76.6, 66.4, 54.9, 50.2, 42.4, 37.0, 30.3, 7.4; MS-ESI m/z: 733.2 [M+Na]+.
4.2.9. Compound 13i
Yield 58%; m.p. 153–155 °C; 1H NMR (CDCl3, 400 MHz) δ: 8.41 (s, 1H, C7-H), 8.23 (d, 1H, J=8.4Hz, C9-H), 7.95(d, 1H, J=8.0Hz, C12-H), 7.84–7.88 (m, 3H, p-ClPh-H, C11-H), 7.69 (t, 1H, J=7.6Hz, C10-H), 7.37 (d, 2H, J=8.8Hz, p-ClPh-H), 7.09–7.12 (m, 3H, p-CH3OPh-H, C14-H), 6.84 (d, 2H, J =8.4 Hz, p-CH3OPh-H), 5.53 (ABq, 2H, J=17.2Hz, C17-H), 5.29 (s, 2H, C5-H), 4.15–4.29 (m, 4H, C23-H, 30-H), 3.76 (s, 3H, p-CH3OPh), 2.05–2.22 (m, 2H, C18-H), 0.94 (t, 3H, J=7.6Hz, C19-H); 13C NMR (100 MHz, DMSO-d6) δ: 167.7, 167.6, 166.9, 158.2, 156.5, 152.2, 147.8, 146.1, 144.7, 142.3, 135.9, 130.3, 130.0, 129.6, 128.9, 128.5, 128.2, 127.9, 127.5, 127.4, 126.0, 119.0, 113.7, 95.0, 76.6, 66.4, 54.9, 50.2, 42.5, 37.0, 30.3, 7.4; MS-ESI m/z: 749.6 [M+Na]+.
4.2.10. Compound 13j
Yield 54%; m.p. 229–231 °C; 1H NMR (CDCl3, 400 MHz) δ: 8.41 (s, 1H, C7-H), 8.18–8.24 (m, 3H, p-NO2Ph-H, C9-H), 8.11 (d, 2H, J=8.8Hz, p-NO2Ph-H), 7.95 (d, 1H, J=8.0Hz, C12-H), 7.86 (t, 1H, J=7.8Hz, C11-H), 7.70 (t, 1H, J=8.0Hz, C10-H), 7.10–7.14 (m, 3H, C14-H, p-CH3OPh-H), 6.85 (d, 2H, J =8.8 Hz, p-CH3OPh-H), 5.53 (ABq, 2H, J=17.2Hz, C17-H), 5.29 (s, 2H, C5-H), 4.16–4.33 (m, 4H, C23-H, C30-H), 3.76 (s, 3H, p-CH3OPh), 2.05–2.26 (m, 2H, C18-H), 0.95 (t, 3H, J=7.2Hz, C19-H); 13C NMR (100 MHz, DMSO-d6) δ: 168.2, 167.7, 167.0, 158.2, 156.5, 152.2, 148.8, 148.4, 147.8, 146.1, 144.7, 130.3, 130.0, 129.6, 128.8, 128.5, 127.9, 127.6, 126.9, 125.6, 123.3, 119.0, 113.6, 95.0, 76.7, 66.4, 54.8, 50.2, 42.6, 37.1, 30.3, 7.4; MS-ESI m/z: 738.3 [M+Na]+.
4.2.11. Compound 13k
Yield 59%; m.p. 151–153 °C; 1H NMR (CDCl3, 400 MHz) δ: 8.40 (s, 1H, C7-H), 8.25 (d, 1H, J=8.4Hz, C9-H), 7.95 (d, 1H, J=8.0Hz, C12-H), 7.83–7.87 (m, 3H, Ts-H, C11-H), 7.69 (t, 1H, J=7.2Hz, C10-H), 7.23 (d, 2H, J=8.4Hz, Ts-H), 7.12–7.14 (m, 3H, C14-H, p-CH3Ph-H), 7.05 (d, 2H, J=8.0Hz, p-CH3Ph-H), 5.52 (ABq, 2H, J=17.2Hz, C17-H), 5.27 (s, 2H, C5-H), 4.14–4.37 (m, 4H, C23-H, C30-H), 2.39 (s, 3H, p-CH3Ph), 2.29 (s, 3H, Ts-CH3), 2.08–2.26 (m, 2H, C18-H), 0.94 (t, 3H, J=7.2Hz, C19-H); 13C NMR (100 MHz, DMSO-d6) δ: 167.8, 166.9, 156.5, 152.2, 147.8, 146.1, 144.6, 141.3, 140.8, 135.8, 131.5, 130.3, 129.6, 128.8, 128.7, 127.9, 127.6, 125.6, 119.1, 95.0, 76.5, 66.4, 50.2, 42.4, 37.4, 30.4, 20.7, 20.5, 7.4; MS-ESI m/z: 691.2 [M+H]+.
4.2.12. Compound 13l
Yield 60%; m.p. 190–192 °C; 1H NMR (CDCl3, 400 MHz) δ: 9.16 (s, 1H, Py-H), 8.70 (d, 1H, J=3.6Hz, Py-H), 8.40 (s, 1H, C7-H), 8.23 (m, 2H, C9-H, Py-H), 7.95 (d, 1H, J=8.4Hz, C12-H), 7.85 (t, 1H, J=7.2Hz, C11-H), 7.68 (t, 1H, J=7.6Hz, C10-H), 7.38 (m, 1H, Py-H), 7.12 (m, 3H, C14-H, p-CH3OPh-H), 6.86 (d, 2H, J=8.8Hz, p-CH3OPh-H), 5.52 (ABq, 2H, J=17.6Hz, C17-H), 5.29 (s, 2H, C5-H), 4.14–4.34 (m, 4H, C23-H, C30-H), 3.76 (s, 3H, p-CH3OPh), 2.08–2.26 (m, 2H, C18-H), 0.94 (t, 3H, J=7.6Hz, C19-H); 13C NMR (100 MHz, DMSO-d6) δ: 167.6, 167.8, 167.6, 166.9, 158.1, 156.4, 152.2, 151.6, 147.8, 146.1, 145.9, 144.6, 139.5, 133.3, 131.4, 130.3, 130.0, 129.6, 128.8, 128.4, 127.9, 127.6, 125.9, 123.1, 118.9, 113.7, 94.9, 76.1, 76.6, 66.3, 54.9, 50.0, 42.5, 37.1, 30.3, 7.4; MS-ESI m/z: 694.2 [M+H]+.
4.2.13. Compound 13m
Yield 51%; m.p. 157–159 °C; 1H NMR (CDCl3, 400 MHz) δ: 8.35 (s, 1H, Nap-H), 8.27 (s, 1H, C7-H), 8.22 (d, 1H, J=8.4Hz, C9-H), 7.90 (d, 1H, J=8.0H, C12-H), 7.76–7.85 (m, 4H, Nap-H), 7.68 (d, 2H, J=8.4Hz, Nap-H), 7.58 (t, 1H, J=6.4Hz, C11-H), 7.52 (t, 1H, J=7.2Hz, C10-H), 7.11 (m, 3H, p-CH3OPh-H, C14-H), 6.82 (d, 2H, J=8.0Hz, p-CH3OPh-H), 5.53 (ABq, 2H, J=17.2Hz, C17-H), 5.10 (ABq, 2H, J=17.2Hz, C5-H), 4.15–4.47 (m, 4H, C23-H, C30-H), 3.74 (s, 3H, p-CH3OPh), 2.05–2.20 (m, 2H, C18-H), 0.93 (t, 3H, J=7.2Hz, C19-H); 13C NMR (100 MHz, DMSO-d6) δ: 167.9, 167.6, 166.9, 157.9, 156.4, 151.9, 147.7, 146.0, 144.8, 140.3, 133.3, 131.2, 131.1, 130.1, 130.0, 129.3, 129.0, 128.5, 128.3, 127.9, 127.8, 127.5, 127.3, 126.6, 126.0, 122.0, 118.7, 113.5,94.9, 76.7, 66.3, 54.7, 50.0, 42.5, 36.8, 7.4; MS-ESI m/z: 765.3 [M+Na]+.
4.2.14. Compound 13n
Yield 48%; m.p. 144–146°C; 1H NMR (CDCl3, 400 MHz) δ: 8.41 (s, 1H, C7-H), 8.21 (d, 1H, J=8.4Hz, C9-H), 7.96 (d, 1H, J=8.4Hz, C12-H), 7.85 (t, 1H, J=7.6Hz, C11-H), 7.69 (t, 1H, J=7.6Hz, C10-H), 7.16 (m, 3H, C14-H, p-CH3OPh-H), 6.85 (d, 2H, J=8.0Hz, p-CH3OPh-H), 5.53 (ABq, 2H, J=17.2Hz, C17-H), 5.30 (s, 2H, C5-H), 4.10–4.33 (m, 4H, C23-H, C30-H), 3.75 (s, 3H, p-CH3OPh), 3.06–3.14 (m, 2H, -CH2CH3), 2.10–2.29 (m, 2H, C18-H), 1.26–1.37 (m, 3H, -CH2CH3), 0.96 (t, 3H, J=7.6Hz, C19-H); 13C NMR (100 MHz, CDCl3) δ: 167.5, 167.2, 166.9, 159.3, 157.2, 152.0, 148.7, 146.5, 145.0, 131.2, 130.7, 129.4, 128.4, 128.1, 124.1, 120.0, 114.7, 95.5, 76.7, 67.1, 55.1, 50.0, 49.3, 42.9, 38.6, 31.8, 8.4, 7.4; MS-ESI m/z: 667.1 [M+Na]+.
4.2.15. Compound 14a
Yield 52%; m.p. 119–121°C; 1H NMR (CDCl3, 400 MHz) δ: 8.42 (s, 1H, C7-H), 8.24 (d, 1H, J=8.4Hz, C9-H), 7.95 (d, 1H, J=8.0Hz, C12-H), 7.84 (m, 3H, C11-H, Ts-H), 7.68 (t, 1H, J=7.2Hz, C10-H), 7.27 (m, 2H, Ts-H), 7.15 (s, 1H, C14-H), 7.01 (d, 2H, J =8.4 Hz, p-CH3OPh-H), 6.74 (d, 2H, J =8.4Hz, p-CH3OPh-H), 5.52 (ABq, 2H, J=17.2Hz, C17-H), 5.31 (s, 2H, C5-H), 4.14 (m, 2H, C30-H), 3.77 (s, 3H, p-CH3OPh), 3.47–3.63 (m, 2H, -CH2CH2NH-), 2.66–2.74 (m, 2H, -CH2CH2NH-), 2.40 (s, 3H, Ts-CH3), 2.05–2.18 (m, 2H, C18-H), 0.95 (t, 3H, J=7.6Hz, C19-H); 13C NMR (100 MHz, CDCl3) δ: 171.1, 167.3, 159.2, 157.2, 152.1, 148.8, 146.4, 145.7, 142.1, 140.7, 131.3, 131.1, 130.8, 129.5, 129.2, 128.4, 128.2, 126.3, 124.4, 119.7, 114.6, 109.7, 95.7, 76.6, 67.0, 55.2, 49.9, 38.7, 37.1, 32.8, 31.5, 21.4, 7.5; MS-ESI m/z: 721.3 [M+H]+.
4.2.16. Compound 14b
Yield 55%; m.p. 112–114 °C; 1H NMR (CDCl3, 400 MHz) δ: 8.41 (s, 1H, C7-H), 8.22 (d, 1H, J=8.8 Hz, C9-H), 7.95 (d, 1H, J=8.0Hz, C12-H), 7.85 (m, 3H, C11-H, Ts-H), 7.69 (t, 1H, J=7.6Hz, C10-H), 7.26 (m, 2H, Ts-H), 7.15 (s, 1H, C14-H), 7.08 (d, 2H, J=8.4Hz, p-CH3OPh-H), 6.85 (d, 2H, J=8.4Hz, p-CH3OPh-H), 5.53 (ABq, 2H, J=17.2Hz, C17-H), 5.29 (s, 2H, C5-H), 4.17 (s, 2H, C30-H), 3.77 (s, 3H, p-CH3OPh), 3.26–3.34 (m, 2H, -CH2CH2CH2NH-), 2.40–2.46 (m, 2H, -CH2CH2CH2NH-), 2.36 (s, 3H, Ts-CH3), 2.08–2.21 (m, 2H, C18-H), 1.75–2.08 (m, 2H, -CH2CH2CH2NH-), 0.95 (t, 3H, J=7.6Hz, C19-H); 13C NMR (100 MHz, CDCl3) δ: 171.9, 167.5, 167.3, 159.2, 157.2, 152.2, 148.8, 146.3, 145.7, 142.0, 140.7, 131.2, 131.1, 130.7, 129.5, 129.1, 128.4, 128.2, 128.0, 126.3, 124.7, 119.9, 114.7, 95.7, 76.1, 67.0, 55.2, 49.9, 40.7, 38.7, 31.6, 30.8, 23.1, 21.4, 7.5; MS-ESI m/z: 735.3 [M+H]+.
4.2.17. Compound 14c
Yield 50%; m.p. 166–168°C; 1H NMR (CDCl3, 400 MHz) δ: 8.02 (d, 1H, J=8.8Hz, C12-H), 7.82 (d, 1H, J=8.0Hz, C11-H), 7.38 (m, 3H, C9-H, Ts-H), 7.23 (d, 2H, J=8.4Hz, Ts-H), 7.08 (s, 1H, C14-H), 7.04 (d, 2H, J =8.4 Hz, p-CH3OPh-H), 6.79 (d, 1H, J= 8.4Hz, p-CH3OPh-H), 5.92 (s, 1H, -C10-OH), 5.51 (ABq, 2H, J=17.2Hz, C17-H), 5.10 (m, 2H, C5-H), 4.38 (dd, 2H, J=5.2, 20.8Hz, C23-H), 4.19 (m, 2H, C30-H), 3.70 (s, 3H, p-CH3OPh), 2.98 (m, 2H, C7-CH2CH3), 2.39 (s, 3H, Ts-CH3), 2.05–2.21 (m, 2H, C18-H), 1.28 (d, 3H, J=7.2Hz, C7-CH2CH3), 0.92 (t, 3H, J=7.2Hz, C19-H); 13C NMR (100 MHz, CDCl3) δ: 167.6, 167.0, 159.3, 157.3, 156.5, 148.5, 147.4, 145.5, 144.4, 143.8, 142.6, 139.9, 131.5, 131.1, 129.3, 128.5, 126.8, 126.4, 123.8, 122.8, 118.8, 114.7, 109.8, 105.4, 95.4, 76.7, 67.0, 55.1, 49.4, 43.2, 38.3, 31.6, 22.9, 21.4, 13.5, 7.4; MS-ESI m/z: 751.5 [M+H]+.
4.2.18. Compound 14d
Yield 52%; m.p. 168–179°C; 1H NMR (CDCl3, 400 MHz) δ: 8.03 (s, 1H, C7-H), 7.98 (d, 1H, J=9.2Hz, C12-H), 7.82 (d, 2H, J=8.0Hz, Ts-H), 7.40 (m, 1H, C11-H), 7.22 (d, 2H, J=8.0Hz, Ts-H), 7.14 (s, 1H, C9-H), 7.10 (s, 1H, C14-H), 7.03 (d, 2H, J=8.4Hz, p-CH3OPh-H), 6.76 (d, 1H, J=8.4Hz, p-CH3OPh-H), 5.99 (s, 1H, -C10-OH), 5.51 (ABq, 2H, J=16.8Hz, C17-H), 5.08 (m, 2H, C5-H), 4.41 (dd, 2H, J=4.8, 19.2Hz, C23-H), 4.16 (m, 2H, C30-H), 3.67 (s, 3H, p-CH3OPh), 2.38 (s, 3H, Ts-CH3), 2.05–2.22 (m, 2H, C18-H), 0.94 (t, 3H, J=7.2Hz, C19-H); 13C NMR (100 MHz, CDCl3) δ: 167.9, 167.0, 159.2, 157.2, 156.5, 148.7, 146.7, 145.7, 143.7, 142.6, 139.9, 131.0, 130.6, 129.7, 129.3, 128.6, 126.4, 124.0, 123.3, 118.7, 114.6, 109.1, 95.5, 76.7, 66.9, 55.0, 50.0, 43.2, 38.3, 31.5, 21.4, 7.5; MS-ESI m/z: 745.2 [M+Na]+.
4.2.19. Compound 14e
Yield 52%; m.p.210–212 °C; 1H NMR (DMSO-d6, 400 MHz) δ: 8.93 (br, 1H, NH), 8.04 (d, 1H, J=9.2Hz, C12-H), 7.90 (s, 1H, C9-H), 7.74 (d, 1H, J=8.4Hz, C11-H), 7.54 (d, 2H, J=7.6Hz, Ts-H), 7.18 (d, 2H, J=8.0Hz, Ts-H), 7.03 (m, 3H, C14-H, p-CH3OPh-H), 6.77 (d, 2H, J=8.4Hz, p-CH3OPh-H), 5.51 (s, 2H, C17-H), 5.31 (s, 2H, C5-H), 4.36 (dd, 1H, J=5.6, 17.6Hz, C23-H), 4.24 (dd, 1H, J=4.8, 17.6Hz, C23-H), 4.00 (s, 2H, C30-H), 3.69 (s, 3H, p-CH3OPh), 3.48 (d, 3H, J=10.8Hz, P=O(OCH3)), 3.11–3.17 (m, 2H, C7-CH2CH3), 2.21 (s, 3H, Ts-CH3), 2.10–2.16 (m, 2H, C18-H), 1.30 (t, 3H, J=6.8Hz, C7-CH2CH3), 0.85 (t, 3H, J=6.8Hz, C19-H); MS-ESI m/z: 845.2 [M+H]+.
4.2.20. Compound 15a
Yield 52%; m.p. 141–143°C; 1H NMR (DMSO-d6, 400 MHz) δ: 9.09 (s, 1H, NH), 8.29 (d, 1H, J=8.3Hz, C9-H), 8.13 (d, 1H, J=8.5 Hz, C12-H), 7.83 (t, 1H, J=7.0Hz, C11-H), 7.73 (t, 1H, J=7.6Hz, C10-H), 7.58 (d, 2H, J=8.2Hz, Ts-H), 7.28 (d, 2H, J=8.7Hz, Ts-H), 7.07 (d, 2H, J=8.2Hz, p-CH3OPh-H), 6.95 (s, 1H, C14-H), 6.79 (d, 2H, J=8.7Hz, p-CH3OPh-H), 5.50 (s, 2H, C17-H), 5.32 (s, 2H, C5-H), 4.80 (m, 1H, C23-H), 4.16 (d, 1H, J=14.2Hz, C30-H), 3.95 (d, 1H, J=14.2Hz, C30-H), 3.66 (s, 3H, p-CH3OPh), 3.23 (q, 2H, J=7.6Hz, C7-CH2CH3), 2.22 (s, 3H, Ts-CH3), 2.04–2.07 (m, 2H, C18-H), 1.46 (d, 3H, J=7.3Hz, L-alanine-CH3), 1.31 (t, 3H, J=7.5Hz, C7-CH2CH3), 0.77 (t, 3H, J=7.4Hz, C19-H); 13C NMR (100 MHz, DMSO-d6) δ: 170.1, 166.8, 166.0, 158.1, 156.5, 151.6, 148.5, 146.6, 145.6, 144.5, 141.3, 140.9, 130.0, 128.8, 127.9, 127.6, 126.9, 126.6, 125.8, 124.1, 119.0, 113.7, 94.6, 76.4, 66.4, 55.0, 49.6, 49.1, 37.1, 30.2, 22.2, 20.8, 16.6, 14.0, 7.4 ; MS-ESI m/z: 749.3 [M+H]+.
4.2.21. Compound 15b
Yield 55%; m.p. 139–141 °C; 1H NMR (DMSO-d6, 400 MHz) δ: 9.18 (s, 1H, NH), 8.29 (d, 1H, J=8.6Hz, C9-H), 8.13 (d, 1H, J=7.8Hz, C12-H), 7.83 (t, 1H, J=7.0Hz, C11-H), 7.73 (t, 1H, J=7.7Hz, C10-H), 7.59 (d, 2H, J=8.2Hz, Ts-H), 7.36 (d, 2H, J=7.3Hz, Ts-H), 7.25 (t, 2H, J=7.1Hz, Ph-H), 7.18 (t, 1H, J=7.2Hz, Ph-H), 7.07 (d, 2H, J=8.0Hz, Ph-H), 6.95 (s, 1H, C14-H), 5.49 (s, 2H, C17-H), 5.33 (s, 2H, C5-H), 4.81 (m, 1H, C23-H), 4.25 (d, 1H, J=14.4Hz, C30-H), 4.03 (d, 1H, J=14.4Hz, C30-H), 3.23 (q, 2H, J=7.2Hz, C7-CH2CH3), 2.21 (s, 3H, Ts-CH3), 2.01–2.08 (m, 2H, C18-H), 1.46 (d, 3H, J=7.3Hz, L-alanine-CH3), 1.31 (t, 3H, J=7.5Hz, C7-CH2CH3), 0.76 (t, 3H, J=7.3Hz, C19-H); 13C NMR (100 MHz, DMSO-d6) δ: 170.1, 166.8, 165.5, 156.5, 151.6, 148.5, 146.6, 145.6, 144.5, 141.4, 140.8, 135.2, 130.0, 128.8, 128.7, 128.3, 127.9, 127.6, 126.6, 125.8, 124.1, 119.0, 94.6, 76.4, 66.4, 49.6, 49.1, 37.9, 30.2, 22.2, 20.8, 16.6, 14.0, 7.4 ; MS-ESI m/z: 719.3 [M+H]+.
4.2.22. Compound 15c
Yield 56%; m.p. 146–148 °C; 1H NMR (DMSO-d6, 400 MHz) δ: 8.88 (s, 1H, NH), 8.28 (d, 1H, J=8.3Hz, C9-H), 8.13 (d, 1H, J=7.9Hz, C12-H), 7.83 (t, 1H, J=7.1Hz, C11-H), 7.71 (t, 1H, J=7.7Hz, C10-H), 7.52 (d, 2H,J= 8.2Hz, Ts-H), 7.18 (d, 2H, J=8.7Hz, Ts-H), 7.08 (s, 1H, C14-H), 7.01 (d, 2H, J=8.0Hz, p-CH3OPh-H), 6.76 (d, 2H, J= 8.7Hz, p-CH3OPh-H), 5.51 (s, 2H, C17-H), 5.32 (s, 2H, C5-H), 4.35 (dd, 1H, J=5.9, 17.9Hz, C23-H), 4.28 (dd, 1H, J=5.8, 17.9Hz, C23-H), 4.00 (s, 2H, C30-H), 3.64 (s, 3H, p-CH3OPh), 3.21–3.23 (m, 2H, C7-CH2CH3), 2.19 (s, 3H, Ts-CH3), 2.10–2.12 (m, 2H, C18-H), 1.30 (t, 3H, J=7.5Hz, C7-CH2CH3), 0.84 (t, 3H, J=7.3Hz, C19-H); 13C NMR (100 MHz, DMSO-d6) δ: 167.7, 167.1, 166.9, 158.0, 156.4, 151.6, 148.4, 146.6, 145.4, 144.6, 141.2, 140.7, 130.0, 129.8, 128.7, 127.8, 127.5, 126.5, 126.3, 125.6, 118.8, 113.6, 94.9, 76.5, 66.3, 54.9, 49.4, 42.3, 36.9, 30.3, 22.1, 20.7, 13.9, 7.4; MS-ESI m/z: 735.8 [M+H]+.
4.2.23. Compound 15d
Yield 54%; m.p. 145–147 °C; 1H NMR (DMSO-d6, 400 MHz) δ: 9.03 (s, 1H, NH), 8.28 (d, 1H, J=8.3Hz, C9-H), 8.13 (d, 1H, J=7.9Hz, C12-H), 7.83 (t, 1H, J=7.5Hz, C11-H), 7.72 (t, 1H, J=7.6Hz, C10-H), 7.52 (d, 2H, J=8.2Hz, Ts-H), 7.26 (d, 2H, J=7.0Hz, Ts-H), 7.15–7.23 (m, 3H, Ph-H), 7.09 (s, 1H, C14-H), 7.00 (d, 2H, J=8.0Hz, Ph-H), 5.51 (s, 2H, C17-H), 5.32 (s, 2H, C5-H), 4.38 (dd, 1H, J=5.8, 18.0Hz, C23-H), 4.26 (dd, 1H, J=5.9, 18.0Hz, C23-H), 4.08 (s, 2H, C30-H), 3.22 (q, 2H, J=7.4Hz, C7-CH2CH3), 2.18 (s, 3H, Ts-CH3), 2.08–2.14 (m, 2H, C18-H), 1.29 (t, 3H, J=7.5Hz, C7-CH2CH3), 0.73 (t, 3H, J=8.3Hz, C19-H); 13C NMR (100 MHz, DMSO-d6) δ: 167.8, 166.9, 166.6, 156.5, 151.7, 148.5, 146.6, 145.5, 144.6, 141.4, 140.7, 134.8, 129.9, 128.8, 128.7, 128.3, 127.8, 127.5, 126.6, 126.5, 125.7, 124.0, 118.9, 94.9, 76.6, 66.4, 49.5, 42.4, 37.8, 30.4, 22.1, 20.7, 13.9, 7.4; MS-ESI m/z: 705.3 [M+H]+.
4.2.24. Compound 15e
Yield 59%; m.p. 132–134 °C; 1H NMR (DMSO-d6, 400 MHz) δ: 8.93 (s, 1H, NH), 8.25 (d, 1H, J=8.2Hz, C9-H), 8.14 (d, 1H, J=8.1Hz, C12-H), 7.85 (t, 1H, J=7.0Hz, C11-H), 7.73 (t, 1H, J=7.4Hz, C10-H), 7.38 (d, 2H, J=8.4Hz, p-CH3OPh-H), 6.98 (s, 1H, C14-H), 6.86 (d, 2H, J=8.4Hz, p-CH3OPh-H), 5.50 (s, 2H, C17-H), 5.33 (s, 2H, C5-H), 4.63 (t, 1H, J=7.1Hz, C23-H), 4.12 (d, 1H, J=14.0Hz, C30-H), 3.93 (d, 1H, J=14.1Hz, C30-H), 3.67 (s, 3H, p-CH3OPh), 3.21 (m, 2H, C7-CH2CH3), 2.92 (s, 3H, Ms-CH3), 2.11 (q, 2H, J=7.1Hz, C18-H), 1.46 (d, 3H, J=7.2Hz, L-alanine-CH3), 1.29 (t, 3H, J=7.2Hz, C7-CH2CH3), 0.82 (t, 3H, J=7.0Hz, C19-H); 13C NMR(100 MHz, DMSO-d6) δ: 170.4, 166.8, 165.7, 158.1, 156.5, 151.6, 148.4, 146.6, 145.6, 144.8, 130.3, 130.0,129.8, 127.9, 127.6, 127.1, 126.6, 124.0, 118.9, 113.8, 113.7, 94.5, 76.5, 66.3, 55.0, 49.5, 49.3, 43.1, 36.9, 30.2, 22.2, 16.4, 13.9, 7.4; MS-ESI m/z: 673.3 [M+H]+.
4.2.25. Compound 15f
Yield 60%; m.p. 126–128 °C; 1H NMR (DMSO-d6, 400 MHz) δ: 8.71 (s, 1H, NH), 8.29 (d, 1H, J=8.4Hz, C9-H), 8.13 (d, 1H, J=7.8Hz, C12-H), 7.85 (t, 1H, J=7.5Hz, C11-H), 7.73 (t, 1H, J=7.6Hz, C10-H), 7.27 (d, 2H, J= 8.7Hz, p-CH3OPh-H), 7.06 (s, 1H, C14-H), 6.83 (d, 2H, J=8.7Hz, p-CH3OPh-H), 5.51 (s, 2H, C17-H), 5.33 (d, 2H, J=4.3Hz, C5-H), 4.30 (dd, 1H, J=6.0, 18.0Hz, C23-H), 4.13 (dd, 1H, J=5.8, 18.0Hz, C23-H), 4.00 (d, 1H, J=15.2Hz, C30-H), 3.97 (d, 1H, J=14.7Hz, C30-H), 3.67 (s, 3H, p-CH3OPh), 3.21 (m, 2H, C7-CH2CH3), 2.85 (s, 3H, Ms-CH3), 2.10–2.15 (m, 2H, C18-H), 1.30 (t, 3H, J=7.5Hz, C7-CH2CH3), 0.88 (t, 3H, J=7.3Hz, C19-H); 13C NMR (100 MHz, DMSO-d6) δ: 167.8, 166.9, 166.6, 158.2, 156.5, 151.6, 148.3, 146.6, 145.6, 145.1, 130.3, 129.7, 128.0, 127.6, 126.6, 124.0, 118.7, 113.8, 94.7, 76.6, 66.2, 55.0, 49.5, 43.2, 42.3, 37.1, 30.2, 22.2, 13.9, 7.5; MS-ESI m/z: 659.3 [M+H]+.
4.2.26. Compound 15g
Yield 60%; m.p. 133–135°C; 1H NMR (DMSO-d6, 400 MHz) δ: 9.27 (s, 1H, NH), 8.29 (d, 1H, J =8.5Hz, C9-H), 8.13 (d, 1H, J=7.9Hz, C12-H), 7.83 (t, 1H, J=7.0Hz, C11-H), 7.72 (t, 1H, J=7.7Hz, C10-H), 7.57 (d, 2H,J=8.2Hz, Ts-H), 7.36 (d, 2H, J=8.5Hz, p-ClPh-H), 7.27 (d, 2H, J=8.5Hz, Ts-H), 7.06 (d, 2H, J=8.1Hz, p-ClPh-H), 6.94 (s, 1H, C14-H), 5.51 (s, 2H, C17-H), 5.33 (s, 2H, C5-H), 4.81 (m, 1H, C23-H), 4.25 (d, 1H, J=14.4Hz, C30-H), 3.98 (d, 1H, J=14.5Hz, C30-H), 3.29 (q, 2H, J=7.6Hz, C7-CH2CH3), 2.22 (s, 3H, Ts-CH3), 2.03–2.08 (m, 2H, C18-H), 1.46 (d, 3H, J=7.3Hz, L-alanine-CH3), 1.31 (t, 3H, J=7.5Hz, C7-CH2CH3), 0.77 (t, 3H, J=7.3Hz, C19-H); 13C NMR (100 MHz, DMSO-d6) δ: 170.0, 166.7, 165.1, 156.5, 151.6, 148.5, 146.6, 145.6, 144.4, 141.4, 140.7, 134.0, 131.4, 130.6, 129.9, 128.7, 128.1, 127.8, 127.6, 126.6, 125.7, 124.0, 119.0, 94.5, 76.4, 66.4, 49.5, 49.2, 37.2, 30.2, 22.2, 20.8, 16.5, 13.9, 7.3; MS-ESI m/z: 753.3 [M+H]+.
4.2.27. Compound 16a
Yield 30%; m.p. 160–162°C; 1H NMR (DMSO-d6, 400 MHz) δ: 9.15 (s, 1H, NH), 8.31 (s, 1H, C7-H), 8.13–8.21 (m, 2H, C9-H, C12-H), 7.86–7.94 (m, 1H, C10-H), 7.68–7.79 (m, 3H, C11-H, Ts-H), 7.50 (d, 2H, J=8.4Hz, Ts-H), 7.29 (s, 1H, C14-H), 7.03–7.07 (m, 2H, p-CH3OPh-H), 6.79 (d, 2H, J=8.8Hz, p-CH3OPh-H), 5.83 (m, 2H, C17-H), 5.55 (m, 2H, C5-H), 4.81 (m, 1H, C23-H), 3.91–4.20 (m, 2H, C30-H), 3.66 (s, 3H, p-CH3OPh), 2.21 (s, 3H, Ts-CH3), 2.02–2.09 (m, 2H, C18-H), 1.46 (d, 3H, J=7.2Hz, L-alanine-CH3), 0.75–0.86 (m, 3H, C19-H); MS-ESI m/z : 774.9 [M+K]+.
4.2.28. Compound 16b
Yield 28%; m.p. 160–162°C; 1H NMR (DMSO-d6, 400 MHz) δ: 8.97 (s, 1H, NH), 8.33(s, 1H, C7-H), 8.12–8.20 (m, 1H, C9-H), 7.86–7.94 (m, 1H, C12-H), 7.72–7.79 (m, 2H, C10-H, C11-H), 7.44–7.56 (m, 3H, C14-H, Ts-H), 7.16–7.24 (m, 2H, Ts-H), 7.01 (d, 2H, J=8.2Hz, p-CH3OPh-H), 6.74–6.76 (m 2H, p-CH3OPh-H), 5.85 (m, 2H, C17-H), 5.53 (m, 2H, C5-H), 4.22–4.24 (m, 2H, C23-H), 3.99 (s, 2H, C30-H), 3.63 (s, 3H, p-CH3OPh), 2.20 (s, 3H, Ts-CH3), 2.12–2.17 (m, 2H, C18-H), 0.81–0.84 (m, 3H, C19-H); MS-ESI m/z: 723.0 [M+H]+.
4.2.29. Compound 16c
Yield 59%; m.p. 162–164 °C; 1H NMR (DMSO-d6, 400 MHz) δ: 9.01 (s, 1H, NH), 8.87(s, 1H, C7-H), 8.43 (d, 1H, J=7.4 Hz, C12-H), 8.38 (t, 1H, J=6.3Hz, C10-H), 7.85 (t, 1H, J=7.6Hz, C11-H), 7.40 (d, 2H, J=8.2Hz, Ts-H), 7.16–7.26 (m, 5H, Ts-H, Ph-H), 7.06 (d, 2H, J=8.0Hz, Ph-H), 7.01 (s, 1H, C14-H), 5.49 (m, 2H, C17-H), 5.28 (m, 2H, C5-H), 4.25 (d, 2H, J=5.8Hz, C23-H), 4.05 (dd, 2H, J=4.0, 14.6Hz, C30-H), 2.23 (s, 3H, Ts-CH3), 2.15–2.18 (m, 2H, C18-H), 0.80–0.88 (m, 3H, J=7.3Hz, C19-H); 13C NMR (100 MHz, DMSO-d6) δ: 167.9, 166.6, 156.3, 154.2, 147.7, 145.3, 143.7, 141.3, 140.7, 138.6, 134.8, 131.6, 128.8, 128.5, 128.2, 126.6, 125.5, 120.8, 95.6, 76.1, 66.7, 50.2, 42.4, 37.7, 30.9, 20.8, 7.3; MS-ESI m/z: 722.2 [M+H]+.
4.2.30. Compound 16d
Yield 55%; m.p. 161–163 °C; 1H NMR (CDCl3, 400 MHz) δ: 8.49 (s,1H,C7-H), 8.16 (d, 1H, J=2.8Hz, C12-H), 814 (d, 1H, J=1.6Hz, C10-H), 7.80 (d, 2H, J=8.4Hz, Ts-H), 7.75 (t, 1H, J=7.6Hz, C11-H), 7.29 (d, 2H, J=8.0Hz, Ts-H), 7.11 (s, 1H, C14-H), 7.09 (d, 2H, J=3.6Hz, p-CH3OPh-H), 6.84 (d, 2H, J=8.6Hz, p-CH3OPh-H), 5.52 (ABq, 2H, J=17.6Hz, C17-H), 5.30 (s, 2H, C5-H), 4.20 (m, 4H, C23-H, C30-H), 3.75 (s, 3H, p-CH3OPh), 2.42 (s, 3H, Ts-CH3), 2.09–2.30 (m, 2H, C18-H), 0.90 (t, 3H, J=7.5Hz, C19-H); 13C NMR (100 MHz, CDCl3) δ: 167.7, 166.9, 166.6, 159.2, 156.9, 154.2, 148.2, 145.4, 144.5, 142.4, 140.1, 139.9, 132.2, 131.4, 131.2, 130.3, 129.2, 128.6, 126.3, 124.6, 124.0, 121.6, 114.7, 114.6, 96.9, 76.7, 67.2, 55.2, 49.9, 43.2, 38.4, 31.8, 21.3, 7.4; MS-ESI m/z: 752.2 [M+H]+.
4.2.31. Compound 16e
Yield 33%; m.p.163–165°C; 1H NMR (CDCl3, 400 MHz) δ: 8.56 (s, 1H, C7-H), 8.15 (m, 2H, C9-H, C11-H), 7.74–7.79 (m, 3H, C10-H, Ts-H), 7.27 (s, 1H, C14-H), 7.01–7.11 (m, 4H, Ts-H, p-CH3OPh-H), 6.85 (m, 2H, p-CH3OPh-H), 5.54 (m, 2H, C17-H), 5.37 (m, 2H, C5-H), 4.12–4.19 (m, 4H, C23-H, C30-H), 3.75 (s, 3H, p-CH3OPh), 2.42 (s, 3H, Ts-CH3), 2.05–2.18 (m, 2H, C18-H), 0.93 (t, 3H, J=7.6Hz, C19-H); MS-ESI m/z: 790.2 [M+K]+.
4.2.32. Compound 16f
Yield 45%; m.p. 131–133°C; 1H NMR (CDCl3, 400 MHz) δ: 8.42 (s, 1H, C7-H), 8.22 d, 1H, J=8.4Hz, C9-H), 7.95 (d, 1H, J=8.0, C12-H), 7.81–7.86 (m, 3H, C11-H, Ts-H), 7.68 (t, 1H, J=7.2Hz, C10-H), 7.23–7.25 (m, 2H, Ts-H), 7.07–7.10 (m, 2H, p-CH3OPh-H), 6.99 (s, 1H, C14-H), 6.83–6.87 (m, 2H, p-CH3OPh-H), 5.76–5.79 (m, 1H, C5-H), 5.43 (ABq, 2H, J=17.6Hz, C17-H), 4.13–4.38 (m, 4H, C23-H, C30-H), 3.76 (s, 3H, p-CH3OPh), 3.64 (s, 3H, 5-OCH3), 2.38 (s, 3H, Ts-H), 2.05–2.23 (m, 2H, C18-H), 0.95 (m, 3H, C19-H); 13C NMR (100 MHz, DMSO-d6) δ: 167.9, 167.2, 166.6, 158.1, 157.2, 150.8, 148.9, 145.4, 144.3, 141.2, 140.8, 131.1, 130.1, 129.6, 129.0, 128.7, 128.1, 126.3, 125.6, 121.8, 121.5, 113.7, 95.3, 89.5, 76.3, 66.3, 57.5, 54.9, 42.3, 37.0, 30.1, 20.7, 7.4; MS-ESI m/z: 759.1 [M+Na]+.
4.2.33. Compound 16g
Yield 48%; m.p. 201–203 °C; 1H NMR (CDCl3, 400 MHz) δ: 8.37 (s, 1H, C7-H), 8.18 (d, 1H, J=8.4Hz, C9-H), 7.93 (d, 1H, J=8.0Hz, C12-H), 7.81–7.86 (m, 3H, C11-H, Ts-H), 7.67 (t, 1H, J=7.6Hz, C10-H), 7.29 (d, 2H, J=7.6Hz, Ts-H), 7.10 (m, 3H, C14-H, CH3OPh-H), 6.84 (d, 2H, J=8Hz, CH3OPh-H), 5.57 (ABq, 2H, J=17.2Hz, C17-H), 5.23 (s, 2H, C5-H), 3.92–4.32 (m, 4H, C30-H, C23-H), 3.74 (s, 3H, p-CH3OPh), 2.75–2.96 (m, 2H, E-ring-H), 2.38 (s, 3H, Ts-CH3), 1.78–2.20 (m, 2H, C18-H), 1.05 (t, 3H, J=7.2Hz, C19-H); 13C NMR (100 MHz, CDCl3) δ: 170.2, 167.4, 167.0, 159.4, 152.3, 152.1, 148.7, 145.7, 142.7, 131.3, 129.3, 128.6, 128.1, 126.6, 126.3, 124.1, 122.5, 114.8, 97.7, 76.7, 62.2, 55.2, 50.3, 43.9, 38.5, 37.5, 29.6, 21.4, 7.4; MS-ESI m/z: 743.2 [M+Na]+.
4.2.34. Compound 16h
Yield 30%; m.p. 130–133 °C; 1H NMR (CDCl3, 400 MHz) δ: 8.17 (s, 1H, C7-H), 8.09 (d, 1H, J=8.8 Hz, C9-H), 7.84 (d, 2H, J =8.0 Hz, Ts-H), 7.73 (t, 1H, J=7.2Hz, C11-H), 7.68 (d, 1H, J=8.4Hz, C12-H), 7.54 (s, 1H, C14-H), 7.50 (t, 1H, J=7.6Hz, C10-H), 7.37 (s, 1H, O=CNH-), 7.26–7.28 (m, 2H, Ts-H), 7.16 (d, 2H, J=8.4Hz, p-CH3OPh-H), 6.87 (d, 2H, J=8.4Hz, p-CH3OPh-H), 6.40 (s, 1H, C20-OH), 5.43 (s, 2H, C17-H), 5.07–5.20 (m, 2H, C5-H), 4.14–4.25 (m, 2H, 30-H), 3.73 (s, 3H, p-CH3OPh), 3.25–3.50 (m, 4H, O=CNHCH2CH2-), 2.38 (s, 3H, Ts-CH3), 2.27–2.33 (m, 2H, C18-H), 2.02 (s, 3H, OCOCH3), 0.96 (t, 3H, J=6.8Hz, C19-H); MS-ESI m/z: 774.2 [M+Na]+.
4.3. Cytotoxicity assays
Cytotoxic activity was determined by the sulforhodamine B (SRB) colorimetric assay as previously described [22]. In brief, the cells (3–5 × 103 cells/well) were seeded in 96-well plates filled with RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) containing various concentrations of samples, and incubated for 72 h. At the end of the exposure period, the attached cells were fixed with cold 50% trichloroacetic acid for 30 min followed by staining with 0.04% SRB (Sigma Chemical Co.) for 30 min. The bound SRB was solubilized in 10 mM Tris-base and the absorbance was measured at 515 nm on a Microplate Reader ELx800 (Bio-Tek Instruments, Winooski, VT) with a Gen5 software. All results were representative of three or more experiments.
4.4. Molecular docking study
The binding modes of two compounds 9 and 15c were investigated using molecular docking modeling in the AutoDock 4.2 software. Before starting the docking process, the protein structure was subjected to optimization step in order to minimize the crystallographic induced bond clashes. The Kollman united atom charges and polar hydrogen was added to the receptor and the crystallographic waters were removed. PyMol was used to construct the 3D structure of the compounds 9 and 15c on the basis of camptothecin in the crystal. Charges of the Gasteiger type were assigned to the new constructed structures in AutoDock. Non-polar hydrogen atoms were merged and rotatable bonds were defined. The grid maps of the protein were calculated using AutoGrid module embedded in AutoDock software. The grid was set in a way to include not only the active site amino acids but also the considerable portions of the surrounding surface. Hence, a grid size of 60×60×60 Å points and 0.375 Å spacing were generated based on the binding position of camptothecin in the protein. Docking simulations were performed using the autodock module of the software. Every docking program was taken out in 250000 energy evaluation with 15 conformations kept and the most favorable pose of each compound was exhibited.
Supplementary Material
HIGHLIGHTS.
20(S)-Sulfonylamidine CPT-derivatives were prepared and tested for cytotoxicity.
Several analogues showed superior cytotoxic activity compared to irinotecan.
Key structural features related to cytotoxicity were identified by SAR analysis.
Compounds 9 and 15c interacted with Topo I-DNA by a different binding mode from CPT.
These compounds are new generation CPT-derived antitumor agents.
Acknowledgments
This work was supported financially by the National Natural Science Foundation of China (30800720, 31371975); Partial financial support was supplied by Xinjiang Production & Construction Corps Key Laboratory of Protection and Utilization of Biological Resources in Tarim Basin (BYRB1306) and the Fundamental Research Funds for the Central Universities (lzujbky-2014-k19). Support was also supplied by NIH grant CA177584 from the National Cancer Institute awarded to K.H. Lee. Thanks are also due to the support of Health and Welfare Surcharge of Tobacco Products, China Medical University Hospital Cancer Research Center of Excellence (MOHW103-TD-B-111-03, Taiwan).
Footnotes
Figure S1. Ternary complex of DNA, Topo I, and 1; Figure S2. Docked 1; Figure S3. Binding mode of 2; Representative NMR spectra for compound 13c.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wall ME, Wani MC, Cook CE, Palmer KH, McPhail AT, Sim GA. Plant antitumor agents I. The isolation and structure of camptothecin, a novel alkaloidal leukemia and tumor inhibitor from Camptotheca acuminata. J Am Chem Soc. 1966;88:3888–90. [Google Scholar]
- 2.Slichenmyer WJ, Rowinsky EK, Donehower RC, Kaufmann SH. The current status of camptothecin analogues as antitumor agents. J Nat Cancer Inst. 1993;85:271–291. doi: 10.1093/jnci/85.4.271. [DOI] [PubMed] [Google Scholar]
- 3.Potmesil M, Pinedo H. Camptothecins: New Anticancer Agents. CRC; Boca Raton: 1995. [Google Scholar]
- 4.Liew ST, Yang LX. Design, synthesis and development of novel camptothecin drugs. Curr Pharm Des. 2008;14:1078–1097. doi: 10.2174/138161208784246180. [DOI] [PubMed] [Google Scholar]
- 5.Li QY, Zu YG, Shi RZ, Yao LP. Review camptothecin: current perspectives. Curr Med Chem. 2006;13:2021–2039. doi: 10.2174/092986706777585004. [DOI] [PubMed] [Google Scholar]
- 6.Zunino F, Dallavalle S, Laccabue D, Beretta G, Merlini L, Pratesi G. Current status and perspectives in the development of camptothecins. Curr Pharm Des. 2002;8:2505–2520. doi: 10.2174/1381612023392801. [DOI] [PubMed] [Google Scholar]
- 7.Adams DJ. The impact of tumor physiology on camptothecin-based drug development. Curr Med Chem - Anti-Cancer Agents. 2005;5:1–13. doi: 10.2174/1568011053352596. [DOI] [PubMed] [Google Scholar]
- 8.Tobin PJ, Rivory LP. Camptothecins and key lessons in drug design. Drug Design Reviews - Online. 2004;1:341–346. [Google Scholar]
- 9.Hatefi A, Amsden B. Camptothecin delivery methods. Pharm Res. 2002;19:1389–1399. doi: 10.1023/a:1020427227285. [DOI] [PubMed] [Google Scholar]
- 10.Onishi H, Machida Y. Macromolecular and nanotechnological modification of camptothecin and its analogs to improve the efficacy. Curr Drug Discov Technol. 2005;2:169–183. doi: 10.2174/1570163054866891. [DOI] [PubMed] [Google Scholar]
- 11.Demarquay D, Huchet M, Coulomb H. The homocamptothecin BN 80915 is a highly potent orally active topoisomerase I poison. Anti-Cancer Drugs. 2001;12:9–19. doi: 10.1097/00001813-200101000-00003. [DOI] [PubMed] [Google Scholar]
- 12.Lavergne O, Lesueur-Ginot L, Pla Rodas F, Kasprzyk PG, Pommier J, Demarquay D, Prévost G, Ulibarri G, Rolland A, Schiano-Liberatore AM, Harnett J, Pons D, Camara J, Bigg DC. Homocamptothecins: Synthesis and antitumor activity of novel E-ring-modified camptothecin analogs. J Med Chem. 1998;41:5410–5419. doi: 10.1021/jm980400l. [DOI] [PubMed] [Google Scholar]
- 13.Zhao H, Lee C, Sai P, Choe YH, Boro M, Pendri A, Guan S, Greenwald RB. 20-O-Acylcamptothecin derivatives: evidence for lactone stabilization. J Org Chem. 2000;65:4601–4606. doi: 10.1021/jo000221n. [DOI] [PubMed] [Google Scholar]
- 14.Liu YQ, Tian X, Yang L, Zhan ZC. First synthesis of novel spin-labeled derivatives of camptothecin as potential antineoplastic agents. Europ J Med Chem. 2008;43:2610–2614. doi: 10.1016/j.ejmech.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 15.Yang L, Zhao CY, Liu YQ. Synthesis and biological evaluation of novel conjugates of camptothecin and 5-flurouracil as cytotoxic agents. J Braz Chem Soc. 2011;22:308–318. [Google Scholar]
- 16.Yang LX, Pan X, Wang HJ. Novel camptothecin derivatives. Part 1: oxyalkanoic acid esters of camptothecin and their in vitro and in vivo antitumor activity. Bioorg Med Chem Lett. 2002;12:1241–1244. doi: 10.1016/s0960-894x(02)00153-1. [DOI] [PubMed] [Google Scholar]
- 17.Cao Z, Harris N, Kozielski A, Vardeman D, Stehlin JS, Giovanella B. Alkyl esters of camptothecin and 9-nitrocamptothecin: synthesis, in vitro pharmacokinetics, toxicity, and antitumor activity. J Med Chem. 1998;41:31–37. doi: 10.1021/jm9607562. [DOI] [PubMed] [Google Scholar]
- 18.De Groot FMH, Busscher GF, Aben RWM, Scheeren HW. Novel 20-carbonate linked prodrugs of camptothecin and 9-aminocamptothecin designed for activation by tumour-associated plasmin. Bioorg Med Chem Lett. 2002;12:2371–2376. doi: 10.1016/s0960-894x(02)00388-8. [DOI] [PubMed] [Google Scholar]
- 19.Lerchen HG, Baumgarten J, dem Bruch K, Lehmann TE, Sperzel M, Kempka G, Fiebig HH. Synthesis of 20-O-linked 20(S)-camptothecin Glycoconjugates: impact of the side chain of the ester-linked amino acid on epimerization during the acylation reaction and on hydrolytic stability of the final glycoconjugates. J Med Chem. 2001;44:4186–4195. [Google Scholar]
- 20.Wang MJ, Liu YQ, Chang LC, Wang CY, Zhao YL, Zhao XB, Qian KD, Nan X, Yang L, Yang XM, Hung HY, Yang JS, Kuo DH, Goto M, Morris-Natschke SL, Pan SL, Teng CM, Kuo SC, Wu TS, Wu YC, Lee KH. Design, Synthesis, Mechanisms of Action, and Toxicity of Novel 20(S)-Sulfonylamidine Derivatives of Camptothecin as Potent Antitumor Agents. J Med Chem. 2014;57:6008–6018. doi: 10.1021/jm5003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoo EJ, Ahlquist M, Bae I, Sharpless KB, Fokin VV, Chang S. Mechanistic studies on the Cu-catalyzed three-component reactions of sulfonyl azides, 1-alkynes and amines, alcohols, or water: dichotomy via a common pathway. J Org Chem. 2008;73:5520–5528. doi: 10.1021/jo800733p. [DOI] [PubMed] [Google Scholar]
- 22.Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, Warren JT, Bokesch H, Kenney S, Boyd MR. New Colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990;82:1107–1112. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- 23.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DSAJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Computational Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RKAJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Computational Chem. 1998;19:1639–1662. [Google Scholar]
- 25.S. C. DeLano Scientific. The PyMOL molecular graphics system. 2002. [Google Scholar]
- 26.Kunimoto T, Nitta K, Tanaka T, Uehara N, Baba H, Takeuchi M, Yokokura T, Sawada S, Miyasaka T, Mutai M. Antitumor activity of 7-ethyl-10-[4-(1-piperidino)-1-piperidino]carbonyloxy-camptothecin, a novel water-soluble derivative of camptothecin, against murine tumors. Cancer Res. 1987;47:5944–5947. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.