Abstract
Background
Persistent hepatitis B virus (HBV) infection is sustained by inadequate immune responses, either natural or acquired. Recent studies have suggested that immune responses to viral infection may be affected by microRNA (miR)-155, via its involvement in immune cell differentiation and maturation. However, little is known on the specific interaction between miR-155 and HBV in host antiviral immunity.
Objectives
This study evaluated the levels of miR-155 in peripheral blood mononuclear cells (PBMCs) of chronic hepatitis B (CHB) patients, relative to that of healthy subjects, and investigated an association between miR-155 levels and HBV DNA or alanine aminotransferase (ALT).
Patients and Methods
Total RNA was extracted from peripheral venous blood samples of 90 treatment-naive patients with chronic HBV infection and 20 healthy volunteers. The levels of miR-155 in the PBMCs were measured by real-time quantitative polymerase chain reaction. Serum HBV DNA and liver enzymes were estimated using standard clinical laboratory methods.
Results
In the HBV-infected patients, the miR-155 levels were significantly lower than in the healthy controls (P = 0.001). Chronic HBV-infected patients with elevated ALT had higher levels of miR-155 compared with patients with normal ALT (P = 0.014). No correlations were found between miR-155 and ALT or HBV DNA.
Conclusions
The miR-155 appeared to be suppressed during HBV infection. The significantly higher miR-155 levels in ALT-elevated patients infected with HBV suggest that miR-155 levels in PBMCs correlate with the immune state of patients with chronic HBV infection.
Keywords: Hepatitis B, Chronic; Immunity; Alanine Transaminase; Leukocytes; Mononuclear; miR-155
1. Background
The hepatitis B virus (HBV) infection remains a threat to global public health, with over 350 million people are chronically infected, of which 20 - 40% will develop liver cirrhosis and hepatocellular carcinoma (1). However, HBV-related liver disease is not caused by direct damage to hepatocytes by the virus, the inadequate immune responses of the host being responsible for the lesions (2, 3). One study reported that the immunological pathogenesis of severe chronic hepatitis B (CHB) is related to significant increases in the levels of CD8+ T cells and nonspecific cytotoxic T lymphocytes and these increases predict that CHB will progress to severe hepatitis (4).
According to the status of interactions between the virus, hepatocytes and host immune system, the natural course of chronic HBV infection may be divided into three phases: immune tolerance, immune clearance and the residual or inactive phases (5). Nevertheless, the exact mechanisms that underlie these interactions, in chronic HBV infection, remain unclear.
MicroRNAs (miRNAs) are small noncoding RNAs, of approximately 22 nucleotides long that are involved in innate and adaptive immunity (6, 7). Among the miRNAs, miR-155 is well characterized, with important research undertaken to investigate its role in the balance between tolerance and immunity. The miR-155 is processed from the B-cell integration cluster (BIC) gene (now designated, MIR155 host gene, or MIR155HG), which appears to be evolutionarily conserved, with relatively high levels of expression in the lymphoid organs and cells of various species (8). Early studies reported that mice deficient in the BIC gene displayed defective adaptive immunity (9). The miR-155 is upregulated in both activated B and T lymphocytes (10, 11), and also in cells of the innate immune system, such as macrophages (12), dendritic cells (13) and natural killer cells (14), in which they can either positively or negatively regulate the inflammatory response in mammals. Due to the essential role of miR-155 in the immune response, more and more studies have focused on its function in viral infection.
Evidence has shown that miR-155 is required for the response of antigen-specific CD8+T cells to viral infection, while viral clearance was impaired in mice deficient in miR-155 (15). Bignami et al. (16) determined that, among the 377 miRNAs in CD4+ T cells studied, only miR-155 could distinguish HIV-1 elite long-term nonprogressors from naive patients. Other studies confirmed that, compared with non-HCV-infected individuals, miR-155 levels were higher in the peripheral blood mononuclear cells (PBMCs), liver tissues and sera of patients with HCV genotypes 1, 2, and 3 (17, 18), while being lower in the liver tissues of patients with HCV genotype 4 (19).
Su et al. (20) studied the function of miR-155 in hepatoma cells and found that miR-155 had a mild anti-HBV effect, by promoting the Janus kinase or signal transducers and activators of transcription pathway and enhancing innate antiviral immunity. Another study reported that higher miR-155 levels were associated with a lower HBV viral load, by targeting CCAAT/enhancer-binding protein-β C/EBP-β (21). Therefore, miR-155 appears to be crucial to the immune response against HBV. However, up to present, miR-155 in the PBMCs of CHB patients has not been investigated.
2. Objectives
The present cross-sectional study evaluated the levels of miR-155 in the PBMCs of CHB patients, relative to that of non-HBV-infected individuals. In addition, the correlation between miR-155 and alanine transaminase (ALT) and the subsequent influence on HBV replication was investigated. The findings should provide new insights into the role of miR-155 in the immune system, during HBV infection.
3. Patients and Methods
The human ethics committee of the third affiliated hospital of Sun Yat-sen university, Guangzhou, China, approved this study. The study was conducted in accordance with the ethical guidelines of the declaration of Helsinki. Every subject provided written informed consent before blood samples were collected.
3.1. Patients and Controls
This cross-sectional study included 90 patients with chronic HBV infection, who underwent routine follow-up at the third affiliated hospital of Sun Yat-sen university, Guangzhou, China. These patients were hepatitis B surface antigen (HBsAg)-seropositive for more than 6 months. The study excluded: all patients confected with the hepatitis viruses A, C, D, E, or HIV; suffering from autoimmune disease, steatohepatitis, drug-induced liver injury, decompensated or compensated liver cirrhosis, or malignant comorbidities within the last 5 years; who had undergone organ transplantation; or who had ever received antiviral or immunomodulatory drugs.
For the control group, 20 healthy volunteers, who came to the hospital for physical check-ups, were recruited. All the healthy controls were negative for HBV, HCV or HIV infection and tested normal for ALT and aspartate aminotransferase (AST), at reference ranges 3 - 35 U/L and 13 - 40 U/L, respectively.
3.2. Peripheral Blood Mononuclear Cells Separation
Peripheral venous blood samples were collected from all subjects into 5 mL tubes, containing EDTA, as the anticoagulant. Within several hours from the collection, PBMCs were separated from the samples, via Ficoll-Hypaque density gradient centrifugation (Axis-Shield PoC AS, Oslo, Norway). Approximately 5 × 106 PBMCs were collected from each sample, processed with 1 mL of TRIzol (Invitrogen, Carlsbad, CA, USA) and stored at -80°C, until analyzed.
3.3. The RNA Extraction
The total RNA from the PBMCs was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), in accordance with the manufacturer’s instructions. The concentration and purity of total RNA were measured, using an ultraviolet spectrophotometer (Tiangen Biotech, Beijing, China). The integrity of the total RNA was determined by gel electrophoresis (Bio-Rad, Richmond, CA, USA).
3.4. Reverse-Transcription and Real-Time Quantitative Polymerase Chain Reaction
Subsequent to the RNA extractions, 5 μL (10 ng) of total RNA were immediately reverse transcribed (RT) in a 15-μL volume, with a TaqMan MicroRNA reverse transcription kit and miRNA-specific stem-loop primers (ID: 002623/001973, Cat. # 4427975, Applied Biosystems, Foster City, CA, USA), in accordance with the manufacturer’s instructions. The U6 small nuclear RNA (U6 snRNA) in the PBMCs was used as a reference to normalize the data. The 15-μL reactions were incubated in an Applied Biosystems 2720 Thermal Cycler, in a 96-well plate, for 30 min, at 16°C, 30 min at 42°C, 5 min at 85°C and then held at 4°C. The products of the RT reactions were stored at -20°C, for quantitative polymerase chain reaction (qPCR) amplification.
Real-time qPCR was performed on a LightCycler 480 system (Roche, Basel, Switzerland). The 20-μL complete qPCR reaction mix included 7.67-μL RNase free water, 10-μL TaqMan Universal PCR Master mix, 1-μL miRNA-specific probes (Invitrogen, Life Technologies, Carlsbad, CA, USA) and 1.33-μL RT product. All the reaction mixtures were incubated in a 96-well plate at 50°C, for 2 minutes, 95°C for 10 minutes, 40 cycles of 95°C for 15 seconds and 60°C for 60 seconds. All reactions were run in triplicate for analysis. No-template controls were employed to evaluate the background signal on a 96-well plate. In accordance with the supplier’s guidelines (Applied Biosystems), the levels of miR-155, normalized to U6 snRNA, were calculated relative to those of the healthy controls, using the Livak 2-ΔΔCt method, where Ct is the cycle threshold (22).
3.5. Laboratory Indices
The quantitative values of the following were tested by Elecsys (Roche Diagnostics GmbH, Mannheim, Germany) with the noted reference ranges (23): HBsAg, < 1.0 cut off index (COI); hepatitis B surface antibody (HBsAb), 0 - 10 IU/L; hepatitis B e antigen (HBeAg), < 1.0 COI; hepatitis B e antibody (HBeAb), > 1.0 COI; and hepatitis B c antibody (HBcAb), > 1.0 COI.
The HBV DNA load in the sera was measured via nucleic acid fluorescent qPCR (DAAN GENE, Guangzhou, China), with the detection sensitivity of quantitation of 100 IU/mL. Biochemical indices were detected using an auto-biochemical analyzer (HITACHI 7180, Tokyo, Japan). Specifically, these were ALT and AST at reference ranges of 3 - 35 U/L and 13 - 40 U/L, respectively.
3.6. Statistical Analysis
All statistical data were analyzed using SPSS 17.0 software for Windows (SPSS Inc., Chicago, IL, USA) and presented as mean ± standard deviation or median (quartile). Multiple comparisons were calculated by nonparametric Kruskal-Wallis test with Bonferroni correction for sub-analyses. Statistical significance between the two groups was determined by the Mann-Whitney U-test. The correlation between the miR-155 levels and clinical parameters was examined using Spearman’s rank correlation. All statistical tests were two-tailed. Differences were considered statistically significant at P < 0.05.
4. Results
4.1. Study Subjects’ Characteristics
The subjects of this study comprised 90 treatment-naive patients, who had been HBsAg-seropositive for more than 6 months, and 20 healthy volunteers with normal ALT and AST. Based on the 2012 Asian-Pacific consensus statement on the management of CHB (5), the 90 HBV-infected patients were further stratified as: immune tolerant (IT) phase (n = 20); immune residual or inactive (IR) phase (n = 20); and immune clearance (IC) phase (n = 50) that comprised HBeAg-positive hepatitis (n = 30) and HBeAg-negative hepatitis (n = 20). Because elevated ALT reflects the host’s immune response to HBV, comparisons were conducted for those with elevated ALT to those with normal ALT (Table 1).
Table 1. Clinical Characteristics of Chronic Hepatitis B Patients, Healthy Controls and Patients With Elevated and Normal Alanine Transaminasea,b.
| Variables | CHB | HC | P Value | ALT Elevated | Normal | P Value |
|---|---|---|---|---|---|---|
| N | 90 | 20 | NA | 50 | 40 | NA |
| Gender | 0.361 | < 0.001 | ||||
| Male | 55 | 10 | 40 | 15 | ||
| Female | 35 | 10 | 10 | 25 | ||
| Age, y | 30.76 ± 9.52 | 24.35 ± 4.49 | < 0.001 | 32.16 ± 10.77 | 29.00 ± 7.44 | 0.104 |
| ALT, U/L | 103.00 (503.50) | 15.50 (7.25) | < 0.001 | 455.50 (708.50) | 22.00 (10.75) | < 0.001 |
| AST, U/L | 66.00 (299.00) | 20.00 (6.00) | < 0.001 | 299.50 (514.75) | 22.00 (7.50) | < 0.001 |
| HBV DNA, log10 IU/mL | 6.01 ± 2.16 | NA | NA | 6.62 ± 1.48 | 5.26 ± 2.61 | 0.005 |
| HBsAg | < 0.001 | NA | NA | NA | ||
| Positive | 90 | 0 | ||||
| Negative | 0 | 20 | ||||
| HBeAg | < 0.001 | 0.343 | ||||
| Positive | 50 | 0 | 30 | 20 | ||
| Negative | 40 | 20 | 20 | 20 | ||
| ALT | < 0.001 | NA | NA | NA | ||
| Elevated | 50 | 0 | ||||
| Normal | 40 | 20 |
Abbreviations: ALT, alanine transaminase; AST, aspartate aminotransferase; CHB, chronic hepatitis B; HBeAg, hepatitis B e antigen; HBsAg, hepatitis B s antigen; HBV, hepatitis B virus; HC, healthy controls; NA, not applicable.
aValues are expressed as mean ± SD for age and HBV DNA ;median (quartile) for ALT and AST.
bHBV DNA < 100 was treated as 100.
4.2. MiR-155 Levels in Peripheral Blood Mononuclear Cells
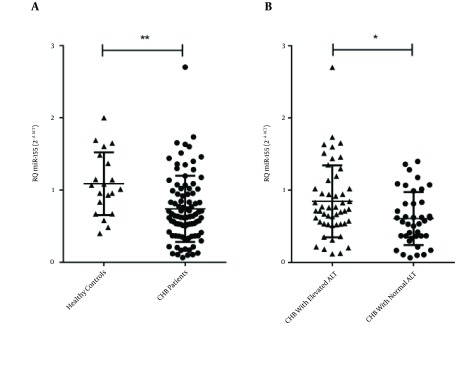
The levels of miR-155 in the PBMC samples were quantified via real-time qPCR (Figure 1). MiRNA levels of U6 snRNA were used to normalize the Ct value. The analysis revealed that the miR-155 levels of the CHB patients were significantly lower than those of the healthy controls (P = 0.001; Figure 1A). This suggested that circulating miR-155 may negatively correlate with chronic HBV infection.
Figure 1. Normalized Levels of MiR-155 in Peripheral Blood Mononuclear Cells of Hepatitis B Virus-Infected Patients and Healthy Controls.
A, levels of miR-155, represented as relative quantitation (RQ), were significantly lower in HBV-infected patients compared with healthy controls (P = 0.001); B, patients with elevated ALT had significantly higher levels of miR-155 than did patients with normal ALT (P = 0.014). Results are expressed as mean of RQ ± standard error of the mean; Abbreviations: ALT, alanine transaminase; CHB, chronic hepatitis B.
We further investigated the association between miR-155 and HBV by stratifying patients according to immune status, that is, IT, IR, or IC. Based on the Kruskal-Wallis tests, there were significant differences among these groups of patients (P = 0.048). MiR-155 levels were higher in IC patients than IT patients (P = 0.042). No difference was observed between IR patients and IT patients (P = 0.946). However, when the Bonferroni correction was done, no differences in miR-155 were observed (data not shown).
The 50 patients in the IC phase were analyzed according to positivity toward the HBeAg, which is a reflection of viral replication in the liver. The difference in miR-155 levels between the HBeAg-positive and -negative IC patients was not significant (P = 0.874, data not shown). However, based on results of the Mann-Whitney U-test, the HBV patients with elevated ALT had significantly higher levels of miR-155 than the patients with normal ALT (P = 0.014, Figure 1B).
4.3. Correlation Between MiR-155 Levels and Clinical Parameters of Chronic Hepatitis B Patients
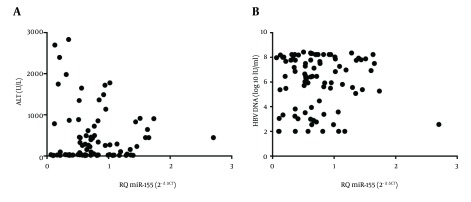
Investigations were conducted upon the correlations between miR-155 levels in PBMCs and patients’ ALT, viral load, age, or gender (Table 1 ; Figure 2). Based on Spearman’s rank correlation analysis, there was no correlation between miR-155 levels and levels of ALT (r = 0.107, P = 0.318; Figure 2 A). Similarly, no correlation was found between HBV DNA and miR-155 levels (r = 0.014, P = 0.899; Figure 2 B).
Figure 2. Correlation of MiR-155 With Alanine Transaminase and Hepatitis B Virus DNA of Hepatitis B Virus-Infected Patients.
Levels of miR-155, presented as relative quantitation (RQ), in PBMCs of HBV-infected treatment-naive patients did not correlate with levels of either (A) ALT (r = 0.107, P = 0.318), or (B) HBV DNA (r = 0.014, P = 0.899). Abbreviations: ALT, alanine transaminase; HBV, hepatitis B virus; PBMCs, peripheral blood mononuclear cells.
In addition, no correlation was shown between miR-155 levels and the ages of either the patients (r = –0.172, P = 0.104) or healthy controls (r = -0.070, P = 0.770), and similarly there was no difference observed in the gender ratios of either the patients (P = 0.107) or healthy control groups (P = 0.496).
5. Discussion
In HBV-infected patients, an efficient immune response will contribute to viral clearance, whereas immune dysfunction can result in persistent infection and chronic inflammatory disease. Numerous miRNAs, including miR-17-92 cluster, miR-150 and miR-155, act as regulatory elements in the immune system, by controlling cellular development, homeostasis and response in highly specific pathways (24). In addition, miRNAs are involved in host-virus interactions and chronic liver inflammation (25, 26). In particular, miR-155 is essential in the maintenance and function of diverse immunological cells and has important roles in both the innate and adaptive immune responses (15, 27-30). The PBMCs represent a crucial component of the immune system that fights infection. In the present study, we chose first to investigate the miR-155 levels in PBMCs from CHB patients, in different phases of the disease.
The present investigation found that the miR-155 levels of PBMCs of treatment-naive HBV-infected patients were significantly lower than those of the non-infected healthy volunteers. This is in agreement with previous data that found that miR-155 levels were significantly lower in both liver biopsy specimens and serum from HBV-infected patients, compared with healthy controls (21). In addition, Tang et al. (31) reported that, compared with healthy volunteers, plasma miRNA-155 was significantly higher in patients with malignant liver and gastric tumors, while it lower in those with benign disease.
However, miR-155 levels in HBV-infected patients differ from that of other virally-infected patients, such as HCV genotypes 1, 2, and 3 in serum or PBMCs (18, 32), Epstein-Barr virus infection in primary human B-cells (33) and HIV infection in macrophages (34). These differences in miR-155 patterns may occur because miR-155 can be induced by various molecules (12). It has tens-to-thousands of targets and its targets, in different diseases, lead to different outcomes. Regarding HBV infection, it has been suggested that lower levels of miR-155 may be due to toll-like receptor 7 suppression (21).
Su et al. (20) showed that overexpression of miR-155 led to suppression of HBV transcription in HepG2 cells, transfected with a pAAV/HBV1.2 plasmid. They examined the expression levels of the HBV X gene (HBx) in vitro, by RT-PCR and found that overexpression of miR-155 may inhibit HBx expression to a certain extent. Sarkar et al. (21) reported that increased miR-155 levels could help reduce the hepatitis B viral load by targeting C/EBP-β in vitro. However, our present results showed no correlation between HBV DNA levels and levels of miR-155 in PBMCs of CHB patients. Based on the present results and the aforementioned studies, it is possible that, in vivo, miR-155 does not inhibit HBV replication, immediately.
MiR-155 has been implicated in driving chronic inflammation (7) and liver enzymes are clinical indicators for activation of the immune response and hepatic inflammation (35). Although, in the present study, no correlation between miR-155 and ALT levels was found, the miR-155 levels in patients with elevated ALT were significantly higher, compared with patients with normal ALT. This suggests that the miR-155 in PBMCs may be associated with chronic HBV infection-related immune pathogenesis.
According to a previous study, miR-155 has both positive and negative roles in the control of the inflammatory response, by regulating cells of the immune system. The different outcomes are likely due to changes in the transcriptome, depending on the cell type and expressions or repressions of the pool of miR-155 direct targets (36). One study observed significant changes in HBV-specific and nonspecific cell-mediated immune responses, after chronic HBV infection deteriorated into severe hepatic disease (4). Therefore, it is important to know what subsets of immune cells are represented in peripheral blood and how PBMC populations differ in distribution and function, from tissue immune cells. Because the pathogenesis of HBV-induced hepatitis is complicated, an in-depth mechanistic study of the function of miR-155 in HBV-related infection is warranted. Further research may lead to novel strategies to treat HBV-infected patients and, perhaps, other related liver diseases. Cieslar-Pobuda et al. (37) recent article noted the adverse effects of liver transplantation, such as HBV infection or reinfection, and that advances in regenerative strategies to treat end-stage liver diseases may avoid these potential problems. A better understanding of the role of miR-155 in HBV infection, after liver transplantation, may lead to new strategies to solve this clinical challenge.
In summary, this is the first report that miR-155 levels in PBMCs of HBV-infected patients are significantly lower, compared with that of healthy control individuals. In addition, our results suggest that miR-155 levels in PBMCs varied among patients. We believe that these findings may contribute to the understanding of the pathogenesis of HBV infection and the development of effective interventions.
Acknowledgments
The authors thank all the HBV-infected individuals and healthy volunteers included in this study and are grateful to all doctors and nurses of third affiliated hospital of Sun Yat-sen university, Guangzhou, China. This manuscript was edited and proofread by Medjaden bioscience limited.
Footnotes
Authors’ Contribution:Study concept and design: Hong Deng, Su-Lin Yu, and Xin-Hua Li; acquisition of data: Su-Lin Yu and Ya-Xin Huang; analysis and interpretation of data: Su-Lin Yu and Ya-Xin Huang; contributed samples: Dong-Ying Xie and Zhi-Liang Gao; statistical analyses: Su-Lin Yu; drafting of the manuscript: Su-Lin Yu; critical revision of the manuscript for important intellectual content: Hong Deng and Xin-Hua Li; study supervision: Hong Deng.
Funding/Support:This study was supported by the science and technology project of Guangdong province (No. 2013B021800209).
References
- 1.Lee WM. Hepatitis B virus infection. N Engl J Med. 1997;337(24):1733–45. doi: 10.1056/NEJM199712113372406. [DOI] [PubMed] [Google Scholar]
- 2.Sugiyama M, Tanaka Y, Kurbanov F, Maruyama I, Shimada T, Takahashi S, et al. Direct cytopathic effects of particular hepatitis B virus genotypes in severe combined immunodeficiency transgenic with urokinase-type plasminogen activator mouse with human hepatocytes. Gastroenterology. 2009;136(2):652–662. e3. doi: 10.1053/j.gastro.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 3.Chisari FV. Viruses, Immunity, and Cancer: Lessons from Hepatitis B. Am J Pathol. 2000;156(4):1117–32. doi: 10.1016/s0002-9440(10)64980-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xibing G, Xiaojuan Y, Zhonghua L, Juanhua W. Alteration in cellular immunity after chronic hepatitis B deteriorated into severe hepatitis and its significance. Hepat Mon. 2011;11(10):810–5. doi: 10.5812/kowsar.1735-143x.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liaw YF, Kao JH, Piratvisuth T, Chan HL, Chien RN, Liu CJ, et al. Asian-Pacific consensus statement on the management of chronic hepatitis B: a 2012 update. Hepatol Int. 2012;6(3):531–61. doi: 10.1007/s12072-012-9365-4. [DOI] [PubMed] [Google Scholar]
- 6.Baltimore D, Boldin MP, O'Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9(8):839–45. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 7.O'Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33(4):607–19. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of Tissue-Specific MicroRNAs from Mouse. Cur Biol. 2002;12(9):735–9. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316(5824):608–11. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van den Berg A, Kroesen BJ, Kooistra K, de Jong D, Briggs J, Blokzijl T, et al. High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin lymphoma. Genes Chromosomes Cancer. 2003;37(1):20–8. doi: 10.1002/gcc.10186. [DOI] [PubMed] [Google Scholar]
- 11.Haasch D, Chen YW, Reilly RM, Chiou XG, Koterski S, Smith M, et al. T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene,. BIC. 2002;217(1-2):78–86. doi: 10.1016/s0008-8749(02)00506-3. [DOI] [PubMed] [Google Scholar]
- 12.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A. 2007;104(5):1604–9. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ceppi M, Pereira PM, Dunand-Sauthier I, Barras E, Reith W, Santos MA, et al. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc Natl Acad Sci U S A. 2009;106(8):2735–40. doi: 10.1073/pnas.0811073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sullivan RP, Fogel LA, Leong JW, Schneider SE, Wong R, Romee R, et al. MicroRNA-155 Tunes Both the Threshold and Extent of NK Cell Activation via Targeting of Multiple Signaling Pathways. The Journal of Immunology. 2013;191(12):5904–13. doi: 10.4049/jimmunol.1301950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gracias DT, Stelekati E, Hope JL, Boesteanu AC, Doering TA, Norton J, et al. The microRNA miR-155 controls CD8+ T cell responses by regulating interferon signaling. Nature Immunology. 2013;14(6):593–602. doi: 10.1038/ni.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bignami F, Pilotti E, Bertoncelli L, Ronzi P, Gulli M, Marmiroli N, et al. Stable changes in CD4+ T lymphocyte miRNA expression after exposure to HIV-1. Blood. 2012;119(26):6259–67. doi: 10.1182/blood-2011-09-379503. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Wei W, Cheng N, Wang K, Li B, Jiang X, et al. Hepatitis C virus-induced up-regulation of microRNA-155 promotes hepatocarcinogenesis by activating Wnt signaling. Hepatology. 2012;56(5):1631–40. doi: 10.1002/hep.25849. [DOI] [PubMed] [Google Scholar]
- 18.Bala S, Tilahun Y, Taha O, Alao H, Kodys K, Catalano D, et al. Increased microRNA-155 expression in the serum and peripheral monocytes in chronic HCV infection. J Transl Med. 2012;10:151. doi: 10.1186/1479-5876-10-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riad SE, El-Ekiaby N, Mekky RY, Ahmed R, El Din MA, El-Sayed M, et al. Expression signature of microRNA-155 in hepatitis C virus genotype 4 infection. Biomed Rep. 2015;3(1):93–7. doi: 10.3892/br.2014.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su C, Hou Z, Zhang C, Tian Z, Zhang J. Ectopic expression of microRNA-155 enhances innate antiviral immunity against HBV infection in human hepatoma cells. Virol J. 2011;8:354. doi: 10.1186/1743-422X-8-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarkar N, Panigrahi R, Pal A, Biswas A, Singh SP, Kar SK, et al. Expression of microRNA-155 correlates positively with the expression of Toll-like receptor 7 and modulates hepatitis B virus via C/EBP-βin hepatocytes. Journal of Viral Hepatitis. 2015;22(10):817–27. doi: 10.1111/jvh.12390. [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Wu ZQ, Tan L, Liu T, Gao ZL, Ke WM. Evaluation of changes of serum hepatitis B surface antigen from a different perspective. World J Gastroenterol. 2015;21(9):2739–45. doi: 10.3748/wjg.v21.i9.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136(1):26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 25.Cullen BR. MicroRNAs as mediators of viral evasion of the immune system. Nat Immunol. 2013;14(3):205–10. doi: 10.1038/ni.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szabo G, Bala S. MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol. 2013;10(9):542–52. doi: 10.1038/nrgastro.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27(6):847–59. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316(5824):604–8. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 29.Yu D. MicroRNAs in Tfh cells: micromanaging inflammaging. Immunity. 2014;41(4):509–11. doi: 10.1016/j.immuni.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Zawislak CL, Beaulieu AM, Loeb GB, Karo J, Canner D, Bezman NA, et al. Stage-specific regulation of natural killer cell homeostasis and response against viral infection by microRNA-155. Proc Natl Acad Sci U S A. 2013;110(17):6967–72. doi: 10.1073/pnas.1304410110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang G, Shen X, Lv K, Wu Y, Bi J, Shen Q. Different normalization strategies might cause inconsistent variation in circulating microRNAs in patients with hepatocellular carcinoma. Med Sci Monit. 2015;21:617–24. doi: 10.12659/MSM.891028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang M, Broering R, Trippler M, Wu J, Zhang E, Zhang X, et al. MicroRNA-155 controls Toll-like receptor 3- and hepatitis C virus-induced immune responses in the liver. J Viral Hepat. 2014;21(2):99–110. doi: 10.1111/jvh.12126. [DOI] [PubMed] [Google Scholar]
- 33.Linnstaedt SD, Gottwein E, Skalsky RL, Luftig MA, Cullen BR. Virally induced cellular microRNA miR-155 plays a key role in B-cell immortalization by Epstein-Barr virus. J Virol. 2010;84(22):11670–8. doi: 10.1128/JVI.01248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swaminathan G, Rossi F, Sierra LJ, Gupta A, Navas-Martin S, Martin-Garcia J. A role for microRNA-155 modulation in the anti-HIV-1 effects of Toll-like receptor 3 stimulation in macrophages. PLoS Pathog. 2012;8(9):e34483. doi: 10.1371/journal.ppat.1002937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liaw YF. Hepatitis flares and hepatitis B e antigen seroconversion: implication in anti-hepatitis B virus therapy. J Gastroenterol Hepatol. 2003;18(3):246–52. doi: 10.1046/j.1440-1746.2003.02976.x. [DOI] [PubMed] [Google Scholar]
- 36.Vigorito E, Kohlhaas S, Lu D, Leyland R. miR-155: an ancient regulator of the immune system. Immunol Rev. 2013;253(1):146–57. doi: 10.1111/imr.12057. [DOI] [PubMed] [Google Scholar]
- 37.Cieslar-Pobuda A, Wiechec E. Research on liver regeneration as an answer to the shortage of donors for liver transplantation. Hepatol Res. 2014;44(9):944–6. doi: 10.1111/hepr.12265. [DOI] [PubMed] [Google Scholar]