Abstract
Dynamic expression of the genome requires coordinated binding of chromatin factors and enzymes that carry out genome-templated processes. Until recently, the molecular mechanisms governing how these factors and enzymes recognize and act on the fundamental unit of chromatin, the nucleosome core particle, have remained a mystery. A small, yet growing set of structures of the nucleosome in complex with chromatin factors and enzymes highlights the importance of multivalency in defining nucleosome binding and specificity. Many such interactions include an arginine anchor motif, which targets a unique acidic patch on the nucleosome surface. These emerging paradigms for chromatin recognition will be discussed, focusing on several recent structural breakthroughs.
The eukaryotic genome is organized in a polymeric complex of protein and nucleic acid called chromatin. The repeating unit of chromatin is the nucleosome core particle separated from each adjacent nucleosome core particle by intervening extranucleosomal linker DNA. The 200 kD nucleosome core particle complex is equally distributed in molecular weight between protein and DNA: the two copies each of histones H2A, H2B, H3 and H4 constitute about 100 kD and the 145-147 bp of DNA the other 100 kD. The nucleosome is a disk shaped complex approximately 100 Å in diameter, but it is far from a featureless disk [1•]. Instead, the nucleosome offers a rich diversity of surfaces for interactions with chromatin proteins and enzymes.
The nucleosome is often depicted in colors to highlight the individual histones and the nucleosomal DNA (Fig. 1a). It can be convenient to consider the nucleosome core particle as three distinct binding surfaces: a central disk of core histones with a diameter of about 65 Å, an outer shell of DNA, and histone tails that splay out from the central histone core (Fig. 1b). Each of these binding surfaces offers possibilities for specific recognition by chromatin proteins and enzymes. The central histone core displays nearly 27,000 Å2 of accessible surface area, replete with nooks and crannies and bulges providing interesting complementary surfaces for interacting proteins. For example, the central histone disk is only 25 Å in thickness at the dyad where two histone H3 molecules come together, but 60 Å in thickness where the H2B αC helices protrude. This surface geometry is further enhanced by a variety of charge distribution on the histone surface. The most prominent charge feature is an acidic patch created by the histone dimer of H2A and H2B (Fig. 1d), the target of the arginine anchor motif described below.
Figure 1.
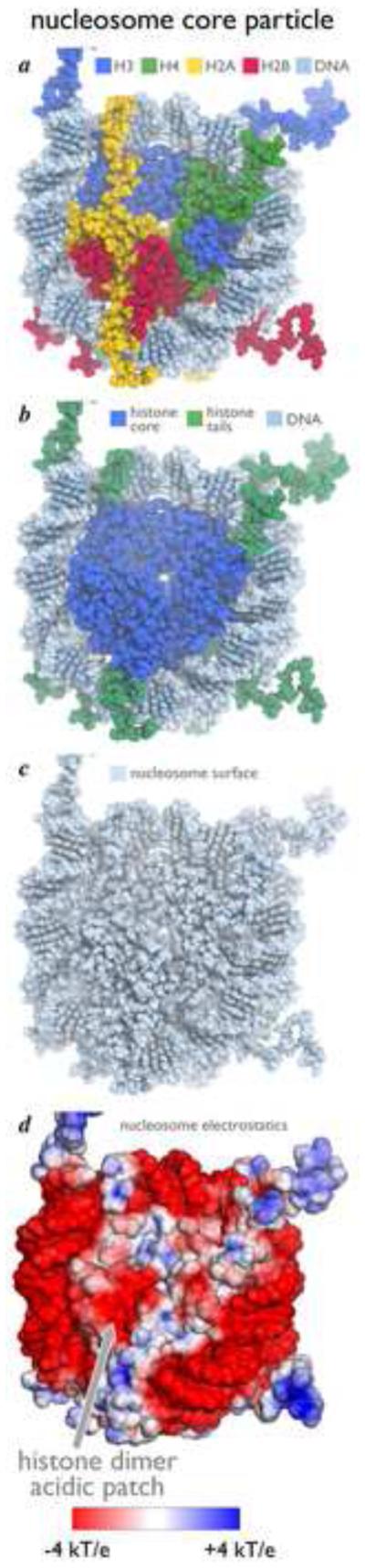
Nucleosome core particle structure. Different surface representations of the nucleosome core particle structure (PDB ID: 1KX5) highlighting (a) individual histone proteins (H3, H4, H2A, and H2B colored blue, green, yellow, and red, respectively) and DNA (light blue), (b) the histone core (blue) and histone tails (green), (c) the nucleosome as single binding surface and (d) electrostatic surface of nucleosome core particle with histone H2A/H2B dimer acidic patch indicated. Electrostatic representation generated with ABPS [58]. All figures in this manuscript were prepared with PyMOL software [57].
The nucleosome was long considered a repressive structure where histone proteins occluded transcription factor binding to DNA. However, much of the DNA on the outside of the nucleosome is solvent accessible and available for binding to chromatin factors. In what is unlikely to be a coincidence, the DNA grooves align across the DNA gyres (Fig. 2 middle panel) [1•, see also Fig. 1.5c in ref. 2], and it would seem probable that at least some transcription factor or chromatin enzyme will bind across the gyres to interact with aligned major or minor grooves. This would increase a transcription factor’s DNA specificity since it would support high affinity binding only when the binding sites on the two gyres were spaced appropriately, and it would also promote binding to the nucleosome over naked DNA.
Figure 2.
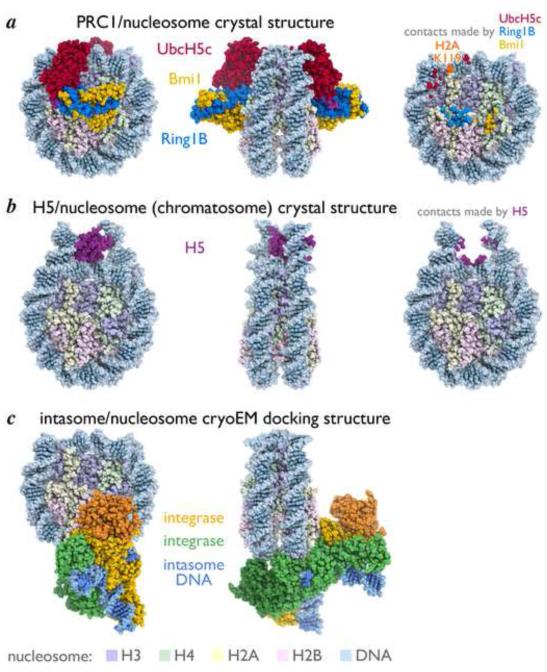
Recent chromatin factor-nucleosome core particle structures. (a) Orthogonal views of PRC1 ubiquitylation module-nucleosome core particle structure, left and center, and footprint of PRC1 on nucleosome surface colored by interacting components of PRC1, right. The Cα atom of the PRC1 targeted residue of H2A (Lys119) is represented by an orange sphere. (b) Similar views of chromatosome structure with the globular domain of H5. (c) Orthogonal views of intasome-nucleosome docking model. Integrase subunits shown in yellow, orange, green and dark green with outer subunits in darker shades. Integrase DNA colored blue. Nucleosome footprint not shown for intasome due to resolution of this model. H3, H4, H2A, H2B and nucleosomal DNA colored light blue, light green, pink, light yellow, and light blue respectively.
The histone tails that extend from the histone core comprise roughly 25% of the mass of the histones and are abundant in post-translational modifications that contribute to epigenetic regulation of gene activity [3,4]. Much has been discovered regarding how chromatin modules can specifically recognize particular post-translational modified histone tail peptides [5,6].
It is also useful to view the nucleosome without coloring to distinguish between the individual components or parts. When so viewed as in Fig. 1c, the nucleosome is a single entity, where histone and DNA atoms cannot be distinguished from each other. This reminds us that chromatin factors and enzymes are unlikely to interact or recognize individual histone or DNA components of the nucleosome. Instead, the architecture of the entire nucleosome is likely to be recognized through binding of a combination of nucleosomal elements. Here we discuss themes in nucleosome recognition by chromatin factors and enzymes and highlight several recent structural breakthroughs.
Multivalency and nucleosome recognition
Despite a wealth of structures detailing the molecular organization of the nucleosome core particle and the structural consequences of histone variants and DNA sequence [7], structures illustrating the mechanisms underlying nucleosome recognition by chromatin factors and enzymes have been more elusive. In fact, nearly a decade passed between the 2.8 Å structure of the nucleosome core particle in 1997 [1•] and the crystal structure of a peptide from the Kaposi’s sarcoma latency-associated nuclear antigen (LANA) anchored to the nucleosome [8•]. Rather than co-crystallization, this first structure of a macromolecule bound to the nucleosome was achieved through soaking of the LANA peptide into preformed nucleosome core particle crystals. The technical challenges associated with co-crystallization of a chromatin protein with the nucleosome core particle were overcome in 2010 with the RCC1 (Regulator of Chromosome Condensation)-nucleosome structure [9•,10]. This was followed by crystal structures of the BAH (Bromo-Associated Homology) domain of the yeast silencing protein Sir3 (Silent information regulator) in complex with the nucleosome [11•] and a peptide from the centromeric protein CENP-C bound to a centromeric nucleosome containing the CENP-A histone variant [12•]. More recently, two more crystal structures have been reported of the first chromatin enzyme, the Polycomb repressive complex 1 (PRC1) E2-E3 ubiquitylation module bound to the nucleosome core particle [13••] as well as that of the chromatosome, containing the nucleosome core, linker DNA, and the globular domain of the linker histone H5 [14••].
Complementary structural techniques have also emerged to add to the current understanding of nucleosome recognition. Methyl-TROSY NMR has allowed the characterization and modeling of the nucleosome in complex with a peptide from HMGN2 (High Mobility Group Nucleosome-Binding Domain-Containing) [15], the globular domain of linker histone H1 [16•], and the PWWP domain from PSIP1/LEDGF (PC4 and SFRS1-interacting protein/lens epithelium-derived growth factor) [17,18]. Such efforts built on the tour de force assignment of methyl resonances from histone Val, Leu, and Ile side chains [15]. More recently, advances in single particle cryo-EM technology have been applied to generate a 7.8 Å structure of the prototype foamy virus intasome bound to the nucleosome core particle [19••].
A central theme emerging from the growing list of structures of the nucleosome bound to chromatin proteins and enzymes is multivalency. Rather than binding to a single component or surface of the nucleosome, chromatin proteins and enzymes often recognize multiple nucleosomal components often across multiple and even non-contiguous nucleosomal surfaces. This allows for specific structural and enzymatic specificities to be distributed to unique nucleosomal locations and emphasizes the importance of studying the nucleosome as an entity, and not simply as the sum of its histone and DNA parts. For example, RCC1 uses its β-propeller loops to bind to both histone and DNA components of the nucleosome [9•,20]. RCC1 anchors to the H2A/H2B acidic patch with its switchback loop and also binds across the major groove of nucleosomal DNA at SHL 6 (superhelical location) with two additional loops. Further contacts are likely made with the nucleosomal DNA at SHL 6.5 by the N-terminal tail of RCC1. [DNA superhelical locations (SHL) identify DNA major groove positions which face the histone octamer, extending from SHL0 at the dyad to SHL±7 near the sites of DNA entry/exit.]
The Sir3 BAH domain binds to all four histones across a large surface of the nucleosome core disk, including both histone tail and histone-fold regions [11•]. Sir3 BAH engages the H4 N-terminal tail organizing an otherwise unstructured region including the H4 basic patch. Additional interactions are made with surfaces of the nucleosome disk contributed by H3, H4 and H2B in the functionally defined LRS (loss of rDNA silencing) region as well as the H2A/H2B acidic patch. Of note, nucleosome affinity is enhanced by N-α acetylation of Sir3, not by direct nucleosome interaction, but indirectly through stabilization of nucleosome binding loops of the Sir3 BAH domain [21,22]. Similar to Sir3, the centromeric protein CENP-C makes multivalent interactions with multiple histone surfaces. A region of CENP-C binds to the H2A/H2B acidic patch while another region interacts with a CENP-A H3 variant-specific hydrophobic surface near the nucleosomal dyad [12•].
The PRC1 ubiquitylation module
PRC1 is a member of the Polycomb group of protein complexes [23]. Similar to other Polycomb complexes, it is a transcriptional repressor of developmentally regulated genes and is commonly misregulated in human cancer [24-26]. Its main molecular functions include ubiquitylation of H2A Lys119 and an intrinsic ability to compact chromatin arrays [27-29]. Rather than being a specific entity, mammalian PRC1 exists as a family of canonical and variant complexes with unique subunit composition [30]. However, despite this complexity all PRC1 family members share a common core, an E3 ligase composed of Ring1B (or Ring1A) and Bmi1 (or one of five other Polycomb group RING finger proteins). This heterodimeric RING-type ubiquitin E3 ligase can pair with one of several E2 ubiquitin-conjugating enzymes including UbcH5c to ubiquitylate H2A, but only in a nucleosomal context [31-33]. While relevant surfaces of Ring1B, Bmi1 and the nucleosome had been identified, the mechanism governing nucleosome specificity remained elusive [32,34].
We have determined the crystal structure of the Ring1B-Bmi1-UbcH5c E3-E2 ubiquitylation module bound to its nucleosome core particle substrate at 3.3 Å resolution [13••]. Unlike other histone modifying enzymes structurally characterized with histone peptides that bind to the targeted amino acid and neighboring residues in the primary sequence, PRC1 binds multiple surfaces of the nucleosome that are distinct from the site of catalysis (Fig. 2a). The Ring1B-Bmi1 heterodimer forms a saddle over the αC helix of H2B (the outermost margin on the nucleosome) anchored on each side by RING domain-histone interactions, the most critical between Ring1B and the H2A/H2B acidic patch. This allows the PRC1 E3 ligase to position its E2 with the E2 active site cleft facing the nucleosome surface near H2A Lys119. Instead of the E2 UbcH5c binding directly to the targeted H2A sequence, it binds to nucleosomal DNA in two places, near the nucleosomal dyad and near the point of DNA entry and exit, with further interactions expected with extranucleosomal linker DNA. By recognizing unique nucleosomal surfaces, PRC1 can efficiently and specifically ubiquitylate H2A without sequence specific recognition of the targeted H2A sequence. A nucleosome substrate is required because the PRC1 ubiquitylation module binds all components of the nucleosome – all four histones and nucleosomal DNA. Moreover, in the context of the nucleosome, the H2A C-terminal tail, which is unstructured in an H2A/H2B dimer, adopts a rigid conformation that presents the targeted lysine to the E2.
The chromatosome
Decades of research support the role of linker histones (denoted H1 or H5) binding to the nucleosome core particle and linker DNA to promote 30 nm fiber formation and resultant chromatin compaction [35]. Linker histones contain a central globular domain flanked by N- and C-terminal extensions, with the globular domain and C-terminal extension contributing to nucleosome binding and function [36-38]. The nucleosome core particle with ~20 bp of extranucleosomal linker DNA bound to a single linker histone is called the chromatosome [39]. Two major models have been proposed for the chromatosome: 1) the symmetric, on-dyad model in which the linker histone globular domain binds the nucleosome on the dyad and interacts with linker DNA on both sides of the nucleosome core, and 2) the asymmetric, off-dyad model in which the linker histone globular domain bind nucleosomal DNA next to the dyad and interacts with linker DNA on one side or both sides of the nucleosome core [40]. A recent crystal structure of the chromatosome assembled with chicken H5 [14••] paired with NMR and cryo-EM investigations using Drosophila H1 and human H1.4 [16•,41•], respectively, suggest that both structures may exist in a linker histone subtype dependent manner and moreover may differentially influence higher order chromatin structure.
Zhou et al. determined the 3.5 Å crystal structure of the chromatosome containing the globular domain of chicken linker histone H5 (most similar to human H1.0) [14••]. The structure shows symmetric, on-dyad binding with the globular domain of H5 interacting with the DNA minor groove at the nucleosomal dyad and minor grooves of extranucleosomal linker DNA on both sides of the nucleosome approximately 1/2 to 1 helical turns from the core particle (Fig. 2b). Nearly all interactions occur with the DNA phosphodiester backbone. Interestingly, asymmetric, off-dyad binding was observed for Drosophila H1 based on methyl-TROSY and spin-labeling NMR characterization [16•]. A similar conclusion was reached using human H1.4 visualized in chromatin arrays by single particle cryo-EM [41•]. Zhou et al. suggest that sequence specific changes in linker histone subtypes dictate binding modes and demonstrate corresponding changes in compaction of chromatin arrays in vitro. Altogether this raises the possibility that deposition of specific linker histone subtypes may differentially influence local chromatin structure in vivo.
The Intasome
Integration of viral DNA into host genomes is performed by a tetrameric complex of integrase bound to viral DNA ends and is called an intasome [42]. The intasome must deform its target DNA to allow phosphodiester bond cleavage and subsequent integration [43]. Surprisingly, many retroviruses integrate into loci within the host genome that are frequently assembled into nucleosomes [44-46]. Moreover, a preferred integration site relative to the nucleosome position is commonly observed. For example, the prototype foamy virus (PFV) intasome preferentially integrates into DNA wrapped into a nucleosome over naked DNA and specifically ±3.5 helical turns from the dyad. However the mechanism governing this nucleosome preference and the resultant site-specificity were unclear until recently.
Maskell et al. used single particle cryo-EM to generate a 7.8 Å map of the intasome nucleosome structure, allowing docking of the intasome and nucleosome atomic structures previously determined by x-ray crystallography [19••] (Fig. 2c). Unlike previous chromatin protein-nucleosome EM structures at resolutions above 20 Å (reviewed in [2]), the resolution of this model allowed direct mechanistic insight into intasome function. The intasome uses three of its four subunits to bind to both histone and DNA components of the nucleosome, binding across both gyres of DNA as well as one adjacent αC helix of H2B. Nucleosome binding results in a 7 Å deformation of one underlying DNA double helix leading to a DNA structure similar to that of the intasome on naked DNA. This may explain the decreased nucleosome binding and integration observed with nucleosomes assembled with Widom 601 nucleosome positioning sequence [47] frequently used in biochemical and structural characterization of chromatin processes in vitro, as this optimized sequence may resist deformation at the target DNA site and therefore integration. By pairing histone interactions with DNA integration, the intasome defines specific sites within the pseudosymmetric nucleosome for integration. The preference for sites occupied by nucleosomes may favor the observed bias toward integration in stable heterochromatic rather that actively transcribed regions of the host genome that are less stably assembled into nucleosomes.
Arginine anchor
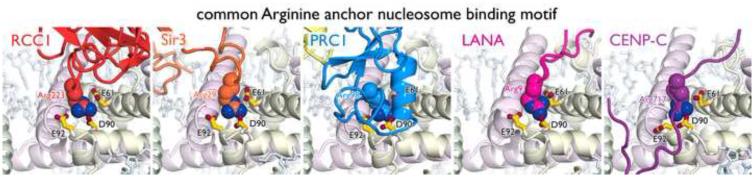
Since limited structural and biochemical data is available detailing nucleosomal recognition by chromatin factors and enzymes, it is remarkable that a clear paradigm for nucleosome binding is emerging. Nearly all of the chromatin factor-nucleosome structures published to date, including the PRC1 ubiquitylation module, RCC1, Sir3, CENP-C and LANA bind the H2A/H2B acidic patch on the nucleosome using a critical arginine side chain, termed the arginine anchor motif [8•,9•,11•,12•,13••]. The H2A/H2B acidic patch is generated by H2A (Glu56, Glu61, Glu64, Asp90, Glu91, Glu92) and H2B (Glu105, Glu113) residues and lies in a groove on the nucleosome disk surface lined on one side by the H2B αC helix and on the other side by the distal ends of the H2A α2 and H2B α1 helices (Fig. 1d). A small ridge separates two sections of the acidic patch groove. The chromatin factor arginine anchor residue projects into the deeper acidic pocket on one side of this central ridge allowing the arginine side chain guanidinium group to make charged interactions with the H2A side chain carboxylates of the Glu61, Asp90, Glu92 acidic triad (Fig. 3). Additional van der Waals interactions exist between the aliphatic region of the arginine anchor side chain and the adjacent H2B αC helix. While the arginine side chain follows a similar trajectory into the acidic patch in each of the aforementioned structures, the main chains surrounding the arginine residue are divergent, suggesting a considerable amount of variability in the types structures that can present an arginine anchor to the nucleosome surface [48,49]. There does not appear to be any sequence conservation beyond the arginine residue itself. Each chromatin factor makes additional contacts in the acidic patch and in many cases elsewhere on the nucleosome surface, but for the PRC1 ubiquitylation module, RCC1, CENP-C and LANA the arginine anchor-acidic patch interaction is among the most essential contacts for nucleosome binding and in the case of PRC1 enzymatic activity. A recently solved unpublished crystal structure of the SAGA (Spt-Ada-Gnc5-Acetyltransferase) complex deubiquitylation module bound to the nucleosome provides yet another example of an arginine anchor interacting with the H2A/H2B acidic patch (Michael Morgan and Cynthia Wolberger, personal communication).
Figure 3.
The arginine anchor motif. Side chains of arginine anchor residues from RCC1, Sir3, PRC1 (Ring1B), LANA, and CENP-C shown in surface representation with side chains from acid triad of H2A in stick representation. Nucleosomal DNA shown as sticks.
Biochemical data suggest additional roles for the acidic patch in other chromatin processes. The acidic patch has been implicated in higher order chromatin structure and is integral to formation of the 30 nm fiber through interaction with a basic patch in the H4 N-terminal tail [1•,50-53]. An NMR-based model of a fragment of the high mobility group protein HMGN2 illustrates another example of an arginine anchor-acidic patch interaction [15]. Similarly to PRC1 subunit Ring1B, BRCA1 likely employs an RING domain arginine anchor to bind to and ubiquitylate nucleosomal H2A [13••]. Another E3 ligase, RNF168, may not bind directly to the H2A/H2B acidic patch, but nonetheless requires an intact H2A/H2B acidic patch for nucleosome and H2A/H2B dimer ubiquitylation [33,34]. In S. cerevisiae, acidic patch mutations disrupt histone modification patterns including H2B K123 ubiquitylation and downstream H2A K4 and K79 methylation [54,55]. It remains to be determined whether arginine anchors are used by these histone modifying enzymes in establishing these marks.
While the arginine anchor-acidic patch interaction is emerging as a paradigm for nucleosome recognition, it is by no means the rule, as seen with recent chromatosome and intasome-nucleosome structures. As the sample size of characterized nucleosome binding proteins is still small, it remains to be discovered how often the acidic patch is used by chromatin factors and what other paradigms exist for nucleosome binding.
Future directions
While much progress had been made over the last five years to reveal molecular details underlying nucleosome recognition, we have only scratched the surface of this complex and fundamental system. The PRC1 ubiquitylation module-nucleosome complex structure presents the first high-resolution depiction of a histone modifying enzyme’s nucleosome recognition. This paves the way for structural characterization of other classes of histone modifying enzymes bound to the nucleosome. Further strides are also needed to understand the molecular details of chromatin binding by other important types of chromatin factors, including chromatin remodelers and readers of epigenetic modifications. The intasome-nucleosome docking model illustrates the power of emerging cryo-EM technology for structural characterization of large nucleosome assemblies to complement existing crystallography and NMR-based methods. Future work in this area will undoubtedly reveal the extent of arginine anchor usage and uncover new paradigms for nucleosome recognition. Ultimately, the combined structural knowledge may inform the design of small molecule or peptidomimetic modulators of chromatin structure and function [56].
Highlights for McGinty and Tan manuscript.
We now possess long-awaited structures of chromatin factors bound to the nucleosome.
The structures highlight the role of multivalency in nucleosome recognition.
The arginine anchor motif is emerging as a paradigm for nucleosome binding.
Acknowledgements
We would like to thank Peter Cherepanov and Allessandro Costa for providing coordinates for their intasome-nucleosome core particle model, and Michael Moore and Cynthia Wolberger for permission to cite their unpublished work. This work was supported by the National Institutes of Health grants GM088236 and GM11165 to S.T. and Damon Runyon Cancer Research Foundation grants DR-2107-12 and DFS-14-15 to R.K.M..
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- •. This is a classic paper reporting the first high-resolution structure of nucleosome core particle.
- 2.McGinty RK, Tan S. Nucleosome structure and function. Chem. Rev. 2015;115:2255–2273. doi: 10.1021/cr500373h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ng MK, Cheung P. A brief histone in time: understanding the combinatorial functions of histone PTMs in the nucleosome context. Biochem. Cell Biol. 2015 doi: 10.1139/bcb-2015-0031. doi:10.1139/bcb-2015-0031. [DOI] [PubMed] [Google Scholar]
- 4.Rothbart SB, Strahl BD. Interpreting the language of histone and DNA modifications. Biochim. Biophys. Acta. 2014;1839:627–643. doi: 10.1016/j.bbagrm.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du J, Patel DJ. Structural biology-based insights into combinatorial readout and crosstalk among epigenetic marks. Biochim. Biophys. Acta. 2014;1839:719–727. doi: 10.1016/j.bbagrm.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel DJ, Wang Z. Readout of epigenetic modifications. Annu. Rev. Biochem. 2013;82:81–118. doi: 10.1146/annurev-biochem-072711-165700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan S, Davey CA. Nucleosome structural studies. Curr. Opin. Struct. Biol. 2011;21:128–136. doi: 10.1016/j.sbi.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, Luger K, Kaye KM. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science. 2006;311:856–861. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]
- •. The authors solve the first crystal structure of a nucleosome core particle bound to a protein fragment, the LANA peptide, which binds the nucleosome using an arginine anchor motif.
- 9.Makde RD, England JR, Yennawar HP, Tan S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature. 2010;467:562–566. doi: 10.1038/nature09321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •. This describes the first structure of a protein bound to the nucleosome. The authors demonstrate that RCC1 binds the H2A/H2B acidic patch using an arginine anchor and also binds to nucleosomal DNA.
- 10.Makde RD, Tan S. Strategies for crystallizing a chromatin protein in complex with the nucleosome core particle. Anal. Biochem. 2013;442:138–145. doi: 10.1016/j.ab.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Armache KJ, Garlick JD, Canzio D, Narlikar GJ, Kingston RE. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 Å resolution. Science. 2011;334:977–982. doi: 10.1126/science.1210915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •. The authors solve the structure of the Sir3 BAH domain bound to the nucleosome core particle. Sir3 binds both the tail of H4 and regions of the core histone disk.
- 12.Kato H, Jiang J, Zhou B-R, Rozendaal M, Feng H, Ghirlando R, Xiao TS, Straight AF, Bai Y. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science. 2013;340:1110–1113. doi: 10.1126/science.1235532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •. The authors present structural studies of a CENP-C peptide bound to a nucleosome core particle assembled with the CENP-A variant. CENP-C recognizes the acidic patch and a histone variant-specific nucleosome surface.
- 13.McGinty RK, Henrici RC, Tan S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature. 2014;514:591–596. doi: 10.1038/nature13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••. This is the first structure of a histone-modifying enzyme bound to the nucleosome core particle. The structure demonstrates that PRC1 binds multiple nucleosome surfaces distinct from the site of catalysis to specifically ubiquitylate H2A.
- 14.Zhou B-R, Jiang J, Feng H, Ghirlando R, Xiao TS, Bai Y. Structural Mechanisms of Nucleosome Recognition by Linker Histones. Mol. Cell. 2015;59:628–638. doi: 10.1016/j.molcel.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••. The authors solve the first crystal structure of the chromatosome. The structure shows on-dyad binding of the globular domain of chicken H5. The authors propose linker histone subtype specific chromatin structural changes.
- 15.Kato H, van Ingen H, Zhou B-R, Feng H, Bustin M, Kay LE, Bai Y. Architecture of the high mobility group nucleosomal protein 2-nucleosome complex as revealed by methyl-based NMR. Proc. Natl. Acad. Sci. U.S.A. 2011;108:12283–12288. doi: 10.1073/pnas.1105848108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou B-R, Feng H, Kato H, Dai L, Yang Y, Zhou Y, Bai Y. Structural insights into the histone H1-nucleosome complex. Proc. Natl. Acad. Sci. U.S.A. 2013;110:19390–19395. doi: 10.1073/pnas.1314905110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •. The authors use methyl-TROSY NMR to demonstrate off-dyad binding of the Drosophila linker histone.
- 17.van Nuland R, van Schaik FM, Simonis M, van Heesch S, Cuppen E, Boelens R, Timmers HM, van Ingen H. Nucleosomal DNA binding drives the recognition of H3K36-methylated nucleosomes by the PSIP1-PWWP domain. Epigenetics Chromatin. 2013;6:12. doi: 10.1186/1756-8935-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eidahl JO, Crowe BL, North JA, McKee CJ, Shkriabai N, Feng L, Plumb M, Graham RL, Gorelick RJ, Hess S, et al. Structural basis for high-affinity binding of LEDGF PWWP to mononucleosomes. Nucleic Acids Res. 2013;41:3924–3936. doi: 10.1093/nar/gkt074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maskell DP, Renault L, Serrao E, Lesbats P, Matadeen R, Hare S, Lindemann D, Engelman AN, Costa A, Cherepanov P. Structural basis for retroviral integration into nucleosomes. Nature. 2015;523:366–369. doi: 10.1038/nature14495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••. The authors report a single-particle cryo-EM model of an intasome bound to the nucleosome at 7.8 Å resolution, the highest resolution cryo-EM structure to date containing the nucleosome. Their model reveals that the intasome recognizes histone and DNA architectures to define a specific nucleosome location for integration of viral DNA.
- 20.England JR, Huang J, Jennings MJ, Makde RD, Tan S. RCC1 Uses a Conformationally Diverse Loop Region to Interact with the Nucleosome: A Model for the RCC1–Nucleosome Complex. J. Mol. Biol. 2010;398:518–529. doi: 10.1016/j.jmb.2010.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang D, Fang Q, Wang M, Ren R, Wang H, He M, Sun Y, Yang N, Xu R-M. Nα-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain. Nat. Struct. Mol. Biol. 2013;20:1116–1118. doi: 10.1038/nsmb.2637. [DOI] [PubMed] [Google Scholar]
- 22.Arnaudo N, Fernández IS, McLaughlin SH, Peak-Chew SY, Rhodes D, Martino F. The N-terminal acetylation of Sir3 stabilizes its binding to the nucleosome core particle. Nat. Struct. Mol. Biol. 2013;20:1119–1121. doi: 10.1038/nsmb.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 24.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 25.Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 2013;20:1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- 26.Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 27.Cao R, Tsukada Y-I, Zhang Y. Role of Bmi-1 and Ring1A in H2A Ubiquitylation and Hox Gene Silencing. Mol. Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 29.Grau DJ, Chapman BA, Garlick JD, Borowsky M, Francis NJ, Kingston RE. Compaction of chromatin by diverse Polycomb group proteins requires localized regions of high charge. Genes Dev. 2011;25:2210–2221. doi: 10.1101/gad.17288211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon JA, Kingston RE. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell. 2013;49:808–824. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buchwald G, van der Stoop P, Weichenrieder O, Perrakis A, van Lohuizen M, Sixma TK. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 2006;25:2465–2474. doi: 10.1038/sj.emboj.7601144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bentley ML, Corn JE, Dong KC, Phung Q, Cheung TK, Cochran AG. Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. EMBO J. 2011;30:3285–3297. doi: 10.1038/emboj.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mattiroli F, Uckelmann M, Sahtoe DD, van Dijk WJ, Sixma TK. The nucleosome acidic patch plays a critical role in RNF168-dependent ubiquitination of histone H2A. Nat. Commun. 2014;5:3291–3291. doi: 10.1038/ncomms4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leung JW, Agarwal P, Canny MD, Gong F, Robison AD, Finkelstein IJ, Durocher D, Miller KM. Nucleosome acidic patch promotes RNF168- and RING1B/BMI1-dependent H2AX and H2A ubiquitination and DNA damage signaling. PLoS Genet. 2014;10:e1004178. doi: 10.1371/journal.pgen.1004178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harshman SW, Young NL, Parthun MR, Freitas MA. H1 histones: current perspectives and challenges. Nucleic Acids Res. 2013;41:9593–9609. doi: 10.1093/nar/gkt700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allan J, Hartman PG, Crane-Robinson C, Aviles FX. The structure of histone H1 and its location in chromatin. Nature. 1980;288:675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- 37.Hendzel MJ, Lever MA, Crawford E, Th'ng JPH. The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J. Biol. Chem. 2004;279:20028–20034. doi: 10.1074/jbc.M400070200. [DOI] [PubMed] [Google Scholar]
- 38.Syed SH, Goutte-Gattat D, Becker N, Meyer S, Shukla MS, Hayes JJ, Everaers R, Angelov D, Bednar J, Dimitrov S. Single-base resolution mapping of H1-nucleosome interactions and 3D organization of the nucleosome. Proc. Natl. Acad. Sci. U.S.A. 2010;107:9620–9625. doi: 10.1073/pnas.1000309107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simpson RT. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978;17:5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- 40.Cutter AR, Hayes JJ. A brief review of nucleosome structure. FEBS Lett. 2015 doi: 10.1016/j.febslet.2015.05.016. doi:10.1016/j.febslet.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song F, Chen P, Sun D, Wang M, Dong L, Liang D, Xu R-M, Zhu P, Li G. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 2014;344:376–380. doi: 10.1126/science.1251413. [DOI] [PubMed] [Google Scholar]
- ••. The authors solve an 11 Å single particle cryo-EM structure of a chromatin array. Their model shows a two-start fiber with off-dyad head-to-head arrangement of the human linker histone H1.4.
- 42.Li M, Mizuuchi M, Burke TR, Craigie R. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 2006;25:1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maertens GN, Hare S, Cherepanov P. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature. 2010;468:326–329. doi: 10.1038/nature09517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pryciak PM, Varmus HE. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell. 1992;69:769–780. doi: 10.1016/0092-8674(92)90289-o. [DOI] [PubMed] [Google Scholar]
- 45.Pruss D, Bushman FD, Wolffe AP. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc. Natl. Acad. Sci. U.S.A. 1994;91:5913–5917. doi: 10.1073/pnas.91.13.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Müller HP, Varmus HE. DNA bending creates favored sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. EMBO J. 1994;13:4704–4714. doi: 10.1002/j.1460-2075.1994.tb06794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 48.Yang D, Arya G. Structure and binding of the H4 histone tail and the effects of lysine 16 acetylation. Phys. Chem. Chem. Phys. 2011;13:2911–2921. doi: 10.1039/c0cp01487g. [DOI] [PubMed] [Google Scholar]
- 49.Kalashnikova AA, Porter-Goff ME, Muthurajan UM, Luger K, Hansen JC. The role of the nucleosome acidic patch in modulating higher order chromatin structure. J. R. Soc., Interface. 2013;10:20121022. doi: 10.1098/rsif.2012.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dorigo B, Schalch T, Bystricky K, Richmond TJ. Chromatin Fiber Folding: Requirement for the Histone H4 N-terminal Tail. J. Mol. Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 51.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent Mediated Interactions in the Structure of the Nucleosome Core Particle at 1.9Å Resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 52.Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 53.Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 54.Nakanishi S, Sanderson BW, Delventhal KM, Bradford WD, Staehling-Hampton K, Shilatifard A. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat. Struct. Mol. Biol. 2008;15:881–888. doi: 10.1038/nsmb.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cucinotta CE, Young AN, Klucevsek KM, Arndt KM. The Nucleosome Acidic Patch Regulates the H2B K123 Monoubiquitylation Cascade and Transcription Elongation in Saccharomyces cerevisiae. PLoS Genet. 2015;11:e1005420. doi: 10.1371/journal.pgen.1005420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.da Silva ITG, de Oliveira PSL, Santos GM. Featuring the nucleosome surface as a therapeutic target. Trends Pharmacol. Sci. 2015;36:263–269. doi: 10.1016/j.tips.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 57. The PyMOL Molecular Graphics System, Version 1.6, Schrodinger, LLC.
- 58.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]