Abstract
Extracellular vesicles (EV) are membranous particles (30–1,000 nm in diameter) secreted by cells. Important biological functions have been attributed to 2 subsets of EV, the exosomes (bud from endosomal membranes) and the microvesicles (MV; bud from plasma membranes). Since both types of particles contain surface proteins derived from their cell of origin, their detection in blood may enable diagnosis and prognosis of disease. We have used an antibody microarray (DotScan) to compare the surface protein profiles of live cancer cells with those of their EV, based on their binding patterns to immobilized antibodies. Initially, EV derived from the cancer cell lines, LIM1215 (colorectal cancer) and MEC1 (B-cell chronic lymphocytic leukaemia; CLL), were used for assay optimization. Biotinylated antibodies specific for EpCAM (CD326) and CD19, respectively, were used to detect captured particles by enhanced chemiluminescence. Subsequently, this approach was used to profile CD19+ EV from the plasma of CLL patients. These EV expressed a subset (~40%) of the proteins detected on CLL cells from the same patients: moderate or high levels of CD5, CD19, CD31, CD44, CD55, CD62L, CD82, HLA-A,B,C, HLA-DR; low levels of CD21, CD49c, CD63. None of these proteins was detected on EV from the plasma of age- and gender-matched healthy individuals.
Keywords: exosomes, microvesicles, luminescence, chronic lymphocytic leukaemia, CD antigen
Extracellular vesicles (EV), comprising exosomes (30–100 nm; formed by inward budding of the endosomal membrane) and microvesicles (MV; 100–1,000 nm; outward budding from plasma membranes; otherwise known as “microparticles,” “ectosomes” or “shed vesicles/particles”), are released by cells and carry proteins, RNA, micro-RNA, and DNA fragments from their cells of origin to other parts of the body via blood and other body fluids (1–3). The biogenesis, purification, contents of proteins and nucleic acids, and functions of these particles have been reviewed (1, 4–9) along with known roles of tumour-derived EV in cancer progression, chemo-resistance, and immune escape (10–14).
Disease-specific EV from blood and other body fluids could provide molecular signatures that aid in diagnosis and prognosis. However, the detection and profiling of disease-specific EV from body fluids has been challenging (15). Due to their small size, EV are difficult to profile by flow cytometry, while the use of mass spectrometry requires purification of the EV subset of interest (e.g. cancer-derived EV) from soluble proteins, protein aggregates, and other EV subsets in plasma and other body fluids.
This article describes the use of an antibody microarray (DotScan) to determine surface protein profiles of EV recovered from the conditioned media of human cancer cell lines, and the application of this method to the detection and analysis of leukaemia-derived EV from the blood of patients with B-cell chronic lymphocytic leukaemia (CLL), the most common form of leukaemia in the Western world (16). We have used CLL as a model to study cancer-derived EV that accumulate in the blood (17). CLL is characterized by the progressive accumulation of mature, monoclonal CD19+/CD5+ B-cells in the peripheral blood, bone marrow, lymph nodes, and spleen (18,19).
DotScan consists of a panel of immobilized antibodies that recognize cluster of differentiation antigens on live cells or EV, which are captured by the corresponding surface antigens. The diagnostic capability of DotScan for the analysis of cells from human blood or bone marrow has been validated with a clinical trial involving 796 leukaemia patients and normal subjects, with a >95% correspondence between the diagnoses made using DotScan alone and diagnoses from the multiple criteria routinely used by pathology laboratories (20). DotScan has also been used to profile the surface proteins of live cells recovered from disaggregated colorectal and melanoma tumours (21–23).
In this study, we have used EV from several human cancer cell lines to optimize DotScan for profiling cancer-derived EV and applied this protocol to analyse human plasma-derived EV. Ultracentrifugation was used to isolate EV from patient plasma, after the removal of platelets and platelet-derived EV. Although the isolation of cell line-derived EV from conditioned medium included the centrifugation at 10,000×g for 20 min (4°C) to deplete MV, this step was omitted when EV were purified from plasma. CLL-derived EV captured on DotScan were profiled by detection with a biotinylated CD19 antibody, without separating exosomes from MV. The strict separation of exosomes and MV is difficult, as their size distributions can overlap significantly (24). Separation based on differential protein expression can also be problematic as different subsets of secreted EV may contain many common markers (25); also expression levels of exosome markers (CD9, CD63, or CD81) may vary or be undetectable (26,27). The relative contributions of exosomes and MV to cancer progression have not yet been defined; however, both subsets of EV can transfer biomolecules from cancer cells to recipient cells (13), and therefore, their combined protein profiles may be informative.
Materials and methods
Human cell lines
LIM1215 (colorectal cancer (28)) cells were obtained from Dr Briony Forbes (University of Adelaide, Adelaide, Australia); MEC1 (B-cell CLL (29)) cells were from the American Type Culture Collection (Manassas, VA, USA). The cells were cultured in a 5% CO2 incubator in RPMI 1640 medium, containing 100 U/ml penicillin/streptomycin (Life Technologies, Mulgrave, VIC, Australia) and 10% foetal calf serum (FCS; In Vitro Technologies, Noble Park North, VIC, Australia). Non-adherent MEC1 cells were harvested by centrifugation at room temperature (400×g, 5 min, 23°C), while adherent LIM1215 cells were collected after 5 min incubation at 37°C with 0.05% trypsin-EDTA (Life Technologies).
Antibodies and other reagents
Antibodies used to make microarrays, with their concentrations, are listed in Supplementary Table 1. Antibody hybridoma clones were selected for their ability to recognize extracellular epitopes of proteins on human cells. Most antibody solutions contained 0.1% (w/v) bovine serum albumin (BSA). However, for some antibodies, BSA reduced the sensitivity of detection, so was omitted as indicated (Supplementary Table 1). Isotype control antibodies (±BSA) were included in the microarray to detect non-specific binding. Biotinylated EpCAM (CD326) and CD19 antibodies (BioLegend, San Diego, CA, USA; catalogue numbers 324216 and 302203, respectively) were used to detect EV derived from LIM1215 and MEC1/CLL cells, respectively, captured on DotScan microarrays. Streptavidin poly-HRP and SuperSignal West Pico Chemiluminescent Substrate were purchased from Thermo Scientific (Rockford, IL, USA). BSA was from Sigma-Aldrich (Castle Hill, NSW, Australia).
Purification of EV from conditioned growth medium
LIM1215 and MEC1 cells were grown to late exponential phase in 175 cm2 flasks. LIM1215 cell monolayers were washed twice with phosphate-buffered saline (PBS) and incubated for 24 h at 37°C with 20 ml of growth medium containing 10% FCS depleted of EV by ultracentrifugation (100,000×g, 16 h, 4°C). This medium showed negative results for EV when tested with DotScan and did not interfere with isolation and analysis of isolated EV. Late exponential phase MEC1 cells were washed twice in PBS by centrifugation (400×g, 5 min, 23°C) and cultured overnight (16 h) in 20 ml of medium with 10% EV-depleted FCS. EV were purified from conditioned medium using a method based on previous protocols (30,31). Briefly, after removal of cells (300×g, 10 min, 23°C), supernatants were centrifuged (1,500×g, 10 min, 23°C, followed by 10,000×g, 20 min, 4°C). EV-containing medium was then concentrated ~4-fold by stirred-cell ultrafiltration on 100 kDa filters (Millipore, North Ryde, NSW, Australia), as previously described (32,33). EV were collected by ultracentrifugation (100,000×g, 16 h, 4°C). Medium supplemented with 10% EV-depleted FCS was used, rather than serum-free medium, to minimize cellular stress effects on EV protein composition (34). EV pellets were stored at −80°C and thawed quickly in a 37°C water bath before resuspension for DotScan analysis and NanoSight analysis.
Purification of cells and EV from blood
Blood samples (10 ml, generally non-fasting) were collected into anti-coagulant tubes (heparin, EDTA or citrate) from CLL patients with progressive disease (35) and normal donors, with local ethics approval and informed patient consent. Three of four CLL blood samples available for this study were collected into heparin and one into EDTA (Supplementary Table 2). Patients were not treated for at least 2 months before blood collection. Blood was processed within 2–4 h of collection. After centrifugation on Histopaque (Sigma-Aldrich, Castle Hill, NSW, Australia; 400×g, 30 min, 23°C, without brake), peripheral blood mononuclear cells (PBMC) and plasma were collected. A protease inhibitor cocktail (Roche, Castle Hill, NSW, Australia) was added to the plasma that was then centrifuged (3 times, 2,500×g, 20 min, 4°C) to deplete platelets (17) before being frozen at −80°C. Frozen clarified plasma (3.5–5 ml) was thawed quickly in a 37°C water bath and diluted to 17 ml with PBS containing 5 mM EDTA, regardless of the anti-coagulant used for collection of the blood sample, in polyallomer centrifuge tubes (16×102 mm; Beckman Coulter, Mount Waverley, VIC, Australia), and EV were pelleted (100,000×g, 16 h, 4°C). Pellets were re-suspended in 200 µl magnetic cell sorting (MACS) buffer (0.5% (w/v) BSA, 20 mM EDTA in PBS, pH 7.2), with thorough disaggregation of EV pellets. Platelet-derived EV were depleted using CD61 antibody-coated magnetic beads (Miltenyi Biotec, Macquarie Park, NSW, Australia). The beads were added to the EV suspension in the ratio of 18 µl beads per ml of original plasma and rotated (10 rpm, 1 h, 4°C), then passed through an LS column (Miltenyi Biotec, Macquarie Park, NSW, Australia) in a strong magnetic field using a QuadroMACS separator (Miltenyi Biotec). The columns were washed with 2 ml of MACS buffer, and the eluent centrifuged (100,000×g, 3.5 h, 4°C). CD61-depleted EV pellets were re-suspended to 300 µl with RPMI-1640 medium and analysed by NanoSight and DotScan without re-freezing. EV captured on the CD61 antibody-coated magnetic beads were also analysed by DotScan after elution from columns by removal of the magnetic field, to confirm that they expressed typical platelet markers (unpublished data).
Nanoparticle tracking analysis (NanoSight)
EV diameters and concentrations were measured over a period of 60 s at 25 frames/s using the NanoSight LM10-HS system with a tuned 405 nm laser (NanoSight Ltd, Amesbury, UK) and nanoparticle tracking analysis software (NTA version 2.3; Malvern Instruments, Malvern, UK). EV samples were diluted in PBS to enable the NanoSight NTA software to detect 108–109 particles/ml using video capture of Brownian movement via the standard CMOS CCD camera of the microscope (camera gain 300; shutter 14.99; frame rate 24.99), with the temperature of the laser unit set at 24°C. To calculate the number of EV used for DotScan, the measured EV concentrations (particles/ml) were adjusted for the dilution factor and the volume applied to the microarrays.
Preparation of DotScan antibody microarrays
Antibodies (10 nl; Supplementary Table 1) were applied to Oncyte nitrocellulose-coated slides (Grace Bio-Labs, Bend, OR, USA) using a PixSys 3200 Aspirate and Dispense System (BioDot, Irvine, CA, USA), as previously described (36). The first section of each microarray consisted of 81 of the original 82 antibodies of the DotScan leukaemia microarray (36). An additional 44 antibodies, against surface proteins on colorectal cancer (CRC) cells with prognostic potential, were added as a “satellite” microarray (24). A third section contained several dilutions of isotype control antibodies and a humanized therapeutic antibody, Mabthera (rituximab; Roche, Castle Hill, NSW, Australia), recognizing CD20. Antibodies were used at concentrations ranging from 50 to 1,000 µg protein/ml, as previously determined for optimal cell capture that correlated with analysis by flow cytometry (20,37). DotScan microarrays were blocked with 5% Diploma skim milk (Fonterra, Macquarie Park, NSW, Australia) in PBS, washed, dried and stored at 4°C with desiccant.
DotScan analysis of cells
Live human cells (3–5×106) were washed in PBS and re-suspended in 300 µl RPMI-1640 medium (no FCS). The cells were applied to DotScan microarrays that had been rehydrated in PBS and then placed in a humidified chamber. The capture of cells on antibody dots of the microarray requires live cells, where CD antigens “cap” to the interface between the cells and dots (38). Longer incubation times are required for large cells (e.g. tumour cell lines) than for small cells (e.g. leukaemia cells) to ensure capture. Microarrays were incubated with cells for 1 h at 37°C (LIM1215), 30 min at 23°C (MEC1) or 12–15 min at 23°C (PBMC from CLL patients). A longer incubation time was required for the capture of the larger LIM1215 cells than for MEC1 cells and CLL cells from patient blood. Unbound cells were gently washed off by immersion in PBS (10–20 s) and bound cells were fixed with 3.7% (w/v) formaldehyde in PBS (2 h, 23°C). Fixative was removed with 3 gentle washes in PBS (total 5 min). While still moist, cell-binding patterns were recorded by optical scanning, using a DotScan DotReader and data analysis software (Medsaic, Darlington, NSW, Australia) (36) that record digital images of cell-binding patterns on microarrays. Visible dots were considered to be positive, with the limit of detection ~100 cells. Dot intensities were quantified using ImageQuant (version 7; GE Healthcare Australia, Rydalmere, NSW, Australia). After background and isotype control subtraction and median centred normalization (39,40), duplicate results were averaged.
DotScan analysis of EV from cell culture medium
EV (108–1011 particles/200 µl FCS-free medium) were captured on DotScan by incubation at 4°C for 16 h, with gentle rocking. A border drawn with a hydrophobic pen (Vector Laboratories, Burlington, CA, USA) was used to restrict liquid samples to the microarray area. Unbound EV were washed off with a single vertical immersion in 20 ml PBS (20 s), and bound EV were fixed to the antibody microarray with 3.7% (w/v) formaldehyde/PBS (2 h, 23°C). Slides were washed by vertical immersion in 20 ml PBS (3 times, 2 min each) and blocked for 20 min at 23°C with 200 µl blocking buffer (1% w/v BSA in PBS) before addition of 200 µl of the following concentrations of biotinylated antibody in blocking buffer: 0.05 µg/ml for CD326 (EpCAM; clone 9C4); 2.5 µg/ml for CD19 (clone HIB19). After 60 min incubation at 23°C and 3×2 min washes in PBS, microarrays were incubated with 200 µl of streptavidin poly-HRP (1.25 ng/ml in blocking buffer) for 30 min at 23°C. After 4 washes, 300 µl of SuperSignal West Pico Chemiluminescent Substrate (prepared according to the manufacturer's instructions) was added. After 5 min at 23°C, excess reagent was drained off and slides were carefully overlaid with a piece of overhead projector plastic and exposed to Amersham Hyperfilm ECL (GE Healthcare, Buckinghamshire, UK) for 5–30 min, as required. A GS-900™ Calibrated Densitometer (Bio-Rad, Hercules, CA, USA) was used for densitometric scanning of ECL films. Visible dots were considered to be positive. Dot luminescence intensities were quantified using ImageQuant (as above). After background and isotype control subtraction and median-centred normalization, duplicate results were averaged.
DotScan analysis of EV from plasma
The protocol was similar to that for DotScan analysis of EV from cell culture medium (above), except EV were profiled in the presence of 2% heat-inactivated human AB serum (Sigma-Aldrich, Castle Hill, NSW, Australia) to block the non-specific Fc receptor binding (41) that was occasionally seen with plasma-derived EV. No EV profiles were detectable in 2% heat-inactivated human serum alone.
Results
Profiling of cancer cell lines and their EV
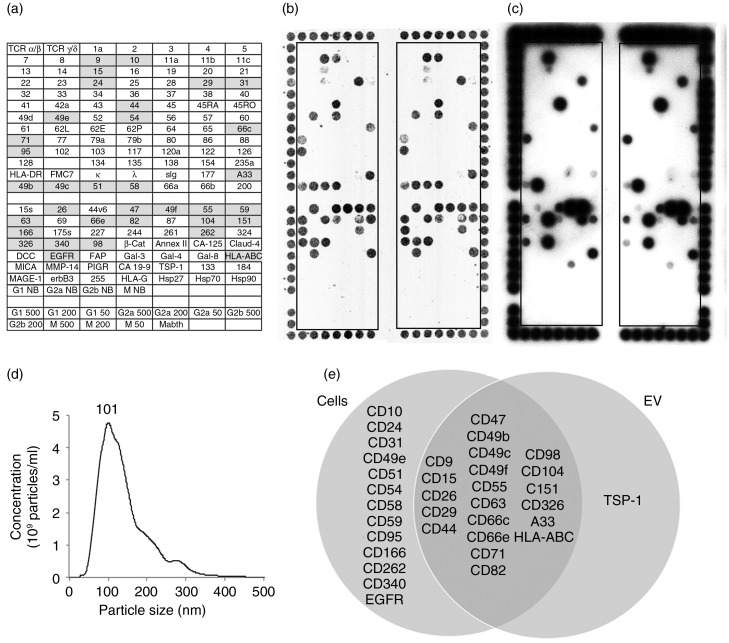
Figure 1 shows surface profiles of live LIM1215 cells acquired using optical detection with a DotScan scanner (Fig. 1b) and those of LIM1215-derived EV (Fig. 1c) using ECL detection. These EV particles, enriched for exosomes by differential centrifugation, ranged in size from 30 to 400 nm by NanoSight analysis (Fig. 1d), with a mode size of 101 nm (the value of the highest point of the peak, i.e. the most frequently occurring size). The Venn diagram (Fig. 1e) shows the antigens co-expressed on cells and EV, and those detected on cells or EV alone. Of the 34 antigens detected on the LIM1215 cells, 24 (70.6%) were also detected on their EV. TSP-1 was detected on the LIM1215 EV, but not the cells.
Fig. 1.
DotScan analysis of LIM1215 cells (b) and their EV (c). The key (a) shows antibody locations, with shaded antibodies indicating cell capture. Duplicate antibody arrays (outlined) are surrounded by a frame of alignment dots consisting of a mixture of CD44/CD29 antibodies. Detection of captured cells was by optical scanning (b). EV (7.8×108 particles derived from 335 µl of LIM1215-conditioned medium) were detected by ECL using biotinylated EpCAM (CD326) antibody, with a 10 min exposure on ECL film (c). NanoSight analysis shows the size distribution of LIM1215 EV (d). The number above the peak represents mode size in nm. A Venn diagram compares surface profiles of LIM1215 cells with their EV (e). TCR, T-cell receptor; κ, λ, immunoglobulin light chains kappa, lambda; sIg, surface immunoglobulin; DCC, deleted in colorectal cancer protein; EGFR, epidermal growth factor receptor; FAP, fibroblast activation protein; HLA-ABC, HLA-DR, human leukocyte antigens A, B, C and DR, respectively; MICA, MHC class I chain-related protein A; MMP-14, matrix metallopeptidase 14; PIGR, polymeric immunoglobulin receptor; TSP-1, thrombospondin-1; Mabthera, chimeric mouse/human anti-CD20. G1, G2a, G2b and M are murine isotype control antibodies IgG1, IgG2a, IgG2b and IgM, respectively. The numbers 500, 200 and 50 refer to isotype control antibody concentrations in µg/ml. NB means no BSA in the antibody solution; these antibodies were at 500 µg/ml. Antibody details are listed in Supplementary Table 1 of Supplementary Material.
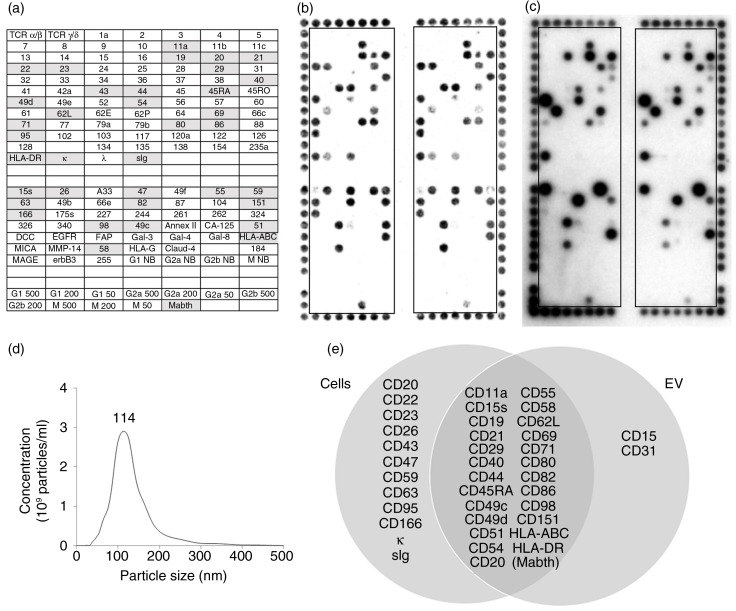
Figure 2 shows a comparison of the surface profiles of MEC1 cells (Fig. 2b) and their EV (Fig. 2c). NanoSight analysis of MEC1 EV (Fig. 2d) showed particles with a mode size of 114 nm. Of the 36 proteins detected on the cells, 25 (69.4%) were also detected on their EV, as summarized in the Venn diagram (Fig. 2e). CD15 and CD31 were detected on the MEC1 EV, but not the cells.
Fig. 2.
DotScan profiling of MEC1 cells (b) and their EV (c). The key (a) shows locations of antibodies (as for Fig. 1), with shaded antibodies indicating cell capture. Detection of captured cells was by optical scanning (b). EV (7.35×1010) were detected by ECL using biotinylated CD19 antibody, with a 5 min exposure on ECL film (c). NanoSight analysis shows the size distribution of MEC1 EV (d). A Venn diagram (e) compares surface profiles of MEC1 cells with their EV. The number above the peak represents mode size in nm.
Profiling of CLL cells and their EV from blood
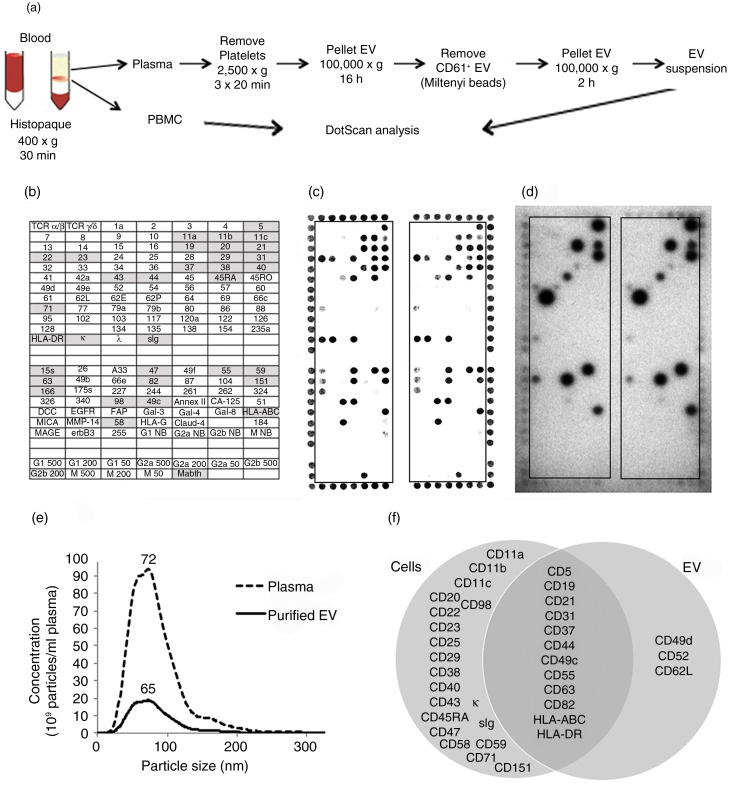
Figure 3a summarizes the protocol for preparation of PBMC and CD61-depleted EV from the blood of CLL patients and healthy controls. Enrichment for CLL-derived EV from plasma was necessary to achieve the required sensitivity for the assay. CLL-derived EV profiles were not clearly detected when plasma (300 µl) was tested directly on DotScan, probably due to interference from plasma proteins and platelet-derived EV. The removal of CD61+ EV by magnetic beads was confirmed by comparing DotScan profiles for CD61-depleted and non-depleted EV using biotinylated CD61 antibody for detection (results not shown); also the CD61+ EV captured on magnetic beads showed distinct platelet-like profiles on DotScan (CD41+, CD42a+, CD61+, CD62P+; unpublished data). Although citrate anti-coagulant has been recommended for proteomic studies because it induces fewer platelet-derived MV ex vivo (42,43), results from 3 independent experiments comparing blood samples collected into different anti-coagulants (citrate, heparin, EDTA) from healthy donors (unpublished data) demonstrated no significant differences (p>0.05 by two-tailed, paired student's t-test) in yield of CD61-depleted EV (determined by NanoSight analysis) or their normalized DotScan profiles (with CD45 detection). However, EV pellets from heparin were stickier than those from EDTA or citrate. The average yields of CD61-depleted EV were almost identical for heparin and citrate, but ~1.5-fold lower for EDTA. Despite this difference in average yield, the intensity of DotScan binding before normalization was similar for CD61-depleted EV from citrate and EDTA, but ~2-fold lower from heparin. Results were also more consistent between duplicate microarray panels for EDTA and citrate.
Fig. 3.
Workflow for preparation of PBMC and CD61-depleted EV from blood (a), with DotScan profiling (b–d). The key (b) shows antibody locations, with shaded antibodies indicating cell capture. DotScan analyses are shown for 3×106 PBMC (c) and CD61-depleted EV from 10 ml of blood (d) from an 87-year-old female CLL patient (Patient 1) with a white blood cell count of 45.3 x109/L. EV were tested in the presence of heat inactivated human AB serum (2%). Detection of captured cells was by optical scanning (c). EV were detected by ECL using biotinylated CD19 antibody, with a 30 min exposure on ECL film (d). NanoSight analysis (e) compares the average size distributions (tested in triplicate) of EV in the plasma and purified CD61-depleted EV from the plasma. The results are shown as average number of particles per ml of plasma before and after enrichment for CD61-depleted EV; the numbers above the peaks represent mode sizes in nm. A Venn diagram (f) compares surface profiles of patient CLL cells and their EV.
DotScan profiles are shown for PBMC (Fig. 3c) and CD19+ EV (Fig. 3d) from CLL Patient 1 with a white blood cell count of 45.3×109/l plasma (Supplementary Table 2). NanoSight analysis comparing total particles in the plasma with the CD61-depleted EV recovered from the plasma (Fig. 3e) shows that ~10% of particles in the plasma were recovered by this procedure, and the size distribution of particles (50–250 nm) was not much altered after EV purification, with mode sizes of 72 and 65 nm, respectively. Although lipoprotein particles may contribute to the EV counts in non-fasting plasma, they are removed by ultracentrifugation (44).
The DotScan profile for the CLL cells of this patient (Fig. 3c) showed a typical CLL profile, as described previously (45), with little or no detection of antigens from normal T-cells (TCRα/β, CD2, CD3, CD4, CD8) or myeloid cells (CD13, CD14, CD15, CD33) that represent relatively small subpopulations in advanced CLL patients with high white blood cell counts. The CD19+ EV profile shown in Fig. 3d (CD5, CD19, CD21, CD31, CD37, CD44, CD49c, CD49d, CD52, CD55, CD62L, CD63, CD82, HLA-DR and HLA-A,B,C) represented a subset of the antigens detected on the corresponding CLL cells (12 of 32; 37.5%), with CD49d, CD52 and CD62L detected on the EV, but not the cells. The co-expression of CD19 (B-cell antigen) and CD5 (aberrantly expressed T-cell antigen) is diagnostic for CLL (46). As shown in the Venn diagram (Fig. 3f), the following antigens detected on the cells were not seen on the corresponding EV: B-cell antigens CD20, CD22 and CD23; integrins CD11a, CD11b, CD11c, CD29; CD38, an important prognostic marker in CLL (47); CD25, CD40, CD43, CD45RA, CD47, CD58, CD59, CD71; and tetraspanin CD151. As for MEC1-derived EV, sIg and κ were not detected. However, these antigens would have been blocked, if present, by immunoglobulin in the human AB serum (added to prevent occasional non-specific Fc receptor binding of EV from human plasma), not required for testing MEC1-derived EV. CD19+ EV from the same volume of blood from age- and gender-matched healthy individuals were not detected.
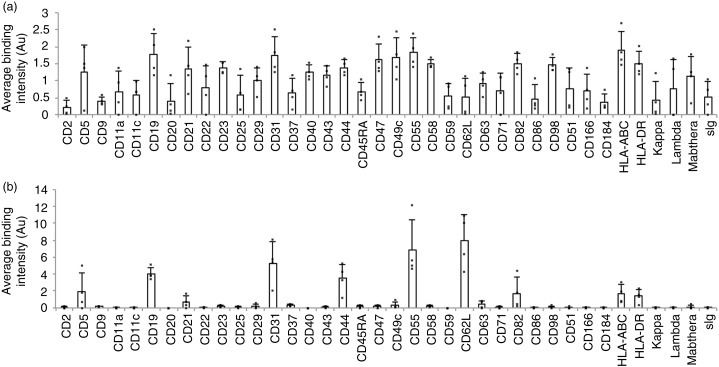
Averaged normalized DotScan data from 4 CLL patients (Supplementary Table 2) are shown in Fig. 4. Of the 38 antigens detected on the cells of 2 or more patients (Fig. 4a), ~33% were also detected on the corresponding CD19+ EV (Fig. 4b), with strong expression of CD19, CD31, CD44, CD55 and CD62L; moderate levels of CD5, CD82, HLA-A,B,C and HLA-DR; and very low levels of CD21, CD49c and CD63. CD5 was not detected on CD19+ EV from Patient 4, whose CLL cells were only weakly positive for CD5 by DotScan. Flow cytometry confirmed that only 19% of this patient's CD19+ leukaemia cells co-expressed CD5 (clinical data). CD19 levels were similar for all 4 EV samples after normalization, confirming that CD19-biotin antibody was suitable for the detection of CLL-derived EV. By contrast, the tetraspanins CD9, CD63 and CD151 were very low or below the limit of detection, while CD62L (L-selectin) was high on EV compared to cells. Compared to CLL cells, EV showed low CD5, CD21, CD82, HLA-ABC and HLA-DR (relative to CD19), and the B-cell antigen CD20 was not detected by either of the CD20 antibodies.
Fig. 4.
Comparison of DotScan surface profiles of PBMC (3×106 cells; a) and CD19+, CD61-depleted EV (b) from the blood of CLL patients (n=4). DotScan analysis was carried out as for Figure 3. After background and isotype control subtraction and median centred normalization, averaged duplicate binding intensities (expressed in arbitrary units, Au) are shown for 38 antigens, each of which was detected on the cells of at least 2 of 4 CLL samples.
Discussion
The results show that DotScan can be used to profile EV derived from cancer cell lines or the plasma of CLL patients, with antibody detection of specific surface markers, such as EpCAM (CD326) for LIM1215 colon cancer, or CD19 for CLL. The activity and specificity of each antibody used in DotScan was previously determined using cell lines and cells from human blood, bone marrow and cancer tissue samples (not shown). As discussed previously (20,37,48), the dot intensities of DotScan data reflect the level of binding of cells (or EV) to the antibody dots and are semi-quantitative. The binding patterns of cells (or EV) on DotScan depend on the proteins expressed on their surfaces at levels above a certain threshold for capture that may vary with the affinity of each antibody and its accessibility to the relevant antigenic epitope. The larger the cell or particle, the more antibody interactions are required for capture. The binding intensities also depend on the number of cells or particles expressing each antigen, until saturation is reached. The sensitivity of the EV assay (i.e. the minimum number of particles required for DotScan detection of a distinctive surface profile for an EV sample) depends on the proportion of EV expressing the antigens recognized by immobilized antibodies, the level of target antigen detected by the biotinylated detection antibody and the ECL exposure time.
CD326 was expressed at high levels on LIM1215 cells and their EV (Fig. 1b, c), and was therefore a suitable detection antigen for this cell line. Other antigens detected strongly on LIM1215 cells and their EV included CD9, CD29, CD44, CD49c, CD49f, CD55, CD63, CD66c, CD66e, CD71, CD98, CD104, CD151 and HLA-A,B,C. Compared to cells, EV showed greatly reduced binding for CD10, CD26, CD49e, CD51, CD54, CD58, CD59, CD82, CD95, CD166, CD262, CD340, A33 and EGFR (Fig. 1b, c), reflecting selective recruitment and compartmentalization of antigens during EV biogenesis (49–53). Interestingly, thrombospondin-1 (TSP-1) was detected on the EV, but not on the cell surface. TSP-1 is secreted by a variety of cells, including LIM1215 (54), and has previously been identified in LIM1215-derived exosomes (www.exocarta.org/gene_summary?gene_id=7057). In colorectal and other cancers, TSP-1 appears to play a complex role in tumour progression (anti-angiogenic and pro-tumorigenic) that has not been fully elucidated (55–58). The role of EV-associated TSP-1 in cancer progression is not known.
EV from the MEC1 cell line were used to optimize DotScan for the subsequent detection of CLL-derived EV from the plasma of CLL patients. Although derived from a CLL patient, MEC1 cells differ from typical CLL cells in their loss of expression of CD5 (29), as confirmed by DotScan (Fig. 2b). B-cell antigens CD19 and CD21 were detected strongly on both MEC1 cells and their EV (Fig. 2b, c). However, the B-cell antigens CD22 and CD23, strongly detected on MEC1 cells, were not detected on their EV (Fig. 2b, c). They were also previously reported to be absent from the exosomes of several B-cell lymphoma cell lines with these antigens (26). Surface immunoglobulin (sIg) with kappa light chain (κ), expressed on the MEC1 cells, was not detected on MEC1-derived EV. CD20 was detected on MEC1 cells and their EV by the Mabthera CD20 antibody (clone MB2 A4). However, the other CD20 antibody (clone H299/B1) captured only cells. This suggests that the Mabthera epitope of CD20 comprising amino acid residues 168–175 (59) remains intact on the surface of MEC1 EV, while the H299/B1 epitope comprising residues 172–178 (59) may be lost or become inaccessible to the immobilized antibody. Although the significance of this difference is not yet understood, it is interesting to note that CD20 antibodies have been classified as Type 1 (e.g. Mabthera) and Type II (e.g. clone H299/B1) based on their different mechanisms of killing B-cells and the epitope position on CD20 (59).
The tetraspanins CD82 and CD151 were detected strongly on both MEC1 cells and their EV (Fig. 2b, c). In contrast, tetraspanins CD9 and CD63, considered to be exosome markers (6), were not detected on MEC1-derived EV (Fig. 2c), although detected strongly on LIM1215 EV (Fig. 1c). CD63 (but not CD9) was detected on MEC1 cells. The absence of CD9 was previously reported for exosomes from the Ramos, Sudhl4 and Sudhl6 B-cell lymphoma lines (26), while lack of CD63 was reported for exosomes derived from stimulated primary B-cells (27). Interestingly, CD15 (Lewis-X antigen) and CD31 (PECAM-1) were detected only on EV. CD15s (Sialyl Lewis X) and CD62L (L-selectin), strongly detected on EV, were only faintly detected on cells. These antigens may be enriched on EV due to the selective recruitment and compartmentalization during EV biogenesis (49–53). Optimal profiles for MEC1-derived (CLL) EV were obtained with ≥8×109 particles using biotinylated CD19 antibody for detection. The limit of detection was approximately 2×106 MEC1 particles per antibody dot (6.5×108/assay).
In this study, our aim was to detect CLL-derived EV in the plasma of patients and define their surface profiles. It has been suggested that CLL-derived EV might be important in the establishment of a pro-survival microenvironment in CLL (60). CLL-exosomes have been shown to switch endothelial and mesenchymal stroma cells into cancer-associated fibroblasts to sustain leukaemic cell survival in vitro (61). Larger circulating EV (100–1,000 nm in diameter) from CLL patients were shown to stimulate bone marrow stromal cells, inducing the production of B-cell survival factor hypoxia-inducible factor-1α (17). CD62L, a homing receptor, is thought to have a pro-survival role in CLL cells (62). Paradoxically, however, low expression of CD62L on CLL cells is associated with poor prognosis (63). In our study, CD62L (L-selectin) was high on CLL EV compared with the corresponding CLL cells (Figs. 2, 3 and 4), suggesting that the reported shedding of CD62L from CLL cells (64) may occur via EV.
The high levels of CD62L detected by DotScan on the surface of CLL-derived EV from 4 patients with progressive disease (Figs. 3 and 4) suggest a possible role for this protein. CD62L could play a role in the homing of these EV to areas of the body where they may offload their protein and miRNA cargo, activating key signalling pathways and modulating gene expression, to suppress immune responses and/or promote disease progression. Such effects could be of particular importance for CLL with the advent of novel protein kinase inhibitors such as ibrutinib (65) and idelalisib (66) that induce clinical remissions accompanied by marked lymphocytosis due to interference with B-cell signalling in the lymph node microenvironment.
It has been suggested that CD20 on exosomes in patients with B-cell malignancies may act as a decoy for rituximab, leading to protection against this therapeutic antibody (67,68). These investigators reported CD20 on exosomes produced in vitro from B-cell lymphoma cell lines and primary CLL or B-cell lymphoma cells. In our study, CD20 was detected with rituximab on EV prepared in vitro from MEC1 CLL cells, but not on CLL EV isolated from the plasma of 4 patients. Using flow cytometry, Caivano et al. (69) found that CD20+ EV were significantly less numerous than CD19+ EV in the plasma of patients with CLL or other B-cell lymphomas. They suggested that CD20 may be excluded from the surface of CLL EV during their generation. The level of expression of CD20 on CLL-derived EV may, in part, depend on the conditions under which they are generated. The exclusion of other surface proteins, such as CD22, CD23, CD40 and CD45RA from CLL EV (Fig. 4), has also been reported for exosomes released from B-cell lymphomas (26).
These preliminary results with CLL have demonstrated the potential for DotScan to identify recognizable disease signatures on EV in the plasma. A related but somewhat different approach was recently described by Jakobsen et al. (70), who used a 37-antibody EV array to profile exosomes from the plasma of non-small cell lung carcinoma patients and control subjects, using a cocktail of CD9, CD63 and CD81 antibodies. This analysis differed from ours in that whole plasma was analysed, yielding profiles of total plasma exosomes, including those derived from platelets and cells involved in inflammation.
Although the methods described in this study allowed the surface profiling of CLL-derived EV from the plasma of advanced CLL patients, higher sensitivity may be required for DotScan profiling of the less abundant subpopulations of EV in blood, for example, to detect early primary tumours or monitor minimal residual disease or recurrence of solid tumours. To improve the yield and quality of EV from plasma, the following points should be considered. The removal of platelets by centrifugation at 2,500×g (20 min, 4°C, 3 times) depletes EV of 100–300 nm diameter, as demonstrated by NanoSight analysis (p<0.05; unpublished data). This centrifugation step should therefore be replaced by centrifuging twice at 1,500×g (20 min, 23°C). In addition, the use of heparin anti-coagulant should be avoided due to the stickiness of EV prepared from heparinized blood, resulting in less consistent DotScan results and reduced sensitivity.
We have shown that EV captured on antibody-coated Miltenyi microbeads (50 nm in diameter) can be profiled directly on DotScan (unpublished data). Positive enrichment for disease-specific EV from plasma using antibody-coated magnetic microbeads may avoid inadvertent CD61-depletion of disease-specific EV that have bound to, or fused with, platelet-derived EV (71) or arise from CD61-expressing cancer cells (72,73). In addition, the sensitivity of the DotScan EV assay could be increased by reducing background luminescence by replacing nitrocellulose-coated slides with glass slides coated with aldehyde silane, poly-L-lysine or aminosilane (74). Although the profiling of CLL cells requires a surface such as nitrocellulose to minimize their tendency to adhere non-specifically (unpublished data), clear slides may provide a better surface for EV analysis.
The International Society for Extracellular Vesicles (ISEV) has provided researchers with a minimal set of biochemical, biophysical and functional criteria for discriminating EV from non-EV components (25). As recommended by ISEV, the EV analysed in our study were prepared from conditioned cell culture medium and body fluids that were collected and treated “gently” to limit cell disruption. Over-confluent growth of cells was avoided during EV production, and the medium was collected and clarified by procedures that minimized damage to cells. Any apoptotic bodies or platelets were removed from plasma before freezing and/or ultracentrifugation, as shown by the absence of particles ≥500 nm by NanoSight analysis of clarified plasma and EV preparations. EV from blood samples processed 4 h after collection gave the same DotScan results as blood processed within 2 h of collection (not shown). Frozen plasma and EV pellets were thawed rapidly at 37°C to minimize disruption to the EV; re-freezing of EV-containing samples was avoided. Although the EV samples used for this study were not analysed by electron microscopy, EV purified previously from conditioned medium and plasma in our laboratory showed the presence of intact EV (not shown). Although the DotScan signals of CD61-depleted EV from the plasma of CLL patients were not compared with depleted plasma signals, they were compared with signals from similarly prepared CD61-depleted EV from the plasma of age- and gender-matched normal healthy individuals and found to be specific for CLL patients.
Tetraspanins such as CD9, CD63 and CD81 are often considered to be markers of exosomes. However, our DotScan results (Figs. 2, 3) support previous reports of exosomes lacking in, or showing variable expression of, CD9 and/or CD63 (26,27), further highlighting the importance of using a cocktail of antibodies for differentiating exosomes from other EV. Although CD81 was not included in the DotScan antibody panel for this study, highly variable CD81 expression levels have been reported for exosomes from B-cell lymphoma cell lines (26) and CD34+ exosomes from the plasma of acute myeloid leukaemia patients (75). To our knowledge, there is currently no single reliable marker for exosomes (76).
Conclusions
DotScan antibody microarrays have been used to compare surface protein profiles of cells and their EV isolated from conditioned growth medium of cell lines, or from the plasma of CLL patients. Further investigations will be required to determine whether the surface CD20 expression levels on CLL-derived EV are influenced by micro-environmental factors during EV secretion and/or have prognostic significance. The clinical significance of the high expression of CD62L detected on MEC1 EV and CLL EV from 4 patients with progressive CLL also requires clarification. Future comparisons of EV from more patients with progressive and stable CLL may further understanding of the mechanisms involved in disease progression.
Supplementary Material
Acknowledgements
We thank Dr. Pauline Y. Huang for preparing the DotScan antibody microarrays.
Conflict of interest and funding
We acknowledge funding support from the George and Elevine Whittaker Research Fellowship and Medsaic Pty Ltd (Darlington, NSW, Australia). RIC owns less than 2% equity in Medsaic Pty Ltd.
References
- 1.D'Souza-Schorey C, Clancy JW. Tumor-derived microvesicles: shedding light on novel microenvironment modulators and prospective cancer biomarkers. Genes Dev. 2012;26:1287–99. doi: 10.1101/gad.192351.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor DD, Gercel-Taylor C. Tumour-derived exosomes and their role in cancer-associated T-cell signalling defects. Br J Cancer. 2005;92:305–11. doi: 10.1038/sj.bjc.6602316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai J, Han Y, Ren H, Chen C, He D, Zhou L, et al. Extracellular vesicle-mediated transfer of donor genomic DNA to recipient cells is a novel mechanism for genetic influence between cells. J Mol Cell Biol. 2013;5:227–38. doi: 10.1093/jmcb/mjt011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi DS, Kim DK, Kim YK, Gho YS. Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics. 2013;13:1554–71. doi: 10.1002/pmic.201200329. [DOI] [PubMed] [Google Scholar]
- 5.Inal JM, Fairbrother U, Heugh S. Microvesiculation and disease. Biochem Soc Trans. 2013;41:237–40. doi: 10.1042/BST20120258. [DOI] [PubMed] [Google Scholar]
- 6.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–20. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–83. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thery C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep. 2011;3:15. doi: 10.3410/B3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25:364–72. doi: 10.1016/j.tcb.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Camussi G, Deregibus MC, Tetta C. Tumor-derived microvesicles and the cancer microenvironment. Curr Mol Med. 2013;13:58–67. [PubMed] [Google Scholar]
- 11.Kucharzewska P, Belting M. Emerging roles of extracellular vesicles in the adaptive response of tumour cells to microenvironmental stress. J Extracell Vesicles. 2013;2:20304. doi: 10.3402/jev.v2i0.20304. http://dx.doi.org/10.3402/jev.v2i0.20304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.An T, Qin S, Xu Y, Tang Y, Huang Y, Situ B, et al. Exosomes serve as tumour markers for personalized diagnostics owing to their important role in cancer metastasis. J Extracell Vesicles. 2015;4:27522. doi: 10.3402/jev.v4.27522. http://dx.doi.org/10.3402/jev.v4.27522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tatischeff I. Cell-derived extracellular vesicles open new perspectives for cancer research. Cancer Res Front. 2015;1:208–24. [Google Scholar]
- 14.Webber J, Yeung V, Clayton A, editors. Extracellular vesicles as modulators of the cancer microenvironment. Sem Cell Dev Biol. 2015;40:27–34. doi: 10.1016/j.semcdb.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 15.Nicholas J. A new diagnostic tool with the potential to predict tumor metastasis. J Natl Cancer Inst. 2013;105:371–2. doi: 10.1093/jnci/djt048. [DOI] [PubMed] [Google Scholar]
- 16.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh AK, Secreto CR, Knox TR, Ding W, Mukhopadhyay D, Kay NE. Circulating microvesicles in B-cell chronic lymphocytic leukemia can stimulate marrow stromal cells: implications for disease progression. Blood. 2010;115:1755–64. doi: 10.1182/blood-2009-09-242719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiorazzi N, Ferrarini M. B cell chronic lymphocytic leukemia: lessons learned from studies of the B cell antigen receptor. Annu Rev Immunol. 2003;21:841–94. doi: 10.1146/annurev.immunol.21.120601.141018. [DOI] [PubMed] [Google Scholar]
- 19.Seifert M, Sellmann L, Bloehdorn J, Wein F, Stilgenbauer S, Durig J, et al. Cellular origin and pathophysiology of chronic lymphocytic leukemia. J Exp Med. 2012;209:2183–98. doi: 10.1084/jem.20120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belov L, Mulligan SP, Barber N, Woolfson A, Scott M, Stoner K, et al. Analysis of human leukaemias and lymphomas using extensive immunophenotypes from an antibody microarray. Br J Haematol. 2006;135:184–97. doi: 10.1111/j.1365-2141.2006.06266.x. [DOI] [PubMed] [Google Scholar]
- 21.Ellmark P, Belov L, Huang P, Lee CS, Solomon MJ, Morgan DK, et al. Multiplex detection of surface molecules on colorectal cancers. Proteomics. 2006;6:1791–802. doi: 10.1002/pmic.200500468. [DOI] [PubMed] [Google Scholar]
- 22.Kaufman KL, Belov L, Huang P, Mactier S, Scolyer RA, Mann GJ, et al. An extended antibody microarray for surface profiling metastatic melanoma. J Immunol Methods. 2010;358:23–34. doi: 10.1016/j.jim.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 23.Zhou J, Belov L, Huang PY, Shin JS, Solomon MJ, Chapuis PH, et al. Surface antigen profiling of colorectal cancer using antibody microarrays with fluorescence multiplexing. J Immunol Methods. 2010;355:40–51. doi: 10.1016/j.jim.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 24.Kanada M, Bachmann MH, Hardy JW, Frimannson DO, Bronsart L, Wang A, et al. Differential fates of biomolecules delivered to target cells via extracellular vesicles. Proc Natl Acad Sci USA. 2015;112:E1433–42. doi: 10.1073/pnas.1418401112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lötvall J, Hill AF, Hochberg F, Buzás EI, Di Vizio D, Gardiner C, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles. 2014;3:26913. doi: 10.3402/jev.v3.26913. http://dx.doi.org/10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oksvold MP, Kullmann A, Forfang L, Kierulf B, Li M, Brech A, et al. Expression of B-cell surface antigens in subpopulations of exosomes released from B-cell lymphoma cells. Clin Ther. 2014;36:847–62 e1. doi: 10.1016/j.clinthera.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 27.Saunderson SC, Schuberth PC, Dunn AC, Miller L, Hock BD, MacKay PA, et al. Induction of exosome release in primary B cells stimulated via CD40 and the IL-4 receptor. J Immunol. 2008;180:8146–52. doi: 10.4049/jimmunol.180.12.8146. [DOI] [PubMed] [Google Scholar]
- 28.Whitehead RH, Macrae FA, John DJBS, Ma J. A colon cancer cell line (LIM1215) derived from a patient with inherited nonpolyposis colorectal cancer. J Natl Cancer Inst. 1985;74:759–65. [PubMed] [Google Scholar]
- 29.Stacchini A, Aragno M, Vallario A, Alfarano A, Circosta P, Gottardi D, et al. MEC1 and MEC2: two new cell lines derived from B-chronic lymphocytic leukaemia in prolymphocytoid transformation. Leuk Res. 1999;23:127–36. doi: 10.1016/s0145-2126(98)00154-4. [DOI] [PubMed] [Google Scholar]
- 30.Logozzi M, De Milito A, Lugini L, Borghi M, Calabro L, Spada M, et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS One. 2009;4:e5219. doi: 10.1371/journal.pone.0005219. doi: http://dx.doi.org/10.1371/journal.pone.0005219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006 doi: 10.1002/0471143030.cb0322s30. Chapter 3:Unit 3.22, doi: http://dx.doi.org/10.1002/0471143030. [DOI] [PubMed] [Google Scholar]
- 32.Colino J, Snapper CM. Dendritic cell-derived exosomes express a Streptococcus pneumoniae capsular polysaccharide type 14 cross-reactive antigen that induces protective immunoglobulin responses against pneumococcal infection in mice. Infect Immun. 2007;75:220–30. doi: 10.1128/IAI.01217-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lai RC, Tan SS, Teh BJ, Sze SK, Arslan F, de Kleijn DP, et al. Proteolytic potential of the MSC exosome proteome: implications for an exosome-mediated delivery of therapeutic proteasome. Int J Proteomics. 2012;2012:971907. doi: 10.1155/2012/971907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, Lee Y, Johansson HJ, Mager I, Vader P, Nordin JZ, et al. Serum-free culture alters the quantity and protein composition of neuroblastoma-derived extracellular vesicles. J Extracell Vesicles. 2015;4:26883. doi: 10.3402/jev.v4.26883. http://dx.doi.org/10.3402/jev.v4.26883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Döhner H, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute–Working Group 1996 guidelines. Blood. 2008;111:5446–56. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belov L, Huang P, Barber N, Mulligan SP, Christopherson RI. Identification of repertoires of surface antigens on leukemias using an antibody microarray. Proteomics. 2003;3:2147–54. doi: 10.1002/pmic.200300599. [DOI] [PubMed] [Google Scholar]
- 37.Belov L, de la Vega O, dos Remedios CG, Mulligan SP, Christopherson RI. Immunophenotyping of leukemias using a cluster of differentiation antibody microarray. Cancer Res. 2001;61:4483–9. [PubMed] [Google Scholar]
- 38.Ellmark P, Woolfson A, Belov L, Christopherson RI. The applicability of a cluster of differentiation monoclonal antibody microarray to the diagnosis of human disease. Methods Mol Biol. 2008;439:199–209. doi: 10.1007/978-1-59745-188-8_14. [DOI] [PubMed] [Google Scholar]
- 39.Hamelinck D, Zhou H, Li L, Verweij C, Dillon D, Feng Z, et al. Optimized normalization for antibody microarrays and application to serum-protein profiling. Mol Cell Proteomics. 2005;4:773–84. doi: 10.1074/mcp.M400180-MCP200. [DOI] [PubMed] [Google Scholar]
- 40.Zhou J, Belov L, Armstrong N, Christopherson RI. Antibody microarrays and multiplexing. Bioinform Hum Proteomics. 2013;3:331–59. [Google Scholar]
- 41.Sedlmayr P, Leitner V, Pilz S, Wintersteiger R, Stewart CC, Dohr G. Species-specific blocking of Fc-receptors in indirect immunofluorescence assays. Laboratory Hematol. 2001;7:81–4. [Google Scholar]
- 42.Lacroix R, Judicone C, Poncelet P, Robert S, Arnaud L, Sampol J, et al. Impact of pre-analytical parameters on the measurement of circulating microparticles: towards standardization of protocol. J Thromb Haemost. 2012;10:437–46. doi: 10.1111/j.1538-7836.2011.04610.x. [DOI] [PubMed] [Google Scholar]
- 43.Mullier F, Bailly N, Chatelain C, Chatelain B, Dogne JM. Pre-analytical issues in the measurement of circulating microparticles: recommendations and pending questions. J Thromb Haemost. 2013;11:693–6. doi: 10.1111/jth.12171. [DOI] [PubMed] [Google Scholar]
- 44.Gardiner C, Ferreira YJ, Dragovic RA, Redman CW, Sargent IL. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. J Extracell Vesicles. 2013;2:19671. doi: 10.3402/jev.v2i0.19671. doi: http://dx.doi.org/10.3402/jev.v2i0.19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang PY, Kohnke P, Belov L, Best OG, Mulligan SP, Christopherson RI. Profiles of surface mosaics on chronic lymphocytic leukemias distinguish stable and progressive subtypes. J Pharm Pharm Sci. 2013;16:231–7. doi: 10.18433/j3f01c. [DOI] [PubMed] [Google Scholar]
- 46.Molica S, Mauro FR, Giannarelli D, Lauria F, Cortelezzi A, Brugiatelli M, et al. Differentiating chronic lymphocytic leukemia from monoclonal B-lymphocytosis according to clinical outcome: on behalf of the GIMEMA chronic lymphoproliferative diseases working group. Haematologica. 2011;96:277–83. doi: 10.3324/haematol.2010.030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dürig J, Naschar M, Schmücker U, Renzing-Köhler K, Hölter T, Hüttmann A, et al. CD38 expression is an important prognostic marker in chronic lymphocytic leukaemia. Leukemia. 2002;16:30–5. doi: 10.1038/sj.leu.2402339. [DOI] [PubMed] [Google Scholar]
- 48.Zhou J, Belov L, Solomon MJ, Chan C, Clarke SJ, Christopherson RI. Colorectal cancer cell surface protein profiling using an antibody microarray and fluorescence multiplexing. J Vis Exp. 2011;55:e3322. doi: 10.3791/3322. doi: http://dx.doi.org/10.3791/3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Jong OG, Verhaar MC, Chen Y, Vader P, Gremmels H, Posthuma G, et al. Cellular stress conditions are reflected in the protein and RNA content of endothelial cell-derived exosomes. J Extracell Vesicles. 2012;1:18396. doi: 10.3402/jev.v1i0.18396. doi: http://dx.doi.org/10.3402/jev.v1i0.18396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Escola JM, Kleijmeer MJ, Stoorvogel W, Griffith JM, Yoshie O, Geuze HJ. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J Biol Chem. 1998;273:20121–7. doi: 10.1074/jbc.273.32.20121. [DOI] [PubMed] [Google Scholar]
- 51.Ji H, Greening DW, Barnes TW, Lim JW, Tauro BJ, Rai A, et al. Proteome profiling of exosomes derived from human primary and metastatic colorectal cancer cells reveal differential expression of key metastatic factors and signal transduction components. Proteomics. 2013;13:1672–86. doi: 10.1002/pmic.201200562. [DOI] [PubMed] [Google Scholar]
- 52.Nazarenko I, Rana S, Baumann A, McAlear J, Hellwig A, Trendelenburg M, et al. Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. 2010;70:1668–78. doi: 10.1158/0008-5472.CAN-09-2470. [DOI] [PubMed] [Google Scholar]
- 53.Perez-Hernandez D, Gutierrez-Vazquez C, Jorge I, Lopez-Martin S, Ursa A, Sanchez-Madrid F, et al. The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J Biol Chem. 2013;288:11649–61. doi: 10.1074/jbc.M112.445304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greening DW, Ji H, Kapp EA, Simpson RJ. Sulindac modulates secreted protein expression from LIM1215 colon carcinoma cells prior to apoptosis. Biochim Biophys Acta. 2013;1834:2293–307. doi: 10.1016/j.bbapap.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 55.Gutierrez LS. The role of thrombospondin 1 on intestinal inflammation and carcinogenesis. Biomark Insights. 2008;3:171–8. doi: 10.4137/bmi.s630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jayachandran A, Anaka M, Prithviraj P, Hudson C, McKeown SJ, Lo PH, et al. Thrombospondin 1 promotes an aggressive phenotype through epithelial-to-mesenchymal transition in human melanoma. Oncotarget. 2014;5:5782–97. doi: 10.18632/oncotarget.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miyata Y, Sakai H. Thrombospondin-1 in urological cancer: pathological role, clinical significance, and therapeutic prospects. Int J Mol Sci. 2013;14:12249–72. doi: 10.3390/ijms140612249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sutton CD, O'Byrne K, Goddard JC, Marshall LJ, Jones L, Garcea G, et al. Expression of thrombospondin-1 in resected colorectal liver metastases predicts poor prognosis. Clin Cancer Res. 2005;11:6567–73. doi: 10.1158/1078-0432.CCR-05-0439. [DOI] [PubMed] [Google Scholar]
- 59.Niederfellner G, Lammens A, Mundigl O, Georges GJ, Schaefer W, Schwaiger M, et al. Epitope characterization and crystal structure of GA101 provide insights into the molecular basis for type I/II distinction of CD20 antibodies. Blood. 2011;118:219–20. doi: 10.1182/blood-2010-09-305847. [DOI] [PubMed] [Google Scholar]
- 60.Haderk F, Hanna B, Richter K, Schnolzer M, Zenz T, Stilgenbauer S, et al. Extracellular vesicles in chronic lymphocytic leukemia. Leuk Lymphoma. 2013;54:1826–30. doi: 10.3109/10428194.2013.796052. [DOI] [PubMed] [Google Scholar]
- 61.Paggetti J, Haderk F, Seiffert M, Janji B, Kim YJ, Diestler U, et al. Chronic lymphocytic leukemia-exosomes switch endothelial and mesenchymal stroma cells into cancer-associated fibroblasts to sustain leukemic cell survival. Blood. 2014;124:2927. doi: 10.1182/blood-2014-12-618025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burgess M, Gill D, Singhania R, Cheung C, Chambers L, Renyolds BA, et al. CD62L as a therapeutic target in chronic lymphocytic leukemia. Clin Cancer Res. 2013;19:5675–85. doi: 10.1158/1078-0432.CCR-13-1037. [DOI] [PubMed] [Google Scholar]
- 63.Zucchetto A, Sonego P, Degan M, Bomben R, Dal Bo M, Bulian P, et al. Surface-antigen expression profiling of B cell chronic lymphocytic leukemia: from the signature of specific disease subsets to the identification of markers with prognostic relevance. J Transl Med. 2006;4:11. doi: 10.1186/1479-5876-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vlad A, Deglesne P-A, Letestu R, Saint-Georges S, Chevallier N, Baran-Marszak F, et al. Down-regulation of CXCR4 and CD62L in chronic lymphocytic leukemia cells is triggered by B-cell receptor ligation and associated with progressive disease. Cancer Res. 2009;69:6387–95. doi: 10.1158/0008-5472.CAN-08-4750. [DOI] [PubMed] [Google Scholar]
- 65.Byrd JC, Brown JR, O'Brien S, Barrientos JC, Kay NE, Reddy NM, et al. Ibrutinib versus ofatumumab in previously treated chronic lymphoid leukemia. N Engl J Med. 2014;371:213–23. doi: 10.1056/NEJMoa1400376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Furman RR, Sharman JP, Coutre SE, Cheson BD, Pagel JM, Hillmen P, et al. Idelalisib and rituximab in relapsed chronic lymphocytic leukemia. N Engl J Med. 2014;370:997–1007. doi: 10.1056/NEJMoa1315226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aung T, Chapuy B, Vogel D, Wenzel D, Oppermann M, Lahmann M, et al. Exosomal evasion of humoral immunotherapy in aggressive B-cell lymphoma modulated by ATP-binding cassette transporter A3. Proc Natl Acad Sci USA. 2011;108:15336–41. doi: 10.1073/pnas.1102855108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paggetti J, Haderk F, Seiffert M, Janji B, Distler U, Ammerlaan W, et al. Exosomes released by chronic lymphocytic leukemia cells induce the transition of stromal cells into cancer-associated fibroblasts. Blood. 2015;126:1106–17. doi: 10.1182/blood-2014-12-618025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Caivano A, Laurenzana I, De Luca L, La Rocca F, Simeon V, Trino S, et al. High serum levels of extracellular vesicles expressing malignancy-related markers are released in patients with various types of hematological neoplastic disorders. Tumor Biol. 2015:1–14. doi: 10.1007/s13277-015-3741-3. [DOI] [PubMed] [Google Scholar]
- 70.Jakobsen KR, Paulsen BS, Bæk R, Varming K, Sorensen BS, Jørgensen MM. Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma. J Extracell Vesicles. 2015;4:26659. doi: 10.3402/jev.v4.26659. doi: http://dx.doi.org/10.3402/jev.v4.26659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tesselaar ME, Romijn FP, Van Der Linden IK, Prins FA, Bertina RM, Osanto S. Microparticle-associated tissue factor activity: a link between cancer and thrombosis? J Thromb Haemost. 2007;5:520–7. doi: 10.1111/j.1538-7836.2007.02369.x. [DOI] [PubMed] [Google Scholar]
- 72.Moreno A, Lucena C, Lopez A, Garrido JJ, Perez de la Lastra JM, Llanes D. Immunohistochemical analysis of ß3 integrin (CD61): expression in pig tissues and human tumors. Histol Histopathol. 2002;17:347–52. doi: 10.14670/HH-17.347. [DOI] [PubMed] [Google Scholar]
- 73.Saalbach A, Wetzel A, Haustein U-F, Sticherling M, Simon JC, Anderegg U. Interaction of human Thy-1 (CD 90) with the integrin αvβ3 (CD51/CD61): an important mechanism mediating melanoma cell adhesion to activated endothelium. Oncogene. 2005;24:4710–20. doi: 10.1038/sj.onc.1208559. [DOI] [PubMed] [Google Scholar]
- 74.Seurynck-Servoss SL, White AM, Baird CL, Rodland KD, Zangar RC. Evaluation of surface chemistries for antibody microarrays. Anal Biochem. 2007;371:105–15. doi: 10.1016/j.ab.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hong CS, Muller L, Boyiadzis M, Whiteside TL. Isolation and characterization of CD34+ blast-derived exosomes in acute myeloid leukemia. PLoS One. 2014;9:e103310. doi: 10.1371/journal.pone.0103310. doi: http://dx.doi.org/10.1371/journal.pone.0103310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Keerthikumar S, Gangoda L, Liem M, Fonseka P, Atukorala I, Ozcitti C, et al. Proteogenomic analysis reveals exosomes are more oncogenic than ectosomes. Oncotarget. 2015;6:15375. doi: 10.18632/oncotarget.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.