Abstract
Bulbophyllum occultum, an epiphytic orchid mainly distributed in the rainforests of (north)eastern Madagascar and La Réunion, represents an interesting model case for testing the effects of anthropogenic vs historical (e.g., climate induced) habitat isolation and long-distance colonization on the genetic structure of plant species with disjunct distributions in the Madagascan region. To this aim, we surveyed amplified fragment length polymorphisms (AFLPs) across 13 populations in Madagascar and nine in La Réunion (206 individuals in total). We found overall high levels of population subdivision (ΦPT=0.387) and low within-population diversity (HE, range: 0.026–0.124), indicating non-equilibrium conditions in a mainly selfing species. There was no impact of recent deforestation (Madagascar) or habitat disturbance (La Réunion) detectable on AFLP diversity. K-means clustering and BARRIER analyses identified multiple gene pools and several genetic breaks, both within and among islands. Inter-island levels of population genetic diversity and subdivision were similar, whereby inter-individual divergence in flower colour explained a significant part of gene pool divergence in La Réunion. Our results suggest that (i) B. occultum persisted across multiple isolated (‘refugial') regions along the eastern rainforest corridor of Madagascar over recent climatic cycles and (ii) populations in La Réunion arose from either single or few independent introductions from Madagascar. High selfing rates and sufficient time for genetic drift likely promoted unexpectedly high population genetic and phenotypic (flower colour) differentiation in La Réunion. Overall, this study highlights a strong imprint of history on the genetic structure of a low-gene-dispersing epiphytic orchid from the Madagascan region.
Introduction
The tropical rainforests of Madagascar represent one of the world's greatest repositories of plant diversity (ca. 12 000 species) and endemism (ca. 70–80% Myers et al., 2000; Goodman and Benstead, 2005) but also rank among the most seriously impacted ecosystems, where human-induced landscape changes occur at an unprecedented rate (Cable, 2011). However, population genetic studies of Madagascan plant species are extremely rare and mostly limited to trees and shrubs (Dawson and Powell, 1999; Amsellem et al., 2000; Voigt et al., 2009; Rivers et al., 2011; Krishnan et al., 2013), with only few having dealt with taxa restricted to the island's eastern rainforest ecosystems (Andrianoelina et al., 2006, 2009 ; Shapcott et al., 2007, 2012,). With their main focus on conservation, these studies have provided some first and important insights into present-day patterns of intraspecific genetic variation, yet suggesting only limited impact of recent loss of vegetation on molecular genetic parameters (Shapcott et al., 2007; Andrianoelina et al., 2009). Moreover, there is a significant gap in our knowledge regarding the historical factors influencing past population dynamics (e.g., habitat fragmentation, colonization) and, in turn, the present-day genetic structure of Madagascan plant species, with implications for evolutionary biology. Somewhat surprisingly, the orchid family (Orchidaceae) has hitherto attracted little attention in this context, despite showing highest species richness (ca. 1000 spp.) and degree of endemism (ca. 85%) among all vascular plant families in Madagascar (Hermans et al., 2007; Cable, 2011; Madagascar Catalogue, 2013).
Because mainly thriving as epiphytes, tropical orchids are generally thought to be less resistant to both anthropogenic and natural changes in the environment when compared with their terrestrial, mostly temperate counterparts (Wiegand et al., 2013). For instance, host tree availability (and possibly specificity) plays a fundamental role in determining the distribution and abundance of epiphytic orchids, often resulting in their patchy distribution across geographic areas and along microenvironments, even within a single tree (Nieder et al., 2000; Winkler et al., 2009; Trapnell et al., 2013; but see Wagner et al., 2015). Furthermore, epiphytic orchids exclusively obtain their moisture and nutrients from air, rain and debris, and thus must be able to withstand periods of extreme water stress between rainfall events (e.g., by development of water-storing pseudobulbs). These intrinsic physiological constraints may further enhance their susceptibility to the genetic consequences of habitat fragmentation, including reduced intra-population diversity and increased population divergence, mainly because of genetic drift and inbreeding (Tremblay et al., 2005; Honnay and Jacquemyn, 2007; Leimu et al., 2010). However, determining these processes remains challenging because levels of genetic diversity of post-fragmentation populations are also likely influenced by other factors, such as physical features of the landscape and species-specific traits, in particular longevity, mating system and dispersal ability (Honnay and Jacquemyn, 2007; Aguilar et al., 2008; Yuan et al., 2012).
This study focuses on the population genetic structure of an epiphytic orchid, Bulbophyllum occultum Thouars (Epidendroideae), which is mainly distributed in the rainforests of northern and eastern Madagascar and the small Mascarene island of La Réunion (2512 km2), ca. 680 km to the East (Figure 1). There are several reasons why this orchid represents an interesting model case to gain insights into the effects of recent (anthropogenic) vs historical (e.g., climate induced) habitat isolation and long-distance colonization on the genetic structure of plant species with a disjunct distribution in the Madagascan region. First, the extant rainforest habitats of B. occultum have been impacted severely by agricultural practices (e.g., ‘slash and burn', livestock production, logging), with only about 10% of the primary vegetation remaining in Madagascar (Burney et al., 2004; Harper et al., 2007; Cable, 2011) and about 25% in La Réunion (Macdonald et al., 1991; Thébaud et al., 2009). Although much of this deforestation occurred only recently (i.e., over the last 100 years), its genetic impact may already have become apparent in species with relatively short generation times (Aguilar et al., 2008; Montoya et al., 2008), such as B. occultum, which may require only about 5 years to reach the reproductive stage as observed for Asian relatives (e.g., B. lobbii) under cultivation (GA Fischer, U Jaros, personal observations). On the other hand, the species' potential for self-pollination (Jacquemyn et al., 2005; Gamisch et al., 2014, 2015, ; see below) and supposedly high dispersal ability (via dust-like, wind-dispersed seeds) may have provided intrinsic buffers to the loss of genetic diversity (Honnay and Jacquemyn, 2007).
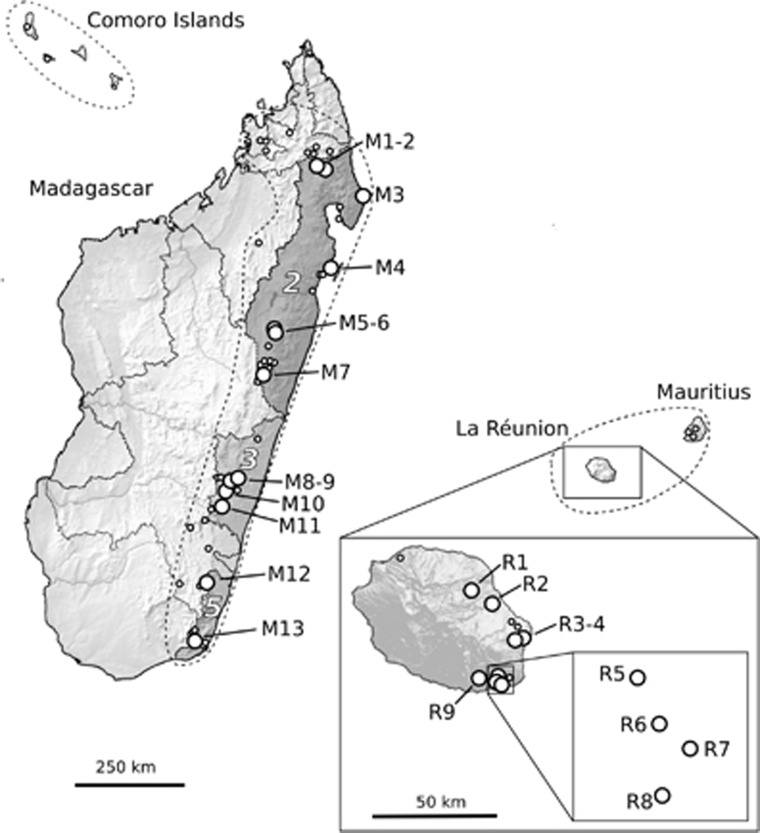
Figure 1.
Geographical distribution of B. occultum in Madagascar, the Mascarenes (La Réunion, Mauritius) and the Comoro Islands (stippled lines) based on the 22 sample locations of this study (large circles) plus 78 geo-referenced herbarium records and own field data (small circles). The dotted lines in Madagascar indicate 12 ‘Centres of Endemism' (COEs sensu Wilmé et al., 2006), out of which three (dark-shaded areas coded 2, 3 and 5) include sample localities for B. occultum (see Table 1 for identification of population codes).
Second, in the more distant past, natural processes of rainforest fragmentation may also have significantly influenced the genetic structure of B. occultum. For example, Quaternary periods of drier and cooler climate (broadly equivalent to glacials at high latitudes; Bennett and Provan, 2008) are believed to have had a major impact on the geographic distribution and within-species genetic structure of Madagascan rainforest biota (Andrianoelina et al., 2006, 2009; Wilmé et al., 2006; Vences et al., 2009; Rakotoarisoa et al., 2012, 2013; Rakotoarinivo et al., 2013). Notably, in Madagascar, the extant range of B. occultum extends across five (out of 12) ‘Centres of Endemism' (COEs) as defined by Wilmé et al. (2006) on the basis of modern land vertebrate records (see Figure 1). These COEs generally circumscribe river catchments in the coastal and montane portions of the island, and have been hypothesized to coincide with glacial refuge locations that fostered allopatric population divergence and speciation of locally endemic taxa (Wilmé et al., 2006; Rakotoarinivo et al., 2013). While recent population genetic studies in lemurs support this hypothesis (Markolf and Kappeler, 2013; Perry et al., 2013), it remains to be established whether COE boundaries significantly contribute to the spatial genetic structure of a plant species as well, and for which B. occultum is a suitable test case along the island's eastern rainforest corridor.
Finally, founder effects may have exerted a strong influence on the distribution of genetic variance among populations of B. occultum in La Réunion. Molecular phylogenetic studies have shown that this species (as represented, however, only by material from Madagascar) firmly nests within a clade of Madagascan congeners (with B. bicoloratum as likely sister; Fischer et al., 2007; Gamisch et al., 2015). Moreover, preliminary chloroplast DNA surveys indicate that La Réunion populations of B. occultum are almost fixed for the most common and widespread haplotype in Madagascan conspecifics (U Jaros, unpublished data). Together, these data suggest that B. occultum colonized La Réunion from Madagascar. In case this occurred relatively recently, we would expect La Réunion populations to be little differentiated from their putative source population(s) in Madagascar, and harbour less diversity relative to the latter (Barrett et al., 1996; Frankham, 1997). On the other hand, recurrent population founding has been shown to minimize bottlenecks created during colonization or even to promote population genetic differentiation (Whitlock and McCauley, 1990; Le Corre and Kremer, 1998; Vandepitte et al., 2012). Notably in this context, La Réunion B. occultum exhibits an unusual flower colour dimorphism, with both red- and yellow-flowered individuals often occurring in sympatry (Cadet, 1989; T Pailler, U Jaros, personal observation). The evolutionary significance of such dimorphisms has been discussed at length (reviewed by Wessinger and Rausher, 2012), but in most instances the mechanism(s) responsible for their maintenance (e.g., various forms of selection or genetic drift) remain unclear. However, for B. occultum, the null hypothesis would be that the genetic consequences of colonization (e.g., founder effects, drift, inbreeding) also affected the distribution of flower colour variation among populations in La Réunion.
In this study, we analysed patterns of genetic structure in populations of B. occultum from Madagascar and La Réunion, using amplified fragment length polymorphisms (AFLPs), a dominant marker system with a wide coverage of the nuclear genome. Our primary aim was to elucidate the extent to which patterns of AFLP variability are shaped by the effects of intrinsic properties (mating system, dispersal ability), drift/migration dynamics (e.g., isolation-by-distance), recent human impact, historical habitat fragmentation and long-distance colonization. More specifically, we addressed the following questions: (1) Are there discernable effects of deforestation or habitat disturbance on levels of within-population genetic diversity in each island? (2) Is there a spatial component to the genetic structure in Madagascar explicable in terms of COE regions and/or glacial refugial isolation? (3) What is the extent of genetic differentiation between Madagascan and La Réunion populations? (4) Which past demographic processes have shaped population genetic diversity and structure in La Réunion (e.g., single or recurrent population founding, founder effects and subsequent genetic drift)? And (5) what is the extent of genetic differentiation between the two flower colour morphs in the latter island? Overall, this study may serve as a first genetic baseline for gaining insights into the population divergence history of an orchid species from the eastern rainforests of Madagascar and with a disjunct distribution in the Mascarenes.
Materials and methods
Study species
B. occultum is a herbaceous perennial orchid, with a diploid chromosome number (2n=36; Pridgeon et al., 2014; M Kiehn, Vienna University, unpublished data) and an epiphytic, or rarely lithophytic way of life (Cadet, 1989; GA Fischer, personal observation). Based on the 22 sample locations of this study (Table 1) plus 78 additional records based on all available geo-referenced herbarium specimens (mostly collected between 1932 and 1989; BR, G, K, MO, P, SZU, WU and www.tropicos.org) and our own field data (all coordinates available upon request), the species is mainly distributed in the (sub)humid coastal and montane forests of northern and eastern Madagascar (0–1500 m; 76 known sites) and (south)eastern La Réunion (0–620 m; 15 sites), with rare occurrences in Mauritius (5) and the Comoro Islands (4) (Figure 1). The conservation status of B. occultum has not been assessed yet by the IUCN (International Union for Conservation of Nature), but there is little doubt that ongoing deforestation activities especially in Madagascar have led to populations becoming progressively more vulnerable and threatened with extinction (GA Fischer, personal observation).
Table 1. Information on sample locations for 22 populations of B. occultum from Madagascar (M1–13) and La Réunion (R1–9) surveyed for AFLP variation.
| Population code | Locality (COE) | Longitude | Latitude | Altitude (m) | LPD/HT | NR/Y |
|---|---|---|---|---|---|---|
| Madagascar | ||||||
| M1 | Andapa 1 (2) | 49.6556 | −14.710 | 560 | 58.8 | — |
| M2 | Andapa 2 (2) | 49.6556 | −14.711 | 534 | 50.2 | — |
| M3 | Ambodirafiana (2) | 50.4582 | −15.269 | 20 | 77.4 | — |
| M4 | Pointe à Larée (2) | 49.7447 | −16.796 | 10 | 70.5 | — |
| M5 | Didy 1 (2) | 48.5583 | −18.133 | 1160 | 15.4 | — |
| M6 | Didy 2 (2) | 48.5569 | −18.069 | 1160 | 14.9 | — |
| M7 | Moramanga (2) | 48.3664 | −19.075 | 1004 | 51.9 | — |
| M8 | Ifanadiana 1 (3) | 47.7086 | −21.338 | 479 | 96.1 | — |
| M9 | Ifanadiana 2 (3) | 47.6192 | −21.326 | 468 | 97.7 | — |
| M10 | Tolongoina (3) | 47.5478 | −21.599 | 336 | 89.9 | — |
| M11 | Ikongo (3) | 47.4575 | −21.852 | 252 | 87.7 | — |
| M12 | Midongy (5) | 47.1289 | −23.499 | 140 | 83.8 | — |
| M13 | Andohahela (5) | 46.8597 | −24.747 | 547 | 18.8 | — |
| La Réunion | ||||||
| R1 | Jardin l'Eden | 55.6271 | −21.026 | 616 | 2 | 1/0 |
| R2 | Ravine Francoise | 55.6970 | −21.085 | 290 | 3 | 0/4 |
| R3 | Bois Blanc 1 | 55.8116 | −21.193 | 217 | 2 | 9/1 |
| R4 | Bois Blanc 2 | 55.8085 | −21.193 | 280 | 2 | 8/0 |
| R5 | Mare Longue 1 | 55.7379 | −21.351 | 288 | 1 | 8/1 |
| R6 | Mare Longue 2 | 55.7398 | −21.356 | 192 | 1 | 4/3 |
| R7 | Mare Longue 4 | 55.7447 | −21.359 | 127 | 1 | 2/4 |
| R8 | Mare Longue 3 | 55.7431 | −21.364 | 36 | 2 | 0/10 |
| R9 | Basse Vallée | 55.7071 | −21.361 | 268 | 1 | 1/5 |
Abbreviations: COEs, ‘Centres of Endemism' in Madagascar (Wilmé et al., 2006), with code numbers given in parentheses (see Figure 1 and text for details); HT, levels of habitat transformation per site in La Réunion, with categories (1–3) according to Strasberg et al. (2005): 1, relatively untransformed (native canopy still pristine and not invaded); 2, moderately degraded (native biota dominated by alien plants); 3, transformed (urban or cultivated areas, and alien secondary vegetation); LPD, log-transformed percentages of deforestation for the period 1950–2005 per site in Madagascar; NR/Y, number of red vs yellow flower-coloured individuals censused per site in La Réunion.
B. occultum produces one or rarely few inflorescences, which arise from pseudobulbs and bear many (ca. 20–40) small flowers (ca. 5 mm). The flower colour appears to be uniformly red in Madagascar (Cribb and Hermans, 2010; GA Fischer, personal observation), but individuals with either red or yellow flowers occur in La Réunion, yet without any other discernable morphological difference (Cadet, 1989; T Pailler, U Jaros, personal observations). Most species of Bulbophyllum are self-compatible and fly (or rarely bee) pollinated (van der Cingel, 2001; Tan et al., 2006; Liu et al., 2010; Humeau et al., 2011). However, direct observations of insect visitors in B. occultum, or related species from the study area, are lacking. In fact, field observations, pollination/bagging experiments and floral-morphological studies in B. occultum indicate that most, if not all, individuals from La Réunion are capable of autonomous within-flower self-pollination (‘auto-pollination' sensu Catling, 1990) due to the lack of the ‘rostellum' (i.e., a modified stigmatic lobe that usually serves as a physical barrier to prevent auto-pollination), whereas both outcrossing and selfing morphs (with and without a rostellum, respectively) occur in Madagascar, albeit at unknown frequencies (Jacquemyn et al., 2005; Gamisch et al., 2014, 2015; U Jaros, personal observation). We therefore anticipated that the genetic data of this study would provide us with some additional clues about the species' mating system, while realizing that dominant AFLPs are less suited for this purpose compared with co-dominant markers.
Plant material and sampling design
In 2008, silica-dried leaf material of B. occultum was collected from 13 populations in Madagascar (M1–13; n=133) and, in 2010, from nine populations in La Réunion (R1–9; n=92), that is, a total of 22 populations and 225 individual plants (see Figure 1 and Table 2). All necessary collecting permits were obtained by the Département des Eaux et Fôrets (Madagascar) and the Parc National de La Réunion (sampling permit code: DIR/I/2010/028). B. occultum has short internodes and thus probably little potential of vegetative propagation through fragmentation of creeping rhizomes (Batygina et al., 2003). Different individuals are easily discriminated by their characteristic growth habit comprising the whole spectrum from small to larger bulbs (GA Fischer, U Jaros, personal observations). Nonetheless, care was taken to collect leaf material from putative genets from different host trees or widely separated branches within a tree. Mean (±s.d.) geographical distances between sampling sites were 331.8 (±213.4) km in Madagascar and 12.4 (±8.2) km in La Réunion. In Madagascar, our sampling of northern (M1–7), central (M8–11) and southern (M12/13) populations covers three (out of five) COE regions in the eastern portion of the island as defined by Wilmé et al. (2006), that is, South Bernarivo/North Mangoro (COE code no. 2), South Mangoro/North Manampatrana (no. 3) and South Mananara (no. 5), respectively (see Figure 1 and Table 1). Within each La Réunion site, we recorded the flower colour of each sampled individual in flower (red or yellow; n=61 in total). Spirit-preserved vouchers of populations analysed in this study are held at the herbarium of Salzburg University (SZU).
Table 2. Summary of AFLP variation for 22 populations (206 individuals) of B. occultum from Madagascar (M1–13) and La Réunion (R1–9).
| Population codea | N | NA | PPF | HE | DW b |
|---|---|---|---|---|---|
| Madagascar | |||||
| M1 | 13 | 1.078 | 36 | 0.095 | 0.422 |
| M2 | 6 | 0.863 | 17 | 0.069 | 0.434 |
| M3 | 11 | 0.951 | 28 | 0.090 | 0.273S |
| M4 | 6 | 0.922 | 21 | 0.091 | 0.466 |
| M5 | 9 | 0.912 | 22 | 0.076 | 0.319 |
| M6 | 10 | 0.892 | 18 | 0.075 | 0.438 |
| M7 | 5 | 0.794 | 13 | 0.061 | 0.539 |
| M8 | 11 | 1.000 | 29 | 0.100 | 0.357 |
| M9 | 11 | 1.020 | 30 | 0.090 | 0.305S |
| M10 | 10 | 0.863 | 18 | 0.045 | 0.363 |
| M11 | 14 | 0.931 | 26 | 0.050 | 0.385 |
| M12 | 14 | 1.029 | 30 | 0.103 | 0.396 |
| M13 | 7 | 0.863 | 19 | 0.079 | 0.492 |
| Regional total | 127 | 1.56 | 71 | 0.115 | — |
| Regional mean | 9.8 (3.0) | 0.932 (0.081) | 24 (6.7) | 0.079 (0.031) | 0.394 (0.069) |
| La Réunion | |||||
| R1 | 10 | 0.941 | 22 | 0.055 | 0.838L |
| R2 | 8 | 0.902 | 21 | 0.059 | 0.425 |
| R3 | 10 | 0.765 | 11 | 0.026 | 0.316S |
| R4 | 9 | 0.755 | 13 | 0.032 | 0.315 |
| R5 | 9 | 0.824 | 16 | 0.039 | 0.379 |
| R6 | 7 | 0.922 | 26 | 0.117 | 0.247S |
| R7 | 8 | 1.010 | 30 | 0.124 | 0.441 |
| R8 | 10 | 0.814 | 13 | 0.044 | 0.750L |
| R9 | 8 | 0.863 | 16 | 0.070 | 0.626L |
| Regional total | 79 | 1.49 | 61 | 0.089 | — |
| Regional mean | 8.8 (1.1) | 0.866 (0.085) | 19 (6.5) | 0.063 (0.026) | 0.482 (0.208) |
| Species total | 206 | 1.84 | 86 | 0.116 | — |
| Species mean | 9.4 (2.4) | 0.91 (0.087) | 22 (6.9) | 0.072 (0.028) | 0.432 (0.147) |
Abbreviations: DW, frequency down-weighted marker values; HE, Nei's (1987) unbiased gene diversity; N, number of individuals assayed; NA, observed number of alleles; PPF, percentage of polymorphic fragments.
Note: Standard deviations are given in parentheses.
Population codes are identified in Table 1.
Significantly small or large DW values are indicated by superscripts 'S' and 'L', respectively.
DNA extraction and AFLP genotyping
For each individual, total genomic DNA was extracted from the dried leaf tissue using the cetyltrimethylammonium bromide method (Doyle, 1991). The AFLP procedure followed the protocol of Vos et al. (1995) with minor modifications that included the use of fluorescent-dye-labelled primers (MWG Biotech, Ebersberg, Germany). An initial primer trial was conducted on eight individuals from two geographically distant populations (M13, R8) using nine selective primer pair combinations with three selective nucleotides (data not shown). Based on this, the following primer combinations that produced the best results with respect to polymorphism and clarity of AFLP profiles were chosen for the complete survey: EcoRI-AGA/MseI-CTG (6-FAM), EcoRI-AGG/MseI-CAG (VIC) and EcoRI-AGC/MseI-CTC (NED). Each PCR plate (48 slots) contained one positive control and one blind sample. The selective PCR products were purified using Sephadex G-50 Superfine (GE Healthcare BioSciences, Uppsala, Sweden) following the manufacturer's instructions. AFLP fragments were separated on a MegaBACE 1000 (GE Healthcare Biosciences, Pittsburgh, PA, USA), using the ET550-R-Rox-MegaBACE sizing standard, and manually scored with the aid of DAX version 8.0 (Van Mierlo Software Consultancy, Eindhoven, The Netherlands). Each marker was coded as 1 or 0 for presence or absence in an individual, resulting in a binary data matrix. We also performed separate test runs with 13 individuals (from 11 different populations) used as replicates. This resulted in a low overall error rate (2%), calculated as the percentage of differently scored loci over all replicates and the three primer pair combinations (Bonin et al., 2004). In total, 206 individuals were finally available for data analysis because 19 individuals failed to amplify in at least one of the three primer combinations. Accordingly, the mean (±s.d.) sample size was 9.4 (±2.4) individuals per population (see Table 2).
Genetic diversity analyses of the sampled populations
In order to assess levels of genetic diversity, we calculated for each population the observed number of alleles (NA), the percentage of polymorphic fragments (PPF) and the number of AFLP phenotypes, using GENALEX version 6.5 (http://biology-assets.anu.edu.au/GenAlEx; Peakall and Smouse, 2006, 2012,). A fragment was considered polymorphic if at least one individual showed a variant pattern. In addition, Nei's (1987) unbiased gene diversity (HE) for small sample sizes and frequency down-weighted marker (DW) values (or ‘rarity-indices') were estimated using the AFLPDAT R package (Ehrich, 2006). Large values of DW imply a high amount of rare AFLP markers as expected for long-term isolated populations, whereas populations affected by genetic drift and/or founder effects exhibit small DW values (Schönswetter and Tribsch, 2005; Skrede et al., 2009). For DW, 95% confidence intervals (CIs) were obtained by 9999 bootstrap replicates over loci. Significantly small or large values of DW (lying outside the 95% CIs) were assessed by a permutation procedure in AFLPDAT using 1000 replicates. Significant differences in mean genetic diversities between Madagascar and La Réunion were tested by an exact Wilcoxon rank-sum test in R version 3.0.3 (R Development Core Team, 2012).
Testing effects of deforestation and habitat disturbance on genetic diversity
For the Madagascan populations, we used linear regression models in R to test for a negative relationship between measures of diversity (NA, PPF, HE, DW), as response variables, and the (log-transformed) percentage of deforestation (LPD) in sampled areas for the period 1950 to 2005, as possible predictor variable (see Table 1). Normality of the distribution of the response variables was confirmed using the Shapiro–Wilk test in R (all P-values >0.1). For estimating LPD, we used forest cover and change data for Madagascar (Harper et al., 2007; https://learning.conservation.org/spatial_monitoring/Pages/data_access.html), including digitalized aerial maps of 1950 (Humbert and Cours Darne, 1965) and Landsat-type images from NASA's GeoCover Project of 2005 (Tucker et al., 2004), to calculate the relative change (i.e., loss) in forest cover within a 10-km radius around each site over this period (1950–2005). As similar data are not available for La Réunion, we classified each population there according to three categories of ‘habitat transformation' (HT) proposed for this island by Strasberg et al. (2005) (see also Lagabrielle et al., 2011): (1) relatively untransformed (native canopy still pristine and not invaded); (2) moderately degraded (native biota dominated by alien plants); and (3) transformed (urban or cultivated areas, and alien secondary vegetation) (see Table 1). Significant differences in genetic diversity parameters among these categories were assessed using a Kruskal–Wallis rank-sum test in R.
Population genetic structure, boundaries, and relationships
To identify the most likely number of genetic groups (K) in the entire AFLP data set, we applied the K-means clustering technique of Hartigan and Wong (1979) using the R script of Arrigo et al. (2010). Based on initial trials, the number of K was restricted to vary from 1 to 10 (i.e., less than the number of populations sampled), first, to avoid unstable groups of small numbers of individuals with increasing numbers of K (set up to 22; data not shown), and, second, to capture the major underlying population structure of the data but without overestimating it (Pritchard et al., 2000). For each value of K, we performed 100 000 independent runs, starting at different random points. To identify the most likely number of groups, we calculated values of ΔK (Evanno et al., 2005) following the method of Arrigo et al. (2010). As a proxy of clustering accuracy, we used the intergroup inertia criterion, that is, the average of the distances between the centroid of each cluster and each sample contained in it (Kergoat and Alvarez, 2008; Espíndola and Alvarez, 2011). We opted for this non-model-based K-means algorithm because, in contrast to alternative model-based algorithms such as implemented in STRUCTURE (Pritchard et al., 2000), it does not assume populations to be in Hardy–Weinberg equilibrium, an assumption probably violated in B. occultum due to its potential capacity of auto-pollination (Gamisch et al., 2014, 2015; see above).
We used the program BARRIER version 2.2 (Manni et al., 2004) to identify geographical locations where major genetic boundaries among populations might occur, based on their pairwise genetic (FST) distances (Supplementary Appendix Table S1). The number of barriers was set to vary from 1 to 10, reflecting their descending order of relative importance (‘priority') for genetic dispersion (Manni et al., 2004). FST was calculated using a fragment–frequency approach in AFLP-SURV version 1.0 (Vekemans et al., 2002). This non-allelic method assumes fixed homozygosity at each locus (e.g., resulting from self-pollination), which may overestimate FST for highly outcrossing species, but is more likely to meet expectations for B. occultum (see above). Statistical support for each barrier was tested by means of 1000 bootstrapped genetic distance matrices generated in AFLP-SURV. Barriers with high (⩾80%) bootstrap support were retained for further analysis.
To visualize the genetic (FST) distances between populations, we performed a principal coordinates analysis (PCoA) in GENALEX. In addition, we constructed a mid-point rooted neighbour-joining phenogram in PHYLIP version 3.69 (Felsenstein, 2005) based on pairwise FST values, with bootstrap support obtained by resampling with replacement over loci (1000 replicates).
Population genetic differentiation and tests of isolation-by-distance
Non-hierarchical analyses of molecular variance (AMOVA) were performed in GENALEX to estimate genetic differentiation among populations (ΦPT), overall and within each island. For each data set, ΦPT values were also calculated in a pairwise fashion. Hierarchical AMOVAs were further used to quantify the partitioning of genetic variance between predefined groups of populations (ΦCT), and between populations within such groups (ΦSC), with populations grouped according to island, areas delimited by genetic (BARRIER) boundaries or COE region (only Madagascar). Significance levels of both Φ-statistics and pairwise ΦPT values were based on 9999 permutations. Effects of isolation-by-distance, as usually caused by restricted gene flow (Daïnou et al., 2010), were tested for each island separately by regressions of ΦPT/(1−ΦPT) against log-transformed geographic distance for all pairs of populations (Rousset, 1997). Significance of regression slopes was evaluated by Mantel tests with 9999 random permutations in GENALEX.
Comparison of AFLP variation and colour morph differentiation in La Réunion
To examine the genetic divergence between red- and yellow-flowered morphs from La Réunion, we computed a PCoA in GENALEX on the AFLP-derived Euclidian distance matrix between individuals and labelled with the K-means groups. Statistical association was further assessed through a contingency table by comparing the occurrence of colour morphs and K-means groups using χ2 tests.
Results
Population genetic diversity
Scoring the sampled material of B. occultum (22 populations, 206 individuals) for three AFLP primer combinations resulted in 102 unambiguous fragments between 75 and 538 bp, of which 88 were polymorphic (PPF=86.3% Table 2). There was some variation in the number of polymorphic fragments per primer pair (‘6-FAM': 36; ‘VIC': 30; ‘NED': 36), with an average of 34.0 (±3.5 s.d.) fragments per combination.
Notably, 139 individuals surveyed had unique AFLP phenotypes (i.e., fragment combinations), while the remainder (67) had redundant ones. However, the great majority of these redundant phenotypes (18 in total) were either scattered across multiple populations (even between islands) or mostly occurred in only two copies if confined to a single population (data not shown). When combined with the species' low capacity for rhizome fragmentation (see Materials and Methods), these patterns strongly suggest that the redundant AFLP phenotypes observed do not result from vegetative propagation but rather reflect low resolution of the AFLP markers in combination with high levels of inbreeding (Dasmahapatra et al., 2008; O'Brien et al., 2014; see also Discussion).
Levels of variation within populations (in terms of NA, PPF and HE) varied among populations but were generally low (Table 2). For example, within-population gene diversity (HE) ranged from 0.026 to 0.124, with both values recorded in La Réunion (pops. R3 vs R8), and averaged 0.072 (±0.028 s.d.) across all populations. The total (species-wide) HE was estimated to be 0.116. Populations in La Réunion displayed on average slightly lower HE values (0.063) than those in Madagascar (0.079), but the differences were not significant (P=0.164), and the same was true for NA and PPF (P=0105 and 0.098, respectively). The mean proportion of rare AFLP markers was of comparable magnitude in Madagascar (DW=0.394±0.069 s.d.) and La Réunion (DW=0.482±0.208 s.d.). However, DW values varied considerably in the latter island, where two populations featured significantly small (R3, R6) and three large (R1, R8, R9) values. In Madagascar, only two populations (M3, M9) had significantly small DW values (Table 2).
Effects of deforestation and habitat disturbance on genetic diversity
In Madagascar, the mean log-transformed percentage of deforestation (LPD) for the period 1950–2005, as estimated across the 13 sample sites, was 63% (±31% s.d.) (see Table 1). However, linear regression models revealed no significant relationship between measures of within-population genetic diversity (NA, PPF, HE) or allelic rarity (DW) and LPD (P=0.131–0.335). In La Réunion, we classified four of the nine sample sites as being ‘untransformed' (R5–7, R9) and the remaining ones as ‘moderately degraded' or ‘transformed' (only R2) (see Table 1). Yet, again, none of the above genetic diversity parameters were significantly correlated with these qualitative measures of habitat transformation (Kruskal–Wallis tests: P=0.134–0.867), whereby results remained unchanged after excluding R2 (P=0.080–0.885).
Population genetic structure, boundaries and relationships
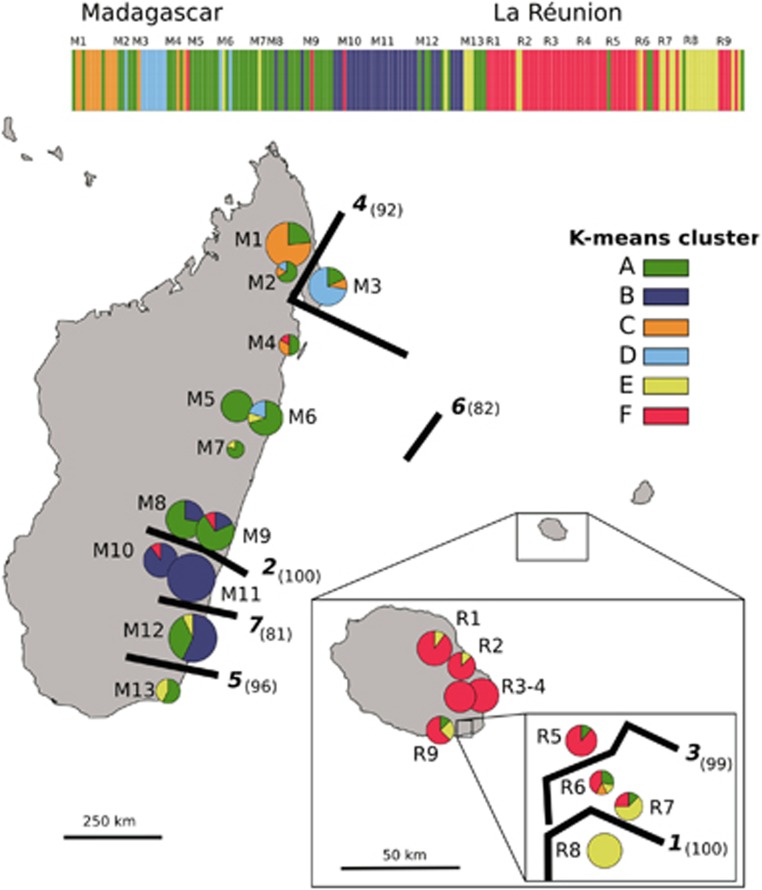
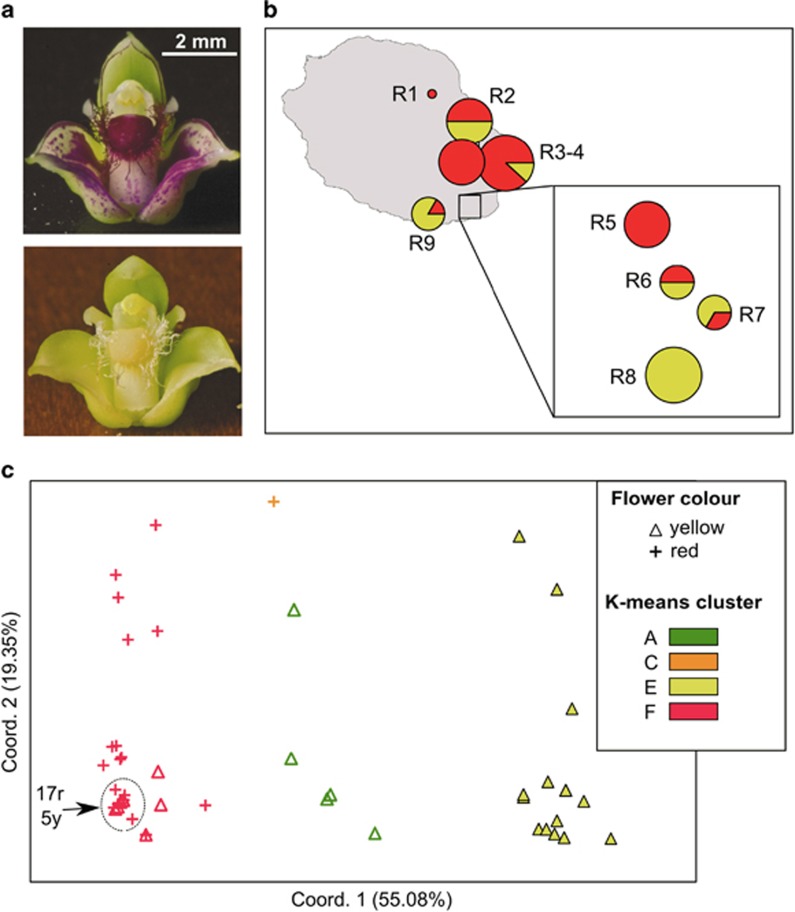
Based on the K-means analysis of the entire data set, K=2 was suggested as the most appropriate number of groups (highest ΔK value), but this grouping identified only certain populations from the South of Madagascar (i.e., M8–12, but excepting M13) as genetically distinct or admixed; by contrast, values of the intergroup inertia (viz. clustering accuracy) progressed to a plateau for levels of K≈6 (see Supplementary Appendix Figure S1). We decided on K=6 as a reasonable solution as the geographical structure captured by this finer grouping also appeared to be associated with several genetic boundaries detected with BARRIER (Figure 2; see below). In detail, all six gene pools (A–F) were present in Madagascar, where most individuals were assigned to groups A (45%) or B (28%), and to a lesser extent C (11%) and D (9%), whereas the remainder (E, F) accounted for ⩽5% of the regional total (Figure 2). Although gene pool A occurred in most populations (except M10/11), it was clearly predominant in North-Central Madagascar, whereas group B was represented in the South (M8–M12). Gene pools C (11%) and D (9%) occurred at relatively high frequency in populations from the North (M2) and Northeast (M3), respectively, and the same applied to group E in the Far South (M13). Four out of the six gene pools (excepting B and D) were also present in La Réunion, but there the majority of individuals were assigned to gene pools F (67%) and E (25%). Individuals of gene pool E group were mainly confined to the South (R6–9), where groups A and C also occurred at low frequencies (⩽8%).
Figure 2.
Distribution of six AFLP gene pool clusters (A–F) within and among 22 populations (206 individuals) of B. occultum from Madagascar (M1–13) and La Réunion (R1–9) as identified by K-means clustering, and with pie charts representing the percentage of individuals. The sizes of the pie charts are proportional to sample size (n), with the smallest circle representing n=5 and the largest representing n=14 (see Table 2). The barplot at the top displays the assignment of individuals to the six K-means clusters, with the smallest vertical bar representing one individual. Significant regions of genetic discontinuity (black lines) identified by BARRIER are numbered (1–7) in descending order of relative importance (‘priority'), with bootstrap values (⩾80%) shown in parentheses (1000 replicates).
The BARRIER analysis identified seven barriers (1–7) of high support (bootstrap support 81–100%) that documented an abrupt rate of change in the genetic profile of populations across the species' range (see Figure 2). In detail, two barriers of upper (1) and high (3) priority were found in the South of La Réunion, and isolated, respectively, a population (R8) fixed for gene pool E, and two nearby ‘admixed' populations (R6, R7) comprised of several gene pools. In Madagascar, the strongest genetic boundary (2) separated North-Central (M1–9) and South (M10–13) populations, whereas barriers of lesser priority marked-down the peripheral populations in the Northeast (4: M3) and Far South (5: M13). Only the sixth boundary separated Madagascan and La Réunion populations, whereas the seventh further isolated a single population (M12) in the South of Madagascar (Figure 2).
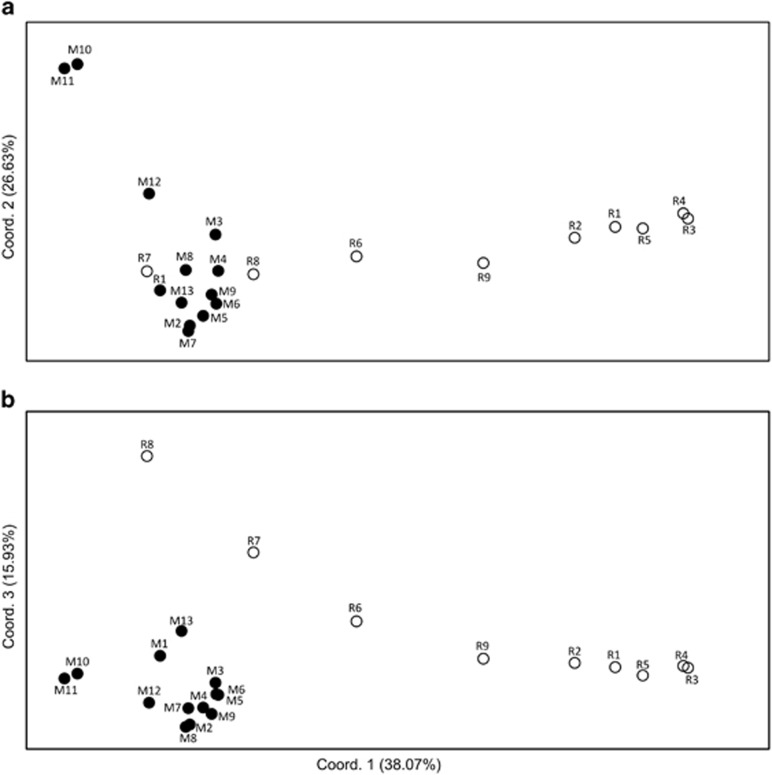
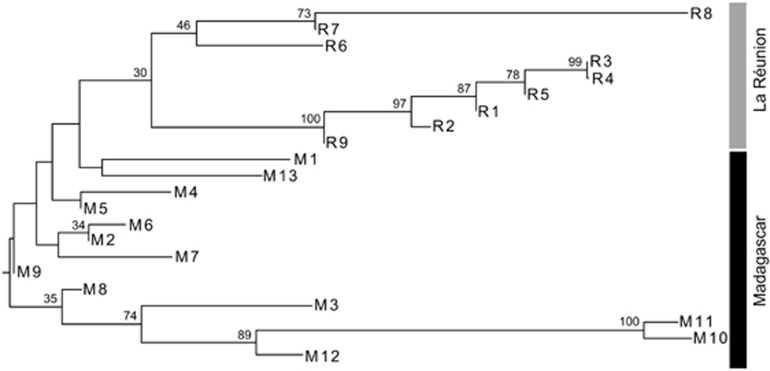
In the two-dimensional PCoA plot (Figure 3a), the three genetically deviant populations from the South of La Réunion (R6–8) were grouped, along the first axis, either among populations from Madagascar (R7, R8) or between the latter and the rest of La Réunion populations (R6). However, taking the third axis into account (Figure 3b), populations R7 and R8 were clearly distinguishable from their Madagascan conspecifics. In the neighbour-joining phenogram (Figure 4), all populations from La Réunion (R1–9) formed a group nested within those from Madagascar, but without receiving strong support bootstrap (30%). Nonetheless, this analysis confirmed (i) the distinct genetic identity of populations M10–12 from South Madagascar in general (89%), and M10/11 in particular (100%); and (ii) the existence of a local cluster of highly differentiated or ‘admixed' populations in the South of La Réunion (R6–8; 46%), different from the one in adjacent areas or further north (R1–5, R9; 100% see also Figure 2).
Figure 3.
Plots of (a) the first and second, and (b) the first and third principal coordinates of 22 populations of B. occultum from Madagascar (M1–13; filled circles) and La Réunion (R1–9; open circles) based on pairwise FST values derived from fragment frequencies of 102 AFLP markers using AFLP-SURV. See Table 1 for explanation of population codes.
Figure 4.
Mid-point rooted neighbour-joining (NJ) phenogram depicting genetic (FST) distances between 22 B. occultum populations from Madagascar (M1–13) and La Réunion (R1–9). Numbers above branches indicate bootstrap values (⩾30%) based on 1000 replicates. See Table 1 for explanation of population codes.
Population genetic differentiation and isolation-by-distance
Non-hierarchical AMOVAs (Table 3) confirmed the high genetic differentiation over all populations of B. occultum (ΦPT=0.387), but further revealed very similar levels of population subdivision in Madagascar and La Réunion (ΦPT=0.341 vs 0.359). At the range-wide scale, 203 of 231 pairwise ΦPT comparisons (88%) had permutation test P-values <0.05, and the number of significant pairwise values was also high within Madagascar (62/78=79%) but lower in La Réunion (24/36=67%) (Supplementary Table S1). When accounting for the species' significant inter-island substructure (ΦCT=0.153, P<0.001), overall levels of population divergence still remained high (ΦSC=0.332) (Table 3). However, the strongest signature of range-wide spatial differentiation was obtained (ΦCT=0.330) when populations were assigned to the eight areas delimited by genetic (BARRIER) boundaries, that is, five in Madagascar and three in La Réunion (Figure 2b). When Madagascan populations were analysed separately, and grouped according to these genetic boundaries, the extent of regional subdivision was much stronger (ΦCT=0.306) compared with the grouping based on COEs (ΦCT=0.073). In each island, there were no detectable effects of isolation-by-distance (Madagascar/La Réunion: P=0.144/0.464).
Table 3. Non-hierarchical and hierarchical AMOVAs for AFLP variation based on several groupings of B. occultum populations from Madagascar and La Réunion.
| Grouping/source of variation | df | Sum of squares | Variance components | Percentage of total variance | Φ-Statistic a |
|---|---|---|---|---|---|
| Total | |||||
| Among populations | 21 | 533.183 | 2.33 | 38.7 | ΦPT=0.387 |
| Within populations | 184 | 677.71 | 3.68 | 61.3 | NC |
| Madagascar | |||||
| Among populations | 12 | 290.335 | 2.05 | 31.9 | ΦPT=0.341 |
| Within populations | 114 | 447.161 | 3.96 | 68.1 | NC |
| La Réunion | |||||
| Among populations | 8 | 143.034 | 1.7 | 35.9 | ΦPT=0.359 |
| Within populations | 70 | 211.953 | 3.03 | 64.1 | NC |
| Madagascar vs La Réunion | |||||
| Among regions | 1 | 118.41 | 1 | 15.3 | ΦCT=0.153 |
| Among populations | 20 | 414.773 | 1.83 | 28.1 | ΦSC=0.332 |
| Within populations | 184 | 677.71 | 3.68 | 56.6 | ΦPT=0.430 |
| Eight barriers (total) b | |||||
| Among regions | 7 | 406.405 | 2.1 | 33 | ΦCT=0.330 |
| Among populations | 14 | 126.778 | 0.6 | 9.3 | ΦSC=0.142 |
| Within populations | 184 | 677.71 | 3.68 | 57.7 | ΦPT=0.420 |
| Five barriers (Madagascar) b | |||||
| Among regions | 4 | 201.057 | 2.03 | 30.6 | ΦCT=0.306 |
| Among populations | 8 | 70.682 | 0.52 | 7.8 | ΦSC=0.110 |
| Within populations | 114 | 465.757 | 4.09 | 61.6 | ΦPT=0.380 |
| Three COEs (Madagascar) c | |||||
| Among regions | 2 | 79.277 | 0.45 | 7.3 | ΦCT=0.073 |
| Among populations | 10 | 192.462 | 1.61 | 26.2 | ΦSC=0.281 |
| Within populations | 114 | 465.757 | 4.09 | 66.5 | ΦPT=0.340 |
Abbreviations: AMOVAs, analyses of molecular variance; df=degrees of freedom; NC=not computed.
All Φ-values were significant (P<0.001) based on 9999 permutations.
Population grouping based on areas defined by the BARRIER analysis (see Figure 2).
Population grouping based on three ‘Centres of Endemism' (COEs; Wilmé et al., 2006) (see Table 1).
Colour morph differentiation in La Réunion and its association with AFLP variation
Across the nine La Réunion populations, we censused 33 individuals that had red flowers and 28 that bore yellow flowers (Figure 5a), based on a sample size range of 1–10 individuals per population (Table 1). Notably, all flowers within a given individual displayed the same colour (U Jaros, personal observation). Among the eight populations surveyed with n⩾4, four were mixed (red/yellow) and two each were uniformly coloured red or yellow, whereby yellow individuals appeared to be more frequent in the South (Figure 5b). A PCoA conducted on the AFLP profiles available for 33 red and 28 yellow coloured morphs (Figure 5c) revealed they were largely distinguishable along the first axis, with only few exceptions. We also tested whether the PCoA results changed as we entered the AFLP data of the remaining 18 individuals from La Réunion with unknown flower colour, but the clustering patterns were essentially the same (Supplementary Appendix Figure S2). As apparent from Figure 5c, red morphs were almost exclusively associated with gene pool F (32 individuals, plus one with C), whereas yellow morphs were mostly assigned to E (15) but also F (8) and A (5). These associations were highly significant when analysed both globally (χ2=35.23, df=3, P<0.001) and for only the predominant gene pools, E and F (χ2=25.50, df=1, P<0.001).
Figure 5.
(a) Red and yellow flower colour morphs of B. occultum from La Réunion. Photo credit: Joseph Vaillant (Parc National de La Réunion). (b) Their distribution (denoted by red vs yellow colour) censused across the nine populations (R1–9) surveyed for AFLP variation in La Réunion. The sizes of the pie charts are proportional to sample size (n), with the smallest circle representing n=1 and the largest n=10 (total n=61; see Table 1). (c) Plot of the first and second principal coordinates of 33 red morphs (crosses) and 28 yellow morphs (triangles) from populations R1–9 based on Euclidian distances derived from AFLP data. Individuals are colour-labelled according to the four AFLP groups detected by K-means clustering (see Figure 2). Numbers refer to densely clustered individuals with red (r) vs yellow (y) flower colour morphs within the dotted circle.
Discussion
In Madagascar, the population divergence history of rainforest-dwelling pant species is still poorly known because of the scarcity of population genetic studies of respective keystone species across the eastern escarpment of the island (see Andrianoelina et al., 2006, 2009; Shapcott et al., 2007, 2012,). This study is the first to provide insights into the patterns of population genetic structure of an epiphytic orchid, B. occultum, which is mainly distributed across the eastern rainforest corridor of Madagascar but also occurs at relatively high frequency in the small Mascarene Island of La Réunion. However, there are potential limitations of this study, including the limited number of polymorphic AFLP fragments scored, and the small sample sizes of some populations (especially in Madagascar). With these caveats in mind, our results nonetheless suggest that the overall genetic structure and diversity of this orchid conforms to expectations for a mainly selfing species with a non-equilibrium (i.e., historically drift-dominated) population system. Rather than revealing effects of recent human impact, this genetic structure contains signatures of long-term habitat isolation in Madagascar and long-distance colonization effects in La Réunion. Somewhat unexpectedly, our data revealed no reduced genetic diversity in the latter island but rather strong population subdivision, which tends to be associated with a divergence in phenotype (flower colour). In the following, we discuss the evidence for these inferences and their wider implication for a general understanding of the species' population history.
Population differentiation and within-population diversity
Our survey of AFLP variation in B. occultum revealed exceptionally high levels of population subdivision (ΦPT), both at the species' range-wide scale (0.387) and within each island (Madagascar/La Réunion: 0.341/0.359; Table 3). These values are at variance with the rather low genetic differentiation among populations reported for orchids in general (e.g., Tremblay et al., 2005) and epiphytic species in particular (e.g., mean FST=0.146 vs 0.109 at allozyme loci; Phillips et al., 2012), which is usually ascribed to extensive long-distance gene flow facilitated by the dust-like and wind-dispersed seeds of these plants (Arditti, 1992; Trapnell and Hamrick, 2004; Vanden Broeck et al., 2014; but see Kartzinel et al., 2013; Pinheiro et al., 2013). In addition, we observed remarkably low levels of genetic diversity within all populations of B. occultum (e.g., HE, range: 0.026–0.124, mean: 0.072; Table 2). These values are much lower than those observed from allozyme analyses in pollinator-dependent Bulbophyllum species from South America (HE, range: 0.391–0.612; Azevedo et al., 2007; see also Ribeiro et al., 2007). Rather, they are strikingly similar to the average value obtained for mainly selfing species screened with dominant marker systems (HE≈HPOP=0.12; Nybom and Bartish, 2000; Nybom, 2004). This is in further agreement with field, experimental and morphological observations that most individuals of B. occultum in La Réunion, and a yet undetermined proportion in Madagascar, are capable of auto-pollination (Jacquemyn et al., 2005; Gamisch et al., 2014, 2015; U Jaros, personal observation).
Taken at face value, the above results suggest that restricted gene flow via pollen and seed, combined with high rates of selfing, are major determinants of the relatively increased population divergence and reduced within-population diversity observed in B. occultum, when compared with most other orchid species (e.g., Tremblay et al., 2005; Phillips et al., 2012; and above). We caution, however, that dominant AFLP markers provide no measure of inbreeding, and it remains to establish whether our inferences on mating system are robust to surveys using co-dominant markers. In addition, our data cannot fully disentangle the relative contributions of contemporary vs historical factors (e.g., gene flow/drift vs range fragmentation/colonization dynamics) on the species' current genetic structure. Remarkably, however, in each island, we did not detect a pattern of isolation-by-distance. When combined with the high overall values of population subdivision (see above), and the findings based on pairwise ΦPT comparisons (Supplementary Table S1), these data suggest that the great majority of B. occultum populations in Madagascar, and still a considerable number of those in La Réunion, are effectively isolated from each other, and that genetic drift, in general, had a larger historical role in shaping population structure compared with limited gene flow alone (Hutchison and Templeton, 1999). Hence, as further discussed below, certain historical events accompanied by drift, must have resulted in non-equilibrium situations that are still detectable in the current genetic structure of this tropical orchid.
Effects of recent habitat fragmentation and disturbance
Barring undetected levels of inbreeding (see above), the consistently low within-population genetic diversity observed in B. occultum could result from recent human-mediated deforestation and increased risks of habitat disturbance. The underlying rationale is that decreasing population size in fragmented or otherwise impacted populations may result in genetic erosion through the loss of alleles by drift, especially rare ones (Young et al., 1996; Erikson and Ehrlén, 2001; Vandepitte et al., 2012). However, we did not find a significantly negative effect of recent (1950–2005) deforestation (LPD) on allelic rarity (DW) in Madagascar, nor was such effect registered by other measures of genetic diversity despite nearly all sites were affected by deforestation to considerable extent (Table 1). Likewise, we did not find an effect of habitat transformation (measured as categorical variable) on any genetic diversity parameter in La Réunion. Comparable to this study, no significant differences in genetic diversity values were recently found between codified ‘fragmented' and ‘non-fragmented' populations of Dalbergia monticola (Fabaceae), an endangered insect-pollinated rosewood tree of the eastern rainforest of Madagascar, using nuclear and chloroplast microsatellites (Andrianoelina et al., 2009). It was suggested that habitat fragmentation has occurred too recently (<100 years) in this tree species relative to the number of generations passed (about two) so that the effects of genetic drift have not become apparent yet (see also Young et al., 1993; Victory et al., 2006; Kramer et al., 2008). A similar explanation, however, appears less likely for B. occultum, given its probably short generation time (see Introduction). Rather, we suspect that the species' capacity for auto-pollination has buffered genetic responses to such recent fragmentation, as reported for other mainly selfing species (Honnay and Jacquemyn, 2007). Hence, based on the present AFLP data, it appears that recent (anthropogenic) habitat fragmentation and degradation had no effects on the genetic diversity characteristics of B. occultum, even though this requires further testing using both an extended population sample and additional genetic markers.
Inferences of population history in Madagascar
Most relevant to inferring the historical population dynamics of B. occultum are the spatial patterns of AFLP diversity revealed by the K-means clustering and BARRIER analyses, which identified remarkably high numbers of gene pools (A–F) and genetic boundaries (1–7) across the species' range (Figure 2). Considering Madagascar first, BARRIER detected five zones of abrupt genetic change across the eastern rainforest corridor, but none of those breaks seems to coincide with apparent landscape features that may act as physical barriers to gene flow (i.e., certain mountain ranges, river systems, etc.). In addition, there was little spatial component to the genetic structure of Madagascan populations when they were grouped according to ‘Centres of Endemism' (COEs; ΦCT=0.073) (Table 3), that is, hypothesized Quaternary glacial refuge locations of endemic vertebrate taxa (Wilmé et al., 2006). This weak concordance may simply reflect that plant and animal species varied in their patterns of distribution over the last glacial cycles, but could also mean that the COEs are too broadly defined in space relative to the more fine-scaled genetic structure identified here. Nonetheless, our data seem to support the hypothesis that Madagascan B. occultum persisted across multiple isolated regions along the eastern rainforest throughout recent climatic cycles, leading to genetic divergence among populations from the Northeast, North-Centre, South and Far South (Figure 2). This scenario of long-term population isolation (or ‘insularization') is also in agreement with the lack of isolation-by-distance at high ΦPT (see above) and the broadly equivalent levels of diversity observed among these populations (Table 2). For example, in case of post-glacial (re-)colonization of the eastern corridor from a single refuge area, such as the one proposed recently on the basis of palaeo-climatic data for palm trees in the Northeast of Madagascar (Rakotoarinivo et al., 2013), we would have expected a latitudinal cline in diversity in the direction of spread (e.g., Hewitt, 2004), but this is not observed (e.g., regression of HE on latitude: R2=0.0104, P=0.741). Even though the primary vicariant factors initiating these multiple genetic boundaries in Madagascan B. occultum remain unclear, it is feasible that they relate to rainforest habitat contraction and isolation events caused by aridification of coastal/lowland sites during glacial maxima (Wilmé et al., 2006; Vences et al., 2009; Rakotoarinivo et al., 2013). This scenario is perhaps best exemplified by the genetic distinctiveness of a coastal population (M3) from the Northeast (Figure 2). However, in contrast to the ‘watershed' hypothesis (Wilmé et al., 2006), the present data predict that during glacial maxima, when the climate conditions were probably cooler and drier in Madagascar (Burney et al., 2004), rainforest species tolerant to water stress, such as epiphytic bulbous orchids (Hermans et al., 2007; Silvera et al., 2009), were able to persist not only in coastal/lowland areas associated with riverine habitats of locally more mesic conditions but also at higher elevation/montane sites along the island's eastern escarpment.
Colonization history and population differentiation in La Réunion
Hierarchical AMOVA (Table 3) suggested moderately strong differentiation between B. occultum populations from Madagascar and La Réunion (ΦCT=0.153), which were also separated by BARRIER (Figure 2). Although the K-means analysis indicated that four out of the six Madagascan gene pools (A–F) were also present in La Réunion (excepting B and D), most individuals there were assigned to gene pools E and F, which otherwise were extremely rare in Madagascar (Figure 2). Finally, the PCoA (Figure 3) highlighted a gradual pattern of inter-island population divergence, while the neighbour-joining analysis (Figure 4) identified all La Réunion populations as a weakly supported group, nested among Madagascan conspecifics. Together, these partly conflicting results are probably best explained by a scenario where La Réunion populations were founded by a single or few independent introductions from Madagascar. However, in case of multiple independent colonizations from genetically different source populations, and/or hybridization with other Bulbophyllum species in La Réunion (ca. 10; Sieder et al., 2007), we would have expected to see increased levels of genetic diversity in this latter island (e.g., Dlugosch and Parker, 2007; Gaudeul et al., 2011), but for which we found no evidence (i.e., inter-island comparisons in all diversity measures were nonsignificant). On the other hand, this latter evidence makes it unlikely that populations in La Réunion arose from a relatively recent, single population introduction, given that founder events almost always generate genetic bottlenecks, at least in the short term (e.g., Frankham, 1997; Dlugosch and Parker, 2007). This leaves us with the alternatives of (i) a relatively ancient single introduction; or (ii) a few independent introductions from genetically similar source populations, whereby, in either instance, mutation and gene flow restored diversity to similar levels as that found in Madagascar. The present data do not allow further distinguishing among these hypotheses. Moreover, we are presently unable to assign any potential source population(s) in Madagascar, neither can we date such putative colonization event(s). These issues will likely be solved only through increased sampling and molecular dating approaches based on DNA sequence data.
Also difficult to interpret is the remarkably high genetic differentiation observed among populations from La Réunion (ΦPT=0.359; Table 3). According to the K-means, BARRIER and neighbour-joining analyses (Figures 2 and 4), this subdivision is largely due to the segregation of two predominant gene pools (E, F), resulting in strong genetic differentiation between certain southern populations (R6–8) and the remainder. In fact, the relevant barriers (1 and 3) are suggestive of a very localized, essentially sympatric transition (‘hybrid') zone transecting one of the least disturbed areas of La Réunion, the Mare Longue Forest-Nature Reserve (Florens et al., 2012). In lieu of evidence for strong founder effects (i.e., reduced genetic diversity), these patterns may be explained by two complementary processes: (i) high selfing rates that increased population genetic structure, viz. the variance among gene pools (i.e., through reductions of inter-population gene flow via pollen; Wright et al., 2013) when the species colonized the island (Whitlock and McCauley, 1990; Giles and Goudet, 1997; García-Verdugo et al., 2010; Horn et al., 2014); and (ii) sufficient time for genetic drift to generate such differentiation since population establishment (see also Gaudeul et al., 2011). Basically the same processes might be responsible for the species' phenotypic variation in flower colour (see below).
Genetic differentiation among flower colour morphs in La Réunion
In general, a flower colour transition as observed in B. occultum from La Réunion, that is, from red to yellow (or white), is common to many animal-pollinated angiosperm taxa, including orchids, and most often involves the loss of floral anthocyanins (Wessinger and Rausher, 2012). In fact, shifts to pale flower colours seem to be common in oceanic islands (Carlquist, 1974; Barrett et al., 1996; Juillet et al., 2010), and this phenomenon has so far been observed also in other species of Bulbophyllum from La Réunion (i.e., B. clavatum, B. densum, B. incurvum, B. variegatum; Cadet, 1989; T Pailler, personal observation) and Madagascar (e.g., B. erectum; A Gamisch, personal communication), but has never been studied in detail. In B. occultum, the relatively high frequency of yellow-flowered variants (ca. 47%) observed in La Réunion (Table 1 and Figure 5b) makes it unlikely that they have been produced by repeated mutations alone (Dormont et al., 2010). Other mechanisms to explain this dimorphism include shifting selection regimes (e.g., in response to new pollinator environments), negative frequency dependent selection or genetic drift (Gigord et al., 2001; Ackerman and Carromero, 2005; Jersáková et al., 2006; Delle-Vedove et al., 2011). Although there is a growing body of investigations directly demonstrating the operation of natural selection on flower colour variation (Wessinger and Rausher, 2012), we may render any form of pollinator-mediated selection (including negative frequency dependent selection) less likely in what appears to be a mainly selfing, pollinator-independent orchid species. Also, it has been suggested that negative frequency dependent selection may not be commonly operational in tropical orchids, given their usually small and scattered distributions (Ackerman and Carromero, 2005). Rather, based on the previous section, we favour the hypothesis that flower colour divergence in B. occultum is maintained by genetic drift or linkage to pleiotropic traits experiencing selection (Wessinger and Rausher, 2012). Intriguingly, our AFLP data provided some first evidence for some genetic differentiation among these red vs yellow morphs (Figure 5c) and their significant association with certain gene pools, especially F and E, respectively. Overall, these results raise the possibility that reproductive isolation factors have accumulated among these sympatric colour morphs, perhaps (in)directly promoted by selfing (Wright et al., 2013) and/or pleiotropic fitness traits associated with morph colour (e.g., seed viability; Jersáková et al., 2006). But evidently, much further field, experimental and genetic work is required to test these hypotheses.
Conclusions
Our AFLP analyses of B. occultum showed that, contrary to what has frequently been observed in other tropical orchids (Phillips et al., 2012), this species has limited gene flow abilities in both Madagascar and La Réunion. Further studies using nuclear and chloroplast markers are required (and currently underway) to infer the relative contributions of gene flow by seed vs pollen in the population genetic structure of this species, along with estimates of inbreeding based on nuclear microsatellite markers. Despite these and other limitations (see above), our results lend support to the hypothesis that two major historical, drift-mediated processes had the greatest effect on spatial patterns of genetic diversity in this study system. First, in Madagascar, we found evidence to support the notion that B. occultum not only exists as largely isolated populations under current climatic conditions but also persisted across multiple isolated (‘refugial') regions along the eastern rainforest corridor over recent climatic cycles. And second, our data suggest that populations of B. occultum in La Réunion arose from either single or few independent introductions from Madagascar. Rather than recent founder effects, high selfing rates and sufficient time for genetic drift to operate are likely synergistic factors explaining the unexpectedly high population genetic and phenotypic (flower colour) differentiation observed in La Réunion. These populations should represent promising candidates to gain further insights into the mechanisms of (incipient) sympatric speciation in small oceanic islands (e.g., Papadopulos et al., 2011; García-Verdugo and Fay, 2014; Mallet et al., 2014). Overall, our study shows that the genetic structure of a low-gene-dispersing epiphytic orchid from the Madagascan region bears a strong imprint of history, tremendous human impact on its rainforest habitat notwithstanding.
Data archiving
Data available from the Dryad Digital Repository: (http://dx.doi.org/10.5061/dryad.1q34p)
Acknowledgments
We like to thank members of the Botanical Garden of Vienna University (Michael Kiehn, Anton Sieder) and the Parc Botanique et Zoologique de Tsimbazaza (Solo Rapanarivo, Jacky Adriantiana) for general support and help with fieldwork in Madagascar; the Département des Eaux et Fôrets (Madagascar) and the Parc National de La Réunion for issuing collecting permits; the curators of herbaria (BR, G, K, MO, P, SZU, WU) for the loan of specimens; Roman Fuchs, Alexander Gamisch, Matthias Affenzeller, Andreas Tribsch (University of Salzburg) and Albin Blaschka (LFZ Raumberg-Gumpenstein) for support in data generation/analysis; and Elisabeth Egger and her team for expert cultivation of Bulbophyllum at HBS. We also gratefully acknowledge insightful comments from three anonymous reviewers on earlier versions of the manuscript. This study was funded by the FWF (Austrian Science Fund) grants P20726-B03 and P17124-B0 to HPC and GAF, respectively.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Ackerman JD, Carromero W. (2005). Is reproductive success related to color polymorphism in a deception pollinated tropical terrestrial orchid? Caribb J Sci 41: 234–242. [Google Scholar]
- Aguilar R, Quesada M, Ashworth L, Herrerias-Diego Y, Lobo J. (2008). Genetic consequences of habitat fragmentation in plant populations: susceptible signals in plant traits and methodological approaches. Mol Ecol 17: 5177–5188. [DOI] [PubMed] [Google Scholar]
- Amsellem L, Noyer JL, Le Bourgeois T, Hossaert-McKey M. (2000). Comparison of genetic diversity of the invasive weed Rubus alceifolius Poir. (Rosaceae) in its native range and in areas of introduction, using amplified fragment length polymorphism (AFLP) markers. Mol Ecol 9: 443–455. [DOI] [PubMed] [Google Scholar]
- Andrianoelina O, Favreau B, Ramamonjisoa L, Bouvet JM. (2009). Small effect of fragmentation on the genetic diversity of Dalbergia monticola, an endangered tree species of the eastern forest of Madagascar, detected by chloroplast and nuclear microsatellites. Ann Bot 104: 1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrianoelina O, Rakotondraoelina H, Ramamonjisoa L, Maley J, Danthu P, Bouvet JM. (2006). Genetic diversity of Dalbergia monticola (Fabaceae) an endangered tree species in the fragmented oriental forest of Madagascar. Biodivers Conserv 15: 1109–1128. [Google Scholar]
- Arditti J. (1992) Fundamentals of Orchid Biology. John Wiley and Sons: New York, USA. [Google Scholar]
- Arrigo N, Felber F, Parisod C, Buerki S, Alvarez N, David J et al. (2010). Origin and expansion of the allotetraploid Aegilops geniculata, a wild relative of wheat. New Phytol 187: 1170–1180. [DOI] [PubMed] [Google Scholar]
- Azevedo MTA, Borba EL, Semir J, Solferini VN. (2007). High genetic variability in neotropical myophilous orchids. Bot J Linn Soc 153: 33–40. [Google Scholar]
- Barrett SCH, Emerson B, Mallet J. (1996). The reproductive biology and genetics of island plants. Philos Trans R Soc B 351: 725–733. [Google Scholar]
- Batygina TB, Bragina EA, Vasilyeva VE. (2003). The reproductive system and germinnation in orchids. Acta Biol Cracoviensia Ser Bot 45/2: 21–34. [Google Scholar]
- Bennett KD, Provan J. (2008). What do we mean by ‘refugia'? Quat Sci Rev 27: 2449–2455. [Google Scholar]
- Bonin A, Bellemain E, Eidesen PB, Pompanon F, Brochmann C, Taberlet P. (2004). How to track and assess genotyping errors in population genetics studies. Mol Ecol 13: 3261–3273. [DOI] [PubMed] [Google Scholar]
- Burney DA, Burney LP, Godfrey LR, Jungers WL, Goodman SM, Wright HT et al. (2004). A chronology for late prehistoric Madagascar. J Hum Evol 47: 25–63. [DOI] [PubMed] [Google Scholar]
- Cable S. (2011) New directions and challenges for the conservation of the flora of Madagascar. In: Bramwell D Caujapé-Castells J (eds). The Biology of Island Floras. Cambridge University Press: Cambridge, UK, pp 425–442. [Google Scholar]
- Cadet J. (1989) Les orchidées de la Réunion. La Nouvelle Imprimerie Dyonysienne: La Réunion.
- Carlquist S. (1974) Island Biology. Columbia University Press: New York, USA. [Google Scholar]
- Catling PM. (1990) Auto-pollination in the Orchidaceae. In: Arditti J (ed), Orchid Biology: Reviews and Perspectives V. Timber Press: Portland, USA, pp 121–158. [Google Scholar]
- Cribb P, Hermans J. (2010) Field Guide to the Orchids of Madagascar. Royal Botanic Gardens: Kew, Richmond, UK. [Google Scholar]
- Dasmahapatra KK, Lacy RC, Amos W. (2008). Estimating levels of inbreeding using AFLP markers. Heredity 100: 286–295. [DOI] [PubMed] [Google Scholar]
- Dawson IK, Powell W. (1999). Genetic variation in the afromontane tree Prunus africana, an endangered medicinal species. Mol Ecol 8: 151–156. [Google Scholar]
- Daïnou K, Bizoux JP, Doucet JL, Mahy G, Hardy OJ, Heuertz M. (2010). Forest refugia revisited: nSSRs and cpDNA sequences support historical isolation in a widespread African tree with high colonization capacity, Milicia excelsa (Moraceae). Mol Ecol 19: 4462–4477. [DOI] [PubMed] [Google Scholar]
- Delle-Vedove R, Juillet N, Bessière J-M, Grison C, Barthes N, Pailler T et al. (2011). Colour-scent associations in a tropical orchid: three colours but two odours. Phytochemistry 72: 735–742. [DOI] [PubMed] [Google Scholar]
- Dlugosch KM, Parker IM. (2007). Founding events in specie invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol Ecol 17: 431–449. [DOI] [PubMed] [Google Scholar]
- Dormont L, Delle-Vedove R, Bessière JM, Hossaert-Mc Key M, Schatz B. (2010). Rare white-flowered morphs increase the reproductive success of common purple morphs in a food-deceptive orchid. New Phytol 185: 300–310. [DOI] [PubMed] [Google Scholar]
- Doyle JJ. (1991) DNA protocols for plants-CTAB total DNA isolation. In: Hewitt GM, Johnston A (eds), Molecular Techniques in Taxonomy. Springer-Verlag: Berlin, Germany, pp 283–293. [Google Scholar]
- Ehrich D. (2006). Aflpdat: a collection of R functions for convenient handling of AFLP data. Mol Ecol Notes 6: 603–604. [Google Scholar]
- Erikson G, Ehrlén J. (2001) Landscape fragmentation and the viability of plant populations. In: Silvertown J, Antonovics J (eds), Integrating Ecology and Evolution in a Spatial Context. Blackwell: Oxford, UK, pp 157–175. [Google Scholar]
- Espíndola A, Alvarez N. (2011). Comparative phylogeography in a specific and obligate pollination antagonism. PLoS One 6: e28662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- Felsenstein J (2005). PHYLIP (Phylogeny Inference Package) Version 3.6. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle..
- Fischer GA, Gravendeel B, Sieder A, Adriantiana J, Heiselmayer P, Cribb PJ et al. (2007). Evolution of resupination of Madagascan species of Bulbophyllum (Orchidaceae). Mol Phylogenet Evol 45: 358–376. [DOI] [PubMed] [Google Scholar]
- Florens FBV, Baider C, Martin GMN, Strasberg D. (2012). Surviving 370 years of human impact: what remains of tree diversity and structure of the lowland wet forests of oceanic island Mauritius? Biodivers Conserv 21: 2139–2167. [Google Scholar]
- Frankham R. (1997). Do island populations have less genetic variation than mainland populations? Heredity 78: 311–327. [DOI] [PubMed] [Google Scholar]
- Gamisch A, Fischer GA, Comes HP. (2014). Recurrent polymorphic mating type variation in Madagascan Bulbophyllum species (Orchidaceae) exemplifies a high incidence of auto-pollination in tropical orchids. Bot J Linn Soc 175: 242–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamisch A, Fischer GA, Comes HP. (2015). Multiple independent origins of auto-pollination in tropical orchids (Bulbophyllum in light of the hypothesis of selfing as an evolutionary dead end. BMC Evol Biol 15: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Verdugo C, Fay MF. (2014). Ecology and evolution on oceanic islands: broadening the botanical perspective. Bot J Linn Soc 174: 271–275. [Google Scholar]
- García-Verdugo C, Forrest AD, Fay MF, Vargas P. (2010). The relevance of gene flow in metapopulation dynamics of an oceanic island endemic, Olea europaea subsp. guanchica. Evolution 64: 3525–3536. [DOI] [PubMed] [Google Scholar]
- Gaudeul M, Giraud T, Kiss L, Shykoff JA. (2011). Nuclear and chloroplast microsatellites show multiple introductions in the worldwide invasion history of common ragweed, Ambrosia artemisiifolia. PLoS One 6: e17658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigord LD, Macnair MR, Smithson A. (2001). Negative frequency-dependent selection maintains a dramatic flower color polymorphism in the rewardless orchid Dactylorhiza sambucina (L.) Soó. Proc Natl Acad Sci USA 98: 6253–6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles BE, Goudet J. (1997). Genetic differentiation in Silene dioica metapopulations: estimation of spatiotemporal effects in a successional plant species. Am Nat 149: 507–526. [Google Scholar]
- Goodman SM, Benstead JP. (2005). Updated estimates of biotic diversity and endemism for Madagascar. Oryx 39: 73–77. [Google Scholar]
- Harper GJ, Steininger MK, Tucker JC, Juhn D, Hawkins F. (2007). Fifty years of deforestation and forest fragmentation in Madagascar. Environ Conserv 34: 1–9. [Google Scholar]
- Hartigan JA, Wong MA. (1979). Algorithm AS 136: a K-means clustering algorithm. J R Stat Soc Ser C Appl Stat 28: 100–109. [Google Scholar]
- Hermans J, Hermans C, Du Puy DJ, Cribb PJ, Bosser J. (2007) Orchids of Madagascar. Royal Botanic Gardens: Kew, Richmond, UK. [Google Scholar]
- Hewitt GM. (2004). Genetic consequences of climatic oscillations in the Quaternary. Philos Trans R Soc B 359: 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnay O, Jacquemyn H. (2007). Susceptibility of common and rare plant species to the genetic consequences of habitat fragmentation. Conserv Biol 21: 823–831. [DOI] [PubMed] [Google Scholar]
- Horn RL, Kuehn R, Drechsel V, Cowley DE. (2014). Discriminating between the effects of founding events and reproductive mode on the genetic structure of Triops populations (Branchiopoda: Notostraca). PLoS One 9: e97473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert H, Cours Darne. (1965). Carte internationale du tapis végétal et des conditions écologiques. 3 coupures au 1/1,000,000 de Madagascar. Travaux de la Section Scientifique et Technique de l'Institut Français de Pondichéry, hors série, 3 maps. Institut français: Pondichéry, France.
- Humeau L, Micheneau C, Jacquemyn H, Gauvin-Bialecki A, Fournel J, Pailler T. (2011). Sapromyiophily in the native orchid, Bulbophyllum variegatum, on Réunion (Mascarene Archipelago, Indian Ocean). J Trop Ecol 27: 591–599. [Google Scholar]
- Hutchison DW, Templeton AR. (1999). Correlation of pairwise genetic and geographic distance measures: inferring the relative influences of gene flow and drift on the distribution of genetic variability. Evolution 53: 1898–1914. [DOI] [PubMed] [Google Scholar]
- Jacquemyn H, Micheneau C, Roberts DL, Pailler T. (2005). Elevational gradients of species diversity, breeding system and floral traits of orchid species on Reunion Island. J Biogeogr 32: 1751–1761. [Google Scholar]
- Jersáková J, Johnson SD, Kindlmann P. (2006). Mechanisms and evolution of deceptive pollination in orchids. Biol Rev 81: 219–235. [DOI] [PubMed] [Google Scholar]
- Juillet N, Delle-Vedove R, Dormont L, Schatz B, Pailler T. (2010). Differentiation in a tropical deceptive orchid: colour polymorphism and beyond. Plant Syst Evol 289: 213–221. [Google Scholar]
- Kartzinel T, Shefferson RP, Trapnell DW. (2013). Relative importance of pollen and seed dispersal across a neotropical mountain landscape for an epiphytic orchid. Mol Ecol 22: 6048–6059. [DOI] [PubMed] [Google Scholar]
- Kergoat GJ, Alvarez N. (2008). Assessing the phylogenetic usefulness of a previously neglected morphological structure through elliptic Fourier analyses: a case study in Bruchus seed-beetles (Coleoptera: Chrysomelidae: Bruchinae). Syst Entomol 33: 289–300. [Google Scholar]
- Kramer AT, Ison JL, Ashley MV, Howe HF. (2008). The paradox of forest fragmentation genetics. Conserv Biol 22: 878–885. [DOI] [PubMed] [Google Scholar]
- Krishnan S, Ranker TA, Davis AP, Rakotomalala JJ. (2013). The study of genetic diversity patterns of Coffea commersoniana, an endangered coffee species from Madagascar: a model for conservation of other littoral forest species. Tree Genet Genomes 9: 179–187. [Google Scholar]
- Lagabrielle E, Rouget M, Le Bourgeois T, Payet K, Durieux L, Baret S et al. (2011). Integrating conservation, restoration and land-use planning in islands—an illustrative case study in Réunion Island (Western Indian Ocean). Landsc Urban Plan 101: 120–130. [Google Scholar]
- Le Corre V, Kremer A. (1998). Cumulative effects of founding events during colonization on genetic diversity and differentiation in an island and stepping-stone model. J Evol Biol 11: 495–512. [Google Scholar]
- Leimu R, Vergeer P, Angeloni F, Ouborg NJ. (2010). Habitat fragmentation, climate change, and inbreeding in plants. Ann NY Acad Sci 1195: 84–95. [DOI] [PubMed] [Google Scholar]
- Liu ZJ, Chen LJ, Liu KW, Li LQ, Rao WH. (2010). A floral organ moving like a caterpillar for pollinating. J Syst Evol 48: 102–108. [Google Scholar]
- Macdonald IAW, Thébaud C, Strahm WA, Strasberg D. (1991). Effects of alien pant invasions on native vegetation remnants on La Réunion (Mascarene Islands, Indian Ocean). Environ Conserv 18: 51–61. [Google Scholar]
- Madagascar Catalogue. (2013) Catalogue of the Vascular Plants of Madagascar. Missouri Botanical Garden: St Louis, USA. Antananarivo, Madagascar. Available at http://www.efloras.org/madagascar (last accessed: October 2013). [Google Scholar]
- Mallet B, Martos F, Blambert L, Pailler T, Humeau L. (2014). Evidence for isolation-by-habitat among populations of an epiphytic orchid species on a small oceanic island. PLoS One 9: e87469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni F, Guérard E, Heyer E, Guerard E. (2004). Geographic patterns of (genetic, morphologic, linguistic) variation: how barriers can be detected by using Monmonier's Algorithm. Ann Hum Biol 76: 173–190. [DOI] [PubMed] [Google Scholar]
- Markolf M, Kappeler PM. (2013). Phylogeographic analysis of the true lemurs (genus Eulemur underlines the role of river catchments for the evolution of micro-endemism in Madagascar. Front Zool 10: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya D, Zavala MA, Rodríguez MA, Purves DW. (2008). Animal versus wind dispersal and the robustness of tree species to deforestation. Science 320: 1502–1504. [DOI] [PubMed] [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. (2000). Biodiversity hotspots for conservation priorities. Nature 403: 853–858. [DOI] [PubMed] [Google Scholar]
- Nei M. (1987) Molecular Evolutionary Genetics. Columbia University Press: New York, USA. [Google Scholar]
- Nieder J, Engwald S, Klawun M, Barthlott W. (2000). Spatial distribution of vascular epiphytes (including Hemiepiphytes) in a lowland Amazonian rain forest (Surumoni Crane Plot) of southern Venezuela. Biotropica 32: 385–396. [Google Scholar]
- Nybom H. (2004). Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13: 1143–1155. [DOI] [PubMed] [Google Scholar]
- Nybom H, Bartish IV. (2000). Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspect Plant Ecol Evol Syst 3: 93–114. [Google Scholar]
- O'Brien EK, Denham AJ, Ayre DJ. (2014). Patterns of genotypic diversity suggest a long history of clonality and population isolation in the Australian arid zone shrub Acacia carneorum. Plant Ecol 215: 55–71. [Google Scholar]
- Papadopulos AST, Baker WJ, Crayn D, Butlin RK, Kynast RG, Hutton I et al. (2011). Speciation with gene flow on Lord Howe Island. Proc Natl Acad Sci USA 108: 13188–13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. (2006). GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6: 288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. (2012). GENALEX 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28: 2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, Louis EE Jr, Ratan A, Bedoya-Reina OC, Burhans RC, Lei R et al. (2013). Aye-aye population genomic analyses highlight an important center of endemism in northern Madagascar. Proc Natl Acad Sci USA 110: 5823–5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips RD, Dixon KW, Peakall R. (2012). Low population genetic differentiation in the Orchidaceae: implications for the diversification of the family. Mol Ecol 21: 5208–5220. [DOI] [PubMed] [Google Scholar]
- Pinheiro F, Cozzolino S, de Barros F, Gouveia TM, Suzuki RM, Fay MF et al. (2013). Phylogeographic structure and outbreeding depression reveal early stages of reproductive isolation in the neotropical orchid Epidendrum denticulatum. Evolution 67: 2024–2039. [DOI] [PubMed] [Google Scholar]
- Pridgeon AM, Cribb PJ, Chase MW, Rasmussen N. (2014) Genera Orchidacearum Vol. 6. Epidendroideae (Part 3). Oxford University Press: Oxford, UK. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. (2012). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Available at http://www.R-project.org/ (last accessed: October 2014).
- Rakotoarinivo M, Blach-Overgaard A, Baker WJ, Dransfield J, Moat J, Svenning J-C. (2013). Palaeo-precipitation is a major determinant of palm species richness patterns across Madagascar: a tropical biodiversity hotspot. P Roy Soc Lond Bio B 280: 20123048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakotoarisoa JE, Raheriarisena M, Goodman SM. (2012). A phylogeographic study of the endemic rodent Eliurus carletoni (Rodentia: Nesomyinae) in an ecological transition zone of northern Madagascar. J Heredity 104: 23–35. [DOI] [PubMed] [Google Scholar]
- Rakotoarisoa JE, Raheriarisena M, Goodman SM. (2013). Late Quaternary climatic vegetational shifts in an ecological transition zone of northern Madagascar: insights from genetic analyses of two endemic rodent species. J Evol Biol 26: 1019–1034. [DOI] [PubMed] [Google Scholar]
- Ribeiro PL, Borba EL, Smidt EC, Lambert SM, Selbach-Schnadelbach A, van den Berg C. (2007). Genetic and morphological variation in the Bulbophyllum exaltatum (Orchidaceae) complex occurring in the Brazilian 'campos rupestres': implications for taxonomy and biogeography. Plant Syst Evol 270: 109–137. [Google Scholar]
- Rivers MC, Brummitt NA, Lughadha EN, Meagher TR. (2011). Genetic variation in Delonix s.l. (Leguminosae) in Madagascar revealed by AFLPs: fragmentation, conservation status and taxonomy. Conserv Genet 12: 1333–1344. [Google Scholar]
- Rousset F. (1997). Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145: 1219–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönswetter P, Tribsch A. (2005). Vicariance and dispersal in the alpine perennial Bupleurum stellatum L. (Apiaceae). Taxon 54: 725–732. [Google Scholar]
- Shapcott A, Quinn J, Rakotoarinivo M, Dransfield J. (2012). Contrasting patterns of genetic diversity between two endangered palms with overlapping distributions, Voanioala gerardii (Arecoideae) and Lemurophoenix halleuxii (Arecoideae), from North-East Madagascar. Conserv Genet 13: 1393–1408. [Google Scholar]
- Shapcott A, Rakotoarinivo M, Smith RJ, Lysaková G, Fay MF, Dransfield J. (2007). Can we bring Madagascar's critically endangered palms back from the brink? Genetics, ecology and conservation of the critically endangered palm Beccariophoenix madagascariensis. Bot J Linn Soc 154: 589–608. [Google Scholar]
- Sieder A, Rainer H, Kiehn M. (2007). CITES Checklist for Bulbophyllum and Allied Taxa (Orchidaceae). Botanical Garden of the University of Vienna. Available at www.cites.org/common/com/NC/tax_ref/Bulbophyllum.pdf (last accessed November 2014).
- Silvera K, Santiago LS, Cushman JC, Winter K. (2009). Crassulacean acid metabolism and epiphytism linked to adaptive radiations in the Orchidaceae. Plant Physiol 149: 1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrede I, Borgen L, Brochmann C. (2009). Genetic structuring in three closely related circumpolar plant species: AFLP versus microsatellite markers and high-arctic versus arctic-alpine distributions. Heredity 102: 293–302. [DOI] [PubMed] [Google Scholar]
- Strasberg D, Rouget M, Richardson DM, Baret S, Dupont J, Cowling RM. (2005). An assessment of habitat diversity and transformation on La Réunion Island (Mascarene Islands, Indian Ocean) as a basis for identifying broad-scale conservation priorities. Biodivers Conserv 14: 3015–3032. [Google Scholar]
- Tan KH, Tan LT, Nishida R. (2006). Floral phenylpropanoid cocktail and architecture of Bulbophyllum vinaceum orchid in attracting fruit flies for pollination. J Chem Ecol 32: 2429–2441. [DOI] [PubMed] [Google Scholar]
- Thébaud C, Warren BH, Strasberg D, Cheke A. (2009) Mascarene Islands, Biology. In: Gillespie RC , Clague DA (eds), Encyclopedia of Islands. University of California Press: Berkeley, USA, pp 612–619. [Google Scholar]
- Trapnell DW, Hamrick JL. (2004). Partitioning nuclear and chloroplast variation at multiple spatial scales in the neotropical epiphytic orchid, Laelia rubescens. Mol Ecol 13: 2655–2666. [DOI] [PubMed] [Google Scholar]
- Trapnell DW, Hamrick JL, Ishibashi C, Kartzinel TR. (2013). Genetic inference of epiphytic orchid colonization; it may only take one. Mol Ecol 22: 3680–3692. [DOI] [PubMed] [Google Scholar]
- Tremblay RL, Ackerman JD, Zimmerman JK, Calvo RN. (2005). Variation in sexual reproduction in orchids and its evolutionary consequences: a spasmodic journey to diversification. Biol J Linn Soc 84: 1–54. [Google Scholar]
- Tucker CJ, Grant DM, Dykstra JD. (2004). NASA's global orthorectified landsat data set. Photogramm Eng Remote Sens 70: 313–322. [Google Scholar]
- van der Cingel NA. (2001) An Atlas of Orchid Pollination: America, Africa, Asia and Australia. A. A. Balkema: Rotterdam, The Netherlands. [Google Scholar]
- Vanden Broeck A, Van Landuyt W, Cox K, De Bruyn L, Gyselings R, Oostermeijer G et al. (2014). High levels of effective long-distance dispersal may blur ecotypic divergence in a rare terrestrial orchid. BMC Ecol 14: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepitte K, Gristina AS, De Hert K, Meekers T, Roldán-Ruiz I, Honnay O. (2012). Recolonization after habitat restoration leads to decreased genetic variation in populations of a terrestrial orchid. Mol Ecol 21: 4206–4215. [DOI] [PubMed] [Google Scholar]
- Vekemans X, Beauwens M, Lemaire M, Roldán-Ruiz I. (2002). Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Mol Ecol 11: 139–151. [DOI] [PubMed] [Google Scholar]
- Vences M, Wollenberg KC, Vieites DR, Lees DC. (2009). Madagascar as a model region of species diversification. Trends Ecol Evol 24: 456–465. [DOI] [PubMed] [Google Scholar]
- Victory ER, Glaubitz JC, Rhodes OE Jr, Woeste KE. (2006). Genetic homogeneity in Juglans nigra (Juglandaceae) at nuclear microsatellites. Am J Bot 93: 118–126. [Google Scholar]
- Voigt FA, Arafeh R, Farwig N, Griebeler EM, Böhning-Gaese K. (2009). Linking seed dispersal and genetic structure of trees: a biogeographical approach. J Biogeogr 36: 242–254. [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M et al. (1995). AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner K, Mendieta-Leiva G, Zotz G. (2015). Host specificity in vascular epiphytes: a review of methodology, empirical evidence and potential mechanisms. AoB PLANTS 7: plu092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessinger CA, Rausher MD. (2012). Lessons from flower colour evolution on targets of selection. J Exp Bot 63: 5741–5749. [DOI] [PubMed] [Google Scholar]
- Whitlock MC, McCauley DE. (1990). Some population genetic consequences of colony formation and extinction: genetic correlations within founding groups. Evolution 44: 1717–1724. [DOI] [PubMed] [Google Scholar]
- Wiegand T, Raventós J, Mújica E, Gonzáles E, Bonet A. (2013). Spatio-temporal analysis of the effects of hurricane Ivan on two contrasting epiphytic orchid species in Guanahacabibes, Cuba. Biotropica 45: 441–449. [Google Scholar]
- Wilmé L, Goodman SM, Ganzhorn JU. (2006). Biogeographic evolution of Madagascar's microendemic biota. Science 312: 1063–1065. [DOI] [PubMed] [Google Scholar]
- Winkler M, Hulber K, Hietz P. (2009). Population dynamics of epiphytic orchids in a metapopulation context. Ann Bot 104: 995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI, Kalisz S, Slotte T. (2013). Evolutionary consequences of self-fertilization in plants. Proc Roy Soc Lond B 280: 20130133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A, Boyle T, Brown T. (1996). The population genetic consequences of habitat fragmentation for plants. Trends Ecol Evol 11: 413–418. [DOI] [PubMed] [Google Scholar]
- Young AG, Merriam HG, Warwick SI. (1993). The effects of forest fragmentation on genetic variation in Acer saccharum Marsh. (sugar maple) populations. Heredity 71: 279–289. [Google Scholar]
- Yuan N, Comes HP, Mao YR, Qi XS, Qiu YX. (2012). Genetic effects of recent habitat fragmentation in the Thousand-Island Lake region of southeast China on the distylous herb Hedyotis chrysotricha (Rubiaceae). Am J Bot 99: 1715–1725. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.