Abstract
Next generation anti-androgen therapies such as enzalutamide and abiraterone have had a profound impact on the management of metastatic castration-resistant prostate cancer (mCRPC). However, mCRPC patients invariably develop resistance to these agents. Here, a series of clonal cell lines were developed from enzalutamide-resistant prostate tumor xenografts to study the molecular mechanism of resistance and test their oncogenic potential under various treatment conditions. Androgen receptor (AR) signaling was maintained in these cell lines which acquired potential resistance mechanisms including expression of AR-variant 7 (AR-v7) and glucocorticoid receptor (GR). BET bromodomain inhibitors were shown previously to attenuate AR signaling in mCRPC; here, we demonstrate the efficacy of BET inhibitors in enzalutamide-resistant prostate cancer models. AR antagonists, enzalutamide and ARN509 exhibit enhanced prostate tumor growth inhibition when combined with BET inhibitors, JQ1 and OTX015, respectively. Taken together, these data provide a compelling pre-clinical rationale to combine BET inhibitors with AR antagonists to subvert resistance mechanisms.
Implications
Therapeutic combinations of BET inhibitors and AR antagonists may enhance the clinical efficacy in the treatment of mCRPC.
Introduction
Metastatic castration-resistant prostate cancer (mCRPC) leads to nearly 30,000 deaths annually in the United States (1). AR is a major driver alteration in mCRPC (2–4) and most tumors continue to rely on androgen receptor signaling (3). Second generation anti-androgen therapies such as abiraterone and enzalutamide have become standard of care treatments for mCRPC (5,6). However, most patients eventually develop resistance to these therapies and, thus alternate approaches to target androgen signaling are needed.
Recently, our group and others have shown that bromodomain and extraterminal (BET) inhibitors block mCRPC growth in animal models (7–9). BET inhibitors are epigenetic therapies that target bromodomain containing proteins BRD2/3/4 and BRDT (10–12). They appear to preferentially affect oncogenic transcription through a super-enhancer based mechanism (13,14). mCRPC is a particularly attractive indication for BET inhibitors due to its reliance on oncogenic transcription factor signaling mediated by AR, ETS fusions, and MYC (7). Several groups including our own have shown that AR signaling is affected by BET inhibitors (7,8,15).
In the current study, we report the efficacy of BET inhibitors in enzalutamide-resistant mCRPC models. We developed enzalutamide-resistant prostate cancer cells from murine xenograft models in an attempt to understand the mechanisms of resistance to enzalutamide. Clonal cell lines derived from distinct enzalutamide-resistant LNCaP-AR and VCAP tumors displayed significantly higher AR expression and signaling relative to controls. Additionally, enzalutamide-resistant CRPC cell lines maintained sensitivity to BET inhibitors, leading to attenuation of AR signaling. Interestingly, AR-v7 which has been reported to be associated with resistance to anti-androgen treatments (16), was specifically elevated in enzalutamide-resistant VCaP cells and was markedly repressed by BET inhibitors. In addition to inhibiting the growth of enzalutamide-resistant CRPC cell lines, BET inhibitors displayed enhanced efficacy in vivo when combined with anti-androgens such as enzalutamide and ARN509.
Materials and methods
Murine Xenografts
Procedures involving mice were approved by the University Committee on Use and Care of Animals at the University of Michigan and conform to all regulatory standards. Please refer to Supplemental Materials and Methods for details.
Cell culture and Viability Assays
Prostate cancer cells were grown in ATCC recommended media. Please refer to Supplemental Materials and Methods for details.
Quantitative Real-Time PCR
Primer sequences are provided in the Table S1. RNA extraction, cDNA synthesis and QPCRs were performed as previously described (7).
Western Blot Analyses
Antibodies used in the study are listed in Table S2. Western blot analysis was performed as described previously (7).
Results and Discussion
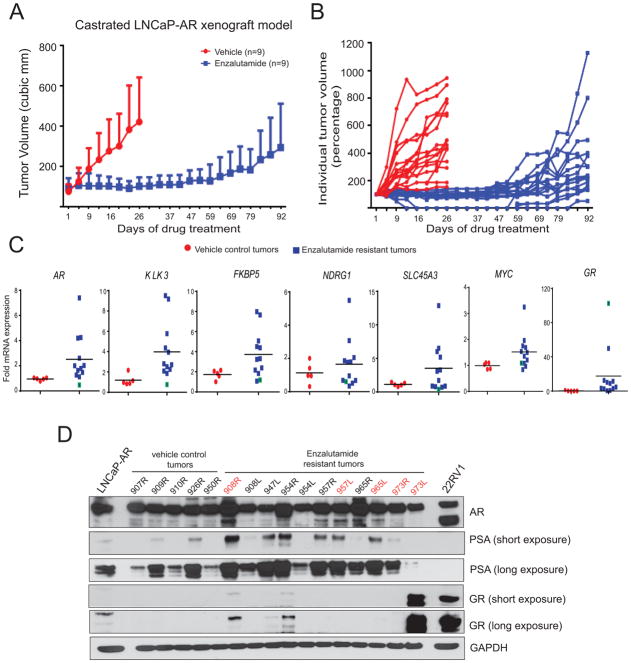
The LNCaP-AR cell line was the key pre-clinical model used in the development of enzalutamide where in castrated male mice, established LNCaP-AR xenograft tumors, regressed upon enzalutamide treatment (17). In order to study the mechanism of resistance to enzalutamide, we performed an in vivo drug efficacy experiment in castrated mice bearing LNCaP-AR tumor xenografts. Consistent with previous studies (17,18), we initially observed robust tumor regression in enzalutamide-treated animals compared to vehicle-treated controls (Fig. 1A and B). However, after about 47days, the tumors became refractory to enzalutamide treatment and tumor growth resumed. Next, we analyzed AR and AR target gene expression in enzalutamide-resistant tumors; as shown in Fig. 1C and D, AR expression was elevated at the transcript and protein levels as well as AR target genes compared to vehicle controls. Interestingly, a few of the tumors displayed high levels of glucocorticoid receptor (GR) transcript and protein expression as reported earlier (18). However, we did not observe high AR signaling, as measured by AR target gene expression, in the GR-overexpressing tumors (Fig. 1C, green boxes- tumor with GR outlier).
Figure 1. Active AR signaling in enzalutamide-resistant xenograft tumors.
A, LNCaP-AR cells were implanted subcutaneously in castrated mice and grown until tumors reached a size of approximately 100mm3. Xenografted mice were randomized and received vehicle or 10mg/kg enzalutamide, 5days a week. Mean tumor volume ±s.d. is shown. B, Individual tumors from A are shown. C, RT-qPCR analysis of AR and AR target genes in LNCaP-AR xenograft tumors Blue box with green boundary indicates tumor sample with outlier GR expression. D, Western blot analysis in tumor. Parental LNCaP-AR and 22RV1 were positive controls. Note tumor sample 973L with outlier GR protein and corresponding PSA levels. The sample number denotes the animal ID.
Furthermore, to expand the model systems used to study the mechanisms of resistance to enzalutamide, we conducted a murine xenograft experiment using VCaP prostate cancer cells that harbor the TMPRSS2:ERG gene fusion and AR amplification, both of which are frequent molecular aberrations in patients with advanced mCRPC. Treatment of VCaP tumor-bearing mice with enzalutamide (30mg/kg) for 5days/week over 5weeks led to a small but significant reduction in tumor volume compared to vehicle controls (Supplementary Fig. 1A). Further, AR and AR-v7 expression levels were elevated in the enzalutamide-treated tumors relative to vehicle controls (Supplementary Fig. 1B), consistent with a previous report that found elevated AR-v7 expression in circulating tumor cells from mCRPC patients with acquired resistance to enzalutamide and abiraterone treatment (16). Additionally, AR downstream targets such as SLC45A3 and MYC were also upregulated in enzalutamide-treated VCaP tumors.
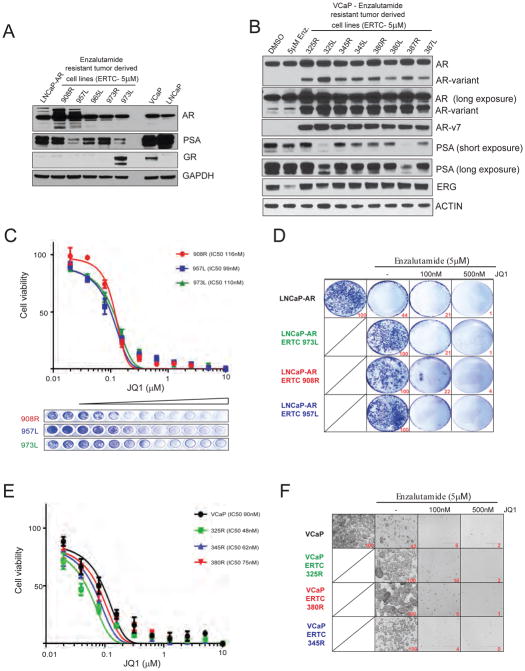
To evaluate the efficacy of BET inhibitors in enzalutamide-refractory disease, we established multiple cell lines from enzalutamide-resistant LNCaP-AR and VCaP tumors that were harvested and cultured in vitro in the presence of 5uM enzalutamide (Fig. 2A and B). In LNCaP-AR enzalutamide-resistant cells, all clones expressed varying levels of AR and PSA while only 1 out 5 clones displayed high GR expression (Fig. 2A). In VCaP enzalutamide-resistant cell lines we observed a significant increase in full length AR and AR-v7 levels with active AR signaling in all clones (Fig. 2B and Supplementary Fig. 1).
Figure 2. Enzalutamide resistant tumor-derived cells maintain BETi sensitivity.
A, Western blot analysis with lysates from enzalutamide-resistant tumor-derived LNCaP-AR cell lines. B, Western blot analysis with lysates from enzalutamide-treated tumor-derived VCaP cell lines (n=8). VCaP cells treated with DMSO or 5μM enzalutamide served as control. Note the overexpression of full-length AR and AR-variant in enzalutamide-resistant VCaP derivatives. C, Cell viability curves for three independent enzalutamide-resistant LNCaP-AR cells treated with JQ1. Crystal violet imaging of the 96 well plate is shown. D, Colony formation assay; cells were cultured in the presence or absence of drugs as indicated for 14days followed by staining. Quantification is shown at bottom right. E, Cell viability curves for three independent enzalutamide-resistant VCaP cancer cells treated with JQ1. Parental VCaP cells served as control. F, Colony formation assay; cells were cultured in the presence or absence of drugs as indicated for 14days followed by imaging. Quantification (relative number of cells) is shown at bottom right.
As these tumor derived cell line clones are enzalutamide-resistant, we asked whether these cells would respond to a BET inhibitor. We first directly compared the efficacies of three different BET inhibitors, JQ1, OTX-015 and I-BET762 in LNCaP vs LNCaP-AR cells and found the first two compounds to be equally efficacious in LNCaP-AR cells with an IC50 value of approximately 65nM, whereas parental LNCaP cells displayed lower sensitivity to the compounds with an IC50 value of approximately 110nM (Supplementary Fig. 2A and B); we chose JQ1 as the representative BET inhibitor in subsequent studies. We treated 3 independent enzalutamide-resistant LNCAP-AR sub-lines with varying concentrations of JQ1 and analyzed for cell viability 4days post-treatment, and found all 3 sub-lines were sensitive to JQ1 with IC50 values of approximately 100nM (Fig. 2C). In order to evaluate the long term effects of JQ1 treatment in the presence of enzalutamide, we performed colony-formation assays. As expected, parental LNCaP-AR cells were sensitive whereas the enzalutamide-resistant clones were insensitive to enzalutamide treatment (Fig. 2D). However, even at low concentrations (100nM) of JQ1, proliferation of enzalutamide-resistant sub-lines were severely inhibited in the long term (Fig. 2D), demonstrating that these AR signaling-positive cells are inherently susceptible to BET inhibition.
In parallel experiments, we tested enzalutamide-resistant VCaP derivatives for sensitivity to BET inhibition; 3 independent enzalutamide-resistant VCaP sub-lines were treated with varying concentrations of JQ1 and analyzed for cell viability 4days post-treatment. All 3 resistant sub-lines along with parental VCaP cells displayed sensitivity to JQ1 with IC50 values of less than 100nM (Fig. 2E). As expected, VCaP parental cells were sensitive to enzalutamide in long term colony formation assays. While enzalutamide-resistant VCaP sub-lines were insensitive to enzalutamide, these cells were sensitive upon JQ1 treatment in long term colony formation assay (Fig. 2F), demonstrating that these AR-amplified, AR-v7-overexpressing cell lines are also susceptible to BET inhibition.
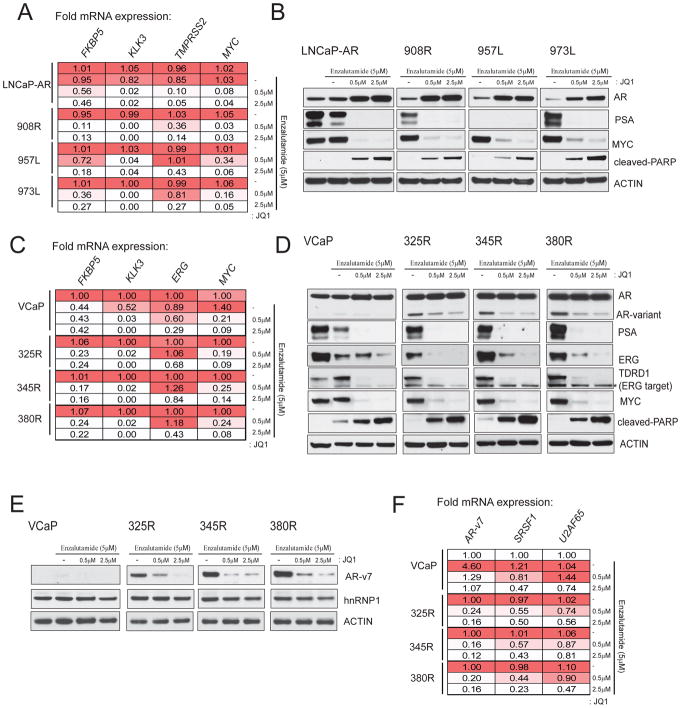
As enzalutamide-resistant clones showed an increase in AR levels but nonetheless remained sensitive to BET inhibitors, we examined whether BET inhibitors would have an effect on AR-mediated gene expression in these cells. Three independent enzalutamide-resistant LNCaP-AR sub-lines were treated with vehicle or two different concentrations of JQ1 and expression of AR target genes was analyzed by qRT-PCR and western blot analysis. As shown in Fig. 3A, FKBP5, KLK3, TMPRSS2 and MYC were transcriptionally down-regulated upon JQ1 treatment in the parental LNCaP-AR as well as in all three enzalutamide-resistant sub-lines. Similarly, PSA and MYC proteins that were expressed in the presence of enzalutamide in the resistant sub-lines were significantly repressed upon JQ1 treatment (Fig. 3B). Interestingly, AR protein levels were elevated with increasing doses of JQ1 in the parental LNCaP-AR and resistant sub-lines (Fig. 3B), suggesting perturbation of a negative feedback loop controlling AR levels.
Figure 3. BETi blocks AR signaling in enzalutamide resistant cells.
A, RT-qPCR analysis of AR target genes in three independent LNCaP-AR ERTCs grown in 5uM enzalutamide, treated with JQ1 for 24hrs. Parental LNCaP-AR cells were treated with either enzalutamide alone and in combination with JQ1. Fold mRNA expression relative to the respective DMSO control is shown. The values are mean of 3 independent readings. B, Western blot analysis of indicated target proteins in the cells from A treated with JQ1 for 48hrs. Cleaved-PARP antibody was used to detect apoptosis. C, As in A with parental VCaP and three independent enzalutamide-resistant VCaP lines. D, As in B. Note the loss of AR-variant band but not the full-length AR band upon JQ1 treatment. TDRD1 is a direct target of ERG and was used as a control for ERG function. Star symbol - non-specific band. E, Western blot with AR-v7 specific antibody using the cell lysates from the same experiment. F, As in C with target specific primers.
Similarly, we sought to determine the effect of JQ1 on AR signaling and in particular, AR-v7 expression in enzalutamide-resistant VCaP sub-lines. Parental VCaP and 3 independent enzalutamide-resistant sub-lines described above were treated with JQ1 followed by qRT-PCR and western blot analysis of AR target genes. FKBP5, KLK3, ERG and MYC were transcriptionally down-regulated upon JQ1 treatment (Fig. 3C); ERG, MYC and PSA protein levels were also reduced (Fig. 3D). Surprisingly, AR-variant protein levels were down-regulated but not full length AR in all 3 resistant sub-lines upon JQ1 treatment (Fig. 3D, top panel); AR-variant was confirmed to be AR-v7 by AR-v7-specific antibody (Fig. 3E). Splicing factors, SRSF1 and U2AF65, reported to be involved in the generation of AR splice variants (19), are subject to down-regulation by JQ1 and would explain the down-regulation of AR-v7 observed here (Fig. 3F). Interestingly, the SRSF1 and U2AF65 promoter regions are enriched for BRD2/3/4 protein displayed reduced levels upon JQ1 treatment (Supplementary Fig. 3). These data suggest that the anti-proliferative effect of JQ1 in the enzalutamide-resistant cells is partly due to its inhibitory effect on AR-v7 generation, which has been proposed as one of the major drivers of resistance to androgen deprivation therapy (ADT) (16).
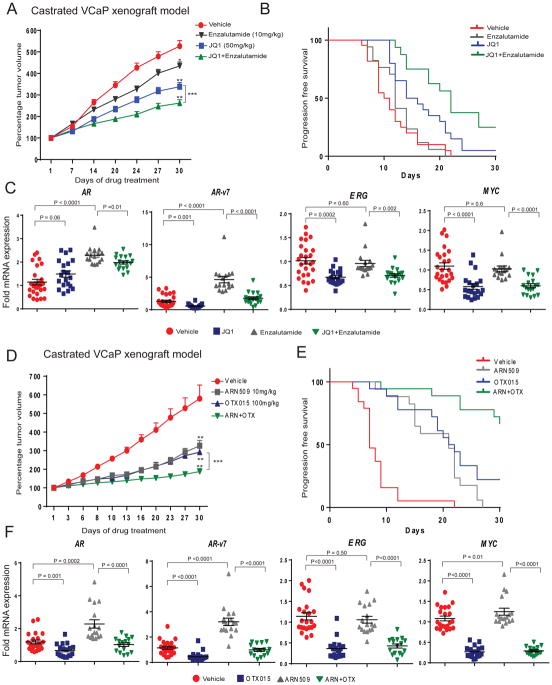
To extend upon the in vitro studies above, we next sought to conduct BET inhibition studies in vivo using the castrated VCaP xenograft mouse model (7). Although VCaP cells are originally derived from a patient with CRPC (20), these cells require androgen supplementation for growth in culture and therefore VCaP tumor xenografts are castration-responsive in mouse models (21). Castrated VCaP-tumor bearing mice were maintained until the tumors reached their original pre-castration volume and then treated with JQ1 (50mg/kg/day), enzalutamide (10mg/kg/day) or a combination. As previously observed (7), JQ1 alone led to approximately 45% reduction in the castration-resistant VCaP tumors compared to vehicle controls at 30days post treatment. However, when JQ1 was combined with enzalutamide tumor growth was inhibited by 62% (Fig. 4A). Further, we assessed the doubling of tumor volume (time interval between initiation of treatment to tumor progression) by generating Kaplan-Meier survival curves and compared the treatment groups using long-rank test. Tumor progression was delayed for the JQ1-treated group (p=0.0008) with a median tumor doubling of 15days whereas tumor doubling for vehicle treated mice was 10.5days, with no significant difference in the enzalutamide alone treatment group (12days; p=0.215). However, the combination treatment group displayed a marked delay in tumor progression (p<0.0001) with a median doubling of 22 days (Fig. 4B). The qRT-PCR analysis of tumors from the various treatment groups showed robust silencing of driver oncogenes, AR-v7, ERG, and MYC in mice treated with JQ1 alone or in combination with enzalutamide whereas treatment with enzalutamide alone showed no change or an increase in AR and AR-v7 expression respectively compared to vehicle controls (Fig. 4C). Additionally, in an AR- and ERG-positive patient-derived xenograft model we observed greater anti-tumor efficacy of JQ1 and enzalutamide in combination than JQ1 alone (Supplementary Fig. 4). Interestingly, JQ1 treatment did not show anti-tumor activity in AR and ERG-negative patient derived xenografts.
Figure 4. BETi in combination with enzalutamide or ARN-509 demonstrates enhanced anti-tumor activity.
A, Castrated mice bearing VCaP CRPC xenograft received vehicle or 10mg/kg enzalutamide or 50mg/kg JQ1 or enza./JQ1 combination as indicated 5days/week. Percentage tumor volume ±s.e.m. is shown. Statistical significance by two-tailed Student’s t-test * P=0.012; ** P<0.0001; *** P=0.005. B, The cumulative incidence plot depicting the percentage of tumors in each treatment group that have doubled in volume as a function of time. C, RT-qPCR analysis of indicated target gene expression in xenograft tumors. Relative fold expression with mean ±s.e.m is shown. D, As in A, except with 10mg/kg ARN-509 or 100mg/kg OTX-015 or ARN-509/OTX-015 combination. Percentage tumor volume ±s.e.m. is shown. Statistical significance by two-tailed Student’s t-test. * P=0.0045; ** P<0.001; *** P=0.0001, P=0.007. E, As in B. F, As in C.
Next, we tested the in vivo efficacy of OTX-015, a BET inhibitor that is being evaluated in a phase Ib clinical trial that includes metastatic prostate cancer, alone or in combination with ARN-509, a second generation anti-androgen related to enzalutamide (22), in the castrated VCaP xenograft mouse model. As shown in Fig. 4D, treatment with a combination of OTX-015 and ARN-509 led to robust anti-tumor activity resulting in 82% growth inhibition in mice, in contrast to 53% and 60% inhibition in mice treated with ARN509 or OTX-015 alone respectively. Likewise, the progression-free survival as measured by tumor doubling was significantly increased for all the treatment groups (P<0.0001) with median doubling of 21 and 21.5 days for ARN509 and OTX-015 respectively compared to 7 days for vehicle-treated group. The median doubling time for the combination treatment group could not be determined over the course of the experiment as the group did not achieve threshold progression (Fig. 4E). Interestingly, ARN-509 alone had improved anti-tumor activity in this model than enzalutamide as was observed in LNCaP-AR model by Clegg et al (23). Furthermore, gene expression analysis of tumors from the OTX-015 and ARN-509 combination treatment groups showed robust silencing of AR-v7, ERG, and MYC while AR and AR-v7 levels were elevated in tumors from mice treated with ARN-509 alone compared to vehicle controls (Fig. 4F). These in vivo data demonstrate enhanced efficacy of combined BET inhibitors and anti-androgens.
Multiple paths leading to primary or acquired resistance to anti-androgen therapies have been reported including overexpression of androgen synthesis enzymes, amplification and point mutations in AR, overexpression of AR splice variants and induction of GR (16,18,24,25). Our cell line and in vivo mouse data recapitulated several of these paths to resistance to enzalutamide and we demonstrated that BET inhibitors could overcome this resistance. The BET inhibitor efficacy data presented here has implications on the design of subsequent clinical trials that will most likely follow the ongoing multiple phase 1 trial of BET inhibitors in castration resistant prostate cancer (www.clinicaltrials.gov). We anticipate that combining BET inhibitors with second generation anti-androgens such as enzalutamide or ARN-509 will result in more durable therapeutic responses.
Supplementary Material
Acknowledgments
We thank A. Palanisamy for technical assistance; M. Cieslik and R. Malik for helpful discussions; J. Athanikar and K. Giles for critically reading the manuscript and submission of documents.
Grant Support
This work was supported by a Movember-Prostate Cancer Foundation Challenge Award and in part by the NCI Prostate SPORE (P50CA186786). A.M.C. is also supported by the American Cancer Society, and A. Alfred Taubman Institute. I.A.A. is supported by NIH pathway to independence (1K99CA187664-01) and a PCF Young Investigator Award.
Footnotes
Disclosure of Potential Conflicts of Interest
A.M.C. and S.W. are co-founders of Oncofusion Therapeutics, which is developing novel BET bromodomain inhibitors. I.A.A. has served as consultant to Oncofusion Therapeutics.
References
- 1.American Cancer Society. Cancer Facts and Figures 2015. 2015 http://wwwcancerorg/acs/groups/content/@editorial/documents/document/acspc-044552pdf.
- 2.Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, et al. Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004;10(1):33–9. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 3.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161(5):1215–28. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Visakorpi T, Hyytinen E, Koivisto P, Tanner M, Keinanen R, Palmberg C, et al. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat Genet. 1995;9(4):401–6. doi: 10.1038/ng0495-401. [DOI] [PubMed] [Google Scholar]
- 5.de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364(21):1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367(13):1187–97. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 7.Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510(7504):278–82. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan SC, Selth LA, Li Y, Nyquist MD, Miao L, Bradner JE, et al. Targeting chromatin binding regulation of constitutively active AR variants to overcome prostate cancer resistance to endocrine-based therapies. Nucleic Acids Res. 2015;43(12):5880–97. doi: 10.1093/nar/gkv262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wyce A, Degenhardt Y, Bai Y, Le B, Korenchuk S, Crouthame MC, et al. Inhibition of BET bromodomain proteins as a therapeutic approach in prostate cancer. Oncotarget. 2013;4(12):2419–29. doi: 10.18632/oncotarget.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478(7370):529–33. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153(2):320–34. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24(6):777–90. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao L, Schwartzman J, Gibbs A, Lisac R, Kleinschmidt R, Wilmot B, et al. Androgen receptor promotes ligand-independent prostate cancer progression through c-Myc upregulation. PLoS One. 2013;8(5):e63563. doi: 10.1371/journal.pone.0063563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371(11):1028–38. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324(5928):787–90. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arora VK, Schenkein E, Murali R, Subudhi SK, Wongvipat J, Balbas MD, et al. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell. 2013;155(6):1309–22. doi: 10.1016/j.cell.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu LL, Xie N, Sun S, Plymate S, Mostaghel E, Dong X. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene. 2014;33(24):3140–50. doi: 10.1038/onc.2013.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korenchuk S, Lehr JE, LMC, Lee YG, Whitney S, Vessella R, et al. VCaP, a cell-based model system of human prostate cancer. In Vivo. 2001;15(2):163–8. [PubMed] [Google Scholar]
- 21.Cai C, Wang H, Xu Y, Chen S, Balk SP. Reactivation of androgen receptor-regulated TMPRSS2:ERG gene expression in castration-resistant prostate cancer. Cancer Res. 2009;69(15):6027–32. doi: 10.1158/0008-5472.CAN-09-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rathkopf DE, Morris MJ, Fox JJ, Danila DC, Slovin SF, Hager JH, et al. Phase I study of ARN-509, a novel antiandrogen, in the treatment of castration-resistant prostate cancer. J Clin Oncol. 2013;31(28):3525–30. doi: 10.1200/JCO.2013.50.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clegg NJ, Wongvipat J, Joseph JD, Tran C, Ouk S, Dilhas A, et al. ARN-509: a novel antiandrogen for prostate cancer treatment. Cancer Res. 2012;72(6):1494–503. doi: 10.1158/0008-5472.CAN-11-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balbas MD, Evans MJ, Hosfield DJ, Wongvipat J, Arora VK, Watson PA, et al. Overcoming mutation-based resistance to antiandrogens with rational drug design. Elife. 2013;2:e00499. doi: 10.7554/eLife.00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitsiades N, Sung CC, Schultz N, Danila DC, He B, Eedunuri VK, et al. Distinct patterns of dysregulated expression of enzymes involved in androgen synthesis and metabolism in metastatic prostate cancer tumors. Cancer Res. 2012;72(23):6142–52. doi: 10.1158/0008-5472.CAN-12-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.