Abstract
Purpose
Head and neck squamous cell carcinoma (HNSCC) remains a devastating disease, and FA gene mutations and transcriptional repression are common. Invasive tumor behaviour is associated with poor outcome, but relevant pathways triggering invasion are poorly understood. There is a significant need to improve our understanding of genetic pathways and molecular mechanisms driving advanced tumor phenotypes, in order to develop tailored therapies. Here we sought to investigate the phenotypic and molecular consequences of FA pathway loss in HNSCC cells.
Experimental Design
Using sporadic HNSCC cell lines with and without FA gene knockdown, we sought to characterize the phenotypic and molecular consequences of FA deficiency. FA pathway inactivation was confirmed by the detection of classical hallmarks of FA following exposure to DNA crosslinkers. Cells were subjected to RNA sequencing with qRT-PCR validation, followed by cellular adhesion and invasion assays in the presence and absence of DNA-PK and Rac1 inhibitors.
Results
We demonstrate that FA loss in HNSCC cells leads to cytoskeletal reorganization and invasive tumor cell behavior in the absence of proliferative gains. We further demonstrate that cellular invasion following FA loss is mediated, at least in part, through NHEJ-associated DNA-dependent protein kinase (DNA-PK) and downstream Rac1 GTPase activity.
Conclusions
These findings demonstrate that FA loss stimulates HNSCC cell motility and invasion, and implicate a targetable DNA-PK/Rac1 signaling axis in advanced tumor phenotypes.
Keywords: Fanconi anemia, head and neck cancer, squamous cell carcinoma, invasion
Introduction
In the general population, HNSCC is associated with smoking, alcohol consumption and HPV infection, and represents the sixth leading type of cancer worldwide, with an annual incidence of 500,000. More than half of sporadic HNSCCs are diagnosed at locally advanced or metastatic stages, and approximately 50% of treated patients relapse with local or distant metastasis, both bearing poor rates of remission (1, 2). Unfortunately, decades of research have not improved HNSCC outcomes significantly, and the classic therapeutic option, a combination of surgery, radiation and chemotherapy, leaves patients permanently disfigured. Thus, there is a need to improve our understanding of the biological processes driving local invasiveness, and develop novel approaches the treatment of late stage tumors. Previous exome sequencing data demonstrated that 11% of HPV+ and HPV− HNSCCs harbor non-synonymous mutations in FA DNA repair genes (3). In addition to classical loss-of-function mutations, transcriptional repression of FANCB, FANCC, FANCF, FANCJ, and FANCM (eg, by promoter methylation) has been noted in dysplastic HN tissue and HNSCC (4, 5). This suggests that loss of FA pathway function can provide a selective advantage to HNSCC cells. However, the underlying biological mechanisms - aside from genome instability - remain poorly understood.
Major advances in understanding how defective DNA repair pathways contribute to tumorigenesis have been achieved by studying DNA repair deficiency syndromes that often manifest during the first years of life (6). Fanconi anemia (FA) is a rare inherited disorder where every cell of an affected individual is exquisitely sensitive to DNA cross-linking agents including mitomycin C or cisplatin (for reviews on mechanisms and disease manifestations see (7, 8), and references within). Clinically, patients with FA are characterized by frequent congenital abnormalities, early progressive bone marrow failure, and a high propensity for developing malignancies, especially acute myeloid leukemia, and squamous cell carcinoma of the head and neck, esophageal and anogenital regions. In fact, FA patients with HNSCCs are usually diagnosed at a young age with advanced tumor stages that have a poor prognosis (9, 10). FA has been associated with recessive mutations in one of 19 FA genes (11), which play a crucial role in triggering and coordinating fundamental mechanisms of DNA repair for the maintenance of genome instability. When intact, the FA core complex, composed of the protein products of eight FA genes including FANCA, is assembled at the site of DNA damage and triggers monoubiquitination of the central and evolutionarily conserved pathway members FANCD2 and FANCI by the E3 ligase FANCL. The activated FANCD2/FANCI dimer then stabilizes the stalled replication fork at the crosslink and coordinates the activity of nucleases, TLS polymerases, and homologous recombination factors. Genes of the homologous recombination (HR) pathway, where bi-allelic mutations clinically cause FA-like syndromes, are BRIP1/FANCJ, BRCA1/FANCS, BRCA2/FANCD1, PALB2/FANCN and RAD51C/FANCO. Cells with bi-allelic mutations in FA genes can compensate for their HR defects by over-activating the error-prone non-homologous end joining (NHEJ) pathway (12, 13), thus triggering inappropriate DNA repair. Individuals with heterozygous loss-of-function germ-line mutations in DNA repair genes also are at an increased risk for tumors, due to the loss of the wild-type allele in the malignant cells (6). Here, heterozygous germ-line defects in the ‘late’ genes of the FA pathway (FANCD1/J/N/O/Q/S) are predominantly associated with the development of hereditary breast/ovarian and pancreatic cancer (14, 15). Furthermore, even sporadic tumors in patients with no family history of cancer frequently harbor mutations in DNA repair genes (3, 16–18).
Several lines of evidence suggest that FA pathway loss in the epidermis, in contrast to the hematopoietic system, promotes growth in the basal stem and progenitor cell compartment. First, in murine models, genetic loss of Fancd2 cooperated with transgenic HPV16 E7 expression targeted to basal epithelial cells to promote the development of HNSCC (19). Importantly, Fancd2 loss alone was already sufficient for a subtle yet consistent increase in basal cell proliferation in E7-negative control mice, thus highlighting a pro-proliferative role for FA pathway defects in the normal epidermis and in an HPV negative environment. Second, using HPV immortalized human keratinocytes, we have previously reported that FANCA or FANCD2 knockdown drive proliferation and HPV E7-dependent hyperplasia in 3D organotypic epithelial raft but not in 2D keratinocyte culture systems (20). Third, we have recently reported that defects in the FA pathway stimulate HPV genome amplification and accumulation of the HPV E7 oncoprotein with concomitant cellular proliferation (21, 22). Forth, FA patient-derived HNSCC cell lines were shown to harbor either similar or increased stem cell populations when compared to sporadic HNSCC lines, using tumor sphere formation, CD44 positivity or ALDH1 status as experimental end points (23, 24).
In order to define the functional effects of acquired FA deficiency in HNSCC cells, we generated isogenic FA HNSCC models using shRNA-mediated stable knockdown and rescue strategies in HPV positive and negative tumor cell lines. While depletion of the key FA pathway components FANCA, FANCD2 and FANCJ induced classical FA phenotypes in these cells when exposed to DNA crosslinkers, minor to no effects on tumor cell growth were observed under standard culture conditions. Surprisingly however, under these same conditions, FA loss caused cytoskeletal reorganization, and dramatic increases in tumor cell invasiveness in the absence of proliferative gains in HPV positive and negative HNSCC cells. Invasive properties were associated, at least in part, with increased activities of the NHEJ associated DNA-dependent protein kinase (DNA-PK) and the Rac1 GTPase, thus linking FA-associated DNA damage responses with cytoskeletal machineries. In summary, these data highlight new and unexpected signaling connections between the FA DNA repair pathway and invasive tumor phenotypes in HNSCC. These might be targeted for the development of novel treatment strategies to lower mortality and improve the prognosis of patients with advanced HNSCCs.
Materials and Methods
Cell culture
The UM-SCC1, UM-SCC6, and UM-SCC47 cell lines were derived and maintained as previously described (25). All cell lines were authenticated regularly by their morphologic characteristics and analysis of corresponding genetic and molecular markers. Nontargeting, FANCD2-, FANCA- and FANCJ (TRCN49915) specific short hairpin RNA (shRNA)-expressing lentiviral vectors were obtained through the Sigma MISSION shRNA program (Sigma-Aldrich) as previously described (20). The MIEG-HPV-16-E7 (GenScript Piscataway, NJ) construct was developed from the pMIEG3 retroviral vector, a kind gift from Dr. David Williams (Children’s Hospital Boston, MA), and has been described earlier. The EcoRI-E7-His(6)-FLAG-XhoI sequence, GAATTCGGCGGCCGCGCCACCATGCATGGAGATACACCTACATTGCATGAATATATGTTAGATTTGCAACCAGAGACAACTGATCTCTACTGTTATGAGCAATTAAATGACAGCTCAGAGGAGGAGGATGAAATAGATGGTCCAGCTGGACAAGCAGAACCGGACAGAGCCCATTACAATATTGTAACCTTTTGTTGCAAGTGTGACTCTACGCTTCGGTTGTGCGTACAAAGCACACACGTAGACATTCGTACTTTGGAAGACCTGTTAATGGGCACACTAGGAATTGTGTGCCCCATCTGTTCAGAAACCAGACTACAAGGACGACGATGACAAGCATCACCATCACCATCACTAACTCGAG, was created by flanking the LXSN-HPV16-E7 gene with primers to introduce EcoRI and and in-frame His(6)-FLAG-Stop-XhoI sequence. The PCR product was produced, cut, and inserted into the EcoRI and XhoI sites in the pMIEG vector by GenScript. RD114 pseudotype retroviruses were produced in 293T cells using an established protocol in the Hanenberg laboratory. Cells were transduced at 30 to 50% confluence for a total of 4 h for retroviruses or 8 h for lentiviruses in a final concentration of 8 μg/ml Polybrene. For MIEG-based vectors, cells were sorted on GFP. Following sorting and replating, cells were then transduced with either non-targeting or FANCD2-specific shRNA. For the lentiviral vectors, cells were selected and carried in 1.25 μg/ml puromycin. A fusion construct between EGFP and FANCD2 cDNA was generated and verified by direct sequencing (H. Hanenberg, unpublished observations). The Rac1 inhibitor NSC23766 was a generous gift by Dr. Yi Zheng.
Cell cycle measurements
Cells were seeded for 24 hours and then either left untreated or treated for 24 hours with 0.25μg/ml or 0.5μg/ml melphalan (Sigma-Aldrich, St. Louis, MO). Cells were trypsinized and 5×105 cells were washed and prepared to assess BrdU incorporation according to manufacturer’s instructions (APC BrdU Flow Kit, BD Pharmingen, San Jose, CA). The cells were pulsed with 10 μM BrdU for 45 minutes. Cell cycle profiles were detected using 7AAD, with samples acquired on a BD FACSCanto instrument (BD Biosciences, San Jose, CA) and the results were analyzed using FlowJo (Treestar).
RNA Sequencing
Individual samples were aligned to the Hg19 genome using TopHat v1.4.1. Gene quantification was performed with Cufflinks v2.0.0 with the “-G -u -b” parameters and the Ensembl gene model. Gene-level quantifications were used throughout. Data was analyzed with GeneSpring 12.6.1 NGS, filtering to remove duplicates, and filtering on post alignment read metrics to remove reads with mapping quality below 40, or with more than one match to genome, or failing vendor QC. Quantification was carried out with DESeq as normalization algorithm and threshold normalized counts to 1, baseline to median of all samples. We filtered to remove genes with fewer than 3 RPKM in at least one sample. Differential expression was determined with the Audic Claverie test (P < 0.05, FC > 1.5). Functional enrichment analysis was carried out with ToppGene and cytoscape figures were made using ToppCluster.
Western blot analysis
Whole-cell protein extracts were harvested and lysed using 1X Laemmli buffer. Blotting was performed as previously described (21). Antibodies used were rabbit polyclonal FANCA (Cascade Bioscience, Winchester, MA), rabbit polyclonal FANCD2 (Novus, Littleton, CO), rabbit polyclonal FANCJ (Novus, Littleton, CO), mouse monoclonal DNA-PKcs (Abcam, Cambridge, MA), rabbit polyclonal phospho-DNA-PKcs (S2056)(Abcam, Cambridge, MA) and actin (Seven Hills Bioresearch, Cincinnati, OH). For detection of HPV16 E7, a primary antibody mix of mouse monoclonal anti-16E7 antibody (1:150, 8C9 (Invitrogen, Grand Island, NY); and a 1:200 dilution of ED17 (Invitrogen, Grand Island, NY)) was used.
Three-dimensional epithelial raft cultures
Organotypic rafts were generated as described previously (20). Briefly, a total of 1×106 UM-SCC1 cells were plated on a collagen matrix with embedded feeder fibroblasts. Exposure to the liquid–air interface resulted in the generation of stratified epithelium with differentiation properties that reflect its natural human counterpart. The tissue was fixed in 2% paraformaldehyde after 16 days of growth, embedded in paraffin, sectioned and morphologically examined by hematoxylin-and-eosin staining.
Immunofluorescence and DIC Imaging
Immunofluorescence was performed as previously described (20) using the Leica DM 5000B. Cells were plated onto coverslips into 6-well plates at equal cell number and collected following 24 h of incubation. Cells were fixed in 4% paraformaldehyde and rinsed in PBS prior to staining with Rhodamine phalloidin (1:200; Invitrogen, Grand Island, NY) and DAPI to stain all nuclei. DIC images were acquired at the time of Phalloidin imaging. Cells on coverslips were treated overnight with 20 μM NSC23766 prior to fixation.
Invasion assays
BioCoat Matrigel transwell invasion assays were performed as per manufacturer’s instructions (BD Bioscience, San Jose, CA). Cells were resuspended in serum free media in the upper chamber and allowed to invade through Matrigel to serum containing media in the bottom chamber. For UMSCC-1, 1.5X105 cells were seeded per transwell while for UMSCC-6 and UMSCC-47 2.5×105 cells were seeded. Invasion was allowed to proceed for 22 hours prior to fixation in methanol and staining with Giemsa. Total numbers of invaded cells were quantified for each transwell using ImageJ.
Migration Assays
Costar transwell migration assay were performed as per manufacturer’s instructions (Corning Incorporated, Charlotte, NC). Briefly, cells were resuspended in serum free media in the upper chamber and allowed to migrate through the polycarbonate membrane to the serum containing media located in the bottom chamber. A total of 1.5 × 105 UM-SCC1 NTsh and D2sh cells were seeded per transwell. Migration was allowed to proceed for 16 hours prior to fixation in methanol and staining with Giemsa. The total number of migrating cells was quantified for each transwell using ImageJ. Six independent migration assay experiments were performed. Graphs were generated using GraphPad Prism 6 and a paired student t-test was applied to determine significance.
Chicken Chorioallantoic Membrane (CAM) assays
Fertilized White Leghorn chicken eggs were incubated while rotating at 37°C in a humidified atmosphere (>60% relative humidity). After 48 hours, 0.5mL of albumin was removed from each egg and the eggs were placed into a non-rotating incubation chamber at 37°C in a humidified atmosphere. After 48 hours, the eggs were windowed to expose the CAM, vasculature and viable embryo. To evaluate tumor cell invasion, a thinly sliced pipette ring was placed on top of the chorion layer of the membrane and a 25uL suspension of 500,000 UM-SCC1 cells that were either FA-deficient or –proficient and embedded in matrigel were pipetted into the ring onto the membrane. Egg shell windows were covered with scotch tape and the eggs were returned to the non-rotating incubator. Following an additional 72 hours of incubation, CAMs with tumor cells were harvested, fixed in 4% paraformaldehyde, processed, embedded in paraffin blocks, and sectioned. 5 micron sections were utilized for standard hematoxylin and eosin staining.
Rac1 activity assay
The Active Rac1 Pull-Down and Detection Kit (Thermo Scientific, Waltham, MA) was used to detect active Rac1 in the absence and presence of DNA-PKcs inhibitor NU7026 (Tocris Bioscience, Bristol, UK). Briefly, cells were plated at equal cell number and allowed to adhere. Cells were treated with either vehicle or 10 μM NU7026 for 24 h. Prior to harvesting, cells were exposed to a 30 min pulse of bleomycin (10 μg/ml). Cells were harvested and lysed as directed by the kit and protein concentrations were determined using the Pierce BCA Protein Assay Kit according to manufacturing instructions (Thermo Scientific, Waltham, MA). Equivalent amounts of protein were loaded onto the column and the protocol outlined within the assay kit was followed. Following elution of activated Rac1 from the column, western blot analysis for Rac1-GTP was performed using the anti-Rac1 mouse monoclonal antibody (1:1000) provided in the assay kit. GTPγS (positive control) and GDP (negative control) were used as controls in the pull-down assays.
Statistics
Statistical significance was determined using two-way analysis of variance (ANOVA) with Sidak post hoc tests using an α value of 0.05 for all calculations using phalloidin projection data. All other significance was determined using a Student t-test. Statistical analyses were performed using GraphPad Prism 6 software.
Results
Classical FA phenotypes result from FA knockdown in HPV positive and negative HNSCC cancer cells
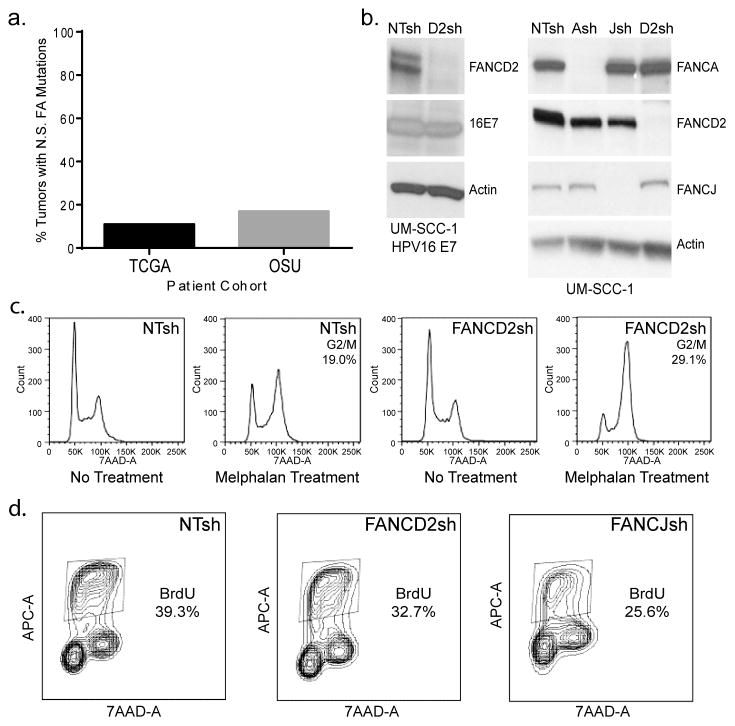
We have recently analyzed whole exome sequencing data of therapy-naïve HNSCCs (26) and found that a significant proportion of sporadic HNSCCs harbor somatic mutations in FA and FA-related genes (3). In order to confirm this with sequencing data from other patient cohorts, we queried The Cancer Genome Atlas (TCGA) (27) (n=306), as well as whole sequencing data from a set of 34 primary human HNSCCs. 11.1% and 17.6%, respectively, of such tumors harbored non-synonymous (N.S.) mutations in the 15 FA genes that had been identified at that time, FANCA, FANCB, FANCC, FANCD1 (BRCA2), FANCD2, FANCE, FANCF, FANCG, FANCI, FANCJ, FANCL, FANCM, FANCN, FANCP, and FANCO (Fig. 1a). In order to explore possible biological effects of FA loss in systems mimicking HPV+ and HPV− HNSCC, we created the following knockdown models using previously published lentiviral shRNA vectors (28). First, an HPV negative UM-SCC1 cell line was transduced with HPV16 E7, and subsequently knocked down for FANCD2. Second, the same cell line was knocked down for FANCA, FANCD2, and FANCJ in the absence of E7. Third, a second HPV− cell line, UM-SCC6, and an HPV+ cell line, UM-SCC47, were similarly knocked down for FANCA and FANCD2. Western blot analyses verified efficient FA protein depletion for all HNSCC cell lines transduced with either FANCA, -D2, and -J shRNA expression vectors (Figures 1b and S1). Cells transduced with the non-targeting (NTsh) control shRNA did not result in FA depletion (as expected). To ensure that FANCD2 depletion induced classical FA phenotypes, DNA crosslinker sensitivity of knockdown versus control cells was quantified. FA lymphoblasts and fibroblasts predictably respond to melphalan exposure with a G2/M cell cycle arrest (28, 29). As expected, FANCD2-deficient UM-SCC1 cancer cells treated with melphalan also displayed an increase in the proportion of cells in G2/M when compared to control NTsh cells (Fig. 1c). FANCJ-deficient UM-SCC1 cells responded to the melphalan treatment in a similar manner when compared to the FANCD2-deficient cancer cells (data not shown). UM-SCC6 and UM-SCC47 HNSCC cell lines depleted for FA proteins were similarly sensitive to melphalan (data not shown); thus FA knockdown induces characteristic hallmarks of FA in HPV positive and negative HNSCC cells. In order to determine the consequences of FA loss on UM-SCC1 cells grown under standard culture conditions, we quantified proliferation rates based on BrdU incorporation (Fig. 1d). FANCD2 and FANCJ loss did not increase the proliferation of UM-SCC1 cells, but either decreased or did not change proliferation (Fig. 4d, S1b). In HPV16 E7 expressing UM-SCC1 cells, FANCD2 loss did not affect cellular proliferation (Fig. S1c). Similar decreases or no effects were observed in UM-SCC6 and UM-SCC47 cell lines (data not shown). Taken together, FA knockdown in HNSCC cell lines did not alter and sometimes reduced cellular growth under standard 2D culture conditions, and conferred classical FA phenotypes in the presence of DNA crosslinkers.
Figure 1. Generation of FA deficient, HPV positive and negative, HNSCC cell models.
(a) The somatic mutations table (MAF file) for head and neck squamous carcinoma (HNSC) samples was obtained from TCGA data portal and from an independent cohort of sporadic HNSCC tumors from Ohio State University (OSU). Analysis of mutational data from 306 (TCGA) and 34 (OSU) sporadic HNSCC tumors determined that 11.1% and 17.6%, respectively, harbored non-synonymous (N.S.) mutations in one of 16 FA genes, respectively. (b) UM-SCC1 HNSCC (HPV16 E7-positive and –negative) cells were knocked down for FANCA, FANCD2 and FANCJ, followed by western blot analysis for verification of protein depletion. (c) FA knockdown by shRNA transduction leads to classical FA phenotypes. UM-SCC1 cells were treated with melphalan, and subjected to flow cytometry based cell cycle analysis. The numbers listed indicate percentages of cells in G2/M following melphalan exposure. FANCD2-deficient cells were increased for the proportion of cells in G2/M when compared to the NTsh control cell population. (d) BrdU incorporation in FANCD2- and FANCJ-deficient compared to control UM-SCC1 cells reveals a slight decrease in proliferation.
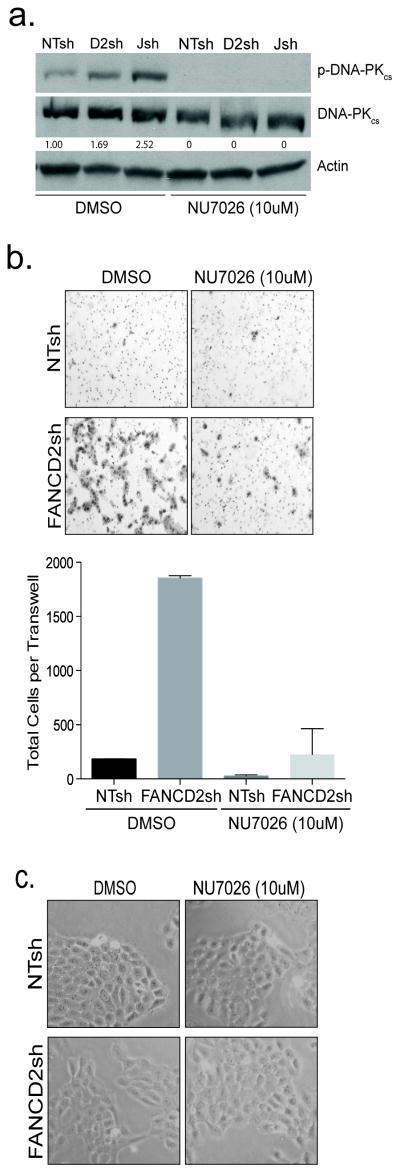
Figure 4. DNA-PK activity promotes cellular invasiveness.
(a) FA loss stimulated DNA-PKcs phosphorylation in UM-SCC1 cells. Western blot analysis shows activation of DNA-PKcs by phosphorylation at Ser2056 following FANCD2 and FANCJ loss. The functionality of the DNA-PKcs inhibitor, NU7026, was verified in the same cells. Cells were treated with either DMSO or NU7026 for 24 hours and exposed to a short 30 minute burst of bleomycin. (b) DNA-PKcs activation contributes to invasion. Cells were control transduced or knocked down for FANCD2, and the number of invasive cells was quantified following overnight incubation. Experiments were carried out in duplicate and standard deviation was calculated. (c) DNA-PKcs inhibition does not affect cell viability as is shown by visual examination of the cells.
FA loss deregulates epithelial HNSCC morphology and cytoskeletal organization
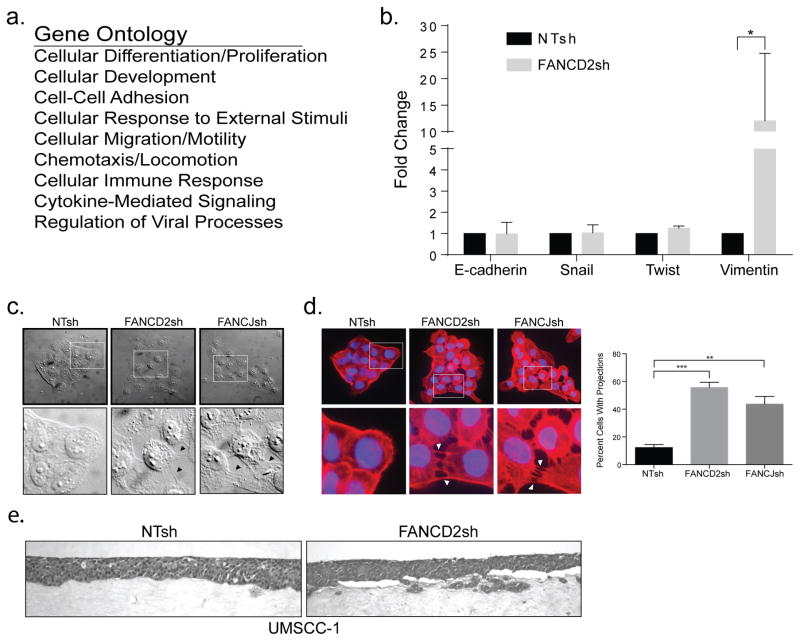
To assess global expression patterns induced by perturbing the FA pathway, we performed RNA sequencing (RNASeq) of the above HPV16 E7 positive UM-SCC1 cells, either depleted for FANCD2 or control transduced (Fig. 1b). A number of genes were differentially expressed in the FANCD2sh compared to the NTsh HNSCC cells (Supplemental Table 1). Following identification of genes with significantly altered expression, ToppGene was used to perform gene ontology analysis and redundancies were eliminated. A large number of gene ontology hits were identified (Supplemental Table 2). Fig. 2a lists top candidate biological processes which – based on gene expression alterations – may be regulated by FANCD2 loss. In line with our previously published data wherein FA pathway loss impaired keratinocyte differentiation in organotypic epithelial raft models, the expression of a number of differentiation-associated genes was reduced by FANCD2 knockdown (Supp. Tables 1 and 2). Importantly, this was not accompanied by the induction of proliferative gene signatures, in agreement with a lack of proliferative gains in the above FA HNSCC cell populations under standard conditions and with previously published data using patient-derived cell lines (Fig. 1d and data not shown) (20). Among the top biological processes were cellular motility and invasion. Increased expression of the intermediate filament vimentin was noted which has already been linked to a number of cancers as a mesenchymal marker for invasive potential (30). Vimentin induction in FA HNSCC cells was validated by qRT-PCR (Fig. 2b). In contrast, the expression of other genes involved in classical epithelial-to-mesenchymal transition (EMT), such as E-cadherin, snail1, or twist1 was not altered. Perhaps related to the observed regulation of genes involved in cellular motility, we observed a marked alteration in the expression of genes involved in cellular adhesion and locomotion (Fig. 2a). Further morphological examination of FA depleted UM-SCC1 cells by differential interference contrast (DIC) microscopy revealed a clear difference in cellular shape and spatial arrangement. FANCD2 and FANCJ knockdown in the UM-SCC1 cell line impaired SCC epithelial morphology: cells physically separated from each other but remained connected by intercellular projections that were largely absent in the control HNSCC cells (Fig. 2c). To further investigate the intercellular projections, the F-actin marker phalloidin was used to stain actin filaments. As shown in Fig. 2d, FANCD2 and FANCJ knockdown cells tended to separate but maintained long intercellular projections, in contrast to control cells which harbored tight epithelial cell-cell contacts. Taken together, FA pathway loss in HNSCC cells de-regulated transcriptomes associated with cellular motility and adhesion, and this was accompanied by morphological and cytoskeletal responses including the formation of intercellular protrusions. In order to assess the ability of FA deficient versus proficient tumor cells to grow as 3D tissues, we engineered 3D organotypic epithelial tumor rafts from NTsh or FANCD2sh UM-SCC1 cells (Fig. 2e). Interestingly, while FA-deficient cells were able to grow and assemble into 3D tissue, we noted their occasional presence in the underlying collagen matrix, a feature which was not shared by the FA-proficient counterparts. We therefore tested the possibility that FA-deficient cells harbored increased invasive properties.
Figure 2. FA loss in HNSCC cells induces cytoskeletal and morphological alterations.
(a) RNA sequencing (RNASeq) and subsequent ToppGene analysis of FANCD2-deficient versus control cells reveals cellular processes with significantly altered gene expression. A list of the top candidate biological processes is shown. (b) Verification of RNA sequencing gene expression results was performed on a select number of genes using qRT-PCR. qRT-PCR also revealed that some classical EMT markers were not uniformly elevated in FA-deficient HNSCCs, with the exception of vimentin. (c) Morphological examination of FA depleted HNSCC cells compared to FA-proficient controls by differential interference contrast (DIC) microscopy shows differences in spatial arrangement and cell shape as well as increased intercellular protrusions in FANCD2- and FANCJ-deficient cells. (d) Intercellular projections were examined further by staining with the F-actin marker phalloidin and subsequent immunofluorescence experiments. Quantification of intercellular projections was performed on three independent experiments and the errors bars represent standard deviation. Asterisks indicate significance at *=p<0.05, **=p<0.001, and ***=p<0.0001 (ANOVA). (e) UM-SCC1 epithelial raft culture shows UM-SCC1 FANCD2sh knockdown cells in the underlying collagen matrix as compared to UM-SCC1 NTsh control cells.
FA pathway loss in HNSCC cells promotes cellular invasion
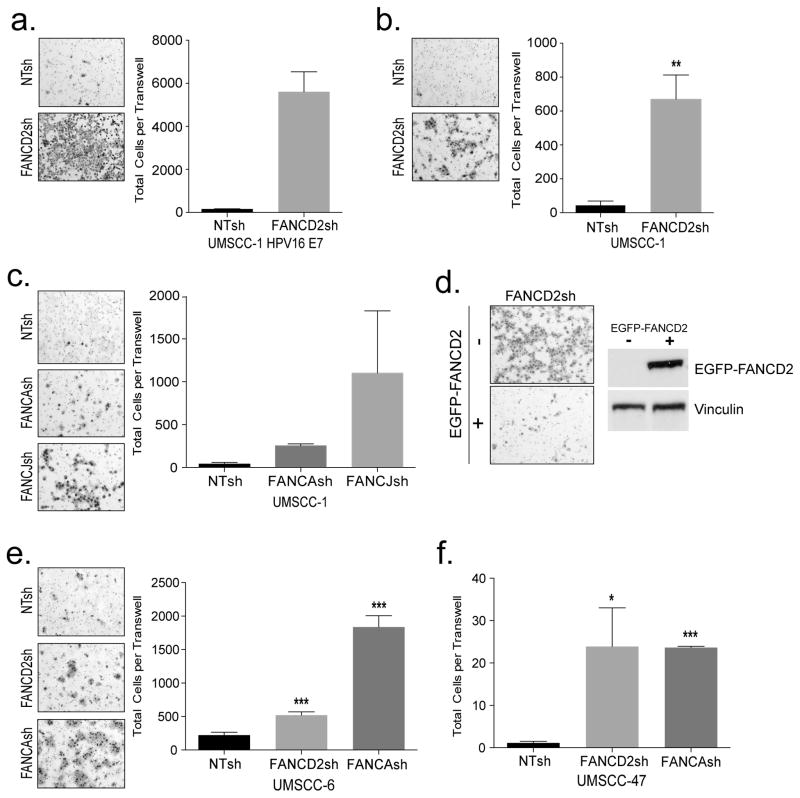
Tumor cell invasiveness was determined directly using Matrigel transwell assays. Both HPV16 E7+ and E7− UM-SCC1 cells were significantly more invasive when knocked down for FANCD2 compared to empty vector controls (Figs. 3a and b). Invasion was not an indirect consequence of increased proliferation (Fig. 1d, data not shown) or increased cellular adhesion (Fig. S2a). Similarly, FANCA and FANCJ knockdown also stimulated invasion in UM-SCC1 cells (Fig. 3c). To rule out any off target effects for the lentiviral knockdown approach, we next expressed a shRNA-resistant FANCD2 construct in FANCD2sh UM-SCC1 cells (Fig. 3d). As predicted, the introduction of EGFP-FANCD2 was sufficient to rescue UM-SCC1 FANCD2 shRNA cells from invasion (Fig. 3d). Expression of the EGFP-FANCD2 fusion protein was confirmed by western blot analysis on the right. Finally, FANCA and FANCD2 knockdown in UM-SCC6 and UM-SCC47 cells also stimulated tumor cell invasion (Figs. 3e and f). As expected, the increased invasiveness of the cancer cells correlated with increased motility seen by standard migration assays (Fig. S2b). In order to further assess the invasive properties of FA-deficient versus -proficient tumor cells, we utilized chicken chorioallantoic membrane (CAM) assays (Fig. S3). Interestingly, FA-deficient cells were occasionally able to invade into the chorion membranes of living chick embryos, and more specifically appeared to invade as cell clusters; however, invasion was never observed with their FA-proficient counterparts. Taken together, FA loss leads to a dramatic increase of invasive capacity and motility in HPV positive and negative HNSCC cells.
Figure 3. FA loss in a panel of HPV negative and positive HNSCC cells promotes invasion.
(a) UM-SCC1 cells expressing HPV16 E7 and knocked down for FANCD2, compared to control NTsh cells were plated to invade through Matrigel coated transwell for 22 hours. Invasive cells were quantified and representative images are shown. (b–c) UM-SCC1 cells knocked down for FANCD2, or for FANCA and FANCJ compared to control NTsh cells were plated to invade through a Matrigel coated transwell for 24 hours. Invasive cells were quantified and representative images are shown. (d) Invasion mediated by FANCD2 depletion could be repressed by the expression of a knockdown resistant eGFP-FANCD2 fusion gene. UM-SCC1 cells were transduced with an eGFP-FANCD2 retroviral expression vector, and sorted for GFP negative cells as a control population, or sorted for cells that express eGFP-FANCD2. Western blot analysis confirms expression of the fusion protein in GFP positive, but not in GFP negative cells. Transwell assays demonstrate EGFP-FANCD2 mediated rescue from invasion that is driven by FANCD2 loss. (e–f) UM-SCC6 and UM-SCC47 knocked down for FANCA and FANCD2 were similarly used for transwell invasion assays compared to controls.
Invasion in response to FA loss requires DNA-PK activity
FA cells exhibit characteristic sensitivity to DNA crosslinkers, and defects in error-free DNA repair by homologous recombination (HR). These defects are accompanied, under some circumstances, by a corresponding increase in the activity of error-prone non-homologous end joining (NHEJ) pathway components (12, 13). NHEJ requires the activation of the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) and subsequent autophosphorylation on serine 2056 (31). In order to probe a possible functional involvement for DNA-PKcs signaling in FA-associated invasion, we first determined whether FANCD2- and FANCJ-deficient UM-SCC1 cells harbored activated DNA-PKcs. We then verified the functionality of a DNA-PK inhibitor NU7026 in this system in order to probe the importance of DNA-PK activity for invasion (Fig. 4a). Control cells and their FANCD2 and FANCJ-depleted counterparts were subjected to a pulse of bleomycin in order to stimulate DNA damage signaling, and the cells were then treated with vehicle or with the DNA-PKcs inhibitor NU7026. Cells were analyzed for activated DNA-PKcs phosphorylated on serine 2056. FANCD2 and particularly FANCJ loss stimulated DNA-PKcs phosphorylation compared to control (Fig. 4a, lanes 1–3), and autophosphorylation was completely eliminated by NU7026. Next, we determined whether DNA-PKcs activation was functionally important for FA-deficient cancer cell invasion. DNA-PKcs inhibition by NU7026 suppressed the invasive phenotype in FANCD2sh UM-SCC1 cells (Fig. 4b), but did not impair cellular viability (Fig. 4c). Importantly, DNA-PKcs inhibition also reduced control cell invasion under these conditions, thus indicating that DNA-PK activation contributes to HNSCC cell invasion. The ability of NU7026 to suppress invasion in FA-deficient cells was also observed in HPV+ FANCD2sh (Fig. S4a) and FANCAsh (Fig. S4b) UM-SCC47 cells.
Invasion in response to FA loss requires Rac1 GTPase activity
Ras-related small GTPase Rho/Rac/CDC42 signaling pathways are key players in cellular morphology and invasion (32). Because the activation of Rac1 was reported to produce intercellular projections similar to the ones noted for FA-deficient cells (Fig. 2) (33, 34), we carried out Rac1-GTP pull downs to first determine whether Rac1 is activated in FA HNSCC cells, and if so, to probe a possible role for DNA-PKcs in its regulation. FANCJsh knockdown cells were chosen based on the observation that DNA-PK activation was maximal in FANCJ when compared to FANCD2 knockdown cells in Fig. 4a. Interestingly, FANCJsh transduced UM-SCC1 cells showed increased Rac1 activity when compared with their NTsh control transduced counterparts (Fig. 5a, compare lanes 1 and 3). Treatment with DNA-PKcs inhibitor did not affect Rac1 activity in the control NTsh cells (Fig. 5a, compare lanes 1 and 2), but significantly lowered Rac1 activity in FANCD2sh and FANCJsh cells (Fig. 5a, compare lanes 3 and 4). Taken together, these results suggest that Rac1 activation downstream from FA loss is, at least in part, dependent upon DNA-PKcs activity. We next sought to examine the requirement for Rac1 GTPase activity in the characteristic cytoskeletal reorganization and invasion of FA-deficient HNSCC cells. We utilized NSC23766, a small molecule that has been shown to specifically inhibit Rac1 structural and functional activity but does not affect the activity of other Rho-related small GTPases (35, 36). Interestingly, NSC23766 reduced the number of intercellular protrusions and stimulated cell-cell adhesion in FANCD2- and FANCJ-deficient compared to control UMSCC1 cells as assessed by DIC (Fig. 5b, top panel) and phalloidin staining (Fig. 5b, bottom panel and quantification below). To determine whether NSC23766 was also capable of suppressing cellular invasion in response to FANCD2 loss, FA-proficient and -deficient HNSCC cells were treated with NSC23766 or vehicle over the course of the transwell assay. Indeed, cellular invasion following FANCD2 loss was repressed by NSC23766 which did not significantly affect control cell invasion (Fig. 5c) or cellular growth (Fig. 5d). NSC23766 could also suppress invasion in FA-deficient HPV+ UM-SCC47 cells (Fig. S4c). Together, these data demonstrate that FA loss and subsequent DNA-PK activation promote Rac1 activity to induce cytoskeletal aberrations and invasive tumor phenotypes in HNSCC cells (see Fig. 5e for a working model).
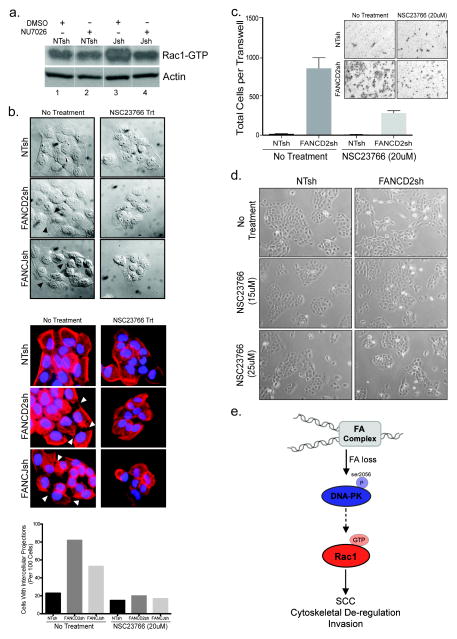
Figure 5. Morphological aberrations and invasive tumor phenotypes triggered by FA loss are dependent upon Rac1 activation.
(a) Rac1-GTP pulldown assays show that DNA-PK inhibition attenuates Rac1 activation in FA-deficient cells. All lanes are from the same western blot. (b) The small molecule Rac1 inhibitor NSC23766 was utilized at 20uM concentrations that did not affect cellular growth. To assess the effects of Rac1 inhibition on aberrant morphology and tumor cell invasion triggered by FA loss, UM-SCC1 cells were plated at equal densities and imaged by DIC microscopy prior to and following an 18 hour exposure to 20uM NSC23766 (left). NSC23766 treated and untreated UM-SCC1 cells were stained for phalloidin and imaged (right). The reduction in intercellular membrane projections following treatment with the Rac1 inhibitor is quantified and included below the images. (c) Invasion of NSC23766 or vehicle treated FA-deficient versus -proficient UM-SCC1 cells was evaluated by transwell assays. Experiments were carried out in duplicate and standard deviation was calculated. (d) UM-SCC1 exposure to NSC23766 at 15μM and 20μM concentrations did not affect cellular fitness and growth for the duration of the invasion assays, 22 hours. (e) A working model depicts the consequences of FA deficiency in HNSCC cells. Loss of FA pathway function results in increased DNA stress and activation of the DNA-PK sensor kinase through autophosphorylation on S2056. DNA-PK activity is directly or indirectly required for downstream Rac1 activation, and subsequent Rac1-dependent SCC cell invasion.
Discussion
Each year, approximately 40,000 new patients are diagnosed with head and neck cancer, predominantly HNSCC, in the United States, and this number continues to rise. Two main causative factors in the majority of oral, oropharyngeal, and laryngeal carcinomas are smoking and alcohol use; however, a growing percentage of these head and neck cancers, approximately 25% at present, have been attributed to HPV infection (37). Cellular and molecular characteristics of HPV–positive and –negative HNSCCs are still being explored, although these are distinct biological and clinical entities (38). Herein we link the loss of the FA DNA repair pathway in HNSCC cells with stimulated invasive potential regardless of HPV status, and uncover novel roles for DNA-PK (unrelated to DNA repair) in promoting Rac1 activation and invasive behavior.
Locoregional dissemination of malignant tumor cells is associated with poor outcome, and is crucially dependent on the migratory and invasive properties of the cancer cell. Tumor cell migration requires the formation of protrusions, leading edge attachment to the surrounding extracellular matrix, contraction of the cell to pull the cell body towards the leading edge, and detachment of the trailing edge. FA deficient HNSCC cells displayed expression changes for genes known to play key roles in cell motility, cell-cell adhesion and locomotion. Furthermore, we observed the presence of pronounced protrusions in FA deficient cells compared to their FA proficient counterparts and noted altered cell morphology. Together with these morphological phenotypes, we showed a significant increase in motility and invasion of FA deficient cells compared their FA proficient counterparts. Invasion was induced by the loss of either upstream (FANCA), central (FANCD2) and downstream (FANCJ) components of the FA pathway, and was reversible by the re-introduction of a knockdown-resistant construct. Together, these data support a scenario whereby the intact FA DNA repair machinery suppresses the transition of transformed to invasive HNSCC phenotypes.
DNA crosslink processing utilizes multiple repair pathways whose coordination appears to be a major function of the FA pathway. Recently, two reports have demonstrated that FA loss engages the DNA damage sensor kinase DNA-PK, a process that is likely followed by aberrant DNA repair by NHEJ and resulting characteristic FA chromosome pathologies. As such, FA proteins might govern the decision to channel double strand breaks (DSBs) into homologous recombination (HR) in favor of the competing DNA-PK associated error-prone non-homologous end-joining (NHEJ) pathway (12, 13). We show that FA loss in HNSCC cells increases the levels of active, auto-phosphorylated DNA-PKcs, and functionally implicate this DNA damage sensor in advanced HNSCC for the first time. While the nuclear role of DNA-PK in DNA repair is well characterized, non-canonical activities have emerged as well. DNA-PK has been implicated in inflammation through the phosphorylation of NF-kB1 (39), and in metabolic gene regulation through interaction with and phosphorylation of USF-1 (40). Furthermore, a number of novel cytoplasmic DNA-PK substrates were published recently that participate in cytoskeletal regulation (41). These include members of the 14-3-3 protein family, vimentin, and desmoplakin. The authors showed that DNA-PK activation decreased motility in melanoma cell lines, in contrast to increased invasion in our HNSCC cells. Thus whereas advanced cancer phenotypes are regulated by DNA-PK in both systems, the direction of the observed regulation may be cell type specific. Such differences might reflect significant complexity in the regulation of cytoskeletal components by DNA-PK. Further analysis identified a novel link between DNA-PKcs and Rac1 signaling in FA HNSCC cells wherein a specific DNA-PK inhibitor decreased active Rac1-GTP protein levels. One possible mechanism might be supported by a previous finding that DNA-PKcs can physically interact with the CDC42/Rac1 guanine exchange factor, ARHGEF6, in ovarian cancer cells (42). In addition, growing evidence supports correlations between DNA damage signaling and Rac1 activity (43–45), although the mechanisms and functional relevance of pathway cross-talk are largely unclear. Whether DNA-PKcs activates Rac1 directly through guanine nucleotide exchange factors or indirectly through signal transduction cascades is currently under investigation.
Rac1 is a well-known regulator of the cellular actin cytoskeleton, adhesion, barrier function and migration. Like other members of the Rho family, Rac1 cycles between GDP-bound inactive and GTP-bound active states (46). These GTPases are controlled by two classes of regulatory molecules: activating guanine nucleotide exchange factors (GEFs), and repressive GTPase-activating proteins (GAPs). Previously published work found that Rac1 was required for Kras-mediated tumorigenesis in skin epithelium and the lung (47, 48). More importantly, Rac1 activity strongly is associated with cell motility and tumor metastasis (49, 50). We found that both cell morphology (protrusion formation) and cellular invasion were dependent upon Rac1 activation, and show a reversion of the FA deficient HNSCC cell phenotype to a more epithelial-like morphology following inhibition of Rac1. We also observed a significant decrease in cancer cell invasion of FA deficient cells following treatment with the Rac1 inhibitor. Taken together, these findings link the loss of the FA pathway with increased Rac1 activation and downstream cytoskeletal aberrations and aggressive invasive potential. Of importance, Rac1 inhibitors are a potential novel therapeutic option for sporadic HNSCC carrying FA mutations, as well as an alternative, non-genotoxic treatment for HNSCC in patients with FA.
Supplementary Material
Statement of Translational Relevance.
Head and neck squamous cell carcinoma (HNSCC) is a devastating cancer type with poor outcomes particularly when diagnosed at advanced, invasive stages. Targetable genes and pathways that stimulate HNSCC progression remain poorly understood. A significant proportion of HNSCCs harbor mutations in FA DNA repair genes. Furthermore, individuals with germline loss of function mutations in FA genes are uniquely predisposed to aggressive HNSCC development. Herein we show that loss of the FA pathway in HNSCC cells stimulate plasma membrane re-organization, and tumor-cell invasiveness in vitro and in vivo – that is reliant upon NHEJ-associated DNA-dependent protein kinase (DNA-PK) and Rac1 GTPase. These findings implicate this important DNA repair pathway in the suppression of advanced tumor phenotypes, and identify new treatment options tailored to FA-deficient HNSCCs.
Acknowledgments
We are grateful to Dr. James Lessard of Cincinnati Children’s Hospital Medical Center (CCHMC) and Seven Hills Bioresearch (Cincinnati, OH) for his gift of the C4 pan-actin monoclonal antibody used in this work. We thank Drs. Stella Davies, Parinda Mehta, and Kasiani Myers of CCHMC and the Cincinnati Children’s Fanconi Anemia Comprehensive Care Center for thoughtful experimental guidance and discussion. This work was supported in part by NIH award RO1 CA102357 (S.I.W.). Helmut Hanenberg is supported by the Lilly Foundation Physician/Scientist initiative.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Jimenez L, Jayakar SK, Ow TJ, Segall JE. Mechanisms of Invasion in Head and Neck Cancer. Arch Pathol Lab Med. 2015 doi: 10.5858/arpa.2014-0498-RA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nature reviews Cancer. 2011;11(1):9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 3.Romick-Rosendale LE, Lui VW, Grandis JR, Wells SI. The Fanconi anemia pathway: repairing the link between DNA damage and squamous cell carcinoma. Mutation research. 2013;743–744:78–88. doi: 10.1016/j.mrfmmm.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith IM, Mithani SK, Mydlarz WK, Chang SS, Califano JA. Inactivation of the tumor suppressor genes causing the hereditary syndromes predisposing to head and neck cancer via promoter hypermethylation in sporadic head and neck cancers. ORL; journal for oto-rhino-laryngology and its related specialties. 2010;72(1):44–50. doi: 10.1159/000292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wreesmann VB, Estilo C, Eisele DW, Singh B, Wang SJ. Downregulation of Fanconi anemia genes in sporadic head and neck squamous cell carcinoma. ORL; journal for oto-rhino-laryngology and its related specialties. 2007;69(4):218–25. doi: 10.1159/000101542. [DOI] [PubMed] [Google Scholar]
- 6.Rosenberg PS, Tamary H, Alter BP. How high are carrier frequencies of rare recessive syndromes? Contemporary estimates for Fanconi Anemia in the United States and Israel. American journal of medical genetics Part A. 2011;155A(8):1877–83. doi: 10.1002/ajmg.a.34087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kee Y, D’Andrea AD. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes & development. 2010;24(16):1680–94. doi: 10.1101/gad.1955310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493(7432):356–63. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kutler DI, Auerbach AD, Satagopan J, Giampietro PF, Batish SD, Huvos AG, et al. High incidence of head and neck squamous cell carcinoma in patients with Fanconi anemia. Archives of otolaryngology--head & neck surgery. 2003;129(1):106–12. doi: 10.1001/archotol.129.1.106. [DOI] [PubMed] [Google Scholar]
- 10.Kutler DI, Singh B, Satagopan J, Batish SD, Berwick M, Giampietro PF, et al. A 20-year perspective on the International Fanconi Anemia Registry (IFAR) Blood. 2003;101(4):1249–56. doi: 10.1182/blood-2002-07-2170. [DOI] [PubMed] [Google Scholar]
- 11.Bogliolo M, Surralles J. Fanconi anemia: a model disease for studies on human genetics and advanced therapeutics. Curr Opin Genet Dev. 2015;33:32–40. doi: 10.1016/j.gde.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Adamo A, Collis SJ, Adelman CA, Silva N, Horejsi Z, Ward JD, et al. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Molecular cell. 2010;39(1):25–35. doi: 10.1016/j.molcel.2010.06.026. [DOI] [PubMed] [Google Scholar]
- 13.Pace P, Mosedale G, Hodskinson MR, Rosado IV, Sivasubramaniam M, Patel KJ. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science. 2010;329(5988):219–23. doi: 10.1126/science.1192277. [DOI] [PubMed] [Google Scholar]
- 14.Friebel TM, Domchek SM, Rebbeck TR. Modifiers of cancer risk in BRCA1 and BRCA2 mutation carriers: systematic review and meta-analysis. Journal of the National Cancer Institute. 2014;106(6):dju091. doi: 10.1093/jnci/dju091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mavaddat N, Peock S, Frost D, Ellis S, Platte R, Fineberg E, et al. Cancer risks for BRCA1 and BRCA2 mutation carriers: results from prospective analysis of EMBRACE. Journal of the National Cancer Institute. 2013;105(11):812–22. doi: 10.1093/jnci/djt095. [DOI] [PubMed] [Google Scholar]
- 16.Hora M, Urge T, Eret V, Stransky P, Klecka J, Kreuzberg B, et al. Tubulocystic renal carcinoma: a clinical perspective. World journal of urology. 2011;29(3):349–54. doi: 10.1007/s00345-010-0614-7. [DOI] [PubMed] [Google Scholar]
- 17.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333(6046):1154–7. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheckenbach K, Baldus SE, Balz V, Freund M, Pakropa P, Sproll C, et al. RAD51C--a new human cancer susceptibility gene for sporadic squamous cell carcinoma of the head and neck (HNSCC) Oral oncology. 2014;50(3):196–9. doi: 10.1016/j.oraloncology.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park JW, Shin MK, Pitot HC, Lambert PF. High incidence of HPV-associated head and neck cancers in FA deficient mice is associated with E7’s induction of DNA damage through its inactivation of pocket proteins. PloS one. 2013;8(9):e75056. doi: 10.1371/journal.pone.0075056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoskins EE, Morris TA, Higginbotham JM, Spardy N, Cha E, Kelly P, et al. Fanconi anemia deficiency stimulates HPV-associated hyperplastic growth in organotypic epithelial raft culture. Oncogene. 2009;28(5):674–85. doi: 10.1038/onc.2008.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoskins EE, Gunawardena RW, Habash KB, Wise-Draper TM, Jansen M, Knudsen ES, et al. Coordinate regulation of Fanconi anemia gene expression occurs through the Rb/E2F pathway. Oncogene. 2008;27(35):4798–808. doi: 10.1038/onc.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoskins EE, Morreale RJ, Werner SP, Higginbotham JM, Laimins LA, Lambert PF, et al. The fanconi anemia pathway limits human papillomavirus replication. Journal of virology. 2012;86(15):8131–8. doi: 10.1128/JVI.00408-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu J, Mu Q, Thiviyanathan V, Annapragada A, Vigneswaran N. Cancer stem cells are enriched in Fanconi anemia head and neck squamous cell carcinomas. International journal of oncology. 2014;45(6):2365–72. doi: 10.3892/ijo.2014.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gammon L, Biddle A, Fazil B, Harper L, Mackenzie IC. Stem cell characteristics of cell sub-populations in cell lines derived from head and neck cancers of Fanconi anemia patients. Journal of oral pathology & medicine : official publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology. 2011;40(2):143–52. doi: 10.1111/j.1600-0714.2010.00972.x. [DOI] [PubMed] [Google Scholar]
- 25.Kimple RJ, Harari PM, Torres AD, Yang RZ, Soriano BJ, Yu M, et al. Development and characterization of HPV-positive and HPV-negative head and neck squamous cell carcinoma tumorgrafts. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19(4):855–64. doi: 10.1158/1078-0432.CCR-12-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–82. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanenberg H, Batish SD, Pollok KE, Vieten L, Verlander PC, Leurs C, et al. Phenotypic correction of primary Fanconi anemia T cells with retroviral vectors as a diagnostic tool. Experimental hematology. 2002;30(5):410–20. doi: 10.1016/s0301-472x(02)00782-8. [DOI] [PubMed] [Google Scholar]
- 29.Chandra S, Levran O, Jurickova I, Maas C, Kapur R, Schindler D, et al. A rapid method for retrovirus-mediated identification of complementation groups in Fanconi anemia patients. Molecular therapy : the journal of the American Society of Gene Therapy. 2005;12(5):976–84. doi: 10.1016/j.ymthe.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 30.Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cellular and molecular life sciences : CMLS. 2011;68(18):3033–46. doi: 10.1007/s00018-011-0735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen BP, Chan DW, Kobayashi J, Burma S, Asaithamby A, Morotomi-Yano K, et al. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. The Journal of biological chemistry. 2005;280(15):14709–15. doi: 10.1074/jbc.M408827200. [DOI] [PubMed] [Google Scholar]
- 32.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420(6916):629–35. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 33.Evers EE, Zondag GC, Malliri A, Price LS, ten Klooster JP, van der Kammen RA, et al. Rho family proteins in cell adhesion and cell migration. European journal of cancer. 2000;36(10):1269–74. doi: 10.1016/s0959-8049(00)00091-5. [DOI] [PubMed] [Google Scholar]
- 34.Caron E, Hall A. Identification of two distinct mechanisms of phagocytosis controlled by different Rho GTPases. Science. 1998;282(5394):1717–21. doi: 10.1126/science.282.5394.1717. [DOI] [PubMed] [Google Scholar]
- 35.Nassar N, Cancelas J, Zheng J, Williams DA, Zheng Y. Structure-function based design of small molecule inhibitors targeting Rho family GTPases. Current topics in medicinal chemistry. 2006;6(11):1109–16. doi: 10.2174/156802606777812095. [DOI] [PubMed] [Google Scholar]
- 36.Gao Y, Dickerson JB, Guo F, Zheng J, Zheng Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(20):7618–23. doi: 10.1073/pnas.0307512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaturvedi AK, Anderson WF, Lortet-Tieulent J, Curado MP, Ferlay J, Franceschi S, et al. Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J Clin Oncol. 2013;31(36):4550–9. doi: 10.1200/JCO.2013.50.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lowy DR, Munger K. Prognostic implications of HPV in oropharyngeal cancer. The New England journal of medicine. 2010;363(1):82–4. doi: 10.1056/NEJMe1003607. [DOI] [PubMed] [Google Scholar]
- 39.Ju J, Naura AS, Errami Y, Zerfaoui M, Kim H, Kim JG, et al. Phosphorylation of p50 NF-kappaB at a single serine residue by DNA-dependent protein kinase is critical for VCAM-1 expression upon TNF treatment. The Journal of biological chemistry. 2010;285(52):41152–60. doi: 10.1074/jbc.M110.158352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong RH, Chang I, Hudak CS, Hyun S, Kwan HY, Sul HS. A role of DNA-PK for the metabolic gene regulation in response to insulin. Cell. 2009;136(6):1056–72. doi: 10.1016/j.cell.2008.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kotula E, Faigle W, Berthault N, Dingli F, Loew D, Sun JS, et al. DNA-PK target identification reveals novel links between DNA repair signaling and cytoskeletal regulation. PloS one. 2013;8(11):e80313. doi: 10.1371/journal.pone.0080313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maiti AK. Gene network analysis of oxidative stress-mediated drug sensitivity in resistant ovarian carcinoma cells. The pharmacogenomics journal. 2010;10(2):94–104. doi: 10.1038/tpj.2009.49. [DOI] [PubMed] [Google Scholar]
- 43.Hajas G, Bacsi A, Aguilera-Aguirre L, Hegde ML, Tapas KH, Sur S, et al. 8-Oxoguanine DNA glycosylase-1 links DNA repair to cellular signaling via the activation of the small GTPase Rac1. Free radical biology & medicine. 2013;61:384–94. doi: 10.1016/j.freeradbiomed.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan Y, Greer PM, Cao PT, Kolb RH, Cowan KH. RAC1 GTPase plays an important role in gamma-irradiation induced G2/M checkpoint activation. Breast cancer research : BCR. 2012;14(2):R60. doi: 10.1186/bcr3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rassool FV, Gaymes TJ, Omidvar N, Brady N, Beurlet S, Pla M, et al. Reactive oxygen species, DNA damage, and error-prone repair: a model for genomic instability with progression in myeloid leukemia? Cancer research. 2007;67(18):8762–71. doi: 10.1158/0008-5472.CAN-06-4807. [DOI] [PubMed] [Google Scholar]
- 46.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nature reviews Molecular cell biology. 2005;6(2):167–80. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 47.Kissil JL, Walmsley MJ, Hanlon L, Haigis KM, Bender Kim CF, Sweet-Cordero A, et al. Requirement for Rac1 in a K-ras induced lung cancer in the mouse. Cancer research. 2007;67(17):8089–94. doi: 10.1158/0008-5472.CAN-07-2300. [DOI] [PubMed] [Google Scholar]
- 48.Samuel MS, Lourenco FC, Olson MF. K-Ras mediated murine epidermal tumorigenesis is dependent upon and associated with elevated Rac1 activity. PloS one. 2011;6(2):e17143. doi: 10.1371/journal.pone.0017143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81(1):53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 50.Parri M, Chiarugi P. Rac and Rho GTPases in cancer cell motility control. Cell communication and signaling : CCS. 2010;8:23. doi: 10.1186/1478-811X-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.