Abstract
The period 2000–2015 brought the advent of high-throughput approaches to protein structure determination. With the overall funding on the order of $2 billion (in 2010 dollars), the structural genomics (SG) consortia established worldwide have developed pipelines for target selection, protein production, sample preparation, crystallization, and structure determination by X-ray crystallography and NMR. These efforts resulted in the determination of over 13,500 protein structures, mostly from unique protein families, and increased the structural coverage of the expanding protein universe. SG programs contributed over 4,400 publications to the scientific literature. The NIH-funded Protein Structure Initiatives (PSI) alone have produced over 2,000 scientific publications, which to date have attracted more than 93,000 citations. Software and database developments that were necessary to handle high-throughput structure determination workflows have led to structures of better quality and improved integrity of the associated data. Organized and accessible data have a positive impact on the reproducibility of scientific experiments. Most of the experimental data generated by the SG centers are freely available to the community and has been utilized by scientists in various fields of research. SG projects have created, improved, streamlined, and validated many protocols for protein production and crystallization, data collection, and functional analysis, significantly benefiting biological and biomedical research.
Introduction: A Brief History of Structural Genomics
The idea of a large-scale, high-throughput approach to protein structure determination arose in the late 1990s, spurred in part by the spectacular success of the Human Genome Project, and rapid progress in DNA sequencing technology [1]. This was accompanied by construction of synchrotron light sources and other structural biology facilities. Several large-scale structural genomics (SG) programs were established at the beginning of the 21st century, with the intention of achieving comprehensive structural coverage of the protein universe. The original goal envisioned making the 3-D structure of most proteins easily available, either directly from the Protein Data Bank (PDB), or through computational modeling methods. At the outset of SG, it was estimated that by solving ~16,000 structures of “carefully selected proteins” it would be possible to obtain structural coverage of “the vast majority of all proteins” (~90%) [2]. However, it was soon realized that this goal might be very challenging due to the continued exponential growth of the number of known protein sequences [3,4]. Later, structural genomics programs refocused on protein families with high biological importance.
In 2000, the US-based National Institutes of Health (NIH) created the Protein Structure Initiative (PSI), “a national effort to assemble a large collection of protein structures in a high-throughput operation” [5]. In its 5-year pilot phase, PSI included nine structure determination centers, with a total funding of 270 million dollars [5]. During this initial phase, it was shown that the high-throughput approach was feasible and could be applied to many protein classes. Several protein production and structure determination pipelines were created using both X-ray crystallography and NMR. In 2005, PSI entered its “production” phase, in which 325 million dollars were budgeted for four large-scale structure determination centers and six specialized centers targeting more specific biology areas [6]. The funding also supported homology modeling centers, a material repository, a knowledgebase, and several synchrotron beamlines. These investments expanded access of the biology community to structural data and models, materials, and technologies and attempted to convert the data and results of PSI into new knowledge. In 2010, PSI evolved into the PSI:Biology program, which associated high-throughput structure determination centers with “High Throughput-Enabled Structural Biology Partnerships.” The price tag for PSI:Biology was 290 million dollars [7] and included funding for four high-throughput centers, nine membrane protein centers, fifteen Biology Partnerships, a material repository, the PSI knowledgebase with homology modeling, and support for several synchrotron beamlines. Before PSI officially ended in July 2015, its combined programs deposited around 7000 protein structures into the PDB. Nearly 90% of these were determined by X-ray crystallography, the rest by NMR.
In 2002, Japan started its own 5-year structural genomics program, Protein 3000, aiming to analyze structural-functional relationship of about three thousand proteins with biomedical importance [8]. The program was implemented by the RIKEN Structural Genomics/Proteomics Initiative (RSGI), with a yearly budget of 45 million dollars, an additional 45 million at collaborating sites [9], and additional funding from partnerships with the pharmaceutical industry [10]. The project set up robotized protein production and sample screening systems and generated a total of 2743 PDB deposits, of which approximately half were determined using NMR, and half using X-ray crystallography.
In 2003, the Structural Genomics Consortium (SGC) was established by researchers from Canada and Great Britain to “focus explicitly on less well-studied areas of the human genome” [11]. At its outset, it secured nearly 100 million dollars funding from the Canadian research organizations, the Wellcome Trust charity, and from pharmaceutical companies. An additional 50 million was committed in 2011 [12]. SGC has grown to include researchers from Sweden, Brazil, Germany, and the US, and so far has contributed around 1400 protein structures to the PDB.
In 2008, the National Institute for Allergy and Infectious Diseases (NIAID) started a structural genomics program targeting the emerging and re-emerging (drug-resistant) infectious human pathogens. The program established two centers, and emphasized target submissions from the wider biology community. NIAID invested $72 million during the first 5 years of the program, and in 2012 budgeted an additional $50 million for the next 5 years [13]. These SG centers took advantage of the technologies, protocols and structure determination pipelines developed by the PSI. As of December 2015, the two NIAID centers have deposited around 1600 protein structures from pathogens into the PDB, including complexes with inhibitors and potential drugs.
There have been other large-scale programs similar to the SG model. A prominent example is the Enzyme Function Initiative (EFI), which operated from 2010 to 2015 and integrated bioinformatics, structural biology, enzymology, genetics, and metabolomics to develop strategies for discovering enzymatic and physiological functions of uncharacterized enzymes [14]. While it was not a SG program itself, the EFI made use of the technologies and protocols developed by the PSI, such as the large-scale pipelines for enzyme production and structure determination, and contributed around 340 deposits to the PDB.
In addition to the major programs described above, there have been around twenty smaller centers in Europe, Asia, and North America that have also contributed PDB deposits classified as “structural genomics.” This classification has been essentially self-assigned by the depositors, and is indicated by the content of the set of fields in the category “pdbx_SG project” [15] in the mmCIF files of the PDB deposits. However, the smaller centers accounted for a relatively small percentage of the SG deposits to the PDB, with the five major initiatives – PSI, Protein 3000, the NIAID centers, SGC, and EFI contributing more than 95% of SG deposits to the PDB. The smaller centers include several programs funded by the European Union and its member countries, the largest of which was the 3-year program SPINE (Structural Proteomics in Europe), which began operations in 2002 and determined over 100 protein structures [16,17]. The Tuberculosis Structural Genomics Consortium, originally formed as part of PSI, has been continuing as an international collaboration comprising almost 500 researchers [18,19]. The “taxonomy” of the all currently living and extinct SG projects is presented in Table 1.
Table 1.
The taxonomy of Structural Genomics: list of the different SG centers, according to the contents of the pdbx_SG_project fields in the mmCIF files in the PDB (curated manually), as of December 31, 2015. Joint deposits for two or more centers are included in the total for each center.
| Center | Country | Funding | # Structures |
|---|---|---|---|
| Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | USA | PSI-2 specialized center | 29 |
| Assembly, Dynamics and Evolution of Cell-Cell and Cell-Matrix Adhesions (CELLMAT) | USA | PSI:Biology partnership | 2 |
| Atoms-to-Animals: The Immune Function Network (IFN) | USA | PSI:Biology partnership | 31 |
| Bacterial targets at IGS-CNRS, France (BIGS) | France | EU | 4 |
| Berkeley Structural Genomics Center (BSGC) | USA | PSI-1 | 103 |
| Center for Eukaryotic Structural Genomics (CESG) | USA | PSI-1,2 | 218 |
| Center for High-Throughput Structural Biology (CHTSB) | USA | PSI-2 specialized center | 6 |
| Center for Structural Genomics of Infectious Diseases (CSGID) | USA | NIAID | 811 |
| Center for Structures of Membrane Proteins (CSMP) | USA | PSI-2 and PSI:Biology specialized center | 28 |
| Chaperone-Enabled Studies of Epigenetic Regulation Enzymes (CEBS) | USA | PSI:Biology partnership | 6 |
| Enzyme Discovery for Natural Product Biosynthesis (NatPro) | USA | PSI:Biology partnership | 66 |
| Enzyme Function Initiative (EFI) | USA | NIH “glue grant” | 338 |
| GPCR Network (GPCR) | USA | PSI:Biology membrane protein center | 48 |
| Integrated Center for Structure and Function Innovation (ISFI) | USA | PSI-2 specialized center | 28 |
| Israel Structural Proteomics Center (ISPC) | Israel | EU | 45 |
| Joint Center for Structural Genomics (JCSG) | USA | PSI-1,2, Biology large scale center | 1601 |
| Marseilles Structural Genomics Program @ AFMB (MSGP) | France | EU | 12 |
| Medical Structural Genomics of Pathogenic Protozoa (MSGPP) | USA | US | 35 |
| Membrane Protein Structural Biology Consortium (MPSBC) | USA | PSI:Biology membrane protein center | 3 |
| Membrane Protein Structures by Solution NMR (MPSbyNMR) | USA | PSI:Biology membrane protein center | 5 |
| Midwest Center for Structural Genomics (MCSG) | USA | PSI-1,2, Biology large scale center | 1872 |
| Mitochondrial Protein Partnership (MPP) | USA | PSI:Biology partnership | 10 |
| Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | Canada | Canadian research organizations | 126 |
| Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | Germany | German government | 36 |
| New York Consortium on Membrane Protein Structure (NYCOMPS) | USA | PSI:Biology membrane protein center | 58 |
| New York SGX Research Center for Structural Genomics (NYSGXRC) | USA | PSI-1,2 large scale center | 1042 |
| New York Structural Genomics Research Consortium (NYSGRC) | USA | PSI:Biology large scale center | 344 |
| Northeast Structural Genomics Consortium (NESG) | USA | PSI-1,2, Biology large scale center | 1212 |
| Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXtals) | USA | PSI:Biology partnership | 5 |
| Ontario Centre for Structural Proteomics (OCSP) | Canada | Canadian research organizations | 35 |
| Oxford Protein Production Facility (OPPF) | UK | EU | 33 |
| Paris-Sud Yeast Structural Genomics (YSG) | France | EU | 6 |
| Partnership for Nuclear Receptor Signaling Code Biology (NHRs) | USA | PSI:Biology partnership | 3 |
| Partnership for Stem Cell Biology (STEMCELL) | USA | PSI:Biology partnership | 20 |
| Partnership for T-Cell Biology (TCELL) | USA | PSI:Biology partnership | 48 |
| Program for the Characterization of Secreted Effector Proteins (PCSEP) | USA | PSI:Biology partnership | 17 |
| Protein Structure Factory (PSF) | Germany | EU | 2 |
| RIKEN Structural Genomics/Proteomics Initiative (RSGI) | Japan | Protein 3000 project | 2743 |
| Scottish Structural Proteomics Facility (SSPF) | UK | EU | 11 |
| Seattle Structural Genomics Center for Infectious Disease (SGCID) | USA | NIAID | 801 |
| Southeast Collaboratory for Structural Genomics (SECSG) | USA | PSI-1 | 121 |
| Structural Genomics Consortium (SGC) | International | Industry, charities, Canadian research organizations | 1404 |
| Structural Genomics Consortium for Research on Gene Expression (SGCGES) | USA | PSI:Biology partnership | 2 |
| Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | USA | PSI-1 | 71 |
| Structural Proteomics in Europe (SPINE) | EU | EU | 119 |
| Structural Proteomics in Europe 2 (SPINE-2) | EU | EU | 8 |
| Structure 2 Function Project (S2F) | USA | US | 54 |
| Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | USA | PSI:Biology partnership | 3 |
| Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | USA | PSI:Biology partnership | 36 |
| TB Structural Genomics Consortium (TBSGC) | International | PSI-1; international | 285 |
| Transcontinental EM Initiative for Membrane Protein Structure (TEMIMPS) | USA | PSI:Biology membrane protein center | 2 |
SG programs, like many other “big science” initiatives, have encountered a variety of often polarized reactions from the scientific community, from a very strong support to, in some cases, strong criticism [8,20–22]. In particular, the target selection process in some SG centers was criticized. The list of “carefully selected proteins” envisioned at the outset [2] has never been actually compiled, and over the years the list of proteins targeted by SG grew to over 330,000 entries. Critics also argued that the funding expended for SG would have been more effective if it were instead directed to traditional, individual programs (e.g. R01). Proponents of structural genomics emphasized that the scientific community at large benefitted from access to publically available technologies, data, structures, materials, software, synchrotron beamlines, NMR facilities etc. developed by the SG programs. Although discussion of such policy and philosophical questions is beyond the scope of our paper, we will briefly summarize the economic aspects of SG.
Although it is difficult to know the budgets of all individual centers, a reasonable estimate of the total costs of the worldwide SG to date is $2.0 billion in 2010 dollars. This amount has been obtained by the summing up the known expenditures of the four major programs (inflation-adjusted to 2010) and extrapolating the result to the other programs. Around 60% of SG funding was provided by the US governmental agencies. While these are significant amounts, they should be put in perspective. The PSI program represented a mere 0.2% of the total NIH budget during the 15 years when PSI was operating. In comparison, the cost of the Human Genome Project was $5.6 billion in 2010 dollars [23,24]. As an investment, the Human Genome Project was a resounding success, driving an estimated $800 billion of worldwide economic impact by the year 2010 [24].
There have been no comparable studies of the economic impact of SG programs and, in our opinion, it would be premature to try to determine it now. It should be emphasized that the two megaprojects were significantly different. The Human Genome Project concentrated its resources on the development of new, focused technologies and methodologies (DNA sequencing and analysis of the data), and the resulting sequence of the human genome was its only end goal and the proof of its success. SG centers have concentrated their resources on a process that is orders of magnitude more complex - from gene sequence to three-dimensional structure. It involved genome analysis and classification of protein families, recombinant technologies for cloning thousands of genes, large-scale production of unique proteins, crystallization, data collection using X-ray crystallography and NMR, de novo structure solution of novel proteins and structural analysis. The integration of many steps and different kind of data creates an enormous amount of complexity. SG programs generated large amounts of experimental data/results produced in a highly reproducible manner; in particular, they cloned over 220,000 protein genes. DNA sequencing programs are not directly comparable to SG efforts due to the differing natures of the problems addressed. Sequencing of large genomes is largely a single step process, whereas structure determination is significantly more complex experimentally, requiring a high level of integration in a multi-step process with a large number of experiments that have to be often replicated.
As of the time of the writing, funding for SG projects has been greatly reduced - only two of the major SG initiatives (NIAID and SGC) are still active. It is an appropriate time to ask: what has been the gain for humanity for the two billion dollars invested in SG so far? In this work, we try to answer this question by discussing the impact of SG in five specific areas: (1) structures and modeling, (2) publications, (3) experimental data, (4) structure determination technologies, and (5) function determination technologies.
Structures, Modeling Leverage and Modeling Methods
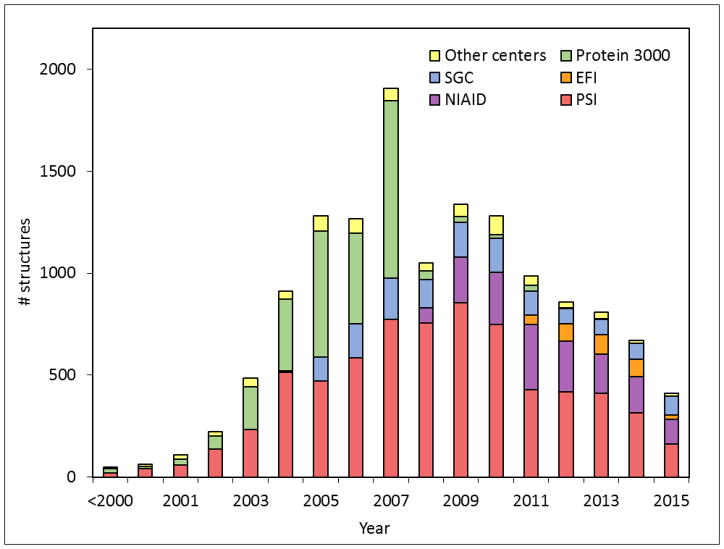
The most often used metric of the outcome of SG is the total number of structures deposited into the PDB. As of December 31, 2015, there were 13,606 PDB deposits (including 11,337 solved by X-ray crystallography, 2267 solved by NMR, 1 solved by electron microscopy, and 1 by combination of NMR and solution scattering) that were attributed to SG centers (as indicated by the mmCIF fields “pdbx_SG_project,” manually curated), constituting 11.8% of all PDB deposits. The PSI alone accounted for slightly over half of all SG deposits. The SG contribution was especially high in terms of “unique” deposits–defined as those for which there was no structure in PDB with ≥ 30% sequence identity at the time of the deposition. As of December 2015, SG contributed around 5100 such structures. The SG contributions constituted about a quarter of all the unique X-ray crystallography deposits in the PDB and nearly a third (29.7%) of the unique NMR deposits.. The growth of SG deposits is shown in Fig. 1.
Fig. 1.
The structural output of SG programs. The vertical axis represents the number of structures deposited to the PDB by SG programs worldwide. Structures were assigned to a particular SG program based on the contents of mmCIF files (field _pdbx_SG_project) with manual curation in the PDB as of December 31, 2015.
These unique deposits are important in terms of “novel modeling leverage,” as they provide templates to build comparatively reliable homology models for domain families that previously had no structural data [25]. Increased structural coverage has also benefited crystallography, as non-redundant structures can be used as templates for structure determination using molecular replacement (MR). This means that crystallography workflows can be simplified and significantly accelerated, facilitating determination of more 3D structures.
SG target selection strategies, particularly at the beginning of SG, focused on selecting targets from the largest structurally uncharacterized families in an effort to maximize the structural coverage of “protein sequence space” [26]. However, the rapid growth in known protein gene sequences due to revolutionary advances in DNA sequencing technology has made this goal more challenging. In 2000, the entire known protein sequence universe consisted of about 300,000 sequences, as measured by the size of the UniProt and TrEMBL databases [27]. In January 2016, the UniProtKB/TrEMBL database had nearly 60 million “non-redundant” entries comprising nearly 20 billion amino acids [28]; the NCBI Reference Sequence (RefSeq) database (release 73) contained a similar number of proteins. The growth of the total number of structures has not been able to match this pace: in recent years, the number of known proteins has been growing exponentially at a rate of about 60% per year, while the number of structures in the PDB has been growing by about 10% per year. Furthermore, the highly skewed, power law distribution of the sizes of protein domain families means that whereas there are a few very large families with many sequences, there are also many “orphan” ones with only one or a few members [29,30].
Despite these challenges, there is evidence to suggest that SG efforts have led to significant progress in increasing the modeling leverage of protein sequence space (the set of proteins for which structures may be obtainable through homology modeling). One study demonstrated that the PSI’s per structure novel coverage rate was four times higher than the equivalent rate for structures solved by non-PSI centers [31]. A more recent study has established that while the rate of growth of sequence databases is still increasing, they are also changing in taxonomic composition (a relative increase in the number of prokaryotic sequences) and are becoming increasingly redundant [32]. Overall, the total structural coverage of the GenBank non-redundant set of protein sequences increased from 30% in 2001 to 40% in 2011, and the increase in the degree of coverage in organisms of greater relevance to human health, such as E.coli, was even greater [33].
To effectively utilize the structural coverage provided by SG, structural and computational biologists developed many comparative modeling methodologies. In particular, PSI laboratories developed modBase–a database of comparative protein structure models [34] and the Protein Model Portal [35], which allows users without expert knowledge to download precomputed models or compute them on demand. The methods of target selection employed by SG, which were designed to maximize structural coverage, were also beneficial to initiatives like the Critical Assessment of Structure Prediction (CASP) [36], in which SG laboratories participated by providing novel structures for blind test cases. The influx of new structures with low sequence similarity to existing models allowed for systematic evaluation of modeling software by challenging them with very difficult comparative and ab initio modeling problems. The CASP competition, which heavily utilized novel SG targets, provided an incentive for software developers to improve both automatic and human-assisted structure prediction.
For some smaller proteomes, SG efforts have succeeded in providing extensive coverage. The proteome of the Thermotoga maritima bacteria consists of 1852 confirmed and predicted proteins (UniProt database release 2016_01, entry up000008183) of which 387 have their 3D structures determined. Nearly 60% of these (227) were first contributed by SG centers. An additional 451 Thermotoga proteins have comparative models available from the Protein Model Portal. Overall, SG provides structures or structural models for close to 37% of the proteome. Successful processing of T. maritima proteome initiated by SG [37] generated enough data to construct a working model of the metabolic networks of this organism [38], which permits simulations of metabolism in the context of whole cell.
The human proteome contains around 20,000 proteins encoded in the genome [39], comprising 11.3 million amino acids (neXtProt database release 2015-09-01) [40]. In the PDB there are currently around 28,500 Homo sapiens entries representing 5485 unique human proteins annotated in neXtProt (3.3 million amino acids) whose structures have been experimentally determined. For nearly a quarter of these (1359 proteins), SG projects have contributed the earliest PDB deposit: RSGI alone determined the first structure for 633 human proteins, the SGC for another 543. The SG contributions correspond to 0.9 million amino acids – 27.9% of the total number of amino acids in human proteins with experimentally determined structures. Structural models for around 14,000 additional human proteins (obtained by automatic modeling methods) have been made available by the Protein Model Portal. Interestingly, nearly all the Gene Ontology (GO) terms occurring in the human proteome have been already structurally characterized, in the sense of having at least one protein with that annotation in the modeling leverage of the PDB [41].
Around 20 to 30% of most eukaryotic and eubacterial proteomes consists of integral membrane proteins (IMPs) [42] for which determination of structure is notoriously challenging. SG efforts have made significant contributions to the progress in this area. Specialized SG centers have targeted over 15,000 IMPs, succeeding in “plucking” a number of “high-hanging” novel membrane targets which increased the structural coverage of membrane proteins by 7% (in terms of providing the first structure for an IMP Pfam-family) [43]. The impact of SG membrane centers, in particular the specialized PSI centers extends beyond the structural coverage: over 2000 purified IMPs have been made available [43]. Combining insights gained from the experimental pipeline and modeling of IMPs, SG has spurred novel approaches to structural characterization of the entire human alpha-helical transmembrane proteome [34].
Publications
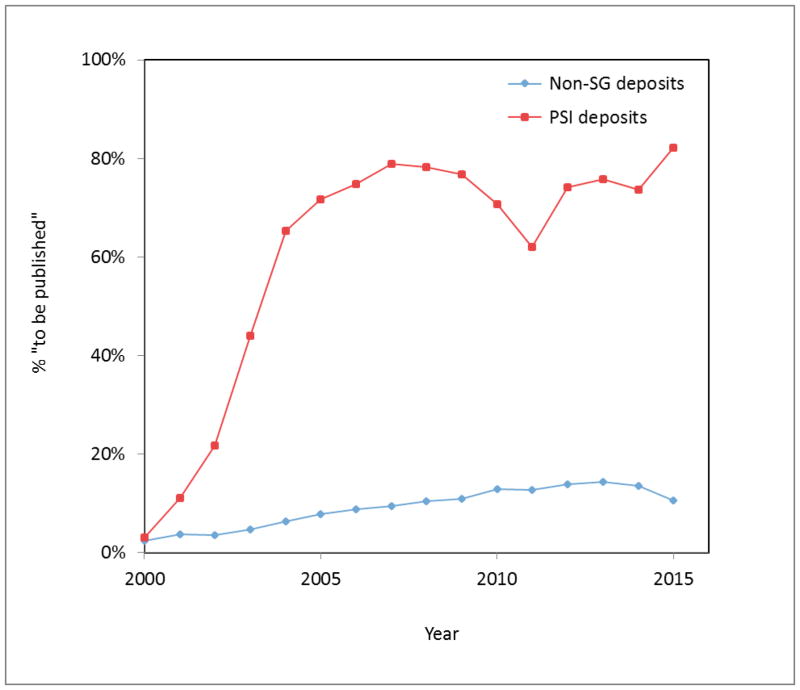
The number of peer-reviewed publications and the number of times they are cited is often used as a measure of the impact of research projects. Until the 1990s, almost any new protein structure warranted a publication, but in the SG model of high volume structure determination, this was simply not possible. All SG consortia were supposed to immediately deposit the structure into the PDB, and not enough time and effort was allotted to obtain “publishable” results such as the determination and validation of function. The difference between the rate of publications associated with SG and non-SG deposits is striking (see Fig. 2). For “traditional” structural biology, the percentage of deposits without a primary citation (marked “to be published” in the PDB deposits) has remained below 15% throughout the last 15 years, though this percentage has been trending upwards. In contrast, around 80% of recent PSI deposits remain “to be published.”
Fig. 2.
Percentage of PDB structures with primary citation “to be published” in the period 2000–2015 in function of the year of deposition for the PSI deposits (red line) and traditional (non-SG) deposits (blue line).
Nevertheless, SG programs have produced a large number of scientific publications. It is difficult to determine the exact numbers of publications for the worldwide SG as a whole, as there are no clear criteria that would allow classifying a particular scientific paper unambiguously as a “SG publication.” As of September, 2015, a total of 4419 scientific publications (the vast majority in peer-reviewed journals) have been reported by the five major initiatives – PSI (2280, [44]), RIKEN Structural Genomic/Proteomics Initiative (1010, [45]), SGC (891, [46]), the two NIAID centers (169, [47,48]), and EFI (69, [49]) Some structural and methodological publications represented very high impact as measured by the number of citations, e.g. [50–54].
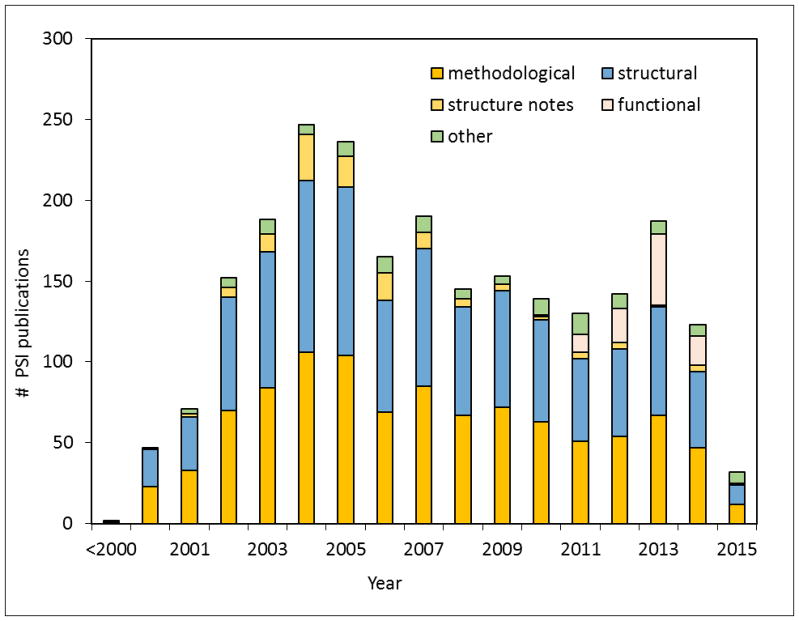
We will focus our analysis on the publications from the PSI, which has set up a database of peer-reviewed publications produced and self-reported by its members ([44], [55]). As of August 24, 2015, it contained 2280 publications credited to the PSI. Of these, 1008 were methodological, 1058 structural, 95 functional, and 119 other (reviews, computational studies, etc.). The growth in the number of PSI publications is shown in Fig. 3. These publications have been cited over 93,000 times in other scientific publications (excluding datasets cited in the Data Citation Index). The average number of citations per publication was around 56 for methodological papers, and around 32 for structural ones (excluding structural notes), which are very respectable numbers.
Fig. 3.
Number of publications produced by the PSI, based on data from the PSI Publications Portal database as of August, 24, 2015.
An often used measure of a set of scientific publications (e.g. a journal) is “cites per document in an n-year window”), defined as the average number of citations received in a given year by papers from the set that were published within the n preceding years. For n=2, this metric is known as the impact factor (a trademark of Thomson Reuters). For the set of all PSI publications, the 2014 impact factor-like parameter was 8.6 (calculated using data from the PSI Publications database). If the set of all PSI publications were a journal, it would have ranked in the top 5% of the more than 1800 journals in the category of Biochemistry, Genetics and Molecular Biology according to data compiled by the SCImago Journal and Country Rank (http:/scimagojr.com).
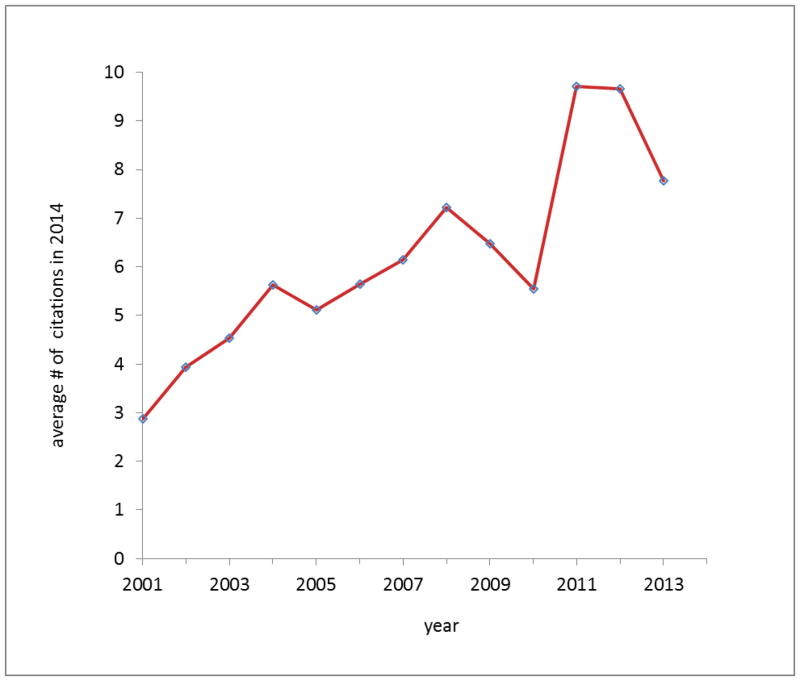
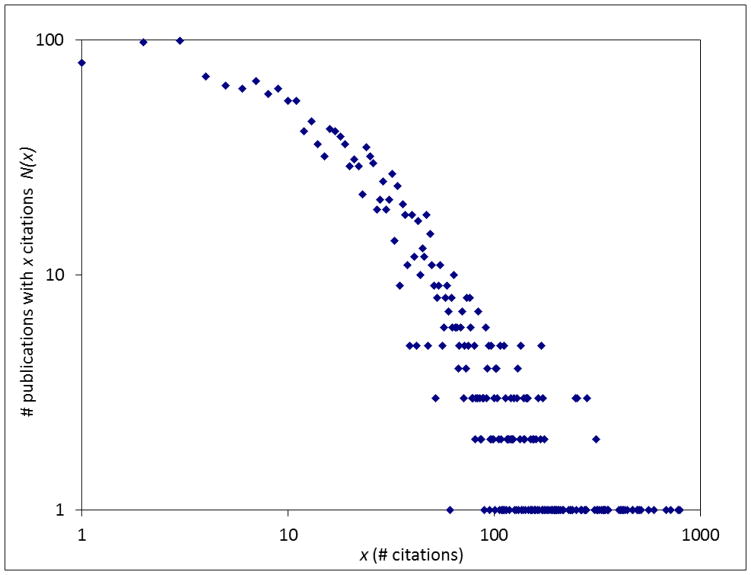
Fig. 4 presents the average number of citations that were received in 2014 by PSI papers as a function of the date of publication. Generally, earlier papers are cited less often than the more recent papers. On average, papers published after 2004 attracted more than 5 citations in 2014. The distribution of N(x), the number of papers with x citations is presented on a log-log scatter plot in Fig. 5. Similarly as for other sets of scientific publications studied in the literature [56] it is characterized by a “heavy tail” of a small number of highly cited papers and the majority of papers with very few citations. Such distributions were suggested to follow a power law distribution, N(x) ~ x-α, with α ~ 3 [56], especially in the region of the most highly cited papers [57,58]. The theoretical model that appears to fit well to the dataset of the PSI publications has a dual character [59], with a direct mechanism describing the papers with few citations, and an indirect mechanism (citing a paper based on citations already existing) describing the highly cited papers. The indirect mechanism yields a non-universal power-law distribution: publications that reach the tipping point where the indirect mechanism dominates tend to become even more highly cited and become “classics”—i.e. “the rich get richer.” Using the method of Clauset and coworkers [60] to analyze the dataset of PSI publications shown in Fig. 5, we found the tipping point to be around 69 citations. Above this tipping point, the citation distribution can be modeled by a power law, with an estimated value of α ~ 2.4. This region includes about 330 (15%) papers that have crossed the tipping point, and thus are potential “classics.”.
Fig. 4.
The average number of citations received in 2014 by PSI paper, as a function of the year of publication.
Fig. 5.
Distribution of the number of citations of PSI publications. The horizontal axis represents the number of citations, and the vertical axis represents number of publications with a given number of citations.
On the other end of the distribution, about 7.6% of PSI publications have not been cited at all to date. While this is potentially a disappointing number, it reflects the policy to concentrate on structures, not on functional experiments. Papers that are “structure notes,” which contain a description of the structure but little or no functional data, attract few citations. However, this problem is not unique to SG, as a significant fraction of papers published in most journals do not attract any citations. For example, Acta Crystallographica Section F publishes short communications on “any aspect of structural biology.” According to the SCImago Journal & Country Rank service, nearly 50% of papers published in this journal have no citations, even after 10 or more years.
Data, Databanks, Databases and Other Resources
From the outset, SG centers have emphasized data management in order to optimize the structure determination pipeline and disseminate results (including negative results). The primary mechanism for reporting experimental data in the PSI (and adopted by other SG centers) was the TargetDB database and its successor TargetTrack [61,55]. Many SG centers developed their own systems for aggregating and reporting experimental data to the TargetTrack system via XML-formatted reports. Many centers also developed laboratory information management systems (LIMS) that provide tools to track many aspects of the structure determination pipeline, from cloning to structure determination. Some of the LIMS originally developed by SG programs were made available to the broader researcher community, such as PiMS/xtalPiMS [62,63] and SESAME [64], and others, such as LabDB [49], are under active development in preparation for public release.
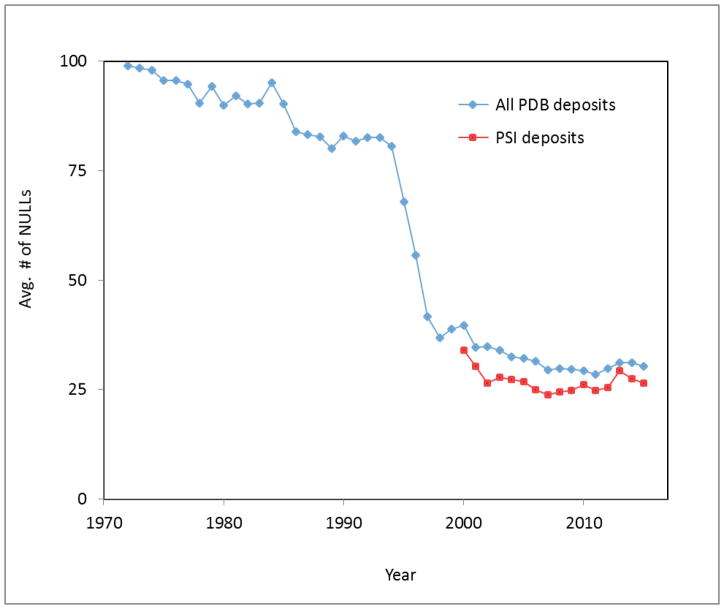
One measure of the completeness of experimental data related to determination of a protein structure is the number of missing data items (“NULLs”) in the headers of PDB deposits. The PDB file format was first specified in 1971 and included around 400 data items [65]. At the outset of the Protein Data Bank, an average PDB deposit header had over 90 data items set to a value of “NULL”. In subsequent years, due to progress in crystallographic software, the average number of NULLs in a given deposit decreased, falling to about 30 in 2006 (Fig. 6). However, since then the average number of NULLs has stayed relatively constant. As not all data items are relevant to all experiments, some NULLs will always be present.
Fig. 6.
The average number of missing data items (“NULLs”) for all PDB deposits using X-ray crystallography (including both traditional structural biology and SG) as a function of the year of deposition from 1972 to 2014 is shown in blue. The average number of NULLs in X-ray crystallography PDB files from the PSI initiative (2000–2014) is shown in red.
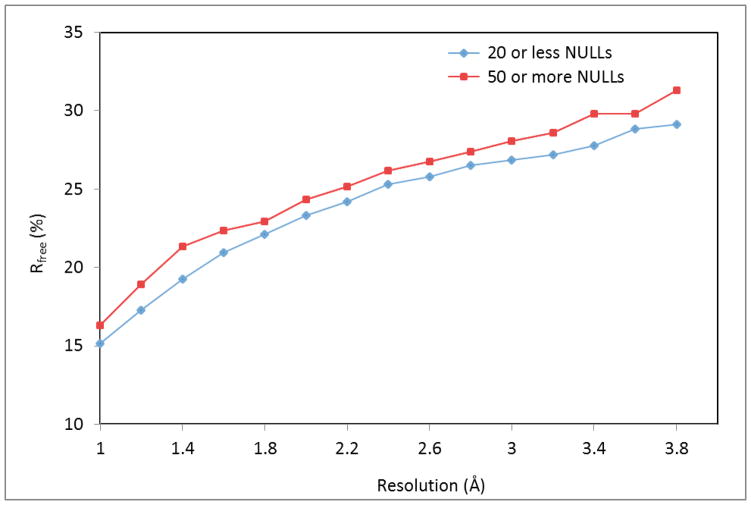
The quality of a crystallographic structure and the number of NULLs in the corresponding PDB deposit appear to be correlated. As shown in Fig. 7, for a wide range of resolutions, the average values of the Rfree factor for “high-completion” deposits (defined as those with 20 NULLs or less) were significantly lower (p=0.05) than the average Rfree values for “low-completion” deposits (defined as those with 50 or more NULLs). One possible explanation for the observed correlation is the “human factor,” namely the experience and knowledge of the crystallographer who determined each structure. An experienced crystallographer presumably is better informed of the procedures to ensure a structure is refined and validated in an optimal way, and also has better knowledge of (and appreciation for) what data items to collect and submit related to the process of solving the structure. Experienced crystallographers are also more likely to be aware of tools, such as PDB_EXTRACT, that can help collect relevant data from pertinent log files.
Fig. 7.
Average Rfree by resolution bin (with a width of 0.2 Å) for X-ray crystallography structures deposited to PDB from January 2001 to March 2015, divided into two groups by the number of missing data items (“NULLs”) in the PDB file. The means for “high-completion” deposits (20 NULLs or less) are shown in blue, and the means for “low-completion” deposits (50 or more NULLs) are shown in red.
SG has been producing large amounts of data and the dissemination of this data has been a long-standing goal of many SG centers. In addition to releasing experimental information in resources such as the PDB and TargetTrack, another approach pursued by several SG initiatives was to find alternative mechanisms to disseminate information about structures that were not accompanied by peer-reviewed papers. Two of these are wiki-like annotation systems—The Open Protein Structure Annotation Network (TOPSAN) [66] and Proteopedia [67]—which permit scientists to collaboratively collect data and annotate structures online. SG centers have also developed novel ways to interactively render and propagate information about structures and their annotations. For example, the Structural Genomics Center (SGC) and the Center for Structural Genomics of Infectious Diseases (CSGID) have developed tools to generate interactive electronic presentations making use of ActiveICM technology (Molsoft LLC). In the case of CSGID these presentations are generated semi-automatically [68].
An important resource presenting SG data is the PSI Structural Biology Knowledgebase [55,69] (SBKB; http://sbkb.org), which makes available protocols created for the purpose of protein structure and function determination, papers highlighting new findings in structural biology work, and recent technology or methodology developments created by SG programs. Recently, a significant fraction of the X-ray diffraction data collected by the US-based structural genomics programs has been made publicly available by the new Integrated Resource for Reproducibility in Macromolecular Crystallography (http://proteindiffraction.org).
SG projects which pursued large-scale NMR studies, such as the RIKEN Structural Genomics/Proteomics Initiative and the Northeast Structural Genomics Consortium (NESGC) have collected large amounts of high-quality protein chemical shift information and other relevant data. RSGI has developed an automated system for annotation of datasets for resonance assignments [70] and their deposition to the BioMagResBank (BMRB) [71]. Some SG centers have been also depositing the original time-domain NMR spectra to BMRB, allowing for recalculation and reinterpretation of structural models. Overall, SG data have significantly increased the quality of the BMRB.
Resources developed by SG programs are not limited to providing data, but also include materials such as expression clones, purified proteins, and chemical probes. In particular, the PSI:Biology Materials Repository was established for storage, annotation, and distribution of genetic constructs designed by PSI centers. As of October 2014, it had nearly 90,000 PSI plasmids containing gene inserts and 130 empty vectors that have been made available to the community [72]. The Seattle Structural Genomics Center for Infectious Disease (SSGCID) has amassed a repository of over 3,000 purified proteins; about half of which have been distributed to requesters [73]. SGC has been developing a library of open access chemical probes – high-quality inhibitors for epigenetic proteins [74].
The emphasis of the SG projects on collecting as much experimental data as possible, focusing on data quality, and inclusion of not only positive but also negative results is of particular importance in the context of recent studies, showing that a significant fraction of experimental results published in the scientific literature cannot be reproduced by groups different from the original researchers [75]. Procedures used by laboratories involved in SG, relying on databases [68] and related tools have been successfully used in identifying problems that may affect reproducibility of experimental results, e.g. mislabeling of a protein sample, contamination with a foreign protein, and other issues [76]. These approaches to detect and correct potential reproducibility problems are directly applicable in many other areas of biomedical research.
Structure determination technologies
To meet their proposed goals, SG centers have had to develop streamlined pipelines expediting processes from target selection, through protein production and crystallization, to structure determination and data management. SG centers have developed techniques for high-throughput cloning, protein expression and purification [77], tools for crystal production, and numerous software packages for X-ray and NMR data collection, structure determination and refinement.
A single protocol can be applied to generate multiple and varied genetic constructs to screen for one that most efficiently expresses the desired protein. SG has improved systems for semiautomated expression and purification of proteins of interest that allow for streamlined testing and searching for the best conditions for large scale production of protein for research applications. The requirement for large quantities of active protein complexes for structural and functional studies was met by the development of efficient pipelines that incorporated numerous technical advances, including tools for co-expression and co-purification of protein partners in E. coli cells [77], as well as eukaryotic expression techniques [78]. Methods for testing the stability of proteins for crystallization experiments have been developed and optimized [79]. Based on their experience crystallizing target proteins, SG centers have developed their own crystallization screens (for example the MCSG suite and JCSG+) [80,81]. The compositions of these screens have been optimized after analysis of all crystallization conditions applied to hundreds of protein targets, and identification of the conditions that produced the most crystals. This would not be possible without extensive data on a large number of targets including both successful and failed crystallographic experiments. The systems developed by SG centers for systematic, wide-scale collection of experimental data described above were crucial for this research.
Parallel to the development of methodologies for efficient protein production and crystallization, bioinformatics tools were also developed for sequence analysis and predicting protein stability, folding, and crystallizability. SG provided bioinformatics servers for the rational design of protein constructs [82] as well as predicting protein folds, interfaces, and protein-protein interactions [83].
SG programs have been a leader in the development of new methodology for structure determination by X-ray crystallography [84,85]. In this century, nearly half of all PDB deposits determined by the single wavelength anomalous dispersion (SAD) method were submitted by SG centers. The large number of diverse protein crystals produced by SG programs created the need for efficient methods of screening for the best diffracting crystal. The unprecedented large number of projects led to the development of programs for fast (albeit not always efficient) data collection and structure determination [51,86–88] and “pushed” protein crystallography synchrotrons to improve both the hardware and the software installed on beamline stations. For example, equipment for sample handling and data collection became more advanced, including the development of robotic equipment for sample mounting [89] and crystal alignment [90]. Microfocus beamlines were developed to permit effective data collection on very small crystals [91]. The technical advances spurred by SG and close proximity of advanced wet laboratories created a basis for interactive crystallography [92]. On many beamlines, data can be collected remotely; however, data quality may occasionally suffer on beamlines that do not allow for remote sample annealing [93,94].
SG projects have also improved technologies for high-throughput NMR data collection and analysis [95]. RSGI has built a robotized NMR pipeline, including both cell-based and cell-free protein expression systems and sample screening [96]. RSGI not only pioneered cell free expression systems but also conducted many workshop to popularize this technique. NESG has developed a protein NMR pipeline, utilizing efficient screening with micro-cryoprobes [97]. This has led to creation of a unique resource that provides experimental data for proteins whose structures have been solved by both NMR and X-ray crystallography [98]. The resource will likely assist further developments of NMR structure determination methods.
Function determination and function determination technologies
Recently, many SG centers pursued the development of methodologies to ascertain a protein’s function, and therefore established a number of computational and bioinformatics strategies to predict possible functions of proteins based on sequence-structure or sequence-function relationships [99,100]. The Fold and Function Assignment Server (FFAS) [101,102], which uses profile-profile algorithms for prediction of fold and function based on sequence, has been consistently ranked as one of the best fold recognition methods.
The mission of the third phase of PSI, i.e. PSI:Biology, was to determine the structures of proteins of biomedical importance and characterize their biological function. Over 100 PSI publications pertain to discovery of protein function (Fig. 3). To understand the structure-function relationships, new methods for functional analysis of proteins with unknown function or methods for analysis of unknown substrates for a protein with determined function have been developed [103,104]. In parallel, tools for protein function prediction via bioinformatics and ligand docking were created to leverage the continuously growing number of 3D structures [105].
The EFI has been pursuing a systematic program of “discovery of in vitro enzymatic and in vivo metabolic and physiological functions of unknown enzymes.” It has focused on large, functionally diverse enzyme superfamilies (amidohydrolases, enolases, glutathione transferases, haloacid dehalogenases and isoprenoid sythethases type I). These efforts contributed to the development of the Structure Function Linkage Database (SFLD) [106], which classifies enzymes by relating chemical/functional features to specific characteristics of sequence and/or structure. Activities of the EFI have resulted in over 60 publications [49].
Conclusions
By providing new methodologies, technologies, training, and knowledge that make determination of difficult and novel structures possible, SG has had a significant impact on structural biology and biomedical sciences. The pipelines for structure determination via X-ray crystallography and NMR has been are a unique resource for the biomedical community worldwide. Before its first quindecennial, SG centers determined more than 13,500 structures. Some members of the crystallographic community may suspect that such productivity is only possible for “low hanging fruit.” In reality, many of these structures were extremely difficult and required the development of advanced molecular biology and structure determination techniques. For that reason, the expertise of the people that built the PSI pipeline has been of benefit to many scientists worldwide for determination of ultra-difficult, biologically important structures. This collected expertise in structure determination as well as other areas of biomedical research (protein production, SAXS, NMR, ligand screening, etc.) has resulted in collaborations with hundreds of laboratories around the world. On the other hand, the initial vision of SG, namely the ability to determine an arbitrary protein structures in an automatic way, is still on the horizon. In particular, coverage of many important pathogens and of the human microbiome is still poor.
At the moment, in light of the extinction of the PSI centers, structural genomics efforts are reduced. It is, however, too early to write an epitaph for SG. Some SG programs continue, most notably the Structural Genomics Consortium and the two programs funded by NIAID. These programs also provide structure determination services to biomedical community. Our observations show that the SG approach is somewhat continued by many individual laboratories, but without taking advantage the economy of the scale, and for obvious reasons, in slightly disorganized fashion. Smaller labs usually cannot afford expensive robotic workstations and rarely can maintain complex pipelines. Unfortunately, some very important projects have been significantly hindered by the end of PSI. We mention only two below—protein production facilities and the development of sophisticated, comprehensive databases that track protein production and related information.
Several centers have developed sophisticated protein production “factories” that could potentially serve as resources for the whole biomedical community [78,107]. In particular, the New York Structural Genomics Research Consortium (NYSGRC) has developed a system for large-scale production of proteins from eukaryotic and anaerobic organisms [108,109]. All details of protein production are harvested into a sophisticated database that is integrated with structural and functional databases into one universal system. The integration of many processes through data management and databases is a key factor for reproducibility of scientific results. Data management in a large modern laboratory has become paramount for coordinating and tracking the vast amount of data generated across multiple experiments, timeframes and centers—not to mention the potential for data mining to extract even more useful and interesting information. Successful data management requires a system with a well-planned, cohesive, and flexible framework. How to best achieve this coordination and level of detail is currently being addressed in different ways, but the measure of success comes back to “data in, information out.” Clearly, gathering details on millions of experiments on thousands of proteins and making them publically available for analysis—even after the projects themselves have ended—may turn out be one of the most important benefits of SG.
The closure of the PSI may result not only in the abandonment of most of the data collected during the 15 years of the initiative, but it may also affect the development of data management tools. The most sophisticated databases developed for PSI have one very uncommon “feature”: the data are harvested automatically and therefore cannot be “censored” by experimenters. We define “censorship” in this case as an omission of unsuccessful experiments, mainly because the researcher did not see the value of negative results. The inclusion of negative results greatly improves the conversion of data into biomedically useful information.
Converting data into information is a general difficulty of modern science. Researchers are swamped in experimental data and extraction of useful information is quite often a Sisyphean task. Addressing this task effectively requires either very substantial manual labor or the implementation of “knowledge-based systems,” with comprehensive tools for efficiently summarizing and mining experimental data, and in some cases, implementation of machine learning methods. Processing structural information, particularly when combined with functional, experimental, and sequential data, while maintaining the context of interaction networks with other bio-macromolecules or bioactive chemical compounds, increasingly requires the use of Big Data paradigms for effective data management, as well as for checking data integrity and accuracy. The absence of documentation regarding which protein preparations were used in functional studies may partly be responsible for reports that a significant number of biomedical experiments cannot be reproduced in other laboratories, even though the changes between protocols seem at first glance to be insignificant [110]. According to a recent study, the annual cost of irreproducibility in life science amounts to a staggering sum of $28 billion for the US alone [111]. By emphasizing and enabling comprehensive data management, SG programs have been directly and indirectly promoting improvement in the reproducibility of research results in structural biology and in biomedical sciences in general. In our opinion, it is likely that the indirect economic benefits of SG stemming only from reducing the irreproducibility in biomedical research have already surpassed the $2 billion investment.
Acknowledgments
We would like to thank Steve Burley, Andrzej Joachimiak, Maks Chruszcz, Gaetano Montelione, Wayne Anderson, Steve Almo, Torsten Schwede, Alex Wlodawer, Zbyszek Dauter and David Cooper for reading the manuscripts and helpful discussion. The authors’ research was supported with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN272201200026C, by NIH grants HG008424, GM093342, GM093324, GM094585 and GM094662.
References
- 1.Terwilliger TC, Waldo G, Peat TS, Newman JM, Chu K, Berendzen J. Class-directed structure determination: foundation for a protein structure initiative. Protein Sci. 1998;7(9):1851–1856. doi: 10.1002/pro.5560070901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vitkup D, Melamud E, Moult J, Sander C. Completeness in structural genomics. Nat Struct Biol. 2001;8(6):559–566. doi: 10.1038/8864088640. pii. [DOI] [PubMed] [Google Scholar]
- 3.Grabowski M, Joachimiak A, Otwinowski Z, Minor W. Structural genomics: keeping up with expanding knowledge of the protein universe. Curr Opin Struct Biol. 2007;17(3):347–353. doi: 10.1016/j.sbi.2007.06.003. S0959-440X(07)00079-6 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levitt M. Nature of the protein universe. Proc Natl Acad Sci U S A. 2009;106(27):11079–11084. doi: 10.1073/pnas.0905029106. 0905029106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.NIGMS. [Accessed November 4, 2015];PSI Pilot Phase Fact Sheet. 2001 https://www.nigms.nih.gov/Research/specificareas/PSI/background/Pages/PilotFacts.aspx.
- 6.NIGMS. [Accessed November 4, 2015];PSI Production Phase Fact Sheet. 2006 http://www.nigms.nih.gov/Research/SpecificAreas/PSI/Background/Pages/PSI2FactSheet.aspx.
- 7.NIGMS. [Accessed November 4, 2015];NIH Grants Will Advance Studies of the Form and Function of Proteins. 2010 http://www.nigms.nih.gov/News/results/Pages/20100930.aspx.
- 8.Yokoyama S, Terwilliger TC, Kuramitsu S, Moras D, Sussman JL. RIKEN aids international structural genomics efforts. Nature. 2007;445(7123):21. doi: 10.1038/445021a. [DOI] [PubMed] [Google Scholar]
- 9.Cassman M, World Technology Evaluation Center. Systems biology : international research and development. Springer; Dordrecht, The Netherlands: 2007. [Google Scholar]
- 10.Tanaka Akiko, Hirai Akimitsu, Harai Daisuke, Nakayama Keitaro, Fujii Atsuko, Yokoyama Shigeyuki. [Accessed November 4, 2015];Intellectual Property Rights Management for Structural Genomics Research. http://www.protein.gsc.riken.jp/Concept/Partnership/partner_eng.htm.
- 11. [Accessed November 4, 2015];SGC Mission and Philosophy. http://www.thesgc.org/about/what_is_the_sgc.
- 12.SGC. [Accessed November 4, 2015];2011 Press release http://www.thesgc.org/sites/default/files/SGC_PhaseIII_PR_FINAL_%20for_release_110928_v110926.pdf.
- 13.O’Connel M. Toxin, Reveal Thyself!-Clues to Deadliest Disease Being Unlocked. Ward Rounds. 2013 http://www.wardrounds.northwestern.edu/summer-fall-2013/features/clues-to-deadliest-diseases/
- 14.Gerlt JA, Allen KN, Almo SC, Armstrong RN, Babbitt PC, Cronan JE, Dunaway-Mariano D, Imker HJ, Jacobson MP, Minor W, Poulter CD, Raushel FM, Sali A, Shoichet BK, Sweedler JV. The Enzyme Function Initiative. Biochemistry. 2011;50(46):9950–9962. doi: 10.1021/bi201312u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.wwPDB. [Accessed August 20, 2015];PDB Exchange Dictionary. 2015 http://mmcif.wwpdb.org/dictionaries/mmcif_pdbx.dic/Items/_pdbx_SG_project.initial_of_center.html.
- 16.Albeck S, Alzari P, Andreini C, Banci L, Berry IM, Bertini I, Cambillau C, Canard B, Carter L, Cohen SX, Diprose JM, Dym O, Esnouf RM, Felder C, Ferron F, Guillemot F, Hamer R, Ben Jelloul M, Laskowski RA, Laurent T, Longhi S, Lopez R, Luchinat C, Malet H, Mochel T, Morris RJ, Moulinier L, Oinn T, Pajon A, Peleg Y, Perrakis A, Poch O, Prilusky J, Rachedi A, Ripp R, Rosato A, Silman I, Stuart DI, Sussman JL, Thierry JC, Thompson JD, Thornton JM, Unger T, Vaughan B, Vranken W, Watson JD, Whamond G, Henrick K. SPINE bioinformatics and data-management aspects of high-throughput structural biology. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 10):1184–1195. doi: 10.1107/S090744490602991X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banci L, Bertini I, Cusack S, de Jong RN, Heinemann U, Jones EY, Kozielski F, Maskos K, Messerschmidt A, Owens R, Perrakis A, Poterszman A, Schneider G, Siebold C, Silman I, Sixma T, Stewart-Jones G, Sussman JL, Thierry JC, Moras D. First steps towards effective methods in exploiting high-throughput technologies for the determination of human protein structures of high biomedical value. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 10):1208–1217. doi: 10.1107/S0907444906029350. [DOI] [PubMed] [Google Scholar]
- 18.Chim N, Habel JE, Johnston JM, Krieger I, Miallau L, Sankaranarayanan R, Morse RP, Bruning J, Swanson S, Kim H, Kim CY, Li H, Bulloch EM, Payne RJ, Manos-Turvey A, Hung LW, Baker EN, Lott JS, James MN, Terwilliger TC, Eisenberg DS, Sacchettini JC, Goulding CW. The TB Structural Genomics Consortium: a decade of progress. Tuberculosis (Edinb) 2011;91(2):155–172. doi: 10.1016/j.tube.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Musa TL, Ioerger TR, Sacchettini JC. The tuberculosis structural genomics consortium: a structural genomics approach to drug discovery. Adv Protein Chem Struct Biol. 2009;77:41–76. doi: 10.1016/S1876-1623(09)77003-8. [DOI] [PubMed] [Google Scholar]
- 20.Cyranoski D. ‘Big science’ protein project under fire. Nature. 2006;443(7110):382. doi: 10.1038/443382a. [DOI] [PubMed] [Google Scholar]
- 21.Petsko GA. An idea whose time has gone. Genome Biol. 2007;8(6):107. doi: 10.1186/gb-2007-8-6-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banci L, Baumeister W, Heinemann U, Schneider G, Silman I, Stuart DI, Sussman JL. An idea whose time has come. Genome Biol. 2007;8(11):408. doi: 10.1186/gb-2007-8-11-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lane E, Ham B. Science policy. The payoff of federal R&D: iPod, Google, and Human Genome Project. Science. 2012;336(6080):433. [PubMed] [Google Scholar]
- 24.Tripp S, Grueber M. [Accessed November 4, 2015];Economic Impact of the Human Genome Project. 2011 http://battelle.org/docs/default-document-library/economic_impact_of_the_human_genome_project.pdf.
- 25.Liu J, Montelione GT, Rost B. Novel leverage of structural genomics. Nat Biotechnol. 2007;25(8):849–851. doi: 10.1038/nbt0807-849. [DOI] [PubMed] [Google Scholar]
- 26.Dessailly BH, Nair R, Jaroszewski L, Fajardo JE, Kouranov A, Lee D, Fiser A, Godzik A, Rost B, Orengo C. PSI-2: structural genomics to cover protein domain family space. Structure. 2009;17(6):869–881. doi: 10.1016/j.str.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Donovan C, Martin MJ, Gattiker A, Gasteiger E, Bairoch A, Apweiler R. High-quality protein knowledge resource: SWISS-PROT and TrEMBL. Brief Bioinform. 2002;3(3):275–284. doi: 10.1093/bib/3.3.275. [DOI] [PubMed] [Google Scholar]
- 28.Uniprot [Accessed November 4, 2015];Current Release Statistics. 2015 http://www.ebi.ac.uk/uniprot/TrEMBLstats.
- 29.Lee D, Grant A, Marsden RL, Orengo C. Identification and distribution of protein families in 120 completed genomes using Gene3D. Proteins. 2005;59(3):603–615. doi: 10.1002/prot.20409. [DOI] [PubMed] [Google Scholar]
- 30.Unger R, Uliel S, Havlin S. Scaling law in sizes of protein sequence families: from superfamilies to orphan genes. Proteins. 2003;51(4):569–576. doi: 10.1002/prot.10347. [DOI] [PubMed] [Google Scholar]
- 31.Nair R, Liu J, Soong TT, Acton TB, Everett JK, Kouranov A, Fiser A, Godzik A, Jaroszewski L, Orengo C, Montelione GT, Rost B. Structural genomics is the largest contributor of novel structural leverage. J Struct Funct Genomics. 2009;10(2):181–191. doi: 10.1007/s10969-008-9055-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khafizov K, Ivanov MV, Glazova OV, Kovalenko SP. Computational approaches to study the effects of small genomic variations. Journal of molecular modeling. 2015;21(10):2794. doi: 10.1007/s00894-015-2794-y. [DOI] [PubMed] [Google Scholar]
- 33.Khafizov K, Madrid-Aliste C, Almo SC, Fiser A. Trends in structural coverage of the protein universe and the impact of the Protein Structure Initiative. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(10):3733–3738. doi: 10.1073/pnas.1321614111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pieper U, Webb BM, Dong GQ, Schneidman-Duhovny D, Fan H, Kim SJ, Khuri N, Spill YG, Weinkam P, Hammel M, Tainer JA, Nilges M, Sali A. ModBase, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2014;42(Database issue):D336–346. doi: 10.1093/nar/gkt1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnold K, Kiefer F, Kopp J, Battey JN, Podvinec M, Westbrook JD, Berman HM, Bordoli L, Schwede T. The Protein Model Portal. Journal of structural and functional genomics. 2009;10(1):1–8. doi: 10.1007/s10969-008-9048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moult J. A decade of CASP: progress, bottlenecks and prognosis in protein structure prediction. Curr Opin Struct Biol. 2005 doi: 10.1016/j.sbi.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 37.Lesley SA, Kuhn P, Godzik A, Deacon AM, Mathews I, Kreusch A, Spraggon G, Klock HE, McMullan D, Shin T, Vincent J, Robb A, Brinen LS, Miller MD, McPhillips TM, Miller MA, Scheibe D, Canaves JM, Guda C, Jaroszewski L, Selby TL, Elsliger MA, Wooley J, Taylor SS, Hodgson KO, Wilson IA, Schultz PG, Stevens RC. Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline. Proc Natl Acad Sci U S A. 2002;99(18):11664–11669. doi: 10.1073/pnas.142413399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Thiele I, Weekes D, Li Z, Jaroszewski L, Ginalski K, Deacon AM, Wooley J, Lesley SA, Wilson IA, Palsson B, Osterman A, Godzik A. Three-dimensional structural view of the central metabolic network of Thermotoga maritima. Science. 2009;325(5947):1544–1549. doi: 10.1126/science.1174671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Omenn GS, Lane L, Lundberg EK, Beavis RC, Nesvizhskii AI, Deutsch EW. Metrics for the Human Proteome Project 2015: Progress on the Human Proteome and Guidelines for High-Confidence Protein Identification. J Proteome Res. 2015;14(9):3452–3460. doi: 10.1021/acs.jproteome.5b00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaudet P, Argoud-Puy G, Cusin I, Duek P, Evalet O, Gateau A, Gleizes A, Pereira M, Zahn-Zabal M, Zwahlen C, Bairoch A, Lane L. neXtProt: organizing protein knowledge in the context of human proteome projects. J Proteome Res. 2013;12(1):293–298. doi: 10.1021/pr300830v. [DOI] [PubMed] [Google Scholar]
- 41.Mizianty MJ, Fan X, Yan J, Chalmers E, Woloschuk C, Joachimiak A, Kurgan L. Covering complete proteomes with X-ray structures: a current snapshot. Acta Crystallogr D Biol Crystallogr. 2014;70(Pt 11):2781–2793. doi: 10.1107/S1399004714019427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7(4):1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kloppmann E, Punta M, Rost B. Structural genomics plucks high-hanging membrane proteins. Curr Opin Struct Biol. 2012;22(3):326–332. doi: 10.1016/j.sbi.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. [Accessed August 24, 2015];PSI Publication Portal. 2015 http://olenka.med.virginia.edu/psi.
- 45.RIKEN Structural/Genomics Proteomics Initiative. Publications. [Accessed August 24 2015];2015 http://www.rsgi.riken.jp/rsgi_e/ResearchResult/index.html.
- 46.SGC. [Accessed August 24, 2015];2015 Publications. www.thesgc.org/publications.
- 47.CSGID Publications. [Accessed August 24 2015];2015 http://csgid.org/publications.
- 48.SSGCID Publications. [Accessed August 24 2015];2015 http://www.ssgcid.org/publications.
- 49.Enzyme Function Initiative Publications. [Accessed November 20, 2015];2015 http://enzymefunction.org/publications.
- 50.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordonez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312(5779):1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 8):859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 52.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318(5854):1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318(5854):1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 54.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gabanyi MJ, Adams PD, Arnold K, Bordoli L, Carter LG, Flippen-Andersen J, Gifford L, Haas J, Kouranov A, McLaughlin WA, Micallef DI, Minor W, Shah R, Schwede T, Tao YP, Westbrook JD, Zimmerman M, Berman HM. The Structural Biology Knowledgebase: a portal to protein structures, sequences, functions, and methods. J Struct Funct Genomics. 2011;12(2):45–54. doi: 10.1007/s10969-011-9106-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Redner S. How popular is your paper? An empirical study of the citation distribution. Eur Phys J B. 1998;4(2):131–134. doi: 10.1007/s100510050359. [DOI] [Google Scholar]
- 57.Albarrán P, Crespo J, Ortuño I, Ruiz-Castillo J. The skewness of science in 219 sub-fields and a number of aggregates. Scientometrics. 2011;88(2):385–397. doi: 10.1007/s11192-011-0407-9. [DOI] [Google Scholar]
- 58.Brzezinski M. Power laws in citation distributions: evidence from Scopus. Scientometrics. 2015;103(1):213–228. doi: 10.1007/s11192-014-1524-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peterson GJ, Pressé S, Dill KA. Nonuniversal power law scaling in the probability distribution of scientific citations. Proceedings of the National Academy of Sciences. 2010;107(37):16023–16027. doi: 10.1073/pnas.1010757107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clauset A, Shalizi C, Newman M. Power-Law Distributions in Empirical Data. SIAM Review. 2009;51(4):661–703. doi: 10.1137/070710111. [DOI] [Google Scholar]
- 61.Chen L, Oughtred R, Berman HM, Westbrook J. TargetDB: a target registration database for structural genomics projects. Bioinformatics. 2004;20(16):2860–2862. doi: 10.1093/bioinformatics/bth300. bth300 [pii] [DOI] [PubMed] [Google Scholar]
- 62.Morris C. PiMS: a data management system for structural proteomics. Methods Mol Biol. 2015;1261:21–34. doi: 10.1007/978-1-4939-2230-7_2. [DOI] [PubMed] [Google Scholar]
- 63.Morris C, Pajon A, Griffiths SL, Daniel E, Savitsky M, Lin B, Diprose JM, da Silva AW, Pilicheva K, Troshin P, van Niekerk J, Isaacs N, Naismith J, Nave C, Blake R, Wilson KS, Stuart DI, Henrick K, Esnouf RM. The Protein Information Management System (PiMS): a generic tool for any structural biology research laboratory. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):249–260. doi: 10.1107/S0907444911007943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zolnai Z, Lee PT, Li J, Chapman MR, Newman CS, Phillips GN, Jr, Rayment I, Ulrich EL, Volkman BF, Markley JL. Project management system for structural and functional proteomics: Sesame. J Struct Funct Genomics. 2003;4(1):11–23. doi: 10.1023/a:1024684404761. [DOI] [PubMed] [Google Scholar]
- 65.Berman HM. The Protein Data Bank: a historical perspective. Acta Crystallogr A. 2008;64(Pt 1):88–95. doi: 10.1107/S0108767307035623. [DOI] [PubMed] [Google Scholar]
- 66.Weekes D, Krishna SS, Bakolitsa C, Wilson IA, Godzik A, Wooley J. TOPSAN: a collaborative annotation environment for structural genomics. BMC bioinformatics. 2010;11:426. doi: 10.1186/1471-2105-11-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prilusky J, Hodis E, Canner D, Decatur WA, Oberholser K, Martz E, Berchanski A, Harel M, Sussman JL. Proteopedia: a status report on the collaborative, 3D web-encyclopedia of proteins and other biomolecules. J Struct Biol. 2011;175(2):244–252. doi: 10.1016/j.jsb.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 68.Zimmerman MD, Grabowski M, Domagalski MJ, Maclean EM, Chruszcz M, Minor W. Data management in the modern structural biology and biomedical research environment. Methods Mol Biol. 2014;1140:1–25. doi: 10.1007/978-1-4939-0354-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gifford LK, Carter LG, Gabanyi MJ, Berman HM, Adams PD. The Protein Structure Initiative Structural Biology Knowledgebase Technology Portal: a structural biology web resource. J Struct Funct Genomics. 2012;13(2):57–62. doi: 10.1007/s10969-012-9133-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kobayashi N, Harano Y, Tochio N, Nakatani E, Kigawa T, Yokoyama S, Mading S, Ulrich EL, Markley JL, Akutsu H, Fujiwara T. An automated system designed for large scale NMR data deposition and annotation: application to over 600 assigned chemical shift data entries to the BioMagResBank from the Riken Structural Genomics/Proteomics Initiative internal database. J Biomol NMR. 2012;53(4):311–320. doi: 10.1007/s10858-012-9641-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Kent Wenger R, Yao H, Markley JL. BioMagResBank. Nucleic Acids Res. 2008;36(Database issue):D402–408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seiler CY, Park JG, Sharma A, Hunter P, Surapaneni P, Sedillo C, Field J, Algar R, Price A, Steel J, Throop A, Fiacco M, LaBaer J. DNASU plasmid and PSI:Biology-Materials repositories: resources to accelerate biological research. Nucleic acids research. 2014;42(Database issue):D1253–1260. doi: 10.1093/nar/gkt1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.SSGCID Available Materials. [Accessed November 23, 2015];2015 http://www.ssgcid.org/available-materials.
- 74.Brown PJ, Muller S. Open access chemical probes for epigenetic targets. Future Med Chem. 2015;7(14):1901–1917. doi: 10.4155/fmc.15.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Collins FS, Tabak LA. Policy: NIH plans to enhance reproducibility. Nature. 2014;505(7485):612–613. doi: 10.1038/505612a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Niedzialkowska E, Gasiorowska O, Handing KB, Majorek KA, Porebski PJ, Shabalin IG, Zasadzinska E, Cymborowski M, Minor W. Protein purification and crystallization artifacts: The tale usually not told. Protein Sci. 2015 doi: 10.1002/pro.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Eschenfeldt WH, Lucy S, Millard CS, Joachimiak A, Mark ID. A family of LIC vectors for high-throughput cloning and purification of proteins. Methods Mol Biol. 2009;498:105–115. doi: 10.1007/978-1-59745-196-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Almo SC, Garforth SJ, Hillerich BS, Love JD, Seidel RD, Burley SK. Protein production from the structural genomics perspective: achievements and future needs. Curr Opin Struct Biol. 2013;23(3):335–344. doi: 10.1016/j.sbi.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ericsson UB, Hallberg BM, Detitta GT, Dekker N, Nordlund P. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Analytical biochemistry. 2006;357(2):289–298. doi: 10.1016/j.ab.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 80.Newman J, Egan D, Walter TS, Meged R, Berry I, Ben Jelloul M, Sussman JL, Stuart DI, Perrakis A. Towards rationalization of crystallization screening for small- to medium-sized academic laboratories: the PACT/JCSG+ strategy. D - 9305878. 2005;61(Pt 10):1426–1431. doi: 10.1107/S0907444905024984. [DOI] [PubMed] [Google Scholar]
- 81.MCSG Suite. [Accessed November 4, 2015];2013 http://www.microlytic.com/content/mcsg-suite.
- 82.Sagemark J, Kraulis P, Weigelt J. A software tool to accelerate design of protein constructs for recombinant expression. Protein expression and purification. 2010;72(2):175–178. doi: 10.1016/j.pep.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 83.Przulj N, Wigle DA, Jurisica I. Functional topology in a network of protein interactions. Bioinformatics. 2004;20(3):340–348. doi: 10.1093/bioinformatics/btg415. [DOI] [PubMed] [Google Scholar]
- 84.NIGMS. [Accessed November 4, 2015];Report of the Protein Structure Initiative Assessment Panel. 2007 https://www.nigms.nih.gov/News/reports/archivedreports2009-2007/Pages/PSIAssessmentPanel2007.aspx.
- 85.NIGMS. [Accessed November 4, 2015];Recommendations for Continued Investment in Structural Biology Following the Sunsetting of the Protein Structure Initiative. 2014 https://www.nigms.nih.gov/News/reports/Documents/NIGMS-FSBC-report2014.pdf.
- 86.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. D - 9305878. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. D - 9305878. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chruszcz M, Domagalski M, Osinski T, Wlodawer A, Minor W. Unmet challenges of structural genomics. Curr Opin Struct Biol. 2010;20(5):587–597. doi: 10.1016/j.sbi.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Snell G, Cork C, Nordmeyer R, Cornell E, Meigs G, Yegian D, Jaklevic J, Jin J, Stevens RC, Earnest T. Automated sample mounting and alignment system for biological crystallography at a synchrotron source. Structure. 2004;12(4):537–545. doi: 10.1016/j.str.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 90.Miller MD, Deacon AM. An X-ray microsource based system for crystal screening and beamline development during synchrotron shutdown periods. Nuclear instruments & methods in physics research Section A, Accelerators, spectrometers, detectors and associated equipment. 2007;582(1):233–235. doi: 10.1016/j.nima.2007.08.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cherezov V, Hanson MA, Griffith MT, Hilgart MC, Sanishvili R, Nagarajan V, Stepanov S, Fischetti RF, Kuhn P, Stevens RC. Rastering strategy for screening and centring of microcrystal samples of human membrane proteins with a sub-10 microm size X-ray synchrotron beam. Journal of the Royal Society, Interface / the Royal Society. 2009;6(Suppl 5):S587–597. doi: 10.1098/rsif.2009.0142.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Advanced Protein Characterization Facility. 2015 http://www.anl.gov/apcf/advanced-protein-characterization-facility.
- 93.Heras B, Martin JL. Post-crystallization treatments for improving diffraction quality of protein crystals. Acta Crystallogr D Biol Crystallogr. 2005;61(Pt 9):1173–1180. doi: 10.1107/S0907444905019451. [DOI] [PubMed] [Google Scholar]
- 94.Krojer T, Pike AC, von Delft F. Squeezing the most from every crystal: the fine details of data collection. Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 7):1303–1313. doi: 10.1107/S0907444913013280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu G, Shen Y, Atreya HS, Parish D, Shao Y, Sukumaran DK, Xiao R, Yee A, Lemak A, Bhattacharya A, Acton TA, Arrowsmith CH, Montelione GT, Szyperski T. NMR data collection and analysis protocol for high-throughput protein structure determination. Proc Natl Acad Sci U S A. 2005;102(30):10487–10492. doi: 10.1073/pnas.0504338102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yokoyama S. Protein expression systems for structural genomics and proteomics. Curr Opin Chem Biol. 2003;7(1):39–43. doi: 10.1016/s1367-5931(02)00019-4. [DOI] [PubMed] [Google Scholar]
- 97.Rossi P, Swapna GV, Huang YJ, Aramini JM, Anklin C, Conover K, Hamilton K, Xiao R, Acton TB, Ertekin A, Everett JK, Montelione GT. A microscale protein NMR sample screening pipeline. J Biomol NMR. 2010;46(1):11–22. doi: 10.1007/s10858-009-9386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Everett JK, Tejero R, Murthy SB, Acton TB, Aramini JM, Baran MC, Benach J, Cort JR, Eletsky A, Forouhar F, Guan R, Kuzin AP, Lee HW, Liu G, Mani R, Mao B, Mills JL, Montelione AF, Pederson K, Powers R, Ramelot T, Rossi P, Seetharaman J, Snyder D, Swapna GV, Vorobiev SM, Wu Y, Xiao R, Yang Y, Arrowsmith CH, Hunt JF, Kennedy MA, Prestegard JH, Szyperski T, Tong L, Montelione GT. A community resource of experimental data for NMR / X-ray crystal structure pairs. Protein Sci. 2015 doi: 10.1002/pro.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Laskowski RA, Watson JD, Thornton JM. ProFunc: a server for predicting protein function from 3D structure. Nucleic acids research. 2005;33(Web Server issue):W89–93. doi: 10.1093/nar/gki414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Watson JD, Laskowski RA, Thornton JM. Predicting protein function from sequence and structural data. Current opinion in structural biology. 2005;15(3):275–284. doi: 10.1016/j.sbi.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 101.Jaroszewski L, Rychlewski L, Li Z, Li W, Godzik A. FFAS03: a server for profile--profile sequence alignments. Nucleic Acids Res. 2005;33(Web Server issue):W284–288. doi: 10.1093/nar/gki418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jaroszewski L, Li Z, Cai XH, Weber C, Godzik A. FFAS server: novel features and applications. Nucleic Acids Res. 2011;39(Web Server issue):W38–44. doi: 10.1093/nar/gkr441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shumilin IA, Cymborowski M, Chertihin O, Jha KN, Herr JC, Lesley SA, Joachimiak A, Minor W. Identification of unknown protein function using metabolite cocktail screening. Structure. 2012;20(10):1715–1725. doi: 10.1016/j.str.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kuhn ML, Majorek KA, Minor W, Anderson WF. Broad-substrate screen as a tool to identify substrates for bacterial Gcn5-related N-acetyltransferases with unknown substrate specificity. Protein science : a publication of the Protein Society. 2013;22(2):222–230. doi: 10.1002/pro.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Watson JD, Sanderson S, Ezersky A, Savchenko A, Edwards A, Orengo C, Joachimiak A, Laskowski RA, Thornton JM. Towards fully automated structure-based function prediction in structural genomics: a case study. J Mol Biol. 2007;367(5):1511–1522. doi: 10.1016/j.jmb.2007.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Akiva E, Brown S, Almonacid DE, Barber AE, 2nd, Custer AF, Hicks MA, Huang CC, Lauck F, Mashiyama ST, Meng EC, Mischel D, Morris JH, Ojha S, Schnoes AM, Stryke D, Yunes JM, Ferrin TE, Holliday GL, Babbitt PC. The Structure-Function Linkage Database. Nucleic Acids Res. 2014;42(Database issue):D521–530. doi: 10.1093/nar/gkt1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Structural Genomics C, China Structural Genomics C, Northeast Structural Genomics C. Graslund S, Nordlund P, Weigelt J, Hallberg BM, Bray J, Gileadi O, Knapp S, Oppermann U, Arrowsmith C, Hui R, Ming J, dhe-Paganon S, Park HW, Savchenko A, Yee A, Edwards A, Vincentelli R, Cambillau C, Kim R, Kim SH, Rao Z, Shi Y, Terwilliger TC, Kim CY, Hung LW, Waldo GS, Peleg Y, Albeck S, Unger T, Dym O, Prilusky J, Sussman JL, Stevens RC, Lesley SA, Wilson IA, Joachimiak A, Collart F, Dementieva I, Donnelly MI, Eschenfeldt WH, Kim Y, Stols L, Wu R, Zhou M, Burley SK, Emtage JS, Sauder JM, Thompson D, Bain K, Luz J, Gheyi T, Zhang F, Atwell S, Almo SC, Bonanno JB, Fiser A, Swaminathan S, Studier FW, Chance MR, Sali A, Acton TB, Xiao R, Zhao L, Ma LC, Hunt JF, Tong L, Cunningham K, Inouye M, Anderson S, Janjua H, Shastry R, Ho CK, Wang D, Wang H, Jiang M, Montelione GT, Stuart DI, Owens RJ, Daenke S, Schutz A, Heinemann U, Yokoyama S, Bussow K, Gunsalus KC. Protein production and purification. Nat Methods. 2008;5(2):135–146. doi: 10.1038/nmeth.f.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bandaranayake AD, Almo SC. Recent advances in mammalian protein production. FEBS Lett. 2014;588(2):253–260. doi: 10.1016/j.febslet.2013.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Almo SC, Love JD. Better and faster: improvements and optimization for mammalian recombinant protein production. Curr Opin Struct Biol. 2014;26:39–43. doi: 10.1016/j.sbi.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Glasziou P, Meats E, Heneghan C, Shepperd S. What is missing from descriptions of treatment in trials and reviews? BMJ. 2008;336(7659):1472–1474. doi: 10.1136/bmj.39590.732037.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Freedman LP, Cockburn IM, Simcoe TS. The Economics of Reproducibility in Preclinical Research. PLoS Biol. 2015;13(6):e1002165. doi: 10.1371/journal.pbio.1002165. [DOI] [PMC free article] [PubMed] [Google Scholar]